Abstract
Many phylogenetically distant animal viruses, including the new coronavirus severe acute respiratory syndrome coronavirus 2, have surface proteins with polybasic sites that are cleaved by host furin and furin-like proteases. Other than priming certain viral surface proteins for fusion, cleavage generates a carboxy-terminal RXXR sequence. This C-end Rule (CendR) motif is known to bind to neuropilin (NRP) receptors on the cell surface. NRPs are ubiquitously expressed, pleiotropic cell surface receptors with important roles in growth factor signaling, vascular biology, and neurobiology, as well as immune homeostasis and activation. The CendR–NRP receptor interaction promotes endocytic internalization and tissue spreading of different cargo, including viral particles. We propose that the interaction between viral surface proteins and NRPs plays an underappreciated and prevalent role in the transmission and pathogenesis of diverse viruses and represents a promising broad-spectrum antiviral target.
Keywords: neuropilin, C-end Rule, virus, internalization
The multitude of viruses with the potential to unleash global outbreaks, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlights the urgent need to understand how these pathogens evolve to gain enhanced virulence and pandemic potential. In particular, identifying the host factors that control viral infection is essential for the development of new antiviral strategies and for the rapid containment of future outbreaks.
To replicate, all viruses must enter a host cell and transfer their genetic material (1). Viruses gain entry by hijacking various host proteins (2, 3). Whereas most virus–receptor interactions involve complex tertiary protein–protein or carbohydrate–protein interactions, virus–host interactions can be in some cases mediated by short (less than seven amino acids) peptide motifs (4). Such elements include viral surface peptides that are involved in cell recruitment, such as positively charged amino acid–containing motifs that engage with negatively charged heparan sulfate polysaccharides on the plasma membrane (5) as well as arginine–glycine–aspartic acid (RGD) and related motifs that bind cell surface integrins (5, 6). Below, we discuss the C-end Rule (CendR) motif–neuropilin (NRP) receptor interaction as an emerging viral peptide–cell receptor system, utilized by SARS-CoV-2, that has potentially broad relevance to the cellular entry and pathogenesis of many viruses (Fig. 1).
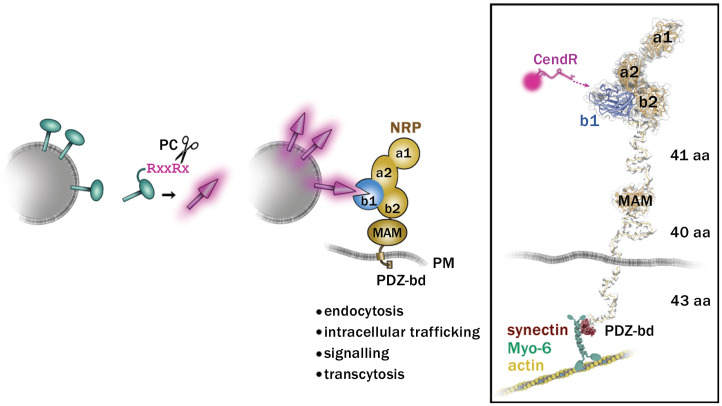
Fig. 1.
Proteolytically activated engagement of viral spike proteins with NRP receptors. (Left) Upon cleavage of R/KXXR/K sequences by furin-like PC, viral spikes (green) are “activated” (magenta spikes) and acquire the ability to bind to NRP receptors. The C-terminal arginine (magenta) of the cleaved viral peptide can interact with the binding pocket in the b1 domain of NRP (blue). On the cytoplasmic side, the postsynaptic density protein (PSD95), Drosophila disc large tumor suppressor (Dlg1), and zonula occludens-1 protein (zo-1) (PDZ)–binding domain (PDZ-bd) of NRP can bind to PDZ-containing proteins and with other receptors, activate different cellular functions, including endocytosis, signaling, intracellular trafficking, and cell-to-cell transport (transcytosis). In Right, the numbers of amino acids (aa) comprising the flexible parts of NRP-1 are indicated on the right. Protein Data Bank ID codes for NRP-1 are 5l73 and 4gz9. Flexible linkers of NRP-1 are modeled using the Coot software. MAM, meprin, A5, receptor protein tyrosine phosphatase mu; PM, plasma membrane.
CendR Peptides: A Position-Dependent Cellular Gateway
In screens for tumor-homing peptides, we discovered a novel peptide motif–receptor system that supports the attachment of various ligands and particles to the cell surface, triggering their endocytic internalization (7, 8). The motif, R/KXXR/K, is recognized by the cognate surface receptors NRP-1 and NRP-2. NRPs are cell surface glycoproteins involved in vascular patterning and angiogenesis, regulation of vascular permeability, modulation of the immune response, and development of the nervous system (9). The R/KXXR/K motif must be carboxy (C) terminal to bind NRPs; therefore, we named the motif CendR. Vascular endothelial growth factor (VEGF)-A165 and other endogenous ligands of NRP-1 and NRP-2 harbor a CendR motif, which binds to a conserved pocket in the extracellular b1 domain of the NRPs (Fig. 1). VEGF-A165 triggers cellular internalization and vascular leakage (10), and short peptides with CendR sequences have similar effects (11), particularly when present in multiple copies on a molecular scaffold or the surface of a nanoparticle (7).
A striking feature of CendR motifs is their ability to penetrate tissue—nanoparticles carrying CendR motifs are transported from the systemic circulation into the extravascular tissue (7, 8). Studies on the internalization of multivalent CendR nanoparticles showed that NRP-1 is an endocytic/sorting receptor. NRP-1 mediates an endocytic pathway similar to macropinocytosis, resulting in the rapid internalization of the ligand–receptor complex along with extracellular solutes and macromolecules (12–14). CendR nanoparticle uptake is independent of dynamin, clathrin, caveolin, and flotillin but depends on actin and on the adaptor protein synectin, which links NRP-1 to the actin motor protein myosin-6 (Fig. 1) (12). Following endocytosis, a yet unidentified sorting signal in NRP-1 mediates intercellular transfer of internalized material, within membrane-enclosed, Rab5-positive vesicles, at cell-to-cell contacts (12). This mechanism is important for the tissue penetration properties of CendR peptides (15).
Whereas early studies on CendR peptides focused on their application for tumor targeting of drugs, the relevance of the CendR peptides as a potential determinant of cellular and tissue tropism of viruses was noted (12).
Furin Convertases and Proteolytic Activation of Pathogenic Viruses
Viral infection often requires the cleavage of viral surface proteins by host proteases. Proteolytic processing can prime these viral fusion proteins (F proteins), enabling penetration into the host cell. The host cell protease furin (also known as paired basic amino-acid-cleaving enzyme and proprotein convertases subtilisin/kexin type 3) (16, 17) is an essential and ubiquitously expressed type I transmembrane serine protease found in the Golgi apparatus, plasma membrane, endosomal compartments, and in soluble form in the extracellular space (18). Furin cleaves after the C-terminal arginine (or lysine) residues of a polybasic target sequence R/KXXR/K, and furin susceptibility of viruses correlates with infectivity and interspecies transmission of different animal viruses (reviewed in ref. 17).
The best-studied case is the influenza virus hemagglutinin (HA) spike glycoprotein, which must be cleaved to allow acid-activated membrane fusion. Low pathogenic avian influenza (LPAI) strains have HAs with a monobasic cleavage site, susceptible to trypsin-like proteases, which have restricted expression in the respiratory and intestinal tracts (19). In contrast, highly pathogenic avian influenza (HPAI) strains have HAs with multiple basic amino acids at the cleavage site, susceptible to furin and the proprotein convertase 6 (PC6) (20). Whereas HPAI strains show rapid dissemination with severe systemic infection, destroying vital organs and frequently causing death, the LPAI strains exhibit limited tissue tropism and cause localized pulmonary and gastrointestinal infections (21). Nevertheless, the insertion of multiple basic amino acids at the cleavage site is not always sufficient to convert LPAI into HPAI (22, 23), and how LPAI viruses evolve into HPAI viruses is incompletely understood (24, 25). Carboxypeptidase B, incorporated in the viral envelope, is capable of cleaving off the C-terminal arginine of HA1 (26), thereby potentially affecting the availability of CendR peptides on the viral surface.
Furin susceptibility is also an important pathogenic determinant for the Newcastle disease virus (NDV). The F protein of avirulent strains of this avian paramyxovirus has a monobasic cleavage site that is processed in the respiratory system. In contrast, the F protein of virulent NDV strains, which spread systemically and cause high rates of mortality in infected birds (27), is cleavable by furin.
The coronavirus spike envelope glycoprotein (S protein) is the main determinant of viral interaction with the host cell surface. Its proteolytic activation at mono- or polybasic sequences is a critical checkpoint that regulates coronavirus cell entry, tissue and cell tropism, host range, and pathogenicity (28). For efficient infection, each subunit of the homotrimeric S protein is cleaved at least once, at the S2’ site. In some coronaviruses (e.g., SARS-CoV-2 and Middle East respiratory syndrome [MERS]), an additional cleavage occurs at the S1/S2 site. Cleavage at S2’ is required to expose the fusogenic portion of the spike (sometimes referred to as the “fusion peptide”), which inserts into the host cell membrane to promote fusion. The S1/S2 site enhances infection in primary cells and in vivo. Interestingly, among lineage B β-coronaviruses, only the SARS-CoV-2 S protein contains a polybasic furin cleavage site RRAR at the junction of S1 and S2 domains, due to insertion of 12 nucleotides in the viral genome (29).
NRPs as Viral Entry Factors
According to the common paradigm, proteolytic cleavage transforms viral surface proteins into a metastable conformation required for productive infection, a molecular “mousetrap” ready to trigger membrane fusion.
However, proteolytic processing of viral proteins by furin has a consequence that has received little attention; it converts polybasic sequence motifs within proteins into CendR peptides that can potentially interact with widely expressed NRP receptors. Indeed, some endogenous substrates of furin, such as class 3 semaphorins, bind to NRPs as their cell surface coreceptors (30–32). Seven viruses to date have been shown to use NRPs as cellular (co-)receptors: the coronavirus SARS-CoV-2 (33, 34), retroviruses human T cell lymphotropic virus-1 (HTLV-1) and HTLV-2 (35, 36), the herpesviruses Epstein-Barr virus (EBV) (37, 38), mouse cytomegalovirus (mCMV) (39), human cytomegalovirus (hCMV) (40), and the arenavirus Lujo virus (LUJV) (41, 42) (Table 1). The spike glycoproteins of all these viruses contain polybasic sites that are cleaved or can potentially be cleaved by a PC. Furin cleavage of viral envelope glycoproteins occurs in phylogenetically distant virus families, including Herpes‐, Corona‐, Flavi‐, Toga‐, Borna‐, Bunya‐, Phyllo‐, Orthomyxo‐, Paramyxo‐, Pneumo‐, Retro-, and the nonenveloped Papillomaviridae (17) (SI Appendix, Table S1). For most of these viruses, furin-mediated cleavages are essential for virus entry and infectivity, and the cleavage products remain associated with the viral particle. We propose that the CendR–NRP interaction could be an underappreciated but critical common pathway used for enhanced infection by PC-activated viruses. In addition to CendR sequences, other regions of the viral spike proteins can interact with NRPs (e.g., the pentamer of hCMV).
Table 1.
Viruses reported to use NRPs as cellular entry factors
| Virus | NRP | Other receptor | Viral NRP-binding protein | Furin cleavage site | Infected cell lines and cells in vivo | Viral family | Genome | Host | Disease | Refs. |
| SARS-CoV-2 | NRP-1 | ACE2 | S | Yes | HEK,* Caco2, Calu-3 | Coronaviridae | +RNA | Human | SARS | 33, 34 |
| HTLV1 | NRP-1 | Glut-1, HSPG | Env | Yes | HeLa, DCs, T cells | Retroviridae | +RNA/DNA | Human | T cell leukemia | 35, 36 |
| EBV | NRP-1 | Ephr-A2 | gB | Yes | Nasopharyngeal epithelial cells | Herpesviridae | DNA | Human | Nasopharyngeal cancer, lymphomas, mononucleosis | 37, 38 |
| mCMV | NRP-1 | n.d. | n.d. | n.d. | Endothelial cells | Herpesviridae | DNA | Mouse | Mostly asymptomatic | 39 |
| hCMV | NRP-2 | CD46 | Pentamer | n.d. | Nasopharyngeal epithelial and endothelial cells | Herpesviridae | DNA | Human | Birth complications, mononucleosis | 40 |
| LUJV | NRP-2 | CD63/LAMP1 | GP | n.d. | Hap1, HEK, endothelial | Arenaviridae | −RNA | Human | Fatal hemorrhagic fever | 41, 42 |
SARS, severe acute respiratory syndrome; DCs, dendritic cells; n.d., not determined.
*HEK cells are susceptible to SARS-CoV-2 only after exogenous expression of ACE2 and TMPRSS2.
In the case of SARS-CoV-2, X-ray crystallography of a peptide corresponding to the furin-cleaved C end of S1 (sequence: SPRRAR) in complex with NRP-1 b1 domain revealed a consensus CendR interaction with the C-terminal arginine of the spike peptide entering the binding pocket of the b1 domain of NRP-1 (34), similar to prototypical NRP-1 ligands (Fig. 1). The binding to NRP-1 appears to be stabilized by additional interaction(s) in other regions of the spike (34).
HTLV-1 and EBV also appear to bind NRP via a canonical CendR interaction, as suggested by competition using full-length VEGFA-165, its C-terminal CendR peptide KPRR, or the soluble b1b2 ectodomain of NRP-1 (35–38). The HTLV-1 interaction with NRP-1 involves additional bridging using heparan sulfate proteoglycans (36), similar to the interactions between NRP-1 and CendR nanoparticles (13), as well as VEGF165 (43). The NRP-1–mediated endocytic mechanism of EBV is dynamin, clathrin, and caveolin independent and based on fluid uptake studies and on small molecule inhibitors sensitivity profiling, resembles macropinocytosis (37). Endocytosed virus–NRP-1 complexes accumulate in intracellular vesicles positive for sorting nexin 5 (SNX5), a protein enriched in macropinosomes (37). Interestingly, VEGFR2 bound to VEGFA165, the isoform that binds to NRP-1, is also internalized by macropinocytosis and accumulates in SNX5 vesicles (44).
hCMV and LUJV use NRP-2 as an entry factor (40–42) in nasopharyngeal epithelial cells.
Physiologically, the two NRP homologs, NRP-1 and NRP-2, bind to both overlapping and distinct members of the VEGF and Sema3 families. For example, whereas Nrp-1 specifically binds the VEGF-A164/5 isoform and Sema3A, NRP-2 binds VEGF-C and Sema3F (9). CendR-binding b1 domains of NRP-1 and NRP-2 show conservation of residues in the binding pocket critical for C-terminal arginine binding and variability within amino acid residues surrounding the interloop cleft (45); this variability and involvement of secondary binding sites are thought to contribute to differences in ligand binding profiles between NRP-1 and NRP-2 (9). NRP-1 and NRP-2 also show isoform-dependent differences in endocytic routes and the intracellular destination of the ligand–partner receptor complexes: recycling endosomes for NRP-1 and late endosomes and lysosomes for NRP-2. NRP-1 guides the VEGFR2/VEGFA complex to the Rab-11 recycling compartment. Depletion of NRP-1 results in redirecting the complex to degradative lysosomes, which results in the termination of signaling (46). On the other hand, NRP-2 has an opposite effect; in the presence of NRP-2, epidermal growth factor receptor (EGFR)–epidermal growth factor (EGF) complexes are targeted for degradation (47). During viral entry, the choice of the NRP isoform may have a profound effect on viral infection, and the preference of one NRP isoform over the other may be dictated by the elements of viral life cycle such as pH or protease required for efficient membrane fusion. In the case of EBV, for instance, NRP-1 enhances entry and infection, whereas NRP-2 is inhibitory (34–37). Importantly, genome delivery of LUJV takes place in late endosomes and lysosomes, supporting the potential contribution of NRP-2 in virus trafficking to these compartments (41, 48).
NRPs are (co-)receptors, and physiological ligands of NRP typically bind a second receptor (e.g., VEGF also binds to VEGFR, and SEMA3A also binds to plexin receptors) (9). Similarly, NRP-interacting viruses seem to use additional receptors for cell binding (e.g., ACE2 for SARS-CoV-2 or Ephrin receptor A2 for EBV) (Table 1). Although the cell surface, particularly the apical membrane of epithelial cells, is rich in proteases, proteolytic CendR activation and NRP engagement may require the high-affinity binding of the viral particle to the primary receptor on the plasma membrane.
NRPs show remarkable structural flexibility and can accommodate ligands and partner receptors of different geometries (Fig. 2), in line with a potential role as multivirus entry/penetration (co-)factors. For a trimeric viral spike, up to three NRP molecules could bind at the same time. A structural comparison of viral spikes that harbor polybasic cleavage sites cleaved, or potentially cleaved, by furin-like convertases shows the presence of water-exposed CendR sequences either on the side of the trimer (e.g., in the case of SARS-CoV-2 and MERS S [Coronaviridae], influenza HA [Orthomyxoviridae], respiratory syncytial virus F [RSV; Pneumoviridae], HIV-1 Env [Retroviridae], and Ebola Zaire virus glycoprotein [GP] [Filoviridae]) or at the forefront of the spike (e.g., in hCMV glycoprotein B [gB] [Herpesviridae] and LUJV GP [Arenaviridae]) (Fig. 2). The flexible parts of the NRP could stretch to reach a CendR peptide exposed on the spike (Fig. 2, center). Following NRP engagement, the force generated by the actin–myosin network coupled to the cytoplasmic PDZ-binding domain of NRP (Fig. 2) could potentially act as a destabilizing factor for the viral spike to facilitate membrane fusion. In line with the role of NRP-1 in SARS-CoV-2 cell–cell fusion (i.e., syncytia formation) (34), the interaction between NRP-1 and S may facilitate the exposure of the second cleavage site (S2’ in SARS-CoV-2 S) (49), bringing this region of the spike to proximity with smaller membrane proteases such as TMPRSS2 (Fig. 2, center). The role of NRP-1 in SARS-CoV-2 infection has not yet been investigated in the epithelial cells of the upper and lower respiratory tracts, where fusion is thought to occur directly at the cell surface rather than in endosomes. The potential facilitation of viral fusion could play an important role in the virus penetration of ciliated polarized epithelial cells, the primary target of SARS-CoV-2. Of note, systemic nanoparticles coated with the CendR peptides show robust microvascular pulmonary accumulation and penetration (7), raising an intriguing possibility of circulation-mediated secondary lung infection. Additional structural and functional studies are required to solve the mechanisms and consequences of NRP engagement with viral spikes.
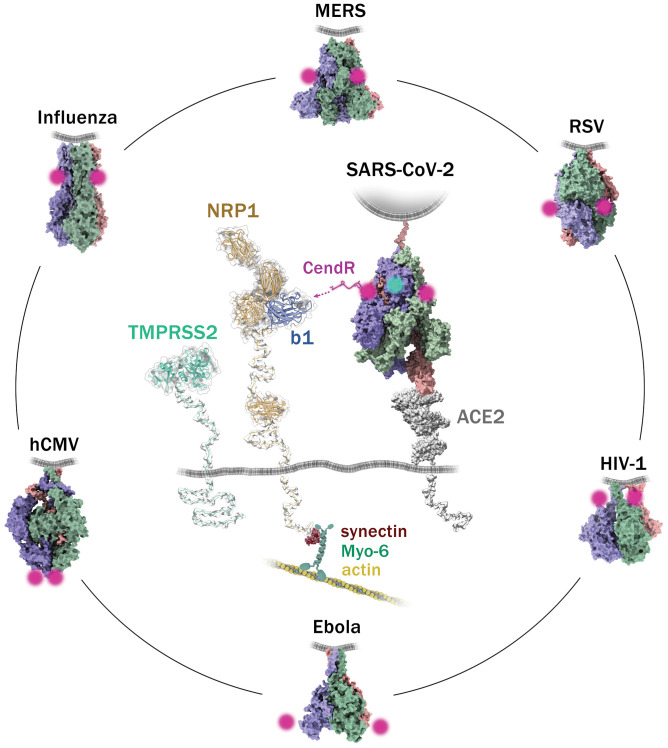
Fig. 2.
Hypothetical interaction model of SARS-CoV-2 spike with entry factors NRP-1 and ACE2 and position of CendR sequences on the spikes of different viruses. The approximate position of the CendR sequence on the trimeric spikes of the respective viruses is indicated by the magenta dots. The three subunits of each trimeric spike are colored in purple, green, and orange. The position of the viral membrane is indicated by the gray line. In the center, the spike of SARS-CoV-2 is binding one ACE2 monomer (gray). The position of the furin-cleaved CendR sequence is indicated in magenta. The b1 domain on NRP is indicated in blue. The virus-activating serine protease TMPRSS2 is shown, and the position of the respective S2’ cleavage site on the spike is indicated by a cyan spot. On the cytoplasmic side, the PDZ-binding domain of NRP-1 binds through synectin to the myosin-6/actin cytoskeleton. All structures were obtained from the Protein Data Bank (PDB) database: SARS-CoV-2 S/ACE2 (7knb, 6m1d), MERS S (7knb), RSV F (6q0s), HIV-1 Env (6mtj), Ebola GP (5jq7), HCMV gB (7kdp), and Influenza HA (2ibx). The long flexible parts between the plasma membrane and NRP-1 meprin, A5, receptor protein tyrosine phosphatase mu (MAM) (PBD ID code 5l73) domain, the MAM and the b1b2a1a2 (PDB ID code 4gz9) domains, as well as the part between the plasma membrane and catalytic domain of TMPRSS2 (PDB ID code 7meq) were modeled using Coot software based on the maximal length of the 40 to 41 (NRP-1) and 42 (TMPRSS2) amino acids in the primary sequence of the respective proteins.
CendR in Viral Infection: The Path Forward
Based on the existing data, we propose that many different viruses ultimately expose CendR peptides to bind NRP, facilitating infection and pathogenesis. As a variation to this rule, some viral spikes, such as the pentamer of hCMV, can bind NRPs in the absence of consensus CendR motif. Binding to NRP (co-)receptors could contribute to viral infectivity and pathogenesis in several ways.
-
1)
Promoting cellular uptake. Interaction of CendR peptides with NRP triggers robust cell internalization, particularly when multiple copies of the CendR peptides are displayed on the surface of virus-sized nanoparticles (7, 12).
-
2)
Enabling tissue spreading. CendR peptides support trans-tissue transport across biological barriers commonly invaded by viruses (e.g., epithelial, vascular, olfactory, and blood–brain barriers) (7, 33, 50).
-
3)
Competing with endogenous ligands. For example, the SARS-CoV-2 S trimer competes with VEGF binding to NRP-1 in pain sensory neurons, causing an analgesic effect (51). Analogous competition with other NRP-1 ligands could modulate signaling pathways to affect virus entry, protein synthesis, and spreading.
-
4)
Suppressing the antiviral immune response. NRPs are important regulators of innate and adaptive immunity with key roles in inflammation, immune homeostasis, and surveillance (52). NRP-1 is expressed in macrophages (53, 54), is part of immunological synapses (55), and regulates the activity of different T cells (56–58). By binding to NRP, viral spikes could interfere with these immune functions.
-
5)
Modulating signaling cascades. NRPs are well-known multiligand hub receptors of important growth factors (e.g., VEGF, PlGF, TGF-β, PDGF-B, FGF, EGF, and HGF) (59). Whereas the cytoplasmic side of NRP has not been associated with kinase activity, virus-bound NRP could, by partnering with and coactivating signaling receptors, modulate signaling cascades involved in remodeling of cytoskeleton, macropinocytosis, cell survival, and protein synthesis.
Research into the role of CendR–NRP in viral infection is needed to address outstanding questions. For instance, do all viruses that contain polybasic processing sites use NRP for enhanced infectivity? Interestingly, for phage displayed peptides, a single C-terminal arginine suffices for NRP binding (7), and split complement proteins with a monobasic C-terminal sequence bind NRP (60).
From the cell perspective, how does the CendR pathway affect the trafficking of viral particles and infection in vivo? How does viral hijacking of the CendR pathway affect physiological functions of NRP, and what are the pathological consequences of viral modulation of the CendR pathway? Moreover, studies on the uptake of multivalent CendR nanoparticles indicated that nutrient starvation and stress conditions enhance NRP-1–dependent particle uptake (12). So, would these conditions enhance infection by CendR-displaying viruses? Finally, is the CendR pathway relevant to zoonotic spread of viruses? Does the conservation of the CendR binding pocket in NRP across vertebrates render furin-activated viruses particularly prone to the transmission from a reservoir population to a new host species?
The answers to these questions, while unlikely to be simple and straightforward, may establish a novel paradigm: NRPs as (co-)receptors shared by a multitude of proteolytically activated pathogenic viruses. This finding could inform broad-spectrum antivirals that act by regulating NRP or CendR convertases in cells or by blocking the conserved CendR peptides on viruses.
Supplementary Material
Acknowledgments
We thank Antti Vaheri, Guido Serini, and Ari Helenius for critical reading of the manuscript.
Footnotes
Competing interest statement: T.T. is the inventor of iRGD and CendR peptides and a shareholder of Cend Therapeutics Inc., a company that holds a license for the iRGD and CendR peptides.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112457118/-/DCSupplemental.
Data Availability
There are no data underlying this work.
References
- 1.Marsh M., Helenius A., Virus entry: Open sesame. Cell 124, 729–740 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercer J., Schelhaas M., Helenius A., Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Ewers H., Helenius A., Lipid-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 3, a004721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagai T., Azia A., Babu M. M., Andino R., Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 7, 1729–1739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cagno V., Tseligka E. D., Jones S. T., Tapparel C., Heparan sulfate proteoglycans and viral attachment: True receptors or adaptation bias? Viruses 11, E596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobhy H., A review of functional motifs utilized by viruses. Proteomes 4, E3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teesalu T., Sugahara K. N., Kotamraju V. R., Ruoslahti E., C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. U.S.A. 106, 16157–16162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugahara K. N., et al. , Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science 328, 1031–1035 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H. F., Vander Kooi C. W., Neuropilin functions as an essential cell surface receptor. J. Biol. Chem. 290, 29120–29126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker P. M., et al. , Neuropilin-1 regulates vascular endothelial growth factor-mediated endothelial permeability. Circ. Res. 96, 1257–1265 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Roth L., et al. , Neuropilin-1 mediates vascular permeability independently of vascular endothelial growth factor receptor-2 activation. Sci. Signal. 9, ra42 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Pang H. B., et al. , An endocytosis pathway initiated through neuropilin-1 and regulated by nutrient availability. Nat. Commun. 5, 4904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pang H. B., Braun G. B., Ruoslahti E., Neuropilin-1 and heparan sulfate proteoglycans cooperate in cellular uptake of nanoparticles functionalized by cationic cell-penetrating peptides. Sci. Adv. 1, e1500821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun G. B., et al. , Etchable plasmonic nanoparticle probes to image and quantify cellular internalization. Nat. Mater. 13, 904–911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugahara K. N., et al. , Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 16, 510–520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun E., Sauter D., Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunology 8, e1073 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izaguirre G., The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses 11, E837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas G., Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3, 753–766 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klenk H. D., Garten W., Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2, 39–43 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Klenk H. D., Rott R., The molecular biology of influenza virus pathogenicity. Adv. Virus Res. 34, 247–281 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaoka Y., Webster R. G., Molecular mechanism of acquisition of virulence in influenza virus in nature. Microb. Pathog. 5, 311–318 (1988). [DOI] [PubMed] [Google Scholar]
- 22.Stech O., et al. , Acquisition of a polybasic hemagglutinin cleavage site by a low-pathogenic avian influenza virus is not sufficient for immediate transformation into a highly pathogenic strain. J. Virol. 83, 5864–5868 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Londt B. Z., Banks J., Alexander D. J., Highly pathogenic avian influenza viruses with low virulence for chickens in in vivo tests. Avian Pathol. 36, 347–350 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Lee D. H., Criado M. F., Swayne D. E., Pathobiological origins and evolutionary history of highly pathogenic avian influenza viruses. Cold Spring Harb. Perspect. Med. 11, a038679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Short K. R., Veldhuis Kroeze E. J., Reperant L. A., Richard M., Kuiken T., Influenza virus and endothelial cells: A species specific relationship. Front. Microbiol. 5, 653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garten W., Bosch F. X., Linder D., Rott R., Klenk H.-D., Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology 115, 361–374 (1981). [DOI] [PubMed] [Google Scholar]
- 27.Nagai Y., Hamaguchi M., Toyoda T., Molecular biology of Newcastle disease virus. Prog. Vet. Microbiol. Immunol. 5, 16–64 (1989). [PubMed] [Google Scholar]
- 28.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S., Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 100, 605–614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coutard B., et al. , The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 176, 104742 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodkin A. L., et al. , Neuropilin is a semaphorin III receptor. Cell 90, 753–762 (1997). [DOI] [PubMed] [Google Scholar]
- 31.He Z., Tessier-Lavigne M., Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 90, 739–751 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Chen H., Chédotal A., He Z., Goodman C. S., Tessier-Lavigne M., Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron 19, 547–559 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Cantuti-Castelvetri L., et al. , Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daly J. L., et al. , Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghez D., et al. , Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 80, 6844–6854 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert S., et al. , HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood 113, 5176–5185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H. B., et al. , Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 6, 6240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., et al. , Ephrin receptor A2 is an epithelial cell receptor for Epstein–Barr virus entry. Nat. Microbiol. 3, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Lane R. K., et al. , Necroptosis-based CRISPR knockout screen reveals Neuropilin-1 as a critical host factor for early stages of murine cytomegalovirus infection. Proc. Natl. Acad. Sci. U.S.A. 117, 20109–20116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Martin N., et al. , An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 174, 1158–1171.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Raaben M., et al. , NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 22, 688–696.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen-Dvashi H., Kilimnik I., Diskin R., Structural basis for receptor recognition by Lujo virus. Nat. Microbiol. 3, 1153–1160 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Teran M., Nugent M. A., Synergistic binding of vascular endothelial growth factor-A and its receptors to heparin selectively modulates complex affinity. J. Biol. Chem. 290, 16451–16462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basagiannis D., et al. , VEGF induces signalling and angiogenesis by directing VEGFR2 internalisation through macropinocytosis. J. Cell Sci. 129, 4091–4104 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Parker M. W., Xu P., Li X., Vander Kooi C. W., Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 287, 11082–11089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballmer-Hofer K., Andersson A. E., Ratcliffe L. E., Berger P., Neuropilin-1 promotes VEGFR-2 trafficking through Rab11 vesicles thereby specifying signal output. Blood 118, 816–826 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Dutta S., et al. , Neuropilin-2 regulates endosome maturation and EGFR trafficking to support cancer cell pathobiology. Cancer Res. 76, 418–428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunz S., de la Torre J. C., Breaking the barrier: Host cell invasion by Lujo virus. Cell Host Microbe 22, 583–585 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Li Z. L., Buck M., Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of Spike protein S1 and S2. Biophys. J. 120, 2828–2837 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayama M., Berger P., Coordination of VEGF receptor trafficking and signaling by coreceptors. Exp. Cell Res. 319, 1340–1347 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Moutal A., et al. , SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain 162, 243–252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy S., et al. , Multifaceted role of neuropilins in the immune system: Potential targets for immunotherapy. Front. Immunol. 8, 1228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson A. M., et al. , Neuropilin-1 expression in adipose tissue macrophages protects against obesity and metabolic syndrome. Sci. Immunol. 3, eaan4626 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Roy S., et al. , Macrophage-derived neuropilin-2 exhibits novel tumor-promoting functions. Cancer Res. 78, 5600–5617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tordjman R., et al. , A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat. Immunol. 3, 477–482 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Sidaway P., Immunotherapy: Neuropilin-1 is required for Treg stability. Nat. Rev. Clin. Oncol. 14, 458 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Leclerc M., et al. , Regulation of antitumour CD8 T-cell immunity and checkpoint blockade immunotherapy by Neuropilin-1. Nat. Commun. 10, 3345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C., et al. , Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. Nat. Immunol. 21, 1010–1021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raimondi C., Ruhrberg C., Neuropilin signalling in vessels, neurons and tumours. Semin. Cell Dev. Biol. 24, 172–178 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Battin C., et al. , Neuropilin-1 acts as a receptor for complement split products. Front. Immunol. 10, 2209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no data underlying this work.