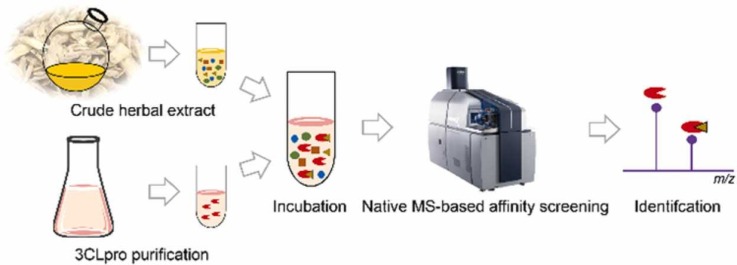
Graphical Abstract
A native MS-based affinity-selection method was developed to screen compounds that target SARS-CoV-2 3CLpro from TCMs crude extracts.
Keywords: SARS-CoV-2, 3CL protease, Native MS, Traditional Chinese medicine, Natural product
Abstract
The 3C-like protease (3CLpro) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is essential to the virus life cycle and is supposed to be a potential target for the treatment of coronaviral infection. Traditional Chinese medicines (TCMs) have played an impressive role in the treatment of COVID-19 in China. The effectiveness of TCM formulations prompts scientists to take continuous effort on searching for bioactive small molecules from the ancient resources. Herein, we developed a native mass spectrometry-based affinity-selection method for rapid screening of active small molecules from crude herbal extracts applied for COVID-19 therapy. Six common herbs named Lonicera japonica, Scutellaria baicalensis, Forsythia suspensa, Glycyrrhiza uralensis, Cirsium japonicum, and Andrographis paniculata were investigated. After preliminary separation of the crude extracts, the fractions were incubated with 3CLpro. A native MS-based affinity screening assay was then conducted to search for the protein-ligand complexes. A UHPLC-Q/TOF-MS with UNIFI data acquisition and data processing software was applied to identify the hit compounds. Standard compounds were used to verify the outcomes. Among the 16 hits, three flavonoids, baicalein, scutellarein and ganhuangenin, were identified as potential noncovalent inhibitors against 3CLpro with IC50 values of 0.94, 3.02, and 0.84 μM, respectively. Their binding affinities were further characterized by native MS, with Kd values being 1.43, 3.85, and 1.09 μM, respectively. Overall, we established an efficient native MS-based strategy for discovering 3CLpro ligands from crude mixtures, which supplies a potential strategy of small molecule lead discovery from TCMs.
1. Introduction
COVID-19, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to a severe pandemic since 2019. To date, more than 260 million cases of this new coronavirus disease have been diagnosed [1]. With the emergence of the mutant coronavirus which exhibits more efficient infection, replication, and competitive fitness, COVID-19 will continuously have a significant global impact [2]. The SARS-CoV-2 3C-like protease (3CLpro) catalyzes the proteolytic processing of polyproteins for viral replication [3], which shows a high degree of conservation across coronaviruses. Since the autocleavage process is essential for viral propagation, 3CLpro is considered a promising target for anti-coronaviral infection [4].
Traditional Chinese medicines (TCMs) have played an impressive role since the outbreak of COVID-19 [5]. Many clinically approved effective TCM formulations contain common herbal medicines such as Scutellaria baicalensis Georgi and Forsythia suspense (Thunb.) Vahl. In our previous study, baicalin and baicalein, two major compounds in S. baicalensis Georgi, showed in vitro 3CLpro inhibition activity [6]. Given the existence of various chemotypes of natural products, effective herbal medicines provide a rich source for drug discovery.
The conventional phytochemistry-pharmacology evaluation approach includes extraction, isolation, purification, identification, and activity screening. Each step is time-consuming, laborious, and more considerably, compound-untargeted. Thus, the development of an effective and selective screening method for active compounds is valuable.
Bio-affinity-based ligand screening has been applied extensively for drug discovery. Due to high sensitivity, low sample consumption and being label-free, bio-affinity-MS (BA-MS) has become an emerging biophysical technique commonly used in screening approaches in recent years [7]. Typical BA-MS methods are classified into the indirect and the direct methods. Indirect methods usually need to isolate the protein-ligand complex first from unbound components using a separation technique such as ultrafiltration [8], solid-phase micro-extraction [9], or size-exclusion chromatography [10], and then analyze the ligands released from the complexes. In contrast, the direct BA-MS method, also called native MS, can provide direct evidence of the existence of the protein-ligand complex as well as the stoichiometric information. Moreover, soft electrospray ionization (ESI) techniques allow solution-phase interactions to be intactly transferred into the gas-phase so that noncovalent binding even with weak affinity can be measured [11], [12], [13]. Recently, an increasing number of studies have shown the feasibility of native MS in natural product-based fragment screenings [14], [15], [16] and protein-compound interaction studies [17]. Therefore, native MS is expected to become a promising screening approach for hit discovery from TCMs.
This work aimed to explore the potential of native MS to capture compounds from complex matrixes, and provide a promising approach for drug discovery. We applied the optimized approach to screen the crude extracts of six TCMs. Guided by the native MS screening data, the compounds bound to the SARS-CoV-2 3CLpro were identified rapidly from the mixtures without time-consuming purification procedures. Then, enzymatic assays and equilibrium dissociation constant (Kd) determinations were performed to validate the screening outcomes. A native MS-based affinity-selection method was finally established to efficiently discover potential noncovalent inhibitors against SARS-CoV-2 3CLpro from herbal extracts.
2. Materials and methods
2.1. Materials
Six common herbs including Lonicera japonica, Scutellaria baicalensis, Forsythia suspensa, Glycyrrhiza uralensis, Cirsium japonicum, and Andrographis paniculata were selected according to the TCM prescriptions [18], [19], [20], and purchased from the Tong-Ren-Tang Company (Beijing, China). All the standard compounds were obtained from Shanghai Standard Technology Co., Ltd. (Shanghai, China). Glutathione (GSH) was obtained from Sigma (Shanghai, China). ODS gel AAG12S50 (12 nm, S-50 µm, YMC Co., Ltd., Japan) was used for column chromatography (CC). All solvents used for CC were of at least analytical grade (Shanghai Chemical Reagents Co., Ltd., Shanghai, China). High-performance liquid chromatography (HPLC)-grade acetonitrile (ACN), methanol (MeOH), acetic acid and formic acid were purchased from Merck (NJ, USA). Dimethyl sulfoxide (DMSO) and ammonium acetate (NH4Ac) were purchased from Sigma Chemical Co. (MO, USA). HPLC-grade water was produced by a Millipore Milli-Q® Advantage A10 Purification System (Millipore, Molsheim, France).
2.2. Expression and purification of 3CLpro
SARS-CoV-2 3CLpro was obtained as previously reported [6]. Briefly, the tagged SARS-CoV-2 3CLpro was expressed in BL21 (DE3) cells. The expressed protein was purified by a Ni-NTA column (GE Healthcare) and then transformed into the cleavage buffer (150 mM NaCl, 25 mM Tris, pH 7.5) containing human rhinovirus 3C protease to remove the additional residues. The resulting 3CLpro was then passed through a size exclusion chromatography (Superdex200, GE Healthcare). The eluted protein samples were stored in a 10 mM Tris buffer (pH 7.5) in a solution (10 mM Tris pH 7.5) for the enzymatic inhibition assay.
2.3. Fraction preparation
The TCM materials were powdered using an electric blender and then extracted with a 10-fold volume of MeOH using ultrasonication (three times, 1 h each). After filtration, the percolates were combined, and the organic solvent was removed by vacuum evaporation at 45 °C. The residue was suspended in water, and separated by ODS column chromatography, eluting with water, MeOH, and acetone, successively. The methanolic fraction (200 mg) was further fractionized using preparative HPLC, running at a flow rate of 30 mL/min with the mobile phases of water/0.1% formic acid (A) and ACN (B). The LC gradient was 0–3 min, B at 10%; 3–50 min, B at 10–80%; 51–60 min, B at 95%. Preparative HPLC was performed on a Waters 2545 Binary Gradient Module instrument with a Waters 2489 UV/Visible Detector using a Waters Sunfire Prep C18 column (5 µm, 30 × 150 mm). The subfractions were collected according to the UV and ELSD responses. Finally, they were powdered with freeze-drying and stored at − 40 °C for use.
2.4. Optimization of mass spectrometry conditions
Native MS screening was performed on a scimaX MRMS system (Bruker Daltonics, Bremen, Germany). The sample solutions were infused into the system with a syringe (250 µL) at a flow rate of 2 µL/min using electrospray ionization in positive ion mode. The source and ion transfer parameters were optimized for protein-ligand complex measurements: Skimmer 1 100 V, Funnel 1 15 V, Funnel RF Amplitude 180 V, collision cell frequency 2.0 MHz, ion transfer frequency 2.0 MHz, time of flight to analyzer cell 2.0 ms with RF amplitude of 350.0 Vpp, source temperature 200 °C. The ion accumulation time was set to 100 ms for optimal S/N of the mass spectrum. Mass spectra were deconvoluted with SNAP2 in Data Analysis 5.2 (Bruker Daltonics, Bremen, Germany).
The stock solution of 1 M NH4Ac was diluted for buffer exchange, and the pH was adjusted using acetic acid. Protein denaturing was obtained using 1% formic acid. Since the compounds were dissolved in organic solvents, different proportions of DMSO, MeOH and ACN were added to 3CLpro solution to evaluate their influence on the spray system.
2.5. Incubation of 3CLpro with crude extracts
The fractions of the crude herbal extracts were dissolved in MeOH to produce a stock solution of 10 mM, assuming an average molecular weight of 500 Da for the constituents. Then the stock solutions were further diluted to afford the working solution before the affinity MS screen. 3CLpro was buffer exchanged with 10 mM NH4Ac 5 times with a Millipore 10k cellulose membrane filter (Merck). The final system was as follows: 3CLpro at 1 µM, mixture of natural products at 10 µM, MeOH at 1%, in 10 mM NH4Ac (pH 6.9). The mixed solution was then incubated for 15 min at room temperature to form the protein-ligand complex before mass spectrometric analysis.
2.6. Hit identification
The fractions of interest were analyzed by ultra high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-Q/TOF-MS) with UNIFI data acquisition and data processing software. High-resolution ESI-MS spectra were recorded on a Waters Synapt G2-Si Q/TOF mass detector. Chromatographic separation was performed on an ACQUITY UPLC HSS T3 column (2.1 × 100 mm, 1.8 µm; Waters) at a temperature of 45 °C using 0.1% formic acid in H2O and 0.1% formic acid in ACN as mobile phases A and B, respectively. The flow rate was 0.4 mL/min and a 1-µL injection volume was used.
The matching and identification of the compounds were performed on the UNIFI informatic platform from Waters Corporation. According to the natural product analytical workflow within UNIFI [21], we established our library by importing the data excerpted from Reaxys® (https://www.reaxys.com/) including compound name, CAS number, and structure. For matching, the m/z accuracy tolerance was set to 10 ppm and spectral matching was performed using the established database. The matching results of high confidence were kept, and the hit compounds were finally identified by comparing the molecular information (molecular weight, retention time, fragment ions) with the standard compounds.
2.7. The inhibition assay of 3CLpro
A fluorescence resonance energy transfer (FRET) protease assay was applied to measure the inhibition of the hit compounds against SARS-CoV-2 3CLpro. The FRET-based protease assay was performed as previously reported [6]. The fluorogenic substrate MCA-AVLQSGFR-Lys(Dnp)-Lys-NH2 was obtained from GenScript (Nanjing, China). SARS-CoV-2 3CLpro (30 nM final concentration) was mixed with different compounds in 80 µL assay buffer (50 mM Tris-HCl, pH 7.3, 1 mM EDTA) and incubated for 10 min. The reaction was triggered by adding fluorogenic substrate (40 µL) at a final concentration of 20 µM. Then, the fluorescence signal at 320 nm (excitation)/405 nm (emission) was measured every 35 s for 3.5 min using a Bio-Tek Synergy4 plate reader.
2.8. Affinity evaluation
Affinity studies between 3CLpro and the hit compounds were conducted under the above-described optimized conditions in 2.4, 2.5. Different concentrations of the hit compounds and 3CLpro (1 μM) were prepared in 10 mM NH4Ac as spray solutions to form differential protein-ligand ratios. The charge states 18+ and 19+ were used to calculate the free protein and protein-ligand complex intensities. Five hundred single scans were added for the final mass spectrum.
The concentration ratios of 3CLpro-natural product complexes (PL) to total 3CLpro (P) were estimated by quantification of their relative MS peak intensity, which was based on the following hypotheses: the noncovalent complexes could be integrally transformed from the solution phase to the gas phase without in-source dissociation; the ionization efficacies of 3CLpro and 3CLpro-natural product complexes were similar. Therefore, the ratio R can be calculated as follows [22]:
The Kd was obtained by plotting the experimentally observed R against the total compound concentration using nonlinear regression in GraphPad Prism 8.2.1. For each compound, at least three independent experiments were performed for the determination.
3. Results and discussion
3.1. Fractionation of the crude extracts
Although native state mass spectrometry allows the direct detection of protein-ligand complexes with weak affinity, the highly abundant components can cover up others in a mixture. As a result, low-affinity ligands in high abundance can be overestimated, while potential active compounds but in low abundance can be overlooked. In addition, for complex samples, ion suppression and signal superposition cannot be completely avoided. Therefore, upfront separation for a mixture is necessary.
In the current study, after preliminary extraction, the six TCM herbs were partitioned through ODS column chromatography with water, MeOH and acetone. To obtain better screening results, the aqueous fractions were discarded to exclude carbohydrates and other highly polar components. Then, the methanolic fractions were further fractionized according to the UV and ELSD responses using preparative HPLC (Figs. S1–S6). A total of 96 fractions were obtained and their weights are shown in Table S1.
3.2. Optimization of the protein solution and spray condition
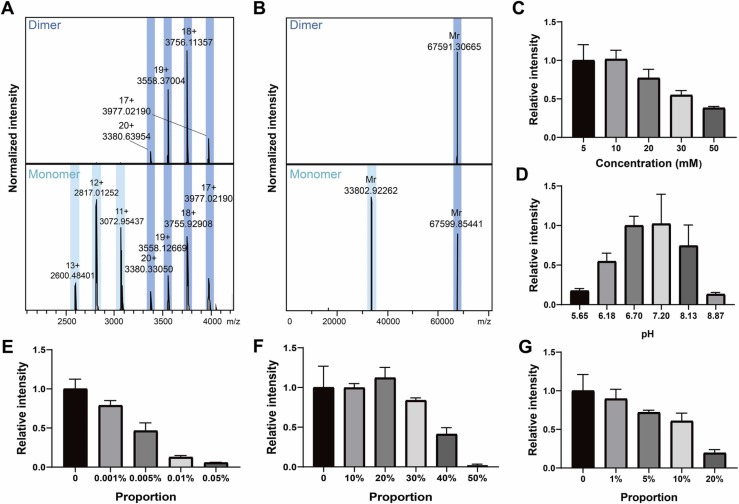
Characterization of 3CLpro was performed on a Magnetic Resonance Mass Spectrometry (MRMS) equipped with an ESI source. We found that 3CLpro remained a homodimer, which is the most common form for the crystal structures of catalytically active 3CLpro [23], and displayed a major 19+ (m/z 3558) charge state together with an 18+ (m/z 3756) charge state species ( Fig. 1A, upper), giving a molecular weight of 67.6 kDa under native condition (Fig. 1B, upper). After adding acid, 3CLpro partially changed into its monomer from a dimer, giving a molecular weight of 33.8 kDa (Fig. 1A, bottom and B, bottom); meanwhile, signals shifted into the lower m/z region implying partial denaturation.
Fig. 1.
Spray condition optimization for 3CLpro. Raw MS spectra (A) and deconvoluted spectra (B) of 3CLpro mainly in dimer (dark blue) or monomer (light blue) states. (C) The intensity of 3CLpro in NH4Ac at different concentrations. (D) The intensity of 3CLpro in 10 mM NH4Ac at different pH values. The influence of different proportions of DMSO (E), MeOH (F), or ACN (G) on the spray system. Experiments were repeated at least three times. Bars represent mean ± SD.
Because the stabilization and aggregation of protein and protein-ligand complexes require appropriate ionic strength and pH, the conditions were then optimized in terms of different NH4Ac concentrations and pH values. The results showed that 3CLpro showed the highest intensity in 10 mM NH4Ac and maintained its native state at pH 6.7–7.2 (Fig. 1C–D). Furthermore, the influence of organic solvents in the spray system was also studied including DMSO, MeOH and ACN (Fig. 1E–G). The results indicated that DMSO had a significant influence on the spray system, which was possibly due to its nonvolatility. Moreover, the spray system had a better tolerance of MeOH than ACN when the proportion was below 20%. Therefore, MeOH was chosen as the solvent for herbal mixture or compound dilution in the following 3CLpro and small molecule interaction studies.
3.3. Native MS-based affinity screening of herbal extracts
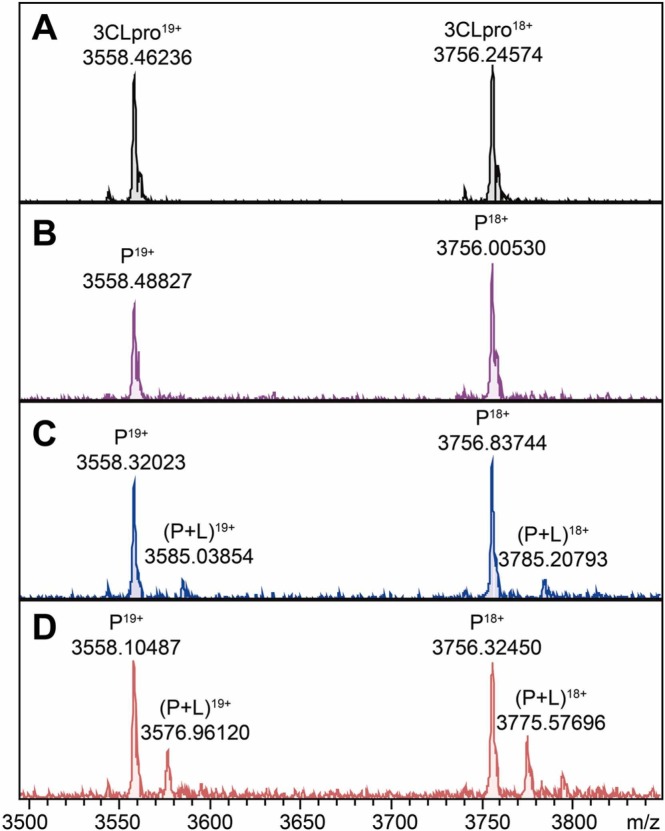
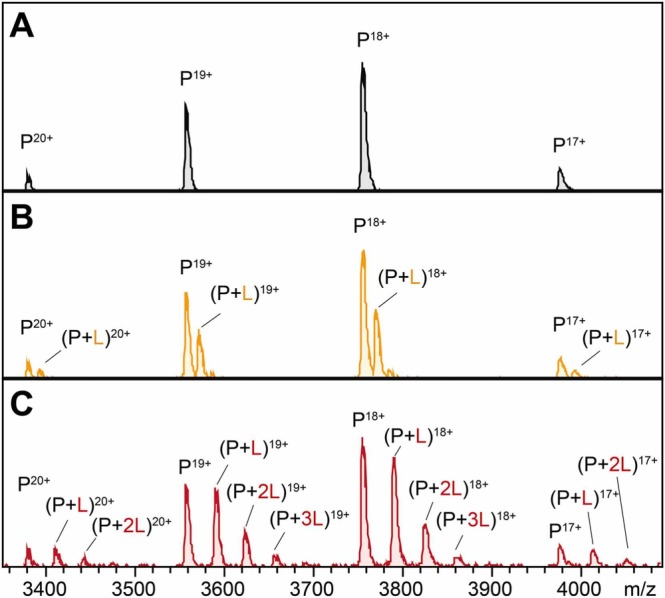
To form the protein-ligand complexes, 3CLpro (1 µM) was incubated with each fraction at the same concentration (10 µM), assuming an average molecular weight of 500 Da for the constituents. Then, a native MS-based affinity screening was conducted. Rapidly, we found the peaks of 3CLpro-natural product complexes. The representative MS spectra are shown in Figs. 2 and S7–S11. The molecular weight of the bound compound was then calculated according to the Δm/z between the protein and protein-ligand complex.
Fig. 2.
Representative MS spectra of native MS-based affinity screening. (A) MS spectrum of apo-3CLpro during the screening process: 3CLpro mainly presents in charge states P19+ and P18+. (B) Representative MS spectrum of a negative result from fraction 1 of Andrographis paniculata. Representative MS spectra of two positive results from fraction 3 (C) and fraction 6 (D) of Andrographis paniculata. The protein-ligand binary complexes (P + L) were found distinctly and the Δm/z was used to calculate the molecular weight of the ligands. Later, andrographoside and andrographolide were identified in fraction 3 and fraction 6 as compounds 12 and 13, respectively.
However, due to the chromatographic peak tail dragging or residue in the fractionation procedures, false positive results could be possible for some fractions. Consequently, more attention should be given to data analysis.
3.4. Identification of the hit compounds
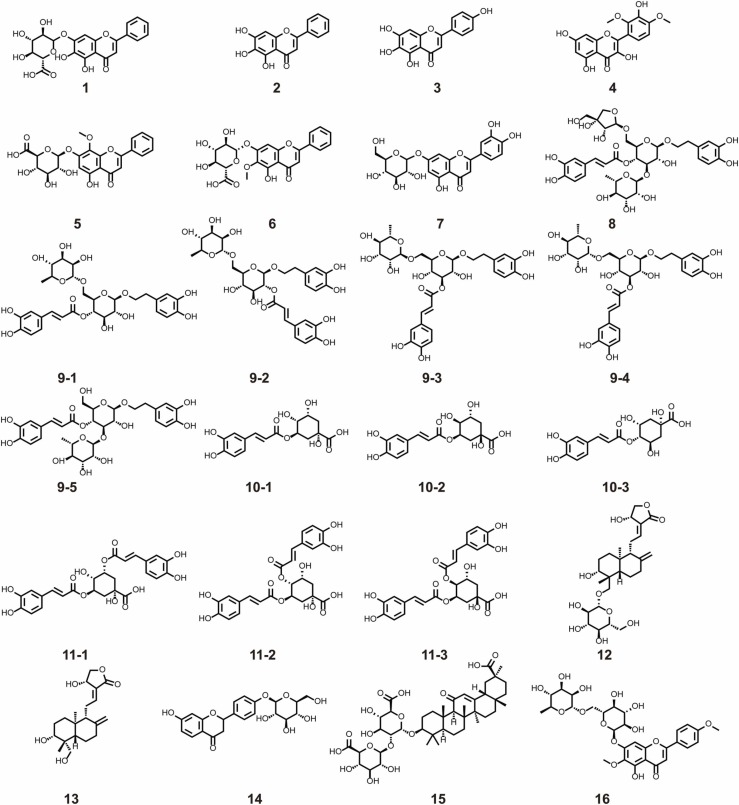
To identify the hit compounds in the fractions, UHPLC-Q/TOF-MS was applied. First, an in-house compound library was set up according to the Reaxys® database for each herb. The exact molecular weight and fragment ions of potential hit compounds were collected and matched to the compound library, and then their structures were proposed. Next, the hit compounds were further confirmed using authentic standards in terms of retention times, MS1 and MS2 mass spectra (Fig. S12). Finally, from 6 herbs, 16 hit compounds were rapidly screened out and identified, including 9 flavonoids, 3 terpenoids and 4 phenylpropanoids ( Table 1, Fig. 3).
Table 1.
The hit compounds identified from the TCMs.
| ID | Name | Chemotype | Source |
|---|---|---|---|
| 1 | Baicalin | Flavonoid | S. baicalensis |
| 2 | Baicalein | Flavonoid | |
| 3 | Scutellarein | Flavonoid | |
| 4 | Ganhuangenin | Flavonoid | |
| 5 | Wogonoside | Flavonoid | |
| 6 | Oroxylin A-7-O-β-D-glucuronide | Flavonoid | |
| 7 | Luteoloside | Flavonoid | |
| 8 | Forsythoside B | Phenylpropanoid | F. suspensa |
| 9-1 | Forsythoside A | Phenylpropanoid | |
| 9-2 | Forsythoside H | Phenylpropanoid | |
| 9-3 | Forsythoside I | Phenylpropanoid | |
| 9-4 | Isoforsythiaside | Phenylpropanoid | |
| 9-5 | Acteoside | Phenylpropanoid | |
| 10-1 | Chlorogenic acid | Phenylpropanoid | L. japonica |
| 10-2 | Neochlorogenic acid | Phenylpropanoid | |
| 10-3 | Cryptochlorogenic acid | Phenylpropanoid | |
| 11-1 | Isochlorogenic acid A | Phenylpropanoid | |
| 11-2 | Isochlorogenic acid B | Phenylpropanoid | |
| 11-3 | Isochlorogenic acid C | Phenylpropanoid | |
| 12 | Andrographoside | Terpenoid | A. paniculata |
| 13 | Andrographolide | Terpenoid | |
| 14 | Liquiritin | Flavonoid | G. uralensis |
| 15 | Glycyrrhizic acid | Terpenoid | |
| 16 | Pectolinarin | Flavonoid | C. japonicum |
Fig. 3.
The structure of the hit compounds.
Remarkably, in our previous studies, compound 2, baicalein, was proven to be the first noncovalent, nonpeptidomimetic inhibitor of SARS-CoV-2 3CLpro that could effectively prevent substrate access to the catalytic dyad within the active site. Moreover, the crystal structure of SARS-CoV-2 3CLpro in complex with baicalein has already been investigated using X-ray protein crystallography [6]. Consistent with the previous results, our current findings indicated that native MS can successfully be applied for the screening of potentially active compounds from the complex mixtures.
3.5. Distinguishment of nonspecific and specific binding
Compared with other methods such as isothermal titration calorimetry (ITC)[24] or surface plasmon resonance spectroscopy (SPR) [25], one great advantage of native MS is that it can provide stoichiometric information. Interestingly, when incubated with the pure compounds, we found that the flavonoids formed fewer binding peaks than other compounds at the same concentration (Fig. S13). A typical example is shown in Fig. 4: compound 2 formed fewer protein-ligand complexes than compound 9-1. It was intriguing to find more than one binding peak because previous study demonstrated that 3CLpro has only one substrate‐binding site, i.e. the active site [23]. Moreover, our previous study demonstrated that compound 2 is bound in the core region of the substrate-binding site [6]. However, more binding peaks of compound 9-1 indicated the presence of nonspecific binding, which meant the binding to other sites. This is probably because the α,β-unsaturated ketone in compound 9-1 acted as a Michael acceptor that could covalently conjugate cysteines in target proteins.
Fig. 4.
Differences in binding selectivity observed by native MS. (A) MS spectrum of apo-3CLpro during the titration assay. 3CLpro mainly presents in charge states P19+ and P18+. (B) MS spectrum of 3CLpro incubated with compound 2. The peaks of protein-ligand complexes were represented by P + L. (C) MS spectrum of 3CLpro incubated with compound 9-1. Nonspecific binding occurred.
To further verify the covalent binding, compounds were incubated with GSH and determined by LC-MS. No adduct with compound 2 was detected after incubation for 24 h, while the adduct with compound 9-1 was obvious. Consistent with previous study [26], these results suggested the covalently nonspecific binding with cysteine. Therefore, the binding selectivity distinguished by native MS indicated the difference between different chemotypes. In addition, stoichiometric information could be used as supplementary evidence to give alerts before putting considerable effort into problematic compounds. Together, native MS analysis allows, in addition to the screening of ligands from the mixture, the distinguishment of different binding modes between protein-ligand complexes.
3.6. Determination of the activity of the hit compounds
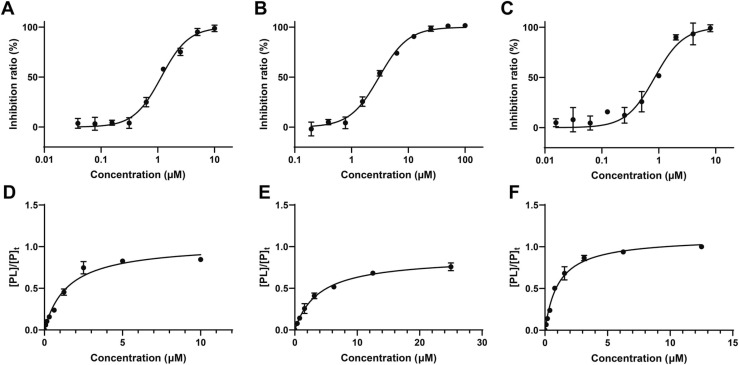
Considering that catechols and Michael acceptors were frequent-hitters in Pan-Assay INterference compoundS (PAINS) [27], the protease assays were conducted only for compounds 2, 3 and 4. The determined IC50 values of the three compounds were 0.94, 3.02 and 0.84 μM, respectively ( Fig. 5A–C and Table 2). Then, based on the optimum tuning condition and the evaluation method mentioned above, a titration experiment using Native MS was applied to measure the binding affinity of 3CLpro-natural product complexes. The titration curves of 3CLpro and the active natural products are shown in Fig. 5D–E and their Kd values were 1.43, 3.85 and 1.09 μM (Table 2). For this chemotype of compound, the results demonstrated that the binding affinities were in keeping with the IC50 values: the stronger the binding, the smaller the IC50 value.
Fig. 5.
Evaluation of the hit compounds. (A–C) Inhibition curves for compounds 2, 3 and 4. (D–F) Binding affinities of compounds 2, 3, and 4 with SARS-CoV-2 3CLpro characterized by native MS. Fraction of ligand bound protein ([PL]/[P]t) obtained under optimized conditions plotted against total ligand concentrations. All data are shown as mean ± SD.
Table 2.
Binding affinity of compound 2, 3 and 4 with SARS-CoV-2 3CLpro determined by MRMS and the inhibition of 3CLpro by the three compounds.
| ID | Name | IC50 (μM) | Kd (μM) | Bmax |
|---|---|---|---|---|
| 2 | Baicalein | 0.94 ± 0.20 | 1.43 ± 0.15 | 1.02 ± 0.02 |
| 3 | Scutellarein | 3.02 ± 0.11 | 3.85 ± 0.47 | 0.88 ± 0.02 |
| 4 | Ganhuangenin | 0.84 ± 0.06 | 1.09 ± 0.14 | 1.11 ± 0.01 |
Although we conjectured that the Michael acceptors were probably promiscuous in the screening and were not considered here, the class of peptidomimetic inhibitors carrying a Michael acceptor has been considered as a potential drug design strategy [28]. More researches thus need to be done in future studies.
4. Conclusions
In this study, we developed a native MS-based affinity-selection method to screen compounds that target SARS-CoV-2 3CLpro from the TCM crude extracts, which resulted in the identification of 16 hit compounds. Directly, protein-ligand complexes and the stoichiometric numbers of the ligands could be visualized using native MS. Although the nonspecific binding interfered with the results of screening to some extent, the binding selectivity of the compounds could be observed directly through mass spectrometry. In addition, potentially problematic compounds could be implied and alerted before investing significant time and effort. Moreover, a FRET protease assay was applied to verify the inhibition of SARS-CoV-2 3CLpro by the three noncovalent-binding compounds and the binding affinities were further measured. Taken together, a native MS-based affinity mass spectrometry method that could rapidly find hits from the complex matrixes was developed in this study. Using this method, the active ingredients from crude herbal extracts could be rapidly screened and identified. The findings support the further use of native MS as a potential tool for ligand screening and evaluation in drug discovery.
CRediT authorship contribution statement
Dafu Zhu: Methodology, Visualization, Writing – original draft. Haixia Su: Validation. Changqiang Ke: Investigation, Data curation. Chunping Tang: Writing – review & editing. Ronald J Quinn AM: Investigation, Writing – review & editing. Matthias Witt: Native MS. Yechun Xu: Resources. Jia Liu: Conceptualization, Writing – review & editing, Supervision. Yang Ye: Resources, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Science and Technology Commission of Shanghai Municipality (20430780300), National Natural Science Foundation of China (21920102003), and Key-Area Research and Development Program of Guangdong Province (2020B0303070002).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jpba.2021.114538.
Appendix A. Supplementary material
Supplementary material.
References
- 1.WHO, Coronavirus disease (COVID-19) outbreak situation, 2021. 〈https://who.sprinklr.com/〉, (Accessed 7 December 2021).
- 2.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Graham R., Edwards C.E., Tse L.V., Okuda K., Markmann A.J., Bartelt L., de Silva A., Margolis D.M., Boucher R.C., Randell S.H., Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Z., Li Y., Zhou L., Zhou X., Xie B., Zhang W., Sun J. Prevention and treatment of COVID-19 using Traditional Chinese Medicine: a review. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao Y., Yan J., Cai B. Label-free bio-affinity mass spectrometry for screening and locating bioactive molecules. Mass Spectrom. Rev. 2021;40(1):53–71. doi: 10.1002/mas.21613. [DOI] [PubMed] [Google Scholar]
- 8.Chen G., Huang B.X., Guo M. Current advances in screening for bioactive components from medicinal plants by affinity ultrafiltration mass spectrometry. Phytochem. Anal. 2018;29(4):375–386. doi: 10.1002/pca.2769. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y.M., Wang L.H., Luo S.F., Wang Q.Q., Moaddel R., Zhang T.T., Jiang Z.J. Magnetic beads-based neuraminidase enzyme microreactor as a drug discovery tool for screening inhibitors from compound libraries and fishing ligands from natural products. J. Chromatogr. A. 2018;1568:123–130. doi: 10.1016/j.chroma.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muckenschnabel I., Falchetto R., Mayr L.M., Filipuzzi I. SpeedScreen: label-free liquid chromatography-mass spectrometry-based high-throughput screening for the discovery of orphan protein ligands. Anal. Biochem. 2004;324(2):241–249. doi: 10.1016/j.ab.2003.09.040. [DOI] [PubMed] [Google Scholar]
- 11.Vivat Hannah V., Atmanene C., Zeyer D., Van Dorsselaer A., Sanglier-Cianferani S. Native MS: an 'ESI' way to support structure- and fragment-based drug discovery. Future Med. Chem. 2010;2(1):35–50. doi: 10.4155/fmc.09.141. [DOI] [PubMed] [Google Scholar]
- 12.Riccardi Sirtori F., Altomare A., Carini M., Aldini G., Regazzoni L. MS methods to study macromolecule-ligand interaction: applications in drug discovery. Methods. 2018;144:152–174. doi: 10.1016/j.ymeth.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Chen G., Fan M., Liu Y., Sun B., Liu M., Wu J., Li N., Guo M. Advances in MS based strategies for probing ligand-target interactions: focus on soft ionization mass spectrometric techniques. Front. Chem. 2019;7:703. doi: 10.3389/fchem.2019.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Y., Feng Y., Di Capua A., Mak T., Buchko G.W., Myler P.J., Liu M., Quinn R.J., Phenotarget A. Approach for identifying an alkaloid interacting with the tuberculosis protein Rv1466. Mar. Drugs. 2020;18(3) doi: 10.3390/md18030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M., Quinn R.J. Fragment-based screening with natural products for novel anti-parasitic disease drug discovery. Expert. Opin. Drug Discov. 2019;14(12):1283–1295. doi: 10.1080/17460441.2019.1653849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vu H., Pedro L., Mak T., McCormick B., Rowley J., Liu M., Di Capua A., Williams-Noonan B., Pham N.B., Pouwer R., Nguyen B., Andrews K.T., Skinner-Adams T., Kim J., Hol W.G.J., Hui R., Crowther G.J., Van Voorhis W.C., Quinn R.J. Fragment-based screening of a natural product library against 62 potential malaria drug targets employing native mass spectrometry. ACS Infect. Dis. 2018;4(4):431–444. doi: 10.1021/acsinfecdis.7b00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B., Qin Q., Chang M., Li S., Shi X., Xu G. Molecular interaction study of flavonoids with human serum albumin using native mass spectrometry and molecular modeling. Anal. Bioanal. Chem. 2018;410(3):827–837. doi: 10.1007/s00216-017-0564-7. [DOI] [PubMed] [Google Scholar]
- 18.Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Li X., Gou C., Li L., Luo X., Zhang C., Zhang Y., Zhang J., Jin A., Li H., Zeng Y., Li T., Wang X. Effect of Jinhua Qinggan granules on novel coronavirus pneumonia in patients. J. Tradit. Chin. Med. 2020;40(3):467–472. doi: 10.19852/j.cnki.jtcm.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang W., Fan Z., Chu Y., Wang H., Yang Y., Wu L., Sun N., Sun G., Shen Y., Lin X., Guo G., Xi S. Chinese patent medicines in the treatment of coronavirus disease 2019 (COVID-19) in China. Front. Pharm. 2020;11:1066. doi: 10.3389/fphar.2020.01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L., Shi A.M., Liu H.Z., Meruva N., Liu L., Hu H., Yang Y., Huang C., Li P., Wang Q. Identification of chemical ingredients of peanut stems and leaves extracts using UPLC-QTOF-MS coupled with novel informatics UNIFI platform. J. Mass Spectrom. 2016;51(12):1157–1167. doi: 10.1002/jms.3887. [DOI] [PubMed] [Google Scholar]
- 22.Pedro L., Van Voorhis W.C., Quinn R.J. Optimization of electrospray ionization by statistical design of experiments and response surface methodology: protein-ligand equilibrium dissociation constant determinations. J. Am. Soc. Mass Spectrom. 2016;27(9):1520–1530. doi: 10.1007/s13361-016-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong M., Su H., Zhao W., Xie H., Shao Q., Xu Y. What coronavirus 3C-like protease tells us: from structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 2021;41(4):1965–1998. doi: 10.1002/med.21783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paketuryte V., Zubriene A., Ladbury J.E., Matulis D. Intrinsic thermodynamics of protein-ligand binding by isothermal titration calorimetry as aid to drug design. Methods Mol. Biol. 1964;2019:61–74. doi: 10.1007/978-1-4939-9179-2_5. [DOI] [PubMed] [Google Scholar]
- 25.Patching S.G. Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta. 2014;1838(1 Pt A):43–55. doi: 10.1016/j.bbamem.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 26.Su H., Yao S., Zhao W., Zhang Y., Liu J., Shao Q., Wang Q., Li M., Xie H., Shang W., Ke C., Feng L., Jiang X., Shen J., Xiao G., Jiang H., Zhang L., Ye Y., Xu Y. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 2021;12(1):3623. doi: 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 28.Wang F., Chen C., Yang K., Xu Y., Liu X., Gao F., Liu H., Chen X., Zhao Q., Liu X., Cai Y., Yang H. Michael acceptor-based peptidomimetic inhibitor of main protease from porcine epidemic diarrhea virus. J. Med. Chem. 2017;60(7):3212–3216. doi: 10.1021/acs.jmedchem.7b00103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.