Abstract
Specific mechanisms must exist to ensure fidelity in selecting the AG dinucleotide that functions as the 3′ splice site during the second transesterification step of splicing. Here we show that the optimal location for this AG is within a narrow distance (19 to 23 nucleotides [nt]) downstream from the branch point sequence (BPS). Contrary to previous expectations, AGs located less than 23 nt from the BPS are always recognized, even when a second AG located more optimally downstream is used in the transesterification reaction. Indeed, the AG closest to the BPS actually dictates the precise location of the AG that engages in the reaction. This mechanism, in which the AG is identified by a general localization step followed by a precise localization step, may be used to achieve fidelity while allowing flexibility in the location of 3′ splice sites.
The first AG dinucleotide located downstream from the branch site is generally chosen as the 3′-splice site during catalytic step II of splicing (reviewed in references 2, 18, and 26). How this AG is selected from among other AGs in the RNA is not known. One proposal is that the AG is selected by a linear scanning mechanism (9, 15, 21, 22). According to this model, a spliceosome component begins at the branch site and scans downstream to the first AG. A related model proposes that the AG is selected via a threading mechanism, in which a 5′-to-3′ search is also involved (1, 3). In Saccharomyces cerevisiae, simple scanning is not thought to occur because distal AGs can outcompete proximal AGs depending on the sequence context (19). In addition, insertion of secondary structure between the branch point sequence (BPS) and the AG does not inhibit step II (8, 13). Finally, step II efficiency is optimal over a narrow BPS-to-AG distance (13 to 22 nucleotides [nt]) (16).
In contrast to that in S. cerevisiae, it is still widely believed that 3′ splice site selection in metazoans involves linear scanning (2). Indeed, despite the high evolutionary conservation of the splicing pathway, it has been proposed that the mechanism of 3′ splice site selection might diverge in yeast and metazoans (1, 3, 21, 22, 26). Among the evidence in favor of the scanning and threading models in metazoans are the observations that the first AG downstream of the BPS is selected in cis-splicing assays (reviewed in reference 26) and that, in a trans-splicing assay, the 5′-most AG on a 3′ RNA substrate is selected (1). Step II is also blocked by hairpin loops inserted between the BPS and AG, consistent with stalling of a scanner on the secondary structure (3, 15, 22). An alternative model to scanning proposes that the 3′ splice site is selected by a BPS-to-AG measuring mechanism. This model is based on the observation that both the sequence and the distance between the BPS and AG affect the efficiency of step II (5). In addition, U5 snRNP proteins contact the intron prior to step II, spanning the distance (∼25 nt) from the BPS to the AG (5). Thus, the AG may be specified by its location relative to the U5 snRNP binding site (5).
The essential splicing factors Slu7 and human Slu7 (hSlu7) have been shown to play important roles in AG selection during step II. In yeast, Slu7 interacts genetically with U5 snRNP and is the last known factor to associate with the 3′ splice site prior to catalysis (11). We recently used pre-mRNAs containing duplicate AGs to analyze AG selection in nuclear extracts lacking hSlu7 (6). This study revealed that hSlu7 is required for selection of the correct AG and defined two steps in AG selection. To gain further insight into the mechanism for AG selection, we have now used normal nuclear extracts to analyze the pre-mRNAs containing duplicate AGs, as well as pre-mRNAs containing single AGs. These data do not support simple scanning or measuring models for AG selection. Instead, the data reveal a complex, multistep mechanism that may be necessary for achieving the high levels of fidelity required for AG selection.
MATERIALS AND METHODS
Plasmids.
Plasmids encoding pre-mRNAs were constructed by inserting oligonucleotides into the HindIII and AccI sites of pAdML (12) to replace the sequences from the BPS to the AG dinucleotide. Beginning with the branch site adenosine (in lowercase), the sequences in this region are the following: 15/18, 5′-aCUUUUUUUCUUUCAGCAG-3′; 15/19, 5′-aCUUUUUUUCUUUCAGUCAG-3′; 15/20, 5′-aCUUUUUUUCUUUCAGUUCAG-3′; 15/21, 5′-aCUUUUUUUCUUUCAGUUUCAG-3′; 16/19, 5′-aCUUUUUUUCUUUUCAGCAG-3′; 16/20, 5′-aCUUUUUUUCUUUUCAGUCAG-3′; 16/21: 5′-aCUUUUUUUCUUUUCAGUUCAG-3′; 16/22, 5′-aCUUUUUUUCUUUUCAGUUUCAG-3′; 17/20, 5′-aCUUUUUUUCUUUUUCAGCAG-3′; 17/21, 5′-aCUUUUUUUCUUUUUCAGUCAG-3′; 17/22, 5′-aCUUUUUUUCUUUUUCAGUUCAG-3′; 17/23, 5′-aCUUUUUUUCUUUUUCAGUUUCAG-3′; 18/21, 5′-aCUUUUUUUCUUUUUUCAGCAG-3′; 18/22, 5′-aCUUUUUUUCUUUUUUCAGUCAG-3′; 18/23, 5′-aCUUUUUUUCUUUUUUCAGUUCAG-3′; 18/24, 5′-aCUUUUUUUCUUUUUUCAGUUUCAG-3′; 19/22, 5′-aCUUUUUUUCUUUUUUUCAGCAG-3′; 20/23, 5′-aCUUUUUUUCUUUUUUUUCAGCAG-3′; 23/26, 5′-aCUUUUUUUCUUUUUUUUUUUCAGCAG-3′; 27/30, 5′-aCUUUUUUUCUUUUUUUUUUUUUUUCAGCAG-3′; 33/36, 5′-aCUUUUUCUUUUUUUCUUUUUUUCUUUUUUUCAGCAG-3′; 15/18/21, 5′-aCUUUUUUUCUUUCAGCAGCAG-3′; 15GG/18/21, 5′-aCUUUUUUUCUUUCGGCAGCAG-3′; 15GA/18/21, 5′-aCUUUUUUUCUUUCGACAGCAG-3′; 15AG, 5′-aCUUUUUUUCUUUCAG-3′; 18AG, 5′-aCUUUUUUUCUUUUUUCAG-3′; 21AG, 5′-aCUUUUUUUCUUUUUUUUUCAG-3′; 24AG, 5′-aCUUUUUUUCUUUUUUUCUUUUCAG-3′; 30AG, 5′-aCUUUUUUUCUUUUUUUCUUUUUUUUUUCAG-3′; and 40AG, 5′-aCUUUUUUUCUUUUUUUCUUUUUUUCUUUUUUUCUUUUUCAG-3′.
Splicing reactions.
32P-labeled pre-mRNAs (10 ng) were incubated in 25-μl splicing reaction mixtures for 1 h unless otherwise indicated. Total RNA was prepared and fractionated on 6.5, 7.5, or 15% denaturing polyacrylamide gels. RNAs were quantitated by phosphorimager (Quantityone; Bio-Rad). The relative use of each AG is indicated in each figure and is also presented in Table 1. The splicing intermediates and products were identified by comigration with known markers on the gels with different percentages of denaturing polyacrylamide.
TABLE 1.
| Pre-mRNA (nt)a | Ratio of distal AG products to proximal AG products |
|---|---|
| 15/18 | 4.2 |
| 15/19 | 1.7 |
| 15/20 | 2.1 |
| 15/21 | 0.1 |
| 15/22 | 0.2 |
| 15/23 | 0b |
| 16/19 | 3.0 |
| 16/20 | 2.4 |
| 16/21 | 1.3 |
| 16/22 | 0b |
| 17/20 | 2.0 |
| 17/21 | 0.6 |
| 17/22 | 0.5 |
| 17/23 | 0b |
| 18/21 | 1.6 |
| 18/22 | 0.3 |
| 18/23 | 0.5 |
| 18/24 | 0b |
| 19/22 | 1.2 |
| 19/23 | 0.2 |
| 23/26 | 0.4 |
| 23/27 | 0b |
Distance of proximal AG downstream from the BPS/distance of distal AG from the BPS.
The levels of the distal lariat were not greater than background.
RESULTS
AG selection depends on the BPS-to-AG and AG-to-AG distances.
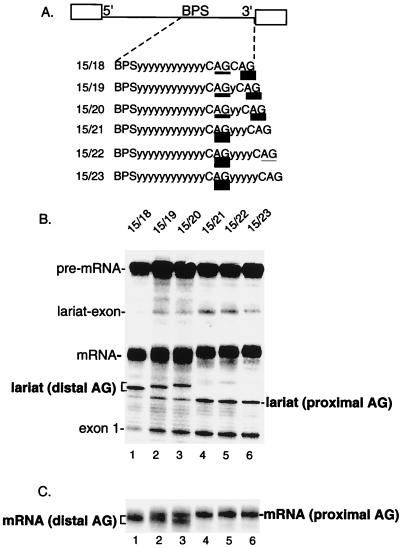
Previous analysis of a pre-mRNA containing duplicated AGs demonstrated that an AG located 11 nt from the BPS is outcompeted by an AG located 15 nt from the BPS (5). To determine whether an AG at 15 nt is optimally located, we asked whether this AG can be outcompeted by distal AGs at various positions downstream (see schematic in Fig. 1A). As shown in Fig. 1B, the proximal AG at 15 nt is efficiently outcompeted by a distal AG located 3 nt downstream (Fig. 1B, lane 1 [compare levels of distal and proximal lariat]; see Fig. 1C, lane 1, for mRNAs). However, as the distal AG is moved further downstream (Fig. 1A), the AG at 15 nt becomes more competitive until it is used almost exclusively (Fig. 1B and C, lanes 2 to 6). The same pattern of AG selection is observed at late time points (90 min) in the splicing reaction (data not shown). We conclude that distal AGs located less than 6 nt downstream from the proximal AG can efficiently compete with the proximal AG.
FIG. 1.
Distal AGs can complete with a proximal AG located 15 nt from the branch site. (A) Schematic of AdML pre-mRNA derivatives. The proximal AG is 15 nt downstream from the BPS, and the distal AG is located between 18 and 23 nt from the BPS. The relative use of the competing AGs is indicated by the thickness of the underlining and is presented in Table 1. y, pyrimidine. (B) Total RNA from splicing reactions was isolated and fractionated on a 7.5% denaturing polyacrylamide gel. The mRNA and lariat introns corresponding to use of the proximal and distal AGs were identified by comigration with known markers on gels with different percentages of polyacrylamide (data not shown). (C) A lighter exposure of the gel in panel B shows the mRNAs generated using proximal versus distal AGs.
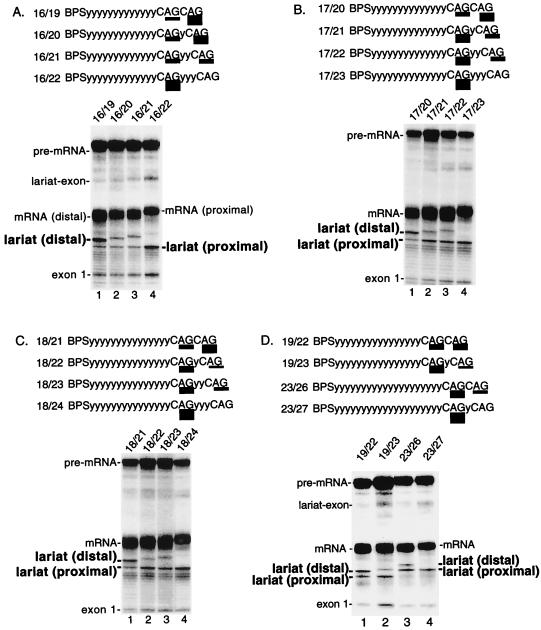
To test whether it is a general rule that distal AGs can compete with proximal AGs, we examined pre-mRNAs containing a proximal AG at increasing BPS-to-AG distances and distal AGs at increasing AG-to-AG distances (Fig. 2). This analysis revealed that distal AGs can indeed compete with proximal AGs located 16, 17, or 18 nt from the BPS (Fig. 2A, B, and C, respectively). Again, the AG-to-AG distance must be less than 6 nt in order for the distal AGs to compete. Within this AG-to-AG distance, distal AGs compete better the shorter the BPS-to-AG distance. For example, a distal AG at an AG-to-AG distance of 5 nt competes optimally when the proximal AG is at 15 nt and does not compete well when the proximal AG is at 17 or 18 nt (Fig. 1 and 2, lanes 3).
FIG. 2.
Competition between duplicated AGs depends on the BPS-to-AG distance and the AG-to-AG distance. Pre-mRNAs contain a proximal AG at 16 nt (A), 17 nt (B), 18 nt (C), and 19 or 23 nt (D) from the BPS. In each panel, a distal AG was located from 3 to 6 nt downstream of the proximal AG. Pre-mRNAs were spliced and analyzed on a 7.5% (A to C) or 6.5% (D) denaturing polyacrylamide gel. Schematics of the pre-mRNAs are shown in each panel. The relative use of each AG is indicated by the thickness of the underlining and is presented in Table 1.
Our assay using duplicated AGs shows that AG selection depends on both the BPS-to-AG distance and the distance between the two adjacent AGs. Moreover, the data indicate that there is probably a maximal BPS-to-AG distance beyond which the proximal AG outcompetes any distal AG. To find this distance, we increased the BPS-to-AG distance to 19 or 23 nt. At a BPS-to-AG distance of 19 nt, the distal AG 3 nt away was still preferentially used (Fig. 2D, lane 1). Efficient use of the proximal AG was observed only when the AG-to-AG distance was further increased to 4 nt (Fig. 2D, lane 2). Significantly, however, when the BPS-to-AG distance was 23 nt, a switch occurred. For the first time, the proximal AG was preferred even when the AG-to-AG distance was only 3 nt (Fig. 2D, lane 3). Moreover, the proximal AG was used exclusively when the AG-to-AG distance was 4 nt (Fig. 2D, lane 4). We conclude that distal AGs can no longer outcompete the proximal AG at a BPS-to-AG distance of 23 nt. Together, these data define the maximal BPS-to-AG distance beyond which a proximal AG can no longer be outcompeted by a distal AG. This distance is between 19 and 23 nt.
An optimal BPS-to-AG distance for step II.
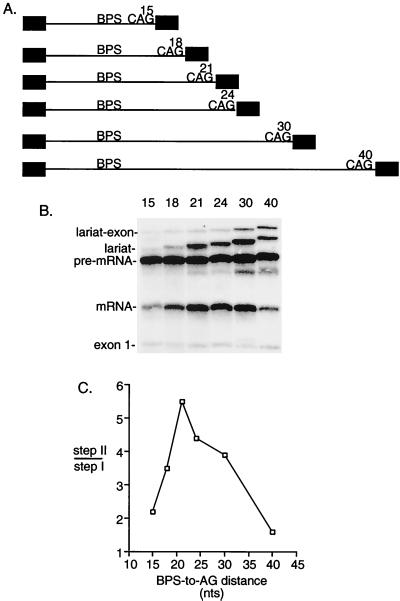
Our assay using duplicated AGs indicated that distal AGs can compete when a proximal AG is located 19 to 23 nt or less from the BPS. The observation that distal AGs can be used at all suggests that proximal AGs are sterically hindered by being too close to the BPS. If this is the case, then we would predict that the efficiency of step II should be impeded when a single AG is located 19 to 23 nt or less from the BPS. To test this possibility, we examined pre-mRNAs containing single AGs with BPS-to-AG distances ranging from 15 to 40 nt (Fig. 3A). As shown in Fig. 3B and C, step II efficiency is indeed impeded when a single AG is located 15 or 18 nt from the BPS (the level of step II was quantitated on a darker exposure of the gel [data not shown]). Significantly, however, when the AG is moved further than 21 nt from the BPS, the efficiency of step II also progressively decreases (Fig. 3B and C). Thus, these data indicate that there is an optimal BPS-to-AG distance of between 19 and 23 nt for step II. Our data with the duplicated AGs indicate that upstream of this distance AGs are sterically hindered by being too close to the BPS. The data with the single AGs indicate that beyond this distance AGs are not used efficiently because they are too far from the BPS. We conclude that step II is optimally efficient over a very narrow BPS-to-AG distance (19 to 23 nt). We note that this distance may vary with other pre-mRNAs due to their different pyrimidine tract sequences. However, given the consistency of our results, it is likely that the distance range is close to what we have observed with this pre-mRNA.
FIG. 3.
A BPS-to-AG distance of 19 to 23 nt is optimal for step II. (A) Schematic of pre-mRNAs containing single AGs located at increasing distances from the BPS. (B) Pre-mRNAs were spliced for 45 min and analyzed on a 15% denaturing polyacrylamide gel. (C) Step II efficiency was calculated as the ratio of spliced products (mRNA and lariat) to splicing intermediates (lariat-exon and exon 1).
Uncoupling two recognition steps for AG selection.
Further insight into the mechanism for AG selection came from comparing the data obtained with the duplicated and single AGs. The single-AG data show that a BPS-to-AG distance between 19 and 23 nt is optimal. Yet with the duplicated AGs, a proximal AG located closer to the BPS was used even when there was a distal AG more optimally located. For example, at a BPS-to-AG distance of 15 nt, the AG at 15 was selected over an AG at 21 nt (Fig. 1). Indeed, the location of the proximal AG, rather than the absolute distance between the BPS and distal AG, determines whether the distal AG is used (Fig. 1 and 2). Specifically, at any given BPS-to-AG distance, distal AGs must be located less than 6 nt from the proximal AG in order to be used. Thus, these data suggest that the proximal AG is recognized, even when it is not used.
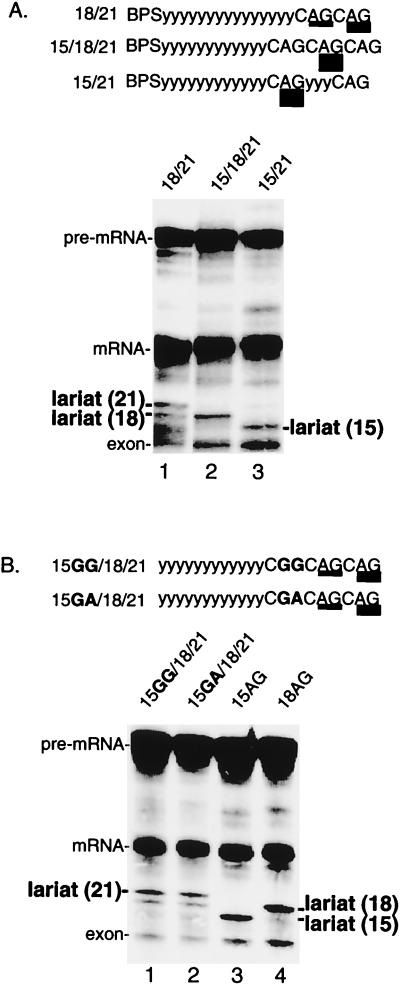
To further test this possibility, we asked whether inserting a third AG upstream of two competing AGs alters the use of the two original AGs (see schematic in Fig. 4A). As shown in Fig. 4A (lane 1), the distal AG at 21 nt is used slightly more efficiently than the proximal AG at 18 nt. When a third AG is inserted into this pre-mRNA, 15 nt from the BPS, this AG is not used (Fig. 4A, lane 2). Importantly, however, the presence of the AG at 15 nt alters the pattern of AG selection, as the AG at 18 nt is now exclusively selected (Fig. 4A, lane 2).
FIG. 4.
A proximal AG determines whether a distal AG is selected. (A) An AG at 15 nt determines selection of distal AGs. Splicing reactions were performed using pre-mRNAs containing duplicate AGs at 18 and 21 nt from the BPS (lane 1) or containing a third AG inserted at 15 nt from the BPS (lane 2). Lane 3 shows splicing of pre-mRNA 15/21, which serves as a marker for the lariat. (B) Other purine dinucleotides at 15 nt do not affect selection of distal AGs. Pre-mRNAs are identical to that shown in panel A (lane 2) except that GG (lane 1) or GA (lane 2) replaces the AG at 15 nt. Pre-mRNAs 15AG and 18AG are markers for the lariats (lanes 3 and 4).
The observation that AG selection can be switched depending on the presence or absence of the AG at 15 nt suggests that the AG at 15 nt is recognized even though it is not used. To further test this possibility, the AG at 15 nt was replaced with either a GG or a GA. Significantly, neither of these purine dinucleotides caused a switch to the AG at 18 nt. We conclude that the AG at 15 nt is specifically recognized even when it is not used. Thus, our assay using multiple AGs suggests that there is an initial step in AG recognition that can be uncoupled from the final selection of the AG.
DISCUSSION
Previous studies showed that the first AG downstream of the BPS is normally used as the 3′ splice site (reviewed in references 2 and 26). This observation is compatible with several models for 3′ splice site selection. In scanning models, the first AG is used because a 5′-to-3′ linear search is conducted. Our analysis does not support a simple scanning model. We show that AGs located closer to the BPS than 19 to 23 nt are not efficiently used, most likely due to steric hindrance from a factor bound at the BPS. Yet, the efficiency of step II also begins to decrease when AGs are located immediately beyond this distance. Thus, assuming that scanning does not become inefficient at the exact point at which it is expected to start (i.e., where steric hindrance is relieved), the AG is not likely to be selected by simple scanning. Our data do not address the previously suggested possibility that scanning is involved in selection of AGs when they are located farther than the optimal distance from the BPS (4, 9, 15, 21, 22). A simple BPS-to-AG measuring mechanism also does not explain our data because this model predicts that step II should always occur at an optimally located AG if one is present. However, our data show that AGs optimally located 19 to 23 nt from the BPS are not always selected if an upstream AG is present (see below).
An assay using duplicate AGs uncouples multiple AG recognition steps.
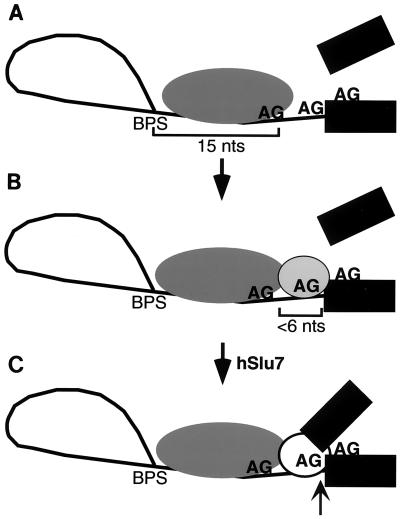
We have used pre-mRNAs containing duplicated AGs as an assay to gain insight into the mechanism of AG selection. Our results show that an AG located closer than the optimal BPS-to-AG distance (19 to 23 nt) is recognized even when it is not used. Indeed, the proximal AG determines whether a distal AG will be selected. Thus, these data reveal two distinct AG recognition events. One possible model to explain our observations, shown in Fig. 5, is that the initial recognition of the proximal AG functions to position the splicing machinery on the RNA downstream of the BPS. The recognition of this positioning AG is relatively flexible, as it can occur when an AG is suboptimally located, only 15 nt from the BPS (Fig. 5A). After this initial positioning of the splicing machinery, selection of the AG that actually functions in the transesterification reaction (the transesterification AG) occurs (Fig. 5B). Our data show that selection of the transesterification AG is more stringent than recognition of the positioning AG. Specifically, the transesterification AG must be located within a narrow range of distance (<6 nt) downstream of the positioning AG. Such a multistep mechanism, which could involve a flexible ruler followed by a stringent ruler, may accommodate the variability in the location of 3′ splice sites while still ensuring high fidelity.
FIG. 5.
Model for AG selection. (A) The proximal AG positions the splicing machinery downstream of the BPS. Recognition of this positioning AG is flexible, as this AG can be located outside the optimal distance for step II (in this example, 15 nt). (B) The AG used in the transesterification reaction is selected in a second step. This transesterification AG must be located within a narrow distance (<6 nt) downstream of the positioning AG. If a second AG is not present within this window, the positioning AG itself is used (but inefficiently). In the absence of hSlu7, the transesterification AG is sequestered, and step II is blocked at this location. (C) In the presence of hSlu7, the sequestered transesterification AG is exposed, and catalysis of step II (arrow) occurs.
In naturally occurring pre-mRNAs, closely clustered AGs are not always present at the 3′ splice site. Nevertheless, our assay using the duplicated AGs uncouples two distinct recognition events that most likely occur when only a single AG is present. It is possible that the two events involve more than one factor or a single factor that undergoes a conformational change. The only proteins known so far to affect AG selection during step II are Slu7 and hSlu7 (in yeast and humans, respectively) (6, 10). We analyzed AG selection in extracts lacking hSlu7 using all of the pre-mRNAs containing the duplicated AGs (6; our unpublished observations). In all cases, the AG that was used in normal extracts was specifically not used in the absence of hSlu7 (whereas incorrect AGs were used). Thus, the hSlu7 study shows that the transesterification AG is selected but is specifically sequestered prior to hSlu7 function. hSlu7 is then required for use of this AG in catalytic step II (Fig. 5C).
One candidate for a factor involved in recognition of the positioning AG is U2AF35, which recognizes the AG early in metazoan spliceosome assembly (17, 27, 30). However, no changes in AG selection were observed in extracts lacking U2AF35, arguing against a role for this protein (K. Chua, R. Reed, and R. Gaur, unpublished data). Previously, it was shown that recognition of a downstream AG was necessary for step I but that a proximal AG was used for step II (14, 29). In this case, it is likely that the first recognition event involved U2AF35. In contrast to these studies, in our constructs the duplicated AGs both have effects on step II.
A candidate factor for recognition of the positioning and/or the transesterification AG is the highly conserved U5 snRNP protein Prp8. In yeast, Prp8 can be UV cross-linked to the YAG, and Prp8 mutants that suppress step II defects caused by mutations in the YAG (7, 20, 23–25, 28) have been identified. These and other observations have led to the proposal that Prp8 may play a role in positioning the AG for step II (7, 20). This possibility is compatible with the observation that hPrp8 can be cross-linked on both sides of the AG at or near the time of step II in metazoans (4). Slu7 is also synthetically lethal with Prp8 in yeast (11). Thus, it will be interesting to determine how these factors function in the different events involved in AG selection.
ACKNOWLEDGMENTS
We are grateful to Z. Zhou and O. Gozani for critical comments on the manuscript.
This work was supported by NIH grant GM 43375-11 to R.R.
REFERENCES
- 1.Anderson K, Moore M J. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 2.Burge C B, Tuschl T H, Sharp P A. Splicing of precursors to mRNA by the spliceosomes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 3.Chen S, Anderson K, Moore M J. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc Natl Acad Sci USA. 2000;97:593–598. doi: 10.1073/pnas.97.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiara M D, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 7.Collins C A, Guthrie C. Allele-specific genetic interactions between Prp8 and RBA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshler J O, Rossi J J. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991;5:1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- 9.Dix I, Russell C S, O'Keefe R T, Newman A J, Beggs J D. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. . (Author's correction, 4:1675–1686.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- 11.Frank D, Patterson B, Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol Cell Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halfter H, Gallwitz D. Impairment of yeast pre-mRNA splicing by potential secondary structure-forming sequences near the conserved branchpoint sequence. Nucleic Acids Res. 1988;16:10413–10423. doi: 10.1093/nar/16.22.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krainer A R, Reed R, Maniatis T. The Robert A. Welch Foundation Conferences on Chemical Research XXIX. In Genetic chemistry: the molecular basis of heredity. Houston, Tex: Welch Foundation; 1985. Mechanisms of human beta-globin pre-mRNA splicing; pp. 353–382. [Google Scholar]
- 15.Liu Z R, Laggerbauer B, Luhrmann R, Smith C W. Crosslinking of the U5 snRNP-specific 116-kDa protein to RNA hairpins that block step 2 of splicing. RNA. 1997;3:1207–1219. [PMC free article] [PubMed] [Google Scholar]
- 16.Luukkonen B G, Seraphin B. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 18.Moore M J, Query C C, Sharp P A. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 19.Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- 20.Siatecka M, Reyes J L, Konarska M M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith C W, Porro E B, Patton J G, Nadal-Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 22.Smith C W J, Chu T T, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teigelkamp S, Newman A J, Beggs J D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umen J G, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umen J G, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 26.Umen J G, Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, Romfo C M, Nilsen T W, Green M R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 28.Wyatt J R, Sontheimer E J, Steitz J A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang Y, Weiner A M. The conserved dinucleotide AG of the 3′ splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene. 1990;90:263–269. doi: 10.1016/0378-1119(90)90189-x. [DOI] [PubMed] [Google Scholar]
- 30.Zorio D A, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]