Abstract
BACKGROUND:
Mechanisms of smell loss in chronic rhinosinusitis (CRS) are still unclear and likely multifactorial. Little attention has been given to olfactory cleft (OC) mucus proteins involved in odorant binding and metabolizing enzymes and their potential role in smell loss.
METHODS:
Mucus from the OC was sampled from patients with CRS (n=20) and controls (n=10). Liquid chromatography and mass spectrometry were performed, followed by data processing so that protein groups could be identified, quantified, and compared. Hierarchical clustering and bioinformatic analysis were performed on significantly different proteins to explore for enrichment in known biologic pathways.
RESULTS:
A total of 2,514 proteins were found in OC mucus from all 30 subjects. Significant differences in protein abundance were found between CRS and controls, including both CRSsNP (n=351 proteins; log2 fold change range: −3.88 to 6.71) and CRSwNP (n=298 proteins; log2 fold change range: −4.00 to −6.13). Significant differences were found between patients with normosmia and those with dysosmia (n=183; log2 fold change range: −3.62 to −2.16) and across groups of interest for a number of odorant binding proteins and metabolizing enzymes.
CONCLUSIONS:
OC mucous in CRS displays a rich and abundant array of proteins, many of which have been implicated in odorant transport and metabolization in animal studies. Significant differences in the olfactory mucus proteome were seen between CRS subtypes and controls, as well as between those with normal and abnormal olfaction. Further study should confirm these findings and explore the role individual proteins play in odorant transport and metabolization.
Keywords: Chronic disease, Mass spectrometry, Proteins, Proteomics, Sinusitis, Smell
INTRODUCTION
Chronic rhinosinusitis (CRS) is a highly prevalent condition that impacts up to 12% of the United States population.1 Loss of smell is a cardinal symptom of CRS, and quantifiable olfactory loss is present in up to 60-80% of patients, which equates to a population prevalence of roughly 30 million Americans.2 Olfactory loss in CRS is generally thought to be a downstream consequence of sinonasal inflammation. Loss of olfaction may result from nasal cavity obstruction, which impairs odorant-containing air from reaching the olfactory cleft. Another possibility is that there is direct inflammation of the olfactory neuroepithelium, disrupting olfactory receptor neuron (ORN) function. Findings from several histologic studies support this mechanism, as well as studies showing inflammation visible in the olfactory cleft (OC) on endoscopy and computed tomography (CT) scan.3–6 However, in order for odorants to reach the ORNs they must first cross the mucus layer overlying the OC neuroepithelium.7 Alterations in OC mucus thus represents an additional possible mechanism through which olfactory dysfunction could occur in patients with CRS, adding to other well-known mechanisms.
The composition of OC mucus has received relatively little attention in patients with CRS, despite its critical role in odorant transport in other animals. Odorants are primarily hydrophobic molecules that must cross the aqueous extra-cellular fluid layer surrounding ORNs.8 Research performed mostly in animal models suggests that odorant-binding proteins (OBPs) are a critical component of olfactory mucus, helping to bind and transport odorants across the mucus layer so they can efficiently interact with ORNs.8 Furthermore, olfactory metabolizing enzymes are highly expressed in mammalian olfactory mucosa. These enzymes are found in the olfactory mucus of nonhuman animals and their catalytic function bio-transforms odorants, facilitating their elimination.9–11 Proteins in the OC mucus thus appear to play a dynamic role promoting sensitivity of the olfactory signal.
Profiling of the human OC mucus proteome is a nascent field, with only 3 prior published studies. The initial study published in 2007 was able to identify 83 unique proteins in healthy controls.12 Using current shot-gun techniques, two additional studies have been reported within the last 2 years, both identifying well over a thousand unique proteins.13,14 However, neither of these studies included patients with CRS. It thus remains unknown whether the proteomic profile of patients with CRS differs from controls without CRS, particularly with regard to odorant binding proteins or metabolizing enzymes, nor is it known whether changes are associated with olfactory function. Considering that at the macroscopic level, thick, discolored mucus is a cardinal diagnostic feature of CRS, we hypothesized that significant differences in OC mucus proteins would exist in patients with CRS compared to controls and could be detected using proteomic methods.
MATERIALS and METHODS
Recruitment
Adult patients with CRS were recruited from the rhinology clinics at the Medical University of South Carolina into a case-control study. All patients met diagnostic criteria for CRS according to the American Academy of Otolaryngology-Head and Neck Surgery, including appropriate symptom criteria and evidence of inflammation on CT scan.15 Patients were excluded if they had cystic fibrosis, primary ciliary dyskinesia, systemic inflammatory disease, or had used systemic corticosteroids in the preceding 30 days. Control subjects were recruited from the community for the purposes of this research protocol. Subjects were excluded if they reported a history of CRS, had symptoms suggestive of CRS, carried a diagnosis of dementia or Parkinson’s disease, or had used systemic corticosteroids in the preceding 30 days. Recruitment was adjusted to ensure that each group was comprised of equal parts normosmic and dysosmic subjects. This study was approved by the MUSC Institutional Review Board and all subjects provided written informed consent per good clinical practice guidelines for medical research in compliance with the Declaration of Helsinki in the study of human subjects.
Clinical assessment
Demographic and comorbidity data was recorded directly from all individuals, supplemented by the medical record. Olfactory testing was performed using Sniffin’ Stick pens (Burghart Messtechnik, Wedel, Germany) which evaluate three separate domains of olfactory function including: odorant threshold (score range: 1-16), odorant discrimination (score range: 0-16), and odorant identification (score range: 0-16).16 Correctly identified threshold (T), discrimination (D), and identification (I) scores, as well as a composite TDI total score, are summarized from item responses (score range: 1-48) with higher scores reflecting superior overall olfactory function.17 For patients with CRS, the 22-item Sinonasal Outcomes Test (SNOT-22) was administered to capture patient-reported disease severity.18
All individuals had sinonasal endoscopy performed using a 3mm rigid telescope (Karl Storz, Tuttlingen, Germany). A cotton ball was used to apply 4% lidocaine with 0.5% oxymetazoline to the head of the inferior turbinate just prior to exam. The endoscopic appearance of the olfactory cleft was graded using the Olfactory Cleft Endoscopy Scale (OCES).19 In those individuals with CRS, the sinonasal cavity was further graded using the Lund Kennedy Endoscopy scale (LKES).20 Patients with CRS were classified as CRS without polyps (CRSsNP) or CRS with polyps (CRSwNP) based on the visible presence of polyps.
Olfactory cleft mucus sampling
Under direct endoscopic visualization, a 1x2 cm Leukosorb filter paper (Pall Scientific, Port Washington, NY) strip was placed directly into the OC of each side. Specifically, the paper strip was placed between the middle turbinate and septum, in a plane posterior to the head of the middle turbinate. After 3 minutes, the Leukosorb was removed and placed into cuvettes that were immediately centrifuged at 4°C for 30 minutes, as described previously.21,22 Mucus from each side was combined, transferred by pipette to a cryotube, and stored at −80°C.
Sample preparation
Mucus samples were lyophilized, resuspended in 100 uL of 0.1% Rapigest (Waters) and boiled at 100°C for 5 minutes. Thirty ug aliquots were diluted to a final volume of 50uL. with Rapigest. Protein was reduced with 5mM. dithiothreitol for 30 minutes and the cysteines alkylated with 15mM. iodoacetamide for 30 minutes in the dark. Samples were diluted with 50uL. of 100mM. ammonium bicarbonate and digested with 300ng. of trypsin (Sigma) overnight at 37°C. The digestion was quenched by adding trifluoroacetic acid (10% v/v) to a final concentration of 0.5% with incubation at 37°C for 45 minutes. Samples were centrifuged at 13,000 RPM for 10 minutes and the supernatant was transferred to a new tube. Peptides were desalted by C18 zip tip (Millipore), dried under vacuum, and stored at −80°C.
Liquid Chromatography and Mass Spectrometry Data Acquisition Parameters
Peptides were resuspended in 7μL. of 2% acetonitrile (ACN), 0.2% formic acid (FA) and 5μL. of this injected. Peptides were separated and analyzed on an EASY 1200 nanoLC (ThermoScientific) in-line with an Orbitrap Fusion Lumos mass spectrometer (ThermoScientific) with instrument control software v. 4.2.28.14. Peptides were pressure loaded at 1,180 bar, and separated on a C18 reversed phase column (Acclaim PepMap RSLC, 75 μm x 50 cm (C18, 2 μm, 100 Å)) (ThermoFisher) using a gradient of 2% to 35% B in 180 min (Solvent A: 0.1% FA; Solvent B: 80% ACN, 0.1% FA) at a flow rate of 300nL/min. The column was thermostated at 45°C. The samples were ionized by electrospray using stainless steel emitters (ThermoScientific). Mass spectra were acquired in data-dependent mode with a high resolution (60,000) FTMS survey scan, mass range of m/z 375-1575, followed by tandem mass spectra (MS/MS) of the most intense precursors with a cycle time of 3 seconds. The automatic gain control target value was 4.0e5 for the survey MS scan. Fragmentation was performed with a precursor isolation window of 1.6m/z, a maximum injection time of 50ms., and HCD collision energy of 35%. Fragment ions were detected in the orbitrap at a 15,000 resolution. Monoisotopic-precursor selection was set to “peptide”. Precursors were dynamically excluded from resequencing for 15 seconds and a mass tolerance of 10ppm. Advanced peak determination was not enabled. Precursor ions with charge states that were undetermined, 1, or > 7 were excluded.
Mass Spectrometry Data Processing
Protein groups were identified, quantified, and normalized using the MaxQuant software platform v.1.6.3.3 with the Andromeda database searching algorithm and label free quantification (LFQ) algorithm.23–25 Data were searched against a human Uniprot reference database UP0000005640 with 74,468 proteins (downloaded August, 2019) and a database of common contaminants. The false discovery rate, determined using a reversed database strategy, was set at 1% at the spectrum, peptide, and protein level. A 4.5ppm tolerance was used for the main search. Fully tryptic peptides with a minimum of 7 residues were required including cleavage between lysine and proline. Two missed cleavages were permitted. Matching between runs was enabled for those features with tandem mass spectra acquired in at least one of the runs. Fast LFQ was enabled with stabilization of large ratios. A minimum ratio count of 2 was required for protein quantification with at least one unique peptide per protein. Parameters included static modification of cysteine with carbamidomethyl and variable N-terminal acetylation and oxidation of methionine.
Statistical and proteome bioinformatics data analysis
The protein groups text file from the MaxQuant search results was processed in Perseus v. 1.6.10.45.26 Identified proteins were filtered to remove proteins only identified by a modified peptide, matches to the reversed database, and potential contaminants. The normalized protein intensities for each biological replicate were log2 transformed. Visualization of the distribution of the data in histograms revealed two samples with insufficient protein for quantitation that were removed from further analysis. Quantitative measurements were required in 80% of biological replicates in at least one of the treatment groups for each binary comparison. For each comparison, Pearson correlation coefficients were evaluated in multi-scatter plots to identify samples that yielded highly variable protein intensities in comparison to other sample and those with correlation coefficients less than 0.8 were removed. Groups of interest were then compared using two-sided Student’s t tests and a permutation-based false discover rate (FDR) was calculated based on 250 randomizations.27 In cases with unequal number of samples per treatment group, a Welch’s t-test was performed. Bioinformatic analysis was performed in the Perseus software environment to test for enrichment of annotations associated with significantly regulated proteins as compared to all the observed proteins.26 The log2 transformed LFQ intensities of proteins with a p-value <0.05 were normalized by z-score and hierarchical clustering was performed using default parameters of Euclidean distance, average linkage, and preprocessing with k-means. Clusters of proteins, visualized in heatmaps, were extracted and enrichment of GO terms was evaluated using a Fisher exact test with a Benjamini-Hochberg FDR<0.02. Categorical annotation was added in Perseus in the form of GO biological process (GOBP), molecular function (GOMF), cellular component (GOCC), and Reactome pathways (downloaded from Uniprot, March 2019). Both unadjusted p-values and FDR are reported throughout and raw data was uploaded to the Proteomics IDEntification (PRIDE) database.
RESULTS
Study population
A total of 30 patients were enrolled, including N=20 with CRS and N=10 controls, with CRS patients equally split between CRSsNP and CRSwNP subgroups. Demographics, comorbidities, and disease severity characteristics are summarized in Table 1. The overall group averaged 48.1 years (SD=17.5, range: 20-77), and included slightly more women than men. Age, gender, and race were similar across groups. As expected based on study design, Sniffin’ stick TDI scores were similar across groups. Patients with CRS had average SNOT-22 and LKES scores typical of cohorts with moderate to severe sinus disease. Patient with CRS were more likely to have comorbid asthma than controls, but other non-CRS comorbidity measures were similar across groups.
Table 1:
Comparison of final study population characteristics.
| Demographics: | CRSsNP Subjects (n=10) | CRSwNP Subjects (n=10) | Control Subjects (n=10) | Test statistic | p-value |
|---|---|---|---|---|---|
| Age in years Median [IQR] | 60.5 [29.0] | 35.0 [32.0] | 52.0 [29.0] | KW=2.6 | 0.277 |
| Males N (%) | 4 (40%) | 2 (20%) | 3 (30%) | χ2=1.0 | 0.621 |
| Females | 6 (60%) | 8 (80%) | 7 (70%) | ||
| White/Caucasian | 10 (100%) | 9 (90%) | 7 (70%) | χ2=4.0 | 0.133 |
| African American | 0 (0%) | 1 (10%) | 3 (30%) | ||
| Hispanic/Latino ethnicity | 1 (10%) | 1 (10%) | 0 (0%) | χ2=1.1 | 0.585 |
| Comorbidity: | |||||
| Nasal polyposis | 0 (0%) | 10 (100%) | 0 (0%) | χ2=30.0 | <0.001 |
| Previous sinus surgery / ESS | 3 (30%) | 3 (30%) | 0 (0%) | χ2=3.8 | 0.153 |
| Asthma | 5 (50%) | 5 (50%) | 0 (0%) | χ2=7.5 | 0.024 |
| Diabetes mellitus (Type I/II) | 1 (10%) | 1 (10%) | 0 (0%) | χ2=1.1 | 0.585 |
| Smoking / tobacco use (current) | 2 (20%) | 0 (0%) | 1 (10%) | χ2=2.2 | 0.329 |
| Alcohol use (current) | 5 (50%) | 7 (70%) | 6 (60%) | χ2=0.8 | 0.659 |
| Allergic rhinitis | 5 (50%) | 5 (50%) | 1 (10%) | χ2=4.6 | 0.101 |
| GERD | 4 (40%) | 2 (20%) | 2 (20%) | χ2=1.4 | 0.506 |
| Clinical Measures of Disease Severity: | |||||
| SNOT-22 total score | 36.0 [51.8] | 72.0 [27.0] | ---- | MWU=3.4 | 0.064 |
| Item: “Sense of smell / taste” | 2.5 [5.0] | 4.5 [1.0] | ---- | MWU=3.6 | 0.058 |
| Lund-Kennedy endoscopy score | 3.0 [2.3] | 8.0 [4.5] | ---- | MWU=11.0 | 0.001 |
| Olfactory cleft endoscopy score | 0.5 [1.8] | 9.0 [8.0] | 0.0 [1.0] | KW=5.6 | 0.060 |
| Measures of Olfactory Function: | |||||
| Sniffin’ Sticks total score | 27.1 [20.0] | 20.3 [24.9] | 28.6 [28.1] | KW= 3.3 | 0.195 |
| Threshold score | 4.5 [4.4] | 4.5 [3.4] | 7.1 [8.8] | KW= 1.5 | 0.465 |
| Discrimination score | 11.0 [7.0] | 8.0 [8.0] | 11.0 10.0] | KW= 1.9 | 0.387 |
| Identification score | 12.5 [10.0] | 7.0 [11.0] | 10.5 [10.0] | KW= 2.9 | 0.239 |
CRSsNP, chronic rhinosinusitis without nasal polyposis; CRSwNP, chronic rhinosinusitis with nasal polyposis; IQR, interquartile range; KW, Kruskall-Wallis test statistic; N, sample size; ESS, endoscopic sinus surgery; GERD, gastroesophageal reflux disease; SNOT-22, 22-item SinoNasal Outcome Test survey.
Across the entire cohort, a total of 2,702 proteins were identified with at least 1 unique peptide (Supplemental Table 1) and 2,514 proteins were identified with at least 2 peptides. A total of 686 proteins were quantified in all 28 LC-MS/MS runs. Comparison of results to other published studies showed that 69 of 84 proteins (82%) detected by Débat et al. were identified in the present study.12 Similarly, 1,075 of 1,117 proteins (96%) detected by Wolf et al. were detected in the present study.13 The overall number of mucus proteins in the current study (N=2,514) was similar to that reported in the OC mucus of non-CRS aging subjects (N=2,586).14 Our samples detected all 87 proteins considered by Yoshikawa to be enriched in olfactory cleft mucus as opposed to nasal cavity mucus, suggesting that our collection technique samples olfactory cleft mucus.
CRSsNP versus controls
To compare samples from patients with CRSsNP to control subjects, protein groups were filtered to retain proteins quantified in 80% of samples in at least one group. Missing values were imputed from a normal distribution with a width of 0.3 that was downshifted from the distribution of quantified proteins by 1.8. To assess for differentially abundant proteins a t test was performed using a p-value <0.05 as the threshold yielding 351 proteins (Supplemental Table 2). This included 235 proteins which were lower in CRSsNP as compared to controls, as well as 116 proteins which were more abundant in patients with CRSsNP (Figure 1). Of these proteins, a total of 61 also had a permutation-based FDR ≤0.05. Next, we wanted to explore specific proteins that have been implicated in odorant transport or metabolism across animal studies.9–11 Lipocalin-15, which has been suggested to function as an odorant-binding protein,14,28,29 was found to be lower in patients with CRSsNP than controls (Table 2). Similarly, we found patients with CRS to have lower levels of glutathione S-transferases (GSTs), alcohol/aldehyde dehydrogenases, and other anti-oxidant proteins (Table 2), all of which have been postulated to impact olfaction by functioning as olfactory metabolizing enzymes in olfactory mucus.7–11 The log2 transformed intensities of regulated proteins were z-scored and hierarchical clustering was performed to probe for enrichment of GO terms and pathways. Significant differences were seen across a number of biological processes as shown in the heat map (Figure 2) and Table 3.
Figure 1:
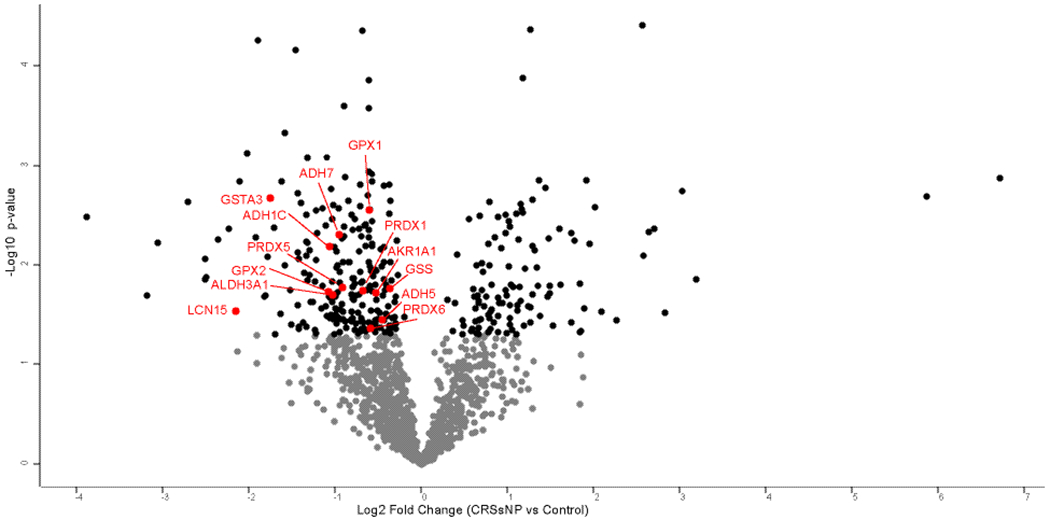
Volcano plot of protein differences between CRSsNP and controls. Black dots present proteins which are significantly different between CRSsNP and controls (p<0.050). Proteins labeled in red correspond to proteins in Table 2.
Table 2:
Proteins hypothesized to play a role in odorant transport and metabolism: CRSsNP versus controls.
| Protein name | Majority protein IDs | Gene name | Log2 Fold Change | p-value* | FDR |
|---|---|---|---|---|---|
| Possible odorant binding protein | |||||
| Lipocalin-15 | Q6UWW0 | LCN15 | −2.15 | 0.030 | 0.130 |
| Glutathione S transferases and synthetase | |||||
| Glutathione S-transferase Mu 2 | P28161;E9PHN7;E9PHN6;F6XZQ7;E9PLF1 | GSTM2 | −1.82 | 0.021 | 0.109 |
| Glutathione S-transferase A3 | Q16772;Q5JW85 | GSTA3 | −1.75 | 0.002 | 0.051 |
| Glutathione synthetase | P48637;A0A2R8Y430;A0A2R8Y5T7 | GSS | −0.36 | 0.017 | 0.102 |
| Alcohol/aldehyde dehydrogenases | |||||
| Alcohol dehydrogenase [NADP(+)] | P14550 | AKR1A1 | −0.52 | 0.022 | 0.110 |
| Alcohol dehydrogenase 1C | P00326 | ADH1C | −1.07 | 0.007 | 0.059 |
| Alcohol dehydrogenase class-3 | P11766 | ADH5 | −0.45 | 0.044 | 0.144 |
| Alcohol dehydrogenase class 4 mu/sigma chain | P40394;A0A0C4DG85;E9PFG0 | ADH7 | −0.95 | 0.005 | 0.054 |
| Aldehyde dehydrogenase, dimeric NADP-preferring | P30838;I3L3I9;A8MYB8;C9JMC5;E9PNN6 | ALDH3A1 | −1.02 | 0.020 | 0.108 |
| Other anti-oxidant enzymes | |||||
| Glutathione peroxidase 1 | A0A087WUQ6;P07203;A0A2R8Y6B6 | GPX1 | −0.60 | 0.003 | 0.048 |
| Glutathione peroxidase 2 | A0A087WTS0;P18283;H0YJK8 | GPX2 | −1.07 | 0.022 | 0.107 |
| Peroxiredoxin-1 | Q06830;A0A0A0MSI0 | PRDX1 | −0.68 | 0.022 | 0.104 |
| Peroxiredoxin-5 | P30044 | PRDX5 | −0.91 | 0.022 | 0.101 |
| Peroxiredoxin-6 | P30041 | PRDX6 | −0.58 | 0.043 | 0.155 |
A two-sided Student’s t test was performed comparing the mean log2 protein intensities from CRSsNP versus controls. CRSsNP, chronic rhinosinusitis without nasal polyposis
=unadjusted p-value; FDR=permutation-based false discovery rate.
Figure 2:
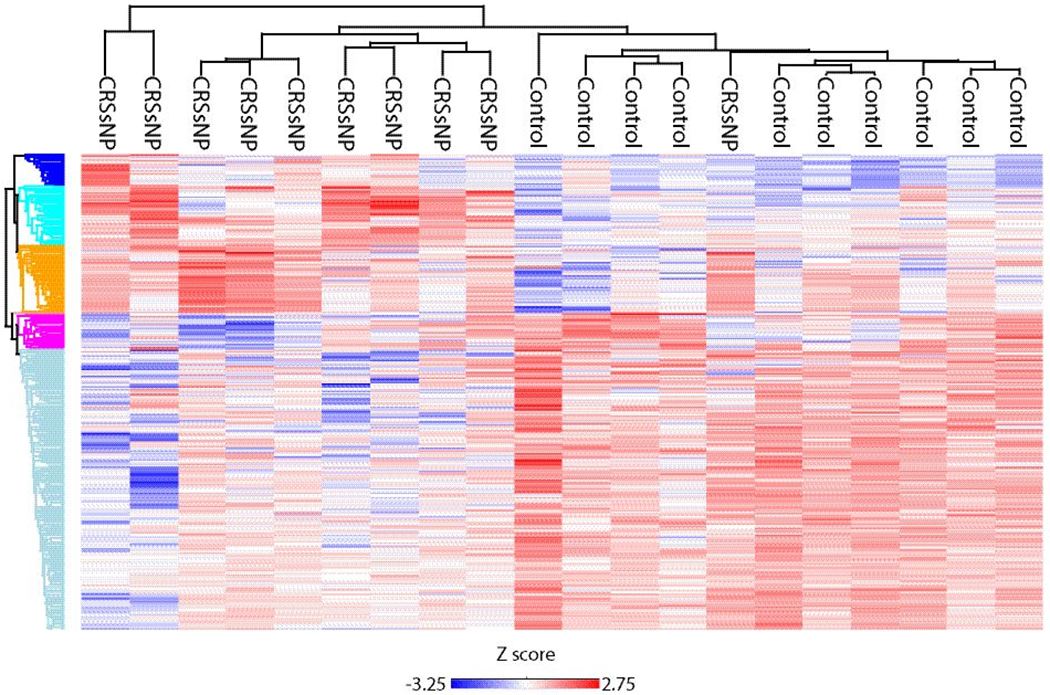
Heat map and dendrogram of significant proteins found in CRSsNP and controls. Red/blue color represents relative abundance of proteins enriched across GO terms and Reactome pathways.
Table 3:
GO and Reactome pathways in CRSsNP versus controls.
| Cluster color from heat map | Gene Ontology and Reactome pathways | Size intersection/Category size | p-value | FDR |
|---|---|---|---|---|
|
| ||||
|
Enriched in CRSsNP
| ||||
| Dark blue | Overall Number of Proteins in Cluster | 42 | 1.44E-08 | 1.73E-08 |
| Blood microparticle [GO:0072562] | 16/84 | 4.30E-10 | 6.27E-07 | |
| Collagen-containing ECM [GO:0062023] | 13/76 | 1.11E-07 | 9.71E-05 | |
| Extracellular region [GO:0005576] | 25/318 | 2.69E-07 | 0.000196 | |
| Extracellular space [GO:0005615] | 20/256 | 1.11E-05 | 0.004388 | |
| Platelet degranulation pathway | 9/51 | 1.07E-05 | 0.011117 | |
| Platelet degranulation [GO:0002576] | 9/46 | 4.42E-06 | 0.018574 | |
|
| ||||
| Aqua | Overall Number of Proteins in Cluster | 68 | 2.34E-17 | 4.67E-17 |
| Actin filament binding [GO:0051015] | 10/38 | 6.66E-06 | 0.018381 | |
|
| ||||
| Orange | Overall Number of Proteins in Cluster | 71 | 7.05E-20 | 2.11E-19 |
| Cytosolic large ribosomal subunit [GO:0022625] | 15/38 | 7.03E-11 | 1.56E-07 | |
| Cytosolic small ribosomal subunit [GO:0022627] | 10/32 | 1.79E-06 | 0.000964 | |
| Focal adhesion [GO:0005925] | 21/140 | 1.99E-06 | 0.000964 | |
| L13a-mediated translational silencing of Ceruloplasmin expression pathway | 25/84 | 1.51E-14 | 3.13E-11 | |
| Membrane [GO:0016020] | 42/329 | 7.14E-11 | 1.56E-07 | |
| Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184] | 25/78 | 2.08E-15 | 1.75E-11 | |
| Nucleolus [GO:0005730] | 15/91 | 2.42E-05 | 0.00755 | |
| Polysomal ribosome [GO:0042788] | 7/19 | 2.17E-05 | 0.007295 | |
| Postsynaptic density [GO:0014069] | 9/36 | 4.43E-05 | 0.012099 | |
| Ribonucleoprotein complex [GO:1990904] | 11/56 | 6.50E-05 | 0.015762 | |
| RNA binding [GO:0003723] | 36/293 | 1.59E-08 | 6.58E-05 | |
| rRNA processing [GO:0006364] | 9/21 | 2.82E-07 | 0.001424 | |
| SRP-dependent cotranslational protein targeting to Membrane [GO:0006614] | 25/68 | 4.64E-17 | 1.17E-12 | |
| Structural constituent of ribosome [GO:0003735] | 25/66 | 1.98E-17 | 1.64E-13 | |
| Synapse [GO:0045202] | 9/11 | 3.49E-05 | 0.010153 | |
| Translation [GO:0006412] | 26/80 | 3.24E-16 | 4.08E-12 | |
| Translational initiation [GO:0006413] | 25/92 | 1.61E-13 | 1.01E-09 | |
|
| ||||
|
Decreased in CRSsNP
| ||||
| Dark pink | Overall Number of Proteins in Cluster | 26 | 2.27E-16 | 3.41E-16 |
| Cytosol [GO:0005829] | 7/892 | 5.18E-05 | 0.013308 | |
| Extracellular space [GO:0005615] | 15/256 | 7.82E-06 | 0.003416 | |
|
| ||||
| Light blue | Overall Number of Proteins in Cluster | 249 | 5.24E-112 | 3.14E-111 |
| Extracellular region [GO:0005576] | 31/318 | 1.12E-06 | 0.000699 | |
| Membrane [GO:0016020] | 30/329 | 9.92E-08 | 9.71E-05 | |
Enrichment of GO terms associated with protein clusters was performed using a Fisher exact test with a Benjamini-Hochberg FDR<0.02. CRSsNP, chronic rhinosinusitis without nasal polyposis; FDR=false discovery rate.
CRSwNP versus controls
Samples from patients with CRSwNP were then compared to controls. Two CRSwNP samples were removed from further analysis based on the low amount of protein. A total of 298 proteins were found to be significantly different between groups using a p-value <0.05 as the threshold (Supplemental Table 3). This included 225 proteins which were lower in CRSwNP as compared with controls, as well 73 proteins which were more abundant in patients with CRSwNP (Figure 3). Of these proteins, a total of 118 also had a permutation-based FDR ≤0.05. Next we explored those proteins that have been implicated in odorant transport or metabolism across other animal studies. We found CRSwNP patients to have decreased abundance of Lipocalin-1 and 15, which have been hypothesized to possibly function in odorant binding.14,28,29 We also found decreased abundance of GSTs, alcohol dehydrogenases, and other oxidizing enzymes that may play a role in odorant metabolism (Table 4).8–11 Log2 protein intensities were z-scored and hierarchical clustering was performed to probe for enrichment of GO terms and Reactome pathways. Highly significant differences were seen across a number of biologic processes as shown in the heat map (Figure 4 and Table 5).
Figure 3:
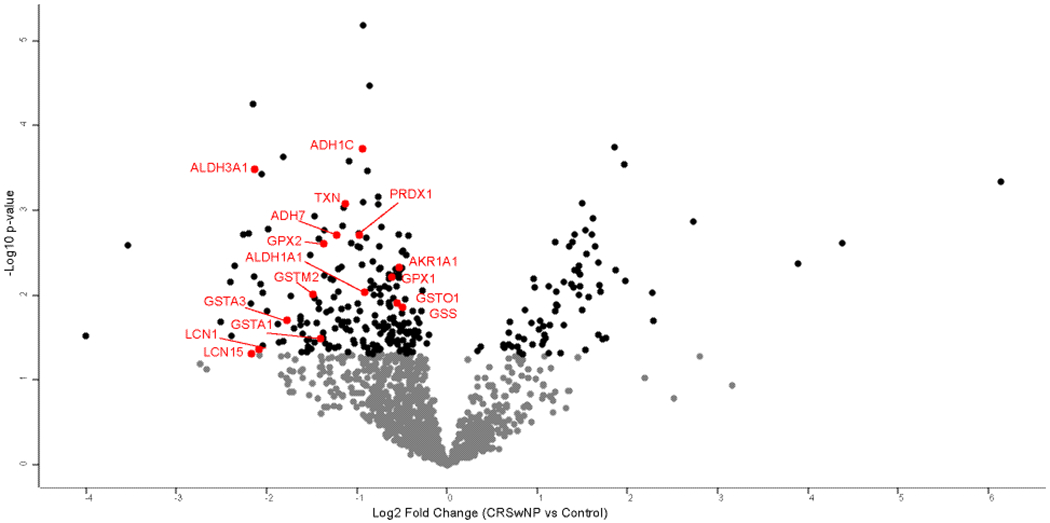
Volcano plot of protein differences between CRSwNP and controls. Black dots present proteins which are significantly different between CRSwNP and controls (p<0.050). Proteins labeled in red correspond to proteins in Table 4.
Table 4:
Proteins hypothesized to play a role in odorant transport and metabolism: CRSwNP versus controls.
| Protein name | Majority protein IDs | Gene name | Log2 Fold Change | p-value* | FDR |
|---|---|---|---|---|---|
| Possible odorant binding proteins | |||||
| Lipocalin-1 | P31025 | LCN1 | −2.16 | 0.049 | 0.177 |
| Lipocalin-15 | Q6UWW0 | LCN15 | −2.08 | 0.043 | 0.121 |
| Glutathione S-transferases and synthetase | |||||
| Glutathione S-transferase A3 | Q16772;Q5JW85 | GSTA3 | −1.77 | 0.020 | 0.067 |
| Glutathione S-transferase Mu 2 | P28161;E9PHN7;E9PHN6;F6XZQ7;E9PLF1 | GSTM2 | −1.49 | 0.010 | 0.053 |
| Glutathione S-transferase A1 | P08263 | GSTA1 | −1.40 | 0.033 | 0.083 |
| Glutathione S-transferase omega-1 | P78417;Q5TA02;Q5TA01 | GSTO1 | −0.55 | 0.012 | 0.046 |
| Glutathione synthetase | P48637;A0A2R8Y430;A0A2R8Y5T7 | GSS | −0.49 | 0.014 | 0.056 |
| Alcohol dehydrogenases | |||||
| Aldehyde dehydrogenase | P30838;I3L3I9;A8MYB8;C9JMC5;E9PNN6 | ALDH3A1 | −2.13 | 0.0003 | 0.003 |
| Alcohol dehydrogenase class 4 mu/sigma chain | P40394;A0A0C4DG85;E9PFG0 | ADH7 | −1.22 | 0.002 | 0.014 |
| Alcohol dehydrogenase 1C | P00326 | ADH1C | −0.94 | 0.0002 | 0.007 |
| Retinal dehydrogenase 1 | P00352 | ALDH1A1 | −0.91 | 0.009 | 0.039 |
| Alcohol dehydrogenase [NADP(+)] | P14550 | AKR1A1 | −0.53 | 0.005 | 0.053 |
| Other oxidizing enzymes | |||||
| Glutathione peroxidase 1 | A0A087WUQ6;P07203;A0A2R8Y6B6 | GPX1 | −0.61 | 0.006 | 0.038 |
| Glutathione peroxidase 2 | A0A087WTS0;P18283;H0YJK8 | GPX2 | −1.36 | 0.002 | 0.009 |
| Peroxiredoxin-1 | Q06830;A0A0A0MSI0 | PRDX1 | −0.97 | 0.002 | 0.015 |
| Thioredoxin | P10599 | TXN | −1.13 | 0.001 | 0.004 |
A two-sided Welch’s t test was performed comparing the mean log2 protein intensities from CRSwNP versus controls. CRSwNP, chronic rhinosinusitis with nasal polyposis;
=unadjusted p-value; FDR=permutation-based false discovery rate.
Figure 4:
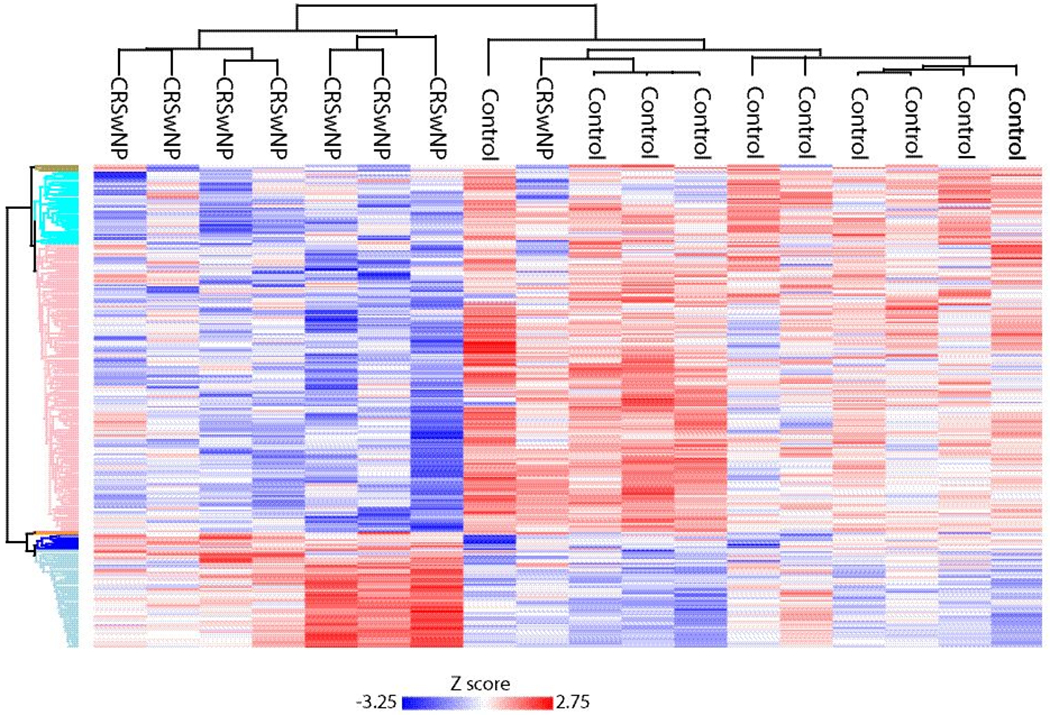
Heat map and dendrogram of significant proteins found in CRSwNP and controls. Red/blue color represents relative abundance of proteins enriched across GO terms and Reactome pathways.
Table 5:
GO and Reactome pathways in CRSwNP versus controls.
| Cluster color from heat map | Gene Ontology and Reactome pathway | Size intersection/Category size | p-value* | FDR |
|---|---|---|---|---|
|
| ||||
|
Enriched in CRSwNP
| ||||
| Blue | Overall Number of Proteins in Cluster | 13 | 0.000336 | 0.000588 |
| Microtubule-based process [GO:0007017] | 4/9 | 6.63E-07 | 0.002286 | |
| Microtubule cytoskeleton organization [GO:0000226] | 4/15 | 6.89E-06 | 0.018143 | |
| Microtubule [GO:0005874] | 5/46 | 3.93E-05 | 0.013999 | |
|
| ||||
| Light blue | Overall Number of Proteins in Cluster | 91 | 1.17E-29 | 4.09E-29 |
| Intrinsic Pathway of Fibrin Clot Formation pathway | 5/5 | 1.33E-06 | 0.00104 | |
| Blood coagulation, intrinsic pathway [GO:0007597] | 6/7 | 5.64E-07 | 0.002286 | |
| Negative regulation of endopeptidase activity [GO:0010951] | 15/26 | 4.13E-12 | 3.98E-08 | |
| Acute-phase response [GO:0006953] | 8/14 | 7.1E-07 | 0.002286 | |
| Serine-type endopeptidase inhibitor activity [GO:0004867] | 17/31 | 3.97E-13 | 3.76E-09 | |
| Heparin binding [GO:0008201] | 10/25 | 1.71E-06 | 0.004046 | |
| Platelet alpha granule lumen [GO:0031093] | 10/28 | 5.65E-06 | 0.00235 | |
| Collagen-containing extracellular matrix [GO:0062023] | 27/77 | 1.97E-14 | 3.27E-11 | |
| Blood microparticle [GO:0072562] | 30/86 | 5.38E-16 | 2.69E-12 | |
| Platelet degranulation [GO:0002576] | 15/44 | 4.15E-08 | 0.000258 | |
| Platelet degranulation pathway | 15/49 | 2.1E-07 | 0.000307 | |
| Regulation of complement activation [GO:0030449] | 12/42 | 8.52E-06 | 0.018985 | |
| Cellular protein metabolic process [GO:0044267] | 13/49 | 8.51E-06 | 0.018985 | |
| Extracellular space [GO:0005615] | 49/257 | 1.37E-14 | 3.27E-11 | |
| Extracellular region [GO:0005576] | 54/317 | 3.66E-14 | 4.56E-11 | |
| Extracellular exosome [GO:0070062] | 67/719 | 3.23E-05 | 0.012402 | |
| Nucleus [GO:0005634] | 11/478 | 1.04E-07 | 7.09E-05 | |
|
| ||||
|
Decreased in CRSwNP
| ||||
| Pink | Overall Number of Proteins in Cluster | 218 | 4.7E-102 | 3.3E-101 |
| Proteasome core complex, alpha-subunit complex [GO:0019773] | 7/7 | 2.87E-06 | 0.001302 | |
| Threonine-type endopeptidase activity [GO:0004298] | 16/17 | 2.32E-12 | 1.1E-08 | |
| Proteasomal ubiquitin-independent protein catabolic process [GO:0010499] | 16/17 | 2.32E-12 | 3.98E-08 | |
| Proteasome core complex [GO:0005839] | 7/7 | 2.32E-12 | 2.31E-09 | |
| Proteasome core complex, beta-subunit complex [GO:0019774] | 9/10 | 6.08E-07 | 0.000337 | |
| Proteasomal protein catabolic process [GO:0010498] | 17/19 | 3.03E-12 | 3.98E-08 | |
| Endopeptidase activity [GO:0004175] | 17/28 | 8.51E-08 | 0.000269 | |
| Proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161] | 19/37 | 5.17E-07 | 0.002286 | |
| Activation of NF-kappaB in B cells pathway | 20/39 | 2.62E-07 | 0.000307 | |
| Proteasome complex [GO:0000502] | 22/44 | 1.14E-07 | 7.09E-05 | |
| Protein deubiquitination [GO:0016579] | 23/54 | 2.03E-06 | 0.005867 | |
| Cytosol [GO:0005829] | 171/871 | 1.76E-06 | 0.000877 | |
|
| ||||
| Aqua | Overall Number of Proteins in Cluster | 64 | 1.43E-23 | 3.33E-23 |
| Epidermis development [GO:0008544] | 7/10 | 4.45E-08 | 0.000258 | |
Enrichment of GO terms associated with protein clusters was performed using a Fisher exact test with a Benjamini-Hochberg FDR<0.02. CRSwNP, chronic rhinosinusitis with nasal polyposis;
=unadjusted p-value; FDR=false discovery rate.
Olfactory cleft mucus proteins and olfaction
Subjects were then classified as normosmic if their TDI score was 31-48 and dysosmic if their TDI score was <31.0. There were 183 proteins whose abundance significantly differed based on olfactory function (Supplemental Table 4). This included 109 proteins that were lower in those with dysosmia, as well as 74 proteins whose abundance was higher in those with dysosmia (Figure 5). None of these proteins had an FDR≤0.05. Once again, we explored proteins that have been implicated in odorant transport or metabolization across other animal studies. We found dysosmic patients to have significantly lower abundance of GSTs, alcohol dehydrogenases, and other oxidizing enzymes that may play a role in odorant metabolism (Table 6). Significant proteins were z scored and clustering was performed as before. Highly significant differences were seen across a number of GO terms and Reactome pathways as shown in the heat map (Figure 6 and Table 7). Of note, dysosmic subjects had enrichment of complement activation pathways, whereas oxidation-reduction pathways were enriched in normosmics. The heat map shows that samples do not appear to cluster based solely on polyp status, suggesting that differences seen are not exclusively driven by disease phenotype.
Figure 5:
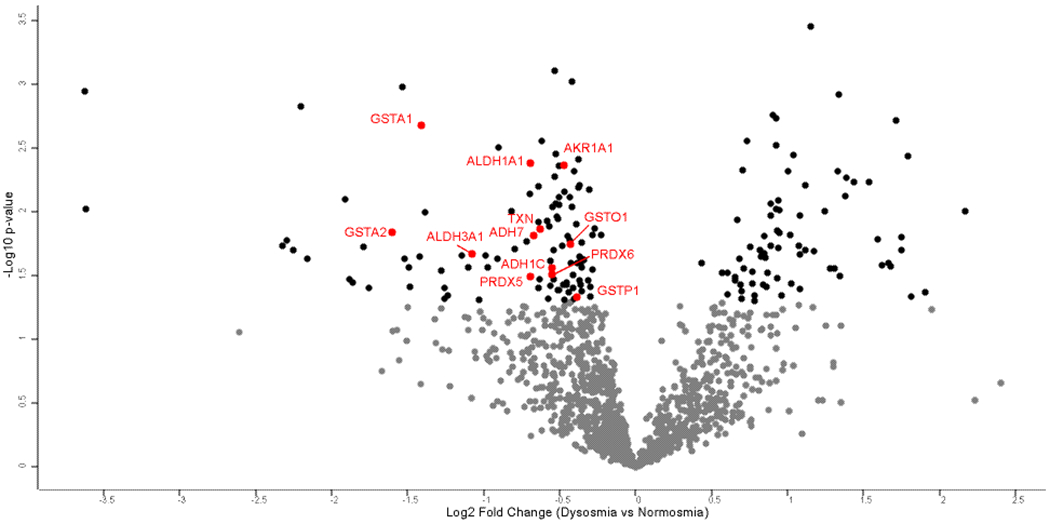
Volcano plot of protein differences between dysosmia and normosmia. Black dots present proteins which are significantly different between dysosmic patients and normosmic patients (p<0.050). Proteins labeled in red correspond to proteins in Table 6.
Table 6:
Proteins hypothesized to play a role in odorant transport and metabolism: normosmic versus dysosmic
| Protein name | Majority protein IDs | Gene name | Log2 Fold Change | p-value* | FDR |
|---|---|---|---|---|---|
| Glutathione S-transferases | |||||
| Glutathione S-transferase A2 | P09210 | GSTA2 | −1.60 | 0.015 | 0.211 |
| Glutathione S-transferase A1 | P08263 | GSTA1 | −1.41 | 0.002 | 0.152 |
| Glutathione S-transferase omega-1 | P78417;Q5TA02;Q5TA01 | GSTO1 | −0.43 | 0.018 | 0.206 |
| Glutathione S-transferase P | P09211;A8MX94 | GSTP1 | −0.39 | 0.047 | 0.278 |
| Alcohol dehydrogenases | |||||
| Aldehyde dehydrogenase, dimeric NADP-preferring | P30838;I3L3I9;A8MYB8;C9JMC5;E9PNN6 | ALDH3A1 | −1.08 | 0.021 | 0.224 |
| Alcohol dehydrogenase class 4 mu/sigma chain | P40394;A0A0C4DG85;E9PFG0 | ADH7 | −0.67 | 0.016 | 0.209 |
| Alcohol dehydrogenase 1C | P00326 | ADH1C | −0.55 | 0.028 | 0.242 |
| Alcohol dehydrogenase [NADP(+)] | P14550 | AKR1A1 | −0.47 | 0.004 | 0.167 |
| Retinal dehydrogenase 1 | P00352 | ALDH1A1 | −0.69 | 0.004 | 0.139 |
| Other oxidative enzymes | |||||
| Peroxiredoxin-5 | P30044 | PRDX5 | −0.69 | 0.033 | 0.242 |
| Peroxiredoxin-6 | P30041 | PRDX6 | −0.55 | 0.031 | 0.226 |
| Thioredoxin | P10599 | TXN | −0.63 | 0.014 | 0.207 |
A two-sided Welch’s t test was performed comparing the mean log2 protein intensities from normosmic versus dysosmic. ID, identifications; FDR=permutation-based false discovery rate.
Figure 6:
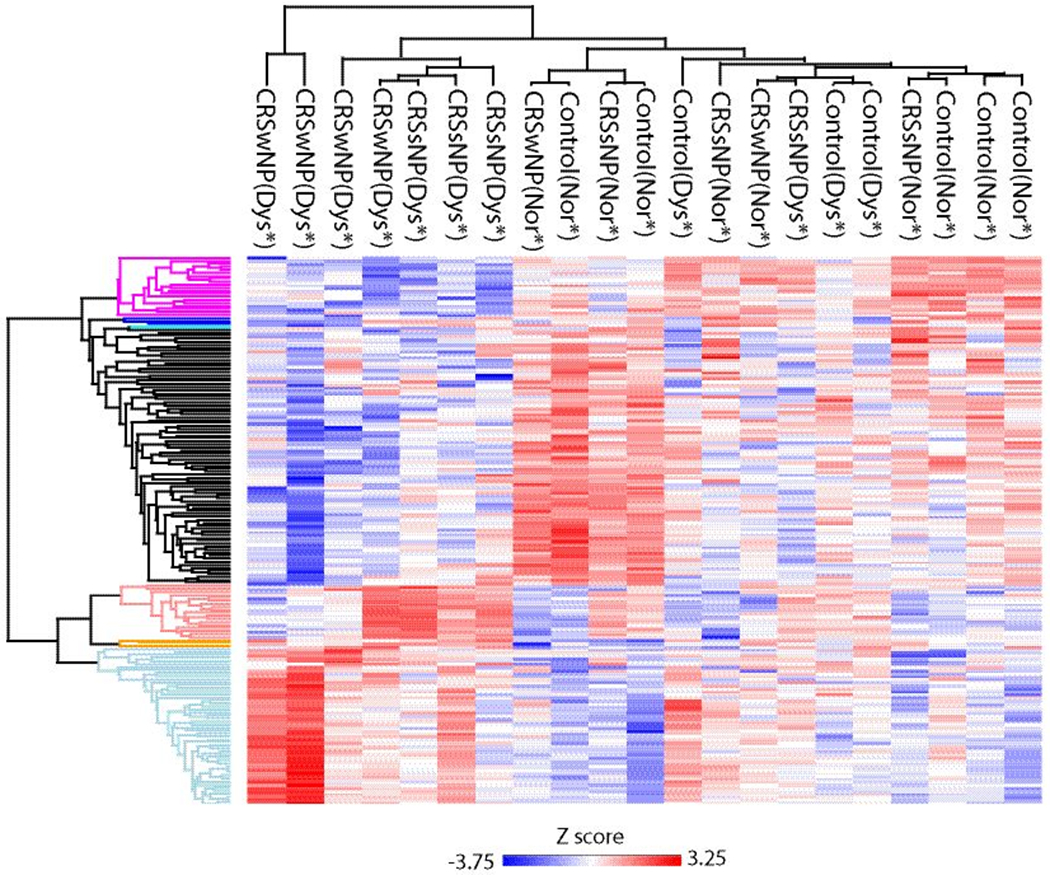
Heat map and dendrogram of significant proteins found in dysosmia and normosmia Red/blue color represents relative abundance of proteins enriched across GO terms and Reactome pathways.
Table 7:
GO and Reactome pathways in normosmic versus dysosmic subjects
| Cluster color from heat map | Gene Ontology and Reactome pathway | Size Intersection/Category size | p-value | FDR |
|---|---|---|---|---|
|
| ||||
|
Enriched in Dysosmic
| ||||
| Peach | Overall Number of Proteins in Cluster | 18 | 1.67E-16 | 2.51E-16 |
| Cytoplasmic translation [GO:0002181] | 7/23 | 4.97E-09 | 1.38E-05 | |
| Polysomal ribosome [GO:0042788] | 5/18 | 1.94E-06 | 0.000639 | |
| Cytosolic large ribosomal subunit [GO:0022625] | 10/37 | 2.99E-12 | 2.14E-09 | |
| Structural constituent of ribosome [GO:0003735] | 14/65 | 3.02E-16 | 2.44E-12 | |
| SRP-dependent cotranslational protein targeting to membrane [GO:0006614] | 14/67 | 4.81E-16 | 1.2E-11 | |
| Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184] | 14/76 | 3.28E-15 | 4.11E-11 | |
| Translation [GO:0006412] | 14/79 | 5.88E-15 | 4.91E-11 | |
| L13a-mediated translational silencing of Ceruloplasmin expression pathway | 14/83 | 1.23E-14 | 2.55E-11 | |
| Translational initiation [GO:0006413] | 14/90 | 4.13E-14 | 2.58E-10 | |
| Nucleolus [GO:0005730] | 7/90 | 8.44E-05 | 0.019025 | |
| Focal adhesion [GO:0005925] | 9/137 | 2.1E-05 | 0.005619 | |
| RNA binding [GO:0003723] | 14/282 | 3.85E-07 | 0.001038 | |
| Membrane [GO:0016020] | 15/323 | 1.89E-07 | 8.1E-05 | |
|
| ||||
| Light Blue | Overall Number of Proteins in Cluster | 53 | 2.11E-49 | 6.32E-49 |
| Positive regulation of cholesterol esterification [GO:0010873] | 4/5 | 1.12E-05 | 0.018638 | |
| Membrane attack complex [GO:0005579] | 5/7 | 1.68E-06 | 0.0006 | |
| Terminal pathway of complement pathway | 5/7 | 1.68E-06 | 0.001157 | |
| Complement activation, alternative pathway [GO:0006957] | 6/9 | 2.37E-07 | 0.00054 | |
| Negative regulation of endopeptidase activity [GO:0010951] | 13/26 | 1.03E-12 | 5.18E-09 | |
| Serine-type endopeptidase inhibitor activity [GO:0004867] | 14/30 | 4.07E-13 | 1.65E-09 | |
| Acute-phase response [GO:0006953] | 6/14 | 7.06E-06 | 0.012625 | |
| Platelet alpha granule lumen [GO:0031093] | 10/26 | 1.41E-08 | 8.6E-06 | |
| Blood microparticle [GO:0072562] | 31/84 | 3.6E-26 | 1.54E-22 | |
| Cellular protein metabolic process [GO:0044267] | 15/48 | 5.54E-11 | 2.31E-07 | |
| Regulation of complement activation [GO:0030449] | 13/42 | 1.53E-09 | 5.47E-06 | |
| Collagen-containing extracellular matrix [GO:0062023] | 23/76 | 1.56E-16 | 1.67E-13 | |
| Platelet degranulation [GO:0002576] | 13/43 | 2.12E-09 | 6.65E-06 | |
| Platelet degranulation pathway | 13/48 | 9.55E-09 | 9.86E-06 | |
| Endoplasmic reticulum lumen [GO:0005788] | 12/48 | 1.04E-07 | 5.56E-05 | |
| Complement activation, classical pathway [GO:0006958] | 11/49 | 1.23E-06 | 0.00257 | |
| Extracellular space [GO:0005615] | 41/252 | 5.76E-21 | 8.22E-18 | |
| Extracellular region [GO:0005576] | 45/307 | 3.04E-22 | 6.51E-19 | |
|
| ||||
|
Decreased in Dysosmic
| ||||
| Dark Pink | Overall Number of Proteins in Cluster | 20 | 2.67E-18 | 5.33E-18 |
| Epidermis development [GO:0008544] | 4/9 | 4.55E-06 | 0.00876 | |
|
| ||||
| Black | Overall Number of Proteins in Cluster | 86 | 8.53E-84 | 5.12E-83 |
| Oxidation-reduction process [GO:0055114] | 11/23 | 3.16E-08 | 7.93E-05 | |
Enrichment of GO terms associated with protein clusters was performed using a Fisher exact test with a Benjamini-Hochberg FDR<0.02. CRSwNP, chronic rhinosinusitis with nasal polyposis;
=unadjusted p-value; FDR=false discovery rate.
DISCUSSION
The composition of OC mucus in the setting of CRS has received relatively little attention. A number of recent studies have focused on the inflammatory cytokine profile of OC mucus;21,30 however, findings from this study suggest that much more than inflammatory proteins are present in mucus collected from the OC. Our hypothesis was that odorant binding proteins and those associated with metabolizing enzymes would be altered in CRS and impact olfaction. We did find notable difference in the proteomic profile of OC mucus between patients with CRS and controls, including differences in proteins thought to act as odorant binding proteins and metabolizing enzymes. These findings suggest that alterations in OC mucus could possibly contribute to olfactory dysfunction in patients with CRS.
CRS-related olfactory dysfunction is most commonly thought to occur via either a conductive pathway, related to impaired airflow, or from direct inflammation of the olfactory neuroepithelium. Findings from this study suggest that alterations in OC mucus are present and could be an additional contributor beyond conductive or neural mechanisms. Research in non-human animals suggests that OC mucus plays a critical and dynamic role in the odorant pathway. Odorants are generally small molecules that can easily evaporate and become airborne. Odorants transported to the OC via airflow then solubilize into the aqueous OC mucus overlying the olfactory neuroepithelium. However, odorants are usually hydrophobic molecules and their solubility is enhanced by interacting with OBPs which help transport odorants through mucus to the ORNs.31 Furthermore, there appear to be a diverse array of metabolizing enzymes that transform odorants within the olfactory mucus, including glutathione s-transferases, alcohol dehydrogenases, and an array of other oxidizing proteins.8 These olfactory metabolizing enzymes can modulate the olfactory signal. An elegant series of experiments by Nagashima and Touhara illustrated how these enzymes impact olfactory function.9 They were able to show that odorants with aldehydes and esters functional groups are targets of metabolic enzymes secreted in the OC mucus in mice. Inhibition of these enzymes blocked glomerular activation patterns and altered mouse food-finding behavior. Enzymatic conversion actually occurred fast enough to affect recognition of the odorant at the levels of ORNs. Similarly, Robert-Hazotte et al. were able to show that glutathione transferases impact newborn rabbit behavioral responsiveness to mammary pheromones, suggesting that olfactory metabolizing enzymes play a role not only in termination but modulation of the olfactory signal.10 Although work in non-human animal models are compelling, most of these experiments have not been extended into humans and thus the role of OBPs and metabolizing enzymes is largely unexplored.
Our current study does provide some insight into proteins that might serve as olfactory binding proteins or metabolizing enzymes. Interestingly, we did not identify OBP2A or OBP2B in any of our samples. This is similar to all 3 prior studies using similar mass spectroscopy techniques. It remains unknown whether these proteins are present, but simply not detected as a consequence of the collection technique or processing. We were able to identify Lipocalin-1 and Lipocalin-15, which have been proposed as proteins that might function as OBPs.28,29 Both of these molecules were less abundant in patients with CRS, suggesting the possibility of dysfunctional odor transport in CRS. Similar to other studies, we did identify a diverse array of proteins which function as olfactory metabolizing enzymes. These included a number of GSTs and alcohol dehydrogenases, both of which have been shown to modulate olfaction in non-human animal studies.9–11 Importantly, these metabolizing enzymes were less abundant in CRS and were less abundant in those with dysosmia. With regard to dysosmia, the oxidation-reduction process pathway [GO:0055114] was significantly different between dysosmic subjects and those with normal smell (p=3.16E-08). Taken together, this data suggests that human OC mucus contains putative OBPs and metabolizing enzymes and that their abundance varies by disease state in patients with CRS. However, this study design does not prove that any specific protein plays a causal role in olfaction and many of these enzymes would impact specific odorants. Future experiments will need to be designed to determine the precise role these proteins play in olfactory dysfunction in CRS that is likely multifactorial.
Considering that thick and discolored mucus is a cardinal feature of CRS, it should not be surprising that significant alterations in the proteomic profile were identified. When evaluating GO terms and pathways, a number of different processes and pathways were identified as shown in Tables 3,5, and 7. Dysosmic subjects were enriched in the complement cascade and platelet degranulation, which could be important with regard to CRS pathophysiology. However, many differences between CRS and controls were in pathways related to cellular function and extracellular space, which may be downstream consequences of the mucosal inflammation as opposed to causal. Recently, Kao et al performed a proteomic analysis of nasal cavity mucus from CRS patients and most differences identified were also in cellular processes and biologic regulation.32
There are several considerations to keep in mind when interpreting results from this study. Although mucus was collected from the olfactory cleft, the exact source of this mucus remains unknown. Yoshikawa et al. simultaneously collected mucus from the OC and anterior nasal cavity in healthy adults using a similar endoscopic-guided technique and were able to show that OC mucus had a unique profile.14 We were able to detect all 87 OC-enriched proteins from the Yoshikawa study which suggests our samples included olfactory mucus. However, a hallmark of CRS is increased sinonasal mucus production. Whether mucus produced in the sinuses can traffic into and mix with olfactory mucus is unknown and cannot be determined with this study design. It is also important to point out that mass spectrometry often fails to detect proteins in small quantities, such as inflammatory cytokines and growth factors which have been found in OC mucus using alternative techniques.3,4,33 This means that complementary techniques would be required not only to confirm these findings, but to obtain a comprehensive understanding of OC mucus in CRS.
Although this is the largest study to perform shotgun proteomics of human olfactory mucus, its sample size is still modest on a clinical basis when one considers the heterogeneity of CRS. Therefore, detailed analysis by CRS disease severity, comorbidities, or age was not feasible. This also limited some of the pair-wise comparisons that could be made across groups defined by disease status and olfactory function. Additionally, one needs to keep in mind that one goal of this study was to test hypotheses related to OBPs and metabolizing enzymes, but another was to explore changes across the entire proteome. Given the large number of statistical tests, the risk of type 1 error increases substantially for any single protein. On the other hand, strict multiple comparisons corrections may inflate Type 2 error given the small sample size. For that reason we reported both unadjusted p-values and a FDR. We would caution that these findings need to be repeated in separate and larger cohorts to ensure they are robust and reproducible. Additionally, mechanistic studies should be performed to confirm that these specific proteins play a role in human olfactory transduction similar to that seen in non-human animals.
CONCLUSIONS
Mucus collected from the OC of patients with CRS displays a rich and abundant array of proteins, many of which have been implicated in odorant transport and metabolization in non-human animal studies. Significant differences in the olfactory mucus proteome were seen between CRS patients and controls, as well as between those with normal smell and those with dysosmia, suggesting olfactory mucus proteins could play a role in CRS-related olfactory dysfunction. Further study will need to confirm these findings in larger sample sizes and explore the role individual proteins play in odorant transport and metabolization.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge technical assistance from the Mass Spectrometry Facility, a University Research Resource, specifically Susana Comte-Walters at the Medical University of South Carolina.
Funding source(s):
ZMS, RJS, JKM, TLS, JCM, and V.R.R. are supported by grants from the National Institute on Deafness and Other Communication Disorders (NIDCD), one of the National Institutes of Health, Bethesda, MD [R01 DC005805; PIs: TLS and ZMS; R03-DC013651 to Z.M.S. and K23DC014747; PI: VRR). This work was also supported by funding mechanisms from Orbitrap Fusion Lumos MS, L.E.B. [S10-OD010731]; Proteomics Core, L.E.B. [GM103542]. These funding organizations did not contribute to the design or conduct of this study; preparation, review, approval or decision to submit this manuscript for publication. There are no relevant financial disclosures for KNC, JRB, or LEB.
Footnotes
Potential conflicts of interest: None of the following consultancy disclosures are affiliated with this study. Z.M.S.: Olympus, OptiNose, Regeneron, Healthy Humming, and Novartis. R.J.S.: OptiNose, Olympus, Stryker, Regeneron, and Healthy Humming. V.R.R.: OptiNose and Medtronic, Inc.
REFERENCES
- 1.Blackwell DL, Lucas JW, Clarke TC. Summary health statistics for U.S. adults: national health interview survey, 2012. Vital Health Stat. 2014; 260:1–161. [PubMed] [Google Scholar]
- 2.Kohli P, Naik AN, Harruff EE, et al. The prevalence of olfactory dysfunction in chronic rhinosinusitis. Laryngoscope. 2017; 127(2):309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yee KK, Pribitkin EA, Cowart BJ, et al. Neuropathology of the olfactory mucosa in chronic rhinosinusitis. Am J Rhinol Allergy. 2010; 24(2):110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kern RC. Chronic sinusitis and anosmia: pathologic changes in the olfactory mucosa. Laryngoscope. 2000; 110:1071–1077. [DOI] [PubMed] [Google Scholar]
- 5.Loftus C, Schlosser RJ, Smith TL, et al. Olfactory cleft and sinus opacification differentially impact olfaction in chronic rhinosinusitis. Laryngoscope. 2020; 130(10):2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soler ZM, Hyer JM, Karnezis TT, Schlosser RJ. Olfactory Cleft Endoscopy Scale correlates with olfactory metrics in patients with chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016; 6(3):293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asakawa M, Fukutani Y, Savangsuksa A, et al. Modification of the response of olfactory receptors to acetophenone by CYP1a2. Sci Rep. 2017; 7(1):10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heydel JM, Coelho A, Thiebaud N, et al. Odorant-binding proteins and xenobiotic metabolizing enzymes: implications in olfactory perireceptor events. Anat Rec (Hoboken). 2013; 296(9):1333–1345. [DOI] [PubMed] [Google Scholar]
- 9.Nagashima A, Touhara K. Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. J Neurosci. 2010; 30(48):16391–16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert-Hazotte A, Faure P, Neiers F, et al. Nasal mucus glutathione transferase activity and impact on olfactory perception and neonatal behavior. Sci Rep. 2019; 9(1):3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heydel JM, Menetrier F, Belloir C, et al. Characterization of rat glutathione transferases in olfactory epithelium and mucus. PloS one. 2019; 14(7):e0220259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debat H, Eloit C, Blon F, et al. Identification of human olfactory cleft mucus proteins using proteomic analysis. J Proteome Res. 2007; 6(5):1985–1996. [DOI] [PubMed] [Google Scholar]
- 13.Wolf A, Liesinger L, Spoerk S, et al. Olfactory cleft proteome does not reflect olfactory performance in patients with idiopathic and postinfectious olfactory disorder: A pilot study. Sci Rep. 2018; 8(1):17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshikawa K, Wang H, Jaen C, et al. The human olfactory cleft mucus proteome and its age-related changes. Sci Rep. 2018; 8(1):17170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, et al. Clinical practice guideline (update): adult sinusitis. Otolaryngolog Head Neck Surg. 2015; 152(2 Suppl):S1–S39. [DOI] [PubMed] [Google Scholar]
- 16.Hummel T, Sekinger B, Wolf SR, Pauli E, Kobal G. ‘Sniffin’ sticks’: olfactory performance assessed by the combined testing of odor identification, odor discrimination and olfactory threshold. Chem Sense. 1997; 22(1):39–52. [DOI] [PubMed] [Google Scholar]
- 17.Hummel T, Kobal G, Gudziol H, Mackay-Sim A. Normative data for the “Sniffin’ Sticks” including tests of odor identification, odor discrimination, and olfactory thresholds: an upgrade based on a group of more than 3,000 subjects. Eur Arch Otorhinolaryngol. 2007; 264:237–243. [DOI] [PubMed] [Google Scholar]
- 18.Hopkins C, Gillett S, Slack R, Lund VJ, Browne JP. Psychometric validity of the 22-item Sinonasal Outcome Test. Clin Otolaryngol. 2009; 34:447–454. [DOI] [PubMed] [Google Scholar]
- 19.Soler ZM, Hyer JM, Karnezis TT, Schlosser RJ. The Olfactory Cleft Endoscopy Scale correlates with olfactory metrics in patients with chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016; 6(3):293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lund VJ, Kennedy DW. Staging for rhinosinusitis. Otolaryngolog Head Neck Surg. 1997; 117:S35–40. [DOI] [PubMed] [Google Scholar]
- 21.Soler ZM, Yoo F, Schlosser RJ, et al. Correlation of mucus inflammatory proteins and olfaction in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2020; 10(3):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo F, Soler ZM, Mulligan JK, et al. Olfactory cleft mucus proteins associated with olfactory dysfunction in a cohort without chronic rhinosinusitis. Int Forum Allergy Rhinol. 2019; 9(10):1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J, Hein MY, Luber CA, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008; 26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- 25.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016; 11:2301–2319. [DOI] [PubMed] [Google Scholar]
- 26.Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016; 13(9):731–740. [DOI] [PubMed] [Google Scholar]
- 27.Tyanova S, Cox J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol Biol. 2018; 1711:133–148. [DOI] [PubMed] [Google Scholar]
- 28.Lacazette E, Gachon AM, Pitiot G. A novel human odorant-binding protein gene family resulting from genomic duplicons at 9q34: differential expression in the oral and genital spheres. Hum Mol Genet. 2000; 9:289–301. [DOI] [PubMed] [Google Scholar]
- 29.Breustedt DA, Schonfeld DL, Skerra A. Comparative ligand-binding analysis of ten human lipocalins. Biochim Biophys Acta. 2006; 1764:161–173. [DOI] [PubMed] [Google Scholar]
- 30.Schlosser RJ, Mulligan JK, Hyer JM, et al. Mucous Cytokine Levels in Chronic Rhinosinusitis-Associated Olfactory Loss. JAMA Otolaryngol Head Neck Surg. 2016; 142:731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Archunan G Odorant Binding Proteins: a key player in the sense of smell. Bioinformation. 2018; 14:36–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih-Teng Kao S, Bassiouni A, Ramezanpour M, et al. Proteomic analysis of nasal mucus samples of healthy patients and patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2020; S0091-6749(20)31071-X. doi: 10.1016/j.jaci.2020.06.037. [DOI] [PubMed] [Google Scholar]
- 33.Turner JH, Liang KL, May L, Lane AP. Tumor necrosis factor alpha inhibits olfactory regeneration in a transgenic model of chronic rhinosinusitis-associated olfactory loss. Am J Rhinol Allergy. 2010; 24:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


