Abstract
Phyllosphere—the harsh foliar plant part exposed to vagaries of environmental and climatic variables is a unique habitat for microbial communities. In the present work, we profiled the phyllosphere microbiome of the rice plants using 16S rRNA gene amplicon sequencing (hereafter termed metabarcoding) and the conventional microbiological methods (culturomics) to decipher the microbiome assemblage, composition, and their functions such as antibiosis and defense induction against rice blast disease. The blast susceptible rice genotype (PRR78) harbored far more diverse bacterial species (294 species) than the resistant genotype (Pusa1602) that showed 193 species. Our metabarcoding of bacterial communities in phyllomicrobiome revealed the predominance of the phylum, Proteobacteria, and its members Pantoea, Enterobacter, Pseudomonas, and Erwinia on the phyllosphere of both rice genotypes. The microbiological culturomic validation of metabarcoding-taxonomic annotation further confirmed the prevalence of 31 bacterial isolates representing 11 genera and 16 species with the maximum abundance of Pantoea. The phyllomicrobiome-associated bacterial members displayed antifungal activity on rice blast fungus, Magnaporthe oryzae, by volatile and non-volatile metabolites. Upon phyllobacterization of rice cultivar PB1, the bacterial species such as Enterobacter sacchari, Microbacterium testaceum, Pantoea ananatis, Pantoea dispersa, Pantoea vagans, Pseudomonas oryzihabitans, Rhizobium sp., and Sphingomonas sp. elicited a defense response and contributed to the suppression of blast disease. qRT-PCR-based gene expression analysis indicated over expression of defense-associated genes such as OsCEBiP, OsCERK1, and phytohormone-associated genes such as OsPAD4, OsEDS1, OsPR1.1, OsNPR1, OsPDF2.2, and OsFMO in phyllobacterized rice seedlings. The phyllosphere bacterial species showing blast suppressive activity on rice were found non-plant pathogenic in tobacco infiltration assay. Our comparative microbiome interrogation of the rice phyllosphere culminated in the isolation and identification of agriculturally significant bacterial communities for blast disease management in rice farming through phyllomicrobiome engineering in the future.
Keywords: antibiosis, blast, defense genes, Magnaporthe oryzae, microbiome, phyllosphere, rice, immunocompetence
Introduction
Microbial communities have an evolutionary association with plant populations where they function as metaorganisms in the natural environment. Here, the microbial activities in total termed microbiomes play a pivotal role in plant development and survival (Hartmann et al., 2008; Bulgarelli et al., 2013; Berg et al., 2016). The microbial communities associated with the plants are called plant microbiome and microbial metagenome that often confer functional flexibility to the plant genome (Sessitsch et al., 2012). The plant microbiomes are presumed to modulate a variety of plant functions. However, the ecological role of the phyllosphere microbial communities on plant functional ecology is among the most understudied and underrated aspects in plant biology.
The microbial life on the foliar niche, the phyllosphere microbiome, is constantly exposed to vagaries of weather events, and other agronomic practices in crop husbandry (Lindow and Leveau, 2002). Currently, the epiphytic phyllosphere microbiomes and their natural functions are increasingly investigated in crops like rice, wheat, maize, and soybean (Andrews and Harris, 2000; Bertani et al., 2016; Compant et al., 2019; Sahu et al., 2020). It is further reported that plant-associated bacteria are prolific for the secretion of primary and secondary metabolites and volatiles for plant growth, developmental regulation, and defense against stresses (Munjal et al., 2016; Rascovan et al., 2016; Sheoran et al., 2016; Gómez Expósito et al., 2017; Eke et al., 2019; Ashajyothi et al., 2020; Vandana et al., 2021).
Rice is the primary staple for the nearly three billion world population and contributes to global food security. Rice production is affected by several biotic and abiotic stresses; among them, blast disease caused by ascomycetous fungus Magnaporthe oryzae (anamorph Pyricularia oryzae Sacc.) is responsible for nearly 30.0% of losses, which can feed 60 million population if prevented preemptively (Dean et al., 2005; Scheuermann et al., 2012; Yasuda et al., 2015; Hashim et al., 2018; Mehta et al., 2019; Prakash et al., 2021). Deployment of fungicides and blast-resistant cultivars are among the blast-combating strategies widely practiced. However, both strategies are under scanner as the chemicals are no longer encouraged due to safety considerations, and the host resistance is not durable (Nalley et al., 2016; Asibi et al., 2019; Sella et al., 2021). Recently, the fungicide residues intercepted on the Indian rice imports have prompted many countries to reject consignments from international trade (Al-Antary et al., 2020). Under this scenario, there is a growing demand among the various stakeholders of the rice production system for an alternative blast mitigation strategy. One promising yet unexplored strategy is the biological control of blast disease by deploying leaf microbiota that shares the same microecological niche with the blast pathogen, M. oryzae. So far, the potential of phyllosphere microbial communities sharing the leaf microhabitat with Magnaporthe—the incitant of blast disease has not yet been harnessed to mitigate the disease in any crop. Hence, the present investigation was carried out to explore the phyllo-microbiome of rice and exploit them for blast disease suppression by microbiome reengineering.
Materials and Methods
Study Location and Sampling for Microbiome Analysis
The 16S rRNA gene amplicon sequencing by NGS (hereafter cited as metabarcoding; Berg et al., 2020), combined with conventional culturomic investigation of phyllomicrobiome, was conducted. For this, we planted rice genotypes in a blast-endemic mountain ecosystem in Palampur, Himachal Pradesh, India (32°6′4.7″N and 76°32′39.79″E) located at an altitude of 1,275 m above mean sea level [weather conditions: mean temperature 22–23°C; precipitation 700–1,000 mm; RH 60.0%; sunshine hours 300–350; source: https://en.climate-data.org; www.worldweatheronline.com]. The rice genotypes were grown during the rice-growing season in August to September 2014. Briefly, rice genotypes PRR78—a blast susceptible variety, and Pusa1602—a near-isogenic line of PRR78 introgressed with Pi2 gene conferring complete resistance to blast disease (Singh et al., 2012), were planted in parallel rows with a spacing of 20 cm by adopting all crop husbandry practices. Leaf samples were excised at 15 and 30 days post-sowing in sterilized falcon tubes and brought to the laboratory in an insulated cool container maintained at a temperature of 4.0 ±1.0°C and processed for microbiome analysis by metabarcoding and culturomic analysis.
Profiling of Phyllomicrobiome by Metabarcoding
Extraction and Isolation of Epiphytic Microbial Community Genomic DNA
Leaf (5.0 g) samples were shaken with 50 ml of sterile phosphate buffer saline [PBS, g L–1 NaCl 8; KCl 0.2; Na2HPO4 1.44; KH2PO4 0.24; pH-7.4] amended with 0.1% Tween-20 (PBS-T) to dislodge the epiphytic microbiome. The leaf epiphytic microbiome was extracted six times serially in 50 ml of PBS-T by agitating for 30 min at 250 rpm and vortexing for 10 s. Thus, the collected epiphytic–microbial suspension (300 ml) was collected aseptically in a presterilized container and centrifuged at 12,000 × g for 60 min at 4.0°C to collect the epiphytic microbial cells. Thus, the obtained pellet was processed to isolate genomic DNA by the cetyltrimethyl ammonium bromide (CTAB) method reported by Moore et al. (2004) with slight modifications like avoidance of phenol in the extraction steps. The quality and quantity of microbial community genomic DNA was determined electrophoretically, spectrophotometrically (Nanodrop 2000, Thermo Scientific, United States), and fluorometrically using Qubit dsDNA BR Assay (Thermo Fisher Scientific Inc., United States).
Preparation of Sequencing Libraries for 2 × 300-bp Run Chemistry
The amplicon libraries were prepared using Nextera XT Index Kit (Illumina Inc.) as per the 16S rRNA Gene Amplicon Sequencing Library Preparation Protocol (Part no. 15044223 Rev. B). PCR primers for the amplification of the 490-bp hypervariable region of V3–V4 of 16S rRNA gene of Eubacteria were designed, synthesized, and used. The sequences of the primers are V3F: 5′CCTACGGGNGGCWGCAG3′ and V4R: 5′GACTACHVGGGTATCTAATCC3′. The target amplicons were generated using the fusion primer that consists of Illumina adaptors and multiplex index sequence as per the instructions of the manufacturer. The amplicon libraries were purified by 1× AMpure XP beads and checked on Agilent High Sensitivity (HS) chip on Bioanalyzer 2100 and quantified on fluorometer by QubitdsDNA HS-Assay kit (Life Technologies, United States). Quality-passed libraries were equimolar pooled and then sequenced using the IlluminaMiSeq platform with 2 × 300-bp paired-end sequencing chemistry following the protocols of the manufacturer (Illumina, San Diego, CA, United States).
Bioinformatic Analyses
Initially, the sequenced raw forward reads (R1) and reverse reads (R2) were scanned and analyzed using the FastQC version (Andrews, 2018) to assess the quality of 16rRNA amplicon reads. Thus, the obtained raw reads were end trimmed and curated using Trimmomatic v0.35 (Bolger et al., 2014) with the following command and settings: (i) remove adapter sequences, (ii) ambiguous reads (reads with unknown nucleotides “N” larger than 5.0%), and (iii) low-quality sequences [reads with more than 10.0% quality threshold (quality value) <20 Phred score]. The resultant quality-passed read pairs were joined using PEAR (Paired-End reAdmergeR) version 0.9.8 (Zhang et al., 2014) with default parameters. The joined paired reads were further processed for downstream taxonomic classification where unpaired reads were discarded. The taxonomic classification of resultant high-quality reads was performed using MG-RAST v4.0, wherein (i) 16S rRNA gene sequence reads were sorted using Sortme RNA, (ii) sorted reads were clustered at ≥97% similarity using CD-HIT method, and (iii) clustered reads were taxonomically classified using SILVA SSU database. The clustered reads and taxon abundance downloaded >100 bases and 90.0% similarity through the best hit classification. Furthermore, PAST v2.17c (Hammer et al., 2001) was used for the determination of α-diversity.
Phyllosphere Microbiome Interrogation by Culturomic Methods
Isolation and Characterization of the Culturable Epiphytic Microbiome
Another set of leaves (500 mg) excised from the rice genotypes were analyzed using the culturomic method on nutrient agar medium [NA, g L–1; peptone 5.0; beef extract 3.0; NaCl 5.0; agar 15.0; pH 7.0 ± 0.2]. Briefly, the leaf was agitated with 50 ml of sterile phosphate buffer saline amended with 0.1% Tween-20 (PBST) for 30 min at 250 rpm followed by vortexing for 10 s; the aliquot, thus, obtained was decimally diluted up to 10–5. An aliquot of 1.0 ml at 10–3, 10–4, and 10–5 from each sample was pour plated in nutrient agar media supplemented with 2,3,5-tetrazolium chloride (50 mg L–1) and incubated at 28 ± 2°C for 72 h for morphotyping the bacterial colonies. The culturable bacterial population and their diversity were assessed and quantified based on morphological traits, such as size, shape, color, texture, and margin. The pure culture of the representative isolates was preserved in −80 and −20°C as glycerol stock (30% V/V) for downstream work.
Identification of Epiphytic Bacterial Species by 16S rRNA Gene Sequencing
Genomic DNA was isolated by the CTAB method described by Moore et al. (2004) with minor modifications as mentioned previously. Isolated and purified genomic DNA was quantitated and quality analyzed as described above. Finally, the genomic DNA reconstituted at 100 ng μl–1 was used as a template in PCR amplification. Box PCR-based DNA fingerprinting was performed for diversity analysis as well as to eliminate the duplicate isolates from the collection (Versalovic et al., 1994); this PCR-based DNA profiling technique specifically amplifies the non-coding conserved sequences in the bacterial genome and is considered a highly discriminatory DNA-fingerprinting technique (Kumar et al., 2004; Eke et al., 2019). Amplicon profiles were resolved in 1.0% agarose gel at 30 V for 10–12 h and imaged (QuantityOne, BioRad, United States). Isolates showing identical amplicon profiles were presumed to be duplicates. PCR amplification of the 16S rRNA gene was performed using primer sets, 27F (27F: 5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (1492R: 5′-GGTTACCTTGTTACGACTT-3′), to amplify the 1,465-bp region (Sheoran et al., 2015; Munjal et al., 2016). Then the PCR amplicons resolved in the agarose gel (1.0 %) were purified and eluted using an elution kit according to the instructions of the manufacturer’ (Promega Corporation, United States). The amplicons were sequenced bidirectionally to achieve maximum coverage of the sequences and analyzed using the nucleotide-Basic Local Alignment Search Tool (BLAST) algorithm at the National Center for Biological Information (NCBI); the bacterial species identity was confirmed by closest match. The diversity analysis of culturable phyllosphere bacterial species was calculated using Shannon diversity indices.
Activity Screening of Epiphytic Phyllospheric Bacteria on Magnaporthe oryzae
Antifungal Activity in vitro
Airborne volatile organic compounds emitted, and diffusible metabolites secreted, during bacterial growth were tested for antifungal activities on M. oryzae. Here, we conducted dual-culture confrontation assays, and mycelial inhibition over mock was calculated as described (Sheoran et al., 2015; Munjal et al., 2016). Additionally, the fungicidal or fungistatic nature of the antifungal activity of volatiles on M. oryzae was also determined. Here, the volatile exposed mycelia of M. oryzae showing complete inhibition were further incubated after replacing bacterial volatile with lid. Depending on the mycelial growth, the bacterial volatile was either categorized as fungicidal or fungistatic on M. oryzae (Sahu et al., 2020). The radial mycelial growth of M. oryzae was measured, and mycelial inhibition (%) over mock was calculated using the following formula
where I = percent inhibition
C = colony diameter in control
T = colony diameter in treatment
Blast-Suppressive Activity in planta
The bacterial isolates showing inhibition of mycelial growth of M. oryzae were selected for in planta blast control assay. Here, blast-susceptible rice genotype, Pusa Basmati 1, was allowed to germinate in the bacterial cell suspension set at three different bacterial densities (∼106, 107, and 108 CFU ml–1) for 5 days. Upon germination, the transplants were further grown in a climate-controlled greenhouse set at a temperature of 28.0 ± 2.0°C, relative humidity of 90.0 ±10.0%, and light/dark cycles of 14/10 h. Thus, the obtained 3-week-old seedlings were foliar sprayed (booster spray) with phyllosphere bacterial suspension (∼106, 107, and 108 CFU ml–1) and challenged with the conidia of M. oryzae-1637 prepared in water (2.0 × 105 conidia ml–1) (Rajashekara et al., 2017). Blast disease severity was determined 7 days post-inoculation on a 0.0–5.0 disease rating scale where 0.0 = no evidence of infection; 1.0 = brown specks <0.5 mm in diameter; 2.0 = brown specks of 0.5–1.0 mm in diameter; 3.0 = round to elliptical lesions of about 1–3 mm in diameter; 4.0 = typical spindle-shaped blast lesion of 3.0 mm or more with little or no coalescence of the lesion; 5.0 = the same as 4.0 but half or more leaves killed by coalescence of lesions. Plants scored 0.0–2.0 were rated resistant, 3.0 as moderately susceptible, and 4.0–5.0 as susceptible (Mackill and Bonman, 1992). The disease severity was calculated using the following formula.
Furthermore, the percent reduction in disease severity compared with control was estimated using the following formula:
C = disease severity in controlT = disease severity in treatment
Phenotyping for Phyllomicrobiome Conferred Immunocompetence
The bacterial isolates showing antifungal activity on M. oryzae were selected for the immunocompetence assay. Here, the germination and phenotypic alterations induced on rice seedlings by bacterial isolates were monitored and scored. In this assay, the blast susceptible Pusa Basmati 1 was subjected to germination for 5 days in the presence of bacterial cells at varying densities such as 106, 107, 108, and 109 (CFU mL–1). Similarly, seeds germinated in sterile double distilled water served as control. The experiment was performed in three replications with 50 seeds in each replication and repeated twice. The seed germination (%) was calculated using the following formula to evaluate the effect of bacterial interaction on germination.
To test the effect of bacterial colonization on the shoot and root growth, five randomly selected seedlings were scored. The percent deviation in shoot and root growth was calculated against the untreated mock seedlings using the following formula. Here, while the negative value indicated growth inhibition, the positive score indicated growth promotion.
where G = percent growth of shoot/root
C = length of shoot/root in control
T = length of shoot/root in the treatment
Assessment of Immunocompetence by qPCR
Having confirmed the blast-suppressive potential of the phyllosphere bacterial species, we performed qPCR experiments to decipher the effect of bacterial supplementation on the expression of genes involved in defense pathways in rice. A total of nine phyllosphere bacterial isolates such as Pantoea vagans OsEp-Plm-30B3, Pantoea ananatis OsEp-Plm-15B6, Enterobacter sacchari OsEp-Plm-15B10, P. ananatis OsEp-Plm-30B17, Pantoea dispersa OsEp-Plm-15B14, Rhizobium sp. OsEp-Plm-30B4, Microbacterium testaceum OsEp-Plm-30B1, Pseudomonas oryzihabitans OsEp-Plm-15B16, and Sphingomonas sp. OsEp-Plm-15B2 that showed blast-suppressive activity on rice was chosen for the study.
Briefly, whole seedlings of Pusa Basmati 1, bacterized with 2 × 107 CFU m–1 and sampled at 24-h interval for three consecutive days were immediately snap frozen in liquid nitrogen to stop all the cellular metabolic activity and then stored instantly at −80°C until further use. The total RNA was isolated using the SV Tool RNA isolation system according to the instructions of the manufacturer (Promega, Madison, WI, United States). The quality and quantity of RNA were assessed spectrophotometrically (NanoDrop 2000, Thermo Scientific, United States) and electrophoretically.
Transcriptional Analysis of Genes Associated With Immunocompetence
Eight rice genes, such as OsCEBiP (Akamatsu et al., 2013), OsCERK1 (Kouzai et al., 2014), OsPAD4 (Ke et al., 2014), OsEDS1 (Ke et al., 2019), OsNPR1 (Sugano et al., 2010), OsPDF2.2 (Thomma et al., 2002), OsFMO1 (Koch et al., 2006; Mishina and Zeier, 2006), and OsPR1.1 (Breen et al., 2017), which were reported to play a role in rice defense, were selected; PCR primers targeting the above defense genes are furnished in Supplementary Tables 1, 2. qPCR was performed in Real-Time Thermal Cycler (LightCycler 96, Roche Life Science, Switzerland) using GoTaq® 1-Step RT-qPCR System (Promega Corporation, United States); qPCR reaction conditions were as follows: one cycle of reverse transcription at 37°C for 15 min followed by reverse transcriptase inactivation step at 95°C for 10 min followed by 30 cycles at 95°C for 10 s, annealing at 58°C for 30 s and extension at 72°C for 30 s followed by three-step melting at 95°C for 10 s, 63°C for 60 s, and 97°C for 1.0 s, and then a final cooling at 37°C for 30 s. Later, cyclic threshold data points were analyzed for the determination of gene expression relative to the reference housekeeping OsActin gene using the software LightCycler®96 Roche. The mean Ct values were considered for the calculation of 2–ΔΔCT to estimate the fold changes in gene expression.
Hypersensitive Reaction on Tobacco
Upon bacterial infiltration on tobacco leaves, potential plant pathogenic bacteria are known to induce hypersensitive reactions (HR) (Klement, 1963); this is considered as a test to ascertain the plant pathogenic nature of bacterial isolates. The best performing nine bacterial isolates for suppression of blast disease were selected for this assay. Tobacco (Nicotiana tabacum) plants were grown under greenhouse conditions at 20°C, 50–60% relative humidity, and 12/12-h light/dark per day. Fully expanded leaves of 2 to 3-month-old plantlets were used in all experiments. Bacterial inoculum (1.0 × 108 CFU ml–1, absorbance at 600 nm = 1.0 OD) was infiltrated onto the leaves using a sterile hypodermal syringe. Thus, treated plants were incubated at 25–30°C under greenhouse conditions (12/12 h of dark/light photoperiods). Similarly, leaves infiltrated with sterile distilled water alone served as a negative control, and a well-known bacterial pathogen, Ralstonia solanacearum, served as a positive control. Plant responses to the bacterial infiltration on tobacco leaves were recorded after 24-h post-inoculation.
Statistical Analysis
All datasets were analyzed using the data analytical tool available in MS Office Excel 2013. The analyzed data obtained were subjected to significance testing by analysis of variance (ANOVA) at a p ≤ 0.05 level of significance. Furthermore, various parameters like the standard error of the mean (SEm), standard error of the difference between two means (SEd), critical difference (CD), and coefficient of variation (CV) were estimated. For figures and tables, the values are represented as the mean of all biological and technical replicates. For the qPCR data analysis, the fold change values calculated for the defense genes were imported into the GraphPad Prism program (https://www.graphpad.com/scientific-software/prism), and two-way ANOVA was conducted using Bonferroni post-hoc test for determining the statistical significance at *p ≤ 0.05, **p = 0.001, and ***p = 0.0001.
Results
16S rRNA Barcode Sequence Read Statistics and Diversity Indices
Phyllomicrobiome profiling of blast-susceptible (PRR78) and blast-resistant rice (Pusa1602) genotypes planted in blast-endemic locations was conducted using integrated 16S rRNA gene amplicon sequencing and microbiological methods. The total curated sequence generated is in the range of 4, 31,222 for Pusa1602 and 1, 81,250 for PRR78 reads (Table 1). The α-diversity indices (Shannon diversity) were 1.25 for Pusa1602 and 1.85 for PRR78. Other diversity indices were also marginally higher for PRR78 than Pusa1602 revealing that the blast-susceptible genotype harbored more diverse microbial species than the resistant type (Table 1).
TABLE 1.
Metabarcoding statistics and diversity indices of phyllosphere microbiome.
| Parameters | Sample origin: Mid Himalayan mountain—Palampur, India | |
|
|
||
| Pusa1602 | PRR78 | |
| MG-RAST accession number* | mgm4619774.3 | mgm4621255.3 |
| Number of base pairs | 201,387,096 | 221,346,634 |
| Total number of sequences | 4,39,681 | 4,68,994 |
| Total number of reads | 431,222 | 181,250 |
| Simpson | 0.6304 | 0.6936 |
| Shannon | 1.253 | 1.851 |
| Evenness | 0.01813 | 0.02166 |
| Fisher-α | 19.03 | 33.71 |
| Berger–Parker | 0.4339 | 0.5018 |
| Chao-1 | 310.4 | 421.7 |
| Observed species | 193 | 294 |
Structure and Composition of Phyllomicrobiome on Blast-Susceptible and Resistant Rice Genotypes
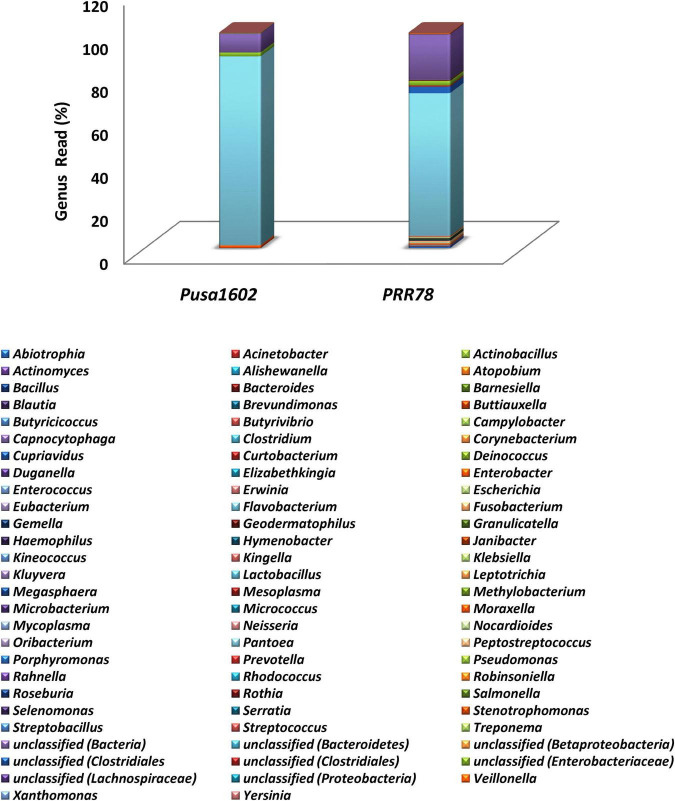
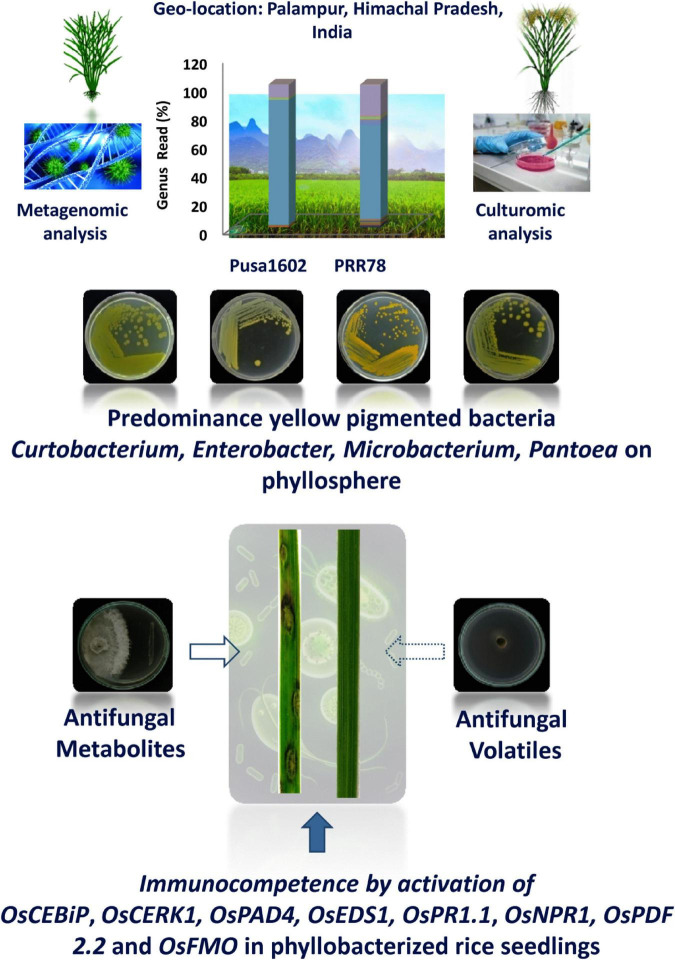
The metabarcoding-assisted taxonomic profiling of the phyllosphere microbiome of two rice genotypes revealed the abundance of bacterial phyla, Proteobacteria (70.7–91.0%), and class Gamma Proteobacteria on both the genotypes (70.4–91.0%). Bacterial communities belong to the order Enterobacteriales (89.2%) followed by Pseudomonadales (1.7%) was found dominant on Pusa1602, while Enterobacteriales (68.0%) followed by Bacteroidales (3.7%) and Pseudomonadales (2.2%) were in high frequency on susceptible genotype, PRR78. Further at the family level, Enterobacteriaceae (89.2%) followed by Pseudomonadaceae (1.7%) in the resistant genotype and Enterobacteriaceae (68.0%) followed by Porphyromonadaceae (3.0%) and Pseudomonadaceae (2.0%) in the susceptible genotype were found overrepresented. Bacterial genus Pantoea (67.3–87.7%) was the most dominant bacteria on both the genotypes. Other dominant genera are Pseudomonas, Enterobacter, Buttiauxella, and Erwinia in the resistant genotype and Porphyromonas, Pseudomonas, Abiotrophia, Enterobacter, and Gemella in the susceptible genotype (Figure 1, Supplementary Figure 1, and Supplementary Table 3).
FIGURE 1.
Genus-level relative abundance of phyllosphere bacterial communities on rice genotypes; refer to Supplementary Figure 1 for other taxonomic hierarchy.
Validation of Metabarcoding Sequence Data by Culturomic Methods
Enumeration of Cultivable Microbiome and Identification of Bacterial Communities
Both the genotypes recorded the nearly identical epiphytic bacterial population (1.55–5.6 log CFU g–1) (Supplementary Table 4). A total of 37 distinct morphotypes of bacterial isolates were enumerated with a diversity index ranging from 1.48 to 1.82 for all the cultured phyllosphere microbiome. The 30-day-old phyllosphere showed more bacterial population (21 morphotypes) and diversity compared with the 15-day seedlings (16 morphotypes). The results of diversity indices further revealed more bacterial diversity in the susceptible genotypes than in the resistant genotypes. The diversity indices of epiphytic bacteria on the rice phyllosphere are presented in Supplementary Table 5. BOX-PCR DNA fingerprinting of all 37 morphotypes culminated in 31 distinct BOX Amplicon Groups (Supplementary Figure 2). Isolates with identical amplicon profiles were considered duplicates, and a representative isolate for each of the BOX groups was retained and subjected to downstream work. The bacterial species identity was established through a 16S rRNA gene sequence analysis. The 31 distinct BOX amplicon groups represented 11 genera and 16 species. We also observed high-frequency occurrence of bacterial species like Acinetobacter (2), Curtobacterium (3), Enterobacter (4), Microbacterium (4), Pantoea (9), Pseudomonas (2), Rhizobium (2), and Sphingomonas (2) on the rice phyllosphere (Table 2 and Supplementary Figures 3, 4a–k). All cultured bacterial genera were also found among the mapped reads in the metabarcoding analysis (Figure 2 and Table 3).
TABLE 2.
Identification of cultivated phyllosphere bacterial isolates by 16S rRNA gene sequencing.
| Sequence ID | Organism | Sequence length (bp) | *Host | GenBank accession |
| OsEp_Plm_15B9 | Acinetobacter calcoaceticus | 1,409 | PRR78 | MT367784 |
| OsEp_Plm_15B13 | Curtobacterium sp. | 1,400 | PRR78 | MT367788 |
| OsEp_Plm_30B20 | Enterobacter ludwigii | 1,422 | PRR78 | MT367806 |
| OsEp_Plm_30B8 | Pantoea ananatis | 1,401 | PRR78 | MT367799 |
| OsEp_Plm_15B16 | Pseudomonas oryzihabitans | 1,403 | PRR78 | MT367791 |
| OsEp_Plm_15B8 | Rhizobium taibaishanense | 1,349 | PRR78 | MT367783 |
| OsEp_Plm_30B9 | Sphingomonas pseudosanguinis | 1,400 | PRR78 | MT367800 |
| OsEp_Plm_15B4 | Acidovorax avenae | 1,400 | PRR78 and Pusa1602 | MT367779 |
| OsEp_Plm_15B15 | Acinetobacter radioresistens | 1,402 | PRR78 and Pusa1602 | MT367790 |
| OsEp_Plm_30B7 | Agrobacterium vitis | 1,400 | PRR78 and Pusa1602 | MT367798 |
| OsEp_Plm_15B3 | Curtobacterium luteum | 1,391 | PRR78 and Pusa1602 | MT367778 |
| OsEp_Plm_15B12 | Curtobacterium luteum | 1,393 | PRR78 and Pusa1602 | MT367787 |
| OsEp_Plm_30B10 | Enterobacter cloacae | 1,403 | PRR78 and Pusa1602 | MT367801 |
| OsEp_Plm_15B10 | Enterobacter sacchari | 1,419 | PRR78 and Pusa1602 | MT367785 |
| OsEp_Plm_15B11 | Enterobacter sp. | 1,402 | PRR78 and Pusa1602 | MT367786 |
| OsEp_Plm_15B7 | Enterococcus faecium | 1,409 | PRR78 and Pusa1602 | MT367782 |
| OsEp_Plm_15B5 | Microbacterium sp. | 1,379 | PRR78 and Pusa1602 | MT367780 |
| OsEp_Plm_15B1 | Microbacterium testaceum | 1,400 | PRR78 and Pusa1602 | MT367776 |
| OsEp_Plm_30B1 | Microbacterium testaceum | 1,398 | PRR78 and Pusa1602 | MT367792 |
| OsEp_Plm_30B5 | Microbacterium testaceum | 1,401 | PRR78 and Pusa1602 | MT367796 |
| OsEp_Plm_15B6 | Pantoea ananatis | 1,405 | PRR78 and Pusa1602 | MT367781 |
| OsEp_Plm_30B2 | Pantoea ananatis | 1,404 | PRR78 and Pusa1602 | MT367793 |
| OsEp_Plm_30B6 | Pantoea ananatis | 1,408 | PRR78 and Pusa1602 | MT367797 |
| OsEp_Plm_30B15 | Pantoea ananatis | 1,412 | PRR78 and Pusa1602 | MT367803 |
| OsEp_Plm_30B17 | Pantoea ananatis | 1,384 | PRR78 and Pusa1602 | MT367804 |
| OsEp_Plm_30B19 | Pantoea ananatis | 1,417 | PRR78 and Pusa1602 | MT367805 |
| OsEp_Plm_15B14 | Pantoea dispersa | 1,410 | PRR78 and Pusa1602 | MT367789 |
| OsEp_Plm_30B3 | Pantoea vagans | 1,409 | PRR78 and Pusa1602 | MT367794 |
| OsEp_Plm_30B14 | Pseudomonas sp. | 1,408 | PRR78 and Pusa1602 | MT367802 |
| OsEp_Plm_30B4 | Rhizobium sp. | 1,360 | PRR78 and Pusa1602 | MT367795 |
| OsEp_Plm_15B2 | Sphingomonas sp. | 1,373 | PRR78 and Pusa1602 | MT367777 |
*Isolated from rice leaf excised from PRR78 and Pusa1602 planted in Palampur, Himachal Pradesh, India.
FIGURE 2.
Microbiological culturomic validation of bacterial species composition in phyllomicrobiome; bacterial species belonging to yellow-pigmented Curtobacterium, Enterobacter, Microbacterium, Pseudomonas, Pantoea, and Sphingomonas were found dominant on the phyllosphere. Data in parentheses represent the number of isolates cultured. *Species identity in Silva Database. #Species identity in GenBank database.
TABLE 3.
Quantification and identification of bacterial population on rice phyllomicrobiome by integrated metabarcoding and culturomic methods.
| Genus | Metabarcoding method |
Culturomic method |
||||
| Blast-resistant Pusa1602 |
Blast-susceptible PRR78 |
Blast-resistant Pusa1602 |
Blast-susceptible PRR78 |
|||
| *Read count | Reads (%) | *Read count | Reads (%) | Log CFU g–1 | Log CFU g–1 | |
| Pantoea | 423,893 | 87.751 | 137,297 | 66.357 | 4.73 | 5.17 |
| Pseudomonas | 8,433 | 1.746 | 4,226 | 2.042 | 3.23 | 4.47 |
| Enterobacter | 4,071 | 0.843 | 1,270 | 0.614 | 4.13 | 5.00 |
| Buttiauxella | 1,210 | 0.250 | − | − | − | − |
| Erwinia | 501 | 0.104 | 264 | 0.128 | − | − |
| Klebsiella | 402 | 0.083 | 769 | 0.372 | − | − |
| Clostridium | 144 | 0.030 | 172 | 0.083 | − | − |
| Salmonella | 141 | 0.029 | 97 | 0.047 | ||
| Bacteroides | 116 | 0.024 | 346 | 0.167 | − | − |
| Enterococcus | 40 | 0.008 | 142 | 0.069 | 3.65 | 2.16 |
| Curtobacterium | 38 | 0.008 | 10 | 0.005 | 2.24 | 4.64 |
| Kineococcus | 35 | 0.007 | 139 | 0.067 | − | − |
| Microbacterium | 27 | 0.006 | 16 | 0.008 | 5.66 | 5.90 |
| Acinetobacter | 22 | 0.005 | − | − | 3.49 | 3.74 |
| Abiotrophia | − | − | 1,522 | 0.736 | − | − |
| Acidovorax | − | − | − | − | 4.54 | 5.37 |
| Agrobacterium | − | − | − | − | 3.87 | 2.47 |
| Butyrivibrio | − | − | 134 | 0.065 | − | − |
| Campylobacter | − | − | 101 | 0.049 | − | − |
| Capnocytophaga | − | − | 518 | 0.250 | − | − |
| Elizabethkingia | − | − | 206 | 0.100 | − | − |
| Escherichia | − | − | 755 | 0.365 | − | − |
| Flavobacterium | − | − | 152 | 0.073 | − | − |
| Fusobacterium | − | − | 1,093 | 0.528 | − | − |
| Gemella | − | − | 1,213 | 0.586 | − | − |
| Granulicatella | − | − | 807 | 0.390 | − | − |
| Haemophilus | − | − | 475 | 0.230 | − | − |
| Kluyvera | − | − | 135 | 0.065 | − | − |
| Leptotrichia | − | − | 220 | 0.106 | − | − |
| Moraxella | − | − | 382 | 0.185 | − | − |
| Neisseria | − | − | 450 | 0.217 | − | − |
| Porphyromonas | − | − | 6,355 | 3.071 | − | − |
| Prevotella | − | − | 1,064 | 0.514 | − | − |
| Rhizobium | − | − | − | − | 5.13 | 5.43 |
| Rothia | − | − | 696 | 0.336 | ||
| Sphingomonas | − | − | − | − | 4.17 | 4.82 |
| Streptobacillus | − | − | 107 | 0.052 | − | − |
| Veillonella | − | − | 608 | 0.294 | − | − |
*Genus with less than 10 reads were not considered.
Activity Screening Against Rice Blast Fungus Magnaporthe oryzae
Screening for Antifungal Activity
Dual-plate confrontation assay showed inhibition of the mycelial growth of M. oryzae by both volatiles and secreted metabolites produced by bacterial species. Among the 31 bacteria evaluated, 11 phyllosphere-associated bacterial isolates displayed over 40.0% inhibition of mycelial growth by their secreted metabolites (Table 4 and Supplementary Figure 5). The antagonistic bacterial isolates represented species, such as Acinetobacter calcoaceticus, Enterobacter cloacae, E. sacchari, P. ananatis, P. vagans, P. oryzihabitans, and Sphingomonas sp. Similarly, a total of 12 of them completely inhibited the growth of M. oryzae (100% inhibition) by bacterial volatile organic compounds (Table 4 and Supplementary Figure 6). The antifungal volatile emitting bacterial isolates represented the species, such as E. sacchari, M. testaceum, P. ananatis, P. dispersa, P. vagans, P. oryzihabitans, and Rhizobium sp. Furthermore, while the volatile of seven bacterial isolates displayed fungicidal activity, the other five bacteria released fungistatic volatiles against M. oryzae as mycelial growth re-emerged upon removal of the volatile exposure (Supplementary Figure 7 and Supplementary Table 6).
TABLE 4.
Antifungal activity of phyllosphere bacteria by secreted metabolites and volatile compounds against Magnaporthe oryzae.
| Bacterial isolates | Mycelial inhibition (%) |
|
| Volatile compounds | Secretory compounds | |
| 1. Enterobacter sacchari OsEp-Plm-15B10 | 100.0 | 57.4 |
| 2. Microbacterium testaceum OsEp-Plm-30B1 | 100.0 | 38.9 |
| 3. Pantoea dispersa OsEp-Plm-15B14 | 100.0 | 35.2 |
| 4. Pantoea ananatis OsEp-Plm-15B6 | 100.0 | 19.4 |
| 5. Pantoea ananatis OsEp-Plm-30B2 | 100.0 | 53.7 |
| 6. Pantoea vagans OsEp-Plm-30B3 | 100.0 | 47.2 |
| 7. Pantoea ananatis OsEp-Plm-30B6 | 100.0 | 45.4 |
| 8. Pantoea ananatis OsEp-Plm-30B8 | 100.0 | 7.4 |
| 9. Pantoea ananatis OsEp-Plm-30B17 | 100.0 | 52.8 |
| 10. Pantoea ananatis OsEp-Plm-30B19 | 100.0 | 8.3 |
| 11. Pseudomonas oryzihabitans OsEp-Plm-15B16 | 100.0 | 41.7 |
| 12. Rhizobium sp. OsEp-Plm-30B4 | 100.0 | 5.6 |
| 13. Sphingomonas sp. OsEp-Plm-15B2 | 62.9 | 53.7 |
| 14. Agrobacterium vitis OsEp-Plm-30B7 | 77.9 | 15.7 |
| 15. Enterobacter cloacae OsEp-Plm-30B10 | 76.4 | 46.3 |
| 16. Pantoea ananatis OsEp-Plm-30B15 | 57.9 | 13.9 |
| 17. Microbacterium testaceum OsEp-Plm-30B5 | 57.1 | 4.6 |
| 18. Enterobacter ludwigii OsEp-Plm-30B20 | 56.4 | 4.6 |
| 19. Curtobacterium luteum OsEp-Plm-15B12 | 48.6 | 8.3 |
| 20. Acidovorax avenae OsEp-Plm-15B4 | 45.7 | 6.5 |
| 21. Acinetobacter radioresistens OsEp-Plm-15B15 | 45.7 | 8.3 |
| 22. Microbacterium testaceum OsEp-Plm-15B1 | 42.9 | 7.4 |
| 23. Curtobacterium sp. OsEp-Plm-15B13 | 38.6 | 7.4 |
| 24. Acinetobacter calcoaceticus OsEp-Plm-15B9 | 37.9 | 44.4 |
| 25. Enterococcus faecium OsEp-Plm-15B7 | 35.0 | 23.2 |
| 26. Sphingomonas pseudosanguinis OsEp-Plm-30B9 | 35.0 | 6.5 |
| 27. Microbacterium sp. OsEp-Plm-15B5 | 34.3 | 8.3 |
| 28. Rhizobium taibaishanense OsEp-Plm-15B8 | 32.9 | 8.3 |
| 29. Enterobacter sp. OsEp-Plm-15B11 | 32.1 | 39.8 |
| 30. Pseudomonas sp. OsEp-Plm-30B14 | 31.4 | 3.7 |
| 31. Curtobacterium luteum OsEp-Plm-15B3 | 29.3 | 11.1 |
| Mock | 0.0 | 0.0 |
| C.D. | 12.0 | 4.8 |
| SE (m) | 4.3 | 1.7 |
| SE (d) | 6.1 | 2.4 |
| C.V. (%) | 13.2 | 12.8 |
| F (calc.) | 52.1 | 130.5 |
| F (tab.) | 1. 6 | 1.6 |
Suppressive Effect of Phyllomicrobiome on Blast Disease
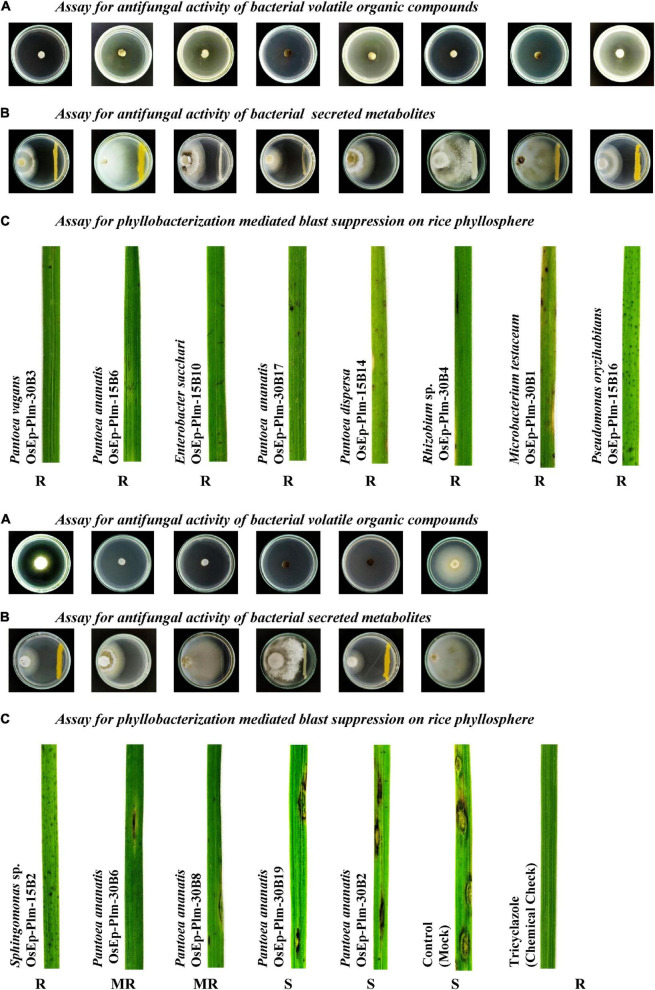
Blast-susceptible rice cultivar, Pusa Basmati 1, was used for evaluating the blast-suppressive effects of rice phyllomicrobiome. A total of 13 bacterial isolates representing Pantoea (eight strains), Enterobacter (one), Microbacterium (one), Pseudomonas (one), Rhizobium (one), and Sphingomonas (one) were evaluated at three different cell densities. Blast incidence and severity were scored as per the blast score chart recommended by Mackill and Bonman (1992). Most of the bacterial isolates were found to reduce the blast disease development in the plants of the susceptible rice cultivar at all tested doses. Maximum reduction in disease severity was shown by P. vagans OsEp-Plm-30B3 (81.9%), P. ananatis OsEp-Plm-15B6 (81.5%), E. sacchari OsEp-Plm-15B10 (78.1%), P. ananatis OsEp-Plm-30B17 (77.7%), P. dispersa OsEp-Plm-15B14 (76.2%), Rhizobium sp. OsEp-Plm-30B4 (69.8%), M. testaceum OsEp-Plm-30B1 (67.5%), P. oryzihabitans OsEp-Plm-15B16 (52.4%), and Sphingomonas sp. OsEp-Plm-15B2 (51.8%) (Figure 3 and Table 5). Nine of the 13 bacterial isolates showed over 50% reduction of blast severity in all bacterial titers. Interestingly, the reduction in blast severity could not be correlated with the bacterial cell densities used for phyllobacterization.
FIGURE 3.
Suppressive effects of phyllosphere bacterial inoculation on Magnaporthe oryzae and the rice blast disease. (A) Phyllosphere bacteria displayed volatile mediated antifungal activity on Magnaporthe oryzae. (B) A few isolates showed secreted metabolite mediated antifungal activity on Magnaporthe oryzae. (C) Nine of the tested 13 bacteria isolates showed blast-suppressive activity on rice leaf.
TABLE 5.
Suppressive effects of phyllobacterization on rice blast disease.
| Bacterial species/isolate | Bacterial dose (0.01 OD at A600 nm = ∼106 cfu per ml) | Bacterial dose (0.1 OD at A600 nm = ∼107cfu per ml) | Bacterial dose (1.0 OD at A600 nm = ∼108 cfu per ml) | Mean | ||||
|
|
|
|
|
|||||
| *Blast severity | **Reduction in severity (%) | *Blast severity | **Reduction in severity (%) | *Blast severity | **Reduction in severity (%) | *Blast severity | **Reduction in severity (%) | |
| Pantoea vagans OsEp-Plm-30B3 | 11.3 | 77.9 | 7.0 | 86.2 | 9.4 | 81.5 | 9.2 | 81.9 |
| Pantoea ananatis OsEp-Plm-15B6 | 8.1 | 84.0 | 13.3 | 73.8 | 6.8 | 86.7 | 9.4 | 81.5 |
| Enterobacter sacchari OsEp-Plm-15B10 | 11.7 | 76.9 | 13.0 | 74.5 | 8.7 | 83.0 | 11.1 | 78.1 |
| Pantoea ananatis OsEp-Plm-30B17 | 15.8 | 68.9 | 12.1 | 76.3 | 6.2 | 87.9 | 11.3 | 77.7 |
| Pantoea dispersa OsEp-Plm-15B14 | 8.0 | 84.3 | 15.5 | 69.5 | 12.8 | 74.9 | 12.1 | 76.2 |
| Rhizobium sp. OsEp-Plm-30B4 | 13.4 | 73.7 | 18.3 | 63.9 | 14.3 | 71.9 | 15.3 | 69.8 |
| Microbacterium testaceum OsEp-Plm-30B1 | 19.0 | 62.6 | 11.4 | 77.5 | 19.1 | 62.5 | 16.5 | 67.5 |
| Pseudomonas oryzihabitans OsEp-Plm-15B16 | 22.4 | 55.9 | 21.9 | 56.9 | 28.3 | 44.4 | 24.2 | 52.4 |
| Sphingomonas sp. OsEp-Plm-15B2 | 20.9 | 58.9 | 24.2 | 52.5 | 28.5 | 43.9 | 24.5 | 51.8 |
| Pantoea ananatis OsEp-Plm-30B6 | 21.5 | 57.7 | 28.9 | 43.2 | 34.8 | 31.6 | 28.4 | 44.2 |
| Pantoea ananatis OsEp-Plm-30B8 | 29.5 | 41.9 | 26.9 | 47.1 | 32.2 | 36.6 | 29.6 | 41.9 |
| Pantoea ananatis OsEp-Plm-30B19 | 41.7 | 18.0 | 42.2 | 17.0 | 35.0 | 31.1 | 39.6 | 22.0 |
| Pantoea ananatis OsEp-Plm-30B2 | 54.6 | −7.5 | 42.9 | 15.7 | 39.0 | 23.2 | 45.5 | 10.5 |
| Control | 50.8 | 0.0 | 50.8 | 0.0 | 50.8 | 0.0 | 50.8 | 0.0 |
| Tricyclazole | 6.8 | 86.7 | 10.1 | 80.1 | 8.1 | 84.0 | 8.33 | 83.6 |
C = disease severity in control.
T = disease severity in treatment.
Phenotyping for Phyllomicrobiome Conferred Immunocompetence
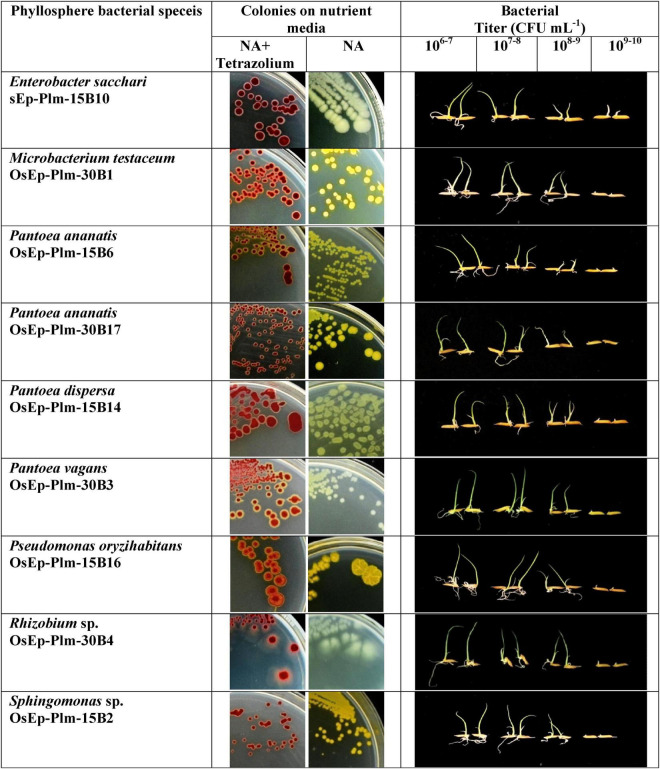
An experiment was conducted to study the potential of phyllosphere bacteria to confer immunocompetence in rice. Seedling growth inhibition due to microbial interactions is touted as a phenotypic marker for the induced immune response in plants. Here, the seed germination and the seedling emergence were monitored during bacterial interaction that showed germination in the range of 40.0–86.7% across various bacterial inoculations. Maximum germination was observed in E. sacchari OsEp-Plm-15B10, while the minimum germination was observed in P. ananatis OsEp-Plm-30B2 (Supplementary Figure 8 and Supplementary Table 7). However, the germinability of rice seeds reduced upon increasing bacterial density with poor germination observed at a titer of 109 CFU ml–1. The bacterial inoculation on seeds and seedlings appeared to trigger the plant phenotypic alteration especially on the shoot and root growth. All nine bacterial isolates representing six genera induced a density-dependent alteration on the shoot and root emergence and growth (Supplementary Tables 8, 9). In particular, the shoot and root growth of rice was found inhibited at high bacterial titer (109 CFU ml–1) (Figure 4).
FIGURE 4.
Assay for phyllomicrobiome-conferred immunocompetence. Seedling growth inhibition as a phenotypic marker of microbiome-conferred immunocompetence, observed with nine blast-suppressive bacterial isolates, represented six genera such as Enterobacter, Microbacterium, Pantoea, Pseudomonas, Rhizobium, and Sphingomonas. The rice phyllosphere bacteria showed various shades of yellow pigmentation; the pink color appearance of the bacterial colony is due to the reduction of tetrazolium dye into insoluble formazan; the inhibition of shoot and root growth—an indicator of innate immunity, can be seen in plantlets interacting with high bacterial titer.
qPCR Assay on Phyllobacterization-Mediated Immunocompetence
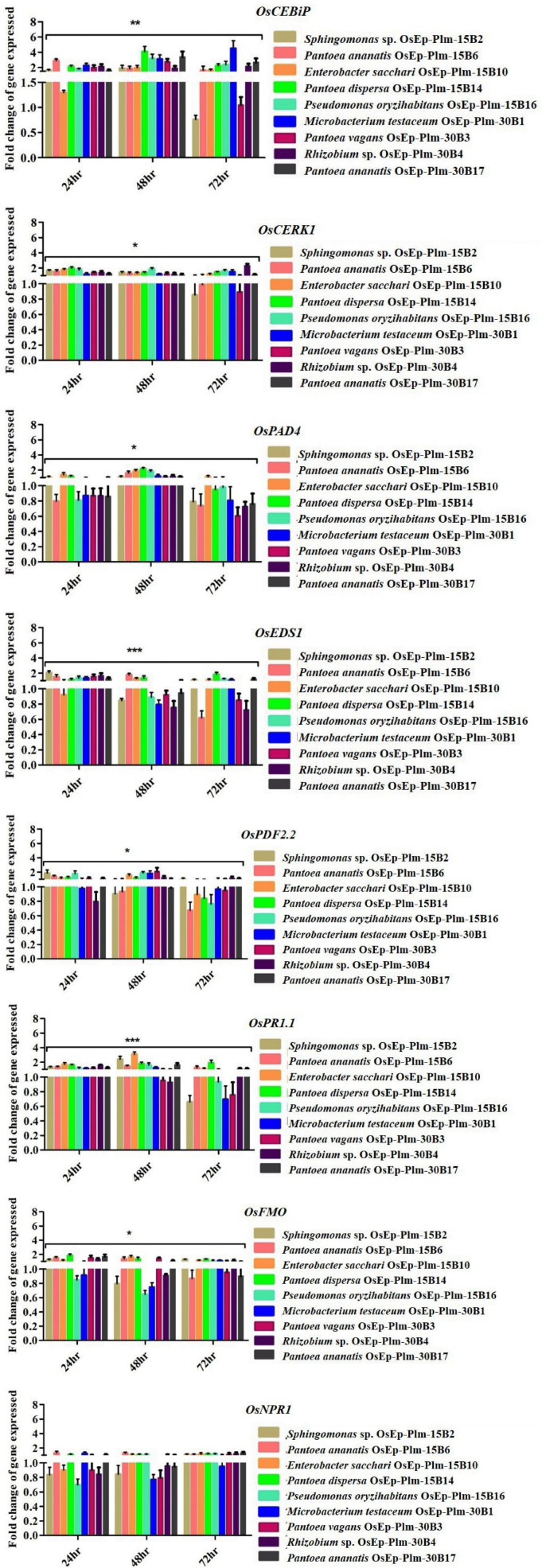
The rice seedlings displaying the altered growth pattern were subjected to transcriptional analysis by qPCR. Defense genes such as OsCEBiP, OsCERK1, OsPAD4, OsNPR1, OsEDS1, OsPDF2.2, OsFMO1, and OsPR1.1 showed marginal to a high level of expression in phyllobacterized rice seedlings compared with the untreated control compared with OsActin used as a reference gene. Interestingly, Sphingomonas sp. OsEp-Plm-15B2, P. ananatis OsEp-Plm-15B6, E. sacchari OsEp-Plm-15B10, P. dispersa OsEp-Plm-15B14, P. oryzihabitans OsEp-Plm-15B16, M. testaceum OsEp-Plm-30B1, P. vagans OsEp-Plm-30B3 Rhizobium sp. OsEp-Plm-30B4, and P. ananatis OsEp-Plm-30B17 sustained the overexpression of OsCEBiP in rice seedlings in all three-time points. P. ananatis OsEp-Plm-15B6 and P. dispersa OsEp-Plm-15B14 showed significant upregulation in almost all the defense-related genes at least for one-time point. E. sacchari OsEp-Plm-15B10 showed sustained upregulation of all the genes for 48 h after bacterial treatment. The epiphytic bacteria-mediated activation of defense genes was more pronounced during the early time points peaking at 48 hpi with a sharp drop at 72 h of bacterial interaction (Figure 5 and Supplementary Table 10).
FIGURE 5.
qPCR-based transcriptional analysis of defense gene expression in rice seedlings upon phyllobacterization. The fold change values obtained for the defense genes were imported into the GraphPad Prism program (https://www.graphpad.com/scientific-software/prism), and two-way ANOVA was conducted using Bonferroni post-hoc test for determining the statistical significance at *p ≤ 0.05, **p = 0.001, and ***p = 0.0001. Refer to Supplementary Table 10 for data pertaining to fold changes of gene expression.
Hypersensitive Reaction on Tobacco
Rice phyllospheric bacteria upon infiltration in tobacco leaf did not trigger any necrotic reactions even after 48 h. The expression of quick necrosis due to the hypersensitive reaction was observed in the pathogen, R. solanacearum, which infiltrated a leaf indicating that the rice phyllosphere-associated bacterial isolates are non-pathogenic on plants (Supplementary Figure 9).
Discussion
The foliar plant niches termed phyllosphere is one of the habitats for a diverse microbiota where the dynamic plant–microbe interactions are believed to impact plant performance in the ecosystem. With assistance from the microbial genome and their metabolic capabilities, the plants are exposed to a plethora of unpredictable, yet competitive or cooperative, microbial interactions (Vorholt, 2012; Hardoim et al., 2015; Reinhold-Hurek et al., 2015; Brader et al., 2017; Lemanceau et al., 2017). In recent years, the impact of microbe–microbe interactions on the host–microbial pathogen interaction outcomes is gaining the attention of researchers. Studies have shown that the microbiome structure, assemblage, and compositions are directly influenced by both micro and macro abiotic and biotic factors (Jacobs et al., 2005). Most of the previous microbiome studies have focused primarily on metagenomic or amplicon sequence surveys, and a few attempts have been made to validate the microbial mNGS datasets using classical microbiological culturomic tools. Strikingly, we have compared the phyllomicrobiome of blast disease-resistant and -susceptible rice genotypes planted in a blast endemic location in India. We recorded marginally high bacterial diversity and species richness on PRR78 that could be attributed to the innate susceptibility of the genotypes owing to the absence of functional NBS-LRR type of receptors termed as R-genes. Besides, the susceptible leaf showing early necrotic symptoms might have paved way for the inevitable microbial succession on the phyllosphere.
In the present work, we not only microbiologically validated the bacterial community structure in the phyllomicrobiome but also functionally characterized them for harnessing their antifungal and defense-inducing potential against rice blast disease. The NGS metabarcoding-based microbiome profiling revealed the predominance of phylum Proteobacteria on the rice phyllosphere. The predominance of Proteobacteria consisting of bacterial communities belonging to Enterobacteriaceae and Pseudomonadaceae is reported on the phyllosphere by many workers (Knief et al., 2012; Ren et al., 2014; Roman-Reyna et al., 2019; Yasmin et al., 2020). At the lower taxonomic genus hierarchy, Pantoea followed by Pseudomonas and Enterobacter were overrepresented on both the rice genotypes. Our findings are in agreement with many other reports that highlighted the ubiquitous occurrence of Pantoea as the most abundant bacterial genera on the phyllosphere (Cottyn et al., 2009; Cother et al., 2010; Kim et al., 2020; Stone and Jackson, 2020; Dhankhar et al., 2021). The microbiological investigation culminated in the isolation of 37 distinct morphotypes on the rice phyllosphere; here, the 4-week-old seedling harbored more bacterial morphotypes compared with the 15-day-old seedlings implying that microbial biomass on plant niches expands with the age of the plants. The morphotypes could be identified as belonging to 31 distinct amplicon groups in BOX-AIR-PCR fingerprinting based on shared amplicon profiles of the isolates. The BOX PCR DNA fingerprinting is one of the widely used molecular tools in bacterial typing and biogeography studies of microbial isolates (Versalovic et al., 1994; Brusetti et al., 2008).
The 16S rRNA gene sequence database search confirmed the identity of cultured bacterial isolates as belonging to Acinetobacter, Enterobacter, Pantoea, Pseudomonas, and Sphingomonas on blast-resistant and -susceptible genotypes. Pantoea, Enterobacter, Microbacterium, and Curtobacterium are recently reported as a member of the core microbiome of the rice leaf endosphere (Kumar et al., 2021). The genus Microbacterium too is frequently observed on the rice phyllosphere and spermosphere by several researchers in the past (Kaku et al., 2000; Leveau and Lindow, 2001; Midha et al., 2016). Characteristically, most of the phyllosphere bacterial genera produced shades of pigmentation as observed in Acinetobacter (pale brown), Curtobacterium (dark yellow), Enterobacter (pale brown to yellow), Microbacterium (yellow), Pantoea (yellow), Pseudomonas (pale brown to yellow), and Sphingomonas (dark yellow) (Supplementary Figures 4a–k). Production of dark pigmentation is one of the adaptive traits of bacteria and other microbes that encounter harsh environmental abiotic stresses like solar radiation and light on the phyllosphere (Green, 1992; Jacobs et al., 2005). Pigmentation on phyllospheric bacteria is believed to protect them from harmful ultraviolet radiation (Sundin and Jacobs, 1999). The rice phyllosphere is considered as the preferred habitat for yellow-pigmented Pantoea and pink-pigmented methylotrophs, which can survive under nutritional and moisture stress as well as can withstand harmful γ-ray radiation (Green, 1992). The effect of solar radiation on the composition and activities of the phyllosphere microbial community is reported by Carvalho and Castillo (2018).
The bacterial isolates showed antifungal activity on M. oryzae by their secreted compounds and volatile organic compounds, while Enterobacter, Microbacterium, Pantoea, Pseudomonas, and Rhizobium showed volatile mediated antifungal activity, and the Acinetobacter and Sphingomonas displayed secretory metabolite-mediated antagonism. The antagonistic potential of these bacterial species has been exploited for combating crop diseases caused by several fungal pathogens. For instance, the antagonistic potential of Acinetobacter baumannii (Liu et al., 2007), Enterobacter sp. (Gong et al., 2019), M. testaceum (Mannaa et al., 2017), P. ananatis (Gasser et al., 2012), P. dispersa (Jiang et al., 2019), P. vagans (Stockwell et al., 2010), P. oryzihabitans (Vagelas and Gowen, 2012; Rariz et al., 2017; Horuz and Aysan, 2018), and Sphingomonas sp. (Innerebner et al., 2011; Wachowska et al., 2013) is reported. Among them, P. vagans strain C9-1, isolated from apple has been registered by Nufarms America Inc., Burr Ridge, IL as “BlightBan C9-1” for the biological control of fire blight of apple caused by Erwinia amylovora. Isolates belonging to Sphingomonas have also been reported to promote plant growth, confer tolerance against abiotic stresses, and offer protection against plant pathogens (Luo et al., 2020; Turner, 2020).
Currently, growing evidence for microbe-induced seedling growth alteration as a phenotypic marker of activated innate immunity is published (Wang et al., 2021). We observed that the phyllosphere bacterial species in a density-dependent manner impacted the seed germination with consequent seedling growth alterations upon seed inoculation. The results are in agreement with the studies of Damodaran et al. (2013) and Zhu et al. (2017) who observed varying effects of endophytic and rhizosphere bacteria on seed germination. The seedling growth assay enabled us to identify the bacterial species that conferred immune competence in rice seedlings. Whereas the seedling growth was found inhibited at a higher dose (109 cells per ml), the lower doses (106–8 cells per ml) showed characteristic non-lethal seedling inhibition presumably owing to a tradeoff between growth and immunity. To confirm this, we performed the qPCR-based temporal transcriptional analysis of defense genes involved in innate immunity on phyllobacterized rice seedlings.
Phyllobacterized rice seedlings showed an elevated expression of defense genes, such as OsCEBiP, OsCERK, OsPR1.1, OsNPR1, OsPDF2.2, OsFMO, and OsPAD4. Significant up-regulation of almost all tested defense-related genes at least for a one-time point was shown by P. ananatis (OsEp-Plm-15B6) and P. dispersa (OsEp-Plm-15B14). Notably, E. sacchari (OsEp-Plm-15B10) showed sustained expression of all the genes 48 hpi. Among the genes, significant expression of OsCEBiP and OsCERK was observed in phyllobacterized rice seedlings. Both OsCEBiP and OsCERK1 are reported to be activating MAMP-triggered immune (MTI) responses in plants upon chitin and peptidoglycan perception (Akamatsu et al., 2013; Kouzai et al., 2014). Defense genes, such as OsPAD4 and OsEDS1 participating in the jasmonic acid-mediated ISR, were also found induced in rice seedlings upon bacterization. Induction of OsPAD4 contributes to the accumulation of rice phytoalexin, mamilactone-A, and contributes to basal resistance (Hasegawa et al., 2010; Ke et al., 2014, 2019). Marginal induction of OsNPR1, OsFMO, OsPDF2.2, and OsPR1.1 was observed in phyllobacterized rice seedlings. Among them, OsNPR1—the key regulator of salicylic acid (SA)-mediated defense signaling is believed to control resource and energy redistribution during the defense reaction (Sugano et al., 2010). Likewise, OsFMO1 is known to modulate systemic acquired resistance in plants against pathogens (Koch et al., 2006; Mishina and Zeier, 2006). While OsPDF2.2 codes for antifungal plant defensin (Thomma et al., 2002), the OsPR1.1 codes for an acidic pathogenesis-related protein to modulate SA-mediated systemic acquired resistance (Brader et al., 2017).
Phyllosphere bacterial species evaluated against rice blast under artificial epiphytotic trial in greenhouse showed a reduction in blast disease (50 % over mock) at all tested bacterial titers. Upon prophylactic foliar application (or phyllobacterization), the species belonging to Pantoea, Enterobacter, Microbacterium, Pseudomonas, Sphingomonas, and Rhizobium showed significant blast suppression at all tested doses. However, we could not observe any dosage response for enhanced blast suppression revealing the sufficiency of bacterial augmentation at 106–7 cells per ml for reducing blast disease. Suppression of blast disease by Bacillus, Streptomyces, Pseudomonas, Pantoea, Paenibacillus, Burkholderia, Enterobacter, Paraburkholderia, and Actinomycetes was earlier reported in the literature (Gómez Expósito et al., 2017; Harsonowati et al., 2017; Schlatter et al., 2017). Rice blast suppression by Microbacterium, Pseudomonas, and Stenotrophomonas are attributed to the direct antifungal antibiosis and the indirect defense activation as evident from the expression of rice defense genes (Ashajyothi et al., 2020; Sahu et al., 2020). Recently, bacterial volatile belonging to pyrazines is reported to modulate defense against blast disease (Patel et al., 2020). Taken together, it is concluded that the enrichment of phyllosphere bacterial communities on the leaf can inflict antifungal antibiosis on M. oryzae and defense elicitation in rice to reduce the incidence and severity of blast disease.
Having confirmed the blast-suppressive potential of phyllosphere bacterial communities, we conducted a tobacco infiltration HR assay to ascertain the biosafety of the phyllosphere bacterial isolate (Klement, 1963). The tobacco HR assay has been recognized to test the pathogenic nature of plant-associated bacterial species (Kucheryava et al., 1999; Medina-Salazar et al., 2020; Sadeghi and Khodakaramian, 2020). None of the phyllosphere bacterial isolates showed any necrotic lesions, while comparing with R. solanacearum served as a positive check. Shades of faded yellowing observed with few phyllosphere bacterial isolates are indicative of activated defense.
In conclusion, the phyllosphere bacterial communities suppressed the blast disease by the dual action of antifungal secreted and volatile metabolites as well as by microbe conferred immunocompetence (Figure 6). The present investigation on phyllosphere microbiome analysis of rice culminated in several potential hitherto unexplored bacterial communities for microbiome-assisted crop protection, especially against rice blast disease.
FIGURE 6.
Phyllosphere microbiome-assisted suppression of rice blast disease. Blast suppressiveness by the predominant and pigmented bacterial isolates of phyllosphere can be attributed to both antifungal activity on Magnaporthe oryzae as well as induced defense in rice as evident from enhanced expression of many defense genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
KS conceptualized the study, developed the methodology, investigated and validated the study, performed the formal analysis, and wrote the original draft. AK conceptualized and investigated the study, provided the resources, wrote, reviewed, and edited the manuscript, and was in charge of the visualization, supervision, and project administration. AP, MK, and NS developed the methodology and validated the study. SM and BR performed the formal analysis. PE and NP validated the study. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer KV declared a shared affiliation with the authors, to the handling editor at the time of the review.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Director, IARI, and Dean, PG School, Indian Council of Agricultural Research–Indian Agricultural Research Institute, New Delhi, for the logistic support and encouragement. We gratefully acknowledge the research grant provided by Genomics-Assisted Crop Improvement and Management (NAHEP/CAAST/2018-19/07), ICAR-IARI, New Delhi.
Funding
KS offers his sincere thanks to the Council of Scientific and Industrial Research (CSIR) for financial support in the form of Junior and Senior Research Fellowships [File No: 09/083(0367)/2016-EMR-I] for the Ph.D. program. KS and AK were grateful to NAHEP-CAAST on “Genomics assisted crop improvement and management” for financial assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.780458/full#supplementary-material
References
- Akamatsu A., Wong H. L., Fujiwara M., Okuda J., Nishide K., Uno K., et al. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13 465–476. 10.1016/j.chom.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Al-Antary T. M., Alawi M. A., Masaedeh M., Haddad N. A. (2020). Multi-Residue Analysis of 405 Pesticides in Agricultral Crops in Middle Governorates of Jordan in 2018 and 2019 Using QuEChERS Method Followed by LC-MS/MS and GC-ECD. Fresenius Environ. Bull. 29 2534–2539. [Google Scholar]
- Andrews J. H., Harris R. F. (2000). The ecology and biogeography of microorganisms on plant surfaces. Annu. Rev. Phytopathol. 38 145–180. 10.1146/annurev.phyto.38.1.145 [DOI] [PubMed] [Google Scholar]
- Andrews S. (2018). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Institute. [Google Scholar]
- Ashajyothi M., Kumar A., Sheoran N., Ganesan P., Gogoi R., Subbaiyan G. K., et al. (2020). Black pepper (Piper nigrum L.) associated endophytic Pseudomonas putida BP25 alters root phenotype and induces defense in rice (Oryza sativa L.) against blast disease incited by Magnaporthe oryzae. Biol. Control 143:104181. 10.1016/j.biocontrol.2019.104181 [DOI] [Google Scholar]
- Asibi A. E., Chai Q., Coulter J. A. (2019). Rice blast: a disease with implications for global food security. Agronomy 9:451. 10.3390/agronomy9080451 [DOI] [Google Scholar]
- Berg G., Rybakova D., Fischer D., Cernava T., Vergès M.-C. C., Charles T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. 10.1186/s40168-020-00875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Rybakova D., Grube M., Köberl M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67 995–1002. 10.1093/jxb/erv466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani I., Abbruscato P., Piffanelli P., Subramoni S., Venturi V. (2016). Rice bacterial endophytes: isolation of a collection, identification of beneficial strains, and microbiome analysis. Environ. Microbiol. Rep. 8 388–398. 10.1111/1758-2229.12403 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G., Compant S., Vescio K., Mitter B., Trognitz F., Ma L.-J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55 61–83. 10.1146/annurev-phyto-080516-035641 [DOI] [PubMed] [Google Scholar]
- Breen S., Williams S. J., Outram M., Kobe B., Solomon P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22 871–879. 10.1016/j.tplants.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Brusetti L., Malkhazova I., Gtari M., Tamagnini I., Borin S., Merabishvili M., et al. (2008). Fluorescent-BOX-PCR for resolving bacterial genetic diversity, endemism, and biogeography. BMC Microbiol. 8:220. 10.1186/1471-2180-8-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Van Themaat E. V. L., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64 807–838. 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Carvalho S. D., Castillo J. A. (2018). Influence of light on plant–phyllosphere interaction. Front. Plant Sci. 9:1482. 10.3389/fpls.2018.01482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Samad A., Faist H., Sessitsch A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19 29–37. 10.1016/j.jare.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cother E., Noble D., Van De Ven R., Lanoiselet V., Ash G., Vuthy N., et al. (2010). Bacterial pathogens of rice in the Kingdom of Cambodia and description of a new pathogen causing a serious sheath rot disease. Plant Pathol. 59 944–953. 10.1111/j.1365-3059.2010.02310.x [DOI] [Google Scholar]
- Cottyn B., Debode J., Regalado E., Mew T., Swings J. (2009). Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J. Appl. Microbiol. 107 885–897. 10.1111/j.1365-2672.2009.04268.x [DOI] [PubMed] [Google Scholar]
- Damodaran T., Sharma D., Mishra V., Jha S., Kannan R., Sah V., et al. (2013). Isolation of Salt Tolerant Endophytic and Rhizospheric Bacteria by Natural Selection and screening for promising plant growth-promoting Rhizobacteria (PGPR) and growth vigour in Tomato under sodic environment. Afr. J. Microbiol. Res. 7 5082–5089. 10.5897/AJMR2013.6003 [DOI] [Google Scholar]
- Dean R. A., Talbot N. J., Ebbole D. J., Farman M. L., Mitchell T. K., Orbach M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434 980–986. 10.1038/nature03449 [DOI] [PubMed] [Google Scholar]
- Dhankhar R., Mohanty A., Gulati P. (2021). “Microbial diversity of Phyllosphere: exploring the unexplored,” in Phytomicrobiome Interactions and Sustainable Agriculture, eds Verma A., Saini J. K., Hesham A. E. L., Singh H. B. (Hoboken, NJ: John Wiley & Sons Ltd; ), 66–90. 10.1002/9781119644798.ch5 [DOI] [Google Scholar]
- Eke P., Kumar A., Sahu K. P., Wakam L. N., Sheoran N., Ashajyothi M., et al. (2019). Endophytic bacteria of desert triangular spurge (Euphorbia antiquorum L.) confer drought tolerance and induce growth promotion in tomato (Solanum lycopersicum L.). Microbiol. Res. 228:126302. 10.1016/j.micres.2019.126302 [DOI] [PubMed] [Google Scholar]
- Gasser F., Cardinale M., Schildberger B., Berg G. (2012). Biocontrol of Botrytis cinerea by successful introduction of Pantoea ananatis in the grapevine phyllosphere. Int. J. Wine Res. 4 53–63. 10.2147/ijwr.s31339 [DOI] [Google Scholar]
- Gómez Expósito R., De Bruijn I., Postma J., Raaijmakers J. M. (2017). Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol. 8:2529. 10.3389/fmicb.2017.02529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong A.-D., Dong F.-Y., Hu M.-J., Kong X.-W., Wei F.-F., Gong S.-J., et al. (2019). Antifungal activity of volatile emitted from Enterobacter asburiae Vt-7 against Aspergillus flavus and aflatoxins in peanuts during storage. Food Control 106:106718. 10.1016/j.foodcont.2019.106718 [DOI] [Google Scholar]
- Green P. (1992). “The genus methylobacterium,” in The Prokaryotes, 2nd Edn, eds Balows A., Trüper H. G., Dworkin M., Harder W., Schleifer K. H. (Berlin: Springer-Verlag; ), 2342–2349. 10.1007/0-387-30745-1_14 [DOI] [Google Scholar]
- Hammer Ø., Harper D. A., Ryan P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4 1–9. [Google Scholar]
- Hardoim P. R., Van Overbeek L. S., Berg G., Pirttilä A. M., Compant S., Campisano A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79 293–320. 10.1128/mmbr.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsonowati W., Astuti R. I., Wahyudi A. T. (2017). Leaf blast disease reduction by rice-phyllosphere actinomycetes producing bioactive compounds. J. Gen. Plant Pathol. 83 98–108. 10.1007/s10327-017-0700-4 [DOI] [Google Scholar]
- Hartmann A., Rothballer M., Schmid M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312 7–14. 10.1007/s11104-007-9514-z [DOI] [Google Scholar]
- Hasegawa M., Mitsuhara I., Seo S., Imai T., Koga J., Okada K., et al. (2010). Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant Microbe Interact. 23 1000–1011. 10.1094/mpmi-23-8-1000 [DOI] [PubMed] [Google Scholar]
- Hashim I., Mamiro D. P., Mabagala R. B., Tefera T. (2018). Smallholder farmers’ knowledge, perception and management of rice blast disease in upland rice production in Tanzania. J. Agric. Sci. 10 137–145. [Google Scholar]
- Horuz S., Aysan Y. (2018). Biological control of watermelon seedling blight caused by Acidovorax citrulli using antagonistic bacteria from the genera Curtobacterium, Microbacterium, and Pseudomonas. Plant Prot. Sci. 54 138–146. 10.17221/168/2016-pps [DOI] [Google Scholar]
- Innerebner G., Knief C., Vorholt J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77 3202–3210. 10.1128/aem.00133-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J., Carroll T., Sundin G. (2005). The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microb. Ecol. 49 104–113. 10.1007/s00248-003-1061-4 [DOI] [PubMed] [Google Scholar]
- Jiang L., Jeong J. C., Lee J.-S., Park J. M., Yang J.-W., Lee M. H., et al. (2019). Potential of Pantoea dispersa as an effective biocontrol agent for black rot in sweet potato. Sci. Rep. 9:16354. 10.1038/s41598-019-52804-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H., Subandiyah S., Ochiai H. (2000). Red stripe of rice [Oryza sativa] is caused by a bacterium Microbacterium sp. J. Gen. Plant Pathol. 66 149–152. 10.1007/pl00012937 [DOI] [Google Scholar]
- Ke Y., Kang Y., Wu M., Liu H., Hui S., Zhang Q., et al. (2019). Jasmonic acid-involved OsEDS1 signaling in Rice-bacteria interactions. Rice 12:25. 10.1186/s12284-019-0283-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y., Liu H., Li X., Xiao J., Wang S. (2014). Rice OsPAD4 functions differently from ArabidopsisAtPAD4 in host-pathogen interactions. Plant J. 78 619–631. 10.1111/tpj.12500 [DOI] [PubMed] [Google Scholar]
- Kim H., Lee K. K., Jeon J., Harris W. A., Lee Y.-H. (2020). Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome 8:20. 10.1186/s40168-020-00805-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement Z. (1963). Rapid detection of the pathogenicity of phytopathogenic pseudomonads. Nature 199 299–300. 10.1038/199299b0 [DOI] [PubMed] [Google Scholar]
- Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6 1378–1390. 10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M., Vorwerk S., Masur C., Sharifi-Sirchi G., Olivieri N., Schlaich N. L. (2006). A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 47 629–639. 10.1111/j.1365-313x.2006.02813.x [DOI] [PubMed] [Google Scholar]
- Kouzai Y., Mochizuki S., Nakajima K., Desaki Y., Hayafune M., Miyazaki H., et al. (2014). Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant Microbe Interact. 27 975–982. [DOI] [PubMed] [Google Scholar]
- Kucheryava N., Fiss M., Auling G., Kroppenstedt R. M. (1999). Isolation and characterization of epiphytic bacteria from the phyllosphere of apple, antagonistic in vitro to Venturia inaequalis, the causal agent of apple scab. Syst. Appl. Microbiol. 22 472–478. 10.1016/s0723-2020(99)80057-5 [DOI] [Google Scholar]
- Kumar A., Sarma Y., Anandaraj M. (2004). Evaluation of genetic diversity of Ralstonia solanacearum causing bacterial wilt of ginger using REP–PCR and PCR–RFLP. Curr. Sci. 87 1555–1561. [Google Scholar]
- Kumar M., Kumar A., Sahu K. P., Patel A., Reddy B., Sheoran N., et al. (2021). Deciphering core-microbiome of rice leaf endosphere: revelation by metagenomic and microbiological analysis of aromatic and non-aromatic genotypes grown in three geographical zones. Microbiol. Res. 246:126704. 10.1016/j.micres.2021.126704 [DOI] [PubMed] [Google Scholar]
- Lemanceau P., Blouin M., Muller D., Moënne-Loccoz Y. (2017). Let the core microbiota be functional. Trends Plant Sci. 22 583–595. 10.1016/j.tplants.2017.04.008 [DOI] [PubMed] [Google Scholar]
- Leveau J. H., Lindow S. E. (2001). Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. U.S.A. 98 3446–3453. 10.1073/pnas.061629598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Leveau J. H. (2002). Phyllosphere microbiology. Curr. Opin. Biotechnol. 13 238–243. 10.1016/s0958-1669(02)00313-0 [DOI] [PubMed] [Google Scholar]
- Liu C., Chen X., Liu T., Lian B., Gu Y., Caer V., et al. (2007). Study of the antifungal activity of Acinetobacter baumannii LCH001 in vitro and identification of its antifungal components. Appl. Microbiol. Biotechnol. 76 459–466. 10.1007/s00253-007-1010-0 [DOI] [PubMed] [Google Scholar]
- Luo Y., Zhou M., Zhao Q., Wang F., Gao J., Sheng H., et al. (2020). Complete genome sequence of Sphingomonas sp. Cra20, a drought-resistant and plant growth-promoting rhizobacteria. Genomics 112 3648–3657. 10.1016/j.ygeno.2020.04.013 [DOI] [PubMed] [Google Scholar]
- Mackill D., Bonman J. (1992). Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82 746–749. 10.1094/phyto-82-746 [DOI] [Google Scholar]
- Mannaa M., Oh J., Kim K. D. (2017). Microbe-mediated control of Aspergillus flavus in stored rice grains with a focus on aflatoxin inhibition and biodegradation. Ann. Appl. Biol. 171 376–392. 10.1111/aab.12381 [DOI] [Google Scholar]
- Medina-Salazar S. A., Rodriguez-Aguilar M., Vallejo-Pérez M. R., Flores-Ramirez R., Marin-Sanchez J., Aguilar-Benitez G., et al. (2020). Biodiversity of epiphytic Pseudomonas strains isolated from leaves of pepper and lettuce. Biologia 75 773–784. 10.2478/s11756-019-00392-y [DOI] [Google Scholar]
- Mehta S., Singh B., Dhakate P., Rahman M., Islam M. A. (2019). “Rice, marker-assisted breeding, and disease resistance,” in Disease Resistance in Crop Plants, ed. Wani S. H. (Cham: Springer; ), 83–111. 10.1007/978-3-030-20728-1_5 [DOI] [Google Scholar]
- Midha S., Bansal K., Sharma S., Kumar N., Patil P. P., Chaudhry V., et al. (2016). Genomic resource of rice seed associated bacteria. Front. Microbiol. 6:1551. 10.3389/fmicb.2015.01551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina T. E., Zeier J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141 1666–1675. 10.1104/pp.106.081257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E., Arnscheidt A., Krüger A., Strömpl C., Mau M. (2004). “Simplified protocols for the preparation of genomic DNA from bacterial cultures,” in Molecular Microbial Ecology Manual, eds Kowalchuk G., De Bruijn F. J., Head I., Akkermans A. D. L., van Elsas J. D. (London: Kluwer Academic Publishers; ), 1–15. 10.1007/978-1-4020-2177-0_101 [DOI] [Google Scholar]
- Munjal V., Nadakkakath A. V., Sheoran N., Kundu A., Venugopal V., Subaharan K., et al. (2016). Genotyping and identification of broad-spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium BP17. Biol. Control 92 66–76. 10.1016/j.biocontrol.2015.09.005 [DOI] [Google Scholar]
- Nalley L., Tsiboe F., Durand-Morat A., Shew A., Thoma G. (2016). Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLoS One 11:e0167295. 10.1371/journal.pone.0167295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Kumar A., Sheoran N., Kumar M., Sahu K. P., Ganeshan P., et al. (2020). Antifungal and defense elicitor activities of pyrazines identified in endophytic Pseudomonas putida BP25 against fungal blast incited by Magnaporthe oryzae in rice. J. Plant Dis. Prot. 128 261–272. 10.1007/s41348-020-00373-3 [DOI] [Google Scholar]
- Prakash G., Patel A., Prakash I., Sahu K. P., Hosahatti R., Kumar A. (2021). “Microconidia: understanding its role in the fungus Magnaporthe oryzae Inciting Rice Blast Disease,” in Blast Disease of Cereal Crops, eds Nayaka S. C., Hosahatti R., Prakash G., Satyavathi C. T., Sharma T. R. (Cham: Springer; ), 143–150. 10.1007/978-3-030-60585-8_10 [DOI] [Google Scholar]
- Rajashekara H., Prakash G., Pandian R., Sarkel S., Dubey A., Sharma P., et al. (2017). An efficient technique for isolation and mass multiplication of Magnaporthe oryzae from blast infected samples. Indian Phytopathol. 69 260–265. [Google Scholar]
- Rariz G., Ferrando L., Echegoyen N., Scavino A. F. (2017). Antagonism between Azospirillum brasilense Az39 and Pseudomonas oryzihabitans, a seed-borne endophyte, in growing rice plants. Rev. Agron. Noroeste Argent. 37 45–56. [Google Scholar]
- Rascovan N., Carbonetto B., Perrig D., Díaz M., Canciani W., Abalo M., et al. (2016). Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep. 6:28084. 10.1038/srep28084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Bünger W., Burbano C. S., Sabale M., Hurek T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53 403–424. 10.1146/annurev-phyto-082712-102342 [DOI] [PubMed] [Google Scholar]
- Ren G., Zhang H., Lin X., Zhu J., Jia Z. (2014). Response of phyllosphere bacterial communities to elevated CO 2 during rice growing season. Appl. Microbiol. Biotechnol. 98 9459–9471. 10.1007/s00253-014-5915-0 [DOI] [PubMed] [Google Scholar]
- Roman-Reyna V., Pinili D., Borjaa F. N., Quibod I., Groen S. C., Mulyaningsih E. S., et al. (2019). The Rice Leaf Microbiome has a Conserved Community Structure Controlled by Complex Host-Microbe Interactions. Available online at: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3382544 (accessed May 2019). [Google Scholar]
- Sadeghi K., Khodakaramian G. (2020). Characteristics and Ice Nucleation Activity of Sugarcane Epiphytic and Endophytic Bacteria and Their Role in Host Frostbite. Sugar Tech 22 291–302. 10.1007/s12355-019-00759-0 [DOI] [Google Scholar]
- Sahu K. P., Kumar A., Patel A., Kumar M., Gopalakrishnan S., Prakash G., et al. (2020). Rice Blast Lesions: an Unexplored Phyllosphere Microhabitat for Novel Antagonistic Bacterial Species Against Magnaporthe oryzae. Microb. Ecol. 81 731–745. 10.1007/s00248-020-01617-3 [DOI] [PubMed] [Google Scholar]
- Scheuermann K. K., Raimondi J. V., Marschalek R., De Andrade A., Wickert E. (2012). “Magnaporthe oryzae genetic diversity and its outcomes on the search for durable resistance,” in The Molecular Basis of Plant Genetic Diversity, ed. Caliskan M. (Rijeka: IntechOpen; ), 331–356. 10.5772/33479 [DOI] [Google Scholar]
- Schlatter D., Kinkel L., Thomashow L., Weller D., Paulitz T. (2017). Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107 1284–1297. 10.1094/phyto-03-17-0111-rvw [DOI] [PubMed] [Google Scholar]
- Sella L., Vu V. V., Quarantin A., Caracciolo R., Govind R., Bolzonello A., et al. (2021). “Sustainable methods to control Pyricularia oryzae, the causal agent of rice blast disease,” in Innovations in Land, Water and Energy for Vietnam’s Sustainable Development, ed. Anderle M. (Berlin: Springer; ), 67–82. 10.1007/978-3-030-51260-6_7 [DOI] [Google Scholar]
- Sessitsch A., Hardoim P., Döring J., Weilharter A., Krause A., Woyke T., et al. (2012). Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microbe Interact. 25 28–36. 10.1094/mpmi-08-11-0204 [DOI] [PubMed] [Google Scholar]
- Sheoran N., Kumar A., Munjal V., Nadakkakath A. V., Eapen S. J. (2016). Pseudomonas putida BP25 alters root phenotype and triggers salicylic acid signaling as a feedback loop in regulating endophytic colonization in Arabidopsis thaliana. Physiol. Mol. Plant Pathol. 93 99–111. 10.1016/j.pmpp.2016.01.008 [DOI] [Google Scholar]
- Sheoran N., Nadakkakath A. V., Munjal V., Kundu A., Subaharan K., Venugopal V., et al. (2015). Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds. Microbiol. Res. 173 66–78. 10.1016/j.micres.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Singh V. K., Singh A., Singh S., Ellur R. K., Choudhary V., Sarkel S., et al. (2012). Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker-assisted backcross breeding. Field Crops Res. 128 8–16. 10.1016/j.fcr.2011.12.003 [DOI] [Google Scholar]
- Stockwell V., Johnson K., Sugar D., Loper J. (2010). Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 100 1330–1339. 10.1094/phyto-03-10-0097 [DOI] [PubMed] [Google Scholar]
- Stone B. W., Jackson C. R. (2020). Seasonal patterns contribute more towards phyllosphere bacterial community structure than short-term perturbations. Microb. Ecol. 81 146–156. 10.1007/s00248-020-01564-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S., Jiang C.-J., Miyazawa S.-I., Masumoto C., Yazawa K., Hayashi N., et al. (2010). Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 74 549–562. 10.1007/s11103-010-9695-3 [DOI] [PubMed] [Google Scholar]
- Sundin G., Jacobs J. (1999). Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.). Microb. Ecol. 38 27–38. 10.1007/s002489900152 [DOI] [PubMed] [Google Scholar]
- Thomma B. P., Cammue B. P., Thevissen K. (2002). Plant defensins. Planta 216 193–202. 10.1007/s00425-002-0902-6 [DOI] [PubMed] [Google Scholar]
- Turner A. D. (2020). Exploring Interactions of Phyllosphere Epiphytes with Plant Pathogenic Bacteria Pseudomonas and Xanthomonas on Tomato. Available online at: https://csuepress.columbusstate.edu/theses_dissertations/395 (accessed September 2, 2021). [Google Scholar]
- Vagelas I., Gowen S. (2012). Control of Fusarium oxysporum and root-knot nematodes (Meloidogyne spp.) with Pseudomonas oryzihabitans. Pak. J. Phytopathol. 24 32–38. [Google Scholar]
- Vandana U. K., Rajkumari J., Singha L. P., Satish L., Alavilli H., Sudheer P. D., et al. (2021). The endophytic microbiome as a hotspot of synergistic interactions, with prospects of plant growth promotion. Biology 10:101. 10.3390/biology10020101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Schneider M., De Bruijn F., Lupski J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5 25–40. 10.1007/978-1-4615-6369-3_34 [DOI] [Google Scholar]
- Vorholt J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10 828–840. 10.1038/nrmicro2910 [DOI] [PubMed] [Google Scholar]
- Wachowska U., Irzykowski W., Jędryczka M., Stasiulewicz-Paluch A. D., Głowacka K. (2013). Biological control of winter wheat pathogens with the use of antagonistic Sphingomonas bacteria under greenhouse conditions. Biocontrol Sci. Technol. 23 1110–1122. [Google Scholar]
- Wang Y., Holland J., Balint-Kurti P. (2021). Development and use of a seedling growth retardation assay to quantify and map loci underlying variation in the maize basal defense response. PhytoFrontiers. 1, 149–159. 10.1094/phytofr-12-20-0038-r [DOI] [Google Scholar]
- Yasmin S., Hakim S., Zaheer A., Mirza B., Mirza M. S. (2020). Metagenomic Analysis of Bacterial Community Associated with Rhizosphere and Phyllosphere of Basmati Rice. bioRxiv [Preprint]. 10.1101/2020.04.09.034009 [DOI] [Google Scholar]
- Yasuda N., Mitsunaga T., Hayashi K., Koizumi S., Fujita Y. (2015). Effects of pyramiding quantitative resistance genes pi21, Pi34, and Pi35 on rice leaf blast disease. Plant Dis. 99 904–909. 10.1094/pdis-02-14-0214-re [DOI] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T., Stamatakis A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30 614–620. 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. L., She X.-P., Wang J. S., Lv H. Y. (2017). Endophytic bacterial effects on seed germination and mobilization of reserves in Ammodendron biofolium. Pak. J. Bot. 49 2029–2035. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.