Abstract
Objective
We observed that the addition of glucose enhanced the expression of sigX and sigM, encoding extra-cytoplasmic function sigma factors in Bacillus subtilis. Several regulatory factors were identified for this phenomenon, including YqfO, CshA (RNA helicase), and YlxR (nucleoid-associated protein). Subsequently, the relationships among these regulators were analyzed. Among them, YqfO is conserved in many bacterial genomes and may function as a metal ion insertase or metal chaperone, but has been poorly characterized. Thus, to further characterize YqfO, we performed RNA sequencing (RNA-seq) analysis of YqfO in addition to CshA and YlxR.
Results
We first performed comparative RNA-seq to detect the glucose-responsive genes. Next, to determine the regulatory effects of YqfO in addition to CshA and YlxR, three pairs of comparative RNA-seq analyses were performed (yqfO/wt, cshA/wt, and ylxR/wt). We observed relatively large regulons (approximately 420, 780, and 180 for YqfO, CshA, and YlxR, respectively) and significant overlaps, indicating close relationships among the three regulators. This study is the first to reveal that YqfO functions as a global regulator in B. subtilis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-021-05869-1.
Keywords: RNA-seq, Glucose, Catabolite regulation, Transcriptome
Introduction
Glucose is used as the most favorable carbon source for many gram-positive bacteria. Hence, bacteria have developed a variety of glucose-responsive systems. For example, the gram-positive model bacterium Bacillus subtilis possesses the transcription factor catabolite control protein A (CcpA) as the primary carbon catabolite regulator [1, 2]. CcpA causes global transcriptional changes, and additional glucose-responsive transcription factor genes are detected in the B. subtilis genome [2]. Several DNA microarray analyses, however, have revealed many glucose-responsive genes in which glucose responses are caused by unknown factors [3–5].
We observed that glucose in the medium enhanced the expression of sigX and sigM, encoding extra-cytoplasmic function sigma factors [6] (Fig. 1A). We identified several regulatory factors for this phenomenon, including YqfO, CshA, and YlxR, and analyzed the relationships among these regulators [7, 8]. YqfO is a conserved protein among the bacteria of the Firmicutes phylum that bears a DUF34/nif3 conserved domain with a suggested function related to transcriptional regulation [9]. The structure of Bacillus cereus YqfO has been resolved, revealing the presence of a dimetal-binding motif [10]. Recently, bioinformatics analysis using data from the determined genome sequences and published reports revealed that YqfO may function as a metal chaperone or metal insertase [11]. We observed that YqfO is under positive control of YlxR [8].
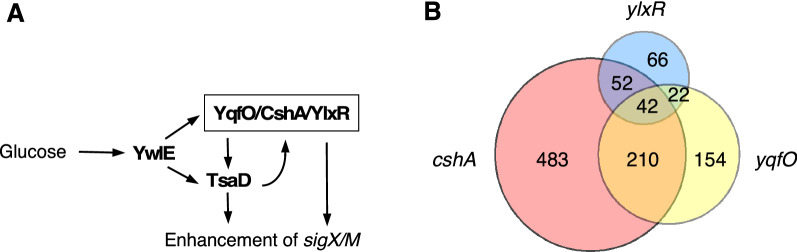
Fig. 1.
Glucose induction of sigX/M and Venn diagram of the detected regulons. A Arrows indicate positive regulation and do not mean direct effects. Details are in the Refs. [16, 21]. B Numbers indicate gene numbers in the sections
CshA, a DEAD-box helicase associates with RNA polymerase (RNAP) [12, 13]. RNAP associated with CshA is involved in the glucose induction (GI) of ylxR [6]. YlxR is a nucleoid-associated protein and regulates the expression of several hundred genes, including tsaD [7]. TsaD is a component of the tRNA modification enzyme TsaEBD, which is thought to be involved in protein translation [14]. TsaD stability is also controlled by the GI of protein arginine phosphatase YwlE, because protein arginine phosphorylation leads to ClpCP-dependent protein degradation [15, 16]. Additionally, the expression of an operon containing a gene encoding protein arginine phosphorylase McsB is under catabolite repression [17]. As shown in Fig. 1A, these regulators are in the feedback regulatory loop, and perhaps CshA-associated RNAP is directly involved in the GI of sigX/M. Based on the previous and RNA-seq analyses, close relationships among these regulatory factors for GI were identified. The RNA-seq analysis of YqfO revealed that YqfO functions as a global regulator in B. subtilis and confirmed that YqfO is involved in the regulatory network for GI composed of CshA, YlxR, and TsaD.
Main text
Methods
Strains and media
The wild-type B. subtilis strain 168 was used for identification of glucose-responsive genes. The strain was grown in 50 ml of the sporulation medium (SM) with or without 2% glucose. For RNA-seq of three regulatory factors, the wild-type (OAM829 bearing amyE::trmK-lacZ) and its derivatives, OAM953 (cshA::Tcr, this study), OAM829 (ylxR::pMUTIN-lacZ::Tcr) [7], and OAM954 (yqfO::pMUTIN-lacZ::Tcr, this study) were used. OAM950 (pftA::pMUTIN-pftA[Emr], this study) and its derivatives OAM951 (yqfO[Emr, lacZ::Tcr], this study) and OAM952 (cshA[Tcr], this study) were constructed as follows. pMUTIN-pftA was constructed by insertion of the PCR product amplified by pMut-pftA-H(5-ATCAAGCTTATGCCGGAACAGAAGATG-3) and pMut-pftA-B(5-ATGGGATCCTTTCTTCACCTCTTTCTCTTTGG-3) after digestion with HindIII and BamHI to pMUTIN treated with the same enzyme pair [8]. OAM955 (fruR::pMUTIN-fruR [Emr], this study) and its derivative OAM956 (yqfO[Emr, lacZ::Tcr], this study) were constructed as follows. pMUTIN-fruR was constructed by insertion of the PCR product amplified by pIS-fruR-E (5-ATCGAATTCCTGGCAGGTTGTATGC-3) and pMut-fruR-BB (5-ATGGGATCCACCATGAACGCGCTTT-3) after digestion with EcoRI and BamHI into pMUTIN treated with the same enzyme pair [8]. The resultant plasmids were used for genetic transformation to generate OAM950 and OAM955, and then each gene disruption was introduced in OAM950 or OAM955.
RNA isolation and RNA-seq analysis
For the experiments shown in Additional file 2: Table S1, 168 was grown in 50 ml of SM with or without 2% glucose and cell culture was sampled at T2 (2 h after the end of the logarithmic growth phase). For the experiments shown in Additional file 2: Tables S2–4, each strain was grown in 50 ml of SM with 2% glucose. Cell culture was sampled at T2. RNA isolation was performed as previously described [7]. Briefly, RNA was isolated from the cells collected by centrifugation using an RNeasy Mini Kit (Qiagen, Germantown, MD, USA). RNA-seq was performed as described in Additional file 1: Supplementary Methods.
β-Gal analysis
β-Gal analysis was performed as described previously [6].
Results and discussion
YqfO, CshA, and YlxR were identified as the controlling factors involved in GI of sigX/M in the early stationary phase cells in a sporulation medium (SM) with 2% glucose [6, 8]. Hence, we first performed comparable RNA-sequencing (RNA-seq) of wild-type B. subtilis 168 cells in SM with or without 2% glucose to detect glucose-responsive genes. We identified 528 (threshold × 10) and 1494 (threshold × 3) of the upshift and downshift genes with glucose, respectively (Additional file 2: Table. S1a, b). This showed the powerful detectability of differentially expressed genes (DEGs) by RNA-seq, because the former analyses using DNA microarray detected 852 (threshold × 2) [3], less than 100 (threshold × 3) [4], and 503 (threshold × 3) [5] DEGs related to glucose addition. CshA- and YlxR-regulated genes have been identified by DNA microarray and RNA-seq, respectively, and are approximately 200 and 400 genes, respectively [7, 13], whereas YqfO-regulated genes have not been characterized. Thus, we performed comparable RNA-seq of the wild-type and its derivatives bearing the disruption of yqfO, cshA, and ylxR. In Additional file 2: Tables S2–4, each DEG (428, 787, and 182) detected by RNA-seq analysis is shown. As a result, we first observed that disruption of the yqfO gene has a broad impact on genome gene expression. YqfO belongs to a large protein superfamily with unknown functions (DUF34), which is conserved in all three domains of life [11]. Although an exact mechanistic analysis was lacking, pleiotropic effects on physiological aspects, including transcription regulation, were observed in the disruptants of the genes encoding DUF34 proteins in many organisms [11]. The Thermus thermophilus DUF34 protein YbgI binds to single-stranded DNA [18], and the Geobacillus stearotherophilus DUF34 protein XynX regulates the xynA gene encoding xylanase through its binding to the xynA promoter [19]. Our RNA-seq analysis for YqfO also revealed more than 400 genes that are under the control of YqfO, suggesting the pleiotropic functions of YqfO in B. subtilis. Notably, the previous study on YqfO revealed only one transcription unit under the control of YqfO [8]. The expression of one of the target operons in RNA-seq, fruRKA (fructose metabolic operon), was confirmed to be affected by yqfO disruption using β-Gal analysis (Fig. 2). B. subtilis YqfO affects the transcription of many genes through unknown mechanisms. It is possible that YqfO may do so through regulation of metal ion homeostasis as the metal chaperone or metal ion insertase, which affects many enzyme activities, because YqfO-His did not bind to the target promoter region in the electromobility shift assay (Additional file 1: Supplementary Methods; Additional file 3: Fig. S1).
Fig. 2.
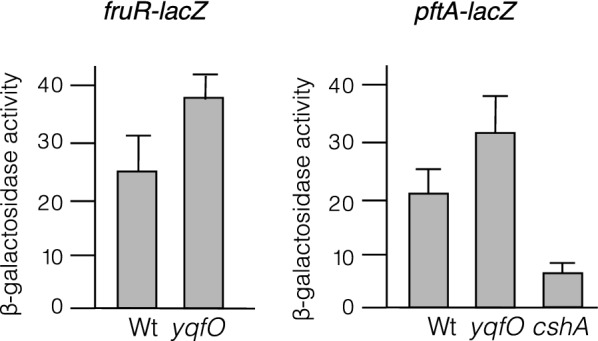
Expression of PfruR-lacZ and PpftA-lacZ in SM with 2% glucose in early stationary phase (T1). Y-axis shows β-Gal activities in Miller units. T-bar shows standard deviation derived from three independent experiments
Compared to the previous report [7], the number of YlxR-regulated genes in this study was relatively small, perhaps because of the different algorithms for DEGs. Compared to the inventory of the glucose-regulated genes, the detected CshA-regulon did not show a particular bias for that inventory (Additional file 2: Tables S1, S3), indicating that many CshA-regulon genes may play other roles in glucose response. It should be noted that the previous studies revealed extreme upregulation of the pftAB (ysbAB, encoding pyruvate transporter) [20] and downregulation of the frl (encoding proteins involved in fructoselysine utilization) operons in the cshA disruptant [13]. Our study detected upregulation of pftAB in the yqfO-disruptant but not in the cshA-disruptant. β-Gal analysis of pftA expression in the yqfO disruptant confirmed our RNA-seq results (Fig. 2). Moreover, we observed downregulation of pftA in the cshA disruptant, which is contrary to the previous results; the reason behind this is unknown. It should be noted that our β-Gal experiments were performed with glucose and different media whereas previous experiments involved the use of LB medium. We observed differential expression of the frl operon among all three disruptants analyzed, especially downregulation in the cshA disruptant (Table 1B). A detailed analysis of the frl operon expression in the ylxR disruptant has been described previously [21]. A previous study indicated a regulatory network including CshA/YlxR/YqfO (Fig. 1A) [6–8]. Overlapping genes in the three regulons are shown in Fig. 1B as Venn diagrams and in Additional file 2: Tables, S2–4. Figure 1B shows that 52% of the YlxR- and 59% of the YqfO-regulon genes overlapped with the CshA-regulon genes, confirming close relationships among the three regulators. A certain group of genes whose expression was independent of YlxR or YqfO was observed in the CshA-regulon. This may be related to the former observation that CshA is a component of RNA degradosome including RNase Y, which controls mRNA abundance of at least 10% of the genes in the genome [13]. We note that in most of the genes regulated by CshA and YqfO, gene disruption effects have the same direction; that is, when cshA disruption affected the expression of particular genes positively, yqfO disruption also did so (see Additional file 2: Table S2). Moreover, it should be noted that this estimation of overlap might be underestimated, because all the genes contained in the specific operon were not always detected in the RNA-seq analysis, as shown in Table 1. These operons in Table 1 encode metabolic genes involved in the biosynthesis of histidine and pyrimidine, and degradation of fructoselysine. The genes commonly detected in the three regulons are listed in Additional file 2: Table S5 and include several metabolic genes, for example, bglH (aryl-phospho-beta-D-glucosidase), ctaD/E/G (cytochrome c oxidase subunits/ assembly factor), rocE/rocF (amino acid permease/arginase), manA (mannose-6-phosphate isomerase), and mtlD (mannitol-1-phosphate 5-dehydrogenase). The RNA-seq analyses of the three regulons confirmed close relationships with each other, suggesting the reliability of the obtained RNA-seq results. Moreover, Venn diagram shows that YlxR and YqfO also have their own regulon genes that are not under the control of other regulators. These results suggest that YlxR and YqfO have distinct roles other than GI of the genes. Hence, these RNA-seq results should be useful for the further research.
Table 1.
Each gene expression of cshA/ylxR/yqfO-regulated operons
| A. his operon | hisZ | hisG | hisD | hisB | hisH | hisA | hisF | hisI | ||
| cshA | R | R | R | R | R | R | R | R | ||
| ylxR | R | R | R | R | R | |||||
| yqfO | R | R | R | R | ||||||
| Glucose | E | E | E | E | E | E | E | E | ||
| B. frl operon | frlB | frlO | frlN | frlM | frlD | yurJ | ||||
| cshA | R | |||||||||
| ylxR | E | E | E | E | E | E | ||||
| yqfO | R | R | ||||||||
| Glucose | R | R | R | R | R | R | ||||
| C. pyr operon | pyrR | pyrP | pyrB | pyrC | pyrAA | pyrAB | pyrK | pyrD | pyrF | pyrE |
| cshA | E | R | R | R | R | R | R | |||
| ylxR | R | R | R | R | R | R | R | |||
| yqfO | R | |||||||||
| Glucose | E | E | E | E | E | E | E | E | E | E |
CcpA-dependent glucose-activation of the pyr operon was reported in Moreno et al. [3]
R repressed in the gene-disruptant or with glucose; E enhanced in the gene-disruptant or with glucose
Operon structure; see Subtiwiki (http://subtiwiki.uni-goettingen.de/)
Limitations
In this study, RNA-seq analysis was performed once per pair for comparison (N = 1); hence the inventory of the genes with marginal fold-change may change if additional RNA-seq experiments are carried out. However, many of the gene lists with critical fold-change will not change, thus, the information in this study would be valuable for researchers studying gene expression in B. subtilis.
Supplementary Information
Additional file 1: Supplementary methods.
Additional file 2: Table S1. a Glucose-regulated genes (high threshold). b Glucose-regulated genes (low threshold). Table S2. YqfO-regulated genes. Table S3. CshA-regulated genes. Table S4. YlxR-regulated genes. Table S5. cshA/ylxR/yqfO-regulated genes.
Additional file 3: Figure S1. Electromobility shift assay of YqfO using the promoter region of the thiL operon.
Acknowledgements
Not applicable.
Abbreviations
- RNA-seq
RNA sequencing
- SM
Sporulation medium
- GI
Glucose induction
- RNAP
RNA polymerase
- DEAD box
Asp-Glu-Ala-Asp box
- PCR
Polymerase chain reaction
- DEG
Differentially expressed gene
Authors’ contributions
YK performed the experiments and data analysis. MO planned the study, performed the experiments and wrote the paper. Both the authors read and approved the final manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Numbers 18K05415 and 21K05349.
Availability of data and materials
The data underlying this article are available in the Sequence Read Archive at https://www.ddbj.nig.ac.jp/dra/index.html. The data for wild-type samples without or with glucose can be accessed with DRR296186 and DRR296187, respectively. The other data of the samples with amyE::trmK-lacZ are as follows; Wt, DRR139003 [W1.fq.gz, W2.fq.gz]; cshA, DRR296184; ylxR, DRR139004 [R1.fq.gz, R2.fq.gz]; and yqfO, DRR296185. All other data generated or analyzed during this study are included in this published article and its Additional files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11(2):87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Fujita Y. Carbon catabolite control of the metabolic network in Bacillussubtilis. Biosci Biotechnol Biochem. 2009;73(2):245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 3.Moreno MS, Schneider BL, Maile RR, et al. Catabolite repression mediated by the CcpA protein in Bacillussubtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2001;39(5):1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 4.Blencke HM, Homuth G, Ludwig H, et al. Transcriptional profiling of gene expression in response to glucose in Bacillussubtilis: regulation of the central metabolic pathways. Metab Eng. 2003;5(2):133–149. doi: 10.1016/S1096-7176(03)00009-0. [DOI] [PubMed] [Google Scholar]
- 5.Vázquez CD, Freyre-González JA, Gosset G, et al. Identification of network topological units coordinating the global expression response to glucose in Bacillussubtilis and its comparison to Escherichiacoli. BMC Microbiol. 2009;9:176. doi: 10.1186/1471-2180-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogura M, Asai K. Glucose induces ECF sigma factor genes, sigX and sigM, independent of cognate anti-sigma factors through acetylation of CshA in Bacillussubtilis. Front Microbiol. 2016;7:1918. doi: 10.3389/fmicb.2016.01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogura M, Kanesaki Y. Newly identified nucleoid-associated-like protein YlxR regulates metabolic gene expression in Bacillussubtilis. mSphere. 2018;3(5):e00501–18. doi: 10.1128/mSphere.00501-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura M, Sato T, Abe K. YlxR which is involved in glucose-responsive metabolic changes, regulates expression of tsaD for protein quality control of pyruvate dehydrogenase. Front Microbiol. 2019;10:923. doi: 10.3389/fmicb.2019.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tascou S, Kang TW, Trappe R, et al. Identification and characterization of NIF3L1 BP1, a novel cytoplasmic interaction partner of the NIF3L1 protein. Biochem Biophys Res Commun. 2003;309(2):440–448. doi: 10.1016/j.bbrc.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Godsey MH, Minasov G, Shuvalova L, et al. The 2.2 A resolution crystal structure of Bacilluscereus Nif3-family protein YqfO reveals a conserved dimetal-binding motif and a regulatory domain. Protein Sci. 2007;16(7):1285–1293. doi: 10.1110/ps.062674007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed CJ, Hutinet G, de Crécy-Lagard V. Comparative genomic analysis of the DUF34 protein family suggests role as a metal ion chaperone or insertase. Biomolecules. 2021;11:1282. doi: 10.3390/biom11091282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delumeau O, Lecointe F, Muntel J, et al. The dynamic protein partnership of RNA polymerase in Bacillussubtilis. Proteomics. 2011;11(15):2992–3001. doi: 10.1002/pmic.201000790. [DOI] [PubMed] [Google Scholar]
- 13.Lehnik-Habrink M, Rempeters L, Kovács ÁT, et al. DEAD-Box RNA helicases in Bacillussubtilis have multiple functions and act independently from each other. J Bacteriol. 2013;195(3):534–544. doi: 10.1128/JB.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiaville PC, El Yacoubi B, Köhrer C, et al. Essentiality of threonylcarbamoyladenosine [t(6)A], a universal tRNA modification, in bacteria. Mol Microbiol. 2015;98(6):1199–1221. doi: 10.1111/mmi.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trentini DB, Suskiewicz MJ, Heuck A, Kurzbauer R, Deszcz L, et al. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539(7627):48–53. doi: 10.1038/nature20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogura M. Glucose-mediated protein arginine phosphorylation/dephosphorylation regulates ylxR encoding nucleoid-associated protein and cell growth in Bacillussubtilis. Front Microbiol. 2020;11:2382. doi: 10.3389/fmicb.2020.590828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii H, Tanaka T, Ogura M. The Bacillussubtilis response regulator gene degU is positively regulated by CcpA and by catabolite-repressed synthesis of ClpC. J Bacteriol. 2013;195(2):193–201. doi: 10.1128/JB.01881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomoike F, Wakamatsu T, Nakagawa N, Kuramitsu S, Masui R. Crystal structure of the conserved hypothetical protein. TTHA1606 from Thermusthermophilus HB8. Proteins Struct Funct Bioinforma. 2009;76(1):244–248. doi: 10.1002/prot.22397. [DOI] [PubMed] [Google Scholar]
- 19.Shulami S, Shenker O, Langut Y, Lavid N, et al. Multiple regulatory mechanisms control the expression of the Geobacillus stearothermophilus gene for extracellular xylanase. J Biol Chem. 2014;289(37):25957–25975. doi: 10.1074/jbc.M114.592873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Esker MH, Kovács ÁT, Kuipers OP. YsbA and LytST are essential for pyruvate utilization in Bacillussubtilis. Environ Microbiol. 2017;19(1):83–94. doi: 10.1111/1462-2920.13454. [DOI] [PubMed] [Google Scholar]
- 21.Ogura M, Shindo K, Kanesaki Y. Bacillussubtilis nucleoid-associated protein YlxR is involved in bimodal expression of the fructoselysine utilization operon (frlBONMD-yurJ) promoter. Front Microbiol. 2020;11:2024. doi: 10.3389/fmicb.2020.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary methods.
Additional file 2: Table S1. a Glucose-regulated genes (high threshold). b Glucose-regulated genes (low threshold). Table S2. YqfO-regulated genes. Table S3. CshA-regulated genes. Table S4. YlxR-regulated genes. Table S5. cshA/ylxR/yqfO-regulated genes.
Additional file 3: Figure S1. Electromobility shift assay of YqfO using the promoter region of the thiL operon.
Data Availability Statement
The data underlying this article are available in the Sequence Read Archive at https://www.ddbj.nig.ac.jp/dra/index.html. The data for wild-type samples without or with glucose can be accessed with DRR296186 and DRR296187, respectively. The other data of the samples with amyE::trmK-lacZ are as follows; Wt, DRR139003 [W1.fq.gz, W2.fq.gz]; cshA, DRR296184; ylxR, DRR139004 [R1.fq.gz, R2.fq.gz]; and yqfO, DRR296185. All other data generated or analyzed during this study are included in this published article and its Additional files.