Abstract
Citrus are among the most prevailing fruit crops produced worldwide. The implementation of effective and reliable breeding programs is essential for coping with the increasing demands of satisfactory yield and quality of the fruit as well as to deal with the negative impact of fast-spreading diseases. Conventional methods are time-consuming and of difficult application because of inherent factors of citrus biology, such as their prolonged juvenile period and a complex reproductive stage, sometimes presenting infertility, self-incompatibility, parthenocarpy, or polyembryony. Moreover, certain desirable traits are absent from cultivated or wild citrus genotypes. All these features are challenging for the incorporation of the desirable traits. In this regard, genetic engineering technologies offer a series of alternative approaches that allow overcoming the difficulties of conventional breeding programs. This review gives a detailed overview of the currently used strategies for the development of genetically modified citrus. We describe different aspects regarding genotype varieties used, including elite cultivars or extensively used scions and rootstocks. Furthermore, we discuss technical aspects of citrus genetic transformation procedures via Agrobacterium, regular physical methods, and magnetofection. Finally, we describe the selection of explants considering young and mature tissues, protoplast isolation, etc. We also address current protocols and novel approaches for improving the in vitro regeneration process, which is an important bottleneck for citrus genetic transformation. This review also explores alternative emerging transformation strategies applied to citrus species such as transient and tissue localized transformation. New breeding technologies, including cisgenesis, intragenesis, and genome editing by clustered regularly interspaced short palindromic repeats (CRISPR), are also discussed. Other relevant aspects comprising new promoters and reporter genes, marker-free systems, and strategies for induction of early flowering, are also addressed. We provided a future perspective on the use of current and new technologies in citrus and its potential impact on regulatory processes.
Keywords: citrus transgenic plants, in vitro regeneration, transformation methods, CRISPR in citrus, reporter and selection markers, cisgenesis and intragenesis, citrus promoters, citrus biotechnology
Introduction
The genus Citrus of the Rutaceae family is one of the most important commercial woody fruit crops from tropical and subtropical areas of the world with a total global production of 124.246 million tons in 2016.1 In 2019, fruit production was 157 million tons worldwide.2 Apart from the fresh fruit and its juice, pectin and essential oils are also important commercialized products of citrus (Fisher and Phillips, 2008). Commercially, several species fall under the term citrus, including lemons, limes, mandarins, satsumas, clementines, common mandarins and tangerines, oranges, grapefruits, and pummelos (Zhong and Nicolosi, 2020). The non-existence of genetic diversity in many commercially cultivated crops (because of monoculture) has made them more susceptible to biotic and abiotic stresses (Esquinas-Alcázar, 2005; Keneni et al., 2012). Citrus trees are susceptible to many pathogens including nematodes, fungi, oomycetes, bacteria, spiroplasmas, phytoplasmas, viruses, and viroids, and the main abiotic stresses affecting these trees are acid, alkaline, and salty soils, flooding and drought, freezing, and high temperatures.
Citrus trees have complex reproductive biology. The apomixis present in citrus, which means that adventitious embryos initiate directly from maternal nucellar cells, limits the development of less vigorous zygotic embryos. They also have long juvenile periods and require at least 5 years for the start of the flowering phase in subtropical areas, and usually, several years more to achieve fully mature characteristics. The complex taxonomic relationships among cultivar groups are another difficulty and one of the reasons for the low-level impact of conventional breeding in citrus genetic improvement (Gmitter and Talon, 2008). Genetic transformation offers an excellent strategy for the genetic enhancement of citrus since it is based on the introduction of specific traits into known genotypes without altering their elite genetic background. Biotechnological tools have assisted in the fast germplasm improvement of current cultivars (Peña and Navarro, 2000) and the development of new varieties.
This review provides insights into the most relevant aspects of genetic transformation of citrus species including explant selection, biological and physical methods for transformation, and dependence on the genotype. We explore the possibilities for the promoter, selection, and reporter systems, and discuss novel and emerging technologies aimed to get more acceptable biotechnological products with no integration of exogenous DNA (“DNA-free”), using cisgenesis, intragenesis, and gene-editing.
Overview
The genus Citrus belongs to the subfamily Aurantoidea. Historically, within this subfamily there have been three genera of economic importance, namely Fortunella, Poncirus, and Citrus; however, more recently, it has been suggested they all belong to Citrus (Mabberley, 2004). Considering an evolutionary perspective, the four taxa identified as the ancestors of most of the cultivated citrus are Citrus medica L. (citron), Citrus reticulata Blanco (mandarin), Citrus maxima (Burm.) Merr. (pummelo), and Citrus micrantha Wester (papeda) (Wu et al., 2018; Ahmed et al., 2019). The secondary species, which result from successive natural hybridizations between the four fundamental species, are Citrus sinensis (L.) Osb. (sweet orange), Citrus aurantium L. (sour orange), Citrus paradisi Macf. (grapefruit), Citrus limon (L.) Burm. (lemon), Citrus jambhiri Lush (rough lemon), and Citrus aurantifolia (Christm.) Swing. (lime) (Wu et al., 2018; Ahmed et al., 2019). Finally, modern commercial cultivars come from artificial hybridizations: the rootstocks Carrizo and Troyer citrange hybrids (sweet orange × Poncirus trifoliata) and Swingle citrumelo (grapefruit × P. trifoliata) (Peña et al., 2008).
Although Poncirus and Citrus genera are the most amenable for in vitro regeneration (Vardi et al., 1982; Kobayashi et al., 1983), all species, hybrids, and economically important cultivars show a high degree of genotype-dependent variability in the efficiency of genetic transformation and regeneration (Bond and Roose, 1998).
Agrobacterium-Based Transformation Methods on Citrus
Citrus crops are not naturally susceptible to Agrobacterium tumefaciens. Although citrus are generally recalcitrant to Agrobacterium-mediated transformation, researchers have successfully achieved the recovery of transgenic plants for many genotypes (Peña et al., 2008). In general, the transformation efficiencies achieved using Agrobacterium can range from 0 to 45% for most citrus cultivars (Febres et al., 2011). The most common disarmed Agrobacterium strains used for the transformation of citrus species and relatives are the octopine strain LBA4404 (Kaneyoshi et al., 1994; Ali et al., 2012) the nopaline strain C58 (Bond and Roose, 1998), and the agropine strains EHA101 or EHA105 (Moore et al., 1992; Peña and Navarro, 2020). A. tumefaciens A281 (the oncogenic ancestor of EHA105) produced the earliest and the highest frequency of tumor formation either in epicotyls or stem segments of Pineapple sweet orange, Mexican lime, Clemenules clementine, Carrizo citrange, P. trifoliata, Fino lemon, Cleopatra mandarin, Citrus macrophylla, sour orange and Mediterranean mandarin (Cervera et al., 1998a,2005; Fagoaga et al., 2005; Peña et al., 2008). Successful transformation of embryogenic calli from Ponkan mandarin and Valencia sweet orange has been also attainable using strain EHA105 (Li et al., 2002; Peña et al., 2008). Transformation efficiency from different strains is mainly attributable to Ti plasmids and specifically to the vir region present on them. The study of Ghorbel et al. (2001) have added extra copies of virG genes from pTiBo542 (Ti plasmid contained in EHA105) to strain C58, which resulted in a significant increment in transformation frequencies of C58 in several citrus genotypes.
Optimizing A. tumefaciens-explant co-cultivation conditions is always essential to enhance citrus transformation efficiency. The main parameters to adjust are bacterial inoculation and co-cultivation time, bacterial concentration, medium composition, and light-darkness conditions (Febres et al., 2011). The Agrobacterium inoculation times range between 5 min (Molinari et al., 2004) and 20 min (Yang et al., 2000; Almeida et al., 2003a). However, incubation periods greater than 10 min have led to an increased number of shoot escapes and a reduction in transformation efficiency (Costa et al., 2002). The bacterial inoculum concentration varies between 4 × 107 (Peña et al., 1995b; Yu et al., 2002) and 5 × 108 cfu/ml (Kaneyoshi et al., 1994; Bond and Roose, 1998; Luth and Moore, 1999) depending on the citrus cultivar. A very low amount of bacteria reduces transformation efficiency but an excess stresses the plant cells (Costa et al., 2002; Yu et al., 2002). The co-cultivation time is usually 2 or 3 days, with an increase in the transformation efficiency with longer co-cultivation periods (Cervera et al., 1998b). Co-cultivation periods of more than 5 days, however, often lead to an overgrowth of Agrobacterium, which decreases the regeneration efficiency. Finally, optimal co-cultivation temperature varies between 19°C (Li et al., 2002, 2003) and 28°C (Luth and Moore, 1999).
Callus cells derived from cambium tissues are the most competent for regeneration (Peña et al., 2004a). Treatments favoring the development of such callus tissue as co-cultivation in a culture medium rich in auxins and incubation of the explants in darkness for the first 2–4 weeks after bacterial inoculation, greatly increased transformation frequencies (Cervera et al., 1998b). The auxin 2,4 dichlorophenoxyacetic acid (2,4-D) in co-cultivation medium led to the highest effects allowing de-differentiation of citrus cells and taking them to a competent state for stable transformation in many cultivars including sour orange, sweet orange, lime, and Troyer citrange (Ghorbel et al., 2000; Peña et al., 2004a,b; Rai, 2006).
Some reports have discussed the need for a pre-culturing step (Spencer and Towers, 1991; Costa et al., 2002). This step, however, has been replaced for the addition of acetosyringone to the bacterial inoculum and the co-cultivation medium, thus promoting transcription of A. tumefaciens virulence genes (Kaneyoshi et al., 1994; Cervera et al., 1998a).
Physical-Based Transformation Methods on Citrus
Biolistic for the Transformation of Citrus Epicotyl Explants
Different methods have been adapted from the original publication by Sanford et al. (1987) to the biolistic-mediated transformation of plant cells. For citrus species, the study of Yao et al. (1996) have reported transformation of tangelo using non-differentiated embryonic callus and therefore demonstrated the integration of transgenes in this species. The study of Bespalhok Filho et al. (2003) performed epicotyl bombardment with GUS as the reporter gene, observing expression in meristematic cambial tissue of Carrizo citrange. In all cases, plant regeneration represented a challenge, since citrus explants are recalcitrant to produce roots, and consequently, to obtain a plant comparable to those established in greenhouses or orchards.
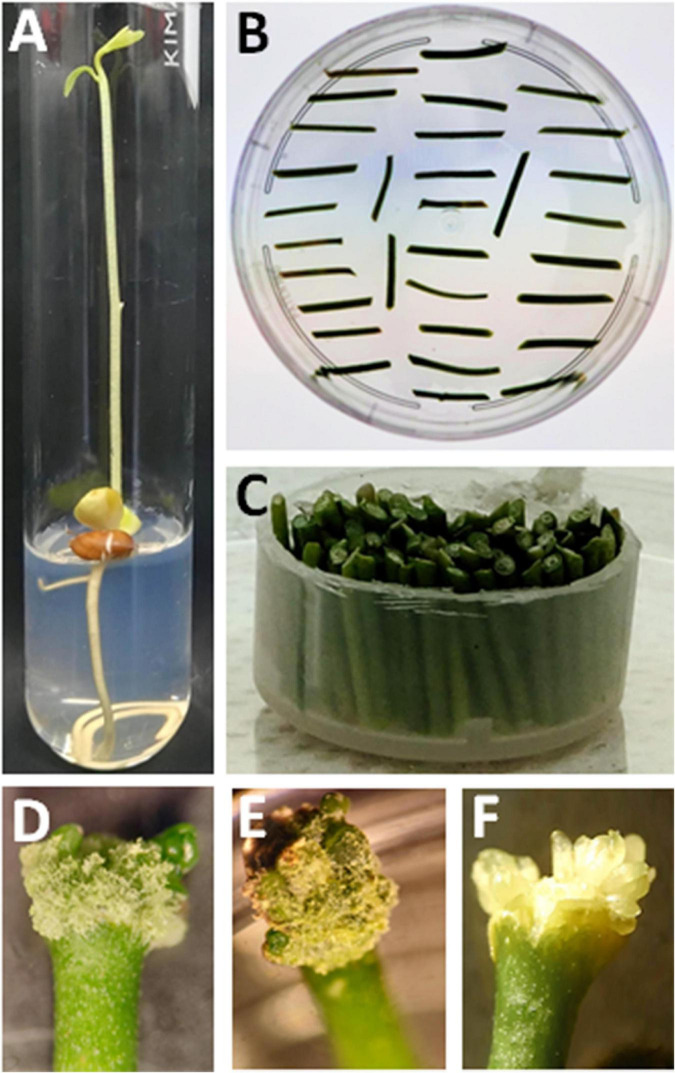
The work of Wu et al. (2016) has described the transformation of epicotyls in Carrizo citrange. In this method, epicotyl explants were bombarded at their apical region with microprojectiles coated with the DNA of interest. The DNA enters the cells, where it is expected to enter the nucleus and can be stably inserted into the genome. Callus formation was developed via tissue culture to obtain regenerated plantlets, which are finally grafted to generate a complete plant. The steps of the biolistic transformation protocol are illustrated in Figure 1.
FIGURE 1.
Bombardment and regeneration of Citrus explants. (A) Germinated seedlings in vitro; (B) Cut of epicotyls. (C) Arrangement of epicotyls in a plastic ring for bombardment. (D,E) Callus in apical regions of the explants produced in the dark after transformation. (F) Photosynthetic somatic embryos with apical dominance were produced after light exposure.
Elongated epicotyl stems are adapted to photoperiod and then used as explants for bombardment with gold or tungsten particles coated with DNA. The decision of using linear or circular DNA (plasmid) will depend on the strategy. For example, if the intention is to induce homologous recombination, the DNA should be linearized (Tomas et al., 1995; Cabrera-Ponce et al., 1997). Otherwise, a circular DNA, either in its supercoiled or relaxed form, can be efficiently transformed. The bombardment of microparticles for citrus tissue has been assayed with pressure from 900 to 2,000 psi but in general, citrus stems bear resistance to the accelerated particle impact. However, the transformation of apical stem tissues improves transformation efficiency. After bombardment, visible photosynthetic tissue emerges from the stem ends as well as from the middle portion of the stem, although in a lower proportion. In a period of 2 months, the embryos can be dissected for further heterografting. The regeneration of plants with a radicular system is a technical challenge. For this reason, grafting of the plantlet onto a vigorous rootstock is advisable. Enhanced transformation efficiency and reduction of escapes were achieved by Wu et al. (2019), with a biolistic strategy based on phosphomannose isomerase (PMI) and mannose selection (see section “Selection Markers, New Reporter Genes, and Marker-Free Systems for Citrus Transformation” for selection markers).
Magnetofection of Pollen With DNA-Functionalized Nanoparticles
Pollen magnetofection is a new technique that has been used to produce transgenic seeds without in vitro plant regeneration. This technique was successfully applied to cotton and tomato pollen-based transformation (Zhao et al., 2017; Zhang et al., 2019; Marcelino-Perez et al., 2021) and consists in the generation of nanoparticles and their functionalization to DNA or RNA encoding for the trait of interest, to generate magnetic nanoparticles (MNPs) which are then introduced into pollen through the application of a magnetic field. This goal is technically efficient since MNP-DNA/RNA complexes interact via electrostatic attractions. Specifically, 1 to 10 million pollen particles can be mixed with the loaded MNPs and placed on a magnetic plate, where they can enter into the pollen via its natural pores. Pollen must be used immediately to pollinate emasculated flowers. It should be considered that pollen viability is measured in hours; therefore, the treated pollen should be used immediately. Seeds obtained through pollination with magnetofected pollen can enclose the transgene of interest. Indeed, exogenous DNA is integrated into the plant genome and is inherited in a Mendelian manner.
Pollen magnetofection could be applied to citrus, thus facilitating the generation of genetically modified plants. Moreover, this technique is potentially useful for gene editing as well. Despite the accessibility in introducing this technique for citrus transformation, a series of considerations must be taken (Xoconostle-Cázares, personal communication). Plants should come from certified orchards and should be maintained in a biosafety greenhouse with full irrigation, fertilization, and a light/dark regime of 18:6 h. Flowering can be naturally or artificially induced and production from 20 to 100 flowers can be achieved in a period of 3 months. C. aurantifolia (Mexican lime) grown in tropical orchards, can produce flowers seven to eight times in a year, while C. sinensis (sweet orange) can flower up to three times in the same period. In a regular magnetofection experiment, 100 flowers can be pollinated. After approximately 3 months, mature fruits would be ready for harvesting. The surface of mature fruits should be smooth. The harvesting of mature fruits will allow the recovery of mature transformed seeds, which can be lately germinated in a seedling nursery at 30°C. Plantlets emerging from the nursery can be then transferred to individual pots for further analyses. An average of five positive plantlets carrying out the transgene in a regular experiment of one hundred pollinated flowers is expected. The transformation efficiency could be influenced by fruit quality. Transgenic plants should be maintained under biosafety conditions.
Polyethylene Glycol-Based Transformation and Electroporation
An alternative strategy for incorporating DNA into plant citrus cells is the transformation of protoplasts. The use of protoplasts, either for cell fusion or for DNA uptake, is a widely employed technology with great potential in the field of citrus genetic improvement (Grosser and Gmitter, 1990, 2011). One of the methods for protoplast transformation is stimulation with polyethylene glycol (PEG), a treatment that induces the agglutination of protoplasts and subsequent incorporation of DNA (or other macromolecules) via endocytosis (Kobayashi and Uchimiya, 1989; Vardi et al., 1990).
In the pioneering early protocols, antibiotic resistance genes were the selection markers of choice. Later, PEG-mediated citrus transformation included the expression of green fluorescent protein (GFP) in order to identify the transformed calli to be subsequently cultured in somatic embryogenesis induction media (Fleming et al., 2000; Olivares-Fuster et al., 2003; Guo and Grosser, 2004; Omar et al., 2007). PEG-mediated transformation is also particularly useful for citrus varieties that offer difficulties for genetic transformation via Agrobacterium, like mandarin hybrids (Dutt et al., 2018a; Omar et al., 2018). This strategy has been recently employed for inducing biallelic and homozygous mutations via genome editing of embryonic protoplast cells (Huang et al., 2020). DNA can also be introduced into protoplasts by electroporation through destabilization of plasma membranes and the formation of pores. This strategy has been employed for citrus genetic transformation of Ponkan mandarin (Hidaka and Omura, 1993) and Hamlin sweet orange (Niedz et al., 2003).
In vitro Regenerations of Transgenic Citrus Plants
Regeneration competence is the first limitation for the production of transgenic plants and many recalcitrant species actually have very low or null regeneration frequencies (Peña et al., 2004a). García-Luis et al. (1999) have proven that citrus genotypes, culture conditions, and medium composition determine the regeneration pathway (García-Luis et al., 1999; Bordón et al., 2000; Moreira-Dias et al., 2000).
Regeneration of whole transgenic citrus plants has been achieved through either organogenesis (direct or indirect) or somatic embryogenesis (Chiancone and Germanà, 2012). In vitro plant organogenesis from epicotyl and internodal stem segments is the chosen strategy which has been applied to several citrus genotypes, including Carrizo citrange, Troyer citrange, sweet orange, Mexican lime, grapefruit, Swingle citrumelo, and P. trifoliata (Peña et al., 2008). The effects of different factors, such as explant orientation, polarity, and cut surface contact with the medium as well as growth regulators treatments, were assessed in different studies in order to improve in vitro regeneration efficiency (Maggon and Deo Singh, 1995; Ghorbel et al., 1998; Pérez-Molphe-Balch and Ochoa-Alejo, 1998; Germanà et al., 2008). Somatic embryogenesis has been used for regeneration of a few transformed species including C. sinensis and C. reticulata (Li et al., 2002; Peña et al., 2008) by using different types of explants such as protoplasts and embryogenic calli (Niedz et al., 1995; Li et al., 2003). The interest in somatic embryogenesis is based on the high regeneration efficiency obtained and in the rare occurrence of somaclonal variation (Henry, 1998).
The culture media used for regeneration by either organogenesis or embryogenesis should contain a series of components to facilitate the formation of calli or buds from transformed explants, but with the minimum of escapes. For that reason, regeneration media normally contain an antibiotic agent for selection (see section “Selection Markers, New Reporter Genes, and Marker-Free Systems for Citrus Transformation” for marker selection), since only a few cells put in contact with the transformation vector are effectively transformed. Peña et al. (1995a,b, 1997) proposed the cultivation of explants in darkness for 2–4 weeks in regeneration/selection medium after co-cultivation with A. tumefaciens for the generation of a higher number of transformed buds (Domínguez et al., 2004; Peña et al., 2004b). Moreover, Moreira-Dias et al. (2000) showed that the addition of the cytokinin 6-benzylaminopurine (BAP) was a requisite for optimal shoot regeneration from Troyer citrange explants, while the influence of auxins appeared to be non-significant. The authors reported no callus and very few bud formations at the apical end of the explants in the absence of BAP and that most of the calli remained quiescent without becoming a shoot. Peña et al. (2004b) also demonstrated that co-cultivation in a BAP-supplemented media promoted a faster differentiation response and multiple bud formation in experiments of indirect organogenesis of Carrizo citrange.
The rooting of shoots and embryos is the most challenging step of citrus transgenic production, strikingly reducing transformation efficiencies. P. trifoliata, grapefruit, and Swingle citrumelo have been successfully rooted in naphthalene-acetic acid (NAA) supplemented media (Kaneyoshi et al., 1994; Luth and Moore, 1999; Molinari et al., 2004). C. macrophylla, Cleopatra mandarin, sour orange, and Mexican lime have been also efficiently rooted in medium supplemented with indole-3-butyric acid (IBA) (Tallón et al., 2012). The use of micrografting or shoot tip grafting to recover plants from transformed shoots or buds, however, has become a routine procedure, with important increases in transformation efficiencies (Peña et al., 1995a,b). This technique involves placement of the shoot tip explant onto a decapitated rootstock, which is generally an etiolated Troyer citrange epicotyl (Figure 2; Navarro et al., 1975; Peña and Navarro, 2000). The regenerated shoot apical end has to be in contact with the vascular ring of the rootstock. Grafted shoots should have previously tested positive for reporter marker activity (GFP or β-D-glucuronidase, GUS, Figure 3), denoting the transgenic nature of the explant. Micrografted shoots or buds are then cultured in a liquid nutrient medium for proper growing. Subsequently, a new grafting of the in vitro-grown plantlets on vigorous rootstocks in the greenhouse allows rapid acclimatization (Peña et al., 2008).
FIGURE 2.
Shoot tip grafting procedure. (A) Decapitated etiolated epicotyls to be used as rootstocks. (B) Micrografting of a transformed shoot using a stereoscopic microscope. INSET: shoot detached from the internodal segment. (C) In vitro growing of the grafted plantlet.
FIGURE 3.
Reporter expression of transformed citrus explants. (A,B) GFP expression of transgenic C. sinensis shoots (green). Red shoot and internodal segments are non-transgenic tissue. (C) GUS expression in a disk section from transgenic shoot base. (D) GUS positive (blue) and negative (green) leaves coming from transformed explants. (E) GUS expression in a disk section from a chimera shoot.
The micrografting methodology has been efficiently used for whole plant generation from shoots derived from organogenesis as well as from germinated somatic embryos (Niedz et al., 2003; Olivares-Fuster et al., 2003). Some of the different citrus genotypes in which this technique was applied include C. sinensis, P. trifoliata, C. aurantifolia, C. aurantium, and grapefruit (Kobayashi et al., 1996; Bond and Roose, 1998; la Malfa et al., 2000; Wong et al., 2001; Januzzi Mendes et al., 2002; Yu et al., 2002; Almeida et al., 2003a,b; Boscariol et al., 2003; Iwanami et al., 2004; Endo et al., 2005; Fagoaga et al., 2005).
Genotype Dependence of Citrus Transformation
The efficiency of genetic transformation and regeneration highly depends on the genotype of all cultivated natural or hybrid species of the genus Citrus and its relatives (Bond and Roose, 1998). As well as for P. trifoliata (Kaneyoshi et al., 1994), the studies by Peña et al. (1995a), Cervera et al. (1998b), and Yu et al. (2002) have developed highly efficient methods for the transformation of modern commercial hybrid cultivars, such as Carrizo and Troyer citrange rootstocks using A. tumefaciens followed by in vitro regeneration and shoot tip grafting.
Other pieces of research have reported biolistic transformations (Wu et al., 2016) and transformation of mature tissues of several varieties including hybrids such as US-942 (C. reticulata × P. trifoliata) and Flying Dragon (Marutani-Hert et al., 2012). Other commercial hybrids currently used, such as swingle citrumelo and tangelo (C. reticulata × C. paradise), are also susceptible to Agrobacterium-mediated transformation (Molinari et al., 2004; De Oliveira et al., 2009).
Among the natural hybrids or secondary species, sweet orange has been extensively studied, and numerous protocols were published mainly based on the Agrobacterium-mediated methodology (Peña et al., 1995c; Bond and Roose, 1998; Yu et al., 2002; Li et al., 2003; Cervera et al., 2005; De Oliveira et al., 2009; Fávero et al., 2012), protoplast transformation (Fleming et al., 2000) and embryogenic suspension cells (Dutt and Grosser, 2010). For sour orange, Gutiérrez-E et al. (1997), as well as Ghorbel et al. (2000), developed an efficient transformation strategy, also based on Agrobacterium co-cultivation. Another natural hybrid used as rootstock is rough lemon, which was genetically transformed by Savita et al. (2011) and Ali et al. (2012). Other crops, such as Mexican lime (Peña et al., 1997; Pérez-Molphe-Balch and Ochoa-Alejo, 1998; Domínguez et al., 2000; De Oliveira et al., 2015), Femminello siracusano lemon (C. limon (L.) Burm. F) (Gentile et al., 2007) and grapefruit (Yang et al., 2000; Costa et al., 2002; Jia et al., 2017) have been efficiently transformed and regenerated.
Agrobacterium-mediated conventional transformation methods, using either juvenile or mature tissue explants showed low efficiency in mandarins and clementines (Cervera et al., 1998b). However, some more recent pieces of research have reported optimized methods (Li et al., 2002; Khawale et al., 2006; Cervera et al., 2008) and alternative strategies based on the use of cell suspensions and protoplast transformation (Dutt et al., 2018a).
To date, several biotechnological developments have been reported for different citrus species and varieties (see detailed information in Table 1).
TABLE 1.
Main biotechnological developments in citrus or relative species.
| Citrus Species/Variety | Trait | Gene of interest | Greenhouse/Field trial | Strategy | References |
| C. sinensis | Resistance to Citrus Canker | CsLOB1 | G | Gene editing | Peng et al., 2017 |
| C. sinensis | WRKY22 | G | Gene editing | Wang et al., 2019 | |
| C. sinensis | hrpN | G | Overexpression | Barbosa-Mendes et al., 2009 | |
| C. paradisi | CsLOB1 | G | Gene editing | Jia et al., 2016, 2017 | |
| C. sinensis | Attacin A | G | Overexpression | Boscariol et al., 2006; Cardoso et al., 2010 | |
| C. sinensis | MdSPDS1 | G | Overexpression | Fu et al., 2011a | |
| C. sinensis | Dermaseptin | G | Overexpression | Furman et al., 2013 | |
| Troyer citrange | Snakin-1 | G | Overexpression | Conti et al., 2020 | |
| C. sinensis | Peroxidase25 | G | Overexpression | Li et al., 2020 | |
| C. sinensis and Carrizo citrange | FLS2 receptor | G | Overexpression | Hao et al., 2016a | |
| C. sinensis | Bs2 | G | Overexpression | Sendín et al., 2017 | |
| W. Murcott mandarin | Xa21 | G | Overexpression | Omar et al., 2018 | |
| Carrizo citrange | Resistance to Citrus Canker and Huanglongbing | M-thionin | G | Overexpression | Hao et al., 2016b |
| C. sinensis | Resistance to Huanglongbing | NPR1 | F | Overexpression | Dutt et al., 2015 |
| C. sinensis | Cecropin B | G | Overexpression | Zou et al., 2017 | |
| C. aurantifolia | β-defensin 2 and Lysozyme | G and F | Overexpression | Guerra-Lupián et al., 2018 | |
| C. sinensis | SAMT1 | G | Overexpression | Zou et al., 2021 | |
| Citrus sp. | SOD2 and SOD7 | G and F | Overexpression | EPA-HQ-OPP-2014-0834, F.-9926-99, 2015 | |
| C. sinensis | Reduced attraction to Diaphorina citri | (E)-β-caryophyllene synthase | G | Overexpression | Alquézar et al., 2021 |
| C. sinensis | Resistance to CPsV | CPsV coat protein | G | RNA silencing | Reyes et al., 2011 |
| C. aurantifolia | Resistance to CTV | CTV p25 coat protein | F | Overexpression | Domínguez et al., 2002 |
| C. aurantifolia | CTV p23 from CTV | F | Overexpression | Soler et al., 2012 | |
| C. aurantifolia | Drought and Salinity Tolerance | CBF3 | G | Overexpression | Romero-Romero et al., 2020 |
| C. sinensis | β-carotene content synthesis | Csβ-CHX | G | RNA silencing | Pons et al., 2014 |
Types of Citrus Explants Used for Genetic Transformation
A variety of explants, such as internodal stem segments, epicotyls, cotyledons, leaf segments, protoplasts, and embryogenic calli from different citrus species and relatives, have been assessed for Agrobacterium-mediated transformation (Moore et al., 1992; Almeida et al., 2003a; Kayim et al., 2004; Khawale et al., 2006; Omar et al., 2007; Chen et al., 2008; De Oliveira et al., 2009, 2015; Ballester et al., 2010; Duan et al., 2010; Fu et al., 2011a,b; Ali et al., 2012). All of them have their intrinsic advantages and drawbacks.
The first report of citrus transformed material was of callus tissue formed from primary explants (Grinblat, 1972; Chatuverdi and Mitra, 1974; Barlass and Skene, 1982; Edriss and Burger, 1984). The addition of growth regulators, mainly the cytokinin BAP, has shown a good response for this kind of explants. However, embryogenic calli lose their regeneration capacity when they are sub-cultured for long periods.
Juvenile internodal stems explants are one of the most prevalent and efficient starting materials for citrus transformation (Orbović and Grosser, 2015). The availability of this kind of explants and the higher regeneration and transformation frequencies are the major advantages over the use of mature tissue explants. The generation of plants using this material, however, would take more than 10 years to flower and fruit, which drastically prolongs the time required to analyze newly introduced traits. The optimization of transformation techniques that bypass the juvenile stage could reduce the time and costs involved in evaluating transgenic new traits (Almeida et al., 2003a; Cervera et al., 2005, 2008). Mature citrus tissue explants help to reduce flowering time but suffer from a considerable decline in regeneration frequency (50–70% less) and transformation potential, therefore it is necessary to carefully select material and adjust tissue culture conditions and media composition (Von Aderkas and Bonga, 2000). As of today, mature materials from sweet orange (Cervera et al., 1998a,2005), sour orange (Ghorbel et al., 2000), lime, and some mandarin genotypes have been successfully transformed and regenerated. A selection of stem pieces from the first flushes of propagated adult buds was the choice for transforming mature tissue. In this regard, transgenic sweet orange plants regenerated from mature tissues flowered and produced fruits 14 months after transferring to the greenhouse (Cervera et al., 2005).
The study of Kaneyoshi et al. (1994) have established the first efficient protocol for the transformation of in vitro seedling material and applied it to the generation of transgenic P. trifoliata plants. By using 1 cm long etiolated epicotyl segments from 20-day-old grown in vitro as starting material for transformation, they demonstrated that these kinds of explants were highly responsive to shoot regeneration, with the extra advantage of not requiring explant disinfection steps. Peña et al. (1995b) used a similar protocol to transform Carrizo citrange and Gutiérrez-E et al. (1997) to transform bitter orange (C. aurantium) and Key lime (C. aurantifolia). Similar protocols were applied to transform epicotyl segments from C. sinensis commercially important cultivars including Washington navel, Valencia, Hamlin, Pera, and Natal (Bond and Roose, 1998; Luth and Moore, 1999; Boscariol et al., 2003), Original protocol from Kaneyoshi et al. (1994) suffered different modifications including longitudinal cuttings of the epicotyl segments (in two halves) to enhance regeneration and transformation frequency (Yu et al., 2002) or cutting of transversally thin layers of about 1–2 mm (Le et al., 1999). All the changed conditions only reduced transformation efficiency compared with the use of 1 cm long explants probably due to Agrobacterium overgrowth and toxicity of such small explants (Molinari et al., 2004). Lately, Costa et al. (2002) and Febres et al. (2003), efficiently transformed C. paradisi cv. Duncan and De Oliveira et al. (2015) also established a similar protocol using 30-day-old epicotyls for the transformation of Mexican lime.
Leaf explants have been also tested for genetic transformation either for direct organogenesis or going through an intermediate process of callus formation (Moore et al., 1992; Khan et al., 2009; Ali et al., 2012). Abundant and rapid accessibility to leaf disks from germinating seedlings and a lower risk of contamination make them a considerable option as starting material for Agrobacterium-mediated transformation. Mature leaf transformation allows introducing new traits without losing the clonal fidelity compared with epicotyls (Sandal et al., 2007). Moore et al. (1992) used citrus leaf disks as explants and compared their organogenic potential with stem segments. They reported that shoot production was much more effective when stems were used as explants compared with leaf segments, probably because organogenesis occurs with higher efficiency from stems (Almeida et al., 2003a).
Sweet orange was the first woody crop in which plant protoplasts were used for regeneration processes (Vardi et al., 1982; Kobayashi et al., 1983). Nowadays, protoplasts are used as starting material for most citrus species and relatives and were applied to produce somatic hybrid plants from more than 150 parental combinations (Grosser and Gmitter, 1990), thus contributing to germplasm expansion and improvement (Grosser et al., 1996, 1998a,b). It is well-known, that protoplast generation is a time-consuming and labor-intensive methodology but the use of this kind of culture as explants has a series of advantages. Protoplast transformation can circumvent the use of antibiotic-resistance genes and antibiotic selection, thus eliminating some public perception problems (Fleming et al., 2000). This system could subsequently be extended to other polyembryonic citrus cultivars, including seedless sweet oranges, lemons, or satsuma mandarins.
Among the citrus species, juvenile tissues from mandarin hybrids, including epicotyls, are the most difficult to infect and transform with A. tumefaciens (Cervera et al., 1998b), which results in low genetic transformation efficiency (Dutt et al., 2010). Direct incorporation of DNA into protoplasts using electroporation (Niedz et al., 2003) or PEG-mediated DNA uptake (Fleming et al., 2000; Omar et al., 2007) is an alternative to bypass Agrobacterium-mediated transformation problems in those genotypes. A clear example is the PEG-mediated transformation of W. Murcott tangor using protoplasts, which allows a considerable increment in the transformation efficiency compared with the conventional epicotyl-mediated Agrobacterium process (Dutt et al., 2018a). The use of protoplasts has been recently taken into consideration again, regarding their amenability for gene editing, with or without the use of DNA molecules, which minimizes the possibility of foreign DNA integration (Huang et al., 2020).
Selection Markers, New Reporter Genes, and Marker-Free Systems for Citrus Transformation
Most of the in vitro citrus regeneration and transformation protocols need selectable marker genes (antibiotic or herbicide resistance) such as the nptII (Neomycin Phosphotransferase II) gene, in combination with kanamycin as a selective agent (reviewed in Peña et al., 2004a). Concomitant to the use of nptII, the product of expression of the uidA gene (GUS), has been widely applied as a co-expressed reporter gene to facilitate the selection of positive transformants (Moore et al., 1992; Peña et al., 1997; Cervera et al., 1998c; Domínguez et al., 2000). GUS has been extensively employed as a reporter gene for citrus genetic transformation (Moore et al., 1992; Peña et al., 1995b,1997; Gutiérrez-E et al., 1997). Its adequate use is important to avoid escapes and the incidence of chimeras (Gutiérrez-E et al., 1997; Yu et al., 2002; Figure 3E). However, Domínguez et al. (2002) have demonstrated that the detection of a high frequency of transformants was possible without nptII/uidA selection. Indeed, once the regeneration of the positive transformants, which are detected by PCR-mediated analysis of regenerated roots, is achieved, antibiotic resistance is no longer necessary. But, in the case of nptII and uidA genes, other than remaining stably integrated into the genome for long term, they do not produce negative effects on crop characteristics (Pons et al., 2012).
To find other selectable marker genes suitable for citrus genetic transformation, Costa et al. (2002) have developed a protocol for grapefruit transformation based on the use of hygromycin as a selective agent. The hpt gene codes for a hygromycin phosphotransferase that, like the protein product of nptII, detoxifies aminoglycoside antibiotics by phosphorylation. Hygromycin selection, however, faces difficulties regarding the screening of transgenic tissues, because of the generation of escapes and chimeras (Padilla and Burgos, 2010). Resistance to phosphinotricine (Basta, Bialaphos, or glufosinate) for transgenic callus selection has been employed in ponkan embryogenic calli overexpressing bar gene (Li et al., 2002). Later on, Zhang Y. Y. et al. (2017) developed a transformation approach for pummelo (C. maxima) based on in planta A. tumefaciens infection and subsequent selective culture using hygromycin, Basta or kanamycin resistance. After PCR-based screening of regenerated shoots, efficiencies achieved were 20.41, 19.37, and 3.21% respectively. Recently, the study of Merritt et al. (2021) reported glyphosate-resistant Duncan grapefruit plants, obtained by inducing native EPSPS mutations.
The use of selectable marker genes for resistance to antibiotics and or herbicides has been a matter of public constraint (Miki and McHugh, 2004) and in some cases has been shown to reduce regeneration capacity (Moore et al., 1992). Therefore, researchers developed alternative methods for screening and selecting transformed tissues. In this regard, GFP and its enhanced derivatives (EGFP) have been extensively employed as reporter genes for the selection of transgenic plant tissues (Chiu et al., 1996; Stewart, 2001) in Agrobacterium-mediated transformation (Ghorbel et al., 1999; Fleming et al., 2000; Yu et al., 2002; Cervera et al., 2008), in protoplast transformation (Guo and Grosser, 2004; Omar and Grosser, 2008), for biolistic assays (Wu et al., 2016) and gene editing (Huang et al., 2020). A novel reporter system based on the measurement of increased anthocyanin accumulation by overexpression of Ruby and VvMYBA1 transcription factors has been used to detect Agrobacterium-mediated Mexican-lime transformed explants (Dutt et al., 2016) or by overexpressing VvMYBA1 in protoplasts, under the control of embryo-specific Dc3 promoter (Dutt et al., 2018b).
Numerous alternative methods have been developed in order to replace the selection systems based on antibiotics resistance. The PMI system (PMI/mannose) is a positive selection strategy based on the expression of the manA gene from Escherichia coli, which is able to metabolize mannose to fructose-6-phosphate. When the only carbon source for explants in the selective culture is mannose, the positive transgenic tissues will be capable of growing, whereas non-transformed tissues will have growth arrest due to carbon starvation (Wang et al., 2000). Boscariol et al. (2003) have used this system for the selection of transformed sweet orange tissues coming from Agrobacterium-mediated transformation of epicotyl segments and achieved transformation efficiencies of 3–23% depending on the variety used. Subsequently, Ballester et al. (2008) achieved higher or similar transformation rates for citrange (30%) and sweet orange (13%) using in vitro-germinated seedlings as a source of epicotyl segments. The PMI/mannose positive selection system combined with EGFP based monitoring of transformed tissues allowed early elimination of escapes in Carrizo explants (Dutt et al., 2010). In biolistic assays, this alternative system has been successfully applied in Carrizo citrange, with efficiencies of 1.9% positive shoots per shot (Wu et al., 2019).
Concomitantly to the use of alternative selection methods that do not rely on antibiotic resistance genes, the possibility of removing the marker gene once the transgenic plants have been recovered is suitable for woody species, where seedless propagation is usually observed, and long juvenility increase time periods required for getting segregation results on transgenic progenies. The multi-auto-transformation (MAT) vector system enables the production of marker-free transgenic plants by a combination of a positive selection mediated by the isopentenyl transferase (ipt) gene and a site-specific DNA recombination tool (Sugita et al., 1999). The enzyme ipt is involved in the production of cytokinins that induce cell division and the overproduction of transgenic shoots. The second component of the MAT system is a site-specific recombinase (R/RS) that removes DNA sequences located between RS recognition sites after transformation. The RS sites flank the ipt marker and the R recombinase transgenes, to facilitate the elimination of the selection marker system after cell transformation. In citrus, the system has been successfully applied in sweet orange, but not in citrange, where a high proportion of chimeras and erroneous sequence recombination occurred (Ballester et al., 2007). The addition of an indolacetamide-hydrolase and tryptophan-monooxygenase (iaaM/H) marker gene and an inducible promoter for controlling the site-specific recombinase R rendered higher efficiency rates (Ballester et al., 2008). Zou et al. (2013) reported the use of other marker-free transformation systems in citrus, based on the expression of Cre/loxP site-recombination coupled to the ipt selectable marker gene. The GFP reporter gene was inserted outside the loxP sequences with the aim of monitoring the rate of transformation and deletion efficiencies. The results obtained demonstrated that Cre/loxP-mediated excision was highly effective and accurate for Jincheng sweet orange. Marker-free transgenic Tarocco blood orange overexpressing antibacterial peptide gene AATCB, which conferred enhanced resistance to citrus canker, was obtained by using a Cre/loxP mediated recombination system combined with ipt positive selection. Transformation efficiency achieved was 21.4% (Peng et al., 2015). Recently, Peng et al. (2021a) described a similar strategy to confer citrus canker resistance but by co-transformation and sequential re-transformation of Tarocco blood orange with the same AATCB gene and another antimicrobial peptide, PR1aCB. They also used Cre/loxP-mediated site-specific recombination system and ipt selection to get marker-free plants and confirmed that double transformants showed enhanced citrus canker resistance.
Promoter Sequences Used for Citrus Transgene Expression
The type of promoter used in the chimeric gene construct for plant transformation is essential to achieve adequate temporal or spatial regulated expression of the desired trait. Although the number of promoter sequences is rather limited, the selection of an adequate promoter is not a trivial issue (reviewed in Smirnova and Kochetov, 2020). Different promoters derived from virus, bacteria, or plant species have been employed for citrus genetic transformation. Table 2 displays a list of promoter sequences used in citrus, considering the species of origin and the regulated gene.
TABLE 2.
Promoters used in citrus genetic transformation.
| Type of expression | Promoter | Source | Transformed plant | Controlled gene | References |
| Constitutive | 35S | CaMV (Cauliflower mosaic virus) | Sour orange | CTV coat protein | Gutiérrez-E et al., 1997 |
| Sour orange | CTV coat protein | Ghorbel et al., 2000 | |||
| Pineapple Sweet orange | GUS | Peña et al., 1995a | |||
| Mexican lime | Peña et al., 1997 | ||||
| Carrizo citrange | Cervera et al., 1998c | ||||
| Pineapple Sweet orange | Cervera et al., 1998a | ||||
| Washington Navel Sweet orange | Bond and Roose, 1998 | ||||
| Citrange | AP1 and LFY | Peña et al., 2001 | |||
| Itaborai Sweet orange | GFP | Fleming et al., 2000 | |||
| Pineapple Sweet orange | CPsV hrps* for 54k, coat protein and 24k | Reyes et al., 2011 | |||
| Mexican lime | CTV hrps* for p20, p23 and p25 | Soler et al., 2012 | |||
| Pineapple Sweet orange | Dermaseptin | Furman et al., 2013 | |||
| Troyer citrange | StSnakin-1 | Conti et al., 2020 | |||
| Duncan grapefruit | PtFT1-scFv | Sinn et al., 2021 | |||
| Wanjincheng Sweet orange | CsSAMT1 | Zou et al., 2021 | |||
| 34S | Duncan grapefruit | CTV coat protein, RdRp and genomic 3′ end RNA | Febres et al., 2003 | ||
| MAS | Agrobacterium tumefaciens | Jincheng and Newhall Navel Sweet orange | Shiva A and Cecropin B | He et al., 2011 | |
| Full length CsCYP, CsGAPC2, and CsEF1 | Sweet orange | Hamlin Sweet orange | GUS | Erpen et al., 2018 | |
| Partial CsCYP, CsGAPC2, and CsEF1 | Nicotiana benthamiana | Corte et al., 2020 | |||
| YAO | Arabidopsis thaliana | Carrizo citrange | PDS | Zhang et al., 2017 | |
| Embryo-specific | Dc3 | Carrot | Hamlin Sweet orange | VvMybA1 | Dutt et al., 2018b |
| Fruit-specific | CitMT45 | Satsuma mandarin | Valencia Sweet orange | GUS | Endo et al., 2007 |
| Pulp and flower-specific | Cl111 | Acid lemon | Acid lemon and acidless lime | Sorkina et al., 2011 | |
| Flower-specific | CitSEP, CitWAX, CitJuSac, CitVO1, and PamMybA | Sweet orange | Micro-tom tomato | Dasgupta et al., 2020 | |
| Seed-specific | CuMFT1 | Satsuma mandarin | Trifoliate orange, satsuma mandarin, Kishu mikan and Arabidopsis thaliana | Nishikawa et al., 2008 | |
| Xylem vessels-specific | CsPP | Madam Vinous orange | Tobacco and Valencia orange | De Azevedo et al., 2006 | |
| Phloem-specific | rolC | Agrobacterium rhizogenes | Mexican lime | GUS | Dutt et al., 2012 |
| RTBV | Rice Tungro Bacilliform Virus | ||||
| RSs1 | Oryza sativa | ||||
| AtSUC2 | Arabidopsis thaliana | ||||
| AtSUC2, AtPP2 | Arabidopsis thaliana | Hamlin, Pera and Valencia Sweet orange | Miyata et al., 2012 | ||
| CsPP2 | Sweet orange | ||||
| AtSUC2, AtPP2 | Arabidopsis thaliana | Hamlin and Valencia Sweet orange | Attacin A | Tavano et al., 2019 | |
| CsPP2 | Sweet orange | Carrizo citrange | CcFT3 | Soares et al., 2020 | |
| AtSUC2 | Arabidopsis thaliana | ||||
| GRP1.8 | Phaseolus vulgaris | Tarocco blood Sweet orange | Cecropin B | Zou et al., 2017 | |
| CsPP2.B1 and CsVTE2 | Sweet orange | Carrizo citrange | GUS | Bezerra et al., 2021 | |
| Pathogen-inducible | gst1 | Potato | Mexican lime | hrpN | Barbosa-Mendes et al., 2009 |
| Pineapple Sweet orange | Bs2 | Sendín et al., 2017 | |||
| Jincheng Sweet orange | GUS | Zou et al., 2014 | |||
| PPP1, hsr203J | Tobacco | ||||
| PR5 | Sweet orange | Troyer citrange | CsMAPK1 | De Oliveira et al., 2013 | |
| Heat shock-inducible | AtHSP70BP | Arabidopsis thaliana | Duncan grapefruit, Valencia Sweet orange, Key lime, Carrizo citrange, Sour orange, and Meiwa kumquat | GUS | Jia and Wang, 2014b |
| Stress-inducible | AtRD29A | Arabidopsis thaliana | Duncan grapefruit and Valencia Sweet orange | CsAP1 and CsLFY | Orbović et al., 2021 |
*hrps: Hairpins to induce RNA silencing.
Over the last years, a lot of work has been done to increase the availability of promoters and other regulatory sequences, including the development of synthetic promoters for citrus genetic transformation and its application to new breeding techniques (reviewed in Ali and Kim, 2019).
Early Flowering Induction to Reduce the Juvenile Phase of Transgenic Citrus
Long juvenile phases (often more than 5 years) are a major constraint to the success of transformation methods based on juvenile tissue explants and of conventional breeding programs. The development and commercial release of new varieties by traditional breeding may require a complete process that can range from 25 to 30 years (Caruso et al., 2020). The discovery of novel genes implicated in citrus precocious flowering is relevant for the improvement of interesting traits, either by transgenesis or by conventional breeding. Some strategies to reduce the long juvenile periods in citrus trees rely on the basis of the knowledge related to flowering pathways in Arabidopsis thaliana.
The first successful approach in citrus plants that was conducted to reduce generation periods consisted of the constitutive overexpression of Arabidopsis LEAFY (LFY) or APETALA1 (AP1) genes in juvenile seedlings (Peña et al., 2001). Both genes are involved in the induction of flowering and their individual expression induced early flowering and fruiting in transgenic citrange rootstocks. AP1, however, was more efficient than LFY, because LFY also induced abnormalities in the vegetative growth. Flowering Locus T (FT) citrus orthologue, CiFT, was ectopically overexpressed in P. trifoliata (Endo et al., 2005) with subsequent shortening of the juvenile period, but again, the vegetative growth and plant architecture were aberrant. Velázquez et al. (2016) developed a viral vector-based tool to induce early flowering: the Citrus Leaf Blotch Virus (CLBV) carrying AtFT or CiFT genes. Within 4–6 months of vector inoculation in different genotypes, flowering was initiated with no other phenotypic abnormalities. Furthermore, Soares et al. (2020) reported a novel strategy to induce precocious flowering by overexpressing the Citrus clementina (CcFT3) orthologue under the control of AtSUC2 phloem-specific promoter in Carrizo citrange rootstocks. This strategy led to plants with normal morphology that flowered 16 months after transformation and, when juvenile scions were grafted, earlier flowering was also induced.
Other alternatives have been employed to reduce the pleiotropic effects derived from constitutive overexpression of early flowering genes. For instance, Sinn et al. (2021) developed transgenic grapefruits expressing P. trifoliata FT1 (PtFT1) as a translational fusion with a single-chain variable fragment antibody. The reduced FT activity rendered transgenic FT chimeras with precocious flowering.
The huge amount of genomic data available for citrus species and phylogenetically related genus would make the discovery of new genes involved in precocious flowering possible. This is the case for Mini-citrus (Fortunella hindsii), wild citrus-related species with dwarf height and early flowering (juvenile period of around 8 months) (Zhu et al., 2019). The discovery of new candidate genes for transgenic or gene editing approaches would speed up the implementation of biotechnological improvements in citrus species (Shimizu, 2020; Rao et al., 2021).
Novel Strategies for Transient and Stable Transformation of Citrus
Generation of stably transformed citrus plants requires arduous and time-consuming procedures, as is the case for most woody species. As mentioned before, the success depends on genotype-associated transformation efficiencies and needs long periods for in vitro regeneration of positive events. Also, the principal characteristics of the candidate genes to introduce before conducting stable transformation of the plants should be previously analyzed. For that purpose, transient gene expression is a useful tool to study the function, subcellular expression patterns, and localization of the genes of interest. This method also allows characterizing novel genes and regulatory sequences in a fast and simple manner (Jones et al., 2009).
In citrus, the study of Figueiredo et al. (2011) has determined the function and subcellular localization of a type III effector AvrGf1 from Xanthomonas citri pv. citri by Agrobacterium-mediated transient expression (agroinfiltration) in grapefruit leaves. In addition, it has been suggested that a X. citri pv. citri treatment before agroinfiltration could significantly enhance transient expression in recalcitrant citrus leaves from different varieties (Jia and Wang, 2014b). The constraints for this pre-treatment are related to the possible side effects derived from the presence of pathogenic bacteria that could interfere with the functional characterization of the gene of interest. Li et al. (2017) have analyzed different factors influencing transient expression efficiency in citrus. They suggested composition of infiltration buffer adequate for an enhanced level of transient expression: 10 mmol L–1 2-(N-morpholino) ethanesulfonic acid (MES), pH 5.6, 10 mmol L–1 MgCl2, and 150 μmol L–1 Acetosyringone. The bacterial suspension density most suitable for transient expression was OD600 = 0.8. The optimal conditions of other parameters, such as temperature, leaf developmental stages, and dependence on genotypes, were also determined. A recent study has shown that an agroinfiltration procedure using a microneedle roller to create abundant little wounds in the leaf surface increased the gene expression efficiency (Acanda et al., 2021). Strategies for transient gene expression based on particle bombardment have been also developed. A held-gene gun system was used for transient transformation of thin epicotyl explants of Carrizo citrange and sweet orange (Bespalhok Filho et al., 2003) as well as for citrus leaves (Levy et al., 2018). The latter system is applicable to a wide variety of genotypes but not all laboratories have the required device. Another novel methodology for transient citrus fruit transgene expression based on fruit immersion in an Agrobacterium suspension and subsequent vacuum infiltration has been reported by Zhang et al. (2021).
A localized expression is a rapid tool allowing testing the correct expression of a transgene. In this regard, Guerra-Lupián et al. (2018) have developed a method for localized expression in stems of Mexican lime by using A. tumefaciens and expression vectors coding for reporter genes and antimicrobial peptides targeted to the vascular tissues (López-Buenfil et al., 2017; Guerra-Lupián et al., 2018). Stem transformation with A. tumefaciens carrying the transgene constructs requires a slight scraping of the corky surface with a scalpel, in order to expose the photosynthetic tissue, and then it is incubated with the bacteria to favor the transformation process (Figure 4). In some instances, the appearance of thick photosynthetic tissue is observed in the injury performed with the scalpel; in general, the scar is lignified and becomes indistinguishable over time (López-Buenfil et al., 2017). Molecular detection in the site of the transformation for evaluation of transgene expression can produce data that are difficult to interpret since Agrobacterium cells can be viable for weeks at the site of inoculation. However, it has been possible to detect reporter gene products systemically, in distant tissue. This fact is very interesting because informational molecules can be mobilized via phloem at long distances in the plant, without having to generate genetically modified plants.
FIGURE 4.
Localized expression procedure. (A) Exposition of photosynthetic tissue by scraping made with a scalpel. (B) Soaking of a cotton swab with Agrobacterium culture. (C) Wrapping of plant tissue. (D) Treated plant, covered with plastic.
The alternative methodology to achieve stable transgene expression by agroinoculation of axillary meristems is very useful to bypass the complexity and time-consuming procedures required for regenerating a whole plant from a single cell. He et al. (2011) have developed transgenic Jincheng orange and Newhall navel orange overexpressing antibacterial Shiva A and Cecropin B proteins by Agrobacterium-transformation of in vitro micrografted mature axillary buds. The transgenic plants subsequently regenerated showed resistance to X. citri pv citri.
An interesting new in planta transformation approach developed by Zhang Y. Y. et al. (2017) consists of an Agrobacterium co-culture with decapitated pummelo seedlings, selective-culture, and dark treatment for inducing bud formation. This strategy resulted in transgene integration with transformation efficiencies of 20.41% when using hygromycin as the selection marker and of 19.37 and 3.21% when using Basta and Kanamycin, respectively.
Other relevant tools for the expression or silencing of citrus genes are the viral-based expression vectors (Folimonov et al., 2007). Citrus Tristeza Virus (CTV)-based engineered constructs expressed the GFP reporter gene for more than 4 years (Folimonov et al., 2007). CTV showed stable expression of transgenes placed within the 3′ region of its genome (El-Mohtar and Dawson, 2014). Velázquez et al. (2016) have reported the use of CLBV for inducing early flowering of juvenile citrus plants, by transient expression of citrus Flowering Locus T gene (CiFT). This viral vector displayed a series of advantages, such as the absence of plant genome integration or recombination, the scarce range of symptoms expressed in most citrus cultivars, its systemic distribution in the plant, and its safety for field trials, because it is not transmissible by insects.
Emerging Technologies: Cis and Intragenesis, Trans-Grafting and Gene Editing in Citrus
In citrus species, as well as in other woody fruit species, breeding programs based on transformation methods using Agrobacterium or biolistic strategies, have been successful in the precise insertion of foreign DNA for the improvement of desired traits without altering the genetic background. However, the presence of genes from other species, the use of selectable markers, and regulatory sequences coming from viruses or bacteria raised considerable public concerns. In this regard, a series of new breeding technologies (NBTs) was developed to modify existing DNA sequences in a plant or to modulate the patterns of endogenous gene expression and were successfully implemented for woody fruit plants in recent years (reviewed in Limera et al., 2017; Poles et al., 2020). Cis and intragenesis, trans-grafting, and gene editing techniques will be considered within this category. Although their application for citrus genetic improvement is still limited, some examples can be mentioned.
Cisgenic and intragenic plants are genetically modified organisms bearing DNA sequences from the species itself (extra copies) or from a closely related species that can be crossed conventionally, in contrast to transgenesis, where genetic material can be mixed between species. In the case of cisgenesis, the natural complete variant includes the promoter, introns, and terminator sequences in the same orientation as the native gene (Lusser and Davies, 2013). In intragenesis, the introduced DNA sequence can be a combination of genes and regulatory sequences (chimeric gene rearrangements) that will lead to different functional versions (Rommens et al., 2007). Recently, several citrus genes that control traits of interest have been cloned and characterized as novel targets for cis and intragenesis approaches. Between them CsSAMT1 (Salicylic acid Carboxyl Methyltransferase 1), which confers tolerance to HLB; CsMADS5, a fruit ripening-associated transcription factor, that positively regulates carotenoid biosynthesis in citrus; a CsMYB96 transcription factor, which enhances citrus fruit resistance against fungal pathogens; CiMADS43, a MADS-Box gene involved in citrus flowering and leaf development and CiNPR4, an NPR1-like gene that enhanced resistance of transgenic citrus plants to HLB (Li et al., 2020; Khan et al., 2021; Lu et al., 2021; Peng et al., 2021b; Zeng et al., 2021; Zhang et al., 2021; Zou et al., 2021).
Cis and intragenic citrus plants must be transformed with citrus-derived DNA sequences. For that purpose, in the case of the Agrobacterium transformation method, a suitable transformation vector system that carries citrus-derived complete T-DNA sequences is desirable. Plant-derived transfer DNA (P-DNA) was already developed to replace bacterial vector T-DNA backbone with a plant DNA sequence (Rommens, 2004). For citrus genetic transformation, An et al. (2013) have developed an intragenic vector system by adding a T-DNA-like sequence from C. clementina, in the correct orientation and with a series of restriction sites for cloning the gene of interest. The empty vector was used to transform A. thaliana and Duncan grapefruit leading to the recovery of positive events under non-selective conditions (3 and 0.67% transformation efficiencies for both species, respectively).
In citrus, the use of reporter genes based on the production of anthocyanin (Dutt et al., 2016, 2018b; Huang et al., 2019) and systems to remove selectable markers have shown positive results (Sugita et al., 1999; Zou et al., 2013). Merritt et al. (2021) developed a citrus DNA glyphosate resistant-selection system by transforming Duncan grapefruit with a glyphosate-resistant mutated version of citrus EPSPS enzyme (TIPS EPSPS). They showed that a glyphosate treatment did not inhibit bud formation and rendered a 40% increment in transformation efficiency. A third requirement is that the native target gene must be linked to a suitable regulatory sequence. For that purpose, several citrus-derived promoters are being characterized (Erpen et al., 2018; Dasgupta et al., 2020; Bezerra et al., 2021).
Trans-grafting is a relevant practice in citrus biotechnological breeding that combines genetic engineering with traditional grafting practices. This method consists of grafting a non-genetically modified scion onto a transgenic rootstock (Kaiser and Dalton, 2001; Haroldsen et al., 2012). Thus, in trans-grafted plants, mobile transgene products can move across the phloem from the transgenic rootstock to the non-transgenic scion, so that the latter can acquire the beneficial trait with no-genetic modification of the final products, for example as in fruits (Song et al., 2015). In citrus crops, non-transgenic scions susceptible to HLB bacterial disease were grafted onto transgenic rootstocks overexpressing a microbial peptide and the resulting plants showed lower rates of infection compared with non-transgrafted plants (Bergey et al., 2015). De Francesco et al. (2020) trans-grafted a transgenic sweet orange interstock overexpressing a hairpin CP-mRNA and observed tolerance to citrus psorosis virus in the non-transgenic scion.
New genome engineering technologies offer encouraging alternatives to create mutations in the citrus genome. For example, the recently developed clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas9 genome editing tool has been successfully applied to citrus species. The efficacy of the CRISPR/Cas9 technique in citrus plants was first studied by targeting the phytoene desaturase (PDS) gene (Jia and Wang, 2014a). The disruption of this gene impairs chlorophyll and carotenoid production resulting in albino or mosaic phenotypes that can be observed visually to estimate the efficacy of the genome modification system (Qin et al., 2007). The study of Jia and Wang (2014a) first reported PDS editing in C. sinensis plants, with a very low editing efficiency of about 3.5%. The work of Zhang F. et al. (2017) has lately developed a higher efficient CRISPR system to edit PDS that relies on (1) Cas9 driven by the Arabidopsis YAO-promoter instead of 35S, and (2) a bifunctional selectable marker used to identify transgenic citrus plants with high expression of Cas9. They obtained albino phenotypes consistent with high mutation frequencies of up to 75% (Zhang F. et al., 2017). The authors also reported that most of the mutations obtained in their study were identified as indels that resulted in a frameshift.
On the other hand, the study of Jia et al. (2017) recently reported editing of the CsLOB1 (C. sinensis Lateral Organ Boundaries) gene. CsLOB1 is a susceptibility gene for citrus canker disease and is induced by the pathogenicity factor PthA4 from X. citri pv. citri. PthA4 binds to the EBEPthA4-CsLOBP to induce CsLOB1 gene expression (Hu et al., 2014). CsLOB1 was targeted for edition both in its promoter and coding sequences, in Valencia sweet orange (C. sinensis) and Duncan grapefruit (C. paradisi Macf.) (Jia et al., 2016, 2017; Peng et al., 2017). The efficiency of recovering mutant plants spanned from 23 to 67% and the transgenic lines with higher mutation rates became resistant to citrus canker. In addition, the generation of homozygous and biallelic canker-resistant Pummelo (C. maxima) plants in the T0 generation was reported (Jia and Wang, 2020). CRISPR edition of CsWRKY22 gene, a marker gene for pathogen-triggered immunity in C. sinensis also reduces susceptibility to X. citri subsp. citri in Wanjincheng orange (Wang et al., 2019).
The LOB1 promoter was also edited in Duncan grapefruit using SaCas9 from Staphylococcus aureus (Jia et al., 2016) instead of the commonly used SpCas9 from Streptococcus pyogenes. SaCas9 (Ran et al., 2015) is a 1053 aa nuclease, much smaller than SpCas9 (1368 aa), that allows easier handling and target cell transformation. No off-targets were observed when using SaCas9 in citrus gene editing neither in transgenic edited tobacco (Kaya et al., 2016). CRISPR-Cas12a from Prevotella and Francisella (a class II/type V CRISPR nuclease), has been also employed to edit PDS or CsLOB1 genes in Duncan grapefruit (Jia et al., 2019) and has been proposed as an alternative system reported having fewer off-targets in relation to Cas9 (Kim et al., 2016; Kleinstiver et al., 2016). The work of Dutt et al. (2020) have recently reported the edition of the PDS gene in C. sinensis plants using embryogenic callus as explants instead of epicotyls. They employed two different constructs: one where the gRNAs were driven by the Arabidopsis U6–26 pol III promoter and another with the RNA processing ability of the Csy4 bacterial endoribonuclease to express several gRNAs (Čermák et al., 2017). All the generated transgenic embryos were completely albino, and no variegated phenotype was observed, which demonstrates a high editing efficiency (Dutt et al., 2020). The embryogenic cell culture mediated transformation system allows a larger population of transformed plants compared with the epicotyl explant mediated system. Furthermore, seedless citrus cultivars and epicotyl transformation recalcitrant cultivars can be easily transformed with this system.
Perspectives
Citrus improvement requires a continuing effort for success. Emerging biotechnologies are providing the research community with new tools that can increase the speed and efficiency of the process. Public perception of transgenic citrus, however, is an actual concern and should be taken into consideration. Even though it could be argued that pathogen-resistant transgenic citrus could improve sustainability by reducing pesticide applications (Caserta et al., 2019), they nevertheless bring other concerns. Some of them are the effect of the modified plants on the environment, the risk of transgene dissemination by pollen, the potential damage to local production and small growers, who cannot adopt the new technology, and potential risks to human health due to the consumption of transgenic citrus fruits, among others.
The expression of genes from citrus origin to obtain cisgenic or intragenic varieties is a promising strategy, considering the issues around the public perception of transgenic plants and the need to address the reduction of the regulation (Holme et al., 2013). When the genetic sequences to be introduced originate in closely related species, instead of phylogenetically distant ones, regulatory processes become easier to achieve. Furthermore, the application of CRISPR/Cas technologies has the potential of generating non-transgenic-edited citrus plants, thus falling within a different regulatory framework (Lema, 2019). In many cases, the result of editing genetic sequences is comparable to that obtained by conventional mutagenesis, if there are no leftovers of Cas9 and gRNA (guide RNA) sequences inserted in the genome. To move toward those alternatives, it is necessary to modify current transformation methods for citrus editing. Promising strategies may include the use of Cas9-ribonucleoprotein complexes for the transitory transformation of either protoplast with PEG or callus explants by biolistic, eluding transgene integration in the plant genome. Therefore, the edited plant can be classified as non-transgenic according to the Cartagena Protocol on Biosafety (Lema, 2019).
Author Contributions
GC and CAR: conceptualization, investigation, supervision, and writing—original draft. BX-C and GM-P: writing—original draft and funding acquisition. HEH: supervision and funding acquisition. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Julia Sabio y García for their critical reading of the manuscript and language revision.
Footnotes
Funding
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (ANPCYT: PICT 2018-02926, PICT 2017-2320, and PICT 2017-1233), and CINVESTAV funding, Mexico.
References
- Acanda Y., Welker S., Orbović V., Levy A. (2021). A simple and efficient agroinfiltration method for transient gene expression in Citrus. Plant Cell Rep. 40 1171–1179. 10.1007/s00299-021-02700-w [DOI] [PubMed] [Google Scholar]
- Ahmed D., Comte A., Curk F., Costantino G., Luro F., Dereeper A., et al. (2019). Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: a case study in diploid and polyploid citrus. Ann. Bot. 123 1231–1251. 10.1093/aob/mcz029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Kim W. C. (2019). A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front. Plant Sci. 10:1433. 10.3389/fpls.2019.01433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Mannan A., El Oirdi M., Waheed A., Mirza B. (2012). Agrobacterium-mediated transformation of rough lemon (Citrus jambhiri Lush) with yeast HAL2 gene. BMC Res. Notes 5:285. 10.1186/1756-0500-5-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida W. A. B., Mourão Filho F. A. A., Mendes B. M. J., Pavan A., Rodriguez A. P. M. (2003a). Agrobacterium-mediated transformation of Citrus sinensis and Citrus limonia epicotyl segments. Sci. Agric. 60 23–29. 10.1590/S0103-90162003000100005 [DOI] [Google Scholar]
- Almeida W. A. B., Mourão Filho F. A. A., Pino L. E., Boscariol R. L., Rodriguez A. P. M., Mendes B. M. J. (2003b). Genetic transformation and plant recovery from mature tissues of Citrus sinensis L. Osbeck. Plant Sci. 164 203–211. 10.1016/S0168-9452(02)00401-6 [DOI] [Google Scholar]
- Alquézar B., Volpe H. X. L., Magnani R. F., de Miranda M. P., Santos M. A., Marques V. V., et al. (2021). engineered orange ectopically expressing the Arabidopsis β-caryophyllene synthase is not attractive to diaphorina citri, the vector of the bacterial pathogen associated to huanglongbing. Front. Plant Sci. 12:641457. 10.3389/fpls.2021.641457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C., Orbović V., Mou Z. (2013). An efficient intragenic vector for generating intragenic and cisgenic plants in citrus. Am. J. Plant Sci. 04 2131–2137. 10.4236/ajps.2013.411265 [DOI] [Google Scholar]
- Ballester A., Cervera M., Peña L. (2007). Efficient production of transgenic citrus plants using isopentenyl transferase positive selection and removal of the marker gene by site-specific recombination. Plant Cell Rep. 26 39–45. 10.1007/s00299-006-0197-3 [DOI] [PubMed] [Google Scholar]
- Ballester A., Cervera M., Peña L. (2008). Evaluation of selection strategies alternative to nptII in genetic transformation of citrus. Plant Cell Rep. 27 1005–1015. 10.1007/s00299-008-0523-z [DOI] [PubMed] [Google Scholar]
- Ballester A., Cervera M., Peña L. (2010). Selectable marker-free transgenic orange plants recovered under non-selective conditions and through PCR analysis of all regenerants. Plant Cell. Tissue Organ Cult. 102 329–336. 10.1007/s11240-010-9737-1 [DOI] [Google Scholar]
- Barbosa-Mendes J. M., Filho F., de A. A. M., Filho A. B., Harakava R., Beer S. V., et al. (2009). Genetic transformation of Citrus sinensis cv. Hamlin with hrpN gene from Erwinia amylovora and evaluation of the transgenic lines for resistance to citrus canker. Sci. Hortic. 122 109–115. 10.1016/j.scienta.2009.04.001 [DOI] [Google Scholar]
- Barlass M., Skene K. G. M. (1982). In vitro plantlet formation from Citrus species and hybrids. Sci. Hortic. 17 333–341. 10.1016/0304-4238(82)90114-5 [DOI] [Google Scholar]
- Bergey D., Dutt M., Barthe G., Irey M., Grosser J. (2015). Genetically modified Citrus rootstocks expressing a synthetic cationic antimicrobial peptide slows down bacterial infection in the non-transgenic scion. Vitr. Cell. Dev. Biol. 51:499. [Google Scholar]
- Bespalhok Filho J. C., Kobayashi A. K., Pereira L. F. P., Galvao R. M., Vieira L. G. E. (2003). Transient gene expression of beta-glucuronidase in citrus thin epicotyl transversal sections using particle bombardment. Braz. Arch. Biol. Technol. 46 1–6. [Google Scholar]
- Bezerra Y. C., dos A., Marques J. P. R., Stipp L. C. L., Attílio L. B., Freitas-Astúa J., et al. (2021). How to drive phloem gene expression? A case study with preferentially expressed citrus gene promoters. Rev. Bras. Frutic. 43 1–12. 10.1590/0100-29452021005 [DOI] [Google Scholar]
- Bond J. E., Roose M. L. (1998). Agrobacterium mediated transformation of the commercially important citrus cultivar Washington navel orange. Plant Cell Rep. 18 229–234. 10.1007/s002990050562 [DOI] [PubMed] [Google Scholar]
- Bordón Y., Guardiola J. L., García-Luis A. (2000). Genotype affects the morphogenic response in vitro of epicotyl segments of Citrus rootstocks. Ann. Bot. 86 159–166. 10.1006/anbo.2000.1177 [DOI] [Google Scholar]
- Boscariol R. L., Almeida W. A. B., Derbyshire M. T. V. C., Mourão Filho F. A. A., Mendes B. M. J. (2003). The use of the PMI/mannose selection system to recover transgenic sweet orange plants (Citrus sinensis L. Osbeck). Plant Cell Rep. 22 122–128. 10.1007/s00299-003-0654-1 [DOI] [PubMed] [Google Scholar]
- Boscariol R. L., Monteiro M., Takahashi E. K., Chabregas S. M., Vieira M. L. C., Vieira L. G. E., et al. (2006). Attacin A gene from Tricloplusia ni reduces susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinesis “Hamlin.”. J. Am. Soc. Hortic. Sci. 131 530–536. 10.21273/jashs.131.4.530 [DOI] [Google Scholar]
- Cabrera-Ponce J. L., Lopez L., Assad-Garcia N., Medina-Arevalo C., Bailey A. M., Herrera-Estrella L. (1997). An efficient particle bombardment system for the genetic transformation of asparagus (Asparagus officinalis L.). Plant Cell Rep. 16 255–260. 10.1007/bf01088276 [DOI] [PubMed] [Google Scholar]
- Cardoso S. C., Barbosa-Mendes J. M., Boscariol-Camargo R. L., Christiano R. S. C., Filho A. B., Vieira M. L. C., et al. (2010). Transgenic sweet orange (Citrus sinensis L. Osbeck) expressing the attacin a gene for resistance to Xanthomonas citri subsp. citri. Plant Mol. Biol. Rep. 28 185–192. 10.1007/s11105-009-0141-0 [DOI] [Google Scholar]
- Caruso M., Smith M. W., Froelicher Y., Russo G., Gmitter F. G. (2020). “Traditional breeding,” in The Genus Citrus, eds Talon M., Caruso M., Gmitter F. (Amsterdam: Elsevier Inc.), 10.1016/B978-0-12-812163-4.00007-3 [DOI] [Google Scholar]
- Caserta R., Teixeira-Silva N. S., Granato L. M., Dorta S. O., Rodrigues C. M., Mitre L. K., et al. (2019). Citrus biotechnology: what has been done to improve disease resistance in such an important crop? Biotechnol. Res. Innov. 3 95–109. 10.1016/j.biori.2019.12.004 [DOI] [Google Scholar]
- Čermák T., Curtin S. J., Gil-Humanes J., Čegan R., Kono T. J. Y., Konečná E., et al. (2017). A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29 1196–1217. 10.1105/tpc.16.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera M., Juárez J., Navarro A., Pina J. A., Durán-Vila N., Navarro L., et al. (1998a). Genetic transformation and regeneration of mature tissues of woody fruit plants bypassing the juvenile stage. Transgenic Res. 7 51–59. 10.1023/A:1008855922283 [DOI] [Google Scholar]
- Cervera M., López M. M., Navarro L., Peña L. (1998b). Virulence and supervirulence of Agrobacterium tumefaciens in woody fruit plants. Physiol. Mol. Plant Pathol. 52 67–78. 10.1006/pmpp.1997.0135 [DOI] [Google Scholar]
- Cervera M., Pina J. A., Juárez J., Navarro L., Peña L. (1998c). Agrobacterium-mediated transformation of citrange: factors affecting transformation and regeneration. Plant Cell Rep. 18 271–278. 10.1007/s002990050570 [DOI] [PubMed] [Google Scholar]
- Cervera M., Juárez J., Navarro L., Peña L. (2005). Genetic transformation of mature citrus plants. Methods Mol. Biol 286 177–188. 10.1385/1-59259-827-7:177 [DOI] [PubMed] [Google Scholar]
- Cervera M., Navarro A., Navarro L., Peña L. (2008). Production of transgenic adult plants from clementine mandarin by enhancing cell competence for transformation and regeneration. Tree Physiol. 28 55–66. 10.1093/treephys/28.1.55 [DOI] [PubMed] [Google Scholar]
- Chatuverdi H. C., Mitra G. C. (1974). Clonal propagation of citrus from somatic callus cultures. Hortic. Sci. 9 118–120. [Google Scholar]
- Chen C., Lyon M. T., O’Malley D., Federici C. T., Gmitter J., Grosser J. W., et al. (2008). Origin and frequency of 2n gametes in Citrus sinensis × Poncirus trifoliata and their reciprocal crosses. Plant Sci. 174 1–8. 10.1016/j.plantsci.2007.08.005 [DOI] [Google Scholar]
- Chiancone B., Germanà M. A. (2012). “Micropropagation of Citrus spp. by organogenesis and somatic embryogenesis,” in Methods in Molecular Biology, ed. Walker J. M. (Totowa, NJ: Humana Press; ), 99–118. 10.1007/978-1-62703-074-8_8 [DOI] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Conti G., Gardella V., Vandecaveye M. A., Gomez C. A., Joris G., Hauteville C., et al. (2020). Transgenic citrange troyer rootstocks overexpressing antimicrobial potato Snakin-1 show reduced citrus canker disease symptoms. J. Biotechnol. 324 99–102. 10.1016/j.jbiotec.2020.09.010 [DOI] [PubMed] [Google Scholar]
- Corte L. E. D., Mendes B. M. J., Filho F. A. A. M., Grosser J. W., Dutt M. (2020). Functional characterization of full-length and 5’ deletion fragments of Citrus sinensis-derived constitutive promoters in Nicotiana benthamiana. Vitr. Cell. Dev. Biol. Plant 56 280–289. 10.1007/s11627-019-10044-0 [DOI] [Google Scholar]
- Costa M. G. C., Otoni W. C., Moore G. A. (2002). An evaluation of factors affecting the efficiency of Agrobacterium-mediated transformation of Citrus paradisi (Macf.) and production of transgenic plants containing carotenoid biosynthetic genes. Plant Cell Rep. 21 365–373. 10.1007/s00299-002-0533-1 [DOI] [Google Scholar]
- Dasgupta K., Hotton S., Belknap W., Syed Y., Dardick C., Thilmony R., et al. (2020). Isolation of novel citrus and plum fruit promoters and their functional characterization for fruit biotechnology. BMC Biotechnol. 20:43. 10.1186/s12896-020-00635-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Azevedo F. A., Mourão Filho F. D. A. A., Schinor E. H., De Paoli L. G., Mendes B. M. J., Harakava R., et al. (2006). GUS gene expression driven by a citrus promoter in transgenic tobacco and “Valencia” sweet orange. Pesqui. Agropecu. Bras. 41 1623–1628. 10.1590/S0100-204X2006001100008 [DOI] [Google Scholar]
- De Francesco A., Simeone M., Gómez C., Costa N., García M. L. (2020). Transgenic Sweet Orange expressing hairpin CP-mRNA in the interstock confers tolerance to Citrus psorosis virus in the non-transgenic scion. Transgenic Res. 29 215–228. 10.1007/s11248-020-00191-1 [DOI] [PubMed] [Google Scholar]
- De Oliveira M. L. P., De Lima Silva C. C., Abe V. Y., Costa M. G. C., Cernadas R. A., Benedetti C. E. (2013). Increased resistance against citrus canker mediated by a citrus mitogen-activated protein kinase. Mol. Plant Microbe Interact. 26 1190–1199. 10.1094/MPMI-04-13-0122-R [DOI] [PubMed] [Google Scholar]
- De Oliveira M. L. P., Febres V. J., Costa M. G. C., Moore G. A., Otoni W. C. (2009). High-efficiency Agrobacterium-mediated transformation of citrus via sonication and vacuum infiltration. Plant Cell Rep. 28 387–395. 10.1007/s00299-008-0646-2 [DOI] [PubMed] [Google Scholar]
- De Oliveira M. L. P., Moore G., Thomson J. G., Stover E. (2015). Agrobacterium-mediated transformation of Mexican lime (Citrus aurantifolia Swingle) using optimized systems for epicotyls and cotyledons. Adv. Biosci. Biotechnol. 6 657–668. 10.4236/abb.2015.611069 [DOI] [Google Scholar]
- Domínguez A., Cervera M., Pérez R. M., Romero J., Fagoaga C., Cubero J., et al. (2004). Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol. Breed. 14:171183. 10.1023/B:MOLB.0000038005.73265.61 [DOI] [Google Scholar]
- Domínguez A., De Mendoza A. H., Guerri J., Cambra M., Navarro L., Moreno P., et al. (2002). Pathogen-derived resistance to Citrus tristeza virus (CTV) in transgenic mexican lime (Citrus arrurantifolia (Christ.) Swing.) plants expressing its p25 coat protein gene. Mol. Breed. 10 1–10. 10.1023/A:1020347415333 [DOI] [Google Scholar]
- Domínguez A., Guerri J., Cambra M., Navarro L., Moreno P., Peña L. (2000). Efficient production of transgenic citrus plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep. 19 427–433. 10.1007/s002990050751 [DOI] [PubMed] [Google Scholar]
- Duan Y. X., Fan J., Guo W. W. (2010). Regeneration and characterization of transgenic kumquat plants containing the Arabidopsis APETALA1 gene. Plant Cell. Tissue Organ Cult. 100 273–281. 10.1007/s11240-009-9646-3 [DOI] [Google Scholar]
- Dutt M., Ananthakrishnan G., Jaromin M. K., Brlansky R. H., Grosser J. W. (2012). Evaluation of four phloem-specific promoters in vegetative tissues of transgenic citrus plants. Tree Physiol. 32 83–93. 10.1093/treephys/tpr130 [DOI] [PubMed] [Google Scholar]
- Dutt M., Barthe G., Irey M., Grosser J. (2015). Transgenic Citrus expressing an arabidopsis NPR1 gene exhibit enhanced resistance against Huanglongbing (HLB; Citrus greening). PLoS One 10:e0137134. 10.1371/journal.pone.0137134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt M., Erpen L., Grosser J. W. (2018a). Genetic transformation of the ‘W Murcott’ tangor: comparison between different techniques. Sci. Hortic. 242 90–94. 10.1016/j.scienta.2018.07.026 [DOI] [Google Scholar]
- Dutt M., Zambon F. T., Erpen L., Soriano L., Grosser J. (2018b). Embryo-specific expression of a visual reporter gene as a selection system for citrus transformation. PLoS One 13:e0190413. 10.1371/journal.pone.0190413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt M., Grosser J. W. (2010). An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of citrus. Plant Cell Rep. 29 1251–1260. 10.1007/s00299-010-0910-0 [DOI] [PubMed] [Google Scholar]
- Dutt M., Lee D. H., Grosser J. W. (2010). Bifunctional selection-reporter systems for genetic transformation of citrus: mannose- and kanamycin-based systems. Vitr. Cell. Dev. Biol. Plant 46 467–476. 10.1007/s11627-010-9300-0 [DOI] [Google Scholar]
- Dutt M., Mou Z., Zhang X., Tanwir S. E., Grosser J. W. (2020). Efficient CRISPR/Cas9 genome editing with Citrus embryogenic cell cultures. BMC Biotechnol. 20:58. 10.1186/s12896-020-00652-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt M., Stanton D., Grosser J. W. (2016). Ornacitrus: development of genetically modified anthocyanin-expressing citrus with both ornamental and fresh fruit potential. J. Am. Soc. Hortic. Sci. 141 54–61. 10.21273/jashs.141.1.54 [DOI] [Google Scholar]
- Edriss M. H., Burger D. W. (1984). In vitro propagation of “Troyer” citrange from epicotyl segments. Sci. Hortic. 23 159–162. 10.1016/0304-4238(84)90019-0 [DOI] [Google Scholar]
- El-Mohtar C., Dawson W. O. (2014). Exploring the limits of vector construction based on citrus tristeza virus. Virology 448 274–283. 10.1016/j.virol.2013.10.017 [DOI] [PubMed] [Google Scholar]
- Endo T., Shimada T., Fujii H., Kobayashi Y., Araki T., Omura M. (2005). Ectopic expression of an FT homolog from Citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res. 14 703–712. 10.1007/s11248-005-6632-3 [DOI] [PubMed] [Google Scholar]
- Endo T., Shimada T., Fujii H., Moriguchi T., Omura M. (2007). Promoter analysis of a type 3 metallothionein-like gene abundant in satsuma mandarin (Citrus unshiu Marc.) fruit. Sci. Hortic. 112 207–214. 10.1016/j.scienta.2006.12.042 [DOI] [Google Scholar]
- EPA-HQ-OPP-2014-0834, F.-9926-99 (2015). Federal Register?:: Defensin Proteins (SoD2 and SoD7) Derived From Spinach (Spinacia oleracea L.) in Citrus Plants; Temporary Exemption From the Requirement of a Tolerance. Environ. Prot. Agency. Available online at: https://www.federalregister.gov/documents/2015/05/06/2015-10486/defensin-proteins-sod2-and-sod7-derived-from-spinach-spinacia-oleracea-l-in-citrus-plants-temporary (accessed August 30, 2021). [Google Scholar]
- Erpen L., Tavano E. C. R., Harakava R., Dutt M., Grosser J. W., Piedade S. M. S., et al. (2018). Isolation, characterization, and evaluation of three Citrus sinensis-derived constitutive gene promoters. Plant Cell Rep. 37 1113–1125. 10.1007/s00299-018-2298-1 [DOI] [PubMed] [Google Scholar]
- Esquinas-Alcázar J. (2005). Protecting crop genetic diversity for food security: political, ethical and technical challenges. Nat. Rev. Genet. 6 946–953. 10.1038/nrg1729 [DOI] [PubMed] [Google Scholar]
- Fagoaga C., Lopez C., Moreno P., Navarro L., Flores R., Peña L. (2005). Viral-like symptoms induced by the ectopic expression of the p23 gene of citrus tristeza virus are Citrus specific and do not correlate with the pathogenicity of the virus strain. Mol. Plant Microbe Interact. 18 435–445. 10.1094/MPMI [DOI] [PubMed] [Google Scholar]
- Fávero P., de Alves Mourão Filho F. A., Stipp L. C. L., Mendes B. M. J. (2012). Genetic transformation of three sweet orange cultivars from explants of adult plants. Acta Physiol. Plant. 34 471–477. 10.1007/s11738-011-0843-4 [DOI] [Google Scholar]
- Febres V. J., Fisher L. C., Khalaf A., Moore G. A. (2011). Citrus transformation: challenges and prospects. Intech 5 101–122. [Google Scholar]
- Febres V. J., Niblett C. L., Lee R. F., Moore G. A. (2003). Characterization of grapefruit plants (Citrus paradisi Macf.) transformed with citrus tristeza closterovirus genes. Plant Cell Rep. 21 421–428. 10.1007/s00299-002-0528-y [DOI] [PubMed] [Google Scholar]
- Figueiredo J. F. L., Römer P., Lahaye T., Graham J. H., White F. F., Jones J. B. (2011). Agrobacterium-mediated transient expression in citrus leaves: a rapid tool for gene expression and functional gene assay. Plant Cell Rep. 30 1339–1345. 10.1007/s00299-011-1045-7 [DOI] [PubMed] [Google Scholar]
- Fisher K., Phillips C. (2008). Potential antimicrobial uses of essential oils in food: is citrus the answer? Trends Food Sci. Technol. 19 156–164. 10.1016/j.tifs.2007.11.006 [DOI] [Google Scholar]
- Fleming G. H., Olivares-Fuster O., Fatta Del-Bosco S., Grosser J. W. (2000). An alternative method for the genetic transformation of sweet orange. Vitr. Cell. Dev. Biol. 36 450–455. [Google Scholar]
- Folimonov A. S., Folimonova S. Y., Bar-Joseph M., Dawson W. O. (2007). A stable RNA virus-based vector for citrus trees. Virology 368 205–216. 10.1016/j.virol.2007.06.038 [DOI] [PubMed] [Google Scholar]
- Fu X. Z., Chen C. W., Wang Y., Liu J. H., Moriguchi T. (2011a). Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: involvement of H2O2production and transcriptional alteration. BMC Plant Biol. 11:55. 10.1186/1471-2229-11-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Z., Khan E. U., Hu S. S., Fan Q. J., Liu J. H. (2011b). Overexpression of the betaine aldehyde dehydrogenase gene from Atriplex hortensis enhances salt tolerance in the transgenic trifoliate orange (Poncirus trifoliata L. Raf.). Environ. Exp. Bot. 74 106–113. 10.1016/j.envexpbot.2011.05.006 [DOI] [Google Scholar]
- Furman N., Kobayashi K., Zanek M. C., Calcagno J., Garcia M. L., Mentaberry A. (2013). Transgenic sweet orange plants expressing a dermaseptin coding sequence show reduced symptoms of citrus canker disease. J. Biotechnol. 167 412–419. 10.1016/j.jbiotec.2013.07.019 [DOI] [PubMed] [Google Scholar]
- García-Luis A., Bordón Y., Moreira-Dias J. M., Molina R. V., Guardiola J. L. (1999). Explant orientation and polarity determine the morphogenic response of epicotyl segments of Troyer citrange. Ann. Bot. 84 715–723. 10.1006/anbo.1999.0972 [DOI] [Google Scholar]
- Gentile A., Deng Z., La Malfa S., Distefano G., Domina F., Vitale A., et al. (2007). Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 126 146–151. 10.1111/j.1439-0523.2007.01297.x [DOI] [Google Scholar]
- Germanà M. A., Macaluso L., Patricolo G., Chiancone B. (2008). Morphogenic response in vitro of epicotyl segments of Citrus macrophylla. Plant Biosyst. 142 661–664. 10.1080/11263500802411395 [DOI] [Google Scholar]
- Ghorbel R., Dominguez A., Navarro L., Peña L. (2000). High efficiency genetic transformation of sour orange (Citrus aurantium) and production of transgenic trees containing the coat protein gene of Citrus tristeza virus. Tree Physiol. 20 1183–1189. 10.1093/treephys/20.17.1183 [DOI] [PubMed] [Google Scholar]
- Ghorbel R., Juárez J., Navarro L., Peña L. (1999). Green fluorescent protein as a screenable marker to increase the efficiency of generating transgenic woody fruit plants. Theor. Appl. Genet. 99 350–358. 10.1007/s001220051244 [DOI] [Google Scholar]
- Ghorbel R., La-Malfa S., López M. M., Petit A., Navarro L., Peña L. (2001). Additional copies of virG from pTiBo542 provide a super-transformation ability to Agrobacterium tumefaciens in citrus. Physiol. Mol. Plant Pathol. 58 103–110. 10.1006/pmpp.2000.0318 [DOI] [Google Scholar]
- Ghorbel R., Navarro L., Duran-Vila N. (1998). Morphogenesis and regeneration of whole plants of grapefruit (Citrus paradisi), sour orange (C. aurantium) and alemow (C. macrophylla). J. Hortic. Sci. Biotechnol. 73 323–327. 10.1080/14620316.1998.11510981 [DOI] [Google Scholar]
- Gmitter F. G., Talon M. (2008). Citrus genomics. Int. J. Plant Genomics 2008:528361. 10.1155/2008/528361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinblat U. (1972). Differentiation of citrus stem in vitro. J. Am. Soc. Hortic. Sci. 97 599–603. [Google Scholar]
- Grosser J. W., Gmitter F. G. (1990). “Protoplast fusion and Citrus improvement,” in Plant Breeding Reviews, eds Grosser J. W., Gmitter F. G. (Portland, OR: Timber Press Inc.), 339–374. 10.1002/9781118061053.ch10 [DOI] [Google Scholar]
- Grosser J. W., Gmitter F. G. (2011). Protoplast fusion for production of tetraploids and triploids: applications for scion and rootstock breeding in citrus. Plant Cell. Tissue Organ Cult. 104 343–357. 10.1007/s11240-010-9823-4 [DOI] [Google Scholar]
- Grosser J. W., Gmitter F. G., Tusa N., Recupero G. R., Cucinotta P. (1996). Further evidence of a cybridization requirement for plant regeneration from citrus leaf protoplasts following somatic fusion. Plant Cell Rep. 15 672–676. 10.1007/BF00231922 [DOI] [PubMed] [Google Scholar]
- Grosser J. W., Jiang J., Louzada E. S., Chandler J. L., Gmitter F. G. (1998a). Somatic hybridization, an integral component of citrus cultivar improvement: II. Rootstock improvement. HortScience 33 1060–1061. 10.21273/hortsci.33.6.1060 [DOI] [Google Scholar]
- Grosser J. W., Jiang J., Mourao-Fo F. A. A., Louzada E. S., Baergen K., Chandler J. L., et al. (1998b). Somatic hybridization, an integral component of citrus cultivar improvement: I. Scion improvement. HortScience 33 1057–1059. 10.21273/hortsci.33.6.1057 [DOI] [Google Scholar]
- Guerra-Lupián M. A., Ruiz-Medrano R., Ramírez-Pool J. A., Ramírez-Ortega F. A., López-Buenfil J. A., Loeza-Kuk E., et al. (2018). Localized expression of antimicrobial proteins mitigates huanglongbing symptoms in Mexican lime. J. Biotechnol. 285 74–83. 10.1016/j.jbiotec.2018.08.012 [DOI] [PubMed] [Google Scholar]
- Guo W. W., Grosser J. W. (2004). Transfer of a potential canker resistance gene into citrus protoplasts using GFP as the selectable marker. Acta Hortic. 632 255–258. 10.17660/ActaHortic.2004.632.32 [DOI] [Google Scholar]
- Gutiérrez-E M. A., Luth D., Moore G. A. (1997). Factors affecting Agrobacterium-mediated transformation in Citrus and production of sour orange (Citrus aurantium L.) plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep. 16 745–753. 10.1007/s002990050313 [DOI] [PubMed] [Google Scholar]
- Hao G., Pitino M., Duan Y., Stover E. (2016a). Reduced susceptibility to xanthomonas citri in transgenic citrus expressing the fls2 receptor from nicotiana benthamiana. Mol. Plant Microbe Interact. 29 132–142. 10.1094/MPMI-09-15-0211-R [DOI] [PubMed] [Google Scholar]
- Hao G., Stover E., Gupta G. (2016b). Overexpression of a modified plant thionin enhances disease resistance to citrus canker and huanglongbing (HLB). Front. Plant Sci. 7:1078. 10.3389/fpls.2016.01078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroldsen V. M., Chi-Ham C. L., Bennett A. B. (2012). Transgene mobilization and regulatory uncertainty for non-GE fruit products of transgenic rootstocks. J. Biotechnol. 161 349–353. 10.1016/j.jbiotec.2012.06.017 [DOI] [PubMed] [Google Scholar]
- He Y., Chen S., Peng A., Zou X., Xu L., Lei T., et al. (2011). Production and evaluation of transgenic sweet orange (Citrus sinensis Osbeck) containing bivalent antibacterial peptide genes (Shiva A and Cecropin B) via a novel Agrobacterium-mediated transformation of mature axillary buds. Sci. Hortic. 128 99–107. 10.1016/j.scienta.2011.01.002 [DOI] [Google Scholar]
- Henry R. J. (1998). “Molecular and biochemical characterization of somaclonal variation,” in Current Plant Science and Biotechnology in Agriculture (PSBA), eds Jain S. M., Brar D. S., Ahloowalia B. S. (Dordrecht: Springer; ), 485–499. 10.1007/978-94-015-9125-6_24 [DOI] [Google Scholar]
- Hidaka T., Omura M. (1993). Transformation of citrus protoplasts by electroporation. J. Jpn. Soc. Hort. Sci 62 371–376. [Google Scholar]
- Holme I. B., Wendt T., Holm P. B. (2013). Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 11 395–407. 10.1111/pbi.12055 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhang J., Jia H., Sosso D., Li T., Frommer W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111 E521–E529. 10.1073/pnas.1313271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Yuan Y., Tang Z., Huang Y., Kang C., Deng X., et al. (2019). Retrotransposon promoter of Ruby1 controls both light- and cold-induced accumulation of anthocyanins in blood orange. Plant Cell Environ. 42 3092–3104. 10.1111/pce.13609 [DOI] [PubMed] [Google Scholar]
- Huang X., Wang Y., Xu J., Wang N. (2020). Development of multiplex genome editing toolkits for citrus with high efficacy in biallelic and homozygous mutations. Plant Mol. Biol. 104 297–307. 10.1007/s11103-020-01043-6 [DOI] [PubMed] [Google Scholar]
- Iwanami T., Shimizu T., Ito T., Hirabayashi T. (2004). Tolerance to citrus mosaic virus in transgenic trifoliate orange lines harboring capsid polyprotein gene. Plant Dis. 88 865–868. 10.1094/PDIS.2004.88.8.865 [DOI] [PubMed] [Google Scholar]
- Januzzi Mendes B. M., Luciana Boscariol R., Mourão Filho F., de A. A., Bastos de Almeida W. A. (2002). Agrobacterium-mediated genetic transformation of “Hamlin” sweet orange. Pesqui. Agropecu. Bras. 37 955–961. 10.1590/s0100-204x2002000700009 [DOI] [Google Scholar]
- Jia H., Orbovic V., Jones J. B., Wang N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4: DCsLOB1.3 infection. Plant Biotechnol. J. 14 1291–1301. 10.1111/pbi.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Orbović V., Wang N. (2019). CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 17 1928–1937. 10.1111/pbi.13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Wang N. (2014b). Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 33 1993–2001. 10.1007/s00299-014-1673-9 [DOI] [PubMed] [Google Scholar]
- Jia H., Wang N. (2014a). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806. 10.1371/journal.pone.0093806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Wang N. (2020). Generation of homozygous canker-resistant citrus in the T0 generation using CRISPR-SpCas9p. Plant Biotechnol. J. 18 1990–1992. 10.1111/pbi.13375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Zhang Y., Orbović V., Xu J., White F. F., Jones J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15 817–823. 10.1111/pbi.12677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H. D., Doherty A., Slarks C. A. (2009). “Transient transformation of plants,” in Methods in Molecular Biology, Plant Genomics, eds Gustafson J. P., Langridge P., Somers D. J. (Totowa, NJ: Humana Press; ), 131–151. 10.1007/978-1-59745-427-8 [DOI] [PubMed] [Google Scholar]
- Kaiser J., Dalton R. (2001). Grafting for transgene containment. Nature 19:1104. [DOI] [PubMed] [Google Scholar]
- Kaneyoshi J., Kobayashi S., Nakamura Y., Shigemoto N., Doi Y. (1994). A simple and efficient gene transfer system of trifoliate orange (Poncirus trifoliata Raf.). Plant Cell Rep. 13 541–545. 10.1007/BF00234507 [DOI] [PubMed] [Google Scholar]
- Kaya H., Mikami M., Endo A., Endo M., Toki S. (2016). Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 6:26871. 10.1038/srep26871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayim M., Ceccardi T. L., Berretta M. J. G., Barthe G. A., Derrick K. S. (2004). Introduction of a citrus blight-associated gene into Carrizo citrange [Citrus sinensis (L.) Osbc. x Poncirus trifoliata (L.) Raf.] by Agrobacterium-mediated transformation. Plant Cell Rep. 23 377–385. 10.1007/s00299-004-0823-x [DOI] [PubMed] [Google Scholar]
- Keneni G., Bekele E., Imtiaz M., Dagne K. (2012). Genetic vulnerability of modern crop cultivars: causes, mechanism and remedies. Int. J. Plant Res. 2 69–79. 10.5923/j.plant.20120203.05 22499009 [DOI] [Google Scholar]
- Khan E. U., Fu X. Z., Wang J., Fan Q. J., Huang X. S., Zhang G. N., et al. (2009). Regeneration and characterization of plants derived from leaf in vitro culture of two sweet orange (Citrus sinensis (L.) Osbeck) cultivars. Sci. Hortic. 120 70–76. 10.1016/j.scienta.2008.10.004 [DOI] [Google Scholar]
- Khan M., Hu J., Dahro B., Ming R., Zhang Y., Wang Y., et al. (2021). ERF108 from Poncirus trifoliata (L.) Raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 108, 705–724. 10.1111/tpj.15465 [DOI] [PubMed] [Google Scholar]
- Khawale R. N., Singh S. K., Garg G., Baranwal V. K., Ajirlo S. A. (2006). Agrobacterium-mediated genetic transformation of Nagpur mandarin (Citrus reticulata Blanco). Curr. Sci. 91 1700–1705. [Google Scholar]
- Kim D., Kim J., Hur J. K., Been K. W., Yoon S. H., Kim J. S. (2016). Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34 863–868. 10.1038/nbt.3609 [DOI] [PubMed] [Google Scholar]
- Kleinstiver B. P., Tsai S. Q., Prew M. S., Nguyen N. T., Welch M. M., Lopez J. M., et al. (2016). Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34 869–874. 10.1038/nbt.3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Nakamura Y., Kaneyoshi J., Higo H., Higo K. (1996). Transformation of kiwifruit (Actinidia chinensis) and trifoliate orange (Poncirus trifoliata) with a synthetic gene encoding the human epidermal growth factor (hEGF). Engei Gakkai zasshi 64 763–769. 10.2503/jjshs.64.763 [DOI] [Google Scholar]
- Kobayashi S., Uchimiya H. (1989). Expression and integration of a foreign gene in orange (Citrus sinensis Osb.) protoplasts by direct DNA transfers). Jpn. J. Genet 64 91–97. [Google Scholar]
- Kobayashi S., Uchimiya H., Ikeda I. (1983). Plant regeneration from “Trovita” orange protoplasts. Japan J. Breed. 33 119–122. [Google Scholar]
- la Malfa S., Gentile A., ZiNiu D., Domina F. (2000). Citrus genetic transformation with a vital reporter gene. Expression of green fluorescent protein in Troyer citrange. Italus Hortus 7 17–21. [Google Scholar]
- Le B. V., Ha N. T., Hong L. T. A., Van K. T. T. (1999). High frequency shoot regeneration from trifoliate orange (Poncirus trifoliata L. Raf.) using the thin cell layer method. Comptes Rendus Academie des Sciences Paris Life Siences 322 1105–1111. [Google Scholar]
- Lema M. A. (2019). Regulatory aspects of gene editing in Argentina. Transgenic Res. 28 147–150. 10.1007/s11248-019-00145-2 [DOI] [PubMed] [Google Scholar]
- Levy A., El-Mochtar C., Wang C., Goodin M., Orbovic V. (2018). A new toolset for protein expression and subcellular localization studies in citrus and its application to citrus tristeza virus proteins. Plant Methods 14 1–11. 10.1186/s13007-017-0270-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. D., Shi W., Deng X. X. (2002). Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep. 21 153–156. 10.1007/s00299-002-0492-6 [DOI] [Google Scholar]
- Li D. D., Shi W., Deng X. X. (2003). Factors influencing Agrobacterium-mediated embryogenic callus transformation of Valencia sweet orange (Citrus sinensis) containing the pTA29-barnase gene. Tree Physiol. 23 1209–1215. 10.1093/treephys/23.17.1209 [DOI] [PubMed] [Google Scholar]
- Li F., Dai S.-m., Deng Z.-n., Li D.-z., Long G.-y., Li N., et al. (2017). Evaluation of parameters affecting Agrobacterium-mediated transient expression in citrus. J. Integr. Agric. 16:572579. 10.1016/S2095-3119(16)61460-0 [DOI] [Google Scholar]
- Li Q., Qin X., Qi J., Dou W., Dunand C., Chen S., et al. (2020). CsPrx25, a class III peroxidase in Citrus sinensis, confers resistance to citrus bacterial canker through the maintenance of ROS homeostasis and cell wall lignification. Hortic. Res. 7 15–17. 10.1038/s41438-020-00415-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limera C., Sabbadini S., Sweet J. B., Mezzetti B. (2017). New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8:1418. 10.3389/fpls.2017.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Buenfil J. A., Ramírez-Pool J. A., Ruiz-Medrano R., Montes-Horcasitas M., del C., Chavarin-Palacio C., et al. (2017). Dynamics of huanglongbing-associated bacterium candidatus liberibacter asiaticus in Citrus aurantifolia Swingle (Mexican lime). Pakistan J. Biol. Sci. 20 113–123. 10.3923/pjbs.2017.113.123 [DOI] [PubMed] [Google Scholar]
- Lu S., Ye J., Zhu K., Zhang Y., Zhang M., Xu Q., et al. (2021). A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot. 72 3028–3043. 10.1093/JXB/ERAB045 [DOI] [PubMed] [Google Scholar]
- Lusser M., Davies H. V. (2013). Comparative regulatory approaches for groups of new plant breeding techniques. N. Biotechnol. 30 437–446. 10.1016/j.nbt.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Luth D., Moore G. (1999). Transgenic grapefruit plants obtained by Agrobacterium tumefaciens-mediated transformation. Plant Cell. Tissue Organ Cult. 57 219–222. 10.1023/A:1006387900496 [DOI] [Google Scholar]
- Mabberley D. J. (2004). Citrus (Rutaceae): a review of recent advances in etymology, systematics and medical applications. Blumea J. Plant Taxon. Plant Geogr. 49 481–498. 10.3767/000651904X484432 [DOI] [Google Scholar]
- Maggon R., Deo Singh B. (1995). Promotion of adventitious bud regeneration by ABA in combination with BAP in epicotyl and hypocotyl explants of sweet orange (Citrus sinensis L. Osbeck). Sci. Hortic. 63 123–128. 10.1016/0304-4238(95)00780-W [DOI] [Google Scholar]
- Marcelino-Perez G., Ruiz-Medrano R., Gallardo-Hernandez S., Xoconostle-Cazares B. (2021). Adsorption of recombinant human β-Defensin 2 and two mutants on mesoporous silica nanoparticles and its effect against Clavibacter michiganensis sbp. michiganensis. Nanomaterials 11:2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani-Hert M., Bowman K. D., McCollum G. T., Mirkov T. E., Evens T. J., Niedz R. P. (2012). a dark incubation period is important for Agrobacterium-mediated transformation of mature internode explants of sweet orange, grapefruit, citron, and a citrange rootstock. PLoS One 7:e47426. 10.1371/journal.pone.0047426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt B. A., Zhang X., Triplett E. W., Mou Z., Orbović V. (2021). Selection of transgenic citrus plants based on glyphosate tolerance conferred by a citrus 5-enolpyruvylshikimate-3-phosphate synthase variant. Plant Cell Rep. 1:3. 10.1007/s00299-021-02760-y [DOI] [PubMed] [Google Scholar]
- Miki B., McHugh S. (2004). Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J. Biotechnol. 107 193–232. 10.1016/j.jbiotec.2003.10.011 [DOI] [PubMed] [Google Scholar]
- Miyata L. Y., Harakava R., Stipp L. C. L., Mendes B. M. J., Appezzato-da-Glória B., de Assis Alves Mourão Filho F. (2012). GUS expression in sweet oranges (Citrus sinensis L. Osbeck) driven by three different phloem-specific promoters. Plant Cell Rep. 31 2005–2013. 10.1007/s00299-012-1312-2 [DOI] [PubMed] [Google Scholar]
- Molinari H. B. C., Bespalhok J. C., Kobayashi A. K., Pereira L. F. P., Vieira L. G. E. (2004). Agrobacterium tumefaciens-mediated transformation of Swingle citrumelo (Citrus paradisi Macf. x Poncirus trifoliata L. Raf.) using thin epicotyl sections. Sci. Hortic. 99 379–385. 10.1016/S0304-4238(03)00111-0 [DOI] [Google Scholar]
- Moore G. A., Jacono C. C., Neidigh J. L., Lawrence S. D., Cline K. (1992). Agrobacterium-mediated transformation of Citrus stem segments and regeneration of transgenic plants. Plant Cell Rep. 11 238–242. [DOI] [PubMed] [Google Scholar]
- Moreira-Dias J. M., Molina R. V., Bordon Y., Guardiola J. L., Garcia-Luis A. (2000). Direct and indirect shoot organogenic pathways in Epicotyl cuttings of Troyer citrange differ in hormone requirements and in their response to light. Ann. Bot. 85 103–110. [Google Scholar]
- Navarro L., Roistacher C. N., Murashige T. (1975). Improvement of shoot-tip grafting in vitro for virus-free citrus. J. Am. Soc. Hortic. Sci. 100:471479. [Google Scholar]
- Niedz R. P., McKendree W. L., Shatters R. G. (2003). Electroporation of embryogenic protoplasts of sweet orange (Citrus sinensis (L.) Osbeck) and regeneration of transformed plants. Vitr. Cell. Dev. Biol. Plant 39 586–594. 10.1079/IVP2003463 [DOI] [Google Scholar]
- Niedz R. P., Sussman M. R., Satterlee J. S. (1995). Green fluorescent protein: an in vivo reporter of plant gene expression. Plant Cell Rep. 14, 403–406. 10.1007/BF00234043 [DOI] [PubMed] [Google Scholar]
- Nishikawa F., Endo T., Shimada T., Fujii H., Shimizu T., Omura M. (2008). Isolation and characterization of a Citrus FT/TFL1 homologue (CuMFT1), which shows quantitatively preferential expression in Citrus seeds. J. Jpn. Soc. Hortic. Sci. 77 38–46. 10.2503/jjshs1.77.38 [DOI] [Google Scholar]
- Olivares-Fuster O., Fleming G. H., Albiach-Marti M. R., Gowda S., Dawson W. O., Grosser J. W. (2003). Citrus tristeza virus (CTV) resistance in transgenic citrus based on virus challenge of protoplasts. Vitr. Cell. Dev. Biol. - Plant 39 567–572. 10.1079/IVP2003432 [DOI] [Google Scholar]
- Omar A. A., Grosser J. W. (2008). Comparison of endoplasmic reticulum targeted and non-targeted cytoplasmic GFP as a selectable marker in citrus protoplast transformation. Plant Sci. 174 131–139. 10.1016/J.PLANTSCI.2007.10.003 [DOI] [Google Scholar]
- Omar A. A., Murata M. M., El-Shamy H. A., Graham J. H., Grosser J. W. (2018). Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 27 179–191. 10.1007/s11248-018-0065-2 [DOI] [PubMed] [Google Scholar]
- Omar A. A., Song W. Y., Grosser J. W. (2007). Introduction of Xa21, a Xanthomonas-resistance, gene from rice, into “Hamlin” sweet orange [Citrus sinensis (L.) Osbeck] using protoplast- GFP co-transformation or single plasmid transformation. J. Hortic. Sci. Biotechnol. 82 914–923. 10.1080/14620316.2007.11512326 [DOI] [Google Scholar]
- Orbović V., Grosser J. W. (2015). “Chapter 20: Citrus transformation using juvenile tissue explants,” in Agrobacterium Protocols, 3rd Edn, Vol. 2 ed. Wang K. (New York, NY: Springer; ), 1–329. 10.1007/978-1-4939-1658-0 [DOI] [PubMed] [Google Scholar]
- Orbović V., Ravanfar S. A., Acanda Y., Narvaez J., Merritt B. A., Levy A., et al. (2021). Stress-inducible Arabidopsis thaliana RD29A promoter constitutively drives Citrus sinensis APETALA1 and LEAFY expression and precocious flowering in transgenic Citrus spp. Transgenic Res. 30 687–699. 10.1007/s11248-021-00260-z [DOI] [PubMed] [Google Scholar]
- Padilla I. M. G., Burgos L. (2010). Aminoglycoside antibiotics: structure, functions and effects on in vitro plant culture and genetic transformation protocols. Plant Cell Rep. 29 1203–1213. 10.1007/s00299-010-0900-2 [DOI] [PubMed] [Google Scholar]
- Peña L., Cervera M., Fagoaga C., Pérez R., Romero J., Juárez J., et al. (2004a). “Agrobacterum-mediated transformation of citrus,” in Transgenic Crops of the World, ed. Curtis I. S. (Dordrecht: Springer Netherlands; ), 145–156. 10.1007/978-1-4020-2333-0_11 [DOI] [Google Scholar]
- Peña L., Pérez R. M., Cervera M., Juárez J. A., Navarro L. (2004b). Early events in Agrobacterium-mediated genetic transformation of citrus explants. Ann. Bot. 94 67–74. 10.1093/aob/mch117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña L., Cervera M., Fagoaga C., Romero J., Ballester A., Soler N., et al. (2008). “Citrus,” in Compendium of Transgenic Crop Plants: Transgenic Tropical and Subtropical Fruits and Nuts, eds Kole C., TC Hall. (Hoboken, NJ: Blackwell Publishing Ltd.). [Google Scholar]
- Peña L., Cervera M., Juárez J., Navarro A., Pina J. A., Durán-Vila N., et al. (1995a). Agrobacterium-mediated transformation of sweet orange and regeneration of transgenic plants. Plant Cell Rep. 14 616–619. 10.1007/BF00232724 [DOI] [PubMed] [Google Scholar]
- Peña L., Cervera M., Juárez J., Ortega C., Pina J. A., Durán-Vila N., et al. (1995b). High efficiency Agrobacterium-mediated transformation and regeneration of citrus. Plant Sci. 104 183–191. 10.1016/0168-9452(94)04021-8 [DOI] [Google Scholar]
- Peña L., Cervera M., Navarro A., Durfin-vila N., Navarro L. (1995c). Agrobacterium-mediated transformation of sweet orange and regeneration of transgenic plants. Plant Cell Rep. 14 616–619. [DOI] [PubMed] [Google Scholar]
- Peña L., Cervera M., Juárez J., Navarro A., Pina J. A., Navarro L. (1997). Genetic transformation of lime (Citrus aurantifolia Swing.): factors affecting transformation and regeneration. Plant Cell Rep. 16 731–737. 10.1007/s002990050311 [DOI] [PubMed] [Google Scholar]
- Peña L., Martín-Trillo M., Juárez J., Pina J. A., Navarro L., Martínez-Zapater J. M. (2001). Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19 263–267. 10.1038/85719 [DOI] [PubMed] [Google Scholar]
- Peña L., Navarro L. (2000). “Transgenic Citrus,” in Transgenic Trees, ed. Bajaj Y. P. S. (Berlin: Springer Berlin Heidelberg; ), 39–54. 10.1007/978-3-642-59609-4_4 [DOI] [Google Scholar]
- Peña L., Navarro L. (2020). “Transgenic trees,” in Forest Products Biotechnology Biotechnology in Agriculture and Forestry, ed. Bajaj Y. P. S. (Berlin: Springer Berlin Heidelberg; ), 297–314. 10.1201/9781482272734-20 [DOI] [Google Scholar]
- Peng A., Chen S., Lei T., Xu L., He Y., Wu L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15 1509–1519. 10.1111/pbi.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A., Xu L., He Y., Lei T., Yao L., Chen S., et al. (2015). Efficient production of marker-free transgenic ‘Tarocco’ blood orange (Citrus sinensis Osbeck) with enhanced resistance to citrus canker using a Cre/loxP site-recombination system. Plant Cell. Tissue Organ Cult. 123 1–13. 10.1007/s11240-015-0799-y [DOI] [Google Scholar]
- Peng A., Zhang J., Zou X., He Y., Xu L., Lei T., et al. (2021a). Pyramiding the antimicrobial PR1aCB and AATCB genes in ‘Tarocco’ blood orange (Citrus sinensis Osbeck) to enhance citrus canker resistance. Transgenic Res. 30 635–647. 10.1007/s11248-021-00245-y [DOI] [PubMed] [Google Scholar]
- Peng A., Zou X., He Y., Chen S., Liu X., Zhang J., et al. (2021b). Overexpressing a NPR1-like gene from Citrus paradisi enhanced huanglongbing resistance in C. sinensis. Plant Cell Rep. 40 529–541. 10.1007/s00299-020-02648-3 [DOI] [PubMed] [Google Scholar]
- Pérez-Molphe-Balch E., Ochoa-Alejo N. (1998). Regeneration of transgenic plants of Mexican lime from Agrobacterium rhizogenes-transformed tissues. Plant Cell Rep. 17 591–596. 10.1007/s002990050448 [DOI] [PubMed] [Google Scholar]
- Poles L., Licciardello C., Distefano G., Nicolosi E., Gentile A., La Malfa S. (2020). Recent advances of in vitro culture for the application of new breeding techniques in citrus. Plants 9 1–25. 10.3390/plants9080938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons E., Alquézar B., Rodríguez A., Martorell P., Genovés S., Ramón D., et al. (2014). Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 12 17–27. 10.1111/pbi.12112 [DOI] [PubMed] [Google Scholar]
- Pons E., Peris J. E., Peña L. (2012). Field performance of transgenic citrus trees: assessment of the long-term expression of uidA and nptII transgenes and its impact on relevant agronomic and phenotypic characteristics. BMC Biotechnol. 12:41. 10.1186/1472-6750-12-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Gu H., Ma L., Peng Y., Deng X. W., Chen Z., et al. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17 471–482. 10.1038/cr.2007.40 [DOI] [PubMed] [Google Scholar]
- Rai M. (2006). Refinement of the citrus tristeza virus resistance gene (Ctv) positional map in Poncirus trifoliata and generation of transgenic grapefruit (Citrus paradisi) plant lines with candidate resistance genes in this region. Plant Mol. Biol. 61 399–414. 10.1007/s11103-006-0018-7 [DOI] [PubMed] [Google Scholar]
- Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. J., Zuo H., Xu Q. (2021). Genomic insights into citrus domestication and its important agronomic traits. Plant Commun. 2:100138. 10.1016/j.xplc.2020.100138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes C. A., Zanek M. C., Velázquez K., Costa N., Plata M. I., Garcia M. L. (2011). Generation of sweet orange transgenic lines and evaluation of citrus psorosis virus-derived resistance against psorosis a and psorosis b. J. Phytopathol. 159 531–537. 10.1111/j.1439-0434.2011.01800.x [DOI] [Google Scholar]
- Romero-Romero J. L., Inostroza-Blancheteau C., Reyes-Díaz M., Matte J. P., Aquea F., Espinoza C., et al. (2020). Increased drought and salinity tolerance in Citrus aurantifolia (Mexican Lemon) plants overexpressing Arabidopsis CBF3 gene. J. Soil Sci. Plant Nutr. 20 244–252. 10.1007/s42729-019-00130-y [DOI] [Google Scholar]
- Rommens C. M. (2004). All-native DNA transformation: a new approach to plant genetic engineering. Trends Plant Sci. 9 457–464. 10.1016/j.tplants.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Rommens C. M., Haring M. A., Swords K., Davies H. V., Belknap W. R. (2007). The intragenic approach as a new extension to traditional plant breeding. Trends Plant Sci. 12 397–403. 10.1016/j.tplants.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Sandal I., Saini U., Lacroix B., Bhattacharya A., Ahuja P. S., Citovsky V. (2007). Agrobacterium-mediated genetic transformation of tea leaf explants: effects of counteracting bactericidity of leaf polyphenols without loss of bacterial virulence. Plant Cell Rep. 26 169–176. 10.1007/s00299-006-0211-9 [DOI] [PubMed] [Google Scholar]
- Sanford J. C., Klein T. M., Wolf E. D., Allen N. (1987). Delivery of substances into cells and tissues using a particle bombardment process. Part. Sci. Technol. 5 27–37. 10.1080/02726358708904533 [DOI] [Google Scholar]
- Savita, Singh B., Virk G. S., Nagpal A. K. (2011). An efficient plant regeneration protocol from callus cultures of Citrus jambhiri Lush. Physiol. Mol. Biol. Plants 17 161–169. 10.1007/s12298-011-0055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendín L. N., Orce I. G., Gómez R. L., Enrique R., Grellet Bournonville C. F., Noguera A. S., et al. (2017). Inducible expression of Bs2 R gene from Capsicum chacoense in sweet orange (Citrus sinensis L. Osbeck) confers enhanced resistance to citrus canker disease. Plant Mol. Biol. 93 607–621. 10.1007/s11103-017-0586-8 [DOI] [PubMed] [Google Scholar]
- Shimizu T. (2020). Genomic Breeding. Amsterdam: Elsevier Inc.. 10.1016/B978-0-12-812163-4.00008-5 [DOI] [Google Scholar]
- Sinn J. P., Held J. B., Vosburg C., Klee S. M., Orbovic V., Taylor E. L., et al. (2021). Flowering Locus T chimeric protein induces floral precocity in edible citrus. Plant Biotechnol. J. 19 215–217. 10.1111/pbi.13463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova O. G., Kochetov A. V. (2020). “Choice of the promoter for tissue and developmental stage-specific gene expression,” in Methods in Molecular Biology, eds Rustgi S., Luo H. (New York, NY: Humana; ), 69–106. 10.1007/978-1-0716-0356-7_4 [DOI] [PubMed] [Google Scholar]
- Soares J. M., Weber K. C., Qiu W., Stanton D., Mahmoud L. M., Wu H., et al. (2020). The vascular targeted citrus FLOWERING LOCUS T3 gene promotes non-inductive early flowering in transgenic Carrizo rootstocks and grafted juvenile scions. Sci. Rep. 10:21404. 10.1038/s41598-020-78417-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N., Plomer M., Fagoaga C., Moreno P., Navarro L., Flores R., et al. (2012). Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol. J. 10 597–608. 10.1111/j.1467-7652.2012.00691.x [DOI] [PubMed] [Google Scholar]
- Song G. Q., Walworth A. E., Loescher W. H. (2015). Grafting of genetically engineered plants. J. Am. Soc. Hortic. Sci. 140 203–213. 10.21273/jashs.140.3.203 [DOI] [Google Scholar]
- Sorkina A., Bardosh G., Liu Y. Z., Fridman I., Schlizerman L., Zur N., et al. (2011). Isolation of a citrus promoter specific for reproductive organs and its functional analysis in isolated juice sacs and tomato. Plant Cell Rep. 30 1627–1640. 10.1007/s00299-011-1073-3 [DOI] [PubMed] [Google Scholar]
- Spencer P. A., Towers G. H. N. (1991). Restricted occurrence of acetophenone signal compounds. Phytochemistry 30 2933–2937. 10.1016/S0031-9422(00)98227-X [DOI] [Google Scholar]
- Stewart J. (2001). The utility of green fluorescent protein in transgenic plants. Plant Cell Rep. 20 376–382. 10.1007/s002990100346 [DOI] [PubMed] [Google Scholar]
- Sugita K., Matsunaga E., Ebinuma H. (1999). Effective selection system for generating marker-free transgenic plants independent of sexual crossing. Plant Cell Rep. 18 941–947. 10.1007/s002990050688 [DOI] [Google Scholar]
- Tallón C. I., Porras I., Pérez-Tornero O. (2012). Efficient propagation and rooting of three citrus rootstocks using different plant growth regulators. Vitr. Cell. Dev. Biol. Plant 48 488–499. 10.1007/s11627-012-9457-9 [DOI] [Google Scholar]
- Tavano E. C., da R., Erpen L., Aluisi B., Harakava R., Lopes J. R. S., et al. (2019). Sweet orange genetic transformation with the attacin A gene under the control of phloem-specific promoters and inoculation with Candidatus Liberibacter asiaticus. J. Hortic. Sci. Biotechnol. 94 210–219. 10.1080/14620316.2018.1493361 [DOI] [Google Scholar]
- Tomas D. T., Ross M. C., Songstad D. D. (1995). “Direct DNA transfer into intact plant cells via microprojectile bombardment,” in Plant Cell, Tissue and Organ Culture (Editorial Springer Lab Manual), eds Gamborg O. L., Phillips G. C. (Berlin: Springer; ), 197–213. [Google Scholar]
- Vardi A., Bleichman S., Aviv D. (1990). Genetic transformation of citrus protoplasts and regeneration of transgenic plants. Plant Sci. 69 199–206. 10.1016/0168-9452(90)90118-8 [DOI] [Google Scholar]
- Vardi A., Spiegel-Roy P., Galun E. (1982). Plant regeneration from Citrus protoplasts: variability in methodological requirements among cultivars and species. Theor. Appl. Genet. 62 171–176. 10.1007/BF00293354 [DOI] [PubMed] [Google Scholar]
- Velázquez K., Agüero J., Vives M. C., Aleza P., Pina J. A., Moreno P., et al. (2016). Precocious flowering of juvenile citrus induced by a viral vector based on Citrus leaf blotch virus: a new tool for genetics and breeding. Plant Biotechnol. J. 14 1976–1985. 10.1111/pbi.12555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Aderkas P., Bonga J. M. (2000). Influencing micropropagation and somatic embryogenesis in mature trees by manipulation of phase change, stress and culture environment. Tree Physiol. 20 921–928. 10.1093/treephys/20.14.921 [DOI] [PubMed] [Google Scholar]
- Wang A. S., Evans R. A., Altendorf P. R., Hanten J. A., Doyle M. C., Rosichan J. L. (2000). A mannose selection system for production of fertile transgenic maize plants from protoplasts. Plant Cell Rep. 19 654–660. 10.1007/s002999900181 [DOI] [PubMed] [Google Scholar]
- Wang L., Chen S., Peng A., Xie Z., He Y., Zou X. (2019). CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 13 501–510. 10.1007/s11816-019-00556-x [DOI] [Google Scholar]
- Wong W. S., Li G. G., Ning W., Xu Z. F., Hsiao W. L. W., Zhang L. Y., et al. (2001). Repression of chilling-induced ACC accumulation in transgenic citrus by over-production of antisense 1-aminocyclopropane-1-carboxylate synthase RNA. Plant Sci. 161 969–977. 10.1016/S0168-9452(01)00505-2 [DOI] [Google Scholar]
- Wu G. A., Terol J., Ibanez V., López-García A., Pérez-Román E., Borredá C., et al. (2018). Genomics of the origin and evolution of Citrus. Nature 554 311–316. 10.1038/nature25447 [DOI] [PubMed] [Google Scholar]
- Wu H., Acanda Y., Canton M., Zale J. (2019). Efficient biolistic transformation of immature citrus rootstocks using phosphomannose-isomerase selection. Plants 8:390. 10.3390/plants8100390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Acanda Y., Jia H., Wang N., Zale J. (2016). Biolistic transformation of Carrizo citrange (Citrus sinensis Osb. × Poncirus trifoliata L. Raf.). Plant Cell Rep. 35 1955–1962. 10.1007/s00299-016-2010-2 [DOI] [PubMed] [Google Scholar]
- Yang Z. N., Ingelbrecht I. L., Louzada E., Skaria M., Mirkov T. E. (2000). Agrobacterium-mediated transformation of the commercially important grapefruit cultivar Rio Red (Citrus paradisi Macf.). Plant Cell Rep. 19 1203–1211. 10.1007/s002990000257 [DOI] [PubMed] [Google Scholar]
- Yao J. L., Wu J. H., Gleave A. P., Morris B. A. M. (1996). Transformation of citrus embryogenic cells using particle bombardment and production of transgenic embryos. Plant Sci. 113 175–183. 10.1016/0168-9452(95)04292-X [DOI] [Google Scholar]
- Yu C., Huang S., Chen C., Deng Z., Ling P., Gmitter F. G. (2002). Factors affecting Agrobacterium-mediated transformation and regeneration of sweet orange and citrange. Plant Cell. Tissue Organ Cult. 71 147–155. 10.1023/A:1019983107509 [DOI] [Google Scholar]
- Zeng R.-F., Zhou H., Fu L.-M., Yan Z., Ye L.-X., Hu S.-F., et al. (2021). Two citrus KNAT-like genes (CsKN1 and CsKN2) are involved in the regulation of sweet orange spring shoot development. J. Exp. Bot. 72 7002–7019. 10.1093/jxb/erab311 [DOI] [PubMed] [Google Scholar]
- Zhang F., LeBlanc C., Irish V. F., Jacob Y. (2017). Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36 1883–1887. 10.1007/s00299-017-2202-4 [DOI] [PubMed] [Google Scholar]
- Zhang M., Wang J., Luo Q., Yang C., Yang H., Cheng Y. (2021). CsMYB96 enhances citrus fruit resistance against fungal pathogen by activating salicylic acid biosynthesis and facilitating defense metabolite accumulation. J. Plant Physiol. 264 153472. 10.1016/j.jplph.2021.153472 [DOI] [PubMed] [Google Scholar]
- Zhang R., Meng Z., Abid M. A., Zhao X. (2019). “Novel pollen magnetofection system for transformation of cotton plant with magnetic nanoparticles as gene carriers,” in Transgenic Cotton: Methods and Protocols, ed. Zhang B. (New York, NY: Humana Press; ), 56–74. 10.1007/978-1-4939-6911-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Y., Zhang D. m., Zhong Y., Chang X. j., Hu M. l., Cheng C. z. (2017). A simple and efficient in planta transformation method for pommelo (Citrus maxima) using Agrobacterium tumefaciens. Sci. Hortic. 214 174–179. 10.1016/j.scienta.2016.11.033 [DOI] [Google Scholar]
- Zhao X., Meng Z., Wang Y., Chen W., Sun C., Cui B., et al. (2017). Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 3 956–964. 10.1038/s41477-017-0063-z [DOI] [PubMed] [Google Scholar]
- Zhong G., Nicolosi E. (2020). “Citrus origin, diffusion, and economic importance,” in The Citrus Genome, eds Gentile A., La Malfa S, Deng Z. (Cham: Springer; ), 5–21. 10.1007/978-3-030-15308-3_2 [DOI] [Google Scholar]
- Zhu C., Zheng X., Huang Y., Ye J., Chen P., Zhang C., et al. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 17 2199–2210. 10.1111/pbi.13132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Jiang X., Xu L., Lei T., Peng A., He Y., et al. (2017). Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to huanglongbing. Plant Mol. Biol. 93 341–353. 10.1007/s11103-016-0565-5 [DOI] [PubMed] [Google Scholar]
- Zou X., Peng A., Xu L., Liu X., Lei T., Yao L., et al. (2013). Efficient auto-excision of a selectable marker gene from transgenic citrus by combining the Cre/loxP system and ipt selection. Plant Cell Rep. 32 1601–1613. 10.1007/s00299-013-1470-x [DOI] [PubMed] [Google Scholar]
- Zou X., Song E., Peng A., He Y., Xu L., Lei T., et al. (2014). Activation of three pathogen-inducible promoters in transgenic citrus (Citrus sinensis Osbeck) after Xanthomonas axonopodis pv. citri infection and wounding. Plant Cell. Tissue Organ Cult. 117 85–98. 10.1007/s11240-013-0423-y [DOI] [Google Scholar]
- Zou X., Zhao K., Liu Y., Du M., Zheng L., Wang S., et al. (2021). Overexpression of salicylic acid carboxyl methyltransferase (CsSAMT1) enhances tolerance to huanglongbing disease in Wanjincheng Orange (Citrus sinensis (L.) Osbeck). Int. J. Mol. Sci. 22:2803. 10.3390/ijms22062803 [DOI] [PMC free article] [PubMed] [Google Scholar]