Abstract
For Drosophila melanogaster flies, sexual fate is determined by the X chromosome number. The basic helix-loop-helix protein product of the X-linked sisterlessB (sisB or scute) gene is a key indicator of the X dose and functions to activate the switch gene Sex-lethal (Sxl) in female (XX), but not in male (XY), embryos. Zygotically expressed sisB and maternal daughterless (da) proteins are known to form heterodimers that bind E-box sites and activate transcription. We examined SISB-Da binding at Sxl by using footprinting and gel mobility shift assays and found that SISB-Da binds numerous clustered sites in the establishment promoter SxlPe. Surprisingly, most SISB-Da sites at SxlPe differ from the canonical CANNTG E-box motif. These noncanonical sites have 6-bp CA(G/C)CCG and 7-bp CA(G/C)CTTG cores and exhibit a range of binding affinities. We show that the noncanonical sites can mediate SISB-Da-activated transcription in cell culture. P-element transformation experiments show that these noncanonical sites are essential for SxlPe activity in embryos. Together with previous deletion analysis, the data suggest that the number, affinity, and position of SISB-Da sites may all be important for the operation of the SxlPe switch. Comparisons with other dose-sensitive promoters suggest that threshold responses to diverse biological signals have common molecular mechanisms, with important variations tailored to suit particular functional requirements.
Cell fate determinations often depend on the ability to recognize and respond to subtle differences in the concentrations of regulatory proteins. The quantitative nature of the problem is particularly clear in Drosophila melanogaster sex determination, for which a twofold difference in the collective concentration of four X-linked gene products ultimately signals sexual fate. Central to the sex determination mechanism are several proteins of the basic helix-loop-helix (bHLH) family. bHLH proteins play important roles in a variety of cellular process, including cell proliferation, blood and muscle development, and neurogenesis (37, 39). These proteins share a bipartite DNA binding and dimerization motif, which consists of a basic α-helix that mediates sequence-specific binding to the consensus E-box sequence CANNTG, and two amphipathic α-helices, which are separated by a loop of variable length, controlling protein dimerization. Although DNA binding by homodimers is not uncommon, most bHLH proteins appear to function as heterodimers. Based on their evolutionary, structural, and DNA-binding characteristics, most bHLH proteins can be grouped into two classes (1, 2). Class A bHLH proteins, including the MyoD family, E12/E47, and the achaete-scute-related proteins, favor E-box sites containing a central GC pair (CAGCTG), while class B proteins, including Myc, Max, and the Hairy-related proteins, prefer E boxes with a central CG pair (CACGTG) (5, 6, 16, 40).
In Drosophila, the class A bHLH protein encoded by the X-linked sisterlessB gene (sisB, also called scute or T4) participates in several highly dose-sensitive transcriptional processes. During sex determination, SISB functions as a direct indicator of X-chromosome number and, along with SISA, is largely responsible for the ability of the fly to discriminate between the XY male and XX female signals (13, 20, 42, 50). During dorsal-ventral fate determination, SISB, and its close relatives in the achaete-scute complex, function to interpret the axis-defining gradient of the dorsal morphogen (23). Later, during neurogenesis, the concentration of SISB (as Scute) determines the capacity of cells to form neural precursors (31, 32). During all these processes, SISB interacts with the ubiquitous product of the daughterless (da) gene to form DNA binding heterodimers (B/Da) that activate the appropriate target genes (8, 15, 17).
The target of the X-chromosome sex determination signal is the regulatory switch gene Sex-lethal (Sxl). Sxl lies at the top of the sex determination and dosage compensation hierarchies and ultimately controls all aspects of somatic sexual development (reviewed in reference 14). In precellular female (XX) embryos, the diplo-X dose of the sisA, sisB, sisC, and runt gene products activates the establishment promoter SxlPe, creating a pulse of Sxl mRNA and protein synthesis that initiates the female developmental program (12, 13, 18, 35, 36, 46). In male (XY) embryos, the haplo-X dose of the four X-counting genes generates too few sisterless and runt proteins to activate SxlPe, and male development follows by default.
While sisB and the other X-linked elements are the true determinants of X-chromosome dose, their action on SxlPe requires a variety of other proteins. These include Da, the maternally supplied dimerization partner of SISB, as well as the positively acting hermaphrodite (her) and Stat92E gene products (13, 14, 34, 45, 46). In addition to these maternal activators, several maternally or zygotically expressed negative regulators, including Groucho, Emc, and Deadpan (Dpn), are needed for proper sex-specific regulation (3, 43, 53).
With the exception of sisC, which encodes a ligand for the JAK-STAT pathway (46), all of the sex signal elements encode transcription factors. SISB and Da, as well as the negative regulator Dpn, are bHLH proteins, while SISA, Her, Stat92E, and Runt are members of the basic-leucine zipper, Zn finger, STAT, and Runt domain families, respectively. How does this diverse collection of transcription factors mediate the on-or-off regulation of SxlPe in response to a twofold difference in SIS and Runt concentrations? While all available evidence suggests that the action of these proteins on SxlPe is direct, little is known about the specific sequences that mediate sex signal element binding. Given the central importance of SISB and Da to sex determination and other dose-sensitive processes, a logical place to begin is with the interactions between B/Da and SxlPe.
Because B/Da is a prototypical bHLH molecule known to bind E boxes in vivo and in vitro, a straightforward prediction would be that the distribution of E-box sites at SxlPe would be well correlated with the major enhancer activities. In fact, only 3 of the 13 class A E-box sequences present in the 3.7-kb region upstream of SxlPe map to regions known to be important for promoter function, and only one maps in the minimal 400-bp segment needed for sex-specific expression (21). We have resolved this paradox in the work reported here. We show that the predominant binding sites for B/Da in the critical region of SxlPe are noncanonical and that many are of relatively low affinity. We show these non-E-box sites play an essential role in Sxl activation, thus providing the first direct evidence that non-E-box sites play important roles in the in vivo functions of class A bHLH proteins. Furthermore, our analysis of binding site affinities along with the deletion analysis of Estes et al. (21) suggests that threshold binding to proximal low-affinity B/Da sites may be critical for the sex-specific response of SxlPe. We propose that once SxlPe is active, higher-affinity distal sites amplify the response, producing the high-level Sxl expression needed to initiate female development.
MATERIALS AND METHODS
Plasmids.
The His6-Da plasmid was based on pRSET-B and encoded C-terminal amino acids 362 to 710. The glutathione S-transferase (GST)-SISB and His6-SISB plasmids carried the C-terminal portion of sisB (residues 82 to 345) in pGEX-2TK and pRSET-B, respectively. Most promoter fusions were derived from an Sxl segment extending from −1.45 kb to +44 bp. This region was PCR amplified from a sequenced genomic plasmid clone with 5′ primer cgaattcgatATCTCTTTTCGCAGCTTCGTA and 3′ primer ggggtaccCAAGATCTCTGAACACAAGTTG, cut with BglII and EcoRI, and inserted into BamHI and EcoRI sites of pGEM-11zf(+) to create pG01 (underlined nucleotides are restriction sites, and lowercase nucleotides are nontemplate bases). Repair-proficient DNA polymerase was used for all PCR experiments. SxlPe-Fluc deletion reporters were made by cloning PCR-amplified SxlPe DNA into SmaI and HindIII cut pGL3-Basic (Promega). Forward primers began at the indicated positions, and the common backward primer extended from −9 to +6 and carried a HindIII site at its 5′ end. Construct −94 bp was made using a forward primer containing 15-base extension GAAAGATCTGAATTC abutting nucleotides −95 to −78 and the common backward primer. The 4X-B/Da site reporters have four copies of the following sequences between the XhoI and EcoRI sites of the −95 bp SxlPe-Fluc plasmid: TGCAGCCGGCA, CGCACCTTGCC; and AACATCTGCCT (underlined nucleotides are B/Da sites). Cell culture sisB and da expression plasmids carried the entire coding regions plus a Kozak sequence in pAct5CPPA (27). Point mutant plasmids were derived from pG01 by site-directed oligonucleotide mutagenesis. All mutations were confirmed by sequencing. The changes were as follows: site 1, GCgaCTTGC; site 2, GCtaGCGGG, site 4, TCtCgagG; sites 5 and 6, GatGCTTGCG CAttaTGCCACGTTCatCC (underlined nucleotides are B/Da sites, and lowercase nucleotides are mismatches). For P-element transformation, the 1.5-kb EcoRI to NotI fragments from pG01 and its [1− 2−], [1− 4−], [1− 5− 6−], [2− 5− 6−], and [4− 5− 6−] derivatives were cloned into the P-element vector pCaSpeR-AUG-βgal with a modified polylinker. Upstream sequences were removed as a 1.1-kb EcoRI to SwaI fragment to create the −390 to +44 bp SxlPe-lacZ transformation vectors. P-element vectors carrying the site 1−, 3−, 4−, and [5− 6−] mutations were made by cloning DraI to BglII (−390 to +44 bp) SxlPe restriction fragments into pCaSpeR-AUG-βgal. The mutated sequences were as follows: site 1, GggCCcTGC; site 3, ACgTCgaC; site 4, TtACgTagC; sites 5 and 6, GgAGCTCGC GaAtaTTGCCgACGTcCCA.
Protein expression and purification.
To produce GST-SISB protein, BL21(DE3) cells carrying the expression vector were grown in Luria broth at 21°C to an optical density at 600 nm of 0.3 and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 to 3 h. Cell pellets were suspended in a 1/40 culture volume of 20 mM HEPES–0.6 M NaCl–0.5 mM EDTA–1% (vol/vol) NP-40–2 mM dithiothreitol (DTT) (pH 7.9) and were sonicated. After a 10-min centrifugation at 10,000 × g, the supernatant was diluted with 1 volume of 20 mM HEPES (pH 7.9), and GST-SISB was purified to homogeneity using glutathione-agarose beads. For His6-tagged proteins, similar procedures were followed except that the lysis buffer contained 1.0 M NaCl and no EDTA or DTT, and a Ni2+-nitrilotriacetic acid affinity resin was used.
DNase I footprinting and electrophoretic mobility shifts.
Binding reaction mixtures (20 μl) contained 15 mM HEPES, 200 mM KCl, 1 mM EDTA, 5 mM DTT, 7.5% (vol/vol) glycerol, 0.1% (vol/vol) NP-40, 1 μg of poly(dI)-poly(dC), 5 μg of bovine serum albumin (BSA) (pH 7.9), and the indicated amounts of premixed SISB and Da proteins. One B/Da unit equaled 0.3 pmol each of GST-SISB (15 ng) and His6-Da (12 ng) proteins. In some experiments with the proximal promoter, His6-SISB was used instead of GST-SISB, with indistinguishable results. For DNase I footprinting, probes were made by PCR amplification with one 32P-end-labeled primer and were gel purified. Between 104 and 105 cpm of probe was incubated with or without B/Da as previously described for electrophoretic mobility shift assays (EMSA). After 30 min at 21°C, 0.05 U of DNase I (Epicentre) was added. Reactions were stopped after 2 min by addition of 80 μl of 0.1 M EDTA–1.0 M NaCl. Samples were phenol-CHCl3 extracted, ethanol precipitated, and dissolved in 80% formamide–0.01 N NaOH–1 mM EDTA, heated to 90°C for 5 min, and loaded on 6% polyacrylamide–8 M urea gels. MspI-cut 32P-labeled pBR322 fragments served as size standards. DNAs shown in Fig. 1 extended (from top to bottom) from −749 to −1013, −229 to −373, and −229 to −16. For EMSA, reaction mixtures were preincubated at 21°C for 10 min before addition of ∼5× 104 cpm of 32P-labeled probe. For competition experiments, unlabeled probes were added immediately after labeled probes. After 30 min, samples were electrophoresed on prerun 0.25× Tris-borate-EDTA–4% polyacrylamide gels at 21°C. Double-stranded probes for gel shifts were prepared from oligonucleotides carrying four extra 5′ bases (GATC) Annealed oligonucleotides were 5′ end labeled with polynucleotide kinase, and then the 5′ overhangs were filled in using Klenow and unlabeled deoxynucleoside triphosphates. Competitor oligonucleotides were filled in but not labeled. Probes used for the experiments in Table 2 but not shown in Table 1 extended from positions −932 to −911, −691 to −673, −613 to −593, −330 to −313, −321 to −291, −271 to −252, −190 to −169, −177 to −150, −131 to −99, −83 to −69, and −70 to −43.
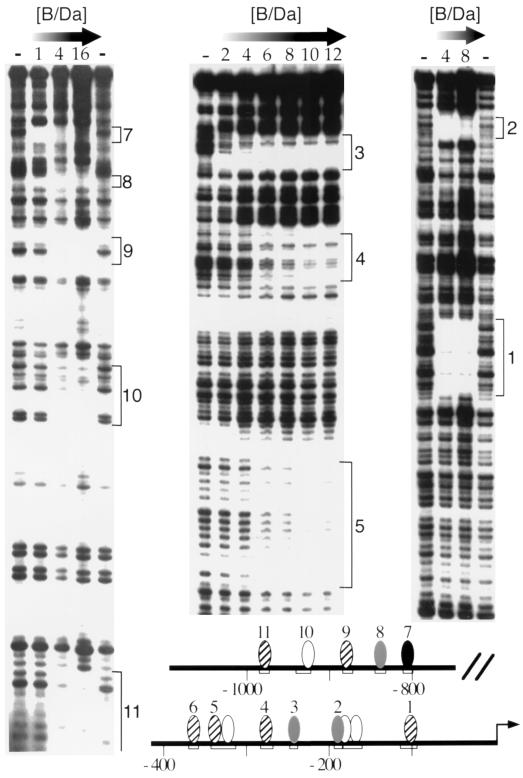
FIG. 1.
B/Da heterodimers bind multiple sites in Sxlpe. DNase I footprinting assays were done at increasing concentrations of GST-tagged SISB and His6-tagged Da proteins. Numbers indicate B/Da footprinting units. One unit equaled 0.3 pmol (15 nM) of each protein. Panels (left to right) illustrate protection from the distal to the proximal region within the 1.4-kb Sxlpe promoter. Site 6 is not shown. Locations of the 11 regions protected by B/Da are summarized in the schematic. Shading represents estimated relative B/Da binding affinity. High-affinity site 7 is black, moderate-affinity sites are gray, low-affinity sites are striped, and putative weak-binding sites are white. Protection of putative sites 2a and 2b was apparent only at 540 nM B/Da (not shown).
TABLE 2.
Naturally occurring nonbinding sequence variants
| Site | Sequencea | Location (bottom strand) |
|---|---|---|
| 6-bp non-E box | ||
| Consensus | CASCCG | |
| Nonbinding | TACCCG | 831, (−381) |
| CASGCG | (−165)b, −121, −110b | |
| CASTCG | (−112), −60 | |
| CACCGG | −305 | |
| CASCCA | (−1339), −309, −184b, −75 | |
| 7-bp non-E box | ||
| Consensus | CASCTTG | |
| Nonbinding | CTGCTTG | −1281 |
| CAACTTG | +32 | |
| CACTTTG | (−1325), −1071 | |
| CAGATTG | (−923)b, −508 | |
| CAGCTTC | −265 | |
| CACCTTT | −326b | |
| E box | ||
| Consensus | CABCTG | |
| Nonbinding | TACCTG | −82 |
| AATCTG | −928, −853, (−503) | |
| CAATTG | −802b, −685 | |
| CATTTG | (−600) | |
| CAGCTC | −1364 | |
| CACCTA | −642 | |
| CATCTT | (−403) | |
| CACCTTt | −326b | |
| CAGCTTc | −265 |
S = G or C; B = G, C, or T.
Maps near footprinted area (no binding in EMSA).
TABLE 1.
Wild-type and mutant B/Da binding sites tested by EMSAa
| Binding sites | Sequence (S = G or C) | EMSA result |
|---|---|---|
| 6-bp non-E-box sites | ||
| Consensus | CASCCG | |
| 2 | GAAATGCAGCCGGCCACC | + |
| 2m1 | GAAATCCAGCCGGCCACC | + |
| 2m2 | GAAAGCCAGCCGTCCACC | + |
| 2m3 | GAAATGCGGCCGGCCACC | − |
| 2m4 | GAAATGCTACCGGCCACC | − |
| 9 | TTTTTCCACCCGGCGTTT | + |
| 9m1 | TTTTTCCACCCGGCGTTT | − |
| 9m2 | TTTTTCCACCCAGCGTTT | − |
| 7-bp non-6-box sites | ||
| Consensus | CASCTTG | |
| 1 | TCGCGCACCTTGCCTCC | + |
| 1m1 | TCGCGGAGCTTGCCTCC | − |
| 1m2 | TCGCGCGACTTGCCTCC | − |
| 4 | AACTATCACCTTGCCG | + |
| 4m1 | AACTATCTCGAGGCCG | − |
| 11 | AACCAAACACCTTGACTGTCTT | + |
| 5 | AACATGCAGCTTGCCACGTTCCAC | + |
| 5m | AACATGCTTATTGCCACGTTCCAC | − |
| 6 | AAAATGCAGCTTGCTTCC | + |
| 6m1 | AAAATGCAGCTTCCTTCC | − |
| 6m2 | AAAATGATGCTTGCTTCC | − |
Sequences of oligonucleotides used in electrophoretic gel mobility shift assays are shown, and their B/Da binding capabilities are indicated as + (binding) or − (nonbinding). Oligonucleotides carried an added GATC at each end. Underlined nucleotides indicate mutations.
Cell culture, P-element transformation, and β-galactosidase staining.
Cultivation, transfection, and assay of Schneider L2 cells were performed according to procedures described by Han et al. (27). One microgram of DNA was used per plate and included 0.1 μg of Fluc reporter, 0.05 μg each of sisB and/or da expression constructs, 0.1 μg of simian virus 40 (SV40) or cytomegalovirus (CMV)-Renilla luciferase reporters to control for transfection efficiency (pRL-SV40 and pRL-CMV; Promega), and carrier DNA. Luciferase activity was determined using a Dual-Luciferase assay kit (Promega) and a Berthold Lumat LB9501 luminometer. P-element transformants were obtained from w1118 flies with a heterozygous Δ2-3 transposase source. Four- to seven-hour-old embryos were stained for β-galactosidase activity as described previously (21). Transcription of wild-type −390-bp SxlPe-lacZ fusions occurs during the normal precellular period, as measured by the production of nascent lacZ nuclear transcripts (19); however, there was a lag in the accumulation of active enzyme, necessitating a later assay.
RESULTS
DNase I footprinting of B/Da binding sites in SxlPe.
Deletion analysis has established that a 1.4-kb region upstream of SxlPe is sufficient to drive uniform high-level female-specific expression of SxlPe-lacZ fusion in a B/Da-dependent manner (21). To identify the B/Da binding sites in SxlPe, we expressed soluble GST-tagged SISB and His6-tagged Da fusion proteins in Escherichia coli and used the purified proteins to systematically footprint the 1.4-kb proximal promoter. In total, we observed 11 protected regions distributed in two clusters. Regions 1 to 6 were located in the proximal 390 bp while regions 7 to 11 mapped between −0.8 and −1.1 kb (Fig. 1). Protected regions 3, 7, and 8 were centered on three type A E boxes, but the sequences in the other eight protected areas lack even the minimal CANNTG E-box consensus (Fig. 2), suggesting that B/Da can bind to non-E-box sites at SxlPe.
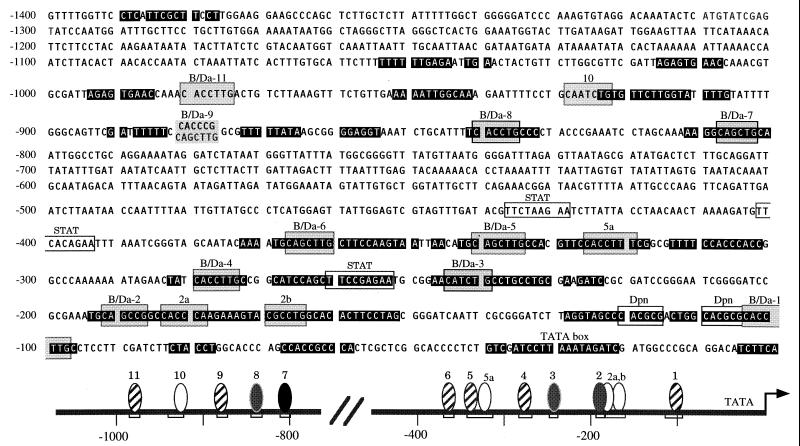
FIG. 2.
Sequence of proximal 1.4-kb Sxlpe. The DNA sequence from −1400 to +1 is shown. Sequence blocks that are identical in D. melanogaster and D. subobscura are shown in white in black boxes. B/Da sites 1 to 9 and 11 are identified by name and number with a shaded block marking each core binding site. The sequence of D. melanogaster B/Da site 9 is shown above the corresponding D. subobscura sequence. Putative sites 2a, 2b, 5a, and 10 are identified by number. Shaded blocks mark the closest matches to the B/Da consensus. The locations of the tandem Dpn binding sites (28), putative STAT sites (34, 46), and TATA box are marked. Schematic is like that in Fig. 1. The numbering scheme differs from those previously published, as in vitro expression and evolutionary analysis place the +1 position 6 bp downstream of that proposed by Keyes et al. (35).
The 11 B/Da sites exhibited a range of apparent binding affinities. Site 7 was protected at lower B/Da concentrations than the other sites, while sites 2, 3, and 8 exhibited intermediate and roughly equivalent affinities (Fig. 1 and data not shown). Although the data varied subtly between experiments, occupancy of sites 1, 4, 5, 6, 9, 10 and 11 generally required higher concentrations of B/Da protein, suggesting that they are of the lowest relative affinity.
For 9 of the 11 protected regions, the size of the footprint (∼10 to 15 bp) was consistent with the binding of a single B/Da heterodimer. For sites 2 and 5, however, a considerably larger region was protected. In the case of site 5, the footprint was approximately 30 bp (Fig. 1). The region 2 footprint was consistent with the presence of a single binding site at low B/Da concentrations (Fig. 1), but the protected region expanded proximally at the highest concentration tested (36 U; data not shown). As discussed below, we were unable to identify more than one binding sequence in either region 2 or 5 and any specific sequence in region 10; accordingly, the hypothetical B/Da sites 2a, 2b, 5a, and 10 are indicated in the schematics as white ovals centered on the closest matches to the binding consensus (Fig. 1 and 2).
CA(G/C)CTTG and CA(G/C)CCG are B/Da binding core sites.
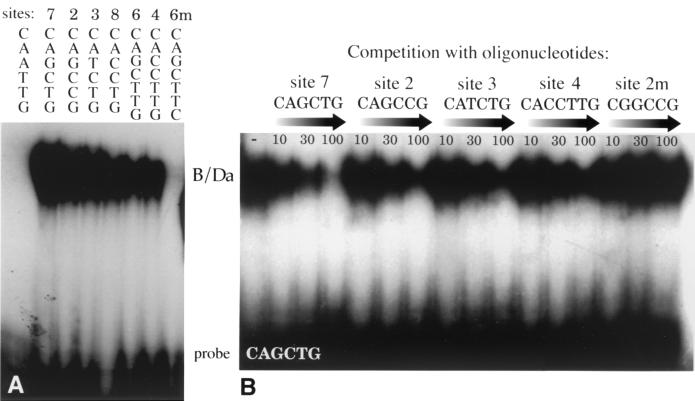
To identify the binding core sequences in the B/Da protected regions, we performed gel mobility shift assays using overlapping oligonucleotide probes from each protected segment. We found that probes from windows 1, 4, and 11 carrying the common sequence CACCTTG were efficiently bound by B/Da in gel mobility shift experiments but that mutations within the common sequence prevented B/Da binding (Fig. 3; Table 1). Windows 5 and 6 carry a common sequence, CAGCTTG, that has previously been suggested as a possible B/Da binding site (28). We found that wild-type, but not mutant, region 5 and 6 oligonucleotides were bound by B/Da in gel mobility shift assays (Fig. 3; Table 1). Taken together, the footprinting and gel mobility shift data suggest that B/Da can bind to a 7-bp core sequence, CA(G/C)CTTG, which is related to the CA(G/C)CTG E box, by the insertion of an internal T residue.
FIG. 3.
EMSA of B/Da DNA binding. (A) B/Da binding to Sxlpe oligonucleotides. B/Da samples (0.1 U) were incubated with 32P-labeled oligonucleotides containing a CAATTG E box or sites 7, 2, 3, 8, 6, 4, and 6m1 (Table 1), and the protein-DNA complexes were resolved on a gel. (B) Binding competition experiments. Complexes were formed between B/Da (0.1 U) and 32P-labeled site 7 and challenged with 10-, 30-, and 100-fold molar excesses of the indicated unlabeled competitor oligonucleotides (Table 1). The site 7 oligonucleotide sequence was GATCAAGGCAGCTGCTATTGGATC.
Sequences within window 2 are protected from DNase I digestion at B/Da concentrations similar to those needed to protect the E-box sites CA(T/C)CTG in windows 3 and 8 (Fig. 1 and data not shown). While window 2 contains neither an E box nor a 7-bp site, it does have the sequence CAGCCG, which differs from the E-box consensus by a single base (underlined). Window 9 carries a similar sequence, CACCCG, suggesting that these may be B/Da binding core sequences. To test these putative sites, we performed gel mobility shift assays. We found that B/Da bound to both wild-type sites and that single-base changes in the core, but not the flanking sequences, prevented binding (Fig. 3A; Table 1).
The assignments of CA(G/C)CTTG and CA(G/C)CTG as specific B/Da binding sites was strengthened by an analysis of similar, but nonbinding, sequences in SxlPe. Altogether, eight non-E-box sequences differing from the 7-bp consensus and 13 differing from the 6-bp consensus by single-base changes were examined in footprinting or gel mobility shift assays (Table 2). Sixteen of these mismatch sequences mapped to areas that were clearly left unprotected in footprinting experiments, indicating that they were not recognized by B/Da. Five mismatch sequences mapped near or within DNase I-protected regions; however, none of these were bound by B/Da in gel mobility shift experiments. This suggests either that they do not form stable complexes under our experimental conditions or that they are not capable of binding independent of other sequence elements.
As expected, oligonucleotides containing the E boxes present in windows 3, 7, and 8 were bound by B/Da in gel retardation assays (Fig. 3A). However, the three other E boxes (CAATTG and CATTTG) in the 1.4-kb promoter were not bound in either assay (Fig. 3A; Table 2).
To ensure that our gel mobility shift results reflected binding by B/Da heterodimers, we also examined the ability of GST-SISB and His6-Da homodimers to bind SxlPe fragments. We found that the individual proteins bound CA(G/C/T)CTG E boxes and that GST-SISB also bound CAGCCG; however, the binding was considerably weaker than that observed with the B/Da heterodimer. Importantly, only heterodimeric DNA binding complexes were detectable in gel mobility shift assays containing both proteins, indicating that B/Da was the predominant active species (data not shown).
Relative affinities of consensus and nonconsensus B/Da binding sites.
To test further the relative affinity of the B/Da sites in SxlPe, we performed binding competition experiments using the gel mobility shift assay. The B/Da heterodimer was incubated with a radiolabeled probe carrying the symmetrical site 7 E box CAGCTG, and increasing concentrations of unlabeled oligonucleotides were added to compete for binding. We found that B/Da could be competed off the consensus site by sites 2, 3, 4, and 7, but not by a mutant site 2 sequence (Fig. 3B and data not shown). The site 7 E box was the strongest binding site, but the nonconsensus site 2 and E-box site 3 oligonucleotides were also reasonably effective competitors. Site 4 CACCTTG was least effective in competing for B/Da binding, consistent with the relative affinities observed in DNase I footprinting experiments (Fig. 1 and data not shown). Based on the relative binding affinities observed in the sum of our footprinting and gel mobility shift experiments, we classify the B/Da sites as being of relatively high (site 7), moderate (sites 2, 3, and 8), or low (sites 1, 4, 5, 6, 9, and 11) DNA binding affinity.
Site distribution and evolutionary conservation of SxlPe structure.
The deletion analysis of Estes et al. (21) suggested that two subsegments within the 1.4-kb promoter account for most SxlPe enhancer activity. An upstream segment (−1.4 to −0.8 kb) contributes to the strength of the promoter but is not essential for sex specificity. A proximal segment, including the start site and 390 upstream base pairs, drives a low-level, nonuniform female-specific expression. The sequence conservation between SxlPe in D. melanogaster and Drosophila subobscura (44) correlates well with the functional analysis (Fig. 2). There is extensive sequence identity in the proximal region, with more limited matches in the distal segment, and no detectable similarity in the similarly sized central spacer segment. Within the proximal 390 bp, the sequences of all six B/Da binding sites are perfectly conserved. In the distal region, E-box sites 7 and 8 are conserved. Interestingly, while the sequence of site 9 is not conserved, another low-affinity B/Da site, CAGCTTG, is present in the equivalent position in D. subobscura (Fig. 2).
Nonconsensus sites can support B/Da-activated transcription in cultured cells.
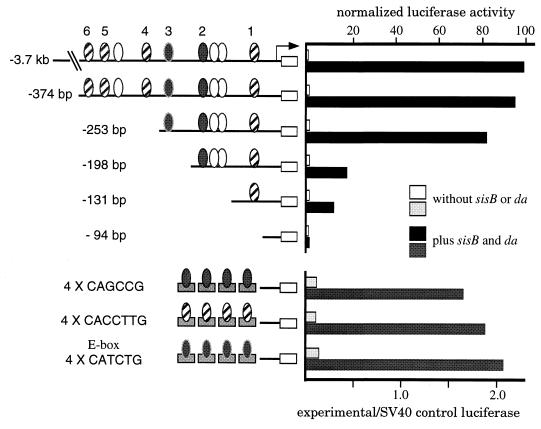
To determine whether the binding sites we identified in vitro can be recognized in vivo, we asked if multimerized sites could support B/Da-mediated transcription in Drosophila Schneider L2 cells. Full-length SISB and Da were expressed under control of the Actin5C promoter and the expression plasmids were introduced into cells along with firefly luciferase (Fluc) reporter plasmids carrying four tandem copies of various B/Da binding sites fused to an otherwise inactive promoter. We found that four copies of the noncanonical CACCTTG or CAGCCG core supported levels of B/Da-activated transcription indistinguishable from those produced by four copies of the canonical site 3 E-box CATCTG (Fig. 4, bottom). It has been shown previously that multimerized fragments containing the sequence CAGCTTG can support B/Da-activated transcription in Kc cells (28), suggesting that this noncanonical core sequence is also recognized by B/Da in vivo.
FIG. 4.
SISB and Da can activate transcription through noncanonical sites in SL2 cell culture. (Top) Activity of a Sxlpe deletion series. Schematics show the structures of the Sxlpe-firefly luciferase plasmids. Only B/Da binding sites in the proximal promoter are shown. The Sxlpe-Fluc plasmids were introduced with full-length sisB and da expression vectors and a CMV-Renilla luciferase (Rluc) plasmid to normalize for transfection efficiency. Normalized F-luc activities are shown in comparison to the 100% active −3.7 kb Sxlpe fusion. Data are the averages of duplicates differing by ≤10%. (Bottom) Activity of multimerized B/Da binding sites. Four copies each (top to bottom) of the site 2, sites 1, 4, and 11, and site 3 E-box core sequences were joined to the −95 bp Sxlpe-Fluc plasmid. The 4 × B/Da site-Fluc plasmids were cotransfected with sisB and da expression vectors and an SV40-Rluc control. Data are the average ratios of experimental Fluc to control Rluc activities. Neither sisB nor da expression vectors alone significantly stimulated Sxlpe or the multimerized site constructs (not shown).
We also addressed the functional importance of the B/Da sites by analyzing a series of SxlPe deletions in L2 cells (Fig. 4, top). We found that sequences in the upstream promoter region had little effect on Pe activity, as a construct with deletion of all upstream elements (−373) behaved similarly to the 3.7-kb promoter that served as the wild-type standard. A further deletion (−253) leaving intact the proximal site 3 E box showed a modest, but reproducible, reduction in Pe activity. The reduction implies that low-affinity sites upstream of the E box can function in the normal sequence context, a conclusion also reached by Hoshijima et al. (28). Deletions extending past the proximal E box caused severe decreases in activity, suggesting that it mediates most of the B/Da-dependent activation observed in cell culture. Several smaller fragments, including the −131 deletion, also responded to B/Da, implying that the CACCTTG sequence in site 1 can be bound in this context (Fig. 4 and data not shown). Deletion to position −95 abolished B/Da-activated transcription.
Mutational analysis of B/Da sites in P-element transgenes.
To determine if the sex-specific response of SxlPe to X-chromosome dose in flies depends on the B/Da sites we identified in vitro, we engineered a variety of inactivating point mutations in the proximal consensus and nonconsensus B/Da binding sites and analyzed their effects in P-element-transformed embryos. We examined the effect of the mutations in the context of the minimal −390 bp promoter because such constructs are expressed sex specifically and are extremely sensitive to changes in the dose of the X-counting elements (21). Accordingly, wild-type and mutant SxlPe promoters were inserted into the P-element transformation vector pCaSpeR-AUG-βgal and injected into flies to create mutant −390 bp SxlPe-lacZ transgenes for in vivo analysis.
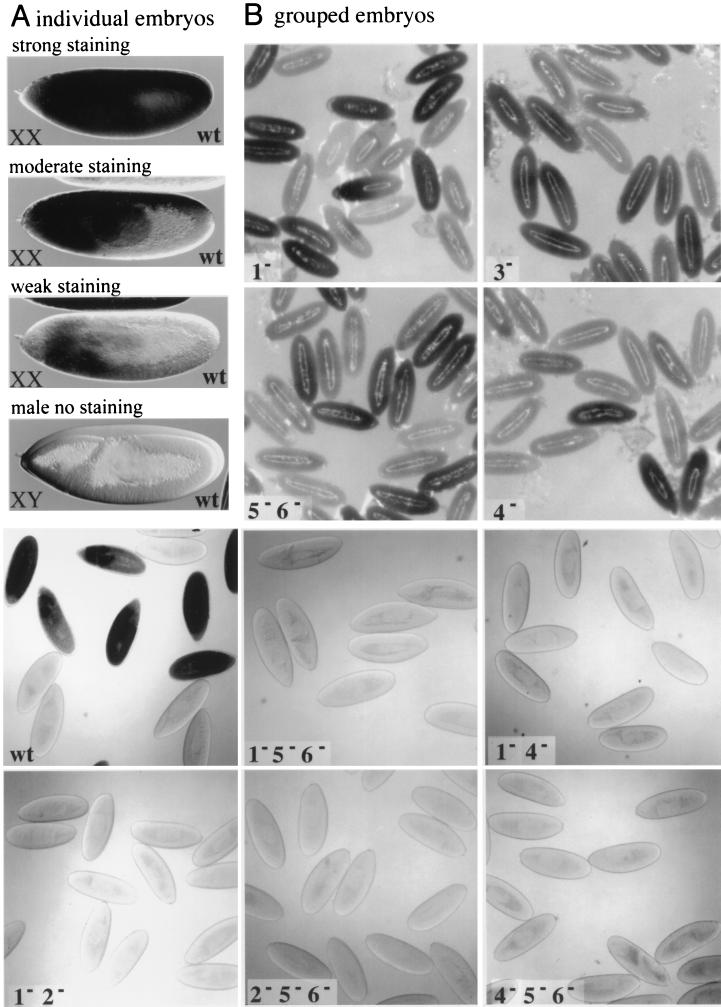
We found that all nine of our wild-type SxlPe-lacZ fusion lines were expressed specifically in females but that promoter activity varied considerably between different transgene lines and even between different females within the same line (Fig. 5A) (also see reference 21). In general, lacZ was most highly expressed in the anterior portions of the embryos, but most lines expressed detectable β-galactosidase activity in all somatic tissues of at least some females. Transgenes carrying mutations in site 1 alone (six lines) or in sites 5, 5a, and 6 (three lines) were indistinguishable from the wild-type lines, indicating that these changes had little effect on SxlPe function (Fig. 5B). Mutations in the site 3 E box (four lines), or in site 4 alone (five lines), appeared to cause modest reductions in lacZ expression. In both cases, this was manifest as a small reduction in the proportion of stained embryos as well as in the intensity with which individual embryos stained (Fig. 5B and data not shown). Variation within the wild-type and mutant lines made precise comparisons impossible, but in general, the effect of the site 4 mutation appeared more severe than that of the E-box change.
FIG. 5.
Noncanonical B/Da sites are needed for Sxlpe expression in P-element-transformed embryos. (A) Female and male embryos carrying wild-type −390 bp Sxlpe-lacZ fusions. Representative 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-stained XX embryos from strongly, moderately, and weakly expressing transformant lines are shown. XY embryos do not express Sxlpe. (B) Wild-type (wt) and mutant Sxlpe-lacZ transgenes stained for β-galactosidase activity. Numbers refer to the B/Da site mutations carried by the transgenes. None of the [1− 5− 6−], [2− 5− 6−], [4− 5− 6−], [1− 2−], or [1− 4−] lines expressed detectable β-galactosidase activity. Other mutant and wild-type lines exhibited a range of expression levels, but all were active. Representatives of moderate strength lines are shown.
Considering that the B/Da sites in the proximal promoter may be functionally redundant, we examined several different mutant site combinations for their effects on Pe. In contrast to the modest effects of the single-site B/Da site mutations, the five mutant site combinations we tested abolished expression of the SxlPe-lacZ transgenes in vivo (Fig. 5B). We examined three independent insertions each for the [2− 5− 6−] and [4− 5− 6−] constructs, four insertions of the [1− 2−] and [1− 4−] constructs, and two insertions of the [1− 5− 6−] transgenes, with identical results. None of the lines exhibited any β-galactosidase activity, even after prolonged staining, suggesting that they were all nonfunctional. To test this conclusion further, we combined two independent insertions of the [2− 5− 6−] mutants and two of the [4− 5− 6−] mutants to see if increasing the mutant transgene copy number from two to four would produce detectable lacZ expression. Even with four copies, the [2− 5− 6−] and [4− 5− 6−] transgenes were inactive (data not shown), confirming that mutations in these noncanonical bHLH binding sites eliminated SxlPe activity.
The P-element transformation data indicate that the noncanonical B/Da sites we identified in vitro are important for the female-specific activation of SxlPe in the embryo. While no individual site mutation prevented SxlPe expression, several combinations containing mutations in low-affinity, or in low- and moderate-affinity, noncanonical sites blocked promoter activity. While SxlPe activity appears to require a minimum number of functional B/Da sites, activity appears not to be a simple function of the number or the affinity of those sites, suggesting that the location of the binding sites and their precise promoter context may be critical for Pe function.
DISCUSSION
Successful dissection of a transcriptional response requires a detailed analysis of the cis elements at promoter targets. A weakness of the Drosophila sex determination system is that the analysis of the Sxl establishment promoter has lagged behind the discovery of its regulators. The identities of most of the important proteins are known, yet with few exceptions, their binding sites at SxlPe are not. In this study, we present evidence that the bHLH heterodimer formed by the sex signal elements SISB and Da activates transcription of Sxl by binding to numerous noncanonical binding sites in SxlPe. Using P-element transformation, we have shown that the noncanonical B/Da sites are required for the activation of SxlPe in embryos. These results suggest that SxlPe utilizes cis elements with different locations and binding affinities to sense the twofold male-female difference in collective SIS protein concentration.
Noncanonical bHLH binding sites.
As measured by footprinting and gel mobility shift assays, the B/Da protein binds to canonical CA(G/C/T)CTG E boxes as well as to noncanonical sites with 6-bp CA(G/C)CCG and 7-bp CA(G/C)CTTG core sequences. These noncanonical sequences can be bound directly by B/Da in vivo, as evidenced by their ability to drive B/Da-dependent transcription from multimerized sites and from truncated versions of SxlPe in cell culture assays. While examples have been found of bHLH proteins that prefer non-E-box sites, such as the Hairy-type repressors (40, 51) and the class C bHLH-PAS proteins (47), the overwhelming majority of bHLH molecules are thought to bind with a strong, if not exclusive, preference for E-box sequences (37, 39). Although so far there have been no other reports of class A proteins binding to non-E-box sites in vivo, there is evidence that noncanonical sequences may be important target sites for the class B Myc-Max heterodimer (24, 25). While the highest-affinity Myc-Max core site is CACGTG, random site selection experiments have demonstrated that the protein also binds efficiently to a number of noncanonical sequences, including CAYGCG, CACGAG, and CACGTTG (4). The importance of this relaxed DNA binding specificity was highlighted by the surprising finding that non-E-box sequences accounted for the majority of genomic Myc-Max sites in immunopurified chromatin (24, 25). Interestingly, if one accounts for the class B preference for CG in the central positions, the 6-bp CANNCG and 7-bp CANNTTG sequences bound by Myc and Max are identical to those we identified as the predominant B/Da sites at SxlPe. This striking similarity in noncanonical DNA sequence recognition by long-diverged representatives of the major bHLH classes (2) suggests that many, if not all, subgroups of bHLH proteins will be found to regulate gene expression through noncanonical sequence elements.
On the other hand, while SISB and its close relatives are considered typical class A molecules, they differ from nearly every other bHLH protein in lacking the usual Arg or Lys residues at the first two positions of the DNA recognition helix (37). If B/Da DNA binding specificity depends on these atypical residues, our findings may be applicable only to achaete-scute family heterodimers. However, even if limited to this family, our results have predictive value for the regulation of other promoters. Indeed, the presence of noncanonical sites in the twist and achaete (ac) promoters may account for two previously paradoxical results. First, the twist promoter lacks E boxes, but expression still depends on synergy between Dorsal and one or more AS-C/Da heterodimers (23). Second, ac-lacZ fusions are still partially regulated by the AS-C even when all the ac E boxes have been mutated (38). Two CAGCCG sequences are present in the D. melanogaster and Drosophila virilis twist promoters (41), and three are in the ac promoter, the most proximal of which abuts the Hairy repressor site (40, 51). Binding of AS-C heterodimers to these noncanonical sites could explain these previously puzzling results. Furthermore, based on promoter structure similarities, our observations may be relevant to the regulation of mammalian homologues. The mammalian achaete-scute homolog MASH-1 promoter has two conserved CAGCCG sequences, one of which is adjacent to the binding site for the mammalian hairy protein HES-1 (11, 52). While speculative, the structural parallel with ac hints that MASH-1 may also be auto- or transregulated by achaete-scute family proteins. If so, the kinds of noncanonical sites we found at SxlPe may be important for a variety of Mash-regulated decisions in vertebrates.
Distribution of B/Da sites at SxlPe—implications for the X-chromosome counting mechanism.
A critical question for sex determination is as follows: how can SxlPe sense the twofold difference in male and female SIS and Runt concentrations and translate that into a strong all-or-nothing response? At some level, SxlPe expression must be related to sex-specific differences in binding site occupancy. This is true whether dose sensitivity arises from cooperative DNA binding, competition with negative regulators, or from the sum of multiple independent interactions between the sex signal elements and the transcription machinery (10).
Based on the deletion analysis of Estes et al. (21), it appears that sex-specific control of SxlPe occurs largely through the activity of two regulatory regions: a central segment located between 1.4 and −0.8 kb, responsible primarily for promoter strength, and a proximal element, −390 to +44 bp, largely responsible for sex specificity. While these regions appear most important, sequences beyond −1.4 kb also contribute to the promoter, as inferred from the stronger lacZ expression from larger promoter fusions and by the ability of upstream sequences to partially substitute for the loss of the central −1.4 to −0.8 kb region (21).
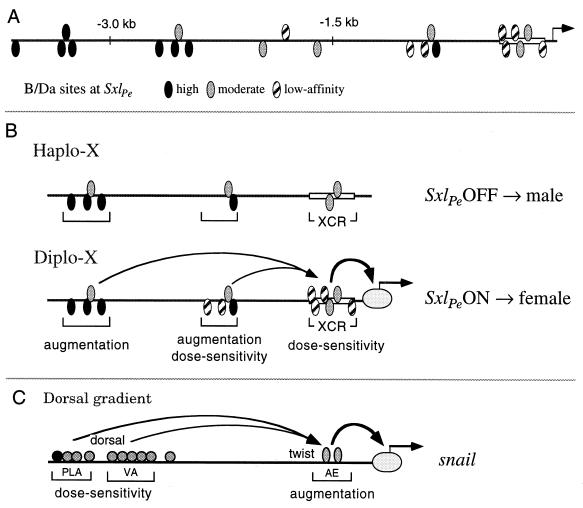
The 10 B/Da sites we identified in vitro are located in the central and proximal promoter elements. In addition, the sequence predicts 11 likely B/Da binding sites of high or moderate binding affinity located in the distal region between −1.6 and −3.7 kb, raising the possibility that there may be 21 or more B/Da sites in the functional SxlPe region (Fig. 6A). Given a 39% GC content, random sequence would predict only 2.7 matches to our B/Da consensus at SxlPe, suggesting that many of these predicted sites are functional binding sequences. Overall, there is a striking positional gradient of predicted binding affinities of the B/Da sites, with the moderate-affinity sites clustered proximally and the highest-affinity sites positioned distally (Fig. 6A). The asymmetric distribution of high- and moderate-affinity sites hints that the distal sites may be occupied at both high and low B/Da concentrations, with full occupancy of the proximal sites occurring only in XX embryos. This suggests a model in which the on or off response of SxlPe to X-chromosome dose occurs primarily within the proximal X-counting region (XCR), with the distal segments providing an augmentation function that enhances transcription only when the female-specific XCR complex forms (Fig. 6B). It is unlikely that the distal high-affinity sites titrate B/Da from the XCR in males, because B/Da is in enormous excess over the Sxl binding sites.
FIG. 6.
A model for Sxlpe function. (A) Distribution of identified and predicted B/Da binding sites from −3.7 kb to +1 in Sxlpe. Note the distal to proximal gradation in relative binding affinities. Predicted high-affinity sites all contain a canonical CAGCTG E box. Of the predicted moderate-affinity sites, the most distal are CATCTG and the third CAGCCG. (B) “Reservoir” model for Sxlpe. Sxlpe is activated in XX embryos as a result of the SIS-directed assembly of a proximal enhancer complex at the XCR. B/Da and other proteins bound to upstream sites function synergistically to augment Sxl transcription when the XCR complex is formed. Sxlpe is left inactive in XY embryos as haplo-X SIS and Runt concentrations are insufficient to drive assembly of the XCR complex. Occupied distal sites cannot stimulate transcription in the absence of a proximal complex. The analogy is to a filled reservoir awaiting release. In XY embryos, the reservoir remains full. In XX embryos, the opening of the proximal promoter causes a flood of Sxl expression. (C) Model for snail regulation in response to dorsal morphogen gradient (30). Dl binds to one high-affinity and nine low-affinity sites in the distal PLA and VA elements. These sites are occupied only in regions of high Dl concentration. The proximal AE contains sites for Twist and other proteins. High-level snail expression requires synergism between the dose sensitivity and augmentation elements.
Structure and function at the Sxl establishment and snail promoters.
The model for SxlPe shares important similarities with that proposed for snail, a target of the dorsal (dl) morphogen (30) (Fig. 6B and C). First, the dose-sensitive control elements are composed of low-affinity sites. For snail, these consist of nine low-affinity D1 binding sites that promote transcription only when D1 concentration is high (Fig. 6C). Second, the dose-sensing regions are not sufficient to promote high-level transcription: separate augmentation elements are required. In the snail promoter, the augmentation element contains two Twist binding sites and functions to increase transcription and refine the boundaries of snail expression (30). Third, the proximal element is necessary for detectable levels of transcription, indicating that the upstream sites function, in a formal genetic sense, through the proximal ones. Thus, both genes may exploit binding site affinity to set the activation threshold and use synergism between distal and proximal regulatory elements to achieve full expression.
While generally similar, snail and SxlPe differ in the relative arrangement of their proposed dose-sensitive and augmentation elements (Fig. 6B and C). In addition, different molecules function as augmentors and as dose determinants at snail, whereas B/Da seems likely to provide, at least part of, both functions at Pe. What is the significance of these differences? Do they offer any clues as to why the promoters are built the way they are?
In order to get strong spatially restricted snail expression, three things must occur: the promoter must be activated at the appropriate Dl concentration, transcription must be enhanced, and the expression boundaries must be refined. The snail AE carries out the latter two functions through synergistic interactions with the Dl sites in the polar and lateral activation (PLA) and ventral activation (VA) elements, thus ensuring that significant transcription occurs only in regions where both Dl and Twist are present (Fig. 6C). If Dl were also used as an augmentor, or if the Dl sites were brought too close to the promoter, the PLA and VA elements might be overly efficient at activation, leading to snail expression in the absence of Twist (33).
In contrast, there are only two requirements at SxlPe: activation at the threshold sis protein concentration and enhancement to ensure high-level expression. Spatial refinement is unnecessary. The Sxl augmentation elements contain high-affinity B/Da sites predicted to be bound even at a low protein concentration. Although bound, their distal location may prevent them from activating transcription unless there is a high enough concentration of B/Da to occupy the lower-affinity binding sites in the proximal promoter. In effect, the promoter appears to be built such that the distal high-affinity B/Da sites could prime SxlPe, allowing for immediate reinforcement of any “on” decision made at the XCR. While proteins other than B/Da could provide the augmentation function, the use of the same molecule for both initiation and enhancement would seem a particularly efficient and effective strategy for Sxl.
Sex specificity and role of negative regulators.
Our model provides an explanation for how the initial decision to turn on may be amplified to generate a strong response, but it does not answer the question of how the critical on or off decision is actually made. While the affinity of the B/Da sites in the XCR may well play an important role, we envision that activators, such as SISA and Runt, as well as maternally provided Stat92E and Her, also contribute to the assembly and function of the XCR complex, as all of the proteins have been shown by genetic means to work through the proximal promoter (21, 34, 36; H. Lu, unpublished data). Intriguingly, no specific Runt binding sites have been identified at SxlPe (36; D. Yang, unpublished data), suggesting that Runt may also bind to sequences of low intrinsic affinity. By analogy, we predict that several low-affinity binding sites for SISA will be present in the XCR and that cooperative binding between various signal proteins will play an important role in determining site occupancy. Direct precedent for these types of protein-protein interactions comes from the rhomboid promoter, where cooperative interactions between ASC/Da and Dl proteins facilitate Dl binding (29, 33, 48). The idea that a large protein complex forms at the XCR is consistent with the number of proteins involved as well as with the extent and relative order of the conserved sequence blocks in the promoter (Fig. 2). Interestingly, the ordering may even extend to the different subunits of heterodimers, as the asymmetric B/Da sites all have their noncanonical half-sites directed proximally. Such local directionality can be important for cooperative binding (7) and is a characteristic of highly structured enhancer complexes, such as those formed at the beta interferon and T-cell receptor α promoters (22, 26, 49).
What might be the roles of negative regulators, such as Dpn and its corepressor Groucho, in Sxl regulation? One possibility is that they directly inhibit the assembly of the XCR complex and that their negative influence is overcome at the high female SIS and Runt concentrations. The close linkage between the paired Dpn binding sites and B/Da site 1 is consistent with competition for DNA binding (Fig. 2). Alternatively, negative regulators may function over a short range to dampen the activation potential of an incompletely assembled male XCR complex or over a long range to prevent proteins bound at the augmentation elements from activating transcription inappropriately. The latter function is consistent with Dpn's function as a long-range repressor (9) and may explain the modest effects of dpn null mutations on Sxl activation in males (3).
ACKNOWLEDGMENTS
We thank Paul Schedl, Dan Kalderon, Jym Mohler, and Tulle Hazelrigg for thoughtful advice during the course of this work and Larry Chasin for valuable assistance with cell culture. L. Chasin, R. Prywes, J. Manley, J. Posakony, and M. Caudy generously provided plasmids and reagents. We are grateful to Teresa Lamb, P. Schedl, and Dan Kalderon for help with the manuscript.
This research was funded by a Searle Community Trust Award and American Cancer Society grant RPG-97-079-01-DB to J.W.E. and by an NIH grant to Paul Schedl (Princeton University).
REFERENCES
- 1.Atchley W R, Fitch W M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atchley W R, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- 3.Barbash D A, Cline T W. Genetic and molecular analysis of the autosomal component of the primary sex determination signal of Drosophila melanogaster. Genetics. 1995;141:1451–1471. doi: 10.1093/genetics/141.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 7.Burz D S, Rivera-Pomar R, Jackle H, Hanes S D. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrera C V, Alonso M C. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai H N, Arnosti D N, Levine M. Long-range repression in the Drosophila embryo. Proc Natl Acad Sci USA. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Thiagalingam A, Chopra H, Borges M W, Feder J N, Nelkin B D, Baylin S B, Ball D W. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cline T W. The Drosophila sex determination signal: how do flies count to two? Trends Genet. 1993;9:385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- 13.Cline T W. Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics. 1988;119:829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cline T W, Meyer B J. Vive la difference: males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 15.Cronmiller C, Cummings C A. The daughterless gene product in Drosophila is a nuclear protein that is broadly expressed throughout the organism during development. Mech Dev. 1993;42:159–169. doi: 10.1016/0925-4773(93)90005-i. [DOI] [PubMed] [Google Scholar]
- 16.Dang C V, Dolde C, Gillison M L, Kato G J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshpande G, Stukey J, Schedl P. scute (sis-b) function in Drosophila sex determination. Mol Cell Biol. 1995;15:4430–4440. doi: 10.1128/mcb.15.8.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson J W, Cline T W. A bZIP protein, SISTERLESS-A, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993;7:1688–1702. doi: 10.1101/gad.7.9.1688. [DOI] [PubMed] [Google Scholar]
- 19.Erickson J W, Cline T W. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development. 1998;125:3259–3268. doi: 10.1242/dev.125.16.3259. [DOI] [PubMed] [Google Scholar]
- 20.Erickson J W, Cline T W. Molecular nature of the Drosophila sex determination signal and its link to neurogenesis. Science. 1991;251:1071–1074. doi: 10.1126/science.1900130. [DOI] [PubMed] [Google Scholar]
- 21.Estes P A, Keyes L N, Schedl P. Multiple response elements in the Sex-lethal early promoter ensure its female-specific expression pattern. Mol Cell Biol. 1995;15:904–917. doi: 10.1128/mcb.15.2.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falvo J V, Parekh B S, Lin C H, Fraenkel E, Maniatis T. Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation. Mol Cell Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Crespo S, Levine M. Interactions between dorsal and helix-loop-helix proteins initiate the differentiation of the embryonic mesoderm and neuroectoderm in Drosophila. Genes Dev. 1993;7:1703–1713. doi: 10.1101/gad.7.9.1703. [DOI] [PubMed] [Google Scholar]
- 24.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 25.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 26.Grosschedl R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 27.Han K, Levine M S, Manley J L. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 28.Hoshijima K, Kohyama A, Watakabe I, Inoue K, Sakamoto H, Shimura Y. Transcriptional regulation of the Sex-lethal gene by helix-loop-helix proteins. Nucleic Acids Res. 1995;23:3441–3448. doi: 10.1093/nar/23.17.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ip Y T, Park R E, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 30.Ip Y T, Park R E, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 31.Jan Y N, Jan L Y. Functional gene cassettes in development. Proc Natl Acad Sci USA. 1993;90:8305–8307. doi: 10.1073/pnas.90.18.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jan Y N, Jan L Y. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 33.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 34.Jinks T M, Polydorides A D, Calhoun G, Schedl P. The JAK/STAT signalling pathway is required for the initial choice of sexual identity in Drosophila melanogaster. Mol Cell. 2000;5:581–587. doi: 10.1016/s1097-2765(00)80451-7. [DOI] [PubMed] [Google Scholar]
- 35.Keyes L N, Cline T W, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. . (Corrigendum, 69:following 572). [DOI] [PubMed] [Google Scholar]
- 36.Kramer S G, Jinks T M, Schedl P, Gergen J P. Direct activation of Sex-lethal transcription by the Drosophila runt protein. Development. 1999;126:191–200. doi: 10.1242/dev.126.1.191. [DOI] [PubMed] [Google Scholar]
- 37.Littlewood T, Evan G I. Helix-loop-helix transcription factors. Oxford, United Kingdom: Oxford University Press; 1998. [Google Scholar]
- 38.Martinez C, Modolell J, Garrell J. Regulation of the proneural gene achaete by helix-loop-helix proteins. Mol Cell Biol. 1993;13:3514–3521. doi: 10.1128/mcb.13.6.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massari M E, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy functions as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 41.Pan D, Valentine S A, Courey A J. The bipartite D. melanogaster twist promoter is reorganized in D. virilis. Mech Dev. 1994;46:41–53. doi: 10.1016/0925-4773(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 42.Parkhurst S M, Bopp D, Ish-Horowicz D. X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell. 1990;63:1179–1191. doi: 10.1016/0092-8674(90)90414-a. . (Corrigendum, 64:following 1046). [DOI] [PubMed] [Google Scholar]
- 43.Paroush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 44.Penalva L O, Sakamoto H, Navarro-Sabate A, Sakashita E, Granadino B, Segarra C, Sanchez L. Regulation of the gene Sex-lethal: a comparative analysis of Drosophila melanogaster and Drosophila subobscura. Genetics. 1996;144:1653–1664. doi: 10.1093/genetics/144.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pultz M A, Baker B S. The dual role of hermaphrodite in the Drosophila sex determination regulatory hierarchy. Development. 1995;121:99–111. doi: 10.1242/dev.121.1.99. [DOI] [PubMed] [Google Scholar]
- 46.Sefton L, Timmer J R, Zhang Y, Beranger F, Cline T W. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405:970–973. doi: 10.1038/35016119. [DOI] [PubMed] [Google Scholar]
- 47.Swanson H I, Chan W K, Bradfield C A. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 48.Szymanski P, Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thanos D. Mechanisms of transcriptional synergism of eukaryotic genes. The interferon-beta paradigm. Hypertension. 1996;27:1025–1029. doi: 10.1161/01.hyp.27.4.1025. [DOI] [PubMed] [Google Scholar]
- 50.Torres M, Sanchez L. The scute (T4) gene acts as a numerator element of the X:A signal that determines the state of activity of Sex-lethal in Drosophila. EMBO J. 1989;8:3079–3086. doi: 10.1002/j.1460-2075.1989.tb08459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Doren M, Bailey A M, Esnayra J, Ede K, Posakony J W. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- 52.Verma-Kurvari S, Savage T, Gowan K, Johnson J E. Lineage-specific regulation of the neural differentiation gene MASH1. Dev Biol. 1996;180:605–617. doi: 10.1006/dbio.1996.0332. [DOI] [PubMed] [Google Scholar]
- 53.Younger-Shepherd S, Vaessin H, Bier E, Jan L Y, Jan Y N. deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell. 1992;70:911–922. doi: 10.1016/0092-8674(92)90242-5. [DOI] [PubMed] [Google Scholar]