Abstract
Background
Papillary thyroid carcinoma (PTC) is the most common type of thyroid cancer (TC), accounting for more than 80% of all cases. Ferroptosis is a novel iron-dependent and Reactive oxygen species (ROS) reliant type of cell death which is distinct from the apoptosis, necroptosis and pyroptosis. Considerable studies have demonstrated that ferroptosis is involved in the biological process of various cancers. However, the role of ferroptosis in PTC remains unclear. This study aims at exploring the expression of ferroptosis-related genes (FRG) and their prognostic values in PTC.
Methods
A ferroptosis-related gene signature was constructed using lasso regression analysis through the PTC datasets of the Cancer Genome Atlas (TCGA). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to investigate the bioinformatics functions of significantly different genes (SDG) of ferroptosis. Additionally, the correlations of ferroptosis and immune cells were assessed through the single-sample gene set enrichment analysis (ssGSEA) and CIBERSORT database. Finally, SDG were test in clinical PTC specimens and normal thyroid tissues.
Results
LASSO regression model was utilized to establish a novel FRG signature with 10 genes (ANGPTL7, CDKN2A, DPP4, DRD4, ISCU, PGD, SRXN1, TF, TFRC, TXNRD1) to predicts the prognosis of PTC, and the patients were separated into high-risk and low-risk groups by the risk score. The high-risk group had poorer survival than the low-risk group (p < 0.001). Receiver operating characteristic (ROC) curve analysis confirmed the signature's predictive capacity. Multivariate regression analysis identified the prognostic signature-based risk score was an independent prognostic indicator for PTC. The functional roles of the DEGs in the TGCA PTC cohort were explored using GO enrichment and KEGG pathway analyses. Immune related analysis demonstrated that the most types of immune cells and immunological function in the high-risk group were significant different with those in the low-risk group. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) verified the SDG have differences in expression between tumor tissue and normal thyroid tissue. In addition, cell experiments were conducted to observe the changes in cell morphology and expression of signature’s genes with the influence of ferroptosis induced by sorafenib.
Conclusions
We identified differently expressed FRG that may involve in PTC. A ferroptosis-related gene signature has significant values in predicting the patients’ prognoses and targeting ferroptosis may be an alternative for PTC’s therapy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-021-02389-7.
Keywords: Papillary thyroid carcinoma, Ferroptosis genes, Gene signature, Prognosis, TCGA
Introduction:
Thyroid cancer (TC) is the most common malignant endocrine tumor and its incidence has steadily increased in many countries over the past several decades [1, 2]. Papillary thyroid carcinoma (PTC) is the most universal type of TC, accounting for more than 80% of all cases, with a high 10-year survival rate over 90% [3, 4]. Although the majority of PTC remain indolent, tumor recurrence and metastasis obstruct the clinical management and survival in certain patients [5].
Ferroptosis describes a novel form of regulated cell death that occurs as a consequence of lethal lipid peroxidation, which is distinct from the apoptosis, necroptosis and pyroptosis [6, 7]. It can be induced by experimental compounds or clinical drugs, such as erastin, ras-selective lethal small molecule 3 (RSL3), or sorafenib, in cancer cells and certain normal cells [8]. It's worth noting that sorafenib was approved by the Food and Drug Administration (FDA) for the treatment of multiple types of advanced cancer, including TC [9–12]. Ferroptosis has been implicated in the pathological cell death and its dysregulation has been associated with carcinogenesis, such as pancreatic cancer, hepatocellular carcinoma, kidney and ovary. Ferroptosis may also have a tumor suppressor function that could be harnessed for cancer therapy [13–16]. In addition, a line of evidence from several researches demonstrate the ferroptotic cells could interact with immune cells, such as CD8 + T cells, by releasing chemotaxis, and then modulating the anticancer immunity [17–21]. Lately, ferroptosis was found to be induced in anaplastic thyroid cancer (ATC) by vitamin C via the inactivation of GPX4, reactive oxygen species (ROS) accumulation and iron sustained lipid peroxidation [22]. Numerous studies have reported that ferroptosis-related genes (FRGs) play a pivotal role in predicting tumor prognosis [23–25], however, the association between FRGs and prognosis in patients with PTC remains to be elucidated.
This study aims at exploring the expression of FRGs and their prognostic value in PTC. Differential expression of FRGs was used to establish a prognostic multigene signature. Additionally, we explored the underlying mechanism of the FRGs signature by a series of analyses to illustrate its prognostic value. Finally, signature genes were tested in clinical PTC specimens and adjacent normal thyroid tissues.
Materials and methods
Acquisition of gene expression and clinical data
The mRNA sequencing (RNA-seq) data and corresponding clinical information of 507 PTC patients were downloaded from the TCGA database (https://portal.gdc.cancer.gov/repository). The dataset included 510 PTC tissue samples and 58 adjacent normal tissue samples. The gene expression profiles were normalized using the Perl language (http://www.perl.org/). The corresponding FRGs were downloaded from FerrDb (http://www.zhounan.org/ferrdb/index.html), a web-based consortium that provided a comprehensive and up-to-date database for ferroptosis markers, regulatory molecules and associated diseases [26]. After eliminating duplicate items from the whole genes (driver: 150; suppressor: 109; marker: 123), we identified 259 FRGs (Additional file 1). The clinical information for 507 patients was presented on Table 1.
Table 1.
Demographic and clinical characteristics of patients in the Cancer Genome Atlas papillary thyroid cancer cohort and validated papillary thyroid cancer cohort
| Clinicopathological features | TCGA(N = 507) | Validated Cohort(N = 75) |
|---|---|---|
| Age (year) | ||
| Mean (SD) | 47.26 (15.78) | 44.09 (12.65) |
| < 55岁 | 340 (67.06%) | 57 (76.00%) |
| ≥ 55岁 | 167 (32.94%) | 18 (24.00%) |
| Gender, n (%) | ||
| Male | 136 (26.82%) | 19 (25.33%) |
| Female | 371 (73.18%) | 56 (74.67%) |
| Stage, n (%) | ||
| Stage I | 285 (56.21%) | 61 (81.33%) |
| Stage II | 52 (10.26%) | 8 (10.67%) |
| Stage III | 114 (22.49%) | 4 (5.33%) |
| Stage IV | 55 (10.85%) | 2 (2.67%) |
| Unknown | 2 (0.39%) | 0 (0.00%) |
| T status, n (%) | ||
| T1 | 144 (28.40%) | 46 (61.33%) |
| T2 | 167 (32.94%) | 5 (6.67%) |
| T3 | 171 (33.73%) | 14 (18.67%) |
| T4 | 23 (4.54%) | 10 (13.33%) |
| TX | 2 (0.39%) | 0 (0.00%) |
| N status, n (%) | ||
| N0 | 231 (45.56%) | 25 (33.33%) |
| N1 | 226 (44.58%) | 50 (66.67%) |
| NX | 50 (9.86%) | 0 (0.00%) |
| M status, n (%) | ||
| MO | 283 (55.82%) | 73 (97.33%) |
| M1 | 9 (1.78%) | 2 (2.67%) |
| MX | 215 (42.41%) | 0 (0.00%) |
Prognostic validity of the gene signature
Differentially expressed genes (DEGs) related to ferroptosis in tumor tissues and adjacent normal tissues in the TCGA PTC cohort were described using the ‘limma’ package in R (version 4.0.4; https://www.r-project.org/), with a false-discovery rate (FDR) < 0.05. FRGs related to overall survival (OS) were identified with univariate Cox regression analysis. Ferroptosis-related DEGs were used to construct a protein–protein interaction network with a minimum required interaction score ≥ 0.15 in the STRING database (version 11.0; https://string-db.org/). Then, we also explored their correlations using the ‘igraph’ package in R. The least absolute shrinkage and selection operator (LASSO) Cox regression model was utilized to construct a multigene signature of the ferroptosis-related DEGs in the TCGA cohort by the the ‘glmnet’ package in R. The formula was established as follows: score = e sum (each gene’s expression × corresponding coefficient). The patients were stratified into high-risk and low-risk groups based on the median value of the risk score. The OS and disease-free survival (DFS) differences between high-risk and low-risk groups were compared by the ‘survival’ and ‘survminer’ package in R. Kaplan–Meier curve was implemented to visualize the survival. The receiver operating characteristic (ROC) analysis was used to examine the sensitivity and specificity of survival prediction using the independent risk factors by the ‘timeROC’ package in R. Univariate and multivariate Cox regression analyses were performed to determine the prognostic value of the risk score and various clinical characteristics. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated. For the clinical stratified analysis, patients were divided into different subgroups according to the clinicopathologic features, include age, gender, stage and TNM stages. Kaplan–Meier survival analysis was presented by the ‘survival’ and ‘survminer’ package in R between high and low risk score groups in different subgroups.
Functional enrichment analysis
The "clusterProfiler" R package was utilized to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses based on the DEGs (|log2FC|≥ 1, FDR < 0.05) between the high-risk and low-risk groups.
Immune-related analysis
Immune infiltration in PTC was investigated by calculating single-sample gene set enrichment analysis (ssGSEA) scores of 16 immune cells and 13 immune-related pathways using the ‘gsva’ package in R. The correlation between the prognostic signature and immune cells was performed by Pearson correlation analysis via CIBERSORT (http://cibersort.stanford.edu/).
Statistical analysis
Data were all analyzed using Bioconductor packages in R software. The significance of the differences in the expression of FRGs between tumor and normal tissues was assessed by the Wilcoxon test. Univariate and multivariate cox regression analyses were used to evaluate the correlation between the affecting factors and patients’ OS or DFS. Mann–Whitney test with P values adjusted by the Benjamini-Hochberg (BH) method was used to compare the ssGSEA scores of immune cells or pathways between the high-risk and low-risk groups. P value < 0.05 was set as statistically significant for all the analyses.
Quantitative real-time PCR
Quantitative Real-Time PCR (qRT-PCR) was performed on 75 pairs of tumor tissues and adjacent normal tissues to validate the mRNA expression levels of the ten signature genes. Consent form was obtained from each patient involved in this study, and the study approved by the Ethics Committee of the First Hospital of China Medical University (Ethical Approval Number: [2021]82). The clinical information for the 75 patients was presented on Table 1. Total RNA was extracted from the sample tissues via RNAiso Plus (TAKARA, China), followed by reverse transcription into cDNA. qRT-PCR was carried out using the TB Green R Premix Ex TaqTM II kit (TAKARA, China). GAPDH served as an internal control. The relative expression levels were quantified by the Ct (2−ΔΔCt) method . The primer sequences were listed (Table 2).
Table 2.
Primer sequences for qRT-PCR
| Primer | Sequence (5′-3′) |
|---|---|
| ANGPTL7-F | TAGAGATGGAGGACTGGGAGG |
| ANGPTL7-R | GTGCACACTTGTCCAAGCAG |
| CDKN2A-F | GGGAGCAGCATGGAGCCG |
| CDKN2A-R | CTGGATCGGCCTCCGACCGTA |
| DPP4-F | AGTGGCGTGTTCAAGTGT |
| DPP4-R | AGTGGCTCATGTGGGTAT |
| DRD4-F | GGGAAGGACAGTGCTTGGATCTG |
| DRD4-R | AGTGCTCTAGATAAGTTGGCAAATGCA |
| ISCU-F | CCAGGTGGATGAAAAGGGGAA |
| ISCU-R | GCAGAGTTCCTTGGCGATGT |
| Srxn1-F | CCCACTGGACCAACTTCTGT |
| Srxn1-R | GTGGCTAGCTCAGACCAAGG |
| TXNRD1-F | AAATTCTTAGGACGGTCGGG |
| TXNRD1-R | AGTCTGCCCTCCTGATAAGC |
| TFRC-F | TCAGTTTCCACCATCTCG |
| TFRC-R | AAGTCTCCAGCACTCCAA |
| TF-F | TCAGCAGAGACCACCGAAGACT |
| TF-R | GACCACACTTGCCCGCTATGTA |
| PGD-F | GTTCCAAGACACCGATGGCAAAC |
| PGD-R | CACCGAGCAAAGACAGCTTCTC |
| GAPDH-F | GTCTCCTCTGACTTCAACAGCG |
| GAPDH-R | ACCACCCTGTTGCTGTAGCCAA |
Cell culture and reagents
Human thyroid cancer cell lines K1(European Collection of Cell Cultures) and KTC-1(Chinese Academy of Sciences Stem Cell Bank) were cultured in RPMI-1640, comprising 10% fetal bovine serum (FBS), 1% non-essential amino acids(NEAA), 1% Glutamax and 1% Sodium Pyruvate at the humidified chamber of 37 °C and 5% CO2. on. Sorafenib (S7397) was obtained from Selleck( Houston, TX, USA).
Cell viability assay
Cells (3000 to 4000/well) were seeded in 96-well plates. After a 24 h culture, cells were treated with different doses of sorafenib for 24 h. The Cell Counting Kit-8(CCK‑8; MedChem Express, China) assay was then carried out to assess the effect of sorafenib on cell viability, and IC50 values were calculated according to the instructions.
Transmission electron microscopy (TEM)
Cell samples were fixed with 2.5% glutaraldehyde, dehydrated in acetone solutions at increasing concentrations, and embedded in an epoxy resin. Then, the Sections (70–90 nm) were stained with lead citrate and uranyl acetate. Ultrastructural images were then captured with a transmission electron microscope (Hitachi H7650, Japan).
Results
Identification of 15 differentially expressed prognostic FRGs
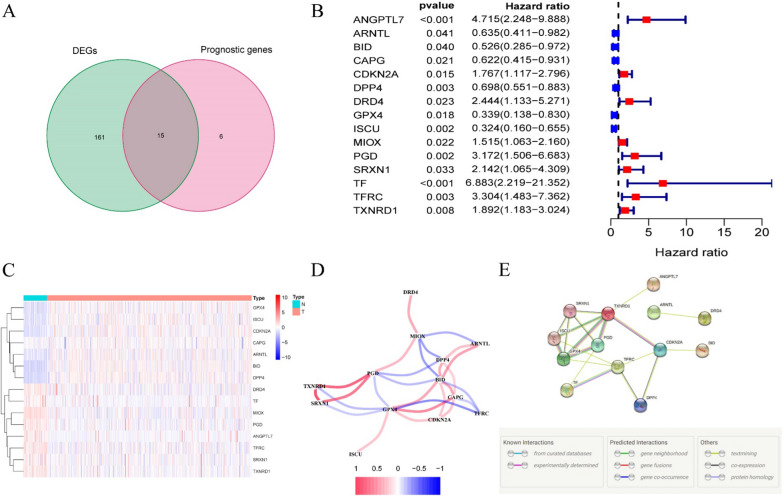
Most of the FRGs (176/259, 68.0%) were differentially expressed between tumor and adjacent normal tissue, and 15 of them were correlated with OS in the univariate Cox regression analysis (Fig. 1A, B). The heatmap exhibited the expression difference of 15 intersect genes between tumor tissues and adjacent nontumorous tissues (Fig. 1C). The correlation network and protein–protein interaction network provided interactive information among these differentially expressed prognostic FRGs (Fig. 1D, E).
Fig. 1.
Identification of differentially expressed genes related to ferroptosis in TCGA papillary thyroid cancer cohort. A 15 out of 21 prognostic ferroptosis-related genes were differentially expressed between tumor tissues and adjacent normal tissues; B univariate Cox proportional regression analysis showed the 15 differentially expressed prognostic ferroptosis-related genes were significantly associated with overall survival; C the heatmap of the expression of 15 differentially expressed prognostic ferroptosis-related genes in tumor and normal thyroid. D the correlation network of 15 differentially expressed prognostic ferroptosis-related genes. Correlation coefficients are represented by different colors. E the PPI network provided interactive information among the 15 differentially expressed prognostic ferroptosis-related genes
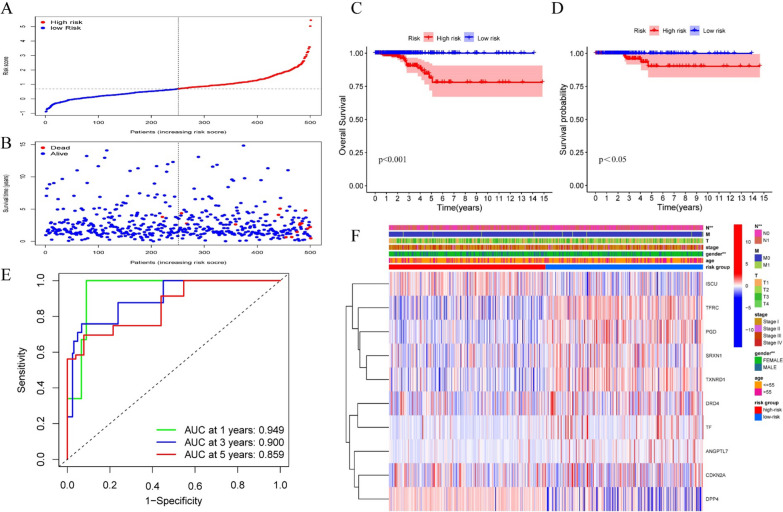
Establishment of a 10-FRGs signature predicting the prognosis of PTC
Lasso Cox regression analysis was utilized based on the 15 differentially expressed prognostic FRGs, and a 10-gene signature was constructed. The risk score for each patient was calculated using the following formula: risk score = ANGPTL7 × 1.52040 + CDKN2A × 0.28426 + DPP4 × (− 0.15329) + DRD4 × 0.56101 + ISCU × (− 0.55270) + PGD × 0.31588 + SRXN1 × 0.29977 + TF × 1.38073 + TFRC × 0.60187 + TXNRD1 × 0.15114. Patients were classified as high risk (n = 250) or low risk (n = 251) based on their signature-based risk scores (Fig. 2A, the median value of the risk score = 0.691487). The scatter diagram showed the patients survival status (Fig. 2B). Kaplan–Meier survival curves confirmed that OS and DFS were significantly worse in high-risk compared with low-risk patients (Fig. 2C, D). When the risk score > 1.72, the patient's mean 5-year survival is < 50% (Additional file 2). Time-dependent ROC curves were used to evaluate the predictive performance of the signature-based risk score for OS. The area under the curve values were: 0.949 for 1-year OS, 0.900 for 3-year OS, and 0.859 for 5-year OS, respectively (Fig. 2E). The heatmap indicated the significant differences in gender and N stage between high- and low-risk groups (Fig. 2F).
Fig. 2.
Establishment of a ferroptosis-related gene signature in TCGA papillary thyroid cancer cohort. A Signature-based risk scores were used to classify patients as high risk or low risk; B survival of high-risk and low-risk patients; C Kaplan–Meier curves for overall survival (P < 0.001); D Kaplan–Meier curves for disease free survival (P < 0.05); E time-dependent receiver operating characteristic curves validated the predictive performance of the signature-based risk score for overall survival; F the heatmap for ferroptosis-related genes prognostic signature and clinicopathological manifestations (*P < 0.05, **P < 0.01, ***P < 0.001)
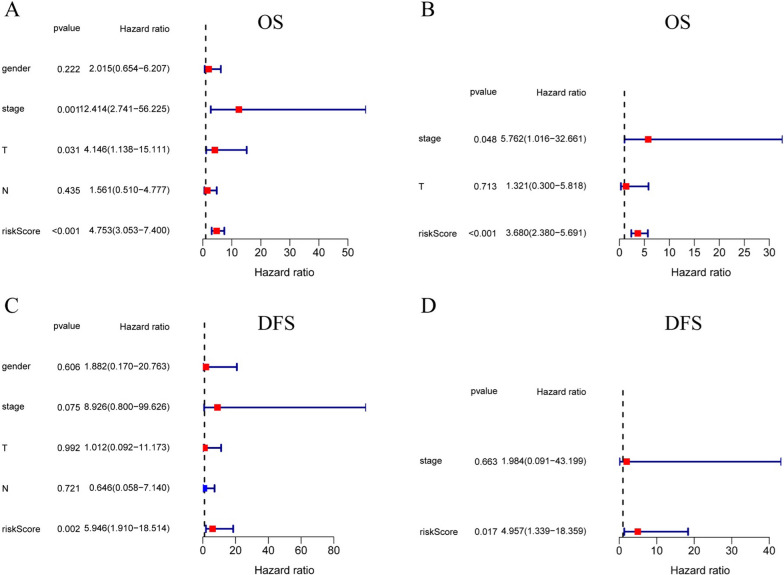
The 10-genes signature is an independent prognostic indicator for OS and DFS
Univariate and multivariate Cox regression analyses were carried out among the available variables to come to the conclusion that the risk score was an independent prognostic predictor for OS (Fig. 3A, B; univariate: HR = 4.753, 95% CI = 3.053–7.400, P < 0.001; multivariate: HR = 3.680, 95% CI = 2.380– 5.691, P < 0.001). Univariate and multivariate Cox regression analyses were carried out to the conclusion that the risk score was significantly associated with DFS (Fig. 3c, D; univariate: HR = 5.946, 95% CI = 1.910–18.514, P < 0.01; multivariate: HR = 4.957, 95% CI = 1.339–18.359, P < 0.05).
Fig. 3.
The association between clinicopathological factors and OS or DFS in patients with papillary thyroid cancer. Univariate Cox regression analyses A and multivariate Cox regression analyses B of the association between clinicopathological factors and OS. Univariate Cox regression analyses C and multivariate Cox regression analyses D of the association between clinicopathological factors and DFS
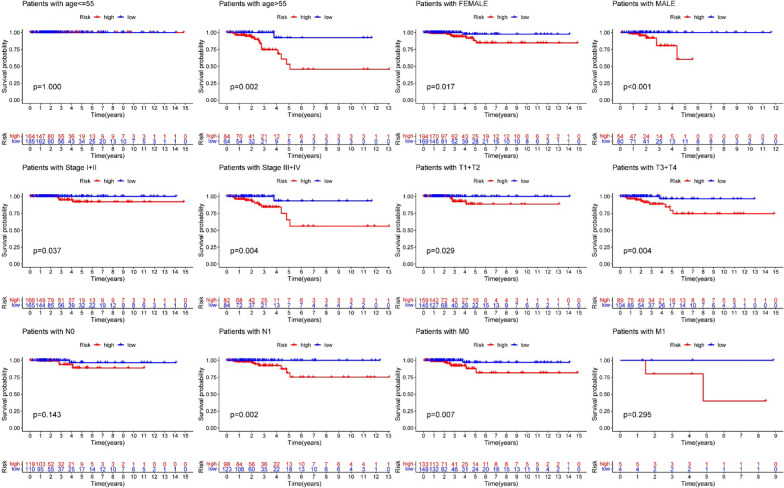
Clinical stratified analysis verifies the signature’s forecasting performance
Clinical stratified analysis was performed to evaluate the forecasting performance of the ten- gene signature (Fig. 4). Although there were no significant difference between high and low risk groups in the OS of three groups (patients with age ≤ 55, N0 and M1), the other 9 clinical groups demonstrated that the high-risk score signified a poorer prognosis in comparison to low-risk score.
Fig. 4.
Stratified analysis for the predictive efficacy of the ferroptosis-related gene signature among TCGA papillary thyroid cancer cohort
GO and KEGG analyses
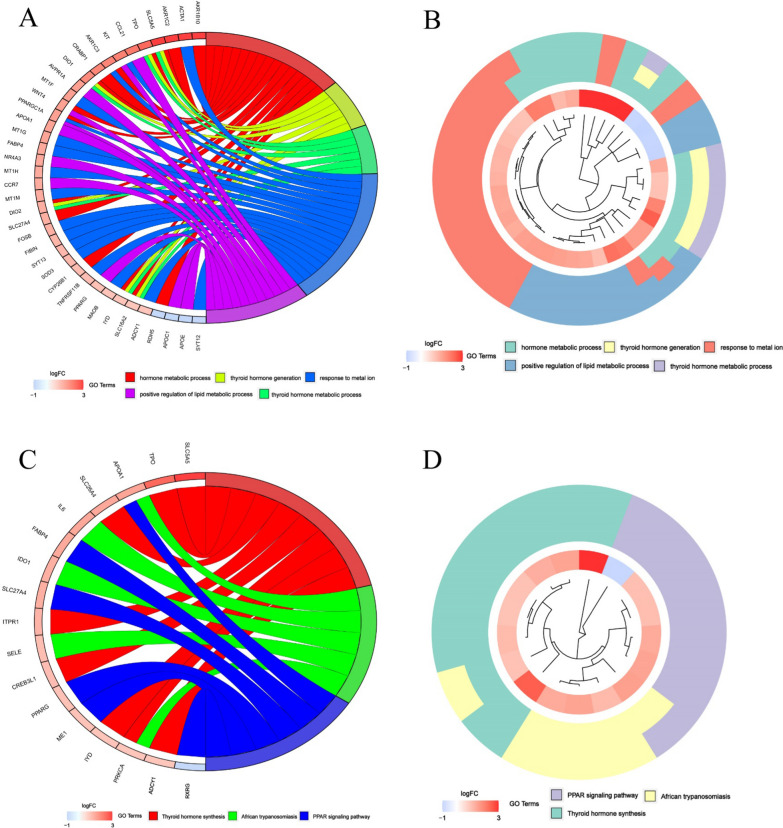
The functional roles of the DEGs in high- and low-risk patients in the TGCA cohort were explored using GO enrichment and KEGG pathway analyses. GO analysis showed that the DEGs were mostly enriched in several hormone metabolic processes, especially the thyroid hormone (Fig. 5A, B). Beyond that, enrichment was also observed in the metabolism of lipid and the activation of receptor (q value < 0.05). KEGG analysis showed that DEGs were mostly enriched in the thyroid hormone synthesis and immune-related pathways such as the PPAR signaling pathway (Fig. 5C, D; q value < 0.05).
Fig. 5.
Enrichment analysis in TCGA papillary thyroid cancer cohort. A, B Gene ontology enrichment. C, D Kyoto Encyclopedia of Genes and Genomes pathway analyses
Immune function analysis found that the signature is closely related to the immune cells
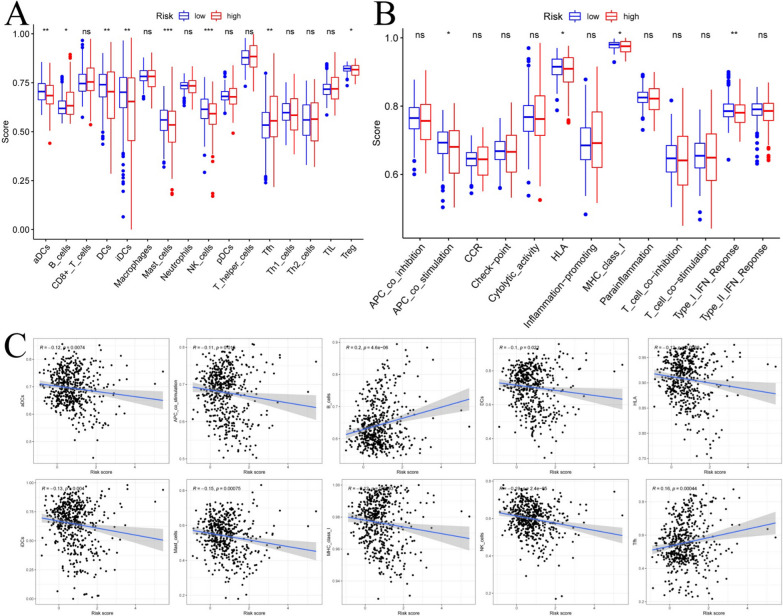
To further explore the correlation between the risk score and immune status, we quantified the enrichment scores of diverse immune cell subpopulations, related functions or pathways with ssGSEA. The results showed the accumulation of various immune cells (such as aDCs, B cells, DCs, iDCs, mast cells, NK cells, Tfh cells and Treg cells) with tumor microenvironment (TME) in the high-risk group were significantly different with those in the low-risk group (Fig. 6A). Moreover, the scores of the immune functions, such as the APC co-stimulation, HLA, MHC class I and Type I IFN response were significantly higher in low-risk group, which indicated anti-tumor immunological function was more active in the low-risk group (Fig. 6B).
Fig. 6.
Immune function analysis of ferroptosis-related gene signature in TCGA papillary thyroid cancer cohort. A, B Comparison of the ssGSEA scores of 13 immune-related functions and 16 immune cells between the high-risk and low-risk groups. C Relations between immune cells and the ferroptosis-related gene signature via CIBERSORT. (ns not significant; *P < 0.05; **P < 0.01; ***P < 0.001)
The CIBERSORT database was used to better understand characteristics of immune cells and FRGs signature (Fig. 6C). The risk score of the 10-gene signature has negative correlations with aDCs (P < 0.01), APC co-stimulation (P < 0.05), DCs (P < 0.05), HLA (P < 0.01), iDCs (P < 0.01), mast cells (P < 0.001), MHC class I (P < 0.01) and NK cells (P < 0.001), while positive correlations with B cell (P < 0.001) and Tfh (P < 0.001).
Verification of the 10 included genes by clinical specimens and cell experiments
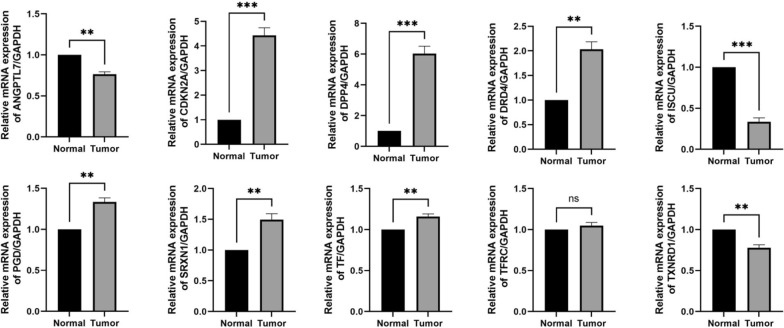
The qRT-PCR was performed on 75 pairs of tumor tissues and adjacent normal tissues to validate the mRNA expression levels of the ten signature genes. The PCR results showed ISCU and TXNRD1 were down-regulated, while the most genes were significantly up-regulated in the PTC tissues. There were no significant differences in the expression of TFRC between the normal and PTC specimens (Fig. 7. *P < 0.05; **P < 0.01, ***P < 0.001, ns: not significant). All results were mainly consistent with the data in TCGA.
Fig. 7.
The relative expression levels of the 10 genes in normal and papillary thyroid cancer tissues by qRT-PCR. (*P < 0.05; **P < 0.01, ***P < 0.001, ns not significant)
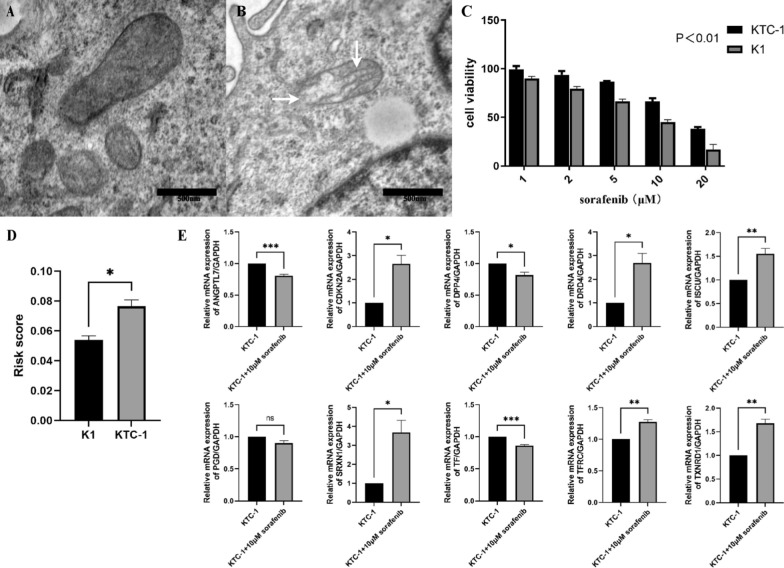
Sorafenib were used in further experiments of thyroid cancer cell lines, as a common inducer of ferroptosis and approved drug for thyroid cancer. TEM showed the ultrastructure of the KTC-1 cell line in control group and 10 μM sorafenib for 24 h. White arrowheads indicate ruptured membrane and the shrunken inner cristae of mitochondria (Fig. 8A, B). K1 and KTC-1 cells were treated with different doses of sorafenib for 24 h, and cell viability was measured by Cell Counting Kit-8. It was found that K1 is more sensitive to sorafenib than KTC-1 (Fig. 8C , p<0.01). And we found that the risk score of KTC-1 was higher than K1 (Fig. 8D). The PCR results showed the expression of most genes were significantly changed under the influence of sorafenib. (Fig. 8E, *P < 0.05; **P < 0.01, ***P < 0.001, ns: not significant).
Fig. 8.
Experiment of thyroid cancer cell line induced by sorafenib. Ultrastructure of the KTC-1 cell line in control group A and 10 μM sorafenib for 24 h (B). White arrowheads indicate ruptured membrane and the shrunken inner cristae of mitochondria. C K1 and KTC-1 cells were treated with different doses of sorafenib for 24 h, and cell growth was measured by Cell Counting Kit-8(P < 0.01). D Risk score of K1 and KTC-1(*P < 0.05). E The relative expression levels of the 10 genes in KTC-1 and KTC-1 + 10 μM sorafenib for 24 h by qRT-PCR. (*P < 0.05; **P < 0.01, ***P < 0.001, ns not significant)
Discussion
Thyroid cancer is the most common endocrine malignancy with a rapidly increasing incidence worldwide [27]. Although the majority of PTC patients have an excellent prognosis, there are also some patients with recurrence, distant metastasis and even other worse ending, affecting the survival time of patients [28, 29]. Therefore, it is particularly important to carry out risk stratification management to optimize the therapy for PTC patients. Ferroptosis has been proved to participate in tumor proliferation and metastasis, and is closely related to TME and tumor immunity [30, 31]. In this study, we developed a FRGs signature to predict the prognosis of PTC.
From the 15 differentially expressed prognostic FRGs, we established a 10-gene signature. The high expression of ANGPTL7, CDKN2A, DRD4, PGD, SRXN1, TF, TFRC, TXNRD1 represent high risk score and worse prognosis, conversely, the high expression of DPP4 and ISCU were associated with lower risk in PTC. ANGPTL7 is the less characterized member of the Angiopoietin-like (ANGPTL) proteins families, who can stimulate human differentiated endothelial cell’s proliferation, motility and invasiveness [32]. Existing research findings implicated ANGPTL7 was a mediator of metastatic progression and a potential target for interference with cancer metastases [33]. Although CDKN2A/p16 is a known tumor suppressor gene, CDKN2A hypermethylation might be a predictive factor for unfavorable prognosis of portion cancer [34]. According to research findings, high level of DRD4 expression has worse survival than low of DRD4 expression in Glioblastomas [35]. DRD4 might regulate chemosensitivity of cancer via inhibiting ferroptosis [36]. Decreasing PGD phosphorylation through EGFR signaling could dramatically attenuate cancer cells’ proliferation, growth and resistance to ionizing radiation [37]. Beyond that PGD inhibition significantly inhibits growth and survival of ATC cells and sensitizes ATC cells to doxorubicin treatment [38]. SRXN1 is a pivotal regulator of the antioxidant response in eukaryotic cells. SRXN1 could promotes tumor growth and metastasis and correlates with worse prognosis and decreased survival in different carcinoma [39, 40]. TF and TFRC are two key molecules in transportation of iron ion during ferroptosis, and both genes were low expressed in tumor tissues. TF could co-work with thyroid hormones in differentiation and other biological processes, and serum TF may also have a further role in stimulating cell proliferation [41, 42]. TFRC Is a specific ferroptosis marker and may could characterize the extent of ferroptosis [43]. The silencing of TFRC gene reduces liver cancer cell growth and survival [44]. Regulation of TXNRD1 could affect the proliferation, invasion and migration of carcinoma [45, 46]. The inhibition of TrxR1 blocks STAT3 activity and induces cancer cell death [47]. DPP4 has been reported as prognostic marker and therapeutic target of PTC [48]. The activity of DPP4 could down-regulation by TP53 to limits erastin-induced ferroptosis [49]. ISCU, a mitochondrial protein, significantly attenuates ferroptosis by regulating iron metabolism, rescuing the mitochondrial function and increasing the level of GSH [50]. Influenced by TP53, the expression of ISCU is related with the maintenance of iron homeostasis in hepatocellular carcinogenesis [51]. It should be noted that the relationship of ten signature’s genes with ferroptosis and thyroid cancer requires further clarification.
Functional analyses showed that the DEGs in the high-risk and low-risk patients identified in the present study were enriched in PPAR signaling pathway, which is existed as three different isoforms (α, γ, and δ) that play major roles in adipose tissue development and lipid metabolism [52, 53]. PPARs are expressed in immune cells and have an emerging critical role in immune cell differentiation and fate commitment [54]. Moreover, previous studies founded that PPARα activity is essential to regulate lipids through promoting ferroptosis [55]. Presented data suggested that PPARγ may promote growth and invasion of cancer cells and play a detrimental role in thyroid cancer [56, 57]. The metabolic processes of thyroid hormone (TH) have a certain influence on the proliferation and anti-apoptosis of thyroid cancer [58]. The existed research shows that the TH effects the differentiation and microenvironment of various tumors [59, 60]. Targeting TH in the thyroid cancer may improve treatment strategies.
There is a tight crosstalk between ferroptosis and antitumor immunity [61]. Different immune cells and mediators can contribute to thyroid cancer development and progression through various functional pathways [62, 63]. This study found that several kinds of tumor-specific cellular immunity and immune-related functions were altered in high-risk compared with low-risk patients. The previous study found that the accumulation of DCs in PTC was possibly correlated with favorable clinical course [64]. iDC were markedly reduced in poorly differentiated thyroid carcinoma(PDTC) and anaplastic thyroid carcinoma(ATC) [65], which partly supported the differences in iDCs between the high and low risk groups in the signature. The increasing of NK cells in the tumor microenvironment has an inversely correlate with advanced stages in patients with PTC [66]. The suppression of NK cell cytotoxicity and differentiation contributed to immune escape of thyroid cancer [67]. Ferritin heavy chain in tumor cells may modulate the expression of MHC class I molecules and influence NK cells, which may explain the association between ferroptosis and NK activation [68]. It has been reported that the exacerbation of ferroptosis via activating IFN-γ/ASK1/JNK signaling pathway [69] revealed the possible connection between the IFN-γ and ferroptosis. Type II IFN response played a pivotal function on cancer immune surveillance, stimulating antitumor immunity and promoting tumor recognition and elimination [70]. The prognostic signature showed a statistically significant negative correlation with most immune cells, at the same time, the immune score of the high-risk group is just below the low-risk group. Previous research demonstrated that mast cells have a contributing to thyroid carcinoma growth and invasiveness [71]. It was reported that mast cells infiltration was correlated with extrathyroidal extension of tumors and tumor invasiveness [72]. But the low numbers of tryptase mast cells were also associated with poor OS and more advanced stage [73]. The role of mast cells in PTC prognosis warrants further research.
The number of bioinformatics articles about ferroptosis is huge, and there are some similarities inevitably. In our manuscript, the signature was not only related to OS, but also related to DFS of thyroid cancer. So, the signature has better predictive performance. In addition, we stimulated thyroid cancer cell by sorafenib, and verified the expression change of the signature genes.
Although we verified the ten signature genes expression in 75 pairs of tumor tissues and paracancerous tissues, as well as PTC cell lines induced by sorafenib, by qRT-PCR and got a result that almost matched the TCGA, the prognostic predictive value of the ten-gene signature is based on a single cohort. Future studies involving larger independent cohorts should be conducted to validate our findings.
Conclusion
In conclusion, here we revealed the expression and prognostic value of FRGs in PTC. Additionally, we developed a risk signature that divided PTC patients into high-risk and low-risk groups with significantly different OS and DFS. These results may allow clinicians to determine individualized treatment for PTC patients with different clinical features and provide crucial evidence supporting further research of the role of ferroptosis in PTC.
Supplementary Information
Additional file 1. A list of 259 ferroptosis-related genes identified from FerrDb.
Additional file 2. Kaplan–Meier curves for overall survival at the best cut-off value of the risk score.
Acknowledgements
Not applicable.
Authors' contributions
JY SHI performed the data analysis and wrote the manuscript. WP helped to accurately process the data. SL, SW and ZH reviewed and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81902726), China Postdoctoral Science Foundation (2018M641739), Shenyang Science and Technology Bureau (213408), Shenyang Department of Education (QNZR2020002).
Availability of data and materials
Gene expression profiles, clinical information and mutation data of THCA in this study are available from the public database (TCGA, https://portal.gdc.cancer.gov/).
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the First Hospital of China Medical University (Shenyang, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.James BC, Mitchell JM, Jeon HD, Vasilottos N, Grogan RH, Aschebrook-Kilfoy B. An update in international trends in incidence rates of thyroid cancer, 1973–2007. Cancer Causes Control. 2018;29(4–5):465–473. doi: 10.1007/s10552-018-1023-2. [DOI] [PubMed] [Google Scholar]
- 2.Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol. 2013;2013:965212. doi: 10.1155/2013/965212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12(11):646–653. doi: 10.1038/nrendo.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito Y, Miyauchi A, Kihara M, Fukushima M, Higashiyama T, Miya A. Overall survival of papillary thyroid carcinoma patients: a single-institution long-term follow-up of 5897 patients. World J Surg. 2018;42(3):615–622. doi: 10.1007/s00268-018-4479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren H, Shen Y, Hu D, He W, Zhou J, Cao Y, Mao Y, Dou Y, Xiong W, Xiao Q, et al. Co-existence of BRAF(V600E) and TERT promoter mutations in papillary thyroid carcinoma is associated with tumor aggressiveness, but not with lymph node metastasis. Cancer Manag Res. 2018;10:1005–1013. doi: 10.2147/CMAR.S159583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, Kang R, Tang D. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabanillas ME, Ryder M, Jimenez C. Targeted therapy for advanced thyroid cancer: kinase inhibitors and beyond. Endocr Rev. 2019;40(6):1573–1604. doi: 10.1210/er.2019-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15(10):599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 11.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 12.Abdelgalil AA, Alkahtani HM, Al-Jenoobi FI. Sorafenib. Profiles Drug Subst Excip Relat Methodol. 2019;44:239–266. doi: 10.1016/bs.podrm.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Dai E, Kang R, Kroemer G, Tang D. The dark side of ferroptosis in pancreatic cancer. Oncoimmunology. 2021;10(1):1868691. doi: 10.1080/2162402X.2020.1868691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie J, Lin B, Zhou M, Wu L, Zheng T. Role of ferroptosis in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144(12):2329–2337. doi: 10.1007/s00432-018-2740-3. [DOI] [PubMed] [Google Scholar]
- 15.Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dancik V, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10(1):1617. doi: 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021 doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Green M, Choi JE, Gijon M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Chen P, Zhai B, Zhang M, Xiang Y, Fang J, Xu S, Gao Y, Chen X, Sui X, et al. The emerging role of ferroptosis in inflammation. Biomed Pharmacother. 2020;127:110108. doi: 10.1016/j.biopha.2020.110108. [DOI] [PubMed] [Google Scholar]
- 19.Rothe T, Gruber F, Uderhardt S, Ipseiz N, Rossner S, Oskolkova O, Bluml S, Leitinger N, Bicker W, Bochkov VN, et al. 12/15-Lipoxygenase-mediated enzymatic lipid oxidation regulates DC maturation and function. J Clin Invest. 2015;125(5):1944–1954. doi: 10.1172/JCI78490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angeli JPF, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 21.Kloditz K, Fadeel B. Three cell deaths and a funeral: macrophage clearance of cells undergoing distinct modes of cell death. Cell Death Discov. 2019;5:65. doi: 10.1038/s41420-019-0146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Xu S, Zhang L, Cheng X, Yu H, Bao J, Lu R. Vitamin C induces ferroptosis in anaplastic thyroid cancer cells by ferritinophagy activation. Biochem Biophys Res Commun. 2021;551:46–53. doi: 10.1016/j.bbrc.2021.02.126. [DOI] [PubMed] [Google Scholar]
- 23.Han F, Li W, Chen T, Yao Y, Li J, Wang D, Wang Z. Ferroptosis-related genes for predicting prognosis of patients with laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2021 doi: 10.1007/s00405-021-06789-3. [DOI] [PubMed] [Google Scholar]
- 24.Jiang P, Yang F, Zou C, Bao T, Wu M, Yang D, Bu S. The construction and analysis of a ferroptosis-related gene prognostic signature for pancreatic cancer. Aging. 2021 doi: 10.18632/aging.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu L, Yang F, Wang L, Dong L, Huang Z, Wang G, Chen G, Li Q. Identification the ferroptosis-related gene signature in patients with esophageal adenocarcinoma. Cancer Cell Int. 2021;21(1):124. doi: 10.1186/s12935-021-01821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford) 2020 doi: 10.1093/database/baaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiltshire JJ, Drake TM, Uttley L, Balasubramanian SP. Systematic review of trends in the incidence rates of thyroid cancer. Thyroid. 2016;26(11):1541–1552. doi: 10.1089/thy.2016.0100. [DOI] [PubMed] [Google Scholar]
- 28.Povoa AA, Teixeira E, Bella-Cueto MR, Melo M, Oliveira MJ, Sobrinho-Simoes M, Maciel J, Soares P. Clinicopathological features as prognostic predictors of poor outcome in papillary thyroid carcinoma. Cancers (Basel) 2020;12(11):3186. doi: 10.3390/cancers12113186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B, Shi Y, Xu Y, Zhang J. The predictive value of coexisting BRAFV600E and TERT promoter mutations on poor outcomes and high tumour aggressiveness in papillary thyroid carcinoma: A systematic review and meta-analysis. Clin Endocrinol (Oxf) 2021;94(5):731–742. doi: 10.1111/cen.14316. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, Chen ZN, Jiang X. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572(7769):402–406. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 32.Parri M, Pietrovito L, Grandi A, Campagnoli S, De Camilli E, Bianchini F, Marchio S, Bussolino F, Jin B, Sarmientos P, et al. Angiopoietin-like 7, a novel pro-angiogenetic factor over-expressed in cancer. Angiogenesis. 2014;17(4):881–896. doi: 10.1007/s10456-014-9435-4. [DOI] [PubMed] [Google Scholar]
- 33.Lim SY, Gordon-Weeks A, Allen D, Kersemans V, Beech J, Smart S, Muschel RJ. Cd11b(+) myeloid cells support hepatic metastasis through down-regulation of angiopoietin-like 7 in cancer cells. Hepatology. 2015;62(2):521–533. doi: 10.1002/hep.27838. [DOI] [PubMed] [Google Scholar]
- 34.Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J, Chen M. The prognostic value of CDKN2A hypermethylation in colorectal cancer: a meta-analysis. Br J Cancer. 2013;108(12):2542–2548. doi: 10.1038/bjc.2013.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dolma S, Selvadurai HJ, Lan X, Lee L, Kushida M, Voisin V, Whetstone H, So M, Aviv T, Park N, et al. Inhibition of dopamine receptor D4 impedes autophagic flux, proliferation, and survival of glioblastoma stem cells. Cancer Cell. 2016;29(6):859–873. doi: 10.1016/j.ccell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Wang ZB, Luo C, Mao XY, Li X, Yin JY, Zhang W, Zhou HH, Liu ZQ. The Prospective value of dopamine receptors on bio-behavior of tumor. J Cancer. 2019;10(7):1622–1632. doi: 10.7150/jca.27780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R, Li W, Tao B, Wang X, Yang Z, Zhang Y, Wang C, Liu R, Gao H, Liang J, et al. Tyrosine phosphorylation activates 6-phosphogluconate dehydrogenase and promotes tumor growth and radiation resistance. Nat Commun. 2019;10(1):991. doi: 10.1038/s41467-019-08921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L, Cheng Q. Inhibiting 6-phosphogluconate dehydrogenase reverses doxorubicin resistance in anaplastic thyroid cancer via inhibiting NADPH-dependent metabolic reprogramming. Biochem Biophys Res Commun. 2018;498(4):912–917. doi: 10.1016/j.bbrc.2018.03.079. [DOI] [PubMed] [Google Scholar]
- 39.Lv X, Yu H, Zhang Q, Huang Q, Hong X, Yu T, Lan H, Mei C, Zhang W, Luo H, et al. SRXN1 stimulates hepatocellular carcinoma tumorigenesis and metastasis through modulating ROS/p65/BTG2 signalling. J Cell Mol Med. 2020;24(18):10714–10729. doi: 10.1111/jcmm.15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barquilha CN, Santos NJ, Moncao CCD, Barbosa IC, Lima FO, Justulin LA, Pertega-Gomes N, Felisbino SL. Sulfiredoxin as a potential therapeutic target for advanced and metastatic prostate cancer. Oxid Med Cell Longev. 2020 doi: 10.1155/2020/2148562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marziali LN, Correale J, Garcia CI, Pasquini JM. Combined effects of transferrin and thyroid hormone during oligodendrogenesis In vitro. Glia. 2016;64(11):1879–1891. doi: 10.1002/glia.23029. [DOI] [PubMed] [Google Scholar]
- 42.Kan M, Yamane I. Effects of ferrous iron and transferrin on cell proliferation of human diploid fibroblasts in serum-free culture. In Vitro. 1984;20(2):89–94. doi: 10.1007/BF02626648. [DOI] [PubMed] [Google Scholar]
- 43.Feng H, Schorpp K, Jin J, Yozwiak CE, Hoffstrom BG, Decker AM, Rajbhandari P, Stokes ME, Bender HG, Csuka JM, et al. Transferrin receptor is a specific ferroptosis marker. Cell Rep. 2020;30(10):3411–3423.e3417. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muhammad JS, Bajbouj K, Shafarin J, Hamad M. Estrogen-induced epigenetic silencing of FTH1 and TFRC genes reduces liver cancer cell growth and survival. Epigenetics. 2020;15(12):1302–1318. doi: 10.1080/15592294.2020.1770917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S, Zhu X, Ke Y, Xiao D, Liang C, Chen J, Chang Y. LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1. Cancer Biol Ther. 2020;21(4):379–387. doi: 10.1080/15384047.2019.1702405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hua S, Quan Y, Zhan M, Liao H, Li Y, Lu L. miR-125b-5p inhibits cell proliferation, migration, and invasion in hepatocellular carcinoma via targeting TXNRD1. Cancer Cell Int. 2019;19:203. doi: 10.1186/s12935-019-0919-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busker S, Qian W, Haraldsson M, Espinosa B, Johansson L, Attarha S, Kolosenko I, Liu J, Dagnell M, Grander D, et al. Irreversible TrxR1 inhibitors block STAT3 activity and induce cancer cell death. Sci Adv. 2020 doi: 10.1126/sciadv.aax7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JJ, Wang TY, Liu CL, Chien MN, Chen MJ, Hsu YC, Leung CH, Cheng SP. Dipeptidyl peptidase IV as a prognostic marker and therapeutic target in papillary thyroid carcinoma. J Clin Endocr Metab. 2017;102(8):2930–2940. doi: 10.1210/jc.2017-00346. [DOI] [PubMed] [Google Scholar]
- 49.Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, Zhong M, Yuan H, Zhang L, Billiar TR, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20(7):1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 50.Du J, Wang TT, Li YC, Zhou Y, Wang X, Yu XX, Ren XY, An YH, Wu Y, Sun WD, et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radical Bio Med. 2019;131:356–369. doi: 10.1016/j.freeradbiomed.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 51.Funauchi Y, Tanikawa C, Lo PHY, Mori J, Daigo Y, Takano A, Miyagi Y, Okawa A, Nakamura Y, Matsuda K. Regulation of iron homeostasis by the p53-ISCU pathway. Sci Rep-Uk. 2015 doi: 10.1038/srep16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahli W, Braissant O, Desvergne B. Peroxisome proliferator activated receptors: transcriptional regulators of adipogenesis, lipid metabolism and more. Chem Biol. 1995;2(5):261–266. doi: 10.1016/1074-5521(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 53.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43(4):527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 54.Christofides A, Konstantinidou E, Jani C, Boussiotis VA. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism. 2021;114:154338. doi: 10.1016/j.metabol.2020.154338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venkatesh D, O'Brien NA, Zandkarimi F, Tong DR, Stokes ME, Dunn DE, Kengmana ES, Aron AT, Klein AM, Csuka JM, et al. MDM2 and MDMX promote ferroptosis by PPARalpha-mediated lipid remodeling. Genes Dev. 2020;34(7–8):526–543. doi: 10.1101/gad.334219.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood WM, Sharma V, Bauerle KT, Pike LA, Zhou Q, Fretwell DL, Schweppe RE, Haugen BR. PPARgamma promotes growth and invasion of thyroid cancer cells. PPAR Res. 2011;2011:171765. doi: 10.1155/2011/171765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toraih EA, Fawzy MS, Abushouk AI, Shaheen S, Hobani YH, Alruwetei AM. Prognostic value of the miRNA-27a and PPAR/RXRalpha signaling axis in patients with thyroid carcinoma. Epigenomics. 2020;12(20):1825–1843. doi: 10.2217/epi-2020-0167. [DOI] [PubMed] [Google Scholar]
- 58.Liu YC, Yeh CT, Lin KH. Molecular functions of thyroid hormone signaling in regulation of cancer progression and anti-apoptosis. Int J Mol Sci. 2019;20(20):4986. [Google Scholar]
- 59.Manka P, Coombes JD, Boosman R, Gauthier K, Papa S, Syn WK. Thyroid hormone in the regulation of hepatocellular carcinoma and its microenvironment. Cancer Lett. 2018;419:175–186. doi: 10.1016/j.canlet.2018.01.055. [DOI] [PubMed] [Google Scholar]
- 60.Cicatiello AG, Ambrosio R, Dentice M. Thyroid hormone promotes differentiation of colon cancer stem cells. Mol Cell Endocrinol. 2017;459:84–89. doi: 10.1016/j.mce.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 61.Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J, Meng Q, Yu X, Shi S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13(1):110. doi: 10.1186/s13045-020-00946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galdiero MR, Varricchi G, Marone G. The immune network in thyroid cancer. Oncoimmunology. 2016;5(6):e1168556. doi: 10.1080/2162402X.2016.1168556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrari SM, Fallahi P, Galdiero MR, Ruffilli I, Elia G, Ragusa F, Paparo SR, Patrizio A, Mazzi V, Varricchi G, et al. Immune and inflammatory cells in thyroid cancer microenvironment. Int J Mol Sci. 2019;20(18):4413. doi: 10.3390/ijms20184413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Batistatou A, Zolota V, Scopa CD. S-100 protein+ dendritic cells and CD34+ dendritic interstitial cells in thyroid lesions. Endocr Pathol. 2002;13(2):111–115. doi: 10.1385/ep:13:2:111. [DOI] [PubMed] [Google Scholar]
- 65.Ugolini C, Basolo F, Proietti A, Vitti P, Elisei R, Miccoli P, Toniolo A. Lymphocyte and immature dendritic cell infiltrates in differentiated, poorly differentiated, and undifferentiated thyroid carcinoma. Thyroid. 2007;17(5):389–393. doi: 10.1089/thy.2006.0306. [DOI] [PubMed] [Google Scholar]
- 66.Gogali F, Paterakis G, Rassidakis GZ, Liakou CI, Liapi C. CD3(−)CD16(−)CD56(bright) immunoregulatory NK cells are increased in the tumor microenvironment and inversely correlate with advanced stages in patients with papillary thyroid cancer. Thyroid. 2013;23(12):1561–1568. doi: 10.1089/thy.2012.0560. [DOI] [PubMed] [Google Scholar]
- 67.Park A, Lee Y, Kim MS, Kang YJ, Park YJ, Jung H, Kim TD, Lee HG, Choi I, Yoon SR. Prostaglandin E2 secreted by thyroid cancer cells contributes to immune escape through the suppression of natural killer (NK) cell cytotoxicity and NK cell differentiation. Front Immunol. 2018 doi: 10.3389/fimmu.2018.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sottile R, Federico G, Garofalo C, Tallerico R, Faniello MC, Quaresima B, Cristiani CM, Di Sanzo M, Cuda G, Ventura V, et al. Iron and ferritin modulate MHC class I expression and NK cell recognition. Front Immunol. 2019;10:224. doi: 10.3389/fimmu.2019.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Jiao W, Cui H, Sun Q, Fan H. Combined exposure of alumina nanoparticles and chronic stress exacerbates hippocampal neuronal ferroptosis via activating IFN-gamma/ASK1/JNK signaling pathway in rats. J Hazard Mater. 2021;411:125179. doi: 10.1016/j.jhazmat.2021.125179. [DOI] [PubMed] [Google Scholar]
- 70.Amorim P, Telch B. A chemotaxis predator-prey model with indirect pursuit-evasion dynamics and parabolic signal. J Math Anal Appl. 2021 doi: 10.1016/j.jmaa.2021.125128. [DOI] [Google Scholar]
- 71.Melillo RM, Guarino V, Avilla E, Galdiero MR, Liotti F, Prevete N, Rossi FW, Basolo F, Ugolini C, de Paulis A, et al. Mast cells have a protumorigenic role in human thyroid cancer. Oncogene. 2010;29(47):6203–6215. doi: 10.1038/onc.2010.348. [DOI] [PubMed] [Google Scholar]
- 72.Visciano C, Liotti F, Prevete N, Cali G, Franco R, Collina F, de Paulis A, Marone G, Santoro M, Melillo RM. Mast cells induce epithelial-to-mesenchymal transition and stem cell features in human thyroid cancer cells through an IL-8-Akt-Slug pathway. Oncogene. 2015;34(40):5175–5186. doi: 10.1038/onc.2014.441. [DOI] [PubMed] [Google Scholar]
- 73.Siiskonen H, Poukka M, Bykachev A, Tyynela-Korhonen K, Sironen R, Pasonen-Seppanen S, Harvima IT. Low numbers of tryptase(+) and chymase(+) mast cells associated with reduced survival and advanced tumor stage in melanoma. Melanoma Res. 2015;25(6):479–485. doi: 10.1097/CMR.0000000000000192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. A list of 259 ferroptosis-related genes identified from FerrDb.
Additional file 2. Kaplan–Meier curves for overall survival at the best cut-off value of the risk score.
Data Availability Statement
Gene expression profiles, clinical information and mutation data of THCA in this study are available from the public database (TCGA, https://portal.gdc.cancer.gov/).