Abstract
OBJECTIVE
Type 1 and type 2 diabetes are associated with gut dysbiosis. However, the relationship between the gut microbiota and latent autoimmune diabetes in adults (LADA), sharing clinical and metabolic features with classic type 1 and type 2 diabetes, remains unclear. Here, we used a multiomics approach to identify the characteristics of the gut microbiota and metabolic profiles in patients with LADA.
RESEARCH DESIGN AND METHODS
This age- and sex-matched case-control study included 30 patients with LADA, 29 patients with classic type 1 diabetes, 31 patients with type 2 diabetes, and 29 healthy individuals. The gut microbiota profiles were identified through the 16S rRNA gene, and fecal and serum metabolites were measured through untargeted liquid chromatography-mass spectrometry.
RESULTS
Patients with LADA had a significantly different structure and composition of the gut microbiota and their metabolites as well as a severe deficiency of short-chain fatty acid–producing bacteria. The gut microbiota structure of the patients with LADA was more similar to that of patients with type 1 diabetes who were positive for GAD antibody. We identified seven serum metabolite modules and eight fecal metabolite modules that differed between the LADA group and the other groups.
CONCLUSIONS
The characteristic gut microbiota and related metabolites of patients with LADA are associated with autoantibodies, glucose metabolism, islet function, and inflammatory factors, which may contribute to the pathogenesis of LADA. Future longitudinal studies should explore whether modulating the gut microbiota and related metabolites can alter the natural course of autoimmune diabetes in the quest for new therapeutics.
Introduction
The recognition of latent autoimmune diabetes in adults (LADA) as adult-onset autoimmune diabetes sharing some genetic, immunologic, metabolic, and clinical features with classic type 1 diabetes (T1D) and type 2 diabetes (T2D) has gradually increased (1–3). The progression of autoimmune β-cell destruction is less intensive in LADA than in classic T1D, and patients are insulin independent for at least 6 months after diagnosis (1–3). Approximately 4–14% of patients with T2D are misdiagnosed and actually have LADA, which, in fact, is more prevalent than classic juvenile-onset T1D in some minority populations (3). In the context of moderate genetic susceptibility to T1D, some environmental factors can trigger islet autoimmunity, leading to β-cell apoptosis and potentially promoting LADA (4). However, the detailed mechanisms underlying the onset of LADA are unclear.
The gut microbiota is an indispensable environmental factor for the development of T1D and T2D (5,6); the structure and composition of the gut microbiota in patients with T1D and T2D differ from those in healthy subjects. Studies in T1D animal models showed that the gut microbiota can regulate toll-like receptor 2/4 signaling, T helper type 17 cells in the intestinal mucosa, sex hormone levels, and the secretion of pancreatic antibacterial peptide, which may modulate the autoimmune targeting of β-cells (7–9). Additionally, in T2D, intestinal dysbiosis can disrupt the gut barrier function and promote chronic metabolic inflammation and the secretion of intestinal hormones, including glucagon-like peptide 1 and peptide YY, affecting insulin sensitivity and secretion (10,11). Importantly, studies have shown that dietary interventions (probiotics, dietary fiber supplements, etc.) and fecal transplants can regulate the gut microbiota; vaccines and various drugs that regulate key factors that affect intestinal barrier function can be used as novel treatment strategies (12). However, no such studies have been conducted in patients with LADA, so the relationships between the gut microbiota, metabolic profile, and LADA remain to be determined.
Here, we compared the gut microbiota and metabolic profiles in patients with LADA, classic T1D, and T2D and healthy subjects. Furthermore, we elucidated the relationships among enterobacterial coabundance groups (CAGs), metabolite modules, and clinical phenotypes.
Research Design and Methods
Study Participants and Recruitment
We consecutively recruited 30 patients with LADA, 29 patients with classic T1D, 31 patients with T2D, and 29 healthy subjects (all age and sex matched, 30–70 years old) at the Henan Provincial People’s Hospital from 3 February 2019 to 30 September 2019; all were of Han origin (Supplementary Diagram). Diabetes was diagnosed according to the criteria recommended by the World Health Organization (13). LADA was diagnosed based on 1) diagnosis age between 30 and 70 years, 2) insulin independence during the initial 6 months, 3) positivity for at least one autoantibody (GAD antibody [GADA], IA-2 antibody [IA-2A], or zinc transporter 8 antibody [ZnT8A]), and 4) no ketosis or ketoacidosis (2,14). Classic T1D was diagnosed based on 1) the onset of acute ketosis or ketoacidosis, 2) the course of insulin replacement therapy, 3) impaired islet function, or 4) positivity for at least one autoantibody (GADA, IA-2A, or ZnT8A). T2D was diagnosed based on 1) the typical history of hyperglycemia, 2) no requirement for immediate insulin treatment, and 3) negativity for islet autoantibodies. All healthy subjects and patients with T2D were negative for GADA, IA-2A, and ZnT8A. All healthy subjects underwent a standard 75-g oral glucose tolerance test (OGTT) to verify their normal blood glucose levels. All patients with diabetes were undergoing treatment with multiple doses of insulin or oral drugs, or both (Supplementary Table 1). The exclusion criteria included the following: secondary diabetes; acute or chronic inflammatory diseases; infectious diseases; pregnancy; malignant tumors; history of steroid or immunosuppressive drug use >7 days; history of treatment with prebiotics, probiotics, antibiotics, or any other medication that could potentially influence the gut microbiota for >3 days in the previous 3 months; gastrointestinal diseases; a history of gastrointestinal surgery in the previous year; and hepatic and renal dysfunction.
The collected demographic/clinical data included age, sex, diabetes duration, height, weight, BMI, waist-to-hip ratio, systolic blood pressure, and diastolic blood pressure. We also collected biochemical determinations, including the 75-g OGTT, C-peptide release test, HbA1c, fasting plasma glucose (FPG), lipid profile, blood cell and hemoglobin counts, liver and renal function, autoantibodies (GADA, IA-2A, ZnT8A), and inflammatory factors. The receiver operating characteristic area under the curve for glucose (AUCGlucose) and C-peptide (AUCC-peptide) during OGTT were also calculated.
All participants completed an interview to determine health status, lifestyle, and medication use. All patients with diabetes received diabetes education and followed a diabetes diet. Dietary intake patterns were determined using a food frequency questionnaire.
All participants provided written informed consent. This study was approved by the Henan Provincial People’s Hospital ethics committee.
Measurement of Autoantibodies
GADA, IA-2A, and ZnT8A were detected as described by Huang et al. (15,16). The cutoff indices of positivity for GADA, IA-2A, and ZnT8A were 18 units/mL (World Health Organization units), 3.3 units/mL, and 0.011 (ZnT8A index), respectively. Positive samples were tested twice for confirmation. According to the Islet Autoantibody Standardization Program 2016, the sensitivity and specificity scores for GADA were 82% and 97.8%; for IA-2A, 76% and 100%; and for ZnT8A, 72% and 100%, respectively.
Measurement of Inflammatory Markers
Peripheral venous blood was collected in the morning after enrollment. Serum samples were stored at −80°C until analysis. The serum levels of interleukin-10 (IL-10), tumor necrosis factor-α (TNF-α), IL-6, and IL-1β were quantified using a custom Human Luminex Discovery Assay (LXSAHM-08; R&D Systems, Minneapolis, MN) according to the manufacturer’s protocol. The detection limits were 4.8–1,162, 9.7–2,359, 4.8–1,154, and 19.5–4,744 pg/mL for IL-10, TNF-α, IL-6, and IL-1β, respectively.
Lipopolysaccharide-Binding Protein Assays
Serum lipopolysaccharide-binding protein (LBP) levels were detected using a commercial ELISA kit (R&D Systems) according to the manufacturer’s instructions (range 4.4–50 ng/mL).
Statistical Analyses
A priori sample size estimation was not possible because of the lack of data on the gut microbiota differences among LADA, classic T1D, T2D, and healthy subjects. For clinical characteristics, serum LBP, and inflammatory factors, relative and absolute frequencies were used as qualitative variables, whereas median and interquartile range values were used as quantitative variables. Differences in quantitative variables among the groups were tested using the nonparametric Mann-Whitney U test or Kruskal-Wallis test. Differences in qualitative variables among the groups were tested using the χ2 test or Fisher exact test. Statistical analyses were performed using SPSS version 19.0 (IBM Corporation, Chicago, IL).
Fecal DNA Extraction and 16S rRNA Gene Sequencing
Fecal samples were collected from all participants according to established procedure and stored at −80°C. Genomic DNA was extracted using the QIAamp PowerFecal Pro DNA Kit (QIAGEN, Hilden, Germany). The fecal microbiota composition was characterized through 16S rRNA gene sequencing. Briefly, PCR targeting the V3–V4 region of the 16S rRNA gene was performed using the following primers: forward, 5′-CCTACGGGNGGCWGCAG-3′, and reverse, 5′-GACTACHVGGGTATCTAATCC-3′ (17). Subsequent amplicon sequencing was performed on a MiSeq platform (Illumina, San Diego, CA) to generate 300-bp paired-end reads; DNA from all 119 fecal samples was included in the same sequencing run.
Sequencing Data Analysis
Sequencing data were demultiplexed using the QIIME2 2019.7 pipeline (18), and forward and reverse reads were trimmed to 268 bp and 194 bp, respectively. An average of 48,972 reads were used as input; after filtering, denoising, merging forward and reverse reads, and removing chimeras, an average of 26,908 reads was recovered. Amplicon sequence variants (ASVs) were identified using the DADA2 plugin (19). Samples were randomly subsampled to equal depths of 20,148 reads before fecal microbiome analysis using the QIIME2 diversity core-metrics-phylogenetic plugin. The generated raw sequencing data are publicly available at the National Center for Biotechnology Information under accession no. SRP272175 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA646610). The phylogenetic tree of the representative ASV sequences was built using the core-metrics-phylogenetic pipeline in QIIME2, and the ASVs were annotated based on the SILVA release 132 database (20). The adonis test using vegan (https://cran.r-project.org/web/packages/vegan/index.html) in R 3.6.1 was performed to calculate the variation in each host factor. The diversity, richness, and evenness of samples were evaluated using QIIME2. Each host physiological factor was calculated according to its explanation rate; P values were generated based on 9,999 permutations. To test the variation in the gut microbiota, Bray-Curtis distances of all samples were calculated in QIIME2. The ASV abundance table was used for the principal coordinate analysis (PCoA) and partial least squares discriminant analysis (PLS-DA) based on the Bray-Curtis distances. PCoA was performed using QIIME2 and visualized using GraphPad Prism 8 (GraphPad Software, San Diego, CA); PLS-DA was performed and plotted using mixOmics in R 3.6.1 (21). Differential ASVs between the groups were identified via linear discriminant analysis effect size (https://huttenhower.sph.harvard.edu/galaxy). Major ASVs were defined as those differing between the LADA group and the healthy, T1D-A (GADA-positive [GADA-P]), T1D-B (GADA-negative [GADA-N]), and T2D groups. CAGs were clustered based on Spearman correlations of the major ASVs using WGCNA (weighted gene coexpression network analysis) with the Ward clustering algorithm in R 3.6.1 (22). The CAG network was visualized in Cytoscape 3.7.2 (https://github.com/cytoscape/cytoscape).
The abundances of ASVs and CAGs in the healthy, T1D-A, T1D-B, and T2D groups were compared with those in the LADA group using the Kruskal-Wallis test with Dunn post hoc analysis in Prism 8.0.1; those in the LADA, T1D-A, T1D-B, and T2D groups were also compared with those in the healthy group, and differences were deemed significant when P < 0.05. Functions of the gut microbiota were predicted from ASVs using PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2). Enriched pathways were determined using the Kyoto Encyclopedia of Genes and Genomes database (https://www.kegg.jp). Wilcoxon rank sum tests (per pathway) between the LADA and the healthy, T1D-A, T1D-B, and T2D groups as well as between the healthy and the T1D-A, T1D-B, and T2D groups were performed using the R package MASS. All correlation analyses were performed by calculating the Spearman correlation coefficient using MATLAB R2019b; the same software was used to design all the heatmaps.
Untargeted Metabolomics Study
The metabolic profiling of fecal and serum samples was performed using the 1290 Infinity LC System (Agilent, Santa Clara, CA) coupled to the TripleTOF 6600 System (SCIEX, Framingham, MA). The electrospray ionization sources were set according to the method described by Zhou et al. (23). Metabolites were characterized via comparison of the retention times, charge/mass ratio values, and fragmentation patterns with those previously reported (24).
Metabolomics Data Analysis
Differential metabolites were identified based on variable importance in the projection generated from the 10-fold cross-validated orthogonal PLS-DA model using a threshold of 1 and then validated with an adjusted P < 0.05 at a univariate level. The metabolites that differed between the LADA and the healthy, T1D-A, T1D-B, and T2D groups were clustered using the R package WGCNA. Serum and fecal metabolites were analyzed separately. Signed, weighted coabundance correlation networks of the metabolites were calculated for all the individuals in the study (24). The soft threshold β = 14, for metabolite correlation, was chosen based on a scale-free topology criterion. Clusters were identified with a dynamic hybrid tree-cutting algorithm using a DeepSplit of 4 and a minimum cluster size of 3 (25). Hub metabolites were defined as those with the highest module membership P value within each module. The heatmap of metabolite modules was plotted using MATLAB R2019b.
Fecal and serum metabolites detected in the positive and negative ion modes among healthy, GADA-P (LADA and T1D-A), and GADA-N (T1D-B) individuals and patients with T2D were subjected to PCoA, performed based on the Bray-Curtis distance. Comparisons were performed using the Kruskal-Wallis test with Dunn post hoc analysis in QIIME2. The abundances of the metabolite modules in the healthy, T1D-A, T1D-B, and T2D groups were compared with those in the LADA group using the Kruskal-Wallis test with Dunn post hoc analysis in Prism 8.0.1; those in the LADA, T1D-A, T1D-B, and T2D groups were also compared with those in the healthy group. The differences were deemed significant when P < 0.05.
Multiomics Correlation Analysis
The Spearman correlations among gut microbiota CAGs, fecal metabolite modules, serum metabolite modules, and clinical parameters were calculated using MATLAB R2019b. The P values were adjusted based on the false discovery rate (FDR) values described by Hochberg and Benjamini (26). Significance was set as FDR <0.05. The visualization of multiomics correlations was performed using Cytoscape 3.7.2.
Results
Anthropometric and Biochemical Measurements
Patient anthropometric and biochemical data are listed in Table 1. The duration of diabetes was shorter in the LADA group than in the other two diabetes groups. The average BMIs of the LADA, T1D groups were lower than those of the T2D and healthy groups. As expected, the FPG and HbA1c levels were significantly higher in patients with LADA and classic T1D and T2D than in healthy subjects; moreover, the fasting C-peptide levels in the LADA and T1D groups were lower than those in the T2D and healthy groups. Blood pressure readings did not differ among the four groups. HDL cholesterol levels were significantly lower in the T2D group than in the other groups. The LADA group had significantly lower IL-10 levels than the other groups. The three diabetes groups had significantly higher TNF-α levels than the healthy group. The levels of IL-6 and IL-1β in the LADA and T1D groups were significantly lower than those in the T2D group and significantly higher than those in the healthy group. The levels of LBP were higher in the three diabetes groups than in the healthy group; LBP levels were the highest in the LADA group.
Table 1.
Anthropometric and biochemical data
| Variable | Healthy controls (n = 29) | Type 2 diabetes patients (n = 31) | LADA patients (n = 30) | Classic type 1 diabetes patients (n = 29) | P |
|---|---|---|---|---|---|
| Male/female | 11/18 | 11/20 | 10/20 | 11/18 | |
| Age (years) | 34.0 (28.50–52.5) | 39.0 (31.00–48.0) | 36.5 (30.5–45.0) | 35.0 (31.0–47.5) | 0.932 |
| Duration of diabetes (months) | 36.0 (12.0–84.0)a | 24.0 (2.5–36.0)b | 36.0 (13.0–108.0)a | <0.001 | |
| BMI (kg/m2) | 22.14(19.99–24.41)a,b | 23.51(21.22–25.40)a | 20.48(18.31–22.29)c | 21.85(20.79–23.02)b,c | 0.002 |
| WHR | 0.86 (0.83–0.92) | 0.92 (0.87–0.94) | 0.88 (0.83–0.94) | 0.90 (0.85–0.93) | 0.221 |
| SBP (mmHg) | 112 (106–120) | 120 (110–130) | 110 (106–125) | 118 (109–130) | 0.130 |
| DBP (mmHg) | 71 (65–77) | 75 (72–80) | 73 (64–80) | 77 (70–84) | 0.060 |
| HbA1c (%) | 5.30 (5.20-5.50)a | 9.00 (7.00-10.60)b | 9.10 (7.08-11.80)b | 8.60 (6.80-9.30)b | <0.001 |
| HbA1c (mmol/mol) | 34(33–37)a | 75(53–92)b | 76(54–105)b | 70(51–78)b | <0.001 |
| FPG (mmol/L) | 4.75 (4.43–5.28)a | 6.90 (6.17–7.40)b | 6.80 (5.70–8.43)b | 7.00 (5.58–8.70)b | <0.001 |
| FCP (ng/ml) | 1.08 (0.86–1.36)a | 0.78 (0.50–1.26)b | 0.37 (0.26–0.50)c | 0.00 (0.00–0.05)d | <0.001 |
| TC (mg/dL) | 4.57 (4.12–4.89) | 4.90 (3.68–5.33) | 4.34 (3.87–5.27) | 4.58 (4.08–5.21) | 0.767 |
| TG (mg/dL) | 1.08 (0.89–1.21) | 1.35 (0.97–2.12) | 0.91 (0.62–1.43) | 1.03 (0.76–1.50) | 0.050 |
| HDL-c (mg/dL) | 1.36 (1.26–1.56)a | 1.15 (0.94–1.27)b | 1.33 (1.10–1.62)a | 1.23 (1.07–1.55)a | 0.004 |
| LDL-c (mg/dL) | 2.56 (2.38–2.78) | 2.99 (2.16–3.27) | 2.33 (1.85–3.03) | 2.34 (2.03–2.84) | 0.185 |
| IL-10 (pg/mL) | 1.63 (1.23–1.93)a | 1.44 (1.21–1.66)a | 1.32 (1.03–1.48)b | 1.44 (1.22–1.77)a | 0.008 |
| TNF-α (pg/mL) | 4.49 (4.06–5.09)a | 5.60 (5.16–6.28)b | 5.44 (4.60–6.74)b | 5.20 (4.26–6.67)b | 0.002 |
| IL-6 (pg/mL) | 1.09 (0.92–1.27)a | 2.69 (1.55–5.68)b | 1.61 (1.25–2.74)c | 1.74 (1.18–3.04)c | <0.001 |
| IL-1β (pg/mL) | 2.19 (2.03–2.64)a | 3.03 (2.47–3.77)b | 2.67 (2.03–2.97)c | 2.47 (2.03–3.18)c | <0.001 |
| LBP (ng/mL) | 26.35(23.58–29.24)a | 32.91(29.41–36.50)b,c | 34.73(29.48–39.89)b | 29.88(26.62–33.44)c | <0.001 |
Data are median (25th–75th percentile) unless otherwise indicated. DBP, diastolic blood pressure; FCP, fasting C-peptide; HDL-C, HDL cholesterol; LDL-C, LDL cholesterol; SBP, systolic blood pressure; TC, total cholesterol; TG, triglyceride; WHR, waist-to-hip ratio. Different superscripted symbols (a, b, c, d) in a row indicate that the medians of the different groups are significantly different (P < 0.05).
Characterization of the Gut Microbiota of Patients With LADA
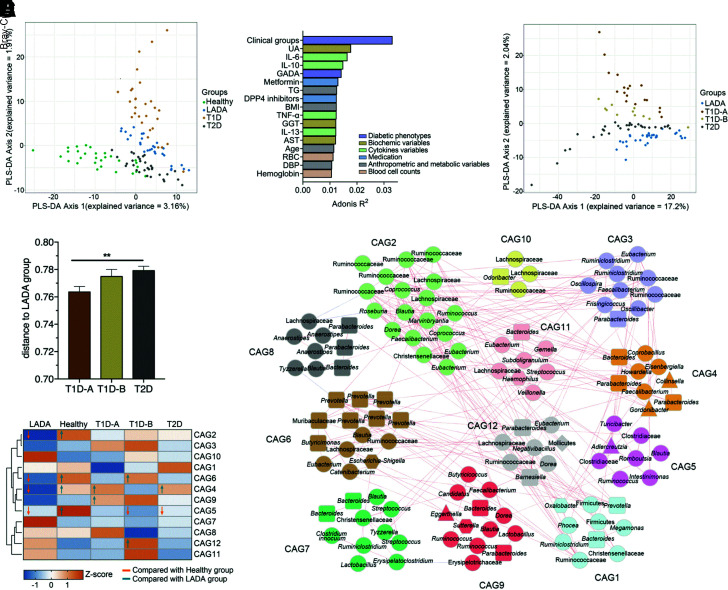
No differences in the richness (observed ASVs), diversity (Shannon index), and evenness (Pielou index) were found among the four groups (Supplementary Fig. 1). However, the PCoA and score plots of the PLS-DA (Fig. 1A and Supplementary Fig. 2) showed that the structure and composition of the gut microbiota differed significantly among the three diabetes groups and healthy group. Furthermore, the structure of the microbiota differed significantly between the LADA group and the other two diabetes groups. Clinical groups, inflammatory factors, autoantibody GADA, and medication use were significantly associated with gut microbial variations (P < 0.1 of permutational MANOVA) (Fig. 1B). As GADA is a very strong explanatory factor for variations in the gut microbiota, the classic T1D group was further divided into T1D-A (GADA-P) and T1D-B (GADA-N) groups. The microbiota structure of the T1D-A group was the most similar to that of the LADA group, whereas that of the T2D group was the most dissimilar to that of the LADA group (Fig. 1C and D). The clinical features of the LADA, T1D-A, and T1D-B groups are shown in Supplementary Table 2. The T1D-B group showed lower BMI, AUCC-peptide, and serum LBP levels than the LADA and T1D-A groups and a longer duration of diabetes than the LADA and T1D-A groups.
Figure 1.
Identification of the major ASV modules in the context of LADA. A: PLS-DA of the gut microbiota composition of healthy subjects and patients with LADA, T1D, and T2D. B: Bar plot revealing the top physiological factors that were significantly associated with variations in the gut microbiota based on Bray-Curtis distances. The colors of the bars represent their clinical categories. Size effects and statistical significance were calculated by adonis/permutational MANOVA. The association was considered significant when P < 0.1. C: PLS-DA of the gut microbiota composition of the patients with LADA, T1D-A, T1D-B, and T2D. D: Between-sample Bray-Curtis distances of the gut microbiota of the T1D-A, T1D-B, and T2D groups compared with those of the LADA group. **P < 0.01. E: Network diagrams of the 12 CAGs. The taxonomy of bacteria phyla identified using the QIIME2 q2-feature-classifier is denoted on the nodes. The rectangular nodes represent Bacteroidetes, the triangular nodes represent Actinobacteria, the round nodes represent Firmicutes, the hexagonal nodes represent Proteobacteria, and the diamond-shaped nodes represent Tenericutes. Lines between nodes represent correlations; only correlations with magnitudes >0.3 are drawn. Red lines mean positive correlations, and blue lines mean negative correlations. F: Z-scores of the abundance of the 12 CAGs across the different groups. Z-scores were transformed by subtracting the average abundance and dividing by the SD of all samples. Blue represents negative Z-scores, and orange represents positive Z-scores. Comparison of the relative abundance of each CAG in the five groups was performed using the Kruskal-Wallis test followed by Dunn post hoc analysis. Significant differences (P < 0.05) are marked with arrows; arrows pointing up represent a significantly higher abundance, and arrows pointing down represent a significantly lower abundance. DBP, diastolic blood pressure; DPP4, dipeptidyl peptidase 4; GGT, glutamyl transpeptidase; RBC, red blood cell; TG, triglyceride; UA, uric acid.
Additionally, we identified 139 ASVs to explore the LADA-specific microbiota (Supplementary Figs. 3 and 4) and constructed 12 CAGs based on Spearman correlation analysis (Fig. 1E). The abundances of CAG2 (i.e., families Ruminococcaceae and Lachnospiraceae), CAG4 (e.g., Parabacteroides species [spp.]), CAG5 (mainly in Clostridiaceae), and CAG6 (including Prevotella spp.) were significantly lower in patients with LADA than in healthy subjects. The abundances of CAG4 and CAG6, CAG6 and CAG12 (i.e., families Ruminococcaceae and Lachnospiraceae), and CAG4 alone were also significantly lower in the LADA group versus the T1D-A, T1D-B, and T2D groups, respectively (Fig. 1F). Collectively, these findings suggest that patients with LADA have a unique gut microbiota structurally and compositionally different from that of healthy subjects and patients with classic T1D and T2D.
For further insight, we also predicted the potentially enriched pathways based on the obtained 16S rRNA gene sequencing data using PICRUSt2 (Supplementary Fig. 5). In the LADA group, amino acid, carbohydrate, and lipid metabolism pathways, including those involved in valine, leucine, and isoleucine degradation, and fatty acid biosynthesis were significantly downregulated, while the secondary bile acid biosynthesis pathway was upregulated in LADA (compared with healthy subjects). Pathways for amino acids, cofactors, and vitamins (e.g., phenylalanine metabolism) were also significantly downregulated in LADA versus T2D. However, in LADA (compared with T1D), no differences were detected.
Considering that all forms of diabetes induced by autoimmune β-cell destruction are included under T1D, patients with LADA and classic T1D were stratified collectively into GADA-P and GADA-N groups. The clinical characteristics of these two groups are shown in Supplementary Table 3. There were no differences in α-diversity among the GADA-P, GADA-N, T2D, and healthy groups (Supplementary Fig. 6). However, the structure and composition of the gut microbiota differed significantly between the GADA-P group and healthy group (Supplementary Figs. 7 and 8).
Characterization of the Fecal and Serum Metabolites in Patients With LADA
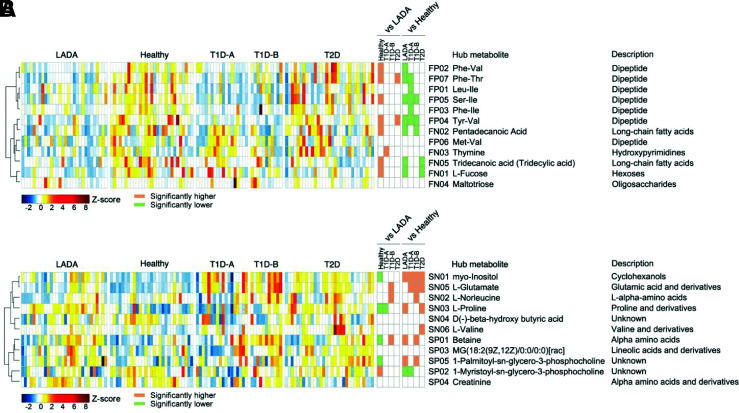
Fecal and serum metabolites were significantly different in patients with diabetes versus healthy subjects. Notably, patients with LADA presented significantly different metabolite profiles from those in healthy subjects and patients with T1D and T2D (Fig. 2). From the orthogonal PLS-DA models (Supplementary Figs. 9 and 10), we identified 422 and 317 metabolites under positive ionic mode and negative ionic modes, respectively, in the feces that were binned into 12 coabundance clusters for all subjects. Additionally, we identified 178 and 147 metabolites (positive and negative modes, respectively) in the serum that were binned into 11 coabundance clusters. Compared with healthy subjects, we found that most fecal metabolite modules (i.e., dipeptide, medium chain fatty acids, hexoses) were less abundant, while most serum metabolite modules (e.g., cyclohexanols, glutamic acid and derivatives, α-amino acids, proline and its derivatives) were more abundant in patients with diabetes. Of note, with respect to fecal metabolites, there was a lower abundance of thymine in the LADA versus T1D-A groups and a higher abundance of dipeptides, including tyrosine, phenylalanine, valine, and isoleucine, in the LADA versus T2D groups. Additionally, concerning serum metabolites, proline and its derivatives were more abundant in the LADA group than in the T1D-A group, while glutamic acid and its derivatives and α-amino acids were less abundant in the LADA group than in the T1D-B group. Interestingly, while PCoA showed that for fecal metabolites, differences were observed only between the three general diabetes groups (GADA-P, GADA-N, T2D) and healthy subjects, for serum metabolites, the differences were between the three diabetes groups and healthy subjects and between the GADA-P and GADA-N groups (Supplementary Figs. 11–15). Thus altogether, these results suggest that patients with diabetes have significantly different metabolite profiles compared with those of healthy subjects and that the metabolite levels differ among patients with LADA and patients with T1D-A, T1D-B, and T2D.
Figure 2.
Identification of major fecal and serum metabolite modules in the context of LADA. Distribution of the 12 fecal metabolite modules (A) and 11 serum metabolite modules (B) among the LADA, healthy, T1D-A, T1D-B, and T2D groups. The abundance profiles were transformed into Z-scores by subtracting the average abundance and dividing by the SD of all samples. Comparison of the abundances of each module in the five groups was performed using the Kruskal-Wallis test followed by Dunn post hoc analysis. P < 0.05 indicates a significant difference.
The Relationships Among the Gut Microbiota, Fecal Metabolites, Serum Metabolites, and Clinical Phenotypes
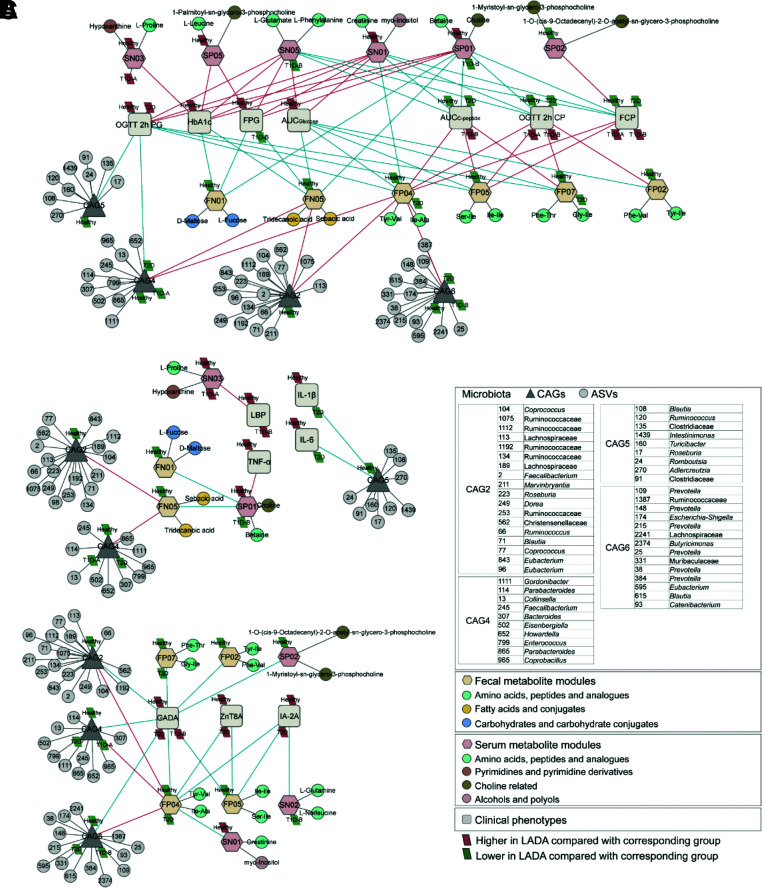
Subsequently, we analyzed the correlations among the gut microbiota profiles, fecal metabolites, serum metabolites, and clinical phenotypes in all participants (Fig. 3). Six fecal metabolite modules, mainly dipeptide-containing branched-chain amino acids (BCAAs) (valine, leucine, and isoleucine) and aromatic amino acids (AAAs) (tyrosine and phenylalanine), were negatively correlated with glucose metabolism–related parameters but positively correlated with islet function–related parameters. Conversely, six serum metabolite modules were positively correlated with glucose metabolism–related parameters and negatively correlated with islet function–related parameters. CAG2, CAG4, and CAG6, less abundant in the LADA group, were associated with glucose metabolism– and islet function–related parameters through fecal metabolites. CAG5 was negatively correlated with IL-6 and IL-1β, whereas CAG2 and CAG4 were associated with TNF-α through fecal and serum metabolites. L-proline and hypoxanthine were positively correlated with LBP. Notably, CAG2, CAG4, CAG6, and the fecal metabolites BCAAs and AAAs were all negatively correlated with GADA; moreover, the fecal metabolites BCAAs and AAAs were also negatively correlated with ZnT8A and IA-2A.
Figure 3.
Correlations among gut microbiota, host fecal or serum metabolites, and host clinical phenotypes in the context of LADA. A: The Spearman correlation network of the gut microbiota, fecal metabolites, serum metabolites, and clinical phenotypes related to blood glucose. B: The Spearman correlation network of the gut microbiota, fecal metabolites, serum metabolites, and clinical phenotypes related to inflammation. C: The Spearman correlation network of the gut microbiota, fecal metabolites, serum metabolites, and autoimmunity antibodies. Red lines indicate positive correlations (FDR <0.05), and blue lines indicate negative correlations (FDR <0.05). 2h CP, 2-h postprandial C-peptide; 2h PG, 2-h postprandial glucose; FCP, fasting C-peptide.
Conclusions
In this study, we found that patients with LADA showed significantly different gut microbiota and metabolite profiles from those of healthy subjects and patients with classic T1D and T2D. Furthermore, we found a correlation among the gut microbiota, fecal metabolites, serum metabolites, and clinical phenotypes.
Remarkably, the gut microbiota of patients with LADA showed distinctive characteristics (e.g., significantly decreased abundance of Faecalibacterium spp., Roseburia spp., and Blautia spp.) compared with the other groups. These are short-chain fatty acid (SCFA)–producing bacteria. SCFA-producing bacteria are known to positively affect glucose metabolism; they strengthen the gut barrier function, reduce chronic inflammation, and modulate intestinal hormones to improve insulin sensitivity and reduce pancreatic autoimmunity (27–29). The structure and composition of the gut microbiota in patients with T1D and T2D are different from that in healthy subjects, with a decrease in the abundance of SCFA-producing bacteria (9,30). Our study found that patients with LADA show a severe deficiency in SCFA-producing bacteria compared not only with healthy subjects but also with patients with classic T1D and T2D. Accordingly, the severe SCFA-producing bacterial deficiency in the guts of patients with LADA may contribute to the occurrence and progression of the disease. However, further studies are needed to identify the key microbiota players and investigate their disease-linked mechanisms of action.
Additionally, we found that the autoantibody GADA strongly associates with the structure and composition of the microbiome but negatively correlates with SCFA-producing bacteria. GADA is one of the most potent autoantigens involved in β-cell–specific autoimmunity. LADA is defined as a heterogeneous disease with respect to susceptibility genes, effects on autoimmunity, and phenotype. The potential causes of LADA involve heterogeneous pathways in the initiation of islet autoimmunity and heterogeneity in cellular responses (31). Interestingly, animal studies found that the SCFAs acetate and butyrate produced by gut microbes protected nonobese diabetic mice from insulitis and slowed the progression of diabetes, whereas butyrate in the diet improved regulatory T cell count and enhanced regulatory T cell function (32). Additionally, several cross-sectional studies have shown that the GADA titer correlates with the phenotypic heterogeneity of autoimmune diabetes, particularly in patients with LADA (33,34). Importantly, in the current study, the findings were similar. Therefore, we hypothesized that the gut microbiota may significantly affect the clinical classification and therapy of autoimmune diabetes. Our understanding of these diseases is insufficient and needs further exploration, and the gut microbiota may provide new insights.
We also identified specific metabolites in the feces and blood, such as BCAAs and AAAs produced by gut bacteria, that were different in patients with LADA versus others with diabetes. Of note, they correlated with glucose metabolism. Other studies have found that BCAAs and AAAs are associated with insulin sensitivity/resistance (35). Large human population studies found that a high intake of dietary BCAAs increases the risk of T2D (36,37). Conversely, animal studies demonstrated that a diet specifically enriched in leucine (BCAA) could improve glucose homeostasis (38). Moreover, lowering dietary BCAAs has been shown to improve insulin sensitivity and increase energy expenditure (39). The mechanism is probably related to the activation of mammalian target of rapamycin, affecting insulin sensitivity (40). Therefore, BCAAs and AAAs might affect glucose metabolism and sensitivity and promote autoantibody expression in patients with LADA.
However, because of the relatively small sample size in this study, multicenter, large-scale trials are required to validate our results. Additionally, since this is a cross-sectional study, the data are correlative but not causal. Future longitudinal studies are essential to explore whether the modulation of the gut microbiota and metabolism can alter the natural course of LADA; the mechanisms involved in immune regulation by commensal bacteria in LADA need further investigation.
In this study, we found unique characteristics of the gut microbiota and metabolic profiles in patients with LADA, distinct from those in healthy subjects and patients with classic T1D and T2D. Additionally, we found that these profiles correlated to glucose metabolism, islet function, inflammatory factors, and the autoimmune status, suggesting an involvement in the onset and progression of LADA. Collectively, the findings of this study may shed new light on autoimmune diabetes.
Article Information
Acknowledgments. The authors thank all the participants.
Funding. This study was supported by National Natural Science Foundation of China grant 81970705.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.F. conceived the study, recruited volunteers, selected all the samples used, performed the statistical analysis and data interpretation, wrote the manuscript, and made key modifications. C.Z. participated in the project design, analyzed the microbiota and metabolomics data, and modified the manuscript. H.S. participated in the study design, recruited volunteers, retained samples, extracted data, carried out the statistical analysis of clinical data, and participated in the writing and modification of the manuscript. W.W. recruited and supervised the participants and performed all clinical procedures. J.S., R.Zhe., L.Y., P.W., J.Y., X.D., Y.Z., S.T., X.S., Y.L., H.Ya., and Q.Y. participated in the recruitment of volunteers and provided the samples used in this study. R.Zha. analyzed the microbiota and metabolomics data. H.Yu. conceived the study, developed the experimental design, wrote the manuscript, and provided critical revision. All authors read and approved the final version of the manuscript. H.Yu. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Y.F., C.Z., and H.S. contributed equally to this work.
Clinical trial reg. no. ChiCTR2000030049, chictr.org
This article contains supplementary material online at https://doi.org/10.2337/figshare.16441407.
References
- 1. Mishra R, Chesi A, Cousminer DL, et al.; Bone Mineral Density in Childhood Study . Relative contribution of type 1 and type 2 diabetes loci to the genetic etiology of adult-onset, non-insulin-requiring autoimmune diabetes. BMC Med 2017;15:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou Z, Xiang Y, Ji L, et al.; LADA China Study Group . Frequency, immunogenetics, and clinical characteristics of latent autoimmune diabetes in China (LADA China study): a nationwide, multicenter, clinic-based cross-sectional study. Diabetes 2013;62:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laugesen E, Østergaard JA, Leslie RD. Corrigendum. Danish Diabetes Academy Workshop and Workshop Speakers. Latent autoimmune diabetes of the adult: current knowledge and uncertainty. Diabet Med 2015;32:1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buzzetti R, Zampetti S, Maddaloni E. Adult-onset autoimmune diabetes: current knowledge and implications for management. Nat Rev Endocrinol 2017;13:674–686 [DOI] [PubMed] [Google Scholar]
- 5. Burcelin R, Serino M, Chabo C, Blasco-Baque V, Amar J. Gut microbiota and diabetes: from pathogenesis to therapeutic perspective. Acta Diabetol 2011;48:257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012;336:1262–1267 [DOI] [PubMed] [Google Scholar]
- 7. Yang Y, Torchinsky MB, Gobert M, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 2014;510:152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol 2016;12:154–167 [DOI] [PubMed] [Google Scholar]
- 9. Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol 2020;17:53–64 [DOI] [PubMed] [Google Scholar]
- 10. Hartstra AV, Nieuwdorp M, Herrema H. Interplay between gut microbiota, its metabolites and human metabolism: dissecting cause from consequence. Trends Food Sci Technol 2016;57:233–243 [Google Scholar]
- 11. Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol 2020;20:40–54 [DOI] [PubMed] [Google Scholar]
- 12. Li X, Watanabe K, Kimura I. Gut microbiota dysbiosis drives and implies novel therapeutic strategies for diabetes mellitus and related metabolic diseases. Front Immunol 2017;8:1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998;15:539–553 [DOI] [PubMed] [Google Scholar]
- 14. Hawa MI, Thivolet C, Mauricio D, et al.; Action LADA Group . Metabolic syndrome and autoimmune diabetes: Action LADA 3. Diabetes Care 2009;32:160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang G, Xiang Y, Pan L, Li X, Luo S, Zhou Z. Zinc transporter 8 autoantibody (ZnT8A) could help differentiate latent autoimmune diabetes in adults (LADA) from phenotypic type 2 diabetes mellitus. Diabetes Metab Res Rev 2013;29:363–368 [DOI] [PubMed] [Google Scholar]
- 16. Huang G, Yin M, Xiang Y, et al. Persistence of glutamic acid decarboxylase antibody (GADA) is associated with clinical characteristics of latent autoimmune diabetes in adults: a prospective study with 3-year follow-up. Diabetes Metab Res Rev 2016;32:615–622 [DOI] [PubMed] [Google Scholar]
- 17. Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012;28:1823–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rohart F, Gautier B, Singh A, Lê Cao KA. mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput Biol 2017;13:e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou X, Liu L, Lan X, et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry 2019;24:1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw 2012;46:i11. [PMC free article] [PubMed] [Google Scholar]
- 25. Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 2008;24:719–720 [DOI] [PubMed] [Google Scholar]
- 26. Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990;9:811–818 [DOI] [PubMed] [Google Scholar]
- 27. Kim CH. Microbiota or short-chain fatty acids: which regulates diabetes? Cell Mol Immunol 2018;15:88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol 2019;15:261–273 [DOI] [PubMed] [Google Scholar]
- 29. Wen L, Wong FS. Dietary short-chain fatty acids protect against type 1 diabetes. Nat Immunol 2017;18:484–486 [DOI] [PubMed] [Google Scholar]
- 30. Wu H, Tremaroli V, Schmidt C, et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab 2020;32:379–390.e3 [DOI] [PubMed] [Google Scholar]
- 31. Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat Rev Endocrinol 2019;15:635–650 [DOI] [PubMed] [Google Scholar]
- 32. Mariño E, Richards JL, McLeod KH, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 2017;18:552–562 [DOI] [PubMed] [Google Scholar]
- 33. Buzzetti R, Di Pietro S, Giaccari A, et al.; Non Insulin Requiring Autoimmune Diabetes Study Group . High titer of autoantibodies to GAD identifies a specific phenotype of adult-onset autoimmune diabetes. Diabetes Care 2007;30:932–938 [DOI] [PubMed] [Google Scholar]
- 34. Hawa MI, Kolb H, Schloot N, et al.; Action LADA Consortium . Adult-onset autoimmune diabetes in Europe is prevalent with a broad clinical phenotype: Action LADA 7. Diabetes Care 2013;36:908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng Y, Li Y, Qi Q, et al. Cumulative consumption of branched-chain amino acids and incidence of type 2 diabetes. Int J Epidemiol 2016;45:1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Isanejad M, LaCroix AZ, Thomson CA, et al. Branched-chain amino acid, meat intake and risk of type 2 diabetes in the Women’s Health Initiative. Br J Nutr 2017;117:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Guo K, LeBlanc RE, Loh D, Schwartz GJ, Yu YH. Increasing dietary leucine intake reduces diet-induced obesity and improves glucose and cholesterol metabolism in mice via multimechanisms. Diabetes 2007;56:1647–1654 [DOI] [PubMed] [Google Scholar]
- 39. Cummings NE, Williams EM, Kasza I, et al. Restoration of metabolic health by decreased consumption of branched-chain amino acids. J Physiol 2018;596:623–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoon MS. The Emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients 2016;8:405. [DOI] [PMC free article] [PubMed] [Google Scholar]