Super-relaxation is a state of myosin in which ATP turnover is very slow, conserving energy in relaxed muscle. This work discusses the molecular interactions underlying super- and hyper-relaxation and how these are modulated in normal muscle physiology and disease.
Abstract
Super-relaxation is a state of muscle thick filaments in which ATP turnover by myosin is much slower than that of myosin II in solution. This inhibited state, in equilibrium with a faster (relaxed) state, is ubiquitous and thought to be fundamental to muscle function, acting as a mechanism for switching off energy-consuming myosin motors when they are not being used. The structural basis of super-relaxation is usually taken to be a motif formed by myosin in which the two heads interact with each other and with the proximal tail forming an interacting-heads motif, which switches the heads off. However, recent studies show that even isolated myosin heads can exhibit this slow rate. Here, we review the role of head interactions in creating the super-relaxed state and show how increased numbers of interactions in thick filaments underlie the high levels of super-relaxation found in intact muscle. We suggest how a third, even more inhibited, state of myosin (a hyper-relaxed state) seen in certain species results from additional interactions involving the heads. We speculate on the relationship between animal lifestyle and level of super-relaxation in different species and on the mechanism of formation of the super-relaxed state. We also review how super-relaxed thick filaments are activated and how the super-relaxed state is modulated in healthy and diseased muscles.
The super-relaxed state of myosin
Animal life is characterized by the constant need for food, which provides the raw materials for making ATP used in the body’s energy-requiring processes. A substantial amount of energy is expended by skeletal muscle, which in humans amounts to 30–40% of body mass (Janssen et al., 2000). During contraction, ATP is rapidly consumed by the molecular motor, myosin II, as it pulls on actin filaments to produce force and movement. But ATP is also used at a significant basal rate even in the resting state. Producing ATP is costly for the cell, so there is a substantial evolutionary advantage to minimizing its waste. Animals have adapted to this requirement by evolving a mechanism that reduces the consumption of ATP to minimal levels in relaxed (RX) muscle. This mechanism is known as super-relaxation or the super-relaxed state (SRX).
SRX is a biochemical state in which ATP turnover by a portion of the myosin heads in the RX state of muscle thick filaments is much slower (∼10 times) than that of myosin II molecules in solution (Stewart et al., 2010; Cooke, 2011; Nag and Trivedi, 2021). This inhibited state, in equilibrium with a faster (RX) state, was suggested by early studies showing that the metabolic rate of live, resting muscle was much less than that expected from the ATP turnover rate of purified myosin (Ferenczi et al., 1978) and similarly that ATPase activity of RX myofibrils was also lower than expected from isolated myosin (Myburgh et al., 1995). A slow rate of ATP turnover was later directly demonstrated in studies of skinned muscle fibers (Stewart et al., 2010). The single ATP turnover rate is typically measured as the rate of binding of fluorescent ATP (mantATP) to, or release from, myosin heads in solution or in skinned fiber bundles (Hooijman et al., 2011; McNamara et al., 2015); conceptually, because Pi release is rate limiting, the apparent rates of ATP binding and ADP release effectively reveal the overall ATP turnover rate. Experimental curves are generally interpreted in terms of two exponentials, one with a slow rate (SRX) and the other with a faster but still slow (RX) rate of ATP use (Cooke, 2011), although additional exponentials can sometimes be resolved (Hooijman et al., 2011; Naber et al., 2011).
The SRX state is ubiquitous and thought to be fundamental to muscle function, acting as a mechanism for parking and switching off—like a car—energy-consuming myosin motors when they are not being used. It has been detected in all muscles where it has been studied: vertebrate (including human) and invertebrate, fast and slow skeletal, and cardiac muscle (Stewart et al., 2010; Cooke, 2011; Hooijman et al., 2011; Naber et al., 2011; McNamara et al., 2015; Phung et al., 2020). A highly inhibited rate of ATP turnover also characterizes the switched-off state of vertebrate smooth muscle and nonmuscle myosin II molecules (Cross et al., 1988). SRX plays a critical role in muscle energetics, conserving ATP in resting as well as contracting muscles and providing a reserve of myosin heads for enhanced contractility in cardiac muscles (Cooke, 2011; Hooijman et al., 2011; Brunello et al., 2020; Ma et al., 2021b).
What are the key features of the SRX state?
The SRX state has two main parameters: (1) the rate of ATP use (i.e., ATP turnover rate, often expressed as its inverse, the ATP turnover time) and (2) the fraction of molecules exhibiting this rate. Both parameters are essential to understanding the significance of SRX in the energy balance of muscle: myosin heads with a very slow rate would be of little consequence if not present in substantial numbers. The rate of ATP use has been attributed to the specific conformation of the myosin head, which can impede, or not, the release of the products of ATP hydrolysis (ADP and Pi; Anderson et al., 2018), and the fraction of heads with this rate in muscle appears to be related to the intra- and intermolecular interactions that the heads undergo in thick filaments, as discussed below.
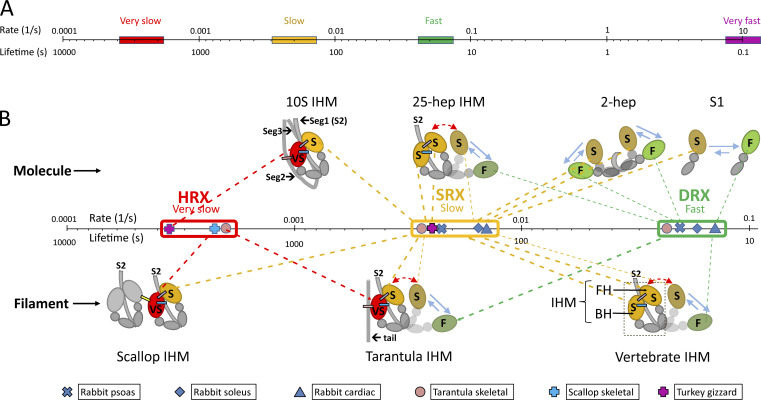
ATP turnover by myosin heads can vary by six orders of magnitude, depending on the muscle and its state of activity (Cooke, 2011; Naber et al., 2011). These fall into four main groups, which, on a logarithmic scale, differ internally by small amounts, but by an order of magnitude or more between groups (Fig. 1 A). The fastest use of ATP is in contracting muscle (rate ≈ 10 s−1), where actin activates the release of hydrolysis products in a process coupled to the power stroke of the myosin head. The advantages of force and movement are paid for by a rapid consumption of ATP. RX heads use ATP at least 100 times more slowly: some at the RX rate, similar to that of purified myosin in solution (<0.1 s−1), and others at an SRX rate, 10 times slower (Fig. 1, A and B). As might be predicted from the two parameters of the SRX, nature has evolved different ways of saving energy in this state: decreasing the ATP turnover rate, increasing the fraction with the low rate, or a combination of the two. These strategies appear adapted to the different systems in which they are found. The SRX rate typically occurs in 50% or more of the heads in a thick filament, the balance being RX heads (Cooke, 2011). In certain muscles, there is an even slower rate (10 times slower than SRX), which we refer to as “hyper-relaxed” (HRX), present together with SRX and RX heads (Naber et al., 2011).
Figure 1.
ATP turnover rates of myosin and proposed relation to head organization. (A) Logarithmic plot of rates, varying from very slow (hyper-relaxed), to slow (super-relaxed), to fast (disordered-relaxed), to very fast (actin-activated). (B) Expanded plot of relaxed rates and proposed relation to head configuration and interactions. The cartoons show the IHM as found in single molecules (upper) and thick filaments (lower); in filaments, only molecules in a single crown are shown. The colored ellipses are the interacting motor domains. The smaller, gray domains are the ELCs and RLCs, and the gray lines are the proximal region of the myosin tail (S2). Molecules with a sufficient length of S2 (25 heptads [25 hep]; ∼250 Å) form an IHM, while single heads and two-headed molecules with only two heptads of tail show no interactions. Molecules in filaments have SRX and DRX turnover rates similar to those of isolated molecules (25-heptad IHM; cf. vertebrate IHM) or more inhibited (HRX) rates. Heads turn over ATP at fast (F; noninteracting), slow (S; interacting), or very slow (VS; more interactions) rates. Slow (attached) FHs of the IHM can detach and sway out (red, double-headed arrows) and, while detached, can equilibrate between the slow and fast conformations, similar to S1 (blue, reversible arrows). The “tail” in tarantula and the uncolored IHM in scallop provide additional, intermolecular interactions leading to the very slow rate; this rate is also seen in 10S myosin through intramolecular interactions with segment 2 of its own tail. The different lengths of the bars in A correspond to the lengths in B, which are drawn to include the range of rates for HRX, SRX, and DRX; the key point is the close clustering of rates within these groups and the large gaps between them.
What is the structural basis of the SRX state?
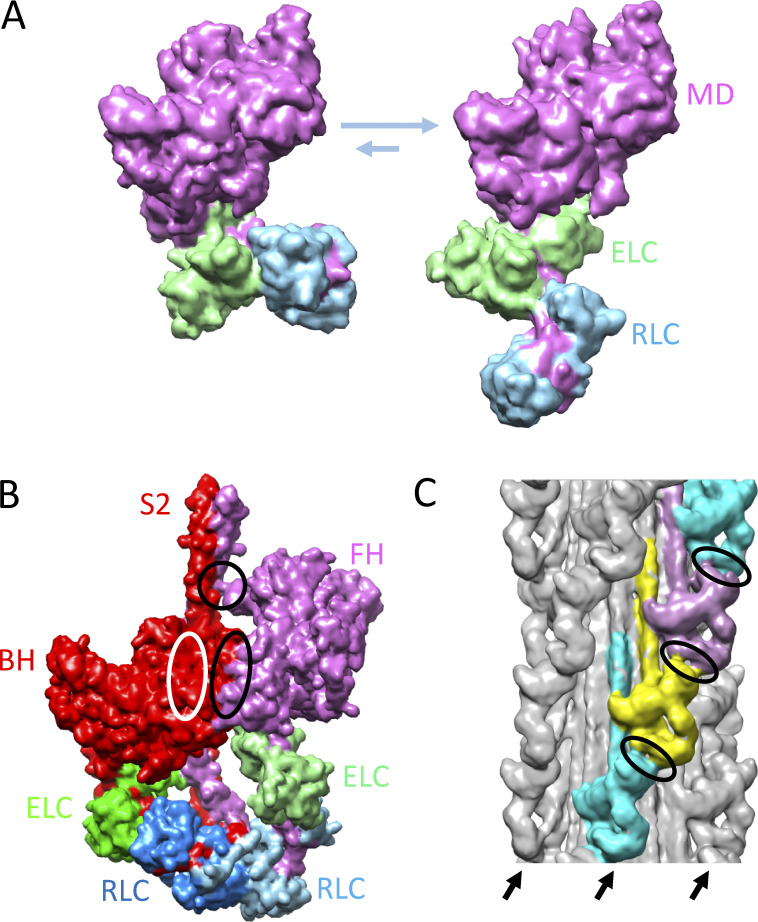
The early finding that myosin filaments in muscle had a slower ATP turnover rate than myosin II molecules in solution led to the idea that interactions of heads possible in the polymer (e.g., with the thick filament backbone), but not in the monomer, inhibited their activity (Myburgh et al., 1995; Stewart et al., 2010; Cooke, 2011). The first study to unequivocally show head organization in a thick filament indeed revealed such interactions—of heads with the backbone, with each other within a molecule, and with each other between molecules (Woodhead et al., 2005). Of particular note was a folding back of the two heads onto the proximal part of the tail (subfragment 2 [S2]), with which they interacted, and intramolecular interaction between the heads through their motor domains. This structure became known as the “interacting-heads motif” (IHM; Fig. 1 B, dotted box; Fig. 2 B; Woodhead et al., 2005; Alamo et al., 2008). Such interaction between heads had first been seen in isolated myosin molecules (Wendt et al., 2001; Burgess et al., 2007) and was thought to inhibit their activity. The two heads were called “blocked” and “free” (BH and FH, respectively), referring to their actin-binding capability (Wendt et al., 2001). In this model, actin binding by the BH would be blocked by interaction of its actin-binding site with the FH. While the FH actin-binding site was exposed, movement of its converter domain, needed for ATP product release, would be inhibited by binding to the BH (Wendt et al., 2001). The overall result would be inhibition of activity of both heads, but by different mechanisms. Interaction of the folded-back heads with S2 would further inhibit the motions required for ATPase activity (Woodhead et al., 2005).
Figure 2.
Structural basis of SRX. (A) Proposed conformations of myosin heads (S1), in equilibrium with each other, underlying SRX (left, bent) and DRX (right, straight) ATP turnover rates (Anderson et al., 2018). MD, motor domain. (B) IHM of cardiac HMM showing BH and FH (based on Protein Data Bank accession no. 5TBY). Ellipses show regions of interaction between BH and FH motor domains (black ellipse), FH and S2 (black circle), and BH and S2 (white ellipse; interaction occurs on rear side of BH). (C) Thick filament (tarantula; EM Data Bank accession no. 1950) showing IHMs lying along four coaxial helices (three on front marked with arrows) creating intermolecular interactions (ellipses) between FH of one IHM and regulatory domain of IHM above. M-line would be at the top of the image. The reconstruction shows the average positions of the heads in the filament; however, the FHs are thought to be dynamic, leaving and returning to the IHM (Fig. 1 B; see text). Models in this figure were created with UCSF Chimera (Pettersen et al, 2004).
The IHMs in RX thick filaments are organized in helical arrays, with intermolecular interactions of IHMs with each other along the helices as well as with the filament backbone (Woodhead et al., 2005; Zoghbi et al., 2008; Zhao et al., 2009; Pinto et al., 2012; Woodhead et al., 2013; Sulbarán et al., 2015). Helical ordering requires the closed conformation of the myosin heads (Xu et al., 2003; Zoghbi et al., 2004; Xu et al., 2009) and is thought to be a signature of the IHM. It can be disrupted in multiple ways (increased salt level, phosphorylation of the myosin regulatory light chains [RLCs], substitution of GTP or ADP for ATP, lowering of temperature), and this is accompanied by reduction of the SRX state in each case (Stewart et al., 2010; Cooke, 2011). Conversely, conditions that enhance the IHM, such as treatment with the myosin inhibitor blebbistatin (Zhao et al., 2008; Xu et al., 2009; Fusi et al., 2015), enhance the SRX (Wilson et al., 2014; Fusi et al., 2015). These correlations led to the view that IHMs in the ADP.Pi prepowerstroke state (Xu et al., 1999), organized in helices and bound to the core of the thick filament, may be the structural basis of the SRX state (Stewart et al., 2010; Cooke, 2011; Wilson et al., 2014; Fusi et al., 2015; Alamo et al., 2016).
Despite this suggestive evidence, however, recent single ATP turnover measurements of myosin constructs have shown that slow turnover of ATP occurs not only in thick filaments but also, to a small extent (∼10–20% of molecules), in isolated myosin heads (S1; Anderson et al., 2018; Rohde et al., 2018; Gollapudi et al., 2021b; the earlier, steady-state solution ATPase observations were not capable of revealing these low rates). Thus, neither the filamentous form of myosin nor the IHM is required for the SRX rate of ATP turnover, as originally thought. It has been suggested, instead, that myosin heads in solution exist in an equilibrium between a strongly bent (“closed”) conformation (in which the lever arm is tilted more toward the prestroke direction similar to that in the IHM structure; 10–20% of molecules) and a more extended (“open”) structure (Anderson et al., 2018) and that inhibition of phosphate release and thus ATP turnover by the closed structure could account for the observation of a small level of SRX in S1 (Figs. 1 B and 2 A). If this is the case, what is the role of the IHM in SRX?
Different degrees of SRX correlate with different levels of head organization
Myosin heads can exist in several structural forms: single heads, two-headed myosin molecules or constructs, and the polymeric myosin filaments found in muscle. Studies show that SRX increases (greater inhibition or greater number of inhibited molecules) as myosin heads are incorporated into more complex structures. As mentioned, ∼10–20% of isolated myosin heads in solution have a slow rate (SRX) and are thought to be in equilibrium with the balance of heads having a turnover rate similar to the conventionally measured ATPase (RX heads; Anderson et al., 2018; Rohde et al., 2018; Gollapudi et al., 2021b). When myosin heads are present in two-headed constructs containing 25 heptads of tail (∼250 Å; Figs. 1 B and 2 B) or full-length S2, similar SRX and RX rates are observed, but the fraction of SRX heads increases to ∼25–30% (at physiological ionic strength; Anderson et al., 2018; Rohde et al., 2018; Gollapudi et al., 2021b), suggesting that the presence of two heads and/or the tail stabilizes the SRX head structure. Importantly, if the tail is short (only two heptads; Fig. 1 B), the rates and fractions are similar to those of S1 (Anderson et al., 2018), demonstrating the importance of the tail in creating the SRX. Modeling and experiments show that 25 heptads (or more) of tail is long enough for molecules to form an IHM with folded-back, interacting FHs and BHs (Figs. 1 B and 2 B; Anderson et al., 2018). Head–head and head–tail interactions would stabilize the heads in their inhibited conformations (Fig. 2 B), consistent with the increase in SRX fraction. The two-heptad-long tail is too short for these interactions (Fig. 1 B), and the molecule does not show evidence of head interactions (Nag et al., 2017): the heads behave like isolated S1 in solution, with only ∼10% having the SRX rate (Fig. 1 B; Anderson et al., 2018). We conclude that the stabilizing interactions in the IHM involving the tail and the two heads increase the number of heads in the SRX conformation.
Studies of whole muscle fibers (vertebrate fast skeletal) show that when myosin molecules are in native thick filaments in their helically ordered IHM conformations (Zoghbi et al., 2008; AL-Khayat et al., 2013), the RX and SRX rates are again similar to those of S1 (Fig. 1 B), but the fraction of SRX now reaches 60–75% (Stewart et al., 2010; Cooke, 2011). A similar, greatly increased fraction of SRX heads is found in slow skeletal and cardiac muscle fibers and myofibrils (Stewart et al., 2010; Hooijman et al., 2011; McNamara et al., 2017; Nelson et al., 2020; Gollapudi et al., 2021a). In filaments, IHMs undergo intermolecular interactions that cannot occur with single molecules (Zoghbi et al., 2008; AL-Khayat et al., 2013). These include interactions between heads of different IHMs along the myosin helices (Fig. 2 C), interaction of heads with tails in the filament backbone, and interaction with myosin-binding protein C (MyBP-C) and titin, lying on the filament surface (Nag et al., 2017). We suggest that these interactions further stabilize the FHs and BHs of the IHMs, without significantly affecting their conformation, leading to the high percentage of SRX in filaments (Anderson et al., 2018).
These observations are consistent with the concept suggested earlier that the SRX rate is a consequence of a specific conformation of myosin heads, which inhibits ATP turnover, and with stabilization of this conformation by intra- and intermolecular interactions of the heads (Anderson et al., 2018). When heads are attached to each other in myosin constructs with a sufficient length of tail, forming the IHM, the inhibited conformation is stabilized by intramolecular interactions of the two heads with each other and with the myosin tail, approximately doubling (25–30%) the SRX fraction found in single heads (10–20%). When IHMs are incorporated into thick filaments, the inhibited heads are further stabilized by intermolecular interactions, with a further doubling of the SRX fraction to 60–75%. Thus, while the IHM is not a requirement for the SRX state, in real muscle, it strongly enhances it.
Are the RX and SRX rates associated with specific heads in the IHM? It is known experimentally that the BH is more stably associated with the IHM than the FH. EM images of smooth muscle heavy meromyosin (HMM; a soluble, proteolytic fragment of myosin containing the two heads and the proximal third of the tail) show molecules where the BH is attached to S2 but the FH has dissociated (Burgess et al., 2007). This and studies of tarantula thick filaments (Brito et al., 2011; Sulbarán et al., 2013) suggest that the FH can dissociate from the IHM and become mobile (“swaying”; Fig. 1 B, red double-headed arrows), with a duty ratio that defines the fraction of time spent in the dissociated state (Alamo et al., 2017). When unconstrained in this way, the FH presumably acts essentially like S1 in solution, equilibrating between its minor SRX (bent) and its predominant RX (straight) rate of ATP turnover (Fig. 1 B, reversible blue arrows). Thus, the more stable BH would account for most of the SRX rate and the dissociated FH for the faster, RX rate. When the FH is docked in the IHM, stabilized in its inhibited form by interactions with the BH and S2 in the IHM, it may slow to approximately the BH SRX rate (Fig. 1 B; Alamo et al., 2016). The BH and docked FH may have similar enough rates that they are not resolved by the two-exponential analysis; there is in fact some evidence that additional exponentials give an improved fit (Hooijman et al., 2011), but this has not been explored in detail. The idea that some heads can move freely for a portion of the time, leading to their disordering in thick filaments, and the correlation of disorder with the RX rate, has led to the concept of the “disordered relaxed state” (DRX; Wilson et al., 2014). Thus, heads in thick filaments are typically referred to as SRX or DRX.
What is the structural basis of the HRX state?
Studies of invertebrate (tarantula) striated muscle show that the SRX state in some species can be enhanced by a further (10 times) slowing of the ATP turnover rate compared with the SRX (Fig. 1) and that this occurs in up to 50% of the heads (Naber et al., 2011). We refer to this as the HRX state. The HRX state has not been observed in vertebrate striated muscle thick filaments or myosin molecules. Is there a structural explanation for the greater inhibition of ATP turnover in tarantula muscle? The cryo-EM reconstruction of tarantula filaments shows that the IHMs in this species are arranged in four coaxial helices, with crowns of four IHMs spaced 145 Å apart axially (Woodhead et al., 2005). Each IHM shows the conventional structure of two heads interacting with each other through their motor domains and with the proximal portion of the myosin tail. Importantly, the reconstruction also suggests an additional key interaction that is apparently absent in vertebrate thick filaments (see below). S2, after leaving its IHM, travels along the filament toward the bare zone, passing near the next IHM along the filament axis, through a groove formed by the BH SRC homology 3 (SH3) and converter domains (Fig. 3 A; and Fig. 4, A and B; Woodhead et al., 2005). This location suggests that S2 could sterically interfere with movement of the BH converter region of this IHM that is required for product release. It could thus act as an additional constraint on the already inhibited conformation of the BH that further inhibits ATP turnover (each BH would thus interact with two S2s, its own and that from the axially adjacent IHM). In this case, we suggest that intermolecular interaction does not simply stabilize the inhibited head conformation but creates a new and additional physical barrier to motions of the BH required for ATPase activity, leading to the highly inhibited HRX turnover rate. The reconstruction shows that only the BHs are affected in this way, which could directly explain the finding that 50% of heads are in the HRX state (Naber et al., 2011).
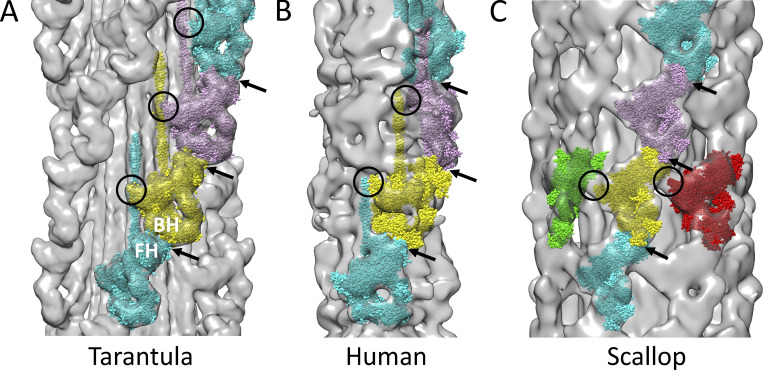
Figure 3.
Comparison of intermolecular interactions in 3-D reconstructions of different thick filaments. Reconstructions are fitted with IHM atomic models (Protein Data Bank accession nos. 3JBH or 5TBY) in each case. All thick filaments (except insect flight muscle; Hu et al., 2016) show interactions between IHMs along the helical tracks (arrows), involving the FH motor domain at one level and the BH lever arm at the next. These interactions are the same at every level in tarantula and scallop (true helical structures) but different at different levels in vertebrates, which are quasi-helical. Additional interactions vary between species. (A) Tarantula shows interaction of S2 from one crown of heads with SH3 and converter domains of the next level (circles; see Fig. 4, A and B). IHM (Protein Data Bank accession no. 3JBH) was fitted to four levels of heads in cryo-EM reconstruction (EMD accession no. 1950; gray). (B) Similar fitting of IHM (Protein Data Bank accession no. 5TBY) to vertebrate (human) cardiac negative stain reconstruction of C-zone (EMD accession no. 2240) shows well-ordered IHMs at two of every three levels of heads (strong map density for pink and cyan IHMs), while the third level (yellow) is poorly ordered (weak map density, suggesting substantial IHM mobility; Zoghbi et al., 2008; AL-Khayat et al., 2013). S2 cannot be fitted unambiguously due to low resolution of map and lack of internal detail with negative stain; within these limitations, there is no obvious S2–head interaction between crowns (circles; see Video 1; cf. Video 2 for 3-D view). (C) In scallop cryo-EM reconstruction, S2 is not resolved, but the tight azimuthal crowding of IHMs around the circumference at each crown suggests potential intermolecular interaction between the SH3 domain of a BH and the motor domain of the neighboring FH (circles). Filaments are oriented with M-line at top; their different symmetries (fourfold, threefold, and sevenfold rotational symmetry, respectively) cause the varying views of the IHMs in the different filaments. All reconstructions are at the same scale. The human has a smaller diameter due to radial shrinkage occurring during negative staining and to the smaller number of molecules (n = 3) at each level. The scallop and tarantula cryo-reconstructions also have different diameters: scallop has seven molecules at each level forming a shell above the filament backbone (Woodhead et al., 2013), while tarantula has four molecules at each level, closer to the backbone (Woodhead et al., 2005), leading to a smaller diameter. Models in this figure were created with UCSF Chimera (Pettersen et al, 2004).
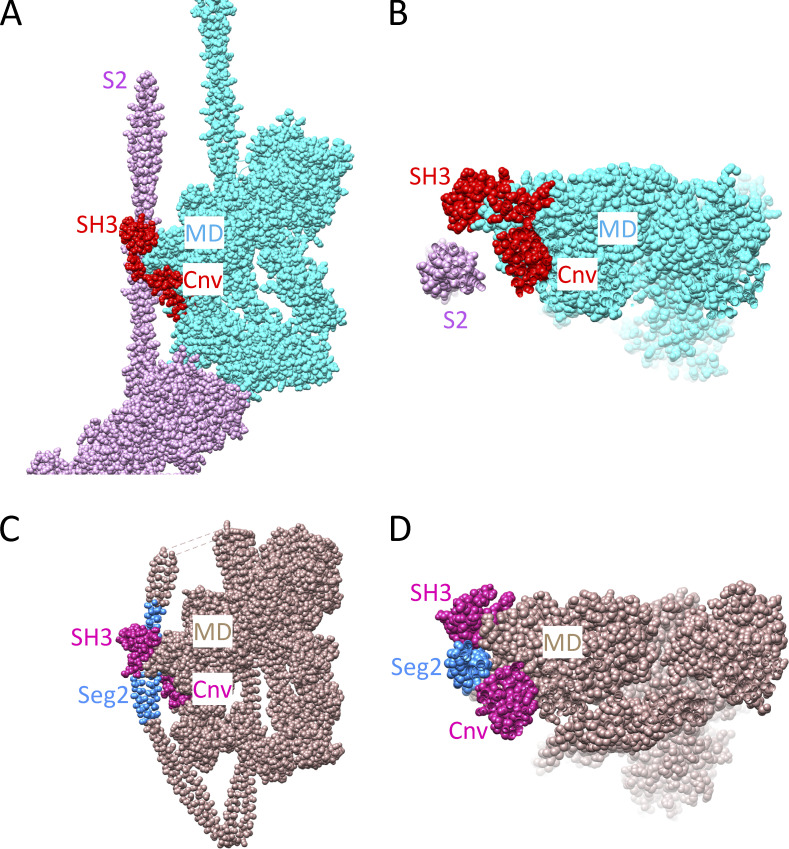
Figure 4.
Comparison of tail interactions with the SH3 and converter domains in atomic models of tarantula thick filaments and 10S smooth muscle myosin. (A and B) Tarantula thick filament (Protein Data Bank accession no. 3JBH). (C and D) 10S myosin (Protein Data Bank accession no. 6XE9). (A and C) Front views at same scale. (B and D) End views (same scale) obtained by rotating A and C 90° around the x axis so that pink IHM in A is closest to viewer (i.e., looking toward M-line). S2 in tarantula filament and segment 2 (Seg2) in 10S myosin both pass through a groove formed by the SH3 and converter (Cnv) domains of the BH motor domain (MD). The looser fit to the groove in tarantula may be due to the lower resolution of the reconstruction used to obtain the atomic model (20 Å resolution; cf. 4.3 Å for 10S myosin). Models in this figure were created with UCSF Chimera (Pettersen et al, 2004).
Several observations support this structural interpretation of the HRX state. Vertebrate thick filaments have a symmetry and organization of myosin heads that is different from those in tarantula. Although no cryo-EM reconstruction of vertebrate filaments is available, two negative stain reconstructions of the C-zone (the middle section of each filament half, containing MyBP-C) clearly show the well-known quasi-helical arrangement of myosin heads characteristic of vertebrates (Zoghbi et al., 2008; AL-Khayat et al., 2013). Fitting of the IHM into the density map reveals well-ordered IHMs at two of every three levels of heads, while the third level is relatively disordered (Zoghbi et al., 2008; AL-Khayat et al., 2013). S2 cannot be fitted with certainty due to the low resolution of the map and ambiguities of negative stain; however, there is no obvious interaction of S2 from one crown with heads in the next (Fig. 3 B; Video 1; cf. Video 2). Correspondingly, vertebrate filaments do not exhibit an HRX state. There is also no HRX state in isolated S1- or S2-headed myosin constructs forming the IHM (Anderson et al., 2018; Rohde et al., 2018). These observations are consistent with the conclusion that in tarantula, intermolecular interaction with S2 from the neighboring crown is responsible for the HRX state. Note that the “ultra-relaxed” state induced in myosin by the inhibitor blebbistatin (Gollapudi et al., 2021a) has a very different origin from the HRX—the former due to internal stabilization of switch 2 in the myosin head in the closed state, inhibiting phosphate release (Zhao et al., 2008), the latter to the external structural constraints on head movements that we have described here.
Video 1.
Human cardiac thick filament reconstruction (EMD accession no. 2240) fitted with human cardiac atomic model (Protein Data Bank accession no. 5TBY) to show apparent absence of interaction between S2 from one level of heads and the SH3 and converter domains (red) of the next level. M-line at top. See also Fig. 3 B. Compare with Video 2.
Video 2.
Tarantula thick filament reconstruction (EMD accession no. 1950) fitted with tarantula IHM atomic model (Protein Data Bank accession no. 3JBH) to show 3-D view of interaction between S2 from one level of heads and the SH3 and converter domains (red) of the next level. M-line at top. See also Fig. 3 A and Fig. 4, A and B.
Additional evidence comes from smooth muscle and nonmuscle myosin molecules, which can exist in a switched-off state in which the ATP turnover rate is similar to that in tarantula—also an HRX state (Cross et al., 1988). Strikingly, these myosins have a folded conformation, in which the two heads form an IHM, and the tail folds into three segments, the middle segment wrapping around the BH (Fig. 1 B; Suzuki et al., 1982; Trybus et al., 1982; Craig et al., 1983; Burgess et al., 2007; Yang et al., 2019). Cryo-EM analysis of this conformation shows intimate contact of the tail with the BH motor as it runs through the groove formed by the BH SH3 and converter domains (Fig. 4, C and D; Scarff et al., 2020; Yang et al., 2020). The regions of tail contact with the BH in these single molecules are very similar to those in the tarantula filament and would sterically impede BH converter movement, contributing to the HRX turnover rate of this inhibited form of the myosin molecule (Fig. 4). Although the inhibitory regions of the tail (S2 in tarantula, the middle segment of the tail in smooth muscle and nonmuscle myosin) and the nature of the interaction (intermolecular in one, intramolecular in the other) are both different, the likely inhibitory effects on BH converter movement and ATPase activity appear to be similar, supporting the importance of this interaction in generating the HRX state. HMM from these myosins lacks the distal two-thirds of the tail and therefore the interaction of the middle segment with the BH. Correspondingly, HMM exhibits an SRX but not an HRX rate (Cross et al., 1988). Together these observations provide strong support for the notion that interaction of the myosin tail with the SH3/converter region of the BH is the structural basis of hyper-relaxation.
Another invertebrate muscle in which a putative HRX state has been observed is the scallop striated adductor. When thick filaments lose their ATP, helical ordering of the heads is lost (we would suggest by straightening of the heads in the apo state, so that the IHM can no longer form). EM shows that this process takes up to 30 min in scallop (Vibert and Craig, 1985), consistent with an HRX turnover rate. Scallop thick filaments differ from tarantula in having sevenfold rather than fourfold rotational symmetry (Vibert and Craig, 1983). Cryo-EM shows that this tightly packs the IHMs around the circumference of each crown, creating intermolecular interactions within crowns involving the SH3 domain of the BH and the motor domain of its neighbor FH (Figs. 1 B and 3 C; Woodhead et al., 2013; the heads in tarantula crowns are more widely spaced and do not show these interactions). While the reconstruction does not provide detail on possible tail interactions between crowns (as in tarantula), these additional intermolecular interactions may constrain the heads within a crown, impeding structural changes of the BH and FH and accounting for the hyper-slow release of products from scallop thick filaments through a mechanism quite different from that in tarantula.
How is the IHM formed?
Formation of the IHM depends on several key properties of myosin II: flexing of the heads and the head–tail junction and interaction of the motor domains with each other and with the proximal region of S2. BH–S2 interaction (where S2 runs over the bent BH) appears to be the primary binding interaction of the IHM (Alamo et al., 2016), as it is observed in smooth muscle HMM even when the FH is not bound to the BH (Burgess et al., 2007); FH–S2 interaction without interaction of the BH and BH–FH interaction without S2 are not observed. Interaction of S2 with the BH can only occur when the BH folds back onto the tail, and then only when the BH is in the strongly bent (Rosenfeld et al., 1994; Alamo et al., 2008, 2016; Scarff et al., 2020; Yang et al., 2020), nucleotide-trapping state, putatively with the SRX rate (Anderson et al., 2018). Thus, we picture the myosin molecule as having flexible heads (in a bent–straight equilibrium biased 90% toward the straight [DRX] conformation), which are flexibly attached to the tail, with the following sequence for formation of the IHM. If a transiently bent head comes in contact with S2, it binds and is stabilized in its bent (SRX) state, thereby becoming a BH (i.e., a precursor IHM; Alamo et al., 2016). The other head, flexing around its head–tail junction and in a similar bent–straight equilibrium, can now be caught (becoming an FH) when its converter region contacts the captured BH motor domain, stabilizing the FH bent conformation (Liu et al., 2003; Alamo et al., 2016; Scarff et al., 2020; Yang et al., 2020). This interaction is strengthened by contact of loops on the FH with S2 (Alamo et al., 2008; Scarff et al., 2020; Yang et al., 2020).
Structural observations show that both heads of the IHM are strongly bent (Wendt et al., 2001; Woodhead et al., 2005; Scarff et al., 2020; Yang et al., 2020), while the bent structure in isolated heads may be uncommon. The sequence proposed above suggests how the IHM can be formed despite these constraints. If the bent structure occurs only rarely (e.g., 10% of the time), the likelihood of two bent heads coming together will be low: this likelihood is greatly increased by initial stabilization of one bent head (the BH) binding to S2 (forming the precursor IHM) and then binding of the second head (the FH) to the already bent BH of the precursor. This kinetic argument would explain why head–head interaction without involvement of the tail has not been observed, consistent with mutational studies (Adhikari et al., 2019). The final IHM is a tripartite structure with multiple weak interactions (BH–FH, BH–S2, and FH–S2) that inhibit ATPase activity as well as actin-binding capability (Scarff et al., 2020; Yang et al., 2020). These interactions are easily broken and in equilibrium with the noninteracting form. The result in isolated sarcomeric myosin soluble fragments at physiological ionic strength is an overall 25% SRX and 75% DRX rate (Anderson et al., 2018; Rohde et al., 2018; Gollapudi et al., 2021b). In filaments, individual IHMs (with the inhibited, bent structure of the individual heads) are stabilized by intermolecular interactions occurring in the polymer (as described earlier), increasing the fraction of SRX heads above 50%.
Is there a relationship of the SRX state with animal lifestyle?
We have suggested that nature uses two ways to enhance the SRX state: increase in fraction of inhibited heads and increase in degree of inhibition. The particular mechanism may be adapted to the lifestyle of the animal (Naber et al., 2011). Tarantulas spend long periods of time immobile and can survive many months without food. Minimizing ATP use during this time by hyper-relaxation of their BHs and SRX of most of their FHs would be an evolutionary advantage (Naber et al., 2011). They would nevertheless be ready for a rapid switch to the active state through the small numbers of swaying FHs that sense thin filament activation when muscle is stimulated (Brito et al., 2011). Scallops can swim quickly for short bursts by jet propulsion, using their striated adductor muscles to rapidly close their shell (e.g., to escape predators), but they remain stationary during extended periods of filter feeding (Speiser and Wilkens, 2016). In this low-activity state, the shell is held partially closed through contraction of the tonic smooth adductor muscle, which enters a catch state, maintaining force with little energy expenditure (Chantler, 2016). The striated adductor is 10 times more massive than its smooth counterpart (Naidu, 1987) and a potential metabolic drain during the long periods when the muscle is not in use: minimizing ATP use during these nonswimming periods through hyper-relaxation of their heads would be advantageous. The folded form of vertebrate smooth muscle and nonmuscle myosin is thought to serve as a storage molecule, which can be activated to form functional filaments as required through phosphorylation of their RLCs (Cross et al., 1988). The complete switching off of ATPase activity through hyper-relaxation when the molecule is not in use would again provide an evolutionary advantage.
Vertebrate striated muscle thick filaments exhibit SRX but not hyper-relaxation. Interaction of IHMs with each other or with the filament backbone, MyBP-C, or titin enhances the fraction of heads in the SRX state but does not significantly increase the level of inhibition of the heads. SRX appears to be developed most strongly in the C-zone (probably in the two crowns of heads showing IHMs in each three-crown repeat), implicating MyBP-C in this inhibition (McNamara et al., 2016, 2019; Nelson et al., 2020). Myosin heads in the D-zone and in the non-IHM crown in the C-zone are less ordered (Zoghbi et al., 2008; AL-Khayat et al., 2013; Brunello et al., 2020) and may be the main source of DRX heads detected by single turnover experiments (Nelson et al., 2020). Vertebrates may make a trade-off of greater (though still low) ATP use in the RX state (SRX with some DRX heads) for the ability to switch on instantly as needed and for fine-tuning of the contractile response through interactions with other proteins such as MyBP-C.
How are thick filaments activated from the SRX state?
We have painted a picture in which most myosin heads in the thick filaments of resting muscle are tacked down on the filament surface in helices of IHMs in an SRX (or HRX) state. However, muscles must be ready for immediate activation from this energy-saving state. The other part of the picture therefore includes a small fraction of heads dissociated from the IHM at any particular moment. These relatively few, mobile, constitutively on (“sentinel”) heads are presumed to constantly explore the interfilament space, able to instantly sense thin filament activation and bind to actin when myosin-binding sites are exposed (by Ca2+-induced tropomyosin movement; Gordon et al., 2000), leading to initial tension development (Linari et al., 2015; Irving, 2017). We suggest that these are the transiently dissociated (swaying) FHs of the IHMs. In vertebrates, the sentinel heads could also include the less well-ordered heads at every third crown of the C-zone (Zoghbi et al., 2008; AL-Khayat et al., 2013) or the disordered heads in the D-zone (Brunello et al., 2020; Nelson et al., 2020). This essential role for a small number of mobile heads as the trigger for thick filament activity, when thin filaments are switched on, is paid for by the increase in ATP consumption over that used by SRX heads. What happens next depends on the type of muscle.
In tarantula, the small amount of thick filament activity in an initial twitch appears to be enhanced in additional twitches by Ca2+-induced activation of myosin light chain kinase, which phosphorylates the FH RLCs, leading to increased release of the FHs from the IHMs, reduction in the HRX fraction and increase in the DRX fraction (Naber et al., 2011), and increased force production (Padrón et al., 2020). Prolonged high Ca2+ (e.g., in a tetanus) subsequently phosphorylates the BHs, resulting in their release and further enhancement of force (Padrón et al., 2020).
Vertebrates have a finely tuned, graded response to activation. X-ray studies show that low-force isotonic contractions use only a small fraction of available heads (Linari et al., 2015). The majority remain helically ordered in their IHM configurations, continuing to save energy—even during contraction—representing a highly efficient use of ATP. As force increases, stress on the thick filaments rises, resulting in release of more heads, which produce greater force, in a positive feedback loop (Linari et al., 2015; Irving, 2017). In this mechanosensing model for thick filament activation, the filament is stretched by ∼1%, which may be sufficient to weaken intermolecular contacts between heads along the helical tracks that help to maintain the IHM conformations (Irving, 2017). Destabilization of the IHMs by stress could thus release the additional heads needed for strong force production. When contraction is switched off by removal of Ca2+ from the cytosol, IHMs rapidly go back to their helically ordered arrangement (Linari et al., 2015; cf. Ma et al., 2019), quickly returning the muscle to its energy-saving SRX state.
In scallop, the thick filaments are directly activated by Ca2+ binding to the essential light chains (ELCs) on the myosin heads rather than through Ca2+ activation of the thin filaments (Szent-Györgyi et al., 1999; Chantler, 2016). Ca2+ binding causes breakage of the IHM intra- and intermolecular interactions, cooperatively releasing heads from their SRX/HRX state (Szent-Györgyi et al., 1999; Zhao and Craig, 2003; Jung et al., 2008; Chantler, 2016). Released Ca2+-activated heads can move freely, similarly to phosphorylated heads of tarantula, interacting with actin to generate force. As this process involves binding of Ca2+ to the myosin heads and not the slower, enzymatic phosphorylation of the light chains, full activation can occur rapidly (Zhao and Craig, 2003), making all heads immediately available for the powerful twitches that produce the strong swimming motions of this species. In the absence of a thin filament switch in scallops, sentinel heads may not be required as the heads directly sense Ca2+ activation.
Interactions between IHMs along their helical tracks are common to all thick filaments in the SRX/HRX state. Reconstructions of tarantula, scallop, and vertebrate thick filaments, representing three distinct mechanisms of activation, all show such interactions, typically between the FH motor domain of one IHM and the BH regulatory domain of its neighbor in the next crown closer to the filament center (Figs. 2 C and 3; Woodhead et al., 2005; Zoghbi et al., 2008; AL-Khayat et al., 2013; Woodhead et al., 2013). As described earlier, these intermolecular interactions appear to stabilize the IHM conformation, enhancing the fraction of SRX heads in filaments. Concomitantly, they may also underlie rapid thick filament activation, as they provide a direct physical path for cooperative disruption of the IHM and thus exit from the SRX state (Stewart et al., 2010; McNamara et al., 2015). Cooperative exit from the SRX state has been experimentally demonstrated in skeletal and cardiac muscle incubated with mantATP chased by ADP (Stewart et al., 2010; Cooke, 2011; Gollapudi et al., 2021b). Cooperative activation is especially well developed in scallop thick filaments (Szent-Györgyi et al., 1999; Chantler, 2016), where it may underlie the rapid Ca2+ activation leading to the strong swimming motions of this species. The intermolecular interactions along helices and around each crown in scallops (Fig. 3 C; Woodhead et al., 2013) connect all heads in an extensive network that could be rapidly disrupted by Ca2+ binding.
How is SRX modulated?
The stability of the IHM and the level of SRX can be modulated in several ways, including myosin RLC phosphorylation and, in vertebrates, phosphorylation of MyBP-C (Stewart et al., 2010; Nag and Trivedi, 2021). RLC phosphorylation in tarantula greatly reduces the fraction of SRX and HRX heads and their ATP turnover times (Naber et al., 2011) while increasing DRX heads. This correlates with a decrease in helical ordering and extension of heads from the filament backbone (suggesting disruption of the IHM), as demonstrated by x-ray diffraction (Padrón et al., 2020) and EM (Craig et al., 1987). Phosphorylation not only activates heads but also maintains a memory of activation following the extensive phosphorylation that occurs in a tetanus; this can greatly potentiate subsequent contraction (Padrón et al., 2020). Disruption of the IHM and extension of heads toward neighboring actin filaments in the RX period following a tetanus presumably enables the stronger and more rapid interaction with actin of a post-tetanic contraction (Padrón et al., 2020). While the phosphorylated RX state that follows a tetanus would temporarily consume more ATP, it could provide a survival benefit by enabling stronger contractions when escaping predators or capturing prey. Following such periods of activity, the RLCs again become dephosphorylated, and the energy-saving SRX/HRX state, with ordered, interconnected IHMs lying along the filament surface, returns (Padrón et al., 2020), characterizing the long periods of inactivity in the life of the tarantula, when ATP savings are critical.
Vertebrate skeletal muscle RLCs can also be phosphorylated, and phosphorylation correlates with post-tetanic potentiation (Sweeney et al., 1993) together with disordering of myosin helices and extension of heads from the filament surface (Levine et al., 1996; Yamaguchi et al., 2016), again suggestive of IHM disruption. As with tarantula, a corresponding reduction in the SRX state with phosphorylation (Stewart et al., 2010; Cooke, 2011; Gollapudi et al., 2021b), with a temporarily greater resting ATP consumption, pays for the greater contractility available following phosphorylation.
Vertebrates have a second means of modulating the SRX state, which may provide finer control of thick filament activation/relaxation than with invertebrates. MyBP-C binds to myosin in the middle one-third of each half of the thick filament (the C-zone; Craig and Offer, 1976; Flashman et al., 2004; Luther et al., 2008), and several lines of evidence demonstrate that the SRX state is more pronounced in these regions (McNamara et al., 2016, 2019; Nelson et al., 2020; Nag and Trivedi, 2021), reaching as high as 90% (Nelson et al., 2020); this correlates with the clearest delineation of IHMs in reconstructions of the thick filament C-zone (Zoghbi et al., 2008; AL-Khayat et al., 2013). Toward the tips of the filament (the distal or D-zone), heads are less ordered (Brunello et al., 2020; R. Craig, unpublished EM data), and the SRX state is diminished (Nelson et al., 2020). In MyBP-C knockout mice, the IHM configuration is weakened or abolished (Zoghbi et al., 2008), and the SRX state is disrupted (McNamara et al., 2016). Thus MyBP-C appears to enhance the SRX state, apparently by stabilizing the IHM. This stabilization is further modulated by phosphorylation of MyBP-C, occurring in the heart in response to β-adrenergic stimulation. Enhancement of cardiac contractility by cardiac MyBP-C (cMyBP-C) phosphorylation may result in part from depression of cMyBP-C’s stabilizing effect on the SRX, which coincides with weakening of the IHM (Kensler et al., 2017; Caremani et al., 2019b; Irving and Craig, 2019; McNamara et al., 2019). These data overall imply that MyBP-C enhances energy saving by stabilizing the IHM structure and that this is modulated in the heart by cMyBP-C phosphorylation (McNamara et al., 2019).
Importantly, in the healthy heart, RLC and MyBP-C phosphorylation are not zero but ∼50% (Chang et al., 2015) and ∼60% of maximum (Previs et al., 2012), respectively. This would suggest a partial weakening of the SRX state (compared with zero phosphorylation) during normal cardiac activity, which may poise myosin heads optimally between sequestration in the IHM (to save energy) and availability for interaction with actin to generate force, with fine-tuning of these levels available upon further phosphorylation. Thus, we assume that the level of SRX of a muscle (degree of IHM formation) is tuned to the physiological needs of the moment, with the goal over time of minimizing energy consumption within these limits. Strikingly, phosphorylation levels of MyBP-C and RLC can both decrease in heart failure (El-Armouche et al., 2007; Toepfer et al., 2013), which would predict a higher fraction of SRX heads. This may reduce the need for energy under these adverse circumstances but may also contribute to the compromised contractility of the failing heart.
Animals can save energy when food supplies are scarce or weather conditions adverse by a reduction in body temperature. This occurs naturally in ectotherms (cold-blooded animals) when exterior temperatures drop and by hibernation in some endotherms (warm-blooded animals), where body temperature and metabolism reduce to low values. Does enhanced SRX play a role in energy conservation under these circumstances? X-ray diffraction of both mammalian and tarantula muscle at low temperatures (10°C) suggests that the number of myosin motors in the helically ordered, IHM conformation decreases substantially compared with temperatures nearer physiological levels (Malinchik et al., 1997; Xu et al., 1997; Caremani et al., 2019a, 2021; Ma et al., 2021a); modeling of tarantula suggests that it is specifically the FHs that are disordered, while the BHs remain ordered (Ma et al., 2021a). This disordering suggests that the SRX state may actually be reduced, rather than enhanced, in cold temperatures. Other factors may contribute to energy conservation by muscle under cold conditions (Caremani et al., 2021; Ma et al., 2021a): myosin heads become refractory to actin binding at low temperature (Caremani et al., 2019a), and the disordered FHs, containing ADP.Pi at the active site, may transition toward the ATP conformation, inhibiting ATP turnover (Xu et al., 1999; Ma et al., 2021a). The impact of torpor (a form of hibernation) on the SRX state has recently been explicitly studied in the 13-lined ground squirrel (Ictidomys tridecemlineatus; Toepfer et al., 2020). During torpor, core body temperature drops to 5°C, metabolic rate to 3% of basal levels, and heart rate from 311 to 6 beats per min; hummingbirds undergo a similar dramatic reduction in metabolic rate to save energy each night (Shankar et al., 2020). The fraction of SRX heads found in cardiac muscle removed from the ground squirrel during torpor was reported to increase from 65 to 75%, contrary to expectation from the low-temperature x-ray studies of mammalian muscle described above that would imply a decrease in the number of IHMs. This discrepancy could be due to performance of the SRX measurements at 21°C (Toepfer et al., 2020) rather than the low temperatures used in the x-ray experiments that revealed helical disordering. The latter would presumably reflect the thick filament structure most accurately at actual torpor temperatures. We speculate that the phenomenon of iguanas (ectotherms) falling from trees and becoming immobile when environmental temperatures are abnormally low (Stroud et al., 2020) may be caused in part by the refractory impact of temperature on the ability of their myosin heads to bind to actin; a similar effect may have contributed to extinction of the dinosaurs during the global cooling that followed the asteroid impact of 66 million years ago (Vellekoop et al., 2014).
Modulation of the SRX and its putative structural correlate, the IHM, may play a critical role in a number of cardiac diseases and their treatment, summarized elsewhere (Alamo et al., 2017; Nag et al., 2017; Yotti et al., 2019; Trivedi et al., 2020; Daniels et al., 2021; Schmid and Toepfer, 2021). Hypertrophic cardiomyopathy (HCM) is an inherited disease caused by mutations in sarcomeric proteins, including myosin, and characterized by hypercontractility and the inability to fully relax during diastole (Ashrafian et al., 2011). Recent studies show that mutations in the myosin heavy chain (MYH7; accounting for ∼40% of HCM cases) strongly cluster in regions of the myosin molecule involved in the interfaces of the IHM (Alamo et al., 2017; Nag et al., 2017), and lead to a substantial decrease in SRX and increased energy use (Anderson et al., 2018; Adhikari et al., 2019; Sarkar et al., 2020). Disruption of the IHM by such mutations, at the head–head or BH–S2 interface, could release more heads for interaction with actin, accounting for the hypercontractility and impaired relaxation observed (Alamo et al., 2017; Anderson et al., 2018; Spudich, 2019; Sarkar et al., 2020). Experimental evidence for this proposal has been obtained (Adhikari et al., 2019; Sarkar et al., 2020). Mutations in MyBP-C leading to HCM also appear to partially disrupt the SRX state of the myosin heads (Toepfer et al., 2019; Nag and Trivedi, 2021), leading to an increase in the number of DRX heads, which may contribute to the observed hypercontractility (McNamara et al., 2017). Depending on the mutation, cMyBP-C may have a reduced affinity for myosin, or the amount of MyBP-C incorporated into the thick filament may be reduced. It has been proposed that either mechanism would reduce the strength of the cMyBP-C–myosin interaction, releasing heads from the filament backbone (DRX heads), which could be partially responsible for the hypercontractile phenotype observed in patients with these mutations (Colson et al., 2007; Toepfer et al., 2019; Nag and Trivedi, 2021).
Impaired relaxation due to HCM mutations in myosin (and other myofibrillar proteins) can be compensated by a recently developed drug, mavacamten, which has been shown to increase the fraction of myosin heads in the SRX state (Anderson et al., 2018; Rohde et al., 2018; Spudich, 2019; Nag and Trivedi, 2021), specifically in the D-zone of thick filaments (Nelson et al., 2020), concomitant with an increased number of molecules folded into the IHM conformation (Anderson et al., 2018; Rohde et al., 2018; Gollapudi et al., 2021b). In thick filaments of intact cardiac muscle, mavacamten increases the degree of quasi-helical ordering of myosin heads, consistent with an increase in the stability or fraction of molecules in the IHM conformation (Anderson et al., 2018), supporting our overall contention that the IHM is the main basis of the SRX state in muscle. Mavacamten inhibits the ATPase activity of isolated S1, specifically by inhibiting phosphate release (Green et al., 2016), suggesting that it enhances the bent (SRX) state of the myosin head, which, based on our earlier reasoning, would lead to the observed increase in the fraction of molecules in the IHM (Anderson et al., 2018).
Future directions
We currently know the structure of the IHM at ∼15 Å resolution in tarantula (Yang et al., 2016) and insect (Hu et al., 2016) filaments and ∼4 Å resolution in single smooth muscle myosin molecules (Scarff et al., 2020; Yang et al., 2020). Improvements in resolution should enhance visualization of the side-chain interactions that stabilize the SRX state in both filament and monomer. For filaments, cryo-EM of tarantula offers the greatest potential, owing to the stability of its helices. To better understand how mutations in the IHM interfaces lead to the hypercontractility of HCM, cryo-EM of vertebrate cardiac thick filaments is required to improve the resolution beyond the current ∼40 Å (obtained with negative staining; Zoghbi et al., 2008; AL-Khayat et al., 2013), a difficult task owing to the lability and quasi-helical symmetry of the vertebrate thick filament head array. Differences in the levels of SRX in the C- and D-zones of the thick filaments (Nelson et al., 2020) suggest differences in IHM stability or interactions, which will need to be analyzed by cryo-EM, again a significant task for the reasons stated above and because of the small size of these zones. The most detailed insights into IHM intramolecular interactions should come from a high-resolution structure of the IHM in isolated cardiac myosin molecules, which is urgently needed. This could potentially be obtained by x-ray crystallography or cryo-EM of IHM constructs (e.g., 25 heptad; Fig. 1; Anderson et al., 2018), although this is likely to be hampered by the relative instability of the structure. Incubation with mavacamten may improve its stability (Anderson et al., 2018). How interaction of MyBP-C with myosin enhances the SRX state is another fertile but challenging area of investigation, which may require cryo-electron tomography of thick filaments or intact myofibrils (Burbaum et al., 2021) or single particle cryo-EM studies of MyBP-C–IHM complexes. Experiments suggest that thick filaments in muscle are in equilibrium with a pool of myosin monomer, which may play a role in thick filament assembly/disassembly during development, hypertrophy, and myosin turnover (Saad et al., 1986; Katoh et al., 1998; Ojima, 2019). Myosin monomers at physiological ionic strength form IHMs with a folded tail structure and slow ATP turnover rate, which may act as a transport form from ribosome to filament (Ankrett et al., 1991; Katoh et al., 1998; Jung et al., 2008). Whether the SRX/IHM structure in thick filaments affects the monomer–polymer equilibrium is an area for future investigation.
Conclusion
The SRX state of thick filaments plays a crucial role in the energy balance of muscle and is ubiquitous across the animal kingdom. It results from a conformation of the myosin head in which ATP turnover is strongly inhibited, minimizing ATP use. This conformation is stabilized by intramolecular interactions when it is incorporated into the IHM and by additional intermolecular interactions when IHMs are assembled into thick filaments, increasing the fraction of energy-saving SRX heads. Thus, we propose that IHMs, helically organized along the thick filament surface in the relaxed state, are the major basis of SRX in living muscle. An HRX state, found in thick filaments of some invertebrates and in the folded (storage) form of smooth muscle and nonmuscle myosin molecules, is also based on the IHM but in these cases results from additional intra/intermolecular interactions. A filament in which every head was in SRX, while maximally saving ATP, would not be useful in contraction: at any time, a small fraction of heads is dissociated (DRX or sentinel heads) and available to sense thin filament activation and generate initial force. The SRX state is down-regulated in situ by RLC and MyBP-C phosphorylation in response to physiological requirements (activation), which disrupt the IHM, releasing heads for interaction with actin. Mutations in myosin or MyBP-C causing HCM disrupt the SRX state, causing hypercontractility, which can be reversed by drugs that stabilize the IHM and thus the SRX.
Online supplemental material
Video 1 shows human cardiac thick filament reconstruction fitted with a human cardiac atomic model to demonstrate an apparent absence of interaction between S2 from one level of heads and the SH3 and converter domains of the next level. Video 2 shows tarantula thick filament reconstruction fitted with a tarantula IHM atomic model to show that in this species there is an interaction between S2 from one level of heads and the SH3 and converter domains of the next level.
Acknowledgments
Henk L. Granzier served as editor.
We thank Dr. Suman Nag for his comments on the manuscript and Dr. Kevin Stokesbury for his advice regarding scallop behavior. Molecular graphics images were produced using UCSF Chimera from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was supported by National Institutes of Health grants AR072036, AR067279, and HL139883. UCSF Chimera is supported by National Institutes of Health grant P41 RR001081.
The authors declare no competing financial interests.
Author contributions: R. Craig and R. Padrón reviewed the literature, analyzed the data, and wrote the paper.
References
- Adhikari, A.S., Trivedi D.V., Sarkar S.S., Song D., Kooiker K.B., Bernstein D., Spudich J.A., and Ruppel K.M.. 2019. β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 10:2685. 10.1038/s41467-019-10555-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Wriggers W., Pinto A., Bártoli F., Salazar L., Zhao F.Q., Craig R., and Padrón R.. 2008. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J. Mol. Biol. 384:780–797. 10.1016/j.jmb.2008.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Qi D., Wriggers W., Pinto A., Zhu J., Bilbao A., Gillilan R.E., Hu S., and Padrón R.. 2016. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J. Mol. Biol. 428:1142–1164. 10.1016/j.jmb.2016.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Ware J.S., Pinto A., Gillilan R.E., Seidman J.G., Seidman C.E., and Padrón R.. 2017. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 6:e24634. 10.7554/eLife.24634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AL-Khayat, H.A., Kensler R.W., Squire J.M., Marston S.B., and Morris E.P.. 2013. Atomic model of the human cardiac muscle myosin filament. Proc. Natl. Acad. Sci. USA. 110:318–323. 10.1073/pnas.1212708110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, R.L., Trivedi D.V., Sarkar S.S., Henze M., Ma W., Gong H., Rogers C.S., Gorham J.M., Wong F.L., Morck M.M., et al. 2018. Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. USA. 115:E8143–E8152. 10.1073/pnas.1809540115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankrett, R.J., Rowe A.J., Cross R.A., Kendrick-Jones J., and Bagshaw C.R.. 1991. A folded (10 S) conformer of myosin from a striated muscle and its implications for regulation of ATPase activity. J. Mol. Biol. 217:323–335. 10.1016/0022-2836(91)90546-I [DOI] [PubMed] [Google Scholar]
- Ashrafian, H., McKenna W.J., and Watkins H.. 2011. Disease pathways and novel therapeutic targets in hypertrophic cardiomyopathy. Circ. Res. 109:86–96. 10.1161/CIRCRESAHA.111.242974 [DOI] [PubMed] [Google Scholar]
- Brito, R., Alamo L., Lundberg U., Guerrero J.R., Pinto A., Sulbarán G., Gawinowicz M.A., Craig R., and Padrón R.. 2011. A molecular model of phosphorylation-based activation and potentiation of tarantula muscle thick filaments. J. Mol. Biol. 414:44–61. 10.1016/j.jmb.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunello, E., Fusi L., Ghisleni A., Park-Holohan S.J., Ovejero J.G., Narayanan T., and Irving M.. 2020. Myosin filament-based regulation of the dynamics of contraction in heart muscle. Proc. Natl. Acad. Sci. USA. 117:8177–8186. 10.1073/pnas.1920632117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbaum, L., Schneider J., Scholze S., Böttcher R.T., Baumeister W., Schwille P., Plitzko J.M., and Jasnin M.. 2021. Molecular-scale visualization of sarcomere contraction within native cardiomyocytes. Nat. Commun. 12:4086. 10.1038/s41467-021-24049-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S.A., Yu S., Walker M.L., Hawkins R.J., Chalovich J.M., and Knight P.J.. 2007. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol. 372:1165–1178. 10.1016/j.jmb.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Caremani, M., Brunello E., Linari M., Fusi L., Irving T.C., Gore D., Piazzesi G., Irving M., Lombardi V., and Reconditi M.. 2019a. Low temperature traps myosin motors of mammalian muscle in a refractory state that prevents activation. J. Gen. Physiol. 151:1272–1286. 10.1085/jgp.201912424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caremani, M., Pinzauti F., Powers J.D., Governali S., Narayanan T., Stienen G.J.M., Reconditi M., Linari M., Lombardi V., and Piazzesi G.. 2019b. Inotropic interventions do not change the resting state of myosin motors during cardiac diastole. J. Gen. Physiol. 151:53–65. 10.1085/jgp.201812196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caremani, M., Fusi L., Linari M., Reconditi M., Piazzesi G., Irving T.C., Narayanan T., Irving M., Lombardi V., and Brunello E.. 2021. Dependence of thick filament structure in relaxed mammalian skeletal muscle on temperature and interfilament spacing. J. Gen. Physiol. 153:e202012713. 10.1085/jgp.202012713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A.N., Battiprolu P.K., Cowley P.M., Chen G., Gerard R.D., Pinto J.R., Hill J.A., Baker A.J., Kamm K.E., and Stull J.T.. 2015. Constitutive phosphorylation of cardiac myosin regulatory light chain in vivo. J. Biol. Chem. 290:10703–10716. 10.1074/jbc.M115.642165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantler, P.D. 2016. Scallop adductor muscles: structure and function. In Scallops: Biology, Ecology, Aquaculture, and Fisheries. Shumway S.E., and Parsons G.J., editors. Elsevier, Amsterdam. 161–218. 10.1016/B978-0-444-62710-0.00004-3 [DOI] [Google Scholar]
- Colson, B.A., Bekyarova T., Fitzsimons D.P., Irving T.C., and Moss R.L.. 2007. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J. Mol. Biol. 367:36–41. 10.1016/j.jmb.2006.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, R. 2011. The role of the myosin ATPase activity in adaptive thermogenesis by skeletal muscle. Biophys. Rev. 3:33–45. 10.1007/s12551-011-0044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, R., and Offer G.. 1976. The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond. B Biol. Sci. 192:451–461. 10.1098/rspb.1976.0023 [DOI] [PubMed] [Google Scholar]
- Craig, R., Smith R., and Kendrick-Jones J.. 1983. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 302:436–439. 10.1038/302436a0 [DOI] [PubMed] [Google Scholar]
- Craig, R., Padrón R., and Kendrick-Jones J.. 1987. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J. Cell Biol. 105:1319–1327. 10.1083/jcb.105.3.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, R.A., Jackson A.P., Citi S., Kendrick-Jones J., and Bagshaw C.R.. 1988. Active site trapping of nucleotide by smooth and non-muscle myosins. J. Mol. Biol. 203:173–181. 10.1016/0022-2836(88)90100-3 [DOI] [PubMed] [Google Scholar]
- Daniels, M.J., Fusi L., Semsarian C., and Naidu S.S.. 2021. Myosin modulation in hypertrophic cardiomyopathy and systolic heart failure: getting inside the engine. Circulation. 144:759–762. 10.1161/CIRCULATIONAHA.121.056324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Armouche, A., Pohlmann L., Schlossarek S., Starbatty J., Yeh Y.H., Nattel S., Dobrev D., Eschenhagen T., and Carrier L.. 2007. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J. Mol. Cell. Cardiol. 43:223–229. 10.1016/j.yjmcc.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Ferenczi, M.A., Homsher E., Simmons R.M., and Trentham D.R.. 1978. Reaction mechanism of the magnesium ion-dependent adenosine triphosphatase of frog muscle myosin and subfragment 1. Biochem. J. 171:165–175. 10.1042/bj1710165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman, E., Redwood C., Moolman-Smook J., and Watkins H.. 2004. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 94:1279–1289. 10.1161/01.RES.0000127175.21818.C2 [DOI] [PubMed] [Google Scholar]
- Fusi, L., Huang Z., and Irving M.. 2015. The conformation of myosin heads in relaxed skeletal muscle: implications for myosin-based regulation. Biophys. J. 109:783–792. 10.1016/j.bpj.2015.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapudi, S.K., Ma W., Chakravarthy S., Combs A.C., Sa N., Langer S., Irving T.C., and Nag S.. 2021a. Two classes of myosin inhibitors, blebbistatin, and mavacamten, stabilize β-cardiac myosin in different structural and functional states. J. Mol. Biol. 433:167295. 10.1016/j.jmb.2021.167295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapudi, S.K., Yu M., Gan Q.F., and Nag S.. 2021b. Synthetic thick filaments: a new avenue for better understanding the myosin super-relaxed state in healthy, diseased, and mavacamten-treated cardiac systems. J. Biol. Chem. 296:100114. 10.1074/jbc.RA120.016506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, A.M., Homsher E., and Regnier M.. 2000. Regulation of contraction in striated muscle. Physiol. Rev. 80:853–924. 10.1152/physrev.2000.80.2.853 [DOI] [PubMed] [Google Scholar]
- Green, E.M., Wakimoto H., Anderson R.L., Evanchik M.J., Gorham J.M., Harrison B.C., Henze M., Kawas R., Oslob J.D., Rodriguez H.M., et al. 2016. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 351:617–621. 10.1126/science.aad3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijman, P., Stewart M.A., and Cooke R.. 2011. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys. J. 100:1969–1976. 10.1016/j.bpj.2011.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z., Taylor D.W., Reedy M.K., Edwards R.J., and Taylor K.A.. 2016. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Å resolution. Sci. Adv. 2:e1600058. 10.1126/sciadv.1600058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving, M. 2017. Regulation of contraction by the thick filaments in skeletal muscle. Biophys. J. 113:2579–2594. 10.1016/j.bpj.2017.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving, T.C., and Craig R.. 2019. Getting into the thick (and thin) of it. J. Gen. Physiol. 151:610–613. 10.1085/jgp.201812307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, I., Heymsfield S.B., Wang Z.M., and Ross R.. 2000. Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr. J Appl Physiol (1985). 89:81–88. 10.1152/jappl.2000.89.1.81 [DOI] [PubMed] [Google Scholar]
- Jung, H.S., Komatsu S., Ikebe M., and Craig R.. 2008. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell. 19:3234–3242. 10.1091/mbc.e08-02-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, T., Konishi K., and Yazawa M.. 1998. Skeletal muscle myosin monomer in equilibrium with filaments forms a folded conformation. J. Biol. Chem. 273:11436–11439. 10.1074/jbc.273.19.11436 [DOI] [PubMed] [Google Scholar]
- Kensler, R.W., Craig R., and Moss R.L.. 2017. Phosphorylation of cardiac myosin binding protein C releases myosin heads from the surface of cardiac thick filaments. Proc. Natl. Acad. Sci. USA. 114:E1355–E1364. 10.1073/pnas.1614020114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, R.J., Kensler R.W., Yang Z., Stull J.T., and Sweeney H.L.. 1996. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys. J. 71:898–907. 10.1016/S0006-3495(96)79293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linari, M., Brunello E., Reconditi M., Fusi L., Caremani M., Narayanan T., Piazzesi G., Lombardi V., and Irving M.. 2015. Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature. 528:276–279. 10.1038/nature15727 [DOI] [PubMed] [Google Scholar]
- Liu, J., Wendt T., Taylor D., and Taylor K.. 2003. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J. Mol. Biol. 329:963–972. 10.1016/S0022-2836(03)00516-3 [DOI] [PubMed] [Google Scholar]
- Luther, P.K., Bennett P.M., Knupp C., Craig R., Padrón R., Harris S.P., Patel J., and Moss R.L.. 2008. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J. Mol. Biol. 384:60–72. 10.1016/j.jmb.2008.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W., Gong H., Kiss B., Lee E.J., Granzier H., and Irving T.. 2019. Response to: Thick filament length changes in muscle have both elastic and structural components. Biophys. J. 116:985–986. 10.1016/j.bpj.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W., Duno-Miranda S., Irving T., Craig R., and Padrón R.. 2021a. Relaxed tarantula skeletal muscle has two ATP energy-saving mechanisms. J. Gen. Physiol. 153:e202012780. 10.1085/jgp.202012780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W., Henze M., Anderson R.L., Gong H., Wong F.L., Del Rio C.L., and Irving T.. 2021b. The super-relaxed state and length dependent activation in porcine myocardium. Circ. Res. 129:617–630. 10.1161/CIRCRESAHA.120.318647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinchik, S., Xu S., and Yu L.C.. 1997. Temperature-induced structural changes in the myosin thick filament of skinned rabbit psoas muscle. Biophys. J. 73:2304–2312. 10.1016/S0006-3495(97)78262-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Dos Remedios C.G., and Cooke R.. 2015. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 7:5–14. 10.1007/s12551-014-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Smith N.J., Lal S., Graham R.M., Kooiker K.B., van Dijk S.J., Remedios C.G.D., Harris S.P., and Cooke R.. 2016. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J. Mol. Cell. Cardiol. 94:65–71. 10.1016/j.yjmcc.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Lal S., Bos J.M., Harris S.P., van der Velden J., Ackerman M.J., Cooke R., and Dos Remedios C.G.. 2017. MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One. 12:e0180064. 10.1371/journal.pone.0180064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Singh R.R., and Sadayappan S.. 2019. Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc. Natl. Acad. Sci. USA. 116:11731–11736. 10.1073/pnas.1821660116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburgh, K.H., Franks-Skiba K., and Cooke R.. 1995. Nucleotide turnover rate measured in fully relaxed rabbit skeletal muscle myofibrils. J. Gen. Physiol. 106:957–973. 10.1085/jgp.106.5.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naber, N., Cooke R., and Pate E.. 2011. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J. Mol. Biol. 411:943–950. 10.1016/j.jmb.2011.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S., and Trivedi D.V.. 2021. To lie or not to lie: super-relaxing with myosins. eLife. 10:e63703. 10.7554/eLife.63703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S., Trivedi D.V., Sarkar S.S., Adhikari A.S., Sunitha M.S., Sutton S., Ruppel K.M., and Spudich J.A.. 2017. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24:525–533. 10.1038/nsmb.3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu, K.S. 1987. Efficiency of meat recovery from Iceland scallops (Chlamys islandica) and sea scallops (Placopecten magellanicus) in the Canadian offshore fishery. J. Northw. Atl. Fish. Sci. 7:131–136. 10.2960/J.v7.a15 [DOI] [Google Scholar]
- Nelson, S.R., Li A., Beck-Previs S., Kennedy G.G., and Warshaw D.M.. 2020. Imaging ATP consumption in resting skeletal muscle: one molecule at a time. Biophys. J. 119:1050–1055. 10.1016/j.bpj.2020.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojima, K. 2019. Myosin: formation and maintenance of thick filaments. Anim. Sci. J. 90:801–807. 10.1111/asj.13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrón, R., Ma W., Duno-Miranda S., Koubassova N., Lee K.H., Pinto A., Alamo L., Bolaños P., Tsaturyan A., Irving T., et al. 2020. The myosin interacting-heads motif present in live tarantula muscle explains tetanic and posttetanic phosphorylation mechanisms. Proc. Natl. Acad. Sci. USA. 117:11865–11874. 10.1073/pnas.1921312117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Phung, L.A., Foster A.D., Miller M.S., Lowe D.A., and Thomas D.D.. 2020. Super-relaxed state of myosin in human skeletal muscle is fiber-type dependent. Am. J. Physiol. Cell Physiol. 319:C1158–C1162. 10.1152/ajpcell.00396.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, A., Sánchez F., Alamo L., and Padrón R.. 2012. The myosin interacting-heads motif is present in the relaxed thick filament of the striated muscle of scorpion. J. Struct. Biol. 180:469–478. 10.1016/j.jsb.2012.08.010 [DOI] [PubMed] [Google Scholar]
- Previs, M.J., Beck Previs S., Gulick J., Robbins J., and Warshaw D.M.. 2012. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 337:1215–1218. 10.1126/science.1223602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, J.A., Roopnarine O., Thomas D.D., and Muretta J.M.. 2018. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc. Natl. Acad. Sci. USA. 115:E7486–E7494. 10.1073/pnas.1720342115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, S.S., Xing J., Rener B., Lebowitz J., Kar S., and Cheung H.C.. 1994. Structural and kinetic studies of the 10 S ↔ 6 S transition in smooth muscle myosin. J. Biol. Chem. 269:30187–30194. 10.1016/S0021-9258(18)43795-7 [DOI] [PubMed] [Google Scholar]
- Saad, A.D., Pardee J.D., and Fischman D.A.. 1986. Dynamic exchange of myosin molecules between thick filaments. Proc. Natl. Acad. Sci. USA. 83:9483–9487. 10.1073/pnas.83.24.9483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S.S., Trivedi D.V., Morck M.M., Adhikari A.S., Pasha S.N., Ruppel K.M., and Spudich J.A.. 2020. The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin. Sci. Adv. 6:eaax0069. 10.1126/sciadv.aax0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarff, C.A., Carrington G., Casas-Mao D., Chalovich J.M., Knight P.J., Ranson N.A., and Peckham M.. 2020. Structure of the shutdown state of myosin-2. Nature. 588:515–520. 10.1038/s41586-020-2990-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., and Toepfer C.N.. 2021. Cardiac myosin super relaxation (SRX): a perspective on fundamental biology, human disease and therapeutics. Biol. Open. 10:bio057646. 10.1242/bio.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar, A., Schroeder R.J., Wethington S.M., Graham C.H., and Powers D.R.. 2020. Hummingbird torpor in context: duration, more than temperature, is the key to nighttime energy savings. J. Avian Biol. 51:e02305. 10.1111/jav.02305 [DOI] [Google Scholar]
- Speiser, D.I., and Wilkens L.A.. 2016. Neurobiology and behaviour of the scallop. In Scallops: Biology, Ecology, Aquaculture, and Fisheries. Shumway S.E., and Parsons G.J., editors. Elsevier, Amsterdam. 219–251. 10.1016/B978-0-444-62710-0.00005-5 [DOI] [Google Scholar]
- Spudich, J.A. 2019. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflugers Arch. 471:701–717. 10.1007/s00424-019-02259-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, M.A., Franks-Skiba K., Chen S., and Cooke R.. 2010. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc. Natl. Acad. Sci. USA. 107:430–435. 10.1073/pnas.0909468107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud, J.T., Mothes C.C., Beckles W., Heathcote R.J.P., Donihue C.M., and Losos J.B.. 2020. An extreme cold event leads to community-wide convergence in lower temperature tolerance in a lizard community. Biol. Lett. 16:20200625. 10.1098/rsbl.2020.0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulbarán, G., Biasutto A., Alamo L., Riggs C., Pinto A., Méndez F., Craig R., and Padrón R.. 2013. Different head environments in tarantula thick filaments support a cooperative activation process. Biophys. J. 105:2114–2122. 10.1016/j.bpj.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulbarán, G., Alamo L., Pinto A., Márquez G., Méndez F., Padrón R., and Craig R.. 2015. An invertebrate smooth muscle with striated muscle myosin filaments. Proc. Natl. Acad. Sci. USA. 112:E5660–E5668. 10.1073/pnas.1513439112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Kamata T., Onishi H., and Watanabe S.. 1982. Adenosine triphosphate-induced reversible change in the conformation of chicken gizzard myosin and heavy meromyosin. J. Biochem. 91:1699–1705. 10.1093/oxfordjournals.jbchem.a133861 [DOI] [PubMed] [Google Scholar]
- Sweeney, H.L., Bowman B.F., and Stull J.T.. 1993. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am. J. Physiol. 264:C1085–C1095. 10.1152/ajpcell.1993.264.5.C1085 [DOI] [PubMed] [Google Scholar]
- Szent-Györgyi, A.G., Kalabokis V.N., and Perreault-Micale C.L.. 1999. Regulation by molluscan myosins. Mol. Cell. Biochem. 190:55–62. 10.1023/A:1006975705724 [DOI] [PubMed] [Google Scholar]
- Toepfer, C., Caorsi V., Kampourakis T., Sikkel M.B., West T.G., Leung M.C., Al-Saud S.A., MacLeod K.T., Lyon A.R., Marston S.B., et al. 2013. Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease. J. Biol. Chem. 288:13446–13454. 10.1074/jbc.M113.455444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepfer, C.N., Wakimoto H., Garfinkel A.C., McDonough B., Liao D., Jiang J., Tai A.C., Gorham J.M., Lunde I.G., Lun M., et al. 2019. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci. Transl. Med. 11:eaat1199. 10.1126/scitranslmed.aat1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepfer, C.N., Garfinkel A.C., Venturini G., Wakimoto H., Repetti G., Alamo L., Sharma A., Agarwal R., Ewoldt J.F., Cloonan P., et al. 2020. Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation. 141:828–842. 10.1161/CIRCULATIONAHA.119.042339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, D.V., Nag S., Spudich A., Ruppel K.M., and Spudich J.A.. 2020. The myosin family of mechanoenzymes: from mechanisms to therapeutic approaches. Annu. Rev. Biochem. 89:667–693. 10.1146/annurev-biochem-011520-105234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus, K.M., Huiatt T.W., and Lowey S.. 1982. A bent monomeric conformation of myosin from smooth muscle. Proc. Natl. Acad. Sci. USA. 79:6151–6155. 10.1073/pnas.79.20.6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellekoop, J., Sluijs A., Smit J., Schouten S., Weijers J.W., Sinninghe Damsté J.S., and Brinkhuis H.. 2014. Rapid short-term cooling following the Chicxulub impact at the Cretaceous–Paleogene boundary. Proc. Natl. Acad. Sci. USA. 111:7537–7541. 10.1073/pnas.1319253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibert, P., and Craig R.. 1983. Electron microscopy and image analysis of myosin filaments from scallop striated muscle. J. Mol. Biol. 165:303–320. 10.1016/S0022-2836(83)80259-9 [DOI] [PubMed] [Google Scholar]
- Vibert, P., and Craig R.. 1985. Structural changes that occur in scallop myosin filaments upon activation. J. Cell Biol. 101:830–837. 10.1083/jcb.101.3.830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt, T., Taylor D., Trybus K.M., and Taylor K.. 2001. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. USA. 98:4361–4366. 10.1073/pnas.071051098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C., Naber N., Pate E., and Cooke R.. 2014. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys. J. 107:1637–1646. 10.1016/j.bpj.2014.07.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead, J.L., Zhao F.Q., Craig R., Egelman E.H., Alamo L., and Padrón R.. 2005. Atomic model of a myosin filament in the relaxed state. Nature. 436:1195–1199. 10.1038/nature03920 [DOI] [PubMed] [Google Scholar]
- Woodhead, J.L., Zhao F.Q., and Craig R.. 2013. Structural basis of the relaxed state of a Ca2+-regulated myosin filament and its evolutionary implications. Proc. Natl. Acad. Sci. USA. 110:8561–8566. 10.1073/pnas.1218462110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., Malinchik S., Gilroy D., Kraft T., Brenner B., and Yu L.C.. 1997. X-ray diffraction studies of cross-bridges weakly bound to actin in relaxed skinned fibers of rabbit psoas muscle. Biophys. J. 73:2292–2303. 10.1016/S0006-3495(97)78261-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., Gu J., Rhodes T., Belknap B., Rosenbaum G., Offer G., White H., and Yu L.C.. 1999. The M.ADP.Pi state is required for helical order in the thick filaments of skeletal muscle. Biophys. J. 77:2665–2676. 10.1016/S0006-3495(99)77101-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., Offer G., Gu J., White H.D., and Yu L.C.. 2003. Temperature and ligand dependence of conformation and helical order in myosin filaments. Biochemistry. 42:390–401. 10.1021/bi026085t [DOI] [PubMed] [Google Scholar]
- Xu, S., White H.D., Offer G.W., and Yu L.C.. 2009. Stabilization of helical order in the thick filaments by blebbistatin: further evidence of coexisting multiple conformations of myosin. Biophys. J. 96:3673–3681. 10.1016/j.bpj.2009.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M., Kimura M., Li Z.B., Ohno T., Takemori S., Hoh J.F., and Yagi N.. 2016. X-ray diffraction analysis of the effects of myosin regulatory light chain phosphorylation and butanedione monoxime on skinned skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 310:C692–C700. 10.1152/ajpcell.00318.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Woodhead J.L., Zhao F.Q., Sulbarán G., and Craig R.. 2016. An approach to improve the resolution of helical filaments with a large axial rise and flexible subunits. J. Struct. Biol. 193:45–54. 10.1016/j.jsb.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Lee K.H., Woodhead J.L., Sato O., Ikebe M., and Craig R.. 2019. The central role of the tail in switching off 10S myosin II activity. J. Gen. Physiol. 151:1081–1093. 10.1085/jgp.201912431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Tiwari P., Lee K.H., Sato O., Ikebe M., Padrón R., and Craig R.. 2020. Cryo-EM structure of the inhibited (10S) form of myosin II. Nature. 588:521–525. 10.1038/s41586-020-3007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotti, R., Seidman C.E., and Seidman J.G.. 2019. Advances in the genetic basis and pathogenesis of sarcomere cardiomyopathies. Annu. Rev. Genomics Hum. Genet. 20:129–153. 10.1146/annurev-genom-083118-015306 [DOI] [PubMed] [Google Scholar]
- Zhao, F.Q., and Craig R.. 2003. Ca2+ causes release of myosin heads from the thick filament surface on the milliseconds time scale. J. Mol. Biol. 327:145–158. 10.1016/S0022-2836(03)00098-6 [DOI] [PubMed] [Google Scholar]
- Zhao, F.Q., Padrón R., and Craig R.. 2008. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys. J. 95:3322–3329. 10.1529/biophysj.108.137067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F.Q., Craig R., and Woodhead J.L.. 2009. Head–head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J. Mol. Biol. 385:423–431. 10.1016/j.jmb.2008.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi, M.E., Woodhead J.L., Craig R., and Padrón R.. 2004. Helical order in tarantula thick filaments requires the “closed” conformation of the myosin head. J. Mol. Biol. 342:1223–1236. 10.1016/j.jmb.2004.07.037 [DOI] [PubMed] [Google Scholar]
- Zoghbi, M.E., Woodhead J.L., Moss R.L., and Craig R.. 2008. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. USA. 105:2386–2390. 10.1073/pnas.0708912105 [DOI] [PMC free article] [PubMed] [Google Scholar]