Abstract
Background
Daphnis nerii cypovirus-23 (DnCPV-23) is a new type of cypovirus and has a lethal effect on the oleander hawk moth, Daphnis nerii which feeds on leave of Oleander and Catharanthus et al. After DnCPV-23 infection, the change of Daphnis nerii responses has not been reported.
Methods
To better understand the pathogenic mechanism of DnCPV-23 infection, 3rd-instar Daphnis nerii larvae were orally infected with DnCPV-23 occlusion bodies and the transcriptional responses of the Daphnis nerii midgut were analyzed 72 h post-infection using RNA-seq.
Results
The results showed that 1979 differentially expressed Daphnis nerii transcripts in the infected midgut had been identified. KEGG analysis showed that protein digestion and absorption, Toll and Imd signaling pathway were down-regulated. Based on the result, we speculated that food digestion and absorption in insect midgut might be impaired after virus infection. In addition, the down-regulation of the immune response may make D. nerii more susceptible to bacterial infections. Glycerophospholipid metabolism and xenobiotics metabolism were up-regulated. These two types of pathways may affect the viral replication and xenobiotic detoxification of insect, respectively.
Conclusion
These results may facilitate a better understanding of the changes in Daphnis nerii metabolism during cypovirus infection and serve as a basis for future research on the molecular mechanism of DnCPV-23 invasion.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-021-01721-x.
Keywords: Daphnis nerii cypovirus-23, Midgut, Transcriptome analysis
Introduction
The oleander hawk moth, Daphnis nerii (D. nerii), belongs to Lepidoptera, Sphingidae family, is a worldwide pest [1]. D. nerii larvae damages leave of Oleander, Catharanthus, Vinca, Adenium, Vitis, Tabernaemontana, Gardenia, Trachelospermum, Amsonia, Asclepias, Carissa, Rhazya, Thevetia, Jasminum and Ipomoea [2, 3], which affect the landscape and the medicinal value of these plants. At present, the chemical pesticide decamethrin is used to control D. nerii [2].
Cypovirus is a member of the Reoviridae family, and is characterized by its single layered capsid [4]. DnCPV-23 was isolated from naturally diseased D. nerii larvae. This was a new type of cypovirus based on different electrophoretic migration patterns and conserved terminal sequences [1, 5, 6]. In addition to Daphnis nerii, it has been found that DnCPV-23 can also induce infection and death in many species of Sphingidae insects, such as Cephonodes hylas Linnaeus, Ampelophaga rubiginosa Bremer & Grey, and Agathia lycaenaria Kollar. The genome of DnCPV-23 consists of ten segments of linear double-stranded RNA, referred to as genomic segments 1 (S1) to 10 (S10), in accordance with the fragments from longest to shortest [7]. Our previous research and unpublished data demonstrated that the virus could successfully replicate on the Sf9 [8] and Manduca sexta cell lines QB-MS 2-2 [9]. However, the molecular mechanism of the interactions between the new type cypovirus and its hosts remains unclear. It is necessary to identify the interactions between the virus and its hosts to achieve an in-depth understanding and reveal the exploitation potential of the virus for future insecticide development.
Recently, many studies in the field have generated large amounts of data using the aforementioned high-throughput approaches, from the silkworms or BmN cells infected with BmCPV, including (1) The possible host’s RNAi response against BmCPV challenge in persistent and pathogenic Bombyx mori model was compared. During the pathogenic infection, it was found that higher level RNAi responses against BmCPV were observed, which further demonstrated the importance of RNAi as an antiviral mechanism [10]. (2) Gene expression profiles [11–19], DNA methylation [20], and lipidomic profile [21] of silkworm midgut or BmN cells after BmCPV infection were analyzed. These results suggested that many genes (for example, genes expressing Calreticulin, FK506-binding protein, and protein kinase c inhibitor gene, microRNAs, and activated protein kinase C) may play important roles in BmCPV replication. In addition, epigenetic regulation may influence silkworm-virus interaction, and BmCPV may modulate the lipid metabolism of cells for their self-interest.
Until now, the molecular mechanism underlying the midgut infection of DnCPV-23 is not clearly understood. Furthermore, since transcriptome analyses regarding D. nerii or DnCPV-23 have not yet been performed, this study aims to fill this gap about the new type cypovirus. The data and analysis presented here provide insights into the possible mechanism of DnCPV-23 infection and host defense and a basis for future DnCPV-23 relevant studies.
Materials and methods
Daphnis nerii larval midgut and virus stock
Newly wild-caught second instar larvae with a similar mass were used in this research investigation for the virus infection. Before infection, the D. nerii were supplied with 12-h day/night cycles under 50 ± 5% relative humidity conditions and were nurtured on oleander leaves at 27 ± 1 ℃ for three days. The midgut tissues were collected from four pathogenically infected larvae at 72 h [13, 15] after feeding with DnCPV-23. The same tissues were also collected from three uninfected control larvae at the same time point. DnCPV was originally isolated from the larvae of D. nerii and propagated in D. nerii larvae [1]. The polyhedra suspension of DnCPV-23 utilized for infecting the D. nerii was stored at 4 °C in the dark.
Virus inoculation
In this study, the DnCPV-23 viral stock was suspended in distilled water at a concentration of 2 × 107 polyhedra/mL. Then, 100 μL of the viral suspension was spread evenly on one piece of oleander leaf measuring approximately 4 cm × 1.5 cm each in size. The leaf was then fed to four D. nerii larvae. The dose of infection was calculated as 2 × 106 polyhedra per larva. In addition, three control larvae were fed the same quantity of leaves treated with only distilled water. After approximately 12 h, fresh oleander leaves were used to feed the inoculated larvae after the DnCPV-23-inoculated leaves had been completely consumed.
Sample preparation
The midguts of both DnCPV-23-infected and control larvae were collected at 72 h post-inoculation by dissecting the larvae on ice. The isolated midgut was then quickly washed in 0.8% diethylpyrocarbonate (DEPC)-treated physiologic saline solution to remove the attached leaf pieces, and then frozen in liquid nitrogen [13, 22].
RNA sequencing
All of the RNA-seq procedures were conducted by the Oebiotech Company (Shanghai, China). The total RNA was extracted from the D. nerii midgut tissue using TRIzol reagent (Invitrogen, USA) according to the manufacture’s protocols. The RNA integrity and concentrations were checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). In addition, seven RNA samples (including three uninfected samples and four infected samples) with RNA integrity were used to construct the libraries. The cDNA libraries were prepared using a TruSeq RNA Sample Preparation Kit (Illumina, USA) according to the manufacturer’s protocols. Thereafter, the obtained cDNA libraries were sequenced on the Illumina HiSeq2500 platform, which generated paired-end raw reads of 125 bp.
De novo assembly and functional annotation
The raw data was pretreated by discarding reads with adaptors and low quality (quality scores < 30). Then, the raw data was assembled using Trinity software with default parameters for de novo transcriptome assembly. Transcripts that were not shorter than 300 bp were used for subsequent analysis. To obtain the functional annotations of predicted protein-coding sequences, we searched against various databases, including the NCBI non-redundant (NR) protein, SwissProt, and euKaryotic Orthologous Groups (KOG) using Blastx with an E-value < 10−5. The top hit was utilized to assign gene names. Whereafter, the Gene Ontology (GO) annotations of the transcripts were then analyzed based on SwissProt annotations, and functional classifications were assigned by WeGO software. In addition, for the purpose of determining the biological pathways involved, the KEGG pathway was annotated based on the KEGG Orthology (KO) identifiers.
Differential gene expression analysis
RNA sequencing results from the two groups were mapped to the assembled transcriptome using bowtie2 [23] and express [24]. The FPKM (fragments per kb per million reads) method [25] was utilized to calculate the expression levels of the unigenes, which eliminated the influencing effects of the different gene lengths and sequencing levels. The differences in the unigene expressions between the two groups were calculated with DESeq [26] and any significant differences were determined with P < 0.05 and an absolute value of log2 fold change > 1.
Real-time quantitative reverse transcription PCR (Real-Time qRT-PCR)
This study utilized qRT-PCR to analyze the expression level of DnCPV-23 S1, S10 genes of transcriptome samples, and verify the DEGs recognized by the RNA-seq. The total RNA was isolated from the samples of the transcriptomic analysis using TRIzol reagent (Life Technologies) and was then treated with DNase I (Fermentas, Glen Burnie, MD, USA). We reversely transcribed 1 μg of the total RNA per sample into complementary DNA (cDNA) using a PrimeScript RT Reagent Kit (Takara). Then, qRT-PCR was performed using Talent qPCR PreMix SYBR Green (Tiangen, China) on a QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems™). One cycle was added for melting curve analysis for all the reactions to verify the product specificity. The expression level of each gene relative to that of the RPL13 gene was calculated using the 2−△△CT method [27]. All of the primers for the aforementioned target genes are listed in Table 1. Results are representative of two to three independent experiments.
Table 1.
Primers used in the qRT-PCR for the the viral RNA detection of transcriptome samples and validation of the RNA-seq
| No | Primer name | Primer sequence (5' to 3') | Tm (°C) | Gene id | Target gene |
|---|---|---|---|---|---|
| S1-RTPCR-F | GTGCTGATGGTCTGCTAA | 49.6 | N/A | DnCPV S1 | |
| S1-RTPCR-R | TGATTGATGACGACATTGAG | 51.5 | |||
| S10-RTPCR-F | GTCCGCCAATACTCTCAG | 52.6 | N/A | DnCPV S10 | |
| S10-RTPCR-R | CGTAGTCCATCGTCAATCA | 51.3 | |||
| 1 | CASP8-F | ACTGGAGAAGACTATGAGGTTA | 51.5 | TRINITY_DN10280_c0_g1_i1_3 | CASP8 |
| 2 | CASP8-R | ACGCTGTCATCTTGGCTAA | 53.7 | ||
| 3 | CYP6AB13-F | GATTCACACCAGCATTCAG | 51.0 | TRINITY_DN11437_c0_g1_i1_6 | CYP6AB13 |
| 4 | CYP6AB13-R | CAGTCGTATATCTCGCCATA | 50.5 | ||
| 5 | CYP6B45-F | GCGATACCGAACCAGAAC | 53.4 | TRINITY_DN12532_c0_g7_i1_1 | CYP6B45 |
| 6 | CYP6B45-R | ATTGGCAGTAAGTGTGAGTT | 51.0 | ||
| 7 | DHRS4-F | TCTTCTATCGCCGCATATCA | 52.8 | TRINITY_DN12896_c1_g2_i3_3 | DHRS4 |
| 8 | DHRS4-R | CACCACCTCATTAGCAATCG | 53.5 | ||
| 9 | PNLIP-F | CACCTCGTAGACTTGGAAGA | 53.5 | TRINITY_DN12381_c0_g2_i1_6 | PNLIP |
| 10 | PNLIP-R | GTTAGCGTTGCCATTGACA | 53.2 | ||
| 11 | PRSS1_2_3-F | CCTGGAAGATGGCGTGTT | 55.4 | TRINITY_DN10836_c0_g5_i1_6 | PRSS1_2_3 |
| 12 | PRSS1_2_3-R | TCGGCGGTAATTCGGTTAT | 53.5 | ||
| 13 | RDH12-F | GTCTAATCGTCCGCTATTGAG | 52.5 | TRINITY_DN14445_c0_g1_i1_3 | RDH12 |
| 14 | RDH12-R | CTGTAGGTGAAGATTGCCATT | 52.2 | ||
| 15 | SCARB1-F | AACACAACAAGAGGCATCAC | 53.0 | TRINITY_DN14140_c0_g1_i1_6 | SCARB1 |
| 16 | SCARB1-R | GTCGTCGGTTCAATATCCATAA | 51.7 | ||
| 17 | SLC46A1-F | TGGAACGACACGACAAGT | 53.7 | TRINITY_DN8071_c0_g1_i2_5 | SLC46A1 |
| 18 | SLC46A1-R | CAACAGAGTGCGAACAGTATA | 51.7 | ||
| 19 | SLC52A3-F | AAGCGATTGTGGAAGATGTC | 52.5 | TRINITY_DN11521_c0_g1_i2_4 | SLC52A3 |
| 20 | SLC52A3-R | CGGCATACACGAGTACGA | 54.4 | ||
| 21 | ABCA3-F | CGATATACGCCGCAAGTAAG | 53.3 | TRINITY_DN12365_c0_g1_i6_2 | ABCA3 |
| 22 | ABCA3-R | GCAGTTCTCTACATTCAGTTGA | 51.8 | ||
| 23 | ABCC4-F | AGTGGATGGAAGGTTGGAAT | 53.3 | TRINITY_DN11997_c1_g1_i24_2 | ABCC4 |
| 24 | ABCC4-R | CGGCTCTTGTGGTATAATTGA | 51.9 | ||
| 25 | CYP6B6-F | GGACTATTGTTGGCGAATC | 50.7 | TRINITY_DN13898_c0_g1_i1_4 | CYP6B6 |
| 26 | CYP6B6-R | TTGTGGAAGAAGACGATGT | 50.5 | ||
| GAPDH-F | TATGTTCGTTGTCGGAGTTA | 50.1 | TRINITY_DN5984_c0_g1_i2_2 | GAPDH | |
| GAPDH-R | TAGCAGTAGTGGCGTGTA | 52.4 | |||
| 27 | LYPLA3-F | ACATCCACGACACAAGACTA | 52.8 | TRINITY_DN10250_c0_g1_i1_1 | LYPLA3 |
| 28 | LYPLA3-R | GACCGATAATGAACTCCTGAAT | 51.5 | ||
| 29 | NTE-F | CAGCCTGGAAGGTAAGTAGT | 53.6 | TRINITY_DN14343_c0_g2_i1_4 | NTE |
| 30 | NTE-R | CTCATAGACGAGCGACAGT | 53.8 | ||
| 31 | UGT-F | GCATTCATTCAAGTCCATCAG | 51.3 | TRINITY_DN14215_c0_g5_i7_5 | UGT |
| 32 | UGT-R | GCCTCCATCAATAATCACCAA | 52.2 | ||
| 33 | DnRPL13-F | GAACTATTGGCATTGCTGTTG | 52 | TRINITY_DN4717_c0_g1_i2_3 | RPL13 |
| 34 | DnRPL13-R | TCCTCCTCATTGGCTTCAC | 54.5 |
Results
Virus infection of the samples
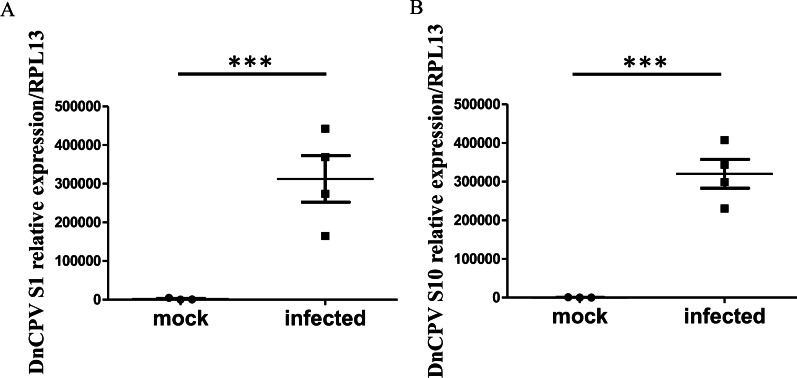
Prior to the transcriptome analysis, qRT-PCR was used to detect the mRNA levels of the DnCPV-23 S1 and S10 genes in the infected and uninfected samples. The results showed that the infected group had been successfully infected based on the high relative expression of the viral gene mRNA compared with uninfected group (Fig. 1).
Fig. 1.
Detection of the viral RNA in transcriptome samples at 72 hpi (hours post infection). After feeding for 72 h, the mRNA levels of DnCPV-23 S1 (A) and S10 (B) in the midgut of D. nerii were detected. The asterisk (***) denotes the presence of a statistically significant difference (p < 0.001) by unpaired Student's t test
Transcriptome sequencing and assembly
The RNA-Seq data from the DnCPV-23-infected and control groups contained 346.39 million reads, and 334.60 million clean reads after trimming, among which 96.17 to 97.39% per sample were determined to be useful. The acquired clean reads were assembled into 31,696 unigenes (> 300 bp). The average length of these unigenes was 1347.61 bp, and the N50 length was 2348 bp; other information about these unigenes were shown in Table 2. This study then assembled 31.696 unigenes ranging from 301 bp to 32,420 bp. The total unigene length was 42,713,980.
Table 2.
Statistics of the assembly results
| Term | All | > = 500 bp | > = 1000 bp | N50 | Total_Length | Max_Length | Min_Length | Average_Length |
|---|---|---|---|---|---|---|---|---|
| Unigene | 31,696 | 20,703 | 12,663 | 2348 | 42,713,980 | 32,420 | 301 | 1347.61 |
Transcriptome annotation
A total of 31,696 assembled unigenes were searched against the public databases, including the NR, Swissprot, KOG, GO, and KEGG databases, among which 16,820 (53.1%) (Fig. 2) unigenes were annotated. The distribution patterns of the unigenes in the different databases were as follows: 16,615 unigenes in the NR database, 11,152 unigenes in the Swissprot database, 10,374 unigenes in the KOG, 10,468 unigenes in the GO, and 5501 unigenes in the KEGG databases (Table 3). Figure 2 shows the degree of overlap between the unigenes annotated in the different databases. It was found that 4353 (13.7%) unigenes overlapped in all five databases, while 12,390 (73.7%) unigenes overlapped in two or more databases.
Fig. 2.
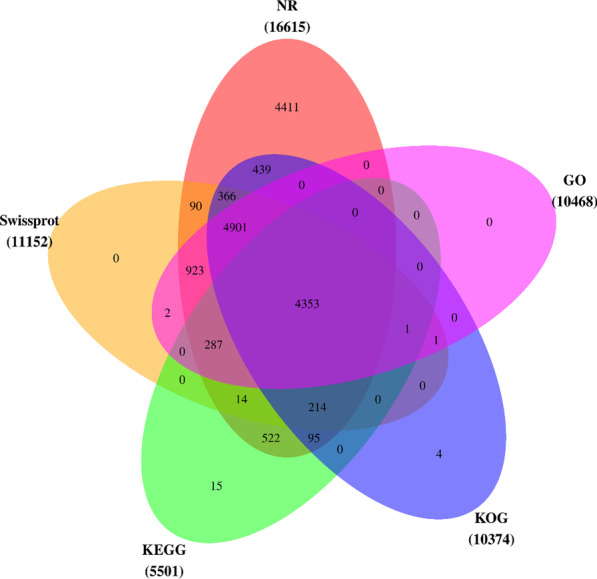
Venn diagram showing the degree of overlapping of the unigenes annotated based on different databases. Numbers in different colors represent the number of unigenes annotated through one or more annotation libraries
Table 3.
Annotation statistics for each database
| Anno_Database | Annotated_Number | 300 < = length < 1000 | Length > = 1000 |
|---|---|---|---|
| NR | 16,615(52.42%) | 6217(19.61%) | 10,398(32.81%) |
| Swissprot | 11,152(35.18%) | 2921(9.22%) | 8231(25.97%) |
| KEGG | 5501(17.36%) | 1694(5.34%) | 3807(12.01%) |
| KOG | 10,374(32.73%) | 2758(8.70%) | 7616(24.03%) |
| eggNOG | 15,249(48.11%) | 5239(16.53%) | 10,010(31.58%) |
| GO | 10,468(33.03%) | 2670(8.42%) | 7798(24.60%) |
| Pfam | 10,594(33.42%) | 2505(7.90%) | 8089(25.52%) |
Significant impacts of the viral infection on the hosts’ transcriptome expressions
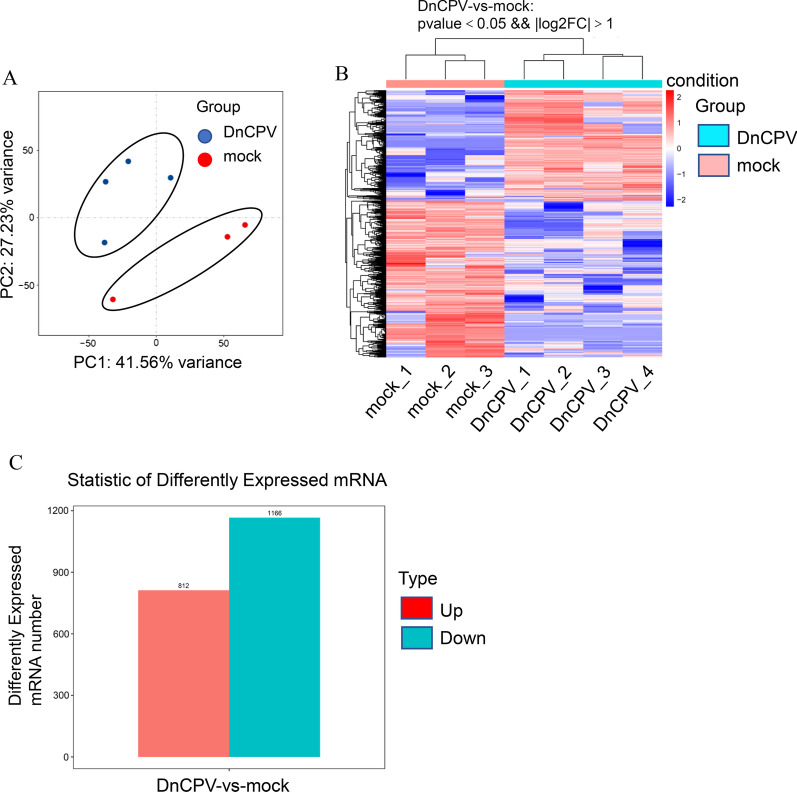
As shown in Fig. 3, the main component PCA1 had reached 41.56%, and the main component PCA2 had reached 27.23%. Therefore, the percentage total of the two was 68.79%, which accounted for a high proportion and represented the overall population to a large extent. This study’s principal component analysis manifested a clear separation of the samples with the two treatments (Fig. 3A), which indicated that the samples had good repeatability. The heat map of the gene expressions is presented in Fig. 3B. The results suggested that these DEGs could distinguish the samples. The results revealed that the viral infection could exert apparent influences on the midgut gene expressions. In addition, the transcriptome results showed that 1166 genes were down-regulated (accounting for 3.68% of the total assembled unigenes) and 812 genes (accounting for 2.56% of the total assembled unigenes) were up-regulated as a response to the DnCPV-23 infection (Fig. 3C).
Fig. 3.
Influence of DnCPV-23 infection on D. nerii transcriptome: A Plot of the 1st and 2nd principal component of the sample variations using the principal component analysis, in which the red dots represent samples without DnCPV-23 infection, and the green dots denote infected samples. B Heat map of 1,978 differently expressed genes (DEGs) in the infected samples and controls. C After infection, 812 genes were up-regulated (red bars) and 1166 genes were down-regulated (blue bars)
Analysis of the differently expressed genes
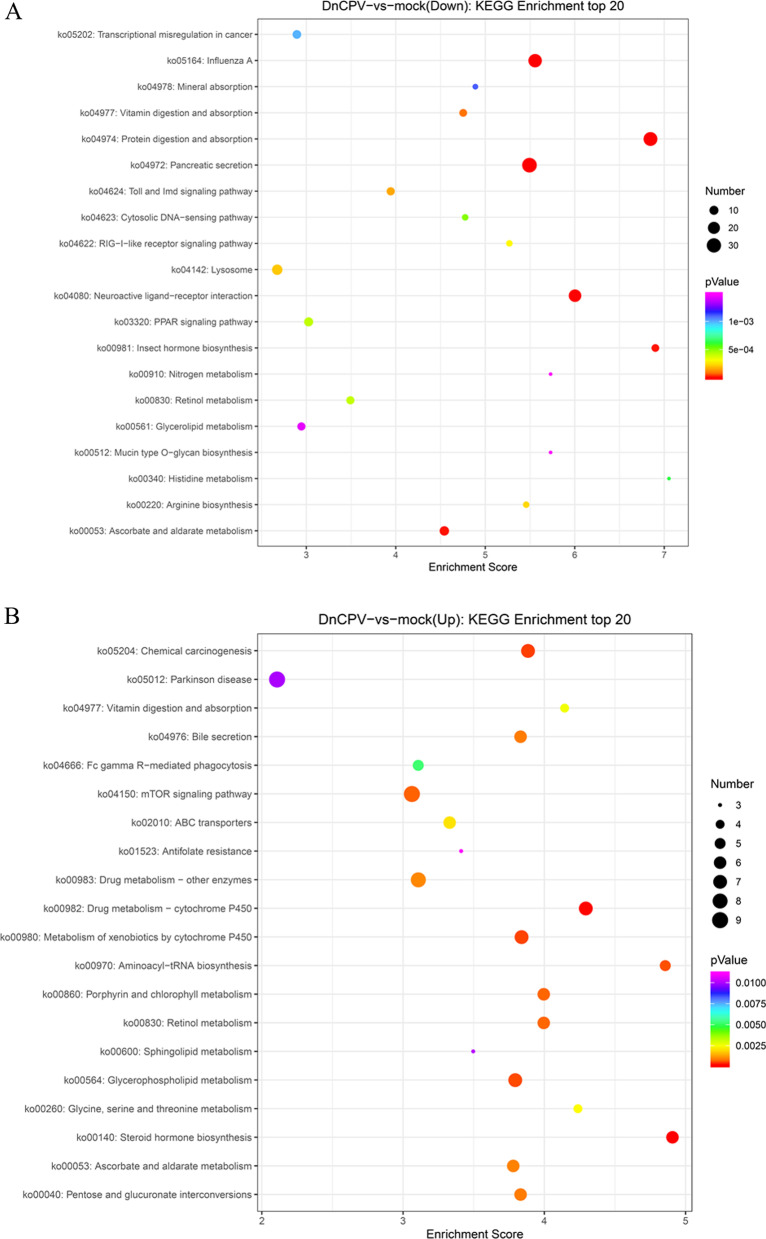
In this study, KEGG function enrichment analysis was performed on the differential genes expressed in the DnCPV-23-infected and uninfected control groups to clarify the relevant biological pathways involved in the differential genes. Among all of the DEGs, 298 DEGs had KEGG annotations, of which 118 were up-regulated genes and 180 were down-regulated genes. According to the pValue of KEGG analysis of up-regulated and down-regulated signal pathways, we identified 20 most significant signal pathways each. These pathways play an important role in insect reproduction, immunity, digestion and absorption and xenobiotic metabolism and so on (Fig. 4).
Fig. 4.
KEGG classifications of DEGs after DnCPV-23 infection (Top 20): A. Down-regulated pathways; B. Up-regulated pathways. Horizontal axis of the figure is the enrichment score. The larger the bubble, the more the number of DEGs. The bubble color changes from purpl E-blu E-green–red, indicating that the smaller the enrichment pValue and the greater the significance
qRT-PCR validation of DEGs
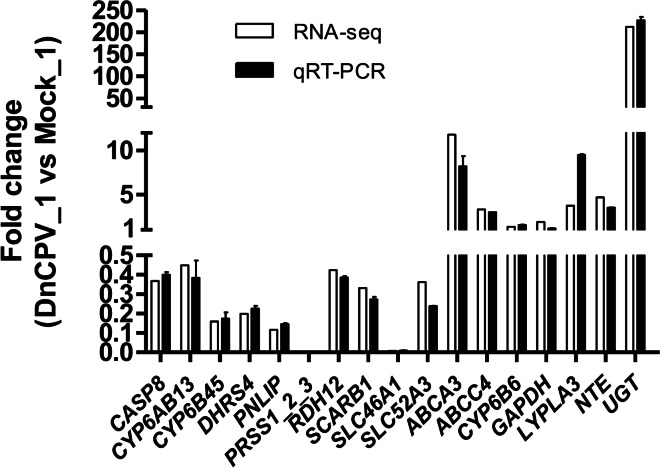
To verify the reliability of the transcriptome data and the DEG results obtained by RNA-seq, seventeen DEGs were selected for qPCR analysis. As shown in Fig. 5, the fold-change values of DnCPV_1 sample vs Mock_1 sample obtained in the qPCR analysis results were consistent with the values obtained by the RNA-seq for all of the selected genes.
Fig. 5.
Validation of RNA-seq profiles by real-time qPCR. To validate the RNA-seq data, the relative mRNA levels of 17 selected DEGs in the DnCPV_1 sample were examined by qPCR; The mRNA levels by qPCR are presented as the fold change compared with the Mock_1 sample after normalization against RPL13. The relative expression levels from the RNA-seq analysis were calculated as RPKM values. Error bars show mean ± SEM
Discussion
This study analyzed the transcriptome of the uninfected D. nerii midgut and the DnCPV-23- infected D. nerii midgut presented unique gene expression profiles induced by DnCPV-23 infection for the first time. In addition, KEGG function enrichment analysis was performed on the differential genes expressed after DnCPV-23 infection. Compared with uninfected D. nerii midgut, the transcriptome profiles of the infected samples displayed universally changed transcript abundances for many pathways.
Based on the pValue of KEGG analysis regarding up-regulated and down-regulated signal pathways, we identified 20 most significant signal pathways each. Among these signal pathways, the retinol metabolism pathway, vitamin digestion, and absorption signal pathway were down-regulated, consistent with the transcriptome study about BmCPV infected midgut vs non-infected midgut [13]. In addition, protein digestion and absorption pathway way was down-regulated in accord with previous research [10]. DnCPV infection may destroy the functions of digestion and the absorption of midguts, which causes the disturbance of protein and amino acid metabolism in D. nerii [13, 28]. Peptidoglycan recognition proteins (PGRPs) are pattern recognition molecules that are conserved from insects to mammals. PGRPs are the first receptors known to recognize, bind, or catalytically cleave the pathogenic microorganisms [29], PGRPs recognize bacteria and their unique cell wall component, eptidoglycan [30, 31]. This study observed nine transcripts of D. nerii isoforms of PGRP genes. Six transcripts were found to be down-regulated in the infected D. nerii midgut. The most highly expressed and most dramatically down-regulated was TRINITY_DN13195_c0_g1_i3_3, which was down-regulated by as much as 51-fold. The down-regulation of PGRP expression can lead to a decrease in the ability of the D. nerii’s innate immune system to recognize bacterial peptidoglycans (PGN), which may lead to D. nerii more susceptible to bacterial infections. In addition, BmPGRP-S2 was up-regulated upon BmCPV infection, overexpression of which can activate the Imd pathway and induce increased AMPs to enhance the antiviral capacity of transgenic silkworm against BmCPV [32]. Moreover, previous study demonstrates [33] that PGRPS2-1 and PGRPS2-2 can prevent BmCPV replication. Based on this work, was speculated that the down-regulation of PGRP was conducive to the replication of DnCPV-23.The gene CASP8 (KEGG gene name: caspase-8, Gene id: TRINITY_DN10280_c0_g1_i1_3) (Dredd in Drosophila) was down-regulated more than two folds, and other caspase genes changed non-significantly. It is predicted to be involved in the cleavage of Relish, the Drosophila homolog of mammalian NF-κB, resulting in activating the immune-deficient pathway (IMD)-induced expression of antimicrobial peptides in response to Gram-negative bacteria [34–36], fungi and viruses [37]. Research performed by Li et al. proved BmDredd interacts with BmSTING to enhance antiviral signaling [38]. The down-regulation of this gene may be very important for DnCPV-23 to escape from the host innate immune system and replicate in the midgut. Our result conflicted with the work by Guo et al. [11]. We speculated the contradiction might be related to the different stages of virus-host interaction or the heterogeneity of different species against virues. The pathways and the genes mentioned above are listed in Table 4 (The expression of genes in each sample is shown in Additional file 1).
Table 4.
The down-regulated pathways focused in the discussion section
| id | Term | pValue | Enrichment_score | gene_id | BaseMean_control_mock | BaseMean_case_DnCPV | FoldChange | pValue | qValue | Regulation | NR annotation | KEGG gene name |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ko04974 | Protein digestion and absorption | 1.33E−17 | 6.845688889 | TRINITY_DN12884_c1_g5_i1_1 | 2843.901036 | 282.1111777 | 0.099198662 | 1.72 E−06 | 0.0007668 | Down | LOW QUALITY PROTEIN: carboxypeptidase B [Bombyx mori] | CPA2 |
| TRINITY_DN13745_c3_g2_i2_4 | 37,228.13358 | 6283.238748 | 0.168776625 | 0.04653249 | 0.7667028 | Down | putative chymotrypsin, partial [Samia ricini] | CELA2 | ||||
| TRINITY_DN13546_c1_g2_i2_1 | 33,763.67895 | 179.8629852 | 0.005327115 | 1.26 E−10 | 2.06 E−07 | Down | RecName: Full = Trypsin, alkaline C; Flags: Precursor | PRSS1_2_3 | ||||
| TRINITY_DN11633_c0_g1_i2_3 | 576.0371804 | 28.38155477 | 0.049270352 | 1.62 E−08 | 1.12 E−05 | Down | trypsin, alkaline C-like [Spodoptera litura] | PRSS1_2_3 | ||||
| TRINITY_DN13619_c0_g2_i1_3 | 701.0350489 | 199.5322761 | 0.28462525 | 0.00871419 | 0.3479086 | Down | sodium/potassium-transporting ATPase subunit alpha isoform X6 [Bombyx mori] | ATP1A | ||||
| TRINITY_DN13597_c0_g2_i2_4 | 683.1424229 | 36.96948382 | 0.054116803 | 0.03730582 | 0.7023823 | Down | serine protease 62 [Mamestra configurata] | PRSS1_2_3 | ||||
| TRINITY_DN10836_c0_g7_i1_6 | 407.2114124 | 52.04490715 | 0.127808076 | 0.00142694 | 0.1232753 | Down | trypsin, partial [Manduca sexta] | PRSS1_2_3 | ||||
| TRINITY_DN7116_c0_g1_i1_5 | 140.0397486 | 41.73985284 | 0.298057182 | 0.02906 | 0.6296715 | Down | Prolylcarboxypeptidase [Danaus plexippus plexippus] | PRCP | ||||
| TRINITY_DN5681_c0_g1_i1_6 | 1202.562374 | 41.63057875 | 0.034618228 | 0.00013096 | 0.0240473 | Down | chymotrypsinogen-like protein 3 [Manduca sexta] | PRSS1_2_3 | ||||
| TRINITY_DN10836_c0_g5_i1_6 | 3539.814633 | 11.52410727 | 0.003255568 | 0.00393418 | 0.2234223 | Down | trypsin, alkaline C [Bombyx mori] | PRSS1_2_3 | ||||
| TRINITY_DN14237_c1_g1_i3_3 | 2139.270665 | 126.8457966 | 0.059293945 | 0.00362815 | 0.2142243 | Down | hypothetical protein B5V51_4161 [Heliothis virescens] | PRSS1_2_3 | ||||
| TRINITY_DN18044_c0_g1_i1_4 | 61.20950346 | 0 | 0 | 0.00262208 | 0.1786659 | Down | RecName: Full = Trypsin, alkaline C; Flags: Precursor | PRSS1_2_3 | ||||
| TRINITY_DN13619_c0_g3_i1_3 | 1098.504638 | 384.681457 | 0.350186466 | 0.02344348 | 0.5771173 | Down | Sodium/potassium-transporting ATPase subunit alpha [Papilio xuthus] | ATP1A | ||||
| TRINITY_DN12770_c1_g2_i2_6 | 33,988.65545 | 2472.057397 | 0.072731838 | 0.03984473 | 0.7217322 | Down | serine protease 62 [Mamestra configurata] | PRSS1_2_3 | ||||
| TRINITY_DN14161_c2_g2_i3_3 | 150,880.9838 | 10,699.40148 | 0.070912856 | 0.027439 | 0.6151748 | Down | trypsin, alkaline C-like [Spodoptera litura] | PRSS1_2_3 | ||||
| TRINITY_DN12929_c2_g1_i1_6 | 9158.048497 | 41.0899202 | 0.004486755 | 0.00143302 | 0.1232753 | Down | trypsin [Manduca sexta] | PRSS1_2_3 | ||||
| TRINITY_DN10836_c0_g1_i4_6 | 157,887.1736 | 53,448.64932 | 0.338524328 | 0.02226533 | 0.5641697 | Down | trypsin, alkaline C-like [Bombyx mori] | PRSS1_2_3 | ||||
| TRINITY_DN12646_c0_g1_i3_6 | 18,799.76899 | 343.0033323 | 0.018245082 | 0.00558226 | 0.271197 | Down | trypsin, alkaline C-like [Spodoptera litura] | PRSS1_2_3 | ||||
| TRINITY_DN3826_c0_g1_i1_3 | 4708.243184 | 116.815754 | 0.024810901 | 0.00083317 | 0.0853474 | Down | serine protease 5 [Mamestra configurata] | PRSS1_2_3 | ||||
| TRINITY_DN12903_c0_g1_i1_6 | 588.272948 | 19.69837276 | 0.03348509 | 7.39 E−10 | 8.85 E−07 | Down | silk gland derived serine protease [Bombyx mori] | PRSS1_2_3 | ||||
| TRINITY_DN14269_c4_g1_i5_4 | 15,015.14212 | 207.7199418 | 0.013834031 | 0.00010037 | 0.019782 | Down | trypsin [Manduca sexta] | PRSS1_2_3 | ||||
| TRINITY_DN8384_c0_g2_i8_4 | 4501.305649 | 35.57861617 | 0.007904066 | 0.00033725 | 0.0475214 | Down | chymotrypsinogen-like protein 3 [Manduca sexta] | PRSS1_2_3 | ||||
| TRINITY_DN17232_c0_g1_i1_5 | 229.0935699 | 80.06529495 | 0.349487308 | 0.03584292 | 0.6919927 | Down | proton-coupled amino acid transporter-like protein CG1139 [Trichoplusia ni] | SLC36A, PAT | ||||
| TRINITY_DN8969_c0_g1_i1_6 | 1070.195299 | 24.35172941 | 0.022754472 | 0.00153816 | 0.1284174 | Down | carboxypeptidase B [Bombyx mori] | CPA2 | ||||
| TRINITY_DN12498_c2_g2_i1_5 | 3577.880426 | 0 | 0 | 0.00308839 | 0.197654 | Down | trypsin CFT-1-like [Trichoplusia ni] | PRSS1_2_3 | ||||
| TRINITY_DN9363_c0_g1_i1_5 | 2166.100895 | 67.76280065 | 0.031283308 | 7.81 E−11 | 1.52 E−07 | Down | trypsin precursor AiD2, partial [Agrotis ipsilon] | PRSS1_2_3 | ||||
| TRINITY_DN7771_c0_g1_i1_1 | 166.8439433 | 24.32627007 | 0.145802536 | 0.01850363 | 0.5173478 | Down | hypothetical protein B5V51_4161 [Heliothis virescens] | PRSS1_2_3 | ||||
| TRINITY_DN1220_c0_g1_i1_5 | 14,636.6041 | 1623.956772 | 0.110951745 | 6.30 E−06 | 0.0020452 | Down | trypsin, alkaline C-like [Spodoptera litura] | PRSS1_2_3 | ||||
| ko04977 | Vitamin digestion and absorption | 8.59 E−05 | 4.753950617 | TRINITY_DN8071_c0_g1_i2_5 | 159.1367963 | 7.893169411 | 0.049599901 | 2.35 E−05 | 0.0062453 | Down | proton-coupled folate transporter isoform X2 [Bombyx mori] | SLC46A1 |
| TRINITY_DN12381_c0_g2_i1_6 | 15,115.15447 | 69.82405532 | 0.004619473 | 1.51 E−05 | 0.0042801 | Down | pancreatic triacylglycerol lipas E−like [Spodoptera litura] | PNLIP, PL | ||||
| TRINITY_DN9781_c0_g1_i1_3 | 75.2460337 | 20.63729453 | 0.274264217 | 0.03100164 | 0.6531949 | Down | scavenger receptor class B type 1 like protein 12 [Bombyx mori] | SCARB1 | ||||
| TRINITY_DN11521_c0_g1_i2_4 | 1196.401288 | 280.0407483 | 0.234069247 | 0.00250876 | 0.1751687 | Down | solute carrier family 52, riboflavin transporter, member 3-B isoform X3 [Trichoplusia ni] | SLC52A3, RFT2 | ||||
| TRINITY_DN14080_c0_g1_i4_5 | 13,236.92182 | 908.4409062 | 0.068629317 | 0.03090689 | 0.6516395 | Down | pancreatic triacylglycerol lipase [Bombyx mori] | PNLIP, PL | ||||
| TRINITY_DN17108_c0_g1_i1_5 | 19.89691746 | 1.317177118 | 0.066200059 | 0.0121168 | 0.4110339 | Down | hypothetical protein B5V51_177 [Heliothis virescens] | SLC46A1 | ||||
| TRINITY_DN14140_c0_g1_i1_6 | 6399.436654 | 1095.005085 | 0.171109606 | 0.00016917 | 0.0286312 | Down | sensory neuron membrane protein 2 [Bombyx mori] | SCARB1 | ||||
| ko04624 | Toll and Imd signaling pathway | 0.00016 | 3.943369176 | TRINITY_DN13195_c0_g1_i3_3 | 48,818.06612 | 740.1059156 | 0.015160492 | 2.79 E−06 | 0.0010722 | Down | peptidoglycan recognition protein 2 [Manduca sexta] | PGRP |
| TRINITY_DN1052_c0_g1_i2_5 | 74.58752535 | 0 | 0 | 0.02832612 | 0.6208957 | Down | Bacteriophage T7 lysozym E−like protein 1 (BTL-LP1) [Bombyx mori] | PGRP | ||||
| TRINITY_DN10280_c0_g1_i1_3 | 1415.480197 | 536.8363029 | 0.379260907 | 0.04150037 | 0.7315422 | Down | caspas E−6 [Manduca sexta] | CASP8 | ||||
| TRINITY_DN14006_c2_g1_i2_4 | 16,714.28346 | 318.7737419 | 0.019071936 | 3.82 E−10 | 5.17 E−07 | Down | peptidoglycan recognition protein 2 [Manduca sexta] | PGRP | ||||
| ko00830 | Retinol metabolism | 0.000409 | 3.492698413 | TRINITY_DN14190_c1_g2_i2_4 | 4018.349256 | 720.63212 | 0.179335362 | 0.04158394 | 0.7316189 | Down | UDP-glucosyltransferase isoform X1 [Bombyx mori] | UGT |
| TRINITY_DN12319_c0_g2_i1_4 | 4465.699274 | 170.972127 | 0.038285634 | 2.79 E−06 | 0.0010722 | Down | UDP-glycosyltransferase UGT340C2 [Bombyx mori] | UGT | ||||
| TRINITY_DN12896_c1_g2_i3_3 | 4251.17249 | 1508.008769 | 0.35472773 | 0.02274633 | 0.5685456 | Down | PREDICTED: RNA-directed DNA polymerase from mobile element jockey-like [Papilio machaon] | DHRS4 | ||||
| TRINITY_DN13518_c1_g1_i6_6 | 745.6825793 | 119.2237632 | 0.159885408 | 0.0001628 | 0.0278557 | Down | UDP-glycosyltransferase UGT340C1 precursor [Bombyx mori] | UGT | ||||
| TRINITY_DN14445_c0_g1_i1_3 | 151.0781746 | 54.27468401 | 0.359249006 | 0.04671659 | 0.7685163 | Down | hypothetical protein B5X24_HaOG201493 [Helicoverpa armigera] | RDH12 | ||||
| TRINITY_DN9738_c0_g1_i1_6 | 438.41149 | 83.17535239 | 0.189719828 | 0.04256225 | 0.7412925 | Down | uncharacterized protein LOC112052352 [Bicyclus anynana] | UGT | ||||
| TRINITY_DN8673_c0_g1_i3_3 | 839.7824168 | 167.2848772 | 0.199200262 | 0.00073535 | 0.0803495 | Down | PREDICTED: UDP-glucuronosyltransferase 2B19-like isoform X6 [Amyelois transitella] | UGT | ||||
| TRINITY_DN17220_c0_g1_i1_4 | 6379.593263 | 3.929259199 | 0.000615911 | 0.00570705 | 0.2744919 | Down | UDP-glycosyltransferase UGT340C1 precursor [Bombyx mori] | UGT |
In this study, the up-regulation of glycerophospholipid metabolism was consistent with Zhang’s research [21]. The up-regulation of this pathway may be related to the viral replication [39, 40]. In addition, Glycine, serine and threonine metabolism were up-regulated in this transcriptome analysis. In the study by Wu et al., two genes related to this signaling pathway were up-regulated and the other down-regulated. In our study, the expression levels of the phosphoserine phosphatase genes were significantly higher in DnCPV-23-infected midgut than in the non-infected group, suggesting that serine metabolism disorders were induced after DnCPV-23 infection. Expression of many UGT genes was up-regulated; UDP-glucuronosyltransferase (UGT) isozymes take endogenic and exogenic toxic substances as substrates, catalyze detoxification of many chemical toxins in our daily diet and environment by conjugation to glucuronic acid or glucose [41, 42]. After DnCPV-23 infection, it was speculated that the D. nerii tended to strengthen the elimination of lipophilic endobiotics such as hormones and xenobiotics including phytoalexins and drugs conjugated by invertebrates and plants mainly with glucose [42] through promoting the transcription of UGTs by regulating the activities of nuclear-receptor family (CAR, PXR, FXR, LXR, and PPAR), the arylhydrocarbon receptor [43] or ubiquitous transcription factors (FOXA1, Sp1, and Cdx2) [44]. However, the interactions between UGT and cypovirus still remain unclear. In Table 5, there were the pathways and genes mentioned above and genes expression of each sample is shown in Additional file 1.
Table 5.
The up-regulated pathways focused in the discussion section
| id | Term | pValue | Enrichment_score | Gene_id | BaseMean_control_mock | BaseMean_case_DnCPV | FoldChange | pValue | qValue | Regulation | NR annotation | KEGG gene name |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ko00564 | Glycerophospholipid metabolism | 0.00046 | 3.794540796 | TRINITY_DN14020_c0_g1_i1_6 | 1066.209777 | 3311.867512 | 3.106206287 | 0.027085 | 0.610813421 | Up | phosphatidate phosphatase LPIN2 isoform X2 [Trichoplusia ni] | LPIN |
| TRINITY_DN14343_c0_g2_i1_4 | 25.87214477 | 100.8782352 | 3.899106012 | 0.02501 | 0.591372647 | Up | hypothetical protein B5V51_748 [Heliothis virescens] | NTE, NRE | ||||
| TRINITY_DN14343_c2_g1_i1_5 | 1212.926019 | 3816.360113 | 3.146407986 | 0.018214 | 0.514696311 | Up | phosphatidate phosphatase LPIN3 isoform X1 [Bombyx mori] | LPIN | ||||
| TRINITY_DN2180_c0_g1_i1_3 | 5.852454693 | 49.91148246 | 8.528298822 | 0.005837 | 0.276535057 | Up | group XV phospholipase A2-like [Trichoplusia ni] | LYPLA3 | ||||
| TRINITY_DN10250_c0_g1_i1_1 | 73.45139125 | 221.6966763 | 3.018277429 | 0.037654 | 0.703084732 | Up | group XV phospholipase A2-like [Trichoplusia ni] | LYPLA3 | ||||
| TRINITY_DN11518_c6_g1_i1_2 | 52.98336083 | 709.749683 | 13.39570899 | 0.008188 | 0.337015221 | Up | Phosphatidylserine decarboxylase [Operophtera brumata] | psd, PISD | ||||
| TRINITY_DN12265_c0_g2_i1_2 | 21.24218102 | 111.7071749 | 5.258743197 | 0.006687 | 0.296214335 | Up | Neuropathy target esterase sws [Papilio xuthus] | NTE, NRE | ||||
| ko00260 | Glycine, serine and threonine metabolism | 0.00232 | 4.238058552 | TRINITY_DN9933_c0_g1_i2_6 | 782.5009178 | 4185.641092 | 5.349055824 | 0.025238 | 0.593696583 | Up | phosphoserine phosphatase isoform X3 [Trichoplusia ni] | serB, PSPH |
| TRINITY_DN7804_c0_g1_i1_2 | 11.1084935 | 91.01956837 | 8.193691462 | 0.032267 | 0.660850357 | Up | glucose dehydrogenase [FAD, quinone] [Bombyx mori] | betA, CHDH | ||||
| TRINITY_DN12220_c1_g1_i9_4 | 107.649078 | 517.8794976 | 4.8108122 | 0.00313 | 0.19793455 | Up | PREDICTED: phosphoserine phosphatase [Amyelois transitella] | serB, PSPH | ||||
| TRINITY_DN10934_c0_g2_i2_1 | 2279.078944 | 10,414.52625 | 4.569620669 | 0.002617 | 0.17866588 | Up | phosphoserine phosphatase isoform X1 [Bombyx mori] | serB, PSPH | ||||
| ko00982 | Drug metabolism—cytochrome P450 | 0.0002 | 4.29382248 | TRINITY_DN11538_c1_g1_i3_2 | 228.6892992 | 2097.050761 | 9.169868326 | 0.023489 | 0.577337157 | Up | hypothetical protein B5V51_11710 [Heliothis virescens] | UGT |
| TRINITY_DN14215_c0_g5_i7_5 | 127.0025656 | 13,180.97268 | 103.7850898 | 0.020886 | 0.5511394 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| TRINITY_DN7938_c0_g2_i1_2 | 3.93470221 | 637.5929402 | 162.0435058 | 6.29 E−05 | 0.013882511 | Up | PREDICTED: uncharacterized protein LOC106102769 [Papilio polytes] | GST, gst | ||||
| TRINITY_DN13727_c0_g2_i1_5 | 185.9951388 | 4205.660038 | 22.61166644 | 0.002663 | 0.180041496 | Up | UDP-glycosyltransferase UGT340C2 [Bombyx mori] | UGT | ||||
| TRINITY_DN13616_c0_g3_i6_5 | 25.95433532 | 8843.383538 | 340.7285692 | 0.014481 | 0.449089688 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| TRINITY_DN11622_c2_g4_i1_2 | 0 | 23.44067936 | Inf | 0.026206 | 0.601380717 | Up | UDP-glucuronosyltransferase 2B15-like isoform X1 [Helicoverpa armigera] | UGT | ||||
| TRINITY_DN11402_c0_g2_i13_2 | 663.9094166 | 6959.047076 | 10.48192254 | 0.000742 | 0.080554531 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| ko00980 | Metabolism of xenobiotics by cytochrome P450 | 0.00043 | 3.839182453 | TRINITY_DN11538_c1_g1_i3_2 | 228.6892992 | 2097.050761 | 9.169868326 | 0.023489 | 0.577337157 | Up | hypothetical protein B5V51_11710 [Heliothis virescens] | UGT |
| TRINITY_DN14215_c0_g5_i7_5 | 127.0025656 | 13,180.97268 | 103.7850898 | 0.020886 | 0.5511394 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| TRINITY_DN7938_c0_g2_i1_2 | 3.93470221 | 637.5929402 | 162.0435058 | 6.29 E− E−05 | 0.013882511 | Up | PREDICTED: uncharacterized protein LOC106102769 [Papilio polytes] | GST, gst | ||||
| TRINITY_DN13727_c0_g2_i1_5 | 185.9951388 | 4205.660038 | 22.61166644 | 0.002663 | 0.180041496 | Up | UDP-glycosyltransferase UGT340C2 [Bombyx mori] | UGT | ||||
| TRINITY_DN13616_c0_g3_i6_5 | 25.95433532 | 8843.383538 | 340.7285692 | 0.014481 | 0.449089688 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| TRINITY_DN11622_c2_g4_i1_2 | 0 | 23.44067936 | Inf | 0.026206 | 0.601380717 | Up | UDP-glucuronosyltransferase 2B15-like isoform X1 [Helicoverpa armigera] | UGT | ||||
| TRINITY_DN11402_c0_g2_i13_2 | 663.9094166 | 6959.047076 | 10.48192254 | 0.000742 | 0.080554531 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| ko00983 | Drug metabolism—other enzymes | 0.00101 | 3.107909605 | TRINITY_DN11538_c1_g1_i3_2 | 228.6892992 | 2097.050761 | 9.169868326 | 0.023489 | 0.577337157 | Up | hypothetical protein B5V51_11710 [Heliothis virescens] | UGT |
| TRINITY_DN14215_c0_g5_i7_5 | 127.0025656 | 13,180.97268 | 103.7850898 | 0.020886 | 0.5511394 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| TRINITY_DN7938_c0_g2_i1_2 | 3.93470221 | 637.5929402 | 162.0435058 | 6.29 E−05 | 0.013882511 | Up | PREDICTED: uncharacterized protein LOC106102769 [Papilio polytes] | GST, gst | ||||
| TRINITY_DN13727_c0_g2_i1_5 | 185.9951388 | 4205.660038 | 22.61166644 | 0.002663 | 0.180041496 | Up | UDP-glycosyltransferase UGT340C2 [Bombyx mori] | UGT | ||||
| TRINITY_DN11728_c0_g1_i4_2 | 1306.667581 | 6467.759766 | 4.949812684 | 0.001549 | 0.128973078 | Up | uridine phosphorylase 1 isoform X2 [Bombyx mori] | udp, UPP | ||||
| TRINITY_DN13616_c0_g3_i6_5 | 25.95433532 | 8843.383538 | 340.7285692 | 0.014481 | 0.449089688 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT | ||||
| TRINITY_DN11622_c2_g4_i1_2 | 0 | 23.44067936 | Inf | 0.026206 | 0.601380717 | Up | UDP-glucuronosyltransferase 2B15-like isoform X1 [Helicoverpa armigera] | UGT | ||||
| TRINITY_DN11402_c0_g2_i13_2 | 663.9094166 | 6959.047076 | 10.48192254 | 0.000742 | 0.080554531 | Up | UDP-glucuronosyltransferase 1-7C-like [Trichoplusia ni] | UGT |
Conclusion
This study revealed substantial differences in the transcriptions of the D. nerii genes related to digestion, immunity, glycerophospholipid metabolism and toxic substances metabolism induced by DnCPV-23 replication. Findings obtained in this research further enriched the understanding of cypovirus-Spodoptera insect interactions in midgut and provided additional basic information for the future exploitation of DnCPV-23.
Supplementary Information
Additional file 1. All the different expression genes in the midgut after DnCPV-23 infection.
Acknowledgements
Thanks to the molecular experiment platform provided by the Institute of Microbiology, Jiangxi Academy of Sciences.
Abbreviations
- DnCPV-23
Daphnis nerii Cypovirus-23
- D. nerii
Daphnis nerii
- PGRP
Peptidoglycan Recognition Protein;
- CASP-8
Caspase-8
Authors' contributions
KW, YC designed and performed the experiments and analysed the data. ZZ was responsible for revising the manuscript. ZZ, GL and CJ collected Daphnis nerii larval. WJ and LJ provided suggestions. KW, YC and JL wrote the manuscript. JL, ZX, and MG supervised the project and revised the manuscript. All Authors have read and approved the final version of the manuscript.
Funding
This research was supported by the Doctoral Research Startup Project of Jiangxi Academy of Sciences (2019-XTPH1-04), the Doctoral Research Startup Fund of Jiangxi University of Traditional Chinese Medicine (2020BSZR012), the Natural Science Foundation of Jiangxi Province (20192ACB20008), the key Research and Development Project of Jiangxi Province (20192BBF60056) and the key Research and Development Project of Jiangxi Province (20202BBFL63050).
Availability of data and materials
The original data of the transcriptome will be released on 2021-10-05 or upon publicationhas, BioProject accession: PRJNA766516.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wendong Kuang, Chenghua Yan and Zhigao Zhan have contributed equally to this work
Contributor Information
Wendong Kuang, Email: kuangwendong@163.com.
Chenghua Yan, Email: yanchenghua23@126.com.
Zhigao Zhan, Email: zgzhan_iom@163.com.
Limei Guan, Email: glmnh@126.com.
Jinchang Wang, Email: wangjinchang75@163.com.
Junhui Chen, Email: 1203854526@qq.com.
Jianghuai Li, Email: 35363052@qq.com.
Guangqiang Ma, Email: maguangqiang@163.com.
Xi Zhou, Email: zhouxi@wh.iov.cn.
Liang Jin, Email: jinliang079@163.com.
References
- 1.Zhan Z, Guan L, Wang J, Liu Z, Guo Y, Xiao Y, Wang H, Jin L. Isolation and genomic characterization of a cypovirus from the oleander hawk moth, Daphnis nerii. J Invertebr Pathol. 2019;163:43. doi: 10.1016/j.jip.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Lei YL, Lin ZG. Bionomics of the oleander hawkmoth, Daphnis nerii. Chin J Appl Entomol. 2010;47:918–922. [Google Scholar]
- 3.Sun Y, Chen C, Gao J, Abbas MN, Kausar S, Qian C, Wang L, Wei G, Zhu BJ, Liu CL. Comparative mitochondrial genome analysis of Daphnis nerii and other lepidopteran insects reveals conserved mitochondrial genome organization and phylogenetic relationships. PLoS ONE. 2017;12:e0178773. doi: 10.1371/journal.pone.0178773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Q, Ren F, Yan J, Zhang Y, Awais M, He J, Sun J. Alkaline phosphatase can promote the replication of Bombyx mori cypovirus 1 by interaction with its turret protein. Virus Res. 2021;292:198261. doi: 10.1016/j.virusres.2020.198261. [DOI] [PubMed] [Google Scholar]
- 5.Green TB, White S, Rao S, Mertens PP, Adler PH, Becnel JJ. Biological and molecular studies of a cypovirus from the black fly Simulium ubiquitum (Diptera: Simuliidae) J Invertebr Pathol. 2007;95:26–32. doi: 10.1016/j.jip.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Qin T, Xiao Y, Qin F, Lei C, Sun X. Genomic and biological characterization of a new cypovirus isolated from Dendrolimus punctatus. PLoS ONE. 2014;9:1. doi: 10.1371/journal.pone.0113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang GB, Yang J, Qin FJ, Xu CR, Wang J, Lei CF, Hu J, Sun XL. A reverse genetics system for cypovirus based on a bacmid expressing T7 RNA polymerase. Viruses Basel. 2019;11. [DOI] [PMC free article] [PubMed]
- 8.Kuang WD, Zhan ZG, Guan LM, Wang JC, Yan CH, Chen JH, Li JH, Zhou X, Jin L. Study on proliferation characteristics of Daphnis nerii cypovirus-23 in Sf9 cells. J. Agric. Biotechnol. 2021;29:772–779 (in Chinese).
- 9.Jiang L, Li GX, Li CY, Robert RG, Gary WB. Growth characteristics and expression of recombinant proteins in three new cell lines from Manduca sexta (Lepidoptera:Sphingidae). Acta Entomol Sin. 2010;53:1227–32 (in Chinese).
- 10.Kolliopoulou A, Van Nieuwerburgh F, Stravopodis DJ, Deforce D, Swevers L, Smagghe G. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS ONE. 2015;10:e0121447. [DOI] [PMC free article] [PubMed]
- 11.Guo R, Wang S, Xue R, Cao G, Hu X, Huang M, Zhang Y, Lu Y, Zhu L, Chen F, et al. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl Microbiol Biotechnol. 2015;99:5175–5187. doi: 10.1007/s00253-015-6634-x. [DOI] [PubMed] [Google Scholar]
- 12.Gao K, Deng XY, Qian HY, Qin GX, Hou CX, Guo XJ. Cytoplasmic polyhedrosis virus-induced differential gene expression in two silkworm strains of different susceptibility. Gene. 2014;539:230–237. doi: 10.1016/j.gene.2014.01.073. [DOI] [PubMed] [Google Scholar]
- 13.Wu P, Wang X, Qin GX, Liu T, Jiang YF, Li MW, Guo XJ. Microarray analysis of the gene expression profile in the midgut of silkworm infected with cytoplasmic polyhedrosis virus. Mol Biol Rep. 2011;38:333–341. doi: 10.1007/s11033-010-0112-4. [DOI] [PubMed] [Google Scholar]
- 14.Gao K, Deng XY, Qian HY, Qin G, Guo XJ. Digital gene expression analysis in the midgut of 4008 silkworm strain infected with cytoplasmic polyhedrosis virus. J Invertebr Pathol. 2014;115:8–13. doi: 10.1016/j.jip.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Jiang L, Peng ZW, Guo YB, Cheng TC, Guo HZ, Sun Q, Huang CL, Zhao P, Xia QY. Transcriptome analysis of interactions between silkworm and cytoplasmic polyhedrosis virus. Sci Rep. 2016; 6. [DOI] [PMC free article] [PubMed]
- 16.Wu P, Han S, Chen T, Qin G, Li L, Guo X. Involvement of microRNAs in infection of silkworm with bombyx mori cytoplasmic polyhedrosis virus (BmCPV). PLoS ONE. 2013; 8:e68209. [DOI] [PMC free article] [PubMed]
- 17.Wu P, Qin G, Qian H, Chen T, Guo X. Roles of miR-278-3p in IBP2 regulation and Bombyx mori cytoplasmic polyhedrosis virus replication. Gene. 2016;575:264–269. doi: 10.1016/j.gene.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Pan ZH, Wu P, Gao K, Hou CX, Qin GX, Geng T, Guo XJ. Identification and characterization of two putative microRNAs encoded by Bombyx mori cypovirus. Virus Res. 2017;233:86–94. doi: 10.1016/j.virusres.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Cao G, Zhu L, Chen F, Zar MS, Wang S, Hu X, Wei Y, Xue R, Gong C. Integrin beta and receptor for activated protein kinase C are involved in the cell entry of Bombyx mori cypovirus. Appl Microbiol Biotechnol. 2017;101:3703–3716. doi: 10.1007/s00253-017-8158-z. [DOI] [PubMed] [Google Scholar]
- 20.Wu P, Jie W, Shang Q, Annan E, Jiang X, Hou C, Chen T, Guo X. DNA methylation in silkworm genome may provide insights into epigenetic regulation of response to Bombyx mori cypovirus infection. Sci Rep. 2017;7:16013. doi: 10.1038/s41598-017-16357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Zhang YS, Shi X, Dai K, Liang Z, Zhu M, Zhang ZY, Shen ZE, Pan J, Wang CL, et al. Characterization of the lipidomic profile of BmN cells in response to Bombyx mori cytoplasmic polyhedrosis virus infection. Dev Comp Immunol. 2021;114. [DOI] [PMC free article] [PubMed]
- 22.Shrestha A, Bao K, Chen W, Wang P, Fei Z, Blissard GW. Transcriptional responses of the Trichoplusia ni Midgut to oral infection by the Baculovirus Autographa californica multiple Nucleopolyhedrovirus. J Virol. 2019;93. [DOI] [PMC free article] [PubMed]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11. [DOI] [PMC free article] [PubMed]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Rubinstein R. Characterization of the proteins and serological relationships of cytoplasmic polyhedrosis virus of Heliothis armigera. J Inverterb Pathol. 1983;42:292–294. doi: 10.1016/0022-2011(83)90078-2. [DOI] [Google Scholar]
- 29.Gao K, Deng XY, Qian HY, Qin GX, Hou CX, Guo XJ. Cloning and expression analysis of a peptidoglycan recognition protein in silkworm related to virus infection. Gene. 2014;552:24–31. doi: 10.1016/j.gene.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Kurz CL, Charroux B, Chaduli D, Viallat-Lieutaud A, Royet J. Peptidoglycan sensing by octopaminergic neurons modulates Drosophila oviposition. Elife. 2017;6. [DOI] [PMC free article] [PubMed]
- 31.Neyen C, Runchel C, Schupfer F, Meier P, Lemaitre B. The regulatory isoform rPGRP-LC induces immune resolution via endosomal degradation of receptors. Nat Immunol. 2016;17:1150–1158. doi: 10.1038/ni.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, Sun Q, Wang B, Wang Y, Xie E, Xia Q. Distinct functions of Bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Front Immunol. 2019;10:776. doi: 10.3389/fimmu.2019.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao P, Xia F, Jiang L, Guo H, Xu G, Sun Q, Wang B, Wang Y, Lu Z, Xia Q. Enhanced antiviral immunity against Bombyx mori cytoplasmic polyhedrosis virus via overexpression of peptidoglycan recognition protein S2 in transgenic silkworms. Dev Comp Immunol. 2018;87:84–89. doi: 10.1016/j.dci.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Cooper DM, Granville DJ, Lowenberger C. The insect caspases. Apoptosis. 2009;14:247–256. doi: 10.1007/s10495-009-0322-1. [DOI] [PubMed] [Google Scholar]
- 35.Ma X, Li X, Dong S, Xia Q, Wang F. A Fas associated factor negatively regulates anti-bacterial immunity by promoting Relish degradation in Bombyx mori. Insect Biochem Mol Biol. 2015;63:144–151. doi: 10.1016/j.ibmb.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci U S A. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manniello MD, Moretta A, Salvia R, Scieuzo C, Lucchetti D, Vogel H, Sgambato A, Falabella P. Insect antimicrobial peptides: potential weapons to counteract the antibiotic resistance. Cell Mol Life Sci. 2021;78:4259–4282. doi: 10.1007/s00018-021-03784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hua X, Li B, Song L, Hu C, Li X, Wang D, Xiong Y, Zhao P, He H, Xia Q, Wang F. Stimulator of interferon genes (STING) provides insect antiviral immunity by promoting Dredd caspase-mediated NF-kappaB activation. J Biol Chem. 2018;293:11878–11890. doi: 10.1074/jbc.RA117.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu LY, Hu XL, Kumar D, Chen F, Feng YJJ, Zhu M, Liang Z, Huang LX, Yu L, Xu J, et al. Both ganglioside GM2 and cholesterol in the cell membrane are essential for Bombyx mori cypovirus cell entry. Dev Comp Immunol. 2018;88:161–168. doi: 10.1016/j.dci.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Nanbo A, Maruyama J, Imai M, Ujie M, Fujioka Y, Nishide S, Takada A, Ohba Y, Kawaoka Y. Ebola virus requires a host scramblase for externalization of phosphatidylserine on the surface of viral particles. PLoS Pathog. 2018;14:e1006848. [DOI] [PMC free article] [PubMed]
- 41.Basu NK, Kovarova M, Garza A, Kubota S, Saha T, Mitra PS, Banerjee R, Rivera J, Owens IS. Phosphorylation of a UDP-glucuronosyltransferase regulates substrate specificity. Proc Natl Acad Sci U S A. 2005;102:6285–6290. doi: 10.1073/pnas.0407872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bock KW. The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: animal-plant arms-race and co-evolution. Biochem Pharmacol. 2016;99:11–17. doi: 10.1016/j.bcp.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Mackenzie PI, Hu DG, Gardner-Stephen DA. The regulation of UDP-glucuronosyltransferase genes by tissue-specific and ligand-activated transcription factors. Drug Metab Rev. 2010;42:99–109. doi: 10.3109/03602530903209544. [DOI] [PubMed] [Google Scholar]
- 44.Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol. 2013;45:1121–1132. doi: 10.1016/j.biocel.2013.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. All the different expression genes in the midgut after DnCPV-23 infection.
Data Availability Statement
The original data of the transcriptome will be released on 2021-10-05 or upon publicationhas, BioProject accession: PRJNA766516.