Supplemental Digital Content is available in the text.
Keywords: autophagy, esophageal adenocarcinoma, long-chain noncoding RNA, overall survival, prognostic model, risk assessment
Abstract
Autophagy-related long-chain noncoding ribonucleic acids play a vital role in the development of esophageal adenocarcinoma. This study aimed to construct a prognostic model of autophagy-related long-chain noncoding ribonucleic acids and identify potential therapeutical targets for esophageal adenocarcinoma. We downloaded 261 long-chain noncoding RNA transcript samples and clinical data of 87 esophageal adenocarcinoma patients from the Cancer Genome Atlas and 307 autophagy-related genes from www.autophagy.com. We performed Kyoto Encyclopedia of Genes and Genomes and Gene Ontology enrichment analyses and Gene Set Enrichment Analysis to determine risk characteristics and bioinformatics functions of signal transduction pathways. Univariate and multivariate Cox regression analyses were used to determine the correlation between autophagy-related long-chain noncoding ribonucleic acids and independent risk factors. The receiver operating characteristic analysis was used to evaluate the feasibility of the prognostic model. Finally, we performed survival analysis, risk analysis and independent prognostic analysis to verify the prognostic model of esophageal adenocarcinoma. We identified 22 autophagic long-chain noncoding ribonucleic acids that were highly correlated with the overall survival of esophageal adenocarcinoma patients. The areas under the receiver operating characteristic curve (0.941) and the calibration curve were significantly similar. Moreover, univariate and multivariate Cox regression analyses indicated that autophagy-related long-chain noncoding ribonucleic acids were independent predictors of esophageal adenocarcinoma. We found that autophagy-related long-chain noncoding ribonucleic acids might affect tumor development and prognosis in esophageal adenocarcinoma patients. The findings indicate that the prognostic model of esophageal adenocarcinoma has potential therapeutic applications in patients with esophageal adenocarcinoma.
Introduction
Esophageal cancer is one of the most common malignant tumors of the upper gastrointestinal tract and is one of the top ten causes of cancer deaths globally. In recent years, the incidence of esophageal cancer has shown an increasing trend globally [1]. The pathological types of esophageal cancer are mainly divided into esophageal squamous cell carcinoma and esophageal adenocarcinoma. Among these types of esophageal cancer, esophageal squamous cell carcinoma is common in China, Japan, South Korea and Southeast Asia, while esophageal adenocarcinoma is more common in Western countries, including those in Europe, America and Australia. However, in recent years, studies have found that in China and other regions in Eastern countries, with changes in the living standards, eating habits and surrounding environment, the incidence of esophageal adenocarcinoma has increased significantly faster than that of esophageal squamous cell carcinoma [2]. In clinical practice, many patients with esophageal adenocarcinoma often lack specific symptoms in the early stage and are not diagnosed in time; the prognosis of patients with advanced esophageal adenocarcinoma is worse during metastasis and recurrence, and the morbidity and mortality of patients with advanced esophageal adenocarcinoma are gradually increasing. Therefore, pretreatment intervention and evaluation will help identify and treat patients with a high risk of recurrence of esophageal adenocarcinoma. In addition, it is important to determine the prognostic indicators to effectively predict overall survival (OS) in patients with esophageal adenocarcinoma.
Long-chain noncoding RNAs (lncRNAs) [4] are defined as RNA sequences greater than 200 nucleotides in length that do not have protein translation functions. LncRNAs participate in cell growth and proliferation and gene replication, including transcription, translation and editing. Recent evidence indicates that under certain conditions, some lncRNAs regulate cell apoptosis through the autophagy pathway [5]. In addition, with the rapid development of biology and genetics, increasing evidence [6,7] shows that autophagy-related lncRNAs play a vital role in the initiation, induction and progression of esophageal adenocarcinoma. The findings suggest that autophagy-related lncRNAs directly or indirectly affect the OS of patients with esophageal adenocarcinoma. Moreover, related studies have also shown that autophagy-related lncRNAs are potential candidate genes as prognostic indicators of esophageal adenocarcinoma and ideal therapeutic targets. Therefore, exploring autophagy-related lncRNAs has important guiding value and broad application prospects for basic research and clinical diagnosis and treatment of esophageal adenocarcinoma. The autophagy-related lncRNA esophageal adenocarcinoma prognosis model proposed in this study could guide targeted therapeutic strategies for patients with esophageal adenocarcinoma [3] to reduce mortality and recurrence rates and improve survival and prognosis of patients.
Materials and methods
Data acquisition and processing
Relevant esophageal adenocarcinoma data [8] were downloaded from The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). These data included 261 samples of lncRNA-related transcription genes and clinical data of 87 patients with esophageal adenocarcinoma. In addition, on 29 October 2020, the lncRNA sequencing data and related clinical information of esophageal adenocarcinoma patients were obtained from TCGA. Moreover, we downloaded a total of 307 autophagy-related genes from www.autuphagy.com. These three sets of data were merged according to the corresponding gene sequence number, and 47 overlapping autophagy-related lncRNAs were screened for further analysis. Among the 47 lncRNA genes, 22 genes related to the prognosis of esophageal adenocarcinoma were identified. In R software, the ‘edgeR’ package (version 4.0, URL: http://bioinf.wehi.edu.au/edgeR https://bioconductor.org/packages/edgeR) was used to identify the DELs of 261 transcribed gene samples and 87 clinical samples from the data downloaded from TCGA; these samples were adjusted according to |logFC|>2 and corrected according to P < 0.05. In the analysis of survival time, 22 prognostic-related lncRNAs were found to have significant differences (Supplementary Fig. 1, Supplemental digital content 1, http://links.lww.com/ACD/A405).
Screening and identification of differential expressed autophagy-related genes
Our study used GEO2R to screen for differentially expressed genes (DEGs) among the autophagy-related lncRNA samples (http://www.ncbi.nlm.gov/geo2r). GEO2R [9,10] is an interactive network tool that allows researchers to compare and analyze two or more data sets in the GEO database to identify DEGs under experimental conditions. Adjusted P values (adj. P) and Benjamini and Hochberg false discovery rates were used to identify statistically significant target genes, correct for false positives and remove probe sets without corresponding gene names or genes with multiple probe sets [11]. The cutoff value was determined by the false discovery rate (FDR) <0.05 and log|FC|>1.
Kyoto Encyclopedia of Genes and Genomes and Gene Ontology enrichment analyses of autophagy-related genes
Kyoto Encyclopedia of Genes and Genomes (KEGG) is an online database resource that is used to evaluate a large number of molecular data generated in high-throughput experiments [12–14]. We used KEGG to evaluate the high-level biological functions of the corresponding lncRNAs. In addition, we used Gene Ontology (GO), an analytical bioinformatics tool, to explain and analyze the biological processes of these genes. We downloaded the relevant corresponding gene IDs, gene expression data and logFC values. Before running the script, we downloaded the ‘colorspace’, ‘string’ and ‘ggplot2’ packages (version: 3.3.3, URL: https://ggplot2.tidyverse.org, https://github.com/tidyverse/ggplot2) and ran the ‘BiocManager’ (version: 1.30.10, Martin Morgan et al) and ‘DOSE’ packages (version: 3.12, URL: https://yulab-smu.top/biomedical-knowledge-mining-book/) in R software. The ‘Enrichplot’ package (version: 3.12, URL: https://yulab-smu.top/biomedical-knowledge-mining-book/) was used to generate visual gene set enrichment maps with P = 0.05 as the filter condition for GO enrichment.
Before generating the GO enrichment bubble chart, we set the width=10, height=8, and −log (adj P value) >3 to display the ID of the corresponding gene [15]. The highest FDR <0.05 was considered to be of research significance. In addition, we used R software to perform KEGG and GO analyses of the differentially expressed autophagy-related lncRNAs to identify the enriched functions and pathways of these lncRNAs.
Screening of prognostic-related autophagy genes
From the 47 autophagy-related lncRNAs obtained through integration and screening by the ‘edgeR’ package (version 4.0, URL: http://bioinf.wehi.edu.au/edgeR https://bioconductor.org/packages/edgeR) of R software, 22 lncRNA genes were found to be related to the prognosis of esophageal adenocarcinoma (including 18 lncRNA genes that were expressed at high levels and 4 lncRNA genes that were expressed at low levels in esophageal adenocarcinoma tumor cells).
Construction of the prognostic model of autophagy-related genes in esophageal adenocarcinoma
Survival analysis
The Kaplan–Meier survival analysis method [16] was used to compare the survival rates between the high-risk and low-risk groups, calculate the risk of patients with esophageal adenocarcinoma and determine their OS [1-year, 3-year and 5-year survival rates].
Receiver operating characteristic analysis
Receiver operating characteristic (ROC) analysis [17] uses independent risk factors to test the sensitivity and specificity of survival predictions. The area under the ROC curve (AUC) ranged from 0.5 to 1.0. The closer the AUC is to 1, the more accurate the predictive ability of the model; a prognostic model has no predictive power if the AUC <0.5.
ROC analysis was performed using the ‘Survival ROC’ package in R software to evaluate the lncRNA signature. The calibration curve was used to evaluate whether the predicted survival time was consistent with the actual survival time, and the AUC was used to verify the prediction performance and accuracy of the prognostic model [18]. Moreover, our study also validated the prognostic model of esophageal adenocarcinoma through single factor and multivariate Cox proportional hazards regression (CPHR) analyses and further explored the independence of 22 lncRNA signatures on the OS of patients with esophageal adenocarcinoma.
Gene set enrichment analysis
Based on the median of the risk scores of the 22 lncRNA signatures, the 261 gene transcript samples and 87 clinical samples downloaded from TCGA were divided into a high-risk group and a low-risk group. We used JavaGSEA software [19] to perform GSEA of the high-risk and low-risk groups [selection criteria (NOM) P value <0.05].
Results
Differential analysis of autophagy-related genes
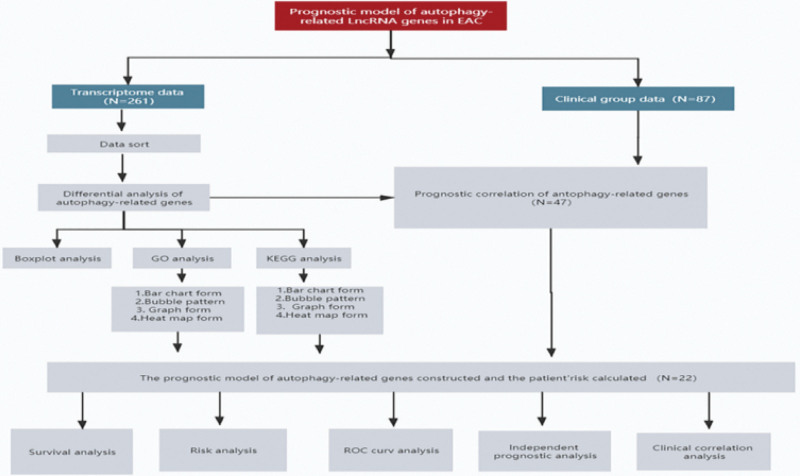
A detailed data flowchart is presented in Fig. 1 to understand the model developed in this study and its verification.
Fig. 1.
Process of designing the lncRNA prognostic model of autophagy-related genes in esophageal adenocarcinoma.
Boxplot
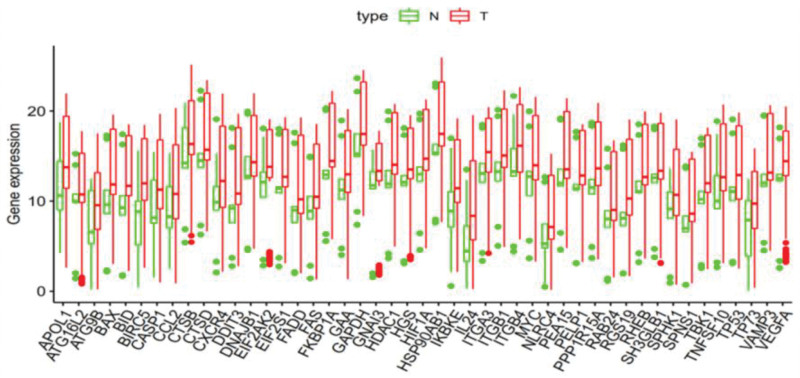
This study analyzed 262 lncRNA transcription samples by box plots [20]. We used the ‘ggpubr’ package in R software to identify the expression levels of differential genes to create autophagy-related lncRNA boxplots (tumor count: 147; normal count: 115) of 47 esophageal adenocarcinoma samples compared with normal tissue samples (Fig. 2). In Fig. 2, the abscissa presents the names of the 47 differentially expressed lncRNA genes, and the ordinate shows the expression of these genes [21]. We found that the 47 differentially expressed autophagy-related lncRNAs were all upregulated in esophageal adenocarcinoma samples.
Fig. 2.
Box plot analysis of the expression of 47 autophagy-related lncRNA genes (red represents tumor samples, and green represents normal samples; the data are from TCGA [https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga] and GEO database [https://www.ncbi.nlm.nih.gov/]).
Kyoto Encyclopedia of Genes and Genomes enrichment analysis
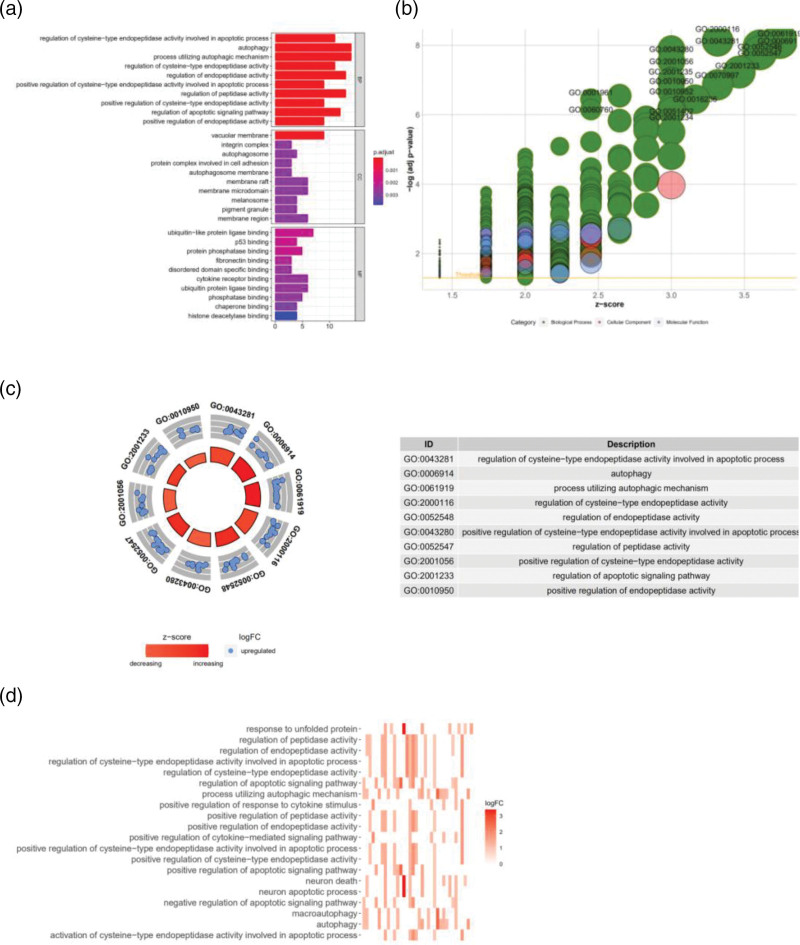
In Fig. 3a, the ordinate of the plot shows the names of GO terms, the abscissa presents the number of genes enriched for each GO term, and the color of the histogram represents the degree of GO enrichment. The GO terms are divided into three categories, including biological process (BP), cellular component (CC) and molecular function (MF) [22]. We found that these genes were mainly concentrated in mechanisms regulating apoptosis and autophagy, including the regulation of cysteine-type endopeptidase activity, regulation of apoptosis signaling pathways, vacuolar membrane integrin complex autophagosomes, cell adhesion, autophagosome membrane rafts, membrane microdomains, melanosome pigment granules and membrane domain protein complexes (Fig. 3a).
Fig. 3.
(a) BarPlot of the GO enrichment analysis of 47 autophagy-related lncRNA genes (red represents signaling pathways that are highly enriched, purple represents signaling pathways that are moderately enriched, and blue represents signaling pathways that are poorly enriched). (b) Bubble plot of the GO enrichment analysis (green represents BP, red represents CC, and blue represents MF). (c) Circle plot of the GO enrichment analysis (count number=10; red indicates upregulated genes, and blue indicates downregulated genes). (d) Heatmap of the enriched signaling pathways (darker colors indicate more intensive signal pathway enrichment). Data processing and analysis were performed by R software (Version 4.0.2). CC, cellular component; BP, biological process; GO, Gene Ontology; MF, molecular function.
In Fig. 3b, the ordinate shows the −log (adj. P value) value, and the abscissa represents the Z-score; a Z-score >0 indicates that the number of upregulated genes enriched in the GO term is large, and a Z-score <0 indicates that the number of upregulated genes enriched in the GO term is small (green represents BP, red represents CC and blue represents MF) [23]. We found that the genes enriched in GO terms were all upregulated, including GO: 0001961, GO0060760 and GO0043280 (Fig. 3b). We identified the GO terms (count number=10) that showed the most significant enrichment of these DEGs, including GO: 0043281, GO: 0006914, GO: 0061919, GO: 2000116, GO: 0052548, GO: 0043280, GO: 0052547, GO: 2001056, GO: 2001233 and GO: 0010950. A darker color indicates more enriched genes. The results of this analysis are consistent with those of the previous BarPlot analysis (Fig. 3c). The genes that we screened out of the GO heat were mainly enriched in responding to unfolded proteins map [24], regulation of the apoptotic signaling pathway, processes utilizing an autophagic mechanism, neuronal death, processes involved in neuronal apoptosis, processes involved in autophagy and others (Fig. 3d).
Kyoto Encyclopedia of Genes and Genomes enrichment analysis
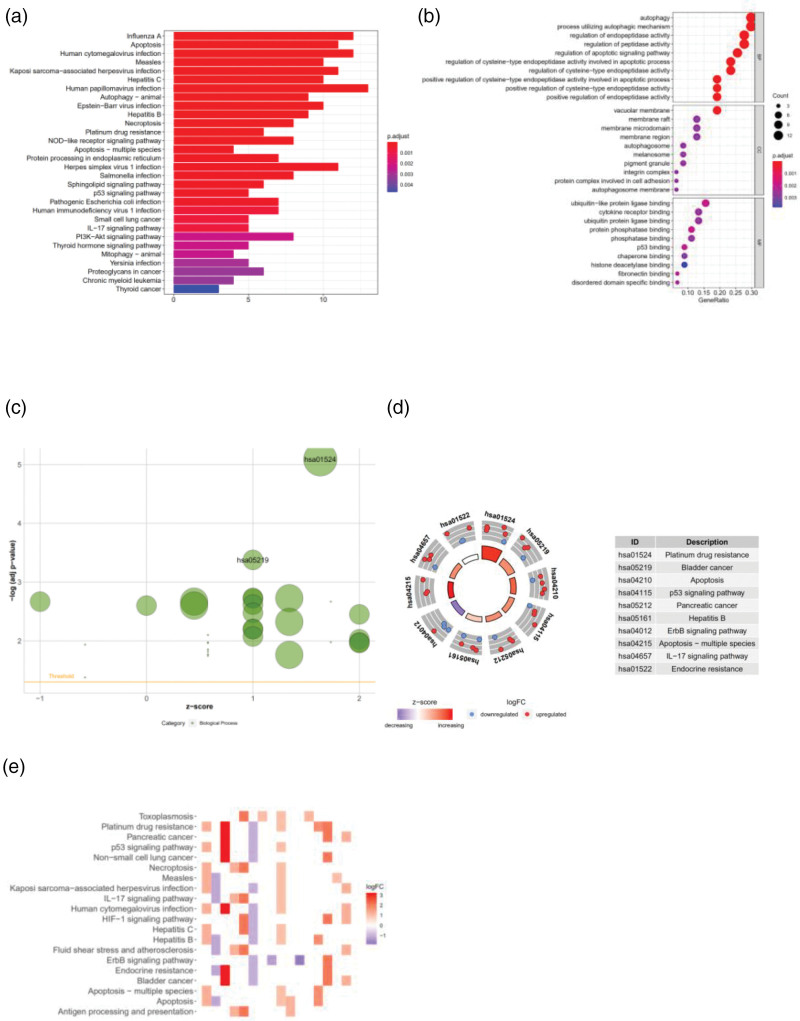
In Fig. 4a, the abscissa shows the number of genes, and the ordinate represents the name of the signaling pathway. The darker the color, the higher the enrichment of the gene. We found that the degree of enrichment was most obvious in KEGG pathways [25], such as autophagy, apoptosis, measles and influenza (Fig. 4a). In Fig 4b, the abscissa shows the number ratio of the genes, the ordinate represents the name of the KEGG signaling pathway and the color of the bubble represents the amount of gene enrichment. We found significant enrichment in KEGG signaling pathways, including autophagy, processes utilizing an autophagic mechanism, regulation of endopeptidase activity and regulation of the apoptotic signaling pathway. Among the pathways, the autophagy pathway was the most enriched pathway, indicating the involvement of the esophageal glands. Cancer-related genes have a certain correlation with the autophagy signal induction pathway (Fig. 4b). We set −log(adj. P value) >3 as the cutoff value for KEGG enrichment of the autophagy-related lncRNA genes. We found that hao05219 and hao01524 were both upregulated in the KEGG enrichment analysis (Fig. 4c). Hao01524 is related to antitumor drug resistance and drug resistance [26]. The KEGG pathways of the upregulated genes were also related to ovarian cancer, lung cancer and bladder cancer, as well as the P53 signaling pathway and the IL-17 signaling pathway (Fig. 4d). The autophagy-related lncRNAs that we downloaded and screened had high expression in pancreatic cancer and bladder cancer and were associated with infection and drug resistance (Fig. 4e).
Fig. 4.
(a) BarPlot of the KEGG enrichment analysis of 47 autophagy-related lncRNA genes (red represents signaling pathways that are highly enriched, purple represents signaling pathways that are moderately enriched, and blue represents signaling pathways that are lowly enriched). (b) Bubble plot of the KEGG enrichment analysis of 47 autophagy-related lncRNA genes. (c) Bubble plot of the enriched signaling pathways (green represents BP, red represents CC, and blue represents MF). (d) Circle plot of the enriched signaling pathways (count number=10; red indicates upregulated genes, and blue indicates downregulated genes). (e) Heatmap of the enriched signaling pathways (darker color indicates greater signal pathway enrichment). Data processing and analysis were performed by R software (Version 4.0.2). CC, cellular component; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function.
Construction and analysis of the prognostic model
Subsistence analysis
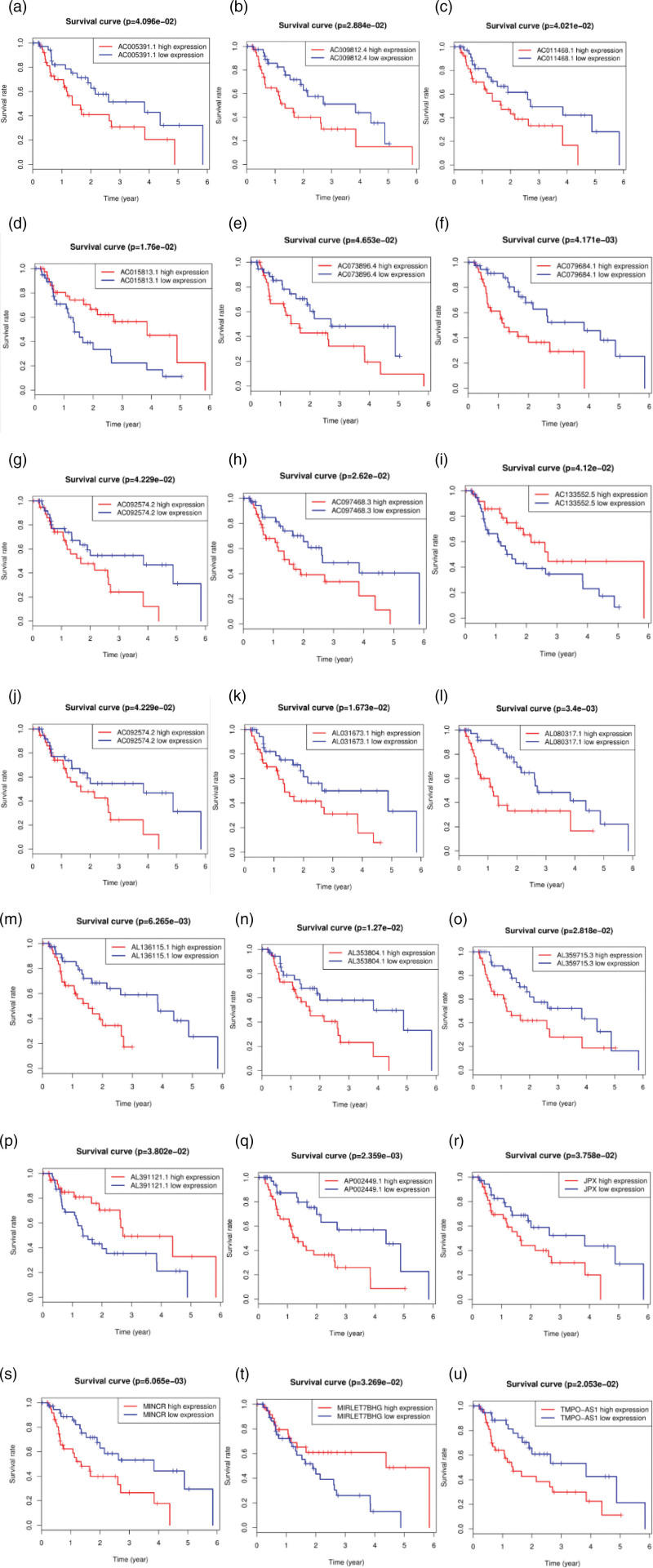
Using R software, we further screened and overlapped the 47 esophageal adenocarcinoma autophagy-related lncRNA genes with the collected clinical OS data (including the 1-year survival rate, 3-year survival rate, and 5-year survival rate) of patients with esophageal adenocarcinoma to construct the prognostic model [27]. Finally, we obtained 22 lncRNAs related to prognosis, including 18 highly expressed lncRNAs and 4 lowly expressed lncRNAs in esophageal adenocarcinoma tumor cells (Fig. 5a–u).
Fig. 5.
Kaplan–Meier survival curves of the overall survival (OS) of patients with esophageal adenocarcinoma (a–u) (red line: high expression of autophagy-related lncRNA genes; blue line: low expression of autophagy-related lncRNA genes). P < 0.05 indicates a significant difference, and P > 0.05 indicates no significant difference.
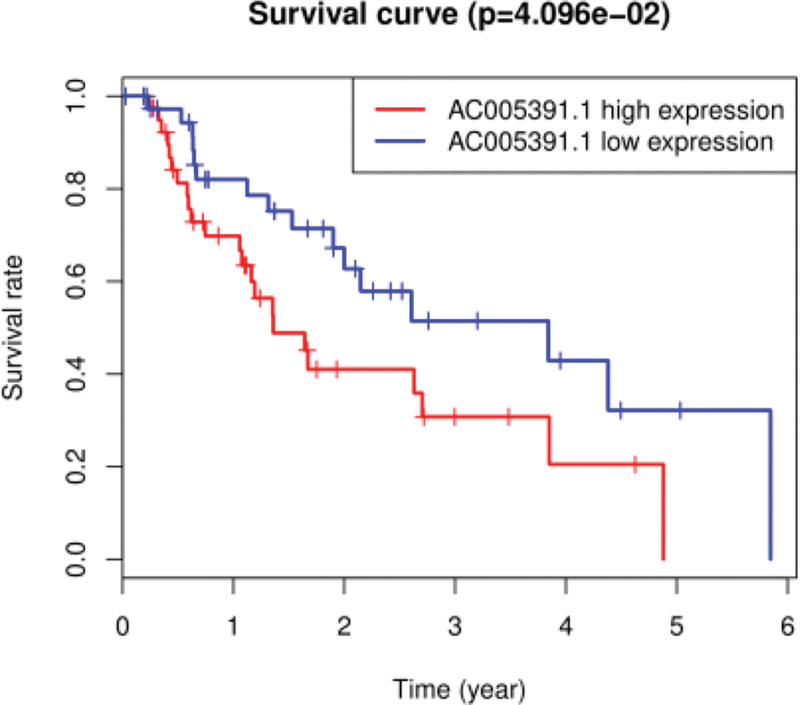
In addition to survival analysis of the 22 prognostic-related lncRNAs, we also performed OS analysis (Fig. 6). We found that lncRNAs were important for the prognosis of esophageal adenocarcinoma patients with a high risk of reduced OS. The specific performances of the lncRNAs [28] are shown in Fig. 6. The survival rates of patients with highly expressed lncRNAs (1-year survival rate: 50%; 3-year survival rate: 0%; 5-year survival rate: 0%) were compared with those of patients with lowly expressed lncRNAs (1-year survival rate: 100%; 3-year survival rate: 90%; 5-year survival rate: 40%), and the difference was statistically significant (P < 0.001).
Fig. 6.
Kaplan–Meier survival curves of patients with esophageal adenocarcinoma (clinical overall survival [OS]). P < 0.05 indicates a significant difference, and P > 0.05 indicates no significant difference.
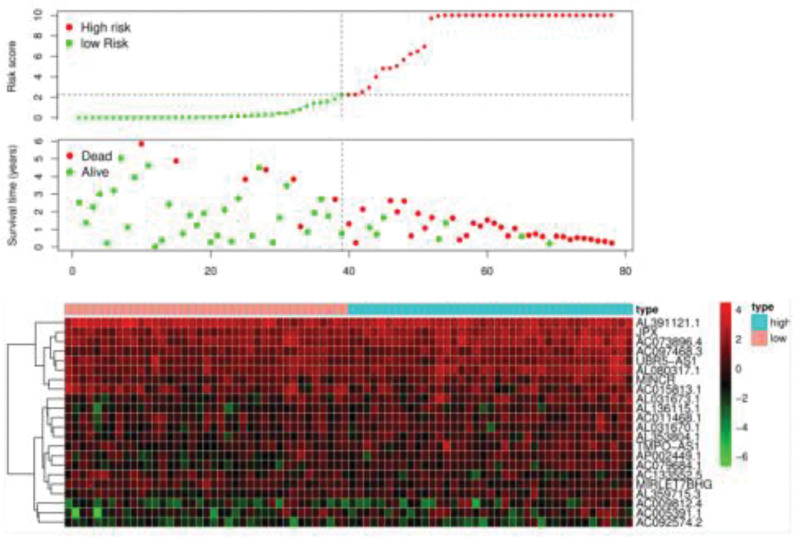
Risk analysis
We performed a risk analysis of the lncRNA prognostic model related to autophagy of esophageal adenocarcinoma; calculated the risk values for each esophageal adenocarcinoma patient, including the risk score, survival time, and risk heat map; and drew a risk curve [29]. In Fig. 7, the abscissa shows the risk value, overall survival, and survival status of each patient (the left shows low-risk patients, and the right shows high-risk patients). The patients’ risk values increased sequentially, and the number of deaths of adenocarcinoma patients also increased sequentially; the finding is in line with the rationality of the prognosis model. We performed a visual analysis of the risk heat maps of these 22 lncRNAs and showed that low-risk lncRNA genes included AL391121.1, AC015813.1, AL031670.1, AP002449.1, AC133552.5, MIRLET7BHG and AC092574.2, and high-risk lncRNA genes included JPX, AC073896.4, AC097468.3, UBR5-AS1, AL080317.1, MINCR, AL031673.1, AL136115.1, AC011468.1, AL353804.1, TMPO-AS1, AC079684.1, AL359715.3, AC009812.4 and AC005391.1.
Fig. 7.
Autophagy-related lncRNA risk curves of 22 esophageal adenocarcinomas obtained after screening.
According to the standard of NOM P value <0.05, 11 KEGG signaling pathway in the high-risk group were found to be significantly changed. KEGG_REGULATION_OF_AUTOPHAGY, as one of the most changed pathways, may promote the occurrence and progression of esophageal adenocarcinoma [30]. Therefore, we only investigated the genes related to autophagy pathways. It was found that the expression levels of AL391121.1, AC015813.1, AL031670.1, AP002449.1, AC133552.5, MIRLET7BHG and AC092574.2 were related to OS in patients with esophageal adenocarcinoma (Fig. 7).
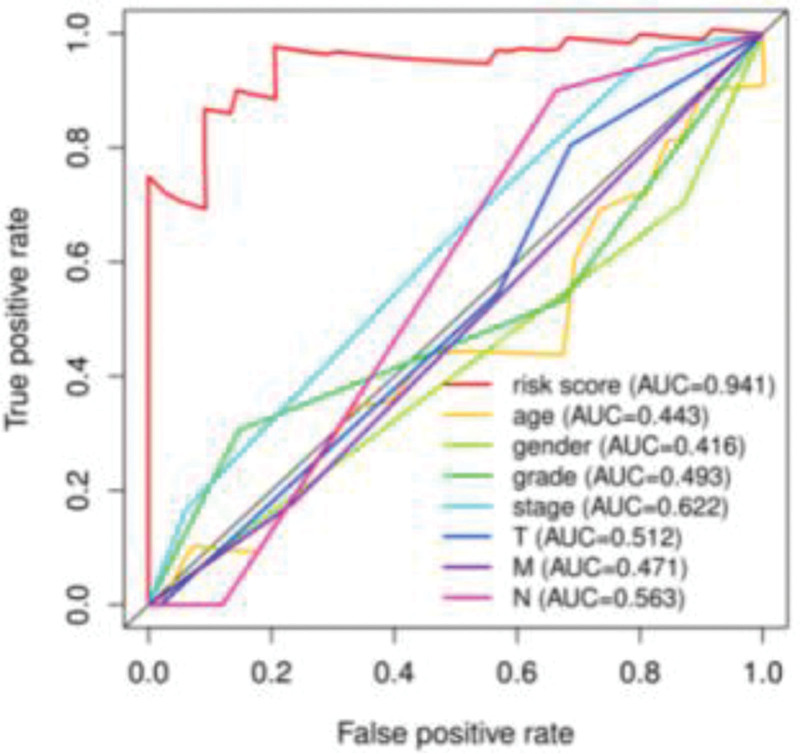
Multi-index receiver operating characteristic curve analysis
In Fig. 8, multiple curves represent various clinical characteristic indexes of esophageal adenocarcinoma, including the risk value, age, sex and tumor stage. Among the curves, the red risk ROC curve drawn using the risk values related to the prognosis of 22 esophageal adenocarcinomas was the main analysis object. The larger the area under the ROC curve of the risk value, the more accurate the established prognostic model. Based on the findings of the risk value (AUC = 0.941), tumor stage (AUC = 0.622), clinical features (AUC > 0.6) and other clinical features of AUC < 0.6, we concluded that the prognosis model of esophageal adenocarcinoma that we constructed using the risk value had an advantage over other clinical features to predict patients’ survival (Fig. 8).
Fig. 8.
Feasibility of the multi-index ROC curve assessment model for patients with esophageal adenocarcinoma. ROC, receiver operating characteristic.
Independent prognostic analysis
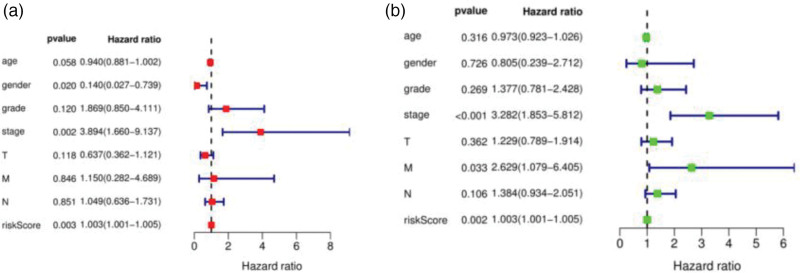
We used the constructed Cox model to analyze the clinical data of patients with esophageal adenocarcinoma. We found that the risk score and stage were independent (P = 0.003 vs. P = 0.002) and multivariate (P = 0.002 vs. P < 0.001) prognostic factors [31]. These two indicators could be used as independent prognostic factors of the esophageal adenocarcinoma model (P < 0.05), while other clinical characteristic indicators could not (P > 0.05). The univariate and multivariate CPHR analyses showed that this feature was an independent predictor of patients with esophageal adenocarcinoma (Fig. 9).
Fig. 9.
Single-factor independent prognostic analysis and multivariate independent prognostic analysis of the esophageal adenocarcinoma model [(a) univariate independent prognostic meta-analysis; (b) multivariate independent prognostic meta-analysis].
Clinical correlation analysis
We downloaded all the clinical data of patients with esophageal adenocarcinoma from TCGA and the GEO database, except for some data without a follow-up, data without specific tumor staging, data without a clear diagnosis and data without complete information, and we finally screened the data to meet the selected conditions. To explore the relationship between the 22 autophagy-related lncRNAs and clinical features of esophageal adenocarcinoma, we used the t test and Kruskal–Wallis test to calculate the correlation [32]. We found that the prognostic model had a certain correlation with the grade and staging of clinical tumors (grade G1-2, P = 0.001; stage I-II, P = 0.02), and the difference was significant (Table 1). In addition, the model was more accurate and valuable for predicting the prognosis of patients with early-stage (stage I-II) esophageal adenocarcinoma (P < 0.05) than that of patients with later-clinical-stage esophageal adenocarcinoma with poor differentiation, high malignancy and metastasis. For patients with advanced esophageal adenocarcinoma, this model was not effective for evaluating their prognosis (P > 0.05).
Table 1.
Correlation analysis of the clinical characteristics of patients with esophageal adenocarcinoma
| Clinical | Group | n | Mean | SD | T | P value |
|---|---|---|---|---|---|---|
| Age | ≤60 | 141 | 1.325 | 1. 186 | −0.768294156 | 0. 443 |
| >60 | 176 | 1.438 | 1.443 | |||
| Sex | Women | 120 | 1.312 | 1.258 | −0.806789468 | 0. 42 |
| Men | 197 | 1.434 | 1.379 | |||
| Grade | Gl-2 | 115 | 1. 103 | 0. 851 | −3.356378317 | 0. 001 |
| G3 | 202 | 1.55 | 1.521 | |||
| Stage | Stage I-II | 143 | 1.204 | 0. 953 | −2.337916733 | 0. 02 |
| Stage III-IV | 174 | 1. 538 | 1.566 | |||
| T | Tl-2 | 78 | 1.264 | 1.073 | −1.076791039 | 0. 283 |
| T3-4 | 239 | 1.428 | 1.408 | |||
| M | MO | 295 | 1.333 | 1. 288 | −2.115479566 | 0. 046 |
| Ml | 22 | 2. 121 | 1.712 | |||
| N | NO | 99 | 1. 199 | 1.001 | −1.946227745 | 0. 053 |
| Nl-3 | 218 | 1.473 | 1.454 |
GSEA
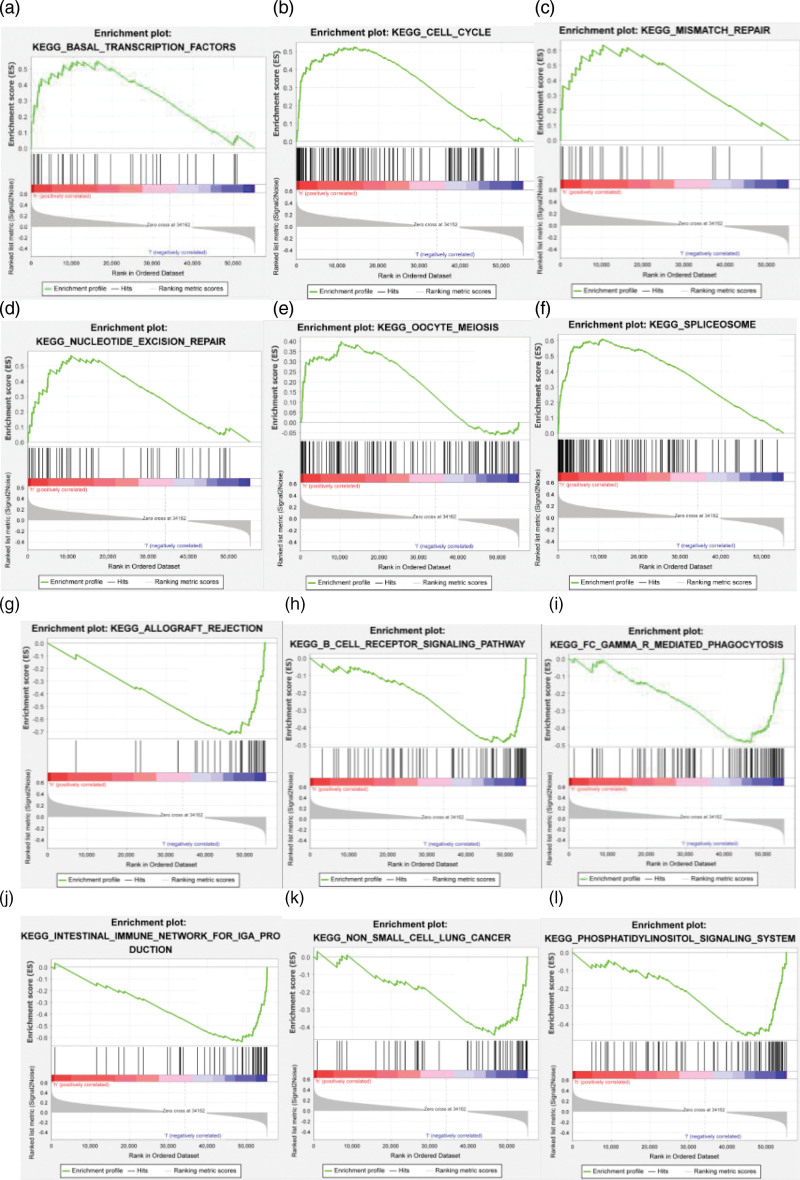
In GSEA, if the peak is on the upper left side, high expression of the gene will promote the corresponding signal transduction pathway. In contrast, if the peak is on the lower right side, high expression of the gene will inhibit the corresponding signal transduction pathway. We found that the high expression of the selected lncRNAs could promote signal transduction pathways, such as transcription-factors, cell-cycle, mismatch-repair, nucleotide-excision-repair, oocyte-meiosis and spliceosome (Fig. 10). However, allograft-rejection, cell receptor signaling pathways, mediated-phagocytosis, immune network for IgA production, phosphatidylinositol signaling system and other pathways were inhibited.
Fig. 10.
GSEA of autophagy-related lncRNA genes in esophageal adenocarcinoma. (a–f) Signaling pathway enrichment regions of the high-risk group; (g–l) signaling pathway enrichment region of the low-risk group.
Discussion
Exosomes, apoptosis, autophagy and enzyme regulation participate in the occurrence and development of tumor cells, and they have become research hotspots in tumor prevention and treatment worldwide. Leidal et al. [33] pointed out that there is a certain correlation between autophagy dysfunction and various diseases, such as infection, tumors, transplantation and metabolic disorders. Autophagy is a double-edged sword in tumorigenesis, and it participates in the growth and metabolism of tumor cells and can promote or inhibit tumor development through signal transduction pathways, regulate tumor cell death or promote tumor cell proliferation. We showed that a total of 22 autophagy-related lncRNAs were involved in the prognostic regulation of esophageal adenocarcinoma. KEGG and GO enrichment analyses demonstrated that almost all of these lncRNAs were involved in autophagy signaling pathways. In addition, we found that among the lncRNAs, AL391121.1, AC015813.1, AL031670.1, AP002449.1, AC133552.5, MIRLET7BHG and AC092574.2 had prognostic value in patients with esophageal adenocarcinoma. Liu et al. [34] demonstrated that MIRLET7BHG polymorphism might be an important predictor of lung cancer related to asbestos exposure, and they used the MIRLET7BHG polymorphism to establish a lung cancer risk assessment model. Consistently, in this study, we found that MIRLET7BHG was also associated with the OS of patients with esophageal adenocarcinoma. Moreover, Yu et al. 35 pointed out that the relationship between autophagy genes and esophageal adenocarcinoma was related to the expression of the corresponding lncRNA.
JPX is the most abundant lncRNA in the human genome, and the mechanisms of its interactions with other factors are largely unknown. Karner et al. [36] found that human JPX and its mouse homolog lncRNA JPX had large differences in their nucleotide sequence and RNA secondary structure. Despite these differences, both lncRNAs showed a strong binding ability to CTCF; the findings support a model of the conservation of the function of lncRNAs independent of their sequence and structural differences. Notably. JPX has mostly been studied in the field of liver cancer, and there are few reports on its mechanism in esophageal adenocarcinoma. Lin et al. [37] found that JPX could induce XIST to inhibit hepatocellular carcinoma by sponging miR-155-5p. Moreover, studies have also indicated that JPX has an inhibitory effect on tumor metabolism signaling. Furthermore, Gong et al. [38] proposed that JPX might be a new tumor-specific prognostic lncRNA marker because there was a certain correlation between JPX and tumor prognosis. Consistently, in this study, the visual analysis of the risk heat maps showed that JPX was one of the high-risk lncRNA genes in esophageal adenocarcinoma.
UBR5-AS1 is recognized as a key lncRNA in many epidemic cancers. Ji et al. [39] explored the relationship between UBR5 expression and cell proliferation, apoptosis and regulatory mechanisms in colon cancer cell lines and proved that UBR5 could degrade P21 by ubiquitination. In addition, Tasaki et al. [40] investigated the relationship between UBR5 and autophagy. They found that loss of UBR5 led to a variety of disorders in autophagy induction and flux, including synthesis and lipidation/activation of the ubiquitin-like protein LC3 and formation of an autophagy double membrane structure. Notably, we found that UBR5-AS1 was one of the high-risk lncRNA genes in esophageal adenocarcinoma. These findings suggest that UBR5 might be a therapeutic target for tumors.
Chen et al. [41] found that MINCR had an inhibitory effect on cell cycle arrest and apoptosis of NSCLC cells, suggesting that this lncRNA could be used as a potential target for NSCLC treatment. In our study, we found that MINCR was positively expressed in patients with esophageal adenocarcinoma. However, to date, the relationship between MINCR and the prognosis of esophageal adenocarcinoma remains unclear. In addition, TMPO-AS1 is a lncRNA that promotes tumorigenesis. TMPO-AS1 mediates the activation of the PI3K and Akt pathways to regulate the cell cycle and proliferation of tumor cells. Although the correlation of TMPO-AS1 and OS is not significant, the increased expression of TMPO-AS1 in esophageal adenocarcinoma is correlated with a poor prognosis. In our study, the expression of TMPO-AS1 in esophageal adenocarcinoma was significantly higher than that in normal control. This result is consistent with that in a previous study by Huang et al. [42], which revealed the effect of TMPO-AS1 in patients with esophageal adenocarcinoma. Thus, high expression of TMPO-AS1 might be related to aggressive tumor behavior of esophageal adenocarcinoma.
In addition to the apoptosis mechanism of cell death, another key mechanism of cell death is autophagy, which involves signal transduction pathways, receptor binding and enzyme decomposition and activation. There is an intricate relationship between autophagy and apoptosis [43]. The interaction between autophagy and apoptosis is mediated by different molecules in the microenvironment of esophageal adenocarcinoma and triggers signal transduction regulation that promotes or inhibits tumor growth. In our study, through KEGG and GO enrichment analyses, we found that most of the lncRNAs related to the prognosis of esophageal adenocarcinoma were mainly enriched in the apoptosis and autophagy pathways. These results are in line with those in the study by Zhu et al. [19], who found that the prognosis of esophageal adenocarcinoma had a certain correlation with autophagy-related lncRNAs. However, the mechanisms of some lncRNAs are still unknown. Further basic experimental investigations are still needed to determine genes related to esophageal adenocarcinoma and autophagy. Moreover, further clinical follow-up studies are also needed to determine prognostic indicators for the OS of patients with esophageal adenocarcinoma. The prognostic model of esophageal adenocarcinoma that we constructed in this study will help more accurately assess the prognosis and explore potential treatment targets in patients with esophageal adenocarcinoma.
Acknowledgements
The authors would like to express their gratitude to AJE for the expert linguistic services provided.
This work was supported by grants from the National Natural Science Foundation of China (81972829), the Science and Technology Innovation Committee of Shenzhen Municipality (Grant No. JCYJ20180228162607111, JCYJ20190809104601662), the Health and Family Planning Commission of Shenzhen Municipality Research Project (Grant No. SZBC2018018), China Scholarship Council (CSC, 201908470124).
X.L. designed the research; J.L. and D.W. guided the research; D.W., Y.Z., X.R. and P.X. collected and downloaded the data of our research; L.W. analyzed the data and wrote the paper.
Our study does not contain data from any individual person or any animals.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Footnotes
Yuzhen Zheng, Jixian Liu and Xiaoqiang Li contributed equally to the writing of this article.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.anti-cancerdrugs.com.
Reference
- 1.Coleman HG, Xie SH, Lagergren J. The epidemiology of esophageal adenocarcinoma. Gastroenterology 2018; 154:390–405. [DOI] [PubMed] [Google Scholar]
- 2.Lee SW, Lien HC, Peng YC, Lin MX, Ko CW, Chang CS. The incidence of esophageal cancer and dysplasia in a Chinese population with nondysplastic Barrett’s esophagus. JGH Open 2018; 2:214–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creemers A, Ebbing EA, Pelgrim TC, Lagarde SM, van Etten-Jamaludin FS, van Berge Henegouwen MI, et al. A systematic review and meta-analysis of prognostic biomarkers in resectable esophageal adenocarcinomas. Sci Rep 2018; 8:13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform 2016; 17:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun 2018; 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janser FA, Adams O, Bütler V, Schläfli AM, Dislich B, Seiler CA, et al. Her2-targeted therapy induces autophagy in esophageal adenocarcinoma cells. Int J Mol Sci 2018; 19:E3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena R, Klochkova A, Murray MG, Kabir MF, Samad S, Beccari T, et al. Roles for autophagy in esophageal carcinogenesis: implications for improving patient outcomes. Cancers (Basel) 2019; 11:E1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weh KM, Howell AB, Kresty LA. Expression, modulation, and clinical correlates of the autophagy protein Beclin-1 in esophageal adenocarcinoma. Mol Carcinog 2016; 55:1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akashi E, Fujihara S, Morishita A, Tadokoro T, Chiyo T, Fujikawa K, et al. Effects of galectin-9 on apoptosis, cell cycle and autophagy in human esophageal adenocarcinoma cells. Oncol Rep 2017; 38:506–514. [DOI] [PubMed] [Google Scholar]
- 10.Worfolk JC, Bell S, Simpson LD, Carne NA, Francis SL, Engelbertsen V, et al. Elucidation of the AGR2 interactome in esophageal adenocarcinoma cells identifies a redox-sensitive chaperone hub for the quality control of MUC-5AC. Antioxid Redox Signal 2019; 31:1117–1132. [DOI] [PubMed] [Google Scholar]
- 11.Adams O, Dislich B, Berezowska S, Schläfli AM, Seiler CA, Kröll D, et al. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget 2016; 7:39241–39255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang JC, Feng YL, Liang X, Cai XJ. Autophagy in 5-fluorouracil therapy in gastrointestinal cancer: trends and challenges. Chin Med J (Engl) 2016; 129:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Chen X, Bajpai M, Verma A, Das KM, Souza RF, et al. Cellular origins and molecular mechanisms of Barrett’s esophagus and esophageal adenocarcinoma. Ann N Y Acad Sci 2013; 1300:187–199. [DOI] [PubMed] [Google Scholar]
- 14.Kresty LA, Weh KM, Zeyzus-Johns B, Perez LN, Howell AB. Cranberry proanthocyanidins inhibit esophageal adenocarcinoma in vitro and in vivo through pleiotropic cell death induction and PI3K/AKT/mTOR inactivation. Oncotarget 2015; 6:33438–33455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Donovan TR, O’Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy 2011; 7:509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong J, Whelan KA, Laczkó D, Dang B, Caro Monroig A, Soroush A, et al. Autophagy levels are elevated in Barrett’s esophagus and promote cell survival from acid and oxidative stress. Mol Carcinog 2016; 55:1526–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams O, Janser FA, Dislich B, Berezowska S, Humbert M, Seiler CA, et al. A specific expression profile of LC3B and p62 is associated with nonresponse to neoadjuvant chemotherapy in esophageal adenocarcinomas. PLoS One 2018; 13:e0197610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Mashed S, O’Donovan TR, Kay EW, Abdallah AR, Cathcart MC, O’Sullivan J, et al. LC3B globular structures correlate with survival in esophageal adenocarcinoma. BMC Cancer 2015; 15:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Dong L, Feng M, Yang F, Jiang W, Huang Z, et al. Profiles of autophagy-related genes in esophageal adenocarcinoma. BMC Cancer 2020; 20:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin RU, Mills JC. The cyclical hit model: how paligenosis might establish the mutational landscape in Barrett’s esophagus and esophageal adenocarcinoma. Curr Opin Gastroenterol 2019; 35:363–370. [DOI] [PubMed] [Google Scholar]
- 21.Roesly HB, Khan MR, Chen HD, Hill KA, Narendran N, Watts GS, et al. The decreased expression of Beclin-1 correlates with progression to esophageal adenocarcinoma: the role of deoxycholic acid. Am J Physiol Gastrointest Liver Physiol 2012; 302:G864–G872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weh KM, Aiyer HS, Howell AB, Kresty LA. Cranberry proanthocyanidins modulate reactive oxygen species in Barrett’s and esophageal adenocarcinoma cell lines. J Berry Res 2016; 6:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blum AE, Venkitachalam S, Guo Y, Kieber-Emmons AM, Ravi L, Chandar AK, et al. RNA sequencing identifies transcriptionally viable gene fusions in esophageal adenocarcinomas. Cancer Res 2016; 76:5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verbeek RE, Siersema PD, Vleggaar FP, Ten Kate FJ, Posthuma G, Souza RF, et al. Toll-like receptor 2 signalling and the lysosomal machinery in Barrett’s esophagus. J Gastrointestin Liver Dis 2016; 25:273–282. [DOI] [PubMed] [Google Scholar]
- 25.Kolenda T, Guglas K, Kopczyńska M, Sobocińska J, Teresiak A, Bliźniak R, Lamperska K. Good or not good: role of miR-18a in cancer biology. Rep Pract Oncol Radiother 2020; 25:808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam F, Gopalan V, Lam AK. RETREG1 (FAM134B): a new player in human diseases: 15 years after the discovery in cancer. J Cell Physiol 2018; 233:4479–4489. [DOI] [PubMed] [Google Scholar]
- 27.Yang ZJ. Letter to the editor: measurement of autophagy-related proteins by immunohistochemistry/tissue microarray to characterize autophagy: problems and considerations. Am J Physiol Gastrointest Liver Physiol 2012; 302:G1356–G1357. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Fan H, Li X, Wu G, Zhao W, Zhang G, et al. Beclin 1 promotes apoptosis and decreases invasion by upregulating the expression of ECRG4 in A549 human lung adenocarcinoma cells. Mol Med Rep 2016; 14:355–360. [DOI] [PubMed] [Google Scholar]
- 29.Hartman KG, Bortner JD, Jr, Falk GW, Ginsberg GG, Jhala N, Yu J, et al. Modeling human gastrointestinal inflammatory diseases using microphysiological culture systems. Exp Biol Med (Maywood) 2014; 239:1108–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang BS, Liu YZ, Yang Y, Zhang Y, Hao JJ, Yang H, et al. Autophagy negatively regulates cancer cell proliferation via selectively targeting VPRBP. Clin Sci (Lond) 2013; 124:203–214. [DOI] [PubMed] [Google Scholar]
- 31.Chueca E, Apostolova N, Esplugues JV, García-González MA, Lanas Á, Piazuelo E. Proton pump inhibitors display antitumor effects in Barrett’s adenocarcinoma cells. Front Pharmacol 2016; 7:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu D, Yang F, He H, Hu J, Lv X, Ma D, Chen YY. Expression of TMEM166 protein in human normal and tumor tissues. Appl Immunohistochem Mol Morphol 2013; 21:543–552. [DOI] [PubMed] [Google Scholar]
- 33.Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol 2018; 20:1338–1348. [DOI] [PubMed] [Google Scholar]
- 34.Liu CY, Stücker I, Chen C, Goodman G, McHugh MK, D’Amelio AM, Jr, et al. Genome-wide gene-asbestos exposure interaction association study identifies a common susceptibility variant on 22q13.31 associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev 2015; 24:1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, Chen X, Cang S. Cancer-related long noncoding RNAs show aberrant expression profiles and competing endogenous RNA potential in esophageal adenocarcinoma. Oncol Lett 2019; 18:4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karner H, Webb CH, Carmona S, Liu Y, Lin B, Erhard M, et al. Functional conservation of LncRNA JPX despite sequence and structural divergence. J Mol Biol 2020; 432:283–300. [DOI] [PubMed] [Google Scholar]
- 37.Lin XQ, Huang ZM, Chen X, Wu F, Wu W. XIST induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med J 2018; 59:816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong J, Jin S, Pan X, Wang G, Ye L, Tao H, et al. Identification of long non-coding RNAs for predicting prognosis among patients with thymoma. Clin Lab 2018; 64:1193–1198. [DOI] [PubMed] [Google Scholar]
- 39.Ji SQ, Zhang YX, Yang BH. UBR5 promotes cell proliferation and inhibits apoptosis in colon cancer by destablizing P21. Pharmazie 2017; 72:408–413. [DOI] [PubMed] [Google Scholar]
- 40.Tasaki T, Kim ST, Zakrzewska A, Lee BE, Kang MJ, Yoo YD, et al. UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc Natl Acad Sci U S A 2013; 110:3800–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Gu T, Lu Z, Qiu L, Xiao G, Zhu X, et al. Roles of MYC-targeting long non-coding RNA MINCR in cell cycle regulation and apoptosis in non-small cell lung Cancer. Respir Res 2019; 20:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang W, Su X, Yan W, Kong Z, Wang D, Huang Y, et al. Overexpression of AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate 2018; 78:1248–1261. [DOI] [PubMed] [Google Scholar]
- 43.D’Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int 2019; 43:582–592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.