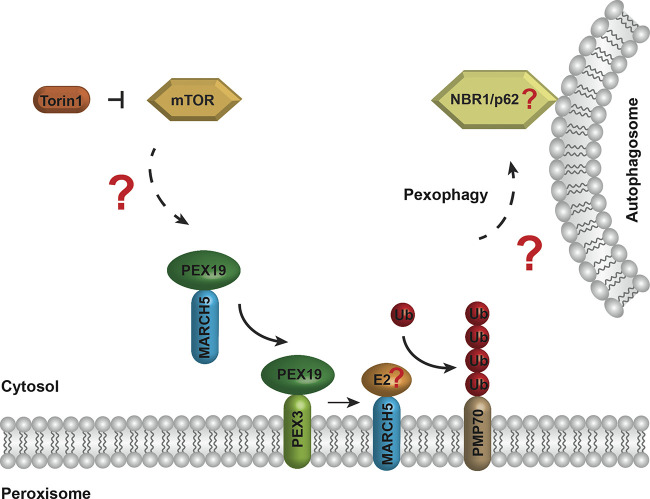
Platta and Erdmann highlight work from Zheng et al. that shows that MARCH5 is recruited to peroxisomes and plays a role in pexophagy regulation.
Abstract
A recent study by Zheng et al. (2021. J. Cell Biol. https://doi.org/10.1083/jcb.202103156) identifies the ubiquitin-protein ligase (E3) MARCH5 as a dual-organelle localized protein that not only targets to mitochondria but also to peroxisomes in a PEX19-mediated manner. Moreover, the authors demonstrate that the Torin1-dependent induction of pexophagy is executed by the MARCH5-catalyzed ubiquitination of the peroxisomal membrane protein PMP70.
Recent research has begun to slowly elucidate the complex processes that underlie selective autophagic degradation of mammalian peroxisomes. The study by Zheng et al. (1) sheds a light on the mechanism underlying pexophagy, which is induced by mTOR (mechanistic target of rapamycin) inactivation (2). The ubiquitously conserved serine/threonine kinase mTOR has a central function in integrating diverse growth signals and orchestrating their physiological effect on a cellular level, while blocking cell growth–restricting mechanisms like the different autophagy pathways (3). Previous work has demonstrated that amino acid starvation could induce mTOR inhibition-dependent peroxisome degradation by up-regulating the activity of the peroxisomal protein ubiquitin (E3) ligase PEX2 (4), which was especially of interest as PEX2 is also required for peroxisomal matrix protein import during the formation of the organelle (5). However, while these data suggested that the dual function of PEX2 might mark it as a point of convergence for the balance of peroxisome formation and degradation, the Zheng et al. study has identified a role for the E3 ligase MARCH5 (membrane-associated RING-CH 5; 1) that aims at a different aspect of peroxisome biology.
Zheng et al. identified the peroxisomal proteins PEX3, PEX19, and PMP70 as close interaction partners of MARCH5 (1). The authors could demonstrate a PEX19-dependent localization of a portion of the MARCH5 population to peroxisomes. Here, MARCH5 can bind and polyubiquitinate the abundant peroxisomal membrane protein PMP70. While it is clear that the increased level of polyubiquitinated PMP70 molecules marks peroxisomes for recognition by ubiquitin-binding autophagy receptors that link the target organelle to the autophagosomal membrane, the identity of the E2 enzyme involved in ubiquitin chain generation as well as the ubiquitin adaptors are unknown (Fig. 1). However, based on published research, NBR1 or p62 are good candidates for the adaptors that engage the autophagy machinery (2). Moreover, the Zheng et al. study demonstrates that MARCH5-mediated polyubiquitination of PMP70 is induced by the mTOR inhibitor Torin1. In return, the described Torin1-induced pexophagy was shown to rely on the peroxisomal localization and activity of the catalytic RING domain of MARCH5 (1).
Figure 1.
The small molecule Torin1 can inhibit the kinase mTOR, resulting in a relief of the mTOR-dependent block of MARCH5 targeting to peroxisomes. MARCH5 is inserted into the peroxisomal membrane in a PEX19- and PEX3-dependent manner. MARCH5 ubiquitinates the abundant peroxisomal membrane protein PMP70 with the help of an unknown ubiquitin (Ub)-conjugating enzyme (E2). The ubiquitinated PMP70 molecules are recognized by ubiquitin-binding autophagy receptors, like NBR1 or p62, that link the organelle to the autophagosome, resulting in the autophagic degradation of the peroxisome via pexophagy.
It is interesting to note that the opponent of pexophagy-linked ubiquitin signals on peroxisomes was already identified as the deubiquitinating enzyme USP30 (6). This combination is even more relevant when considering that MARCH5 and USP30 were described as an antagonizing enzyme pair that regulates the autophagic degradation of mitochondria via mitophagy (6). The function of MARCH5 is also linked to other mitochondrial ubiquitination factors, like the E3 ligase Parkin. While both enzymes can contribute to mitophagy induction by ubiquitinating proteins of the outer mitochondrial membrane, they can also modify each other. MARCH5 ubiquitinates Parkin in order to restrict the number of Parkin molecules during mitophagy and to prevent Parkin-mediated cell death (7).
After mitophagy induction, Parkin can ubiquitinate MARCH5, which results in the p97-mediated membrane extraction of MARCH5 and a PEX3/PEX16-dependent redistribution of MARCH5 to peroxisomes (8). This mechanism was assumed to rescue MARCH5 from degradation by mitophagy. It will be important to elucidate if there is mechanistic overlap between the Parkin-mediated (8) and the Torin1-dependent (1) targeting of MARCH5. Moreover, it will be interesting to determine if MARCH5 is also engaged in an interplay with the peroxisomal E3 ligases PEX2, PEX10, PEX12, or TRIM37.
Mitochondria and peroxisomes share basic components of their fission machineries. Both organelles use the membrane proteins FIS1 and mitochondrial fission factor for the targeting of the membrane-constricting GTPase DRP1 (DLP1; 9). In the case of the mitochondria, MARCH5 can ubiquitinate DRP1 and FIS1 for proteasomal degradation in order to limit mitochondrial However, other data indicate the existence of a feedback mechanism, as DRP1 can also negatively influence MARCH5 activity. In addition, MARCH5 not only limits mitochondrial fission, but also represents a basic requirement for this process. This complex relationship of MARCH5 with mitochondrial fission proteins suggests that it performs a central role in the fine-tuning of the basic regulatory aspects of mitochondrial division (10). Therefore, future studies might not only establish a potential role of MARCH5 in peroxisomal fission but might also uncover aspects that could enable further insights into the related process in mitochondria.
The different roles of MARCH5 in organelle fission and autophagic degradation could possibly be interconnected in one bipartite reaction sequence. Mitochondrial fission is crucial for mitophagy and enables the removal of damaged sections of mitochondria or the limitation of organelle size for a more efficient engulfment by autophagosomal membranes (11). Therefore, both processes can be functionally interconnected. Interestingly, it has been shown that fission also precedes pexophagy in yeast cells (12), which are thought to use organelle-specific adaptors instead of ubiquitin as a degradation tag. However, these observations suggest that MARCH5 might coordinate peroxisomal fission with pexophagy even in mammalian peroxisomes.
In summary, the Zheng et al. study not only identifies a central mechanistic module required for the turnover of mammalian peroxisomes (1) but also raises many interesting questions that will result in further studies dealing with the interplay of the peroxisomal ubiquitination factors, the crosstalk between mitochondria and peroxisomes, and the organization and regulation of the peroxisomal fission machinery as well as the convergence of peroxisomal fission and pexophagy pathways.
Acknowledgments
We thank Jana Tomaschewski for technical assistance in figure preparation.
This work was funded by grants of the Deutsche Forschungsgemeinschaft (FOR 1905).
The authors declare no competing financial interests.
References
- 1.Zheng, J., et al. 2021. J. Cell Biol. 10.1083/jcb.202103156 [DOI] [Google Scholar]
- 2.Li, J., and Wang W.. 2021. Cells. 10.3390/cells10051094 [DOI] [Google Scholar]
- 3.Boutouja, F., and Platta H.W.. 2019. Cells. 10.3390/cells8010018 [DOI] [Google Scholar]
- 4.Sargent, G., et al. 2016. J. Cell Biol. 10.1083/jcb.201511034 [DOI] [Google Scholar]
- 5.Walter, T., and Erdmann R.. 2019. Protein J. 10.1007/s10930-019-09835-6 [DOI] [PubMed] [Google Scholar]
- 6.Eldeeb, M.A., et al. 2020. Trends Cell Biol. 10.1016/j.tcb.2020.04.007 [DOI] [PubMed] [Google Scholar]
- 7.Shiiba, I., et al. 2021. EMBO Rep. 10.15252/embr.201949097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyano, F., et al. 2019. EMBO Rep. 10.15252/embr.201947728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello, J.L., et al. 2018. Subcell. Biochem. 10.1007/978-981-13-2233-4_17 [DOI] [Google Scholar]
- 10.Shiiba, I., et al. 2020. Int. J. Mol. Sci. 10.3390/ijms21113781 [DOI] [Google Scholar]
- 11.Toyama, E.Q., et al. 2016. Science. 10.1126/science.aab4138 [DOI] [Google Scholar]
- 12.Mao, K., et al. 2014. Autophagy. 10.4161/auto.27852 [DOI] [Google Scholar]