Significance
The transport of amino acids across the plasma membrane plays a central role in physiology. The heteromeric amino acid transporters (HATs) of neutral amino acids (LAT1/CD98hc, LAT2/CD98hc, and Asc1/CD98hc) participate in a variety of processes such as modulation of glutamatergic neurotransmission and synaptic plasticity, auditory function, and promotion of brain development and tumor growth by supporting mTORC1 activity. We identify substrate specificity determinants of neutral amino acids HATs within the substrate-binding cavity and in a nearby region that holds the conformation of the substrate-binding site. LAT2 mutations in this scaffold region are associated with human diseases, and our results pave the way to understand the molecular mechanisms of these pathologies.
Keywords: amino acid transporters, HATs, hLAT2, substrate selectivity, structure
Abstract
Despite having similar structures, each member of the heteromeric amino acid transporter (HAT) family shows exquisite preference for the exchange of certain amino acids. Substrate specificity determines the physiological function of each HAT and their role in human diseases. However, HAT transport preference for some amino acids over others is not yet fully understood. Using cryo–electron microscopy of apo human LAT2/CD98hc and a multidisciplinary approach, we elucidate key molecular determinants governing neutral amino acid specificity in HATs. A few residues in the substrate-binding pocket determine substrate preference. Here, we describe mutations that interconvert the substrate profiles of LAT2/CD98hc, LAT1/CD98hc, and Asc1/CD98hc. In addition, a region far from the substrate-binding pocket critically influences the conformation of the substrate-binding site and substrate preference. This region accumulates mutations that alter substrate specificity and cause hearing loss and cataracts. Here, we uncover molecular mechanisms governing substrate specificity within the HAT family of neutral amino acid transporters and provide the structural bases for mutations in LAT2/CD98hc that alter substrate specificity and that are associated with several pathologies.
Amino acids play a central role in cellular metabolism. Dysregulation of both intra- and extracellular amino acid concentrations is associated with pathological conditions (1). Amino acid transfer across the plasma membrane is mediated by specific transporters that bind and transport these molecules from the extracellular medium into the cell or vice versa.
Heteromeric amino acid transporters (HATs) are a family of amino acid transporters comprised by a heavy subunit and a light subunit, linked by a conserved disulfide bridge (2). Heavy subunits (SLC3 family) are ancillary proteins required for trafficking the holotransporter to the plasma membrane (2), whereas the light subunits (LATs; SLC7 family) transport amino acids and confer substrate specificity to the heterodimer (2). HATs are amino acid exchangers that harmonize amino acid concentrations at each side of the plasma membrane and as such they play a critical role in amino acid homeostasis (1, 3).
The physiological relevance of HATs is highlighted by their role in cancer and several inherited diseases (4–8). HAT neutral amino acid transporters in particular are gaining momentum as several mutations linked to human diseases have recently been identified, and new physiological roles for this group of transporters have been uncovered using knockout mouse models (8–13). Several loss-of-function mutations in human LAT2/CD98hc (SLC7A8/SLC3A2) are associated with age-related hearing loss (ARHL) (9) and cataracts (10). Also, some coding variants are linked to an increased risk of autism spectrum disorder (14). In addition, hLAT2/CD98hc overexpression in pancreatic cancer cells sustains glutamine-dependent mTOR activation to promote glycolysis and chemoresistance (15). This observation thus points to hLAT2/CD98hc as a potential pharmacological target in this particular type of cancer. On the other hand, LAT1/CD98hc (SLC7A5/SLC3A2), which is also linked to cancer (4, 7), participates in brain development and autism spectrum disorder (12). Finally, Asc1/CD98hc (SLC7A10/SLC3A2) is considered a target to regulate glutamatergic neurotransmission in some cognitive disorders, such as schizophrenia (16, 17), and a relevant player in adipocyte lipid storage, obesity, and insulin resistance (18).
Several atomic structures of HATs (19–24) and LATs (25) have recently been described, thus paving the way for the dissection of the molecular transport mechanisms. The substrate-binding site of LATs determined in complex with a substrate or competitive inhibitors shows a conserved design consisting of two unwound segments of transmembrane (TM) 1 and TM6, which contain residues that recognize the α-amino and carboxyl groups of the substrate (21–25). Each member of the HAT family displays a preference for transporting a certain set of substrates (2). LAT2/CD98hc, LAT1/CD98hc, and Asc1/CD98hc transport neutral amino acids but of different sizes. LAT1 is specialized in large neutral amino acids but it is inefficient for L-glutamine, and it does not transport small amino acids. LAT2 transports both large and small neutral amino acids, and it is highly efficient for L-glutamine. Finally, Asc1 mediates the preferential uptake of small neutral amino acids, including D-isomers, particularly D-serine (26–28).
Despite recent advances in resolving the structure of several HATs, the molecular mechanisms explaining why each member of the family shows exquisite preference for certain substrates but not others are mostly unknown. Here, we addressed the structural bases of substrate specificity in the HAT family. To this end, we used cryo–electron microscopy (cryo-EM) to determine the structure of human LAT2/CD98hc in inward-facing open and apo conformation. We used this structure to study substrate-binding determinants by combining Protein Energy Landscape Exploration (PELE) and molecular dynamics (MD), together with mutational and functional studies. We reveal that a few residues present in the substrate-binding pocket and nearby regions determine substrate preference, and we demonstrate how the substrate preference of several HATs can be interconverted. In addition, a region located at a certain distance of the substrate cavity but whose structure critically influences the conformation of the substrate-binding site also regulates substrate preference. This region accumulates mutations associated with ARHL and cataracts that alter hLAT2 substrate specificity.
Our work uncovers key structural determinants that govern, by different mechanisms, the differences in substrate specificity found within HAT members of neutral amino acid transporters. It also provides the structural bases for mutations in LAT2/CD98hc associated with deafness and cataracts.
Results
Cryo-EM of hLAT2/CD98hc.
We used single-particle cryo-EM to determine the structure of hLAT2/CD98hc in its apo form and identified the structural basis underlying substrate-size selectivity in the human LAT subfamily of neutral amino acid transporters. After expression in HEK293-6E cells, hLAT2/CD98hc was purified by double affinity chromatography and a final purification step using size-exclusion chromatography (SI Appendix, Fig. S1 A–C). The purified complex was functionally active in transport assays using proteoliposomes (SI Appendix, Fig. S1D).
The peak fraction of the size-exclusion chromatography was applied to holey grids, vitrified, and observed using a 300 kV Titan Krios electron microscope (Thermo Scientific) (SI Appendix, Fig. S2A). Reference-free two-dimensional (2D) averages of the extracted particles revealed that most of the ∼140-kDa complex was made of well-defined TM helices inserted within the detergent micelle, whereas an ectodomain was placed outside the micelle (SI Appendix, Fig. S2B). Extracted particles were subjected to several rounds of 2D and three-dimensional (3D) classification, and 176,132 were selected to reconstruct a high-resolution volume of hLAT2/CD98hc, as described in Materials and Methods (SI Appendix, Fig. S3).
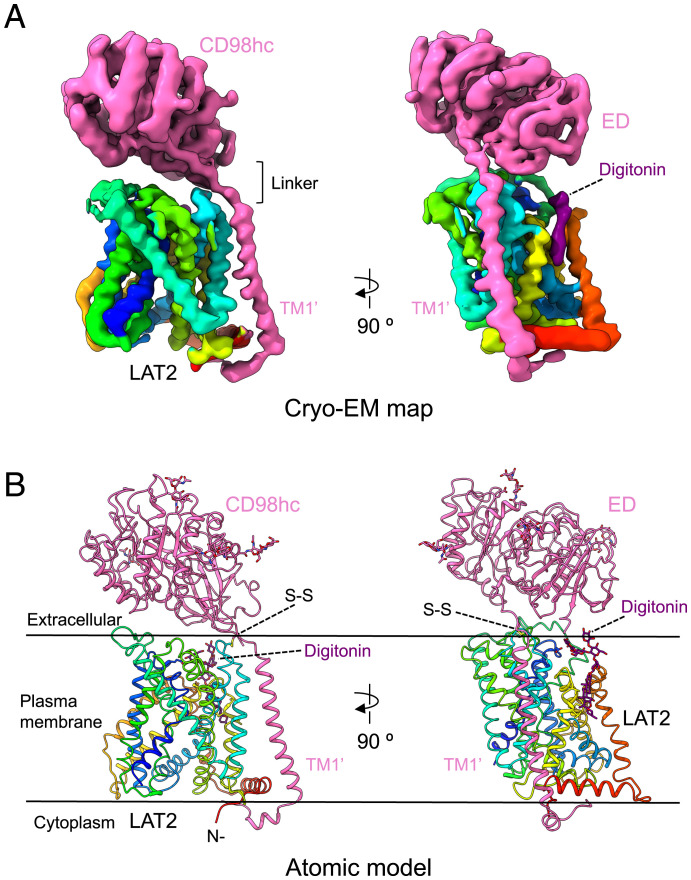
The cryo-EM map of hLAT2/CD98hc revealed the extracellular ectodomain of CD98hc sitting on top of LAT2 and anchored to the membrane by a TM helix (TM1’) that reaches the cytoplasmic side and interacts with hLAT2 (Fig. 1 A and B). This structure is in agreement with previous low-resolution models of hLAT2/CD98hc (29, 30) and similar to other recently solved human HATs (19–23). Indeed, human LAT2/CD98hc apo structure is very similar (rmsd for all backbone atoms of 1.36 Å) to the reported structure of this transporter bound to L-tryptophan (23) (SI Appendix, Fig. S4A). Average resolution estimates of the hLAT2/CD98hc cryo-EM map reported here were 3.9 and 3.7 Å using Fourier Shell Correlation (FSC) in RELION (31) and ResMap (32), respectively (SI Appendix, Fig. S2 C and D). These estimations included the effect of the micelle and the disparity of resolution between the ectodomain and the rigid hLAT2 protein. However, most of hLAT2 was resolved at resolutions between 2.5 and 3.0 Å (SI Appendix, Fig. S2E and Table S1), thereby allowing the unambiguous assignment of residues in each helical element during modeling of the atomic structure of hLAT2 (SI Appendix, Fig. S5). The lower resolution of the CD98hc ectodomain suggested some flexibility with respect to the hLAT2 subunit in the membrane region.
Fig. 1.
Cryo-EM volume and atomic structure of hLAT2/CD98hc. (A) Two views of the cryo-EM map of hLAT2/CD98hc. CD98hc is shown in pink, and the location of its TM helix (TM1’) is indicated. hLAT2 structure is color coded as a rainbow (dark blue, cyan, green, yellow, and red) from the N to the C terminus. A density assigned to digitonin is shown in dark purple. (B) Two views of the atomic model of hLAT2/CD98hc using a color coding as in A. The position of the N terminus of CD98hc and the disulfide bridge linking the two subunits in hLAT2/CD98hc are shown.
In addition, an elongated density near TM helices 3, 9, 10, and 12 was observed and interpreted as digitonin (Fig. 1 A and B). The planar shape of this density was more elongated than that of cholesterol, and it extended toward a disordered density in the extracellular region. A cholesterol molecule in this position might play a physiological role, as also proposed for hLAT1 (33).
hLAT2/CD98hc Apo Structure.
We modeled the complete structure of the hLAT2/CD98hc heterodimer in the absence of substrate (Fig. 1B), except for N-terminal residues 1 to 60 for CD98hc and 1 to 40 for hLAT2 for which we could not identify any density (hLAT2 and CD98hc isoform f numbering used in this study). With the exception of these areas, the sequences of hLAT2 and CD98hc were fully resolved. Like other members of the LAT subfamily, hLAT2 adopts the APC superfamily fold (19–25). TM1–TM5 and TM6–TM10 are related by a pseudo twofold symmetry axis within the plane of the membrane. TM1 and TM6 are unwound in the center, forming two discontinuous helices named 1a, 1b, and 6a, 6b (see details in Unwound Regions in TM1 and TM6 Form the hLAT2 Substrate-Binding Site). hLAT2 and CD98hc are linked by a disulfide bridge between Cys-154 (hLAT2) and Cys-109 (CD98hc) and by a variable number of interactions between TM1’ and the TM portion of hLAT2 (Fig. 1B and SI Appendix, Fig. S6). TM1' crosses the lipid bilayer next to the hLAT2 hash domain (i.e., TMs 3, 4, 8, and 9) forming helix–helix contacts with residues in TM4 and the extracellular end of TM3 in hLAT2 (Fig. 1B and SI Appendix, Fig. S6). These contacts are mediated by hydrophobic residues Phe-88’, Trp-89’ and 92’, Leu-96’, Ala-99’, and Ile-102’ and 103’ in CD98hc TM1’, and Leu-147, Leu-150, and Phe-151 in TM3, and Leu-163, 164, 171, Ile-167, and Trp-174 in TM4 in LAT2 (SI Appendix, Fig. S6). Interestingly, Trp-174, which is conserved only in hLAT2 and hAsc1, faces Phe-88’ and Trp-89’ in TM1’, forming an aromatic triad in the TM1’–TM4 interface (SI Appendix, Fig. S6). Nevertheless, mutation of Trp-174 to alanine (the corresponding residue in hLAT1) had no effect on protein expression, plasma membrane localization, or [3H] L-alanine uptake.
Unwound Regions in TM1 and TM6 Form the hLAT2 Substrate-Binding Site.
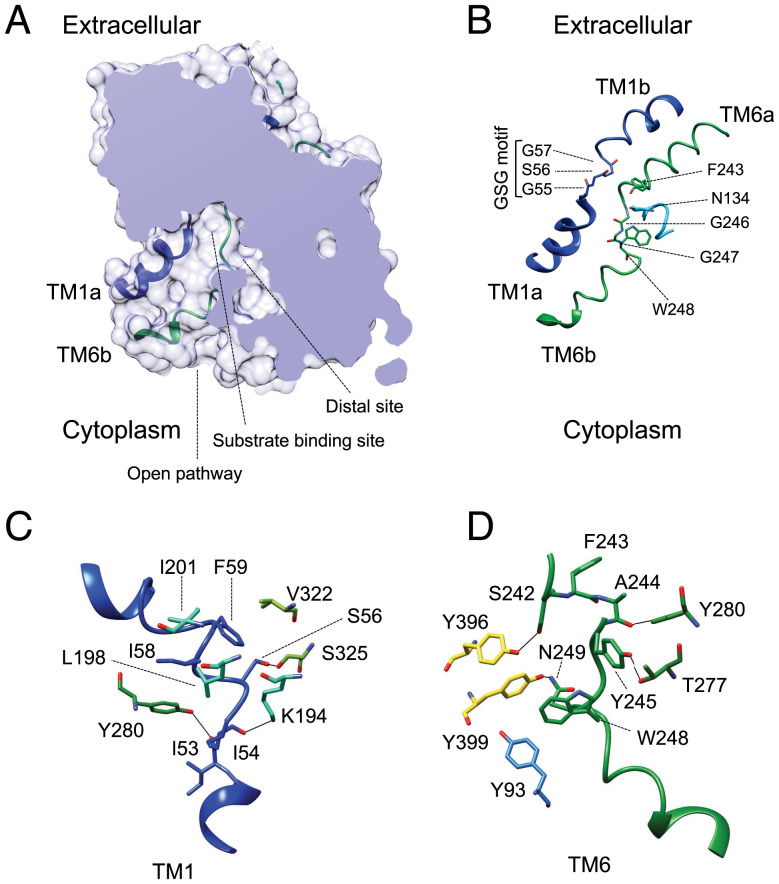
In the absence of substrates, the hLAT2/CD98hc structure is in an inward (cytoplasmic)-facing open conformation with TM1a and TM6b tilted to open a vestibule connecting the cytoplasm to the center of the transporter (Fig. 2A). At the end of this passage, a particular spatial conformation of unwound regions connecting TM1a with TM1b and TM6b with TM6a forms an empty space that, together with residues in TM1a, TM6b, TM3, and TM8, define the substrate-binding site, described before for several amino acid transporters with APC fold (21–23, 25, 34, 35). In particular, in all the transporters in the LAT subfamily, TM1a contains a 55G(S/T)G57 motif with the amide nitrogen atoms of Gly-55, Ser-56, and Gly-57 oriented toward the empty space, providing the possibility of hydrogen bonding with the carboxyl group of substrates. In addition, the unwound region connecting TM6a and TM6b of hLAT2 contains Gly-246 with its carbonyl group facing the empty space formed by TM1 and TM6, and Trp-248 facing residues in the adjacent TM2 and TM10 (Fig. 2B). In addition, Asn-134 from TM3 locates in the vicinity, and this residue was found to be important for the mechanism of substrate selectivity (see Determinants of Substrate Specificity in the LAT Subfamily of Transporters). This substrate-binding cavity is connected to another small vestibule in hLAT2 (Fig. 2A). This open space was first discovered in hLAT1 and interpreted as a distal substrate-binding site (22). In this inward-facing conformation, access of the central cavity to the extracellular space is blocked by several hydrophobic and polar residues, which form a barrier for substrates (Fig. 2A).
Fig. 2.
Unwound regions in TM1 and TM6 form the substrate-binding site opened to the cytosol. (A) Structure of hLAT2, represented as a surface, with a section to show the central cavity harboring the substrate-binding site opened to the cytoplasm but without access to the extracellular space. The TM1a and TM6b helical regions that open the vestibule are shown superimposed. A distal cavity connects to the central vestibule. (B) TM1 and TM6 forming the substrate-binding site are shown as a cartoon with key residues highlighted. The contribution of N134 from the adjacent helix TM3 to the substrate binding is shown. (C) The conformation of TM1 is maintained by interactions with residues in the vicinity. (D) Interactions between TM6 and neighboring regions of the structure. Color codes for hLAT2 helices and residues in all the panels are as used in Fig. 1. Oxygen atoms are shown in red and nitrogen atoms in blue.
The conformation of the unwound regions in TM1 and TM6 is critical to define an open space that can accommodate substrates and to configure an orientation of residues facing this space and ready to interact with substrates. In hLAT2/CD98hc, these requirements are achieved by a set of interactions between residues in TM1 and TM6 with neighboring regions of the structure (Fig. 2 C and D). A hydrophobic cluster formed by Ile-58 and Phe-59 in TM1, Leu-198 and Ile-201 in TM5, and Val-322 in TM8 stabilizes the unwound region of TM1 (Fig. 2C). In addition, hydrogen bonds between the hydroxyl oxygen atoms of Ser-56 in TM1 and Ser-325 in TM8, the carbonyl oxygen atom of Ile-53 in TM1 and the hydroxyl oxygen atom of Tyr-280 in TM7, and the epsilon nitrogen atom of Lys-194 in TM5 and the carbonyl oxygen atom of Ile-54 in TM1 are also established (Fig. 2C).
Similarly, the unwound region of TM6 establishes various interactions with residues from TMs 2, 7, and 10 (Fig. 2D). Indeed, the oxygen atoms of the side chains of residues Ser-242, Tyr-245, and Asn-249 in TM6 form a hydrogen bond network with the oxygen atoms of hydroxyl groups of Tyr-396 in TM10, Thr-277 in TM7, and Tyr-399 in TM10, respectively (Fig. 2D). Moreover, Trp-248 in TM6 establishes aromatic interactions with Tyr-399 in TM10 and Tyr-93 in TM2, as well as hydrophobic contacts with residues in TM2. Finally, as is also the case of TM1, the hydroxyl oxygen atom of Tyr-280 in TM7 interacts with the carbonyl oxygen atom of Ala-244 in TM6 (Fig. 2D).
Structural Basis for Substrate Specificity in hLAT2.
A unique feature of hLAT2 within the LAT subfamily is its substrate specificity toward both large and small neutral amino acids, including L-glutamine (26–28). Very recently, cryo-EM structures of hLAT2/CD98hc bound to L-leucine and L-tryptophan were solved (23); however, they did not shed light on the molecular mechanisms underlying small neutral amino acid and L-glutamine selectivity in hLAT2.
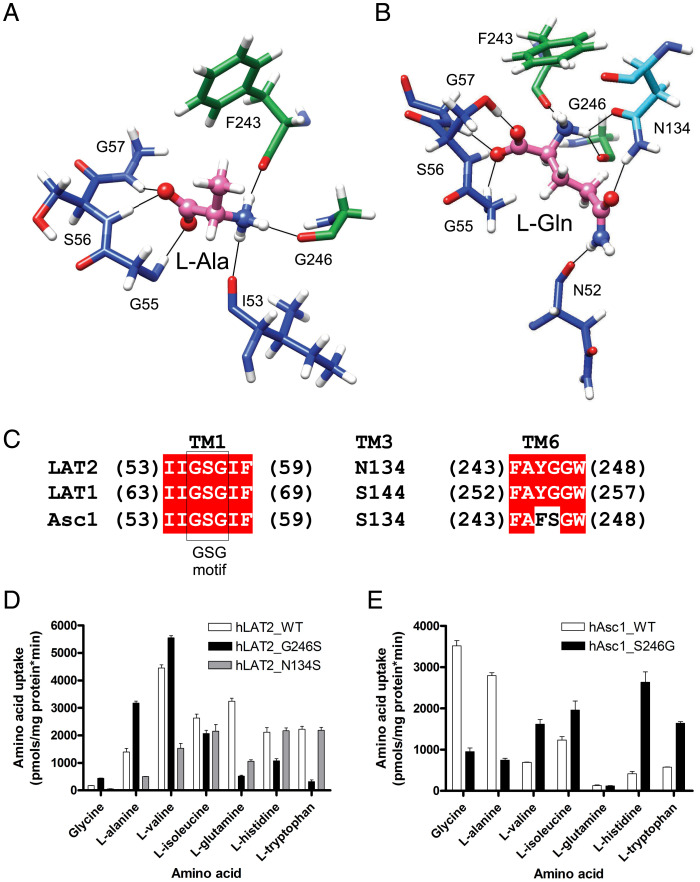
To characterize the structural bases for substrate specificity in hLAT2, we performed molecular docking and PELE studies using a set of hLAT2 amino acid substrate ligands (Gly, L-Ala, L-Trp, and L-Gln) in the apo hLAT2/CD98hc structure. Substrate docking experiments and PELE analysis predicted a minimal energy-binding mode for all the substrates tested (SI Appendix, Fig. S7). This binding mode corresponded to the wild-type (also referred to as canonical) pose within backbone atoms of unwound regions of TM1 and TM6, as reported for other SLC7 transporters [BasC (25), GkApcT (34), human b0+AT (21), LAT1 (22), and LAT2 (23)]. Indeed, the calculated pose of L-tryptophan in hLAT2 is very similar to the cryo-EM structure of hLAT2/CD98hc bound to this substrate, which is compatible with two positions of L-tryptophan (23) (SI Appendix, Fig. S4 B–D). In the calculated poses, one of the α-carboxyl oxygen atoms of glycine, L-alanine, L-glutamine, and L-tryptophan establishes H-bonds with the nitrogen atom of Gly-55, Ser-56, and Gly-57 residues in the GSG motif in the unwound segment of TM1 (Fig. 3 A and B and SI Appendix, Fig. S8). In contrast, the contacts established by the α-amino group of these different substrates with the unwound region of TM6 were substantially more diverse among the tested substrates. Nevertheless, for all the substrates, the hydrogen bond between the carbonyl oxygen atom of Gly-246 (Fig. 2B) and the α-amino nitrogen atom of the substrate was conserved (Fig. 3 A and B and SI Appendix, Fig. S8).
Fig. 3.
Structural determinants of the substrate-binding site in the LAT subfamily of transporters. (A and B) Selected poses in the wild-type binding site from the PELE analysis for L-alanine and L-glutamine substrates, respectively. Carbon atoms of the indicated residues follow the color codes for hLAT2 helices and residues as used in Fig. 1. C atoms of the substrates are shown in pink, whereas O and N atoms of substrates and residues are shown in red and blue, respectively. Black lines connect atoms located at H-bond distance. (C) hLAT2 sequence alignment with human LAT1 and Asc1. Unwound regions of TM1 and TM6, as well as Asn-134, are shown. (D) Transport of 10-µM radiolabeled (3H) glycine, L-alanine, L-valine, L-isoleucine, L-glutamine, L-histidine, and L-tryptophan in HeLa cells by human wild-type LAT2 (white bars) and hLAT2 mutants G246S (black bars) and N134S (gray bars). Data are expressed as mean ± SEM from at least three independent experiments run in quadruplicate. (E) Transport of 10-µM radiolabeled [3H] glycine, L-alanine, L-valine, L-isoleucine, L-glutamine, L-histidine, and L-tryptophan in HeLa cells by human wild-type Asc1 (white bars) and hAsc1 mutant S246G (black bars). Data are expressed as in D.
In addition, other residues contributed to the establishment of additional interactions specific to each substrate. The side chain of L-alanine showed hydrophobic contacts with the aromatic ring of Phe-243 (Fig. 3A), whereas the side chain of L-glutamine interacted via a H-bond with the amide nitrogen atom of the side chain of residue Asn-134 in TM3 and the carbonyl oxygen atom of Asn-52 in TM1 (Fig. 3B). Finally, the side chain of L-tryptophan presented hydrophobic stacking with Gly-246 and Gly-247 in TM6 and H-bond with the amide oxygen atom of the side chain of Asn-134 in TM3 (SI Appendix, Fig. S8B). The hydrophobic staking of L-tryptophan with Gly-246 and Gly-247 was also observed in the cryo-EM structure of hLAT2/CD98hc in complex with L-tryptophan (23) (SI Appendix, Fig. S4 B and D).
A second cavity connected to the substrate-binding pocket has been suggested to form a distal binding site capable of accommodating large side chains (22, 23) (Fig. 2A). Our PELE analysis did not predict any substrate occupying this distal site. Moreover, the recognition of the substrate α-amino and carboxyl groups by the GSG motif and Gly-246 seemed incompatible with the occupancy of the distant “distal site” in hLAT2 in the inward-facing conformation, even for amino acids with large side chains such as L-tryptophan (SI Appendix, Fig. S8B).
Determinants of Substrate Specificity in the LAT Subfamily of Transporters.
Despite similar structures, the members of the LAT subfamily of transporters show important differences in substrate specificity, the structural basis of which is still unknown. Although recognition of amino acid substrates by the cytoplasmic face of the transporter is defined mainly by interactions with backbone atoms of hLAT2, sequence alignment revealed some significant differences at the core of the binding site of hLAT1, hLAT2, and hAsc1 (Fig. 3C). We characterized these differences to identify the molecular bases of the different substrate selectivity reported for neutral amino acid LAT transporters (26–28). Two of the substrate-interacting residues predicted by the PELE analysis (Gly-246 and Asn-134) are not fully conserved in hLAT2, hLAT1, and hAsc1 (Fig. 3C). Gly-246 in hLAT2, which is conserved in hLAT1, became a serine residue in hAsc1. We tested the relevance of this residue in determining hAsc1 as a poor transporter for large neutral amino acids, in contrast to hLAT2 and hLAT1 (26–28). Interestingly, mutation G246S in hLAT2 caused a dramatic decrease in the uptake of large substrates ([3H] L-tryptophan, [3H] L-glutamine, and [3H] L-histidine) but increased the uptake of small neutral amino acids ([3H] glycine and [3H] L-alanine) (Fig. 3D and SI Appendix, Table S2). Conversely, the reverse mutation in hAsc1 (S246G) resulted in a large decreased uptake of the smallest substrates ([3H] glycine and [3H] L-alanine) and in an increased uptake of the large substrates, with the exception of [3H] L-glutamine (Fig. 3E and SI Appendix, Table S2).
PELE analysis predicted that the hydroxyl group of Ser-246 in the hLAT2 G246S mutant provided an additional H-bond with the α-amino nitrogen atom of the substrates (SI Appendix, Fig. S9), resulting in an improved energy of substrate binding in the canonical mode for all the substrates studied (SI Appendix, Fig. S7). Interestingly, mutation G246S in hLAT2 induced an alternative-binding mode facilitated by the H-bond of the hydroxyl oxygen atom of Ser-246 with the carbonyl oxygen atom of Phe-243 due to the rotation of the Cα of Ser-246. This additional binding mode might facilitate escape from (or hinder binding to) the wild-type pose, slightly shifting the substrate side chain out of the main wild-type site/location (SI Appendix, Fig. S9). Notice that the intraprotein hydrogen bond between the mutated Ser-246 and Phe-243 hampers the formation of wild-type substrate interactions. While the interaction between the hydroxyl group of Ser-246 in the hLAT2 G246S mutant with the α-amino nitrogen atom of the substrates was found for all the substrates, the shifted pose was observed only in those amino acids with larger side chains that allowed them to establish interactions with additional residues such as Asn-134 (SI Appendix, Fig. S9). This differential behavior between substrates with small and large side chains might explain the increased uptake of small substrates and the reduced transport activity of large substrates in the G246S LAT2 mutant and also in part in hAsc1.
Kinetic analysis of the [3H] amino acid uptake by the hLAT2 mutant G246S revealed different mechanisms underlying transporter substrate selectivity (SI Appendix, Fig. S10 and Table S3). Indeed, L-alanine uptake showed a decreased extracellular Km, suggesting that the improved binding energy calculated for this substrate in the G246S mutant at the cytosolic side may apply for its binding at the extracellular side. For L-glutamine and L-tryptophan, the external kinetics showed decreased Vmax (∼50%) (SI Appendix, Fig. S10 and Table S3). PELE analysis indicated alternative energetically favorable poses for both substrates at the internal side of the transporter. If this operates at the external side, these alternative poses may be nonproductive, reducing Vmax. Additionally, hLAT2 G246S mutant increased L-glutamine extracellular Km, suggesting that G246S impairs one or several transport-limiting steps affecting Km. Whether alternative poses at the substrate-binding site of L-glutamine predicted by PELE affect these transport-limiting step(s) remains to be established.
In contrast to hLAT2, hLAT1 does not mediate the uptake of small neutral amino acids, and it is a low affinity L-glutamine transporter (26, 27). Moreover, glutamine is not a good intracellular exchange substrate for LAT1 (36). Interestingly, residue Asn-134 in hLAT2 is a serine residue in hLAT1 (Fig. 3C). When we mutated this residue in hLAT2, the substrate specificity of the N134S mutant changed dramatically, showing reduced [3H] L-glutamine, [3H] glycine, and [3H] L-alanine uptake (Fig. 3D and SI Appendix, Table S2), thus mimicking hLAT1. This observation thus points to a key role for Asn-134 in the particular substrate selectivity of hLAT2 compared to that of hLAT1. PELE transporter-substrate structural analysis showed that Asn-134 was a potential interacting residue in the wild-type binding mode for L-glutamine and L-tryptophan but not for small substrates (Fig. 3 A and B and SI Appendix, Fig. S8). In addition, the energy profile for the substrate binding in wild-type hLAT2 compared to that of the N134S mutant showed a different substrate-binding energy landscape (SI Appendix, Fig. S11). The N134S mutant favors the exploration of alternative-binding sites by L-alanine, even though with a worse binding energy than the wild-type binding. External kinetics of the [3H] L-alanine transport showed a clear increase in the extracellular Km without impact on Vmax (SI Appendix, Fig. S10 and Table S3). Whether this increase of the external Km is a consequence of altered binding or to defective non–transport-limiting step(s) in the transport cycle that increases Km remains to be established.
This mutation also reduces the range of binding energy of L-glutamine poses within the wild-type site (SI Appendix, Fig. S11), which should result in reduced entropy and binding affinity. In this regard, [3H] L-glutamine uptake kinetic analysis in the N134S mutant showed an increased extracellular Km. However, Vmax was also reduced by ∼50%, suggesting that Asn-134 governs both L-glutamine translocation and external Km (SI Appendix, Fig. S10 and Table S3). In contrast, the mutation does not cause significant alterations in the landscape of energy binding for L-tryptophan, which would explain why the transport of this substrate is not altered in the N134S mutant (SI Appendix, Fig. S10 and Table S3). Therefore, a change in the residue that occupies the position of Asn-134 in LAT2 explains some of the differences in substrate selectivity in hLAT1 and hLAT2. Our PELE analysis did not predict changes in the energy landscape that might explain the reduced transport of glycine caused by N134S (SI Appendix, Fig. S11). The role of Asn-134 in the transport of glycine could be due to their participation during conformational transitions of the transporter whose structures are not yet available.
ARHL Mutations Map a Region that Regulates Substrate Specificity.
ARHL or presbycusis is one of the most prevalent chronic medical conditions associated with aging. Although ARHL is multifactorial, loss-of-function mutations in hLAT2 transporter protein gene (SLC7A8) have been associated with this condition (9). Two of the four hLAT2 mutations described (V460E and T402M) show a nearly complete reduction of both L-alanine and L-tyrosine transport activity (9). Val-460 is located at the cytoplasmic face of TM12 with its side chain facing toward the aliphatic chain of membrane lipids. In hLAT2/CD98hc, TM12 comprises ∼8 turns of amino acid residues composed mostly of hydrophobic side chains that shield the polypeptide backbone whose H-bond donor and acceptor groups face against the interior of the lipid membrane. Therefore, charged residues within the TM helix would either reduce the stability of the helix in the membrane or inhibit its insertion into the lipid bilayer. Supporting this hypothesis, the hLAT2 V460E mutant showed reduced protein expression and impaired plasma membrane localization, the protein being retained in the endoplasmic reticulum (ER) (9). Also, a prediction of the apparent free energy difference (ΔG) for insertion of the wild-type and V460E hLAT2 protein sequences (−0.29 and 1.36 Kcal/mol, respectively) into the ER membrane suggested that the Sec61 translocon-mediated ER membrane insertion was impaired in the ARHL mutant (37).
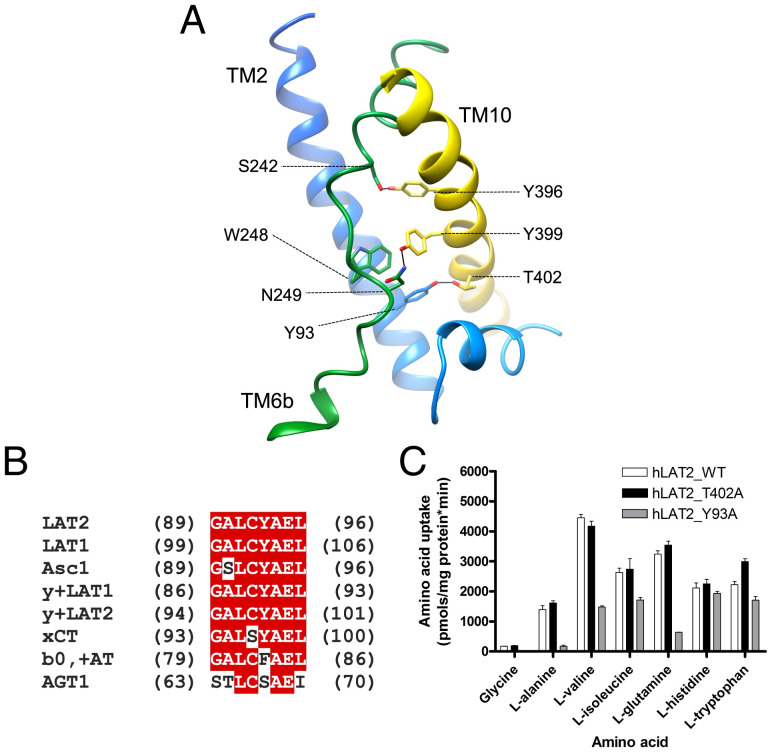
The underlying molecular bases for the transport defect of mutation T402M is unknown. Thr-402 places at a region where the cytoplasmic ends of TM10 and TM2 establish multiple contacts (Fig. 4A). In addition, the unwound region of TM6 contains several residues that interact with TM10, such as Ser-242 with Tyr-396 and Asn-249 with Try 399. Also, Trp-248 presents π–π bonds with Tyr-93 (TM2) and Tyr-399 (TM10). Tyr-93 is part of a GALCYAEL motif [89G (A/s) L (C/s) (Y/f) A E (L/i)96; large letter indicates the most frequent residue; hLAT2 numbering] located in TM2 and which is conserved in human LATs (Fig. 4B). Together, TM2, TM10, and TM6 form a well-interconnected region, and we hypothesized that this contributes to maintaining the correct conformation of TM6 and that changes in this connectivity affect the conformation of TM6 and substrate-binding site. To test this hypothesis, we studied the potential influence of interactions between Tyr-93 and Thr-402, residues that are located at a distance compatible with hydrogen bonding connecting TM2 and TM10. To this end, we generated mutations to Ala that would disrupt this putative bond, and we then analyzed the effects on transport. The T402M ARHL mutant shows reduced uptake of alanine (9), whereas the T402A mutant (the equivalent residue in hLAT1) had a minimal effect on the substrate activity and selectivity profile (Fig. 4C and SI Appendix, Table S2), suggesting that Thr-402–Tyr-93 hydrogen bond was not determinant for hLAT2 transport function. In contrast, the Y93A mutant resulted in a nearly complete reduction of [3H] glycine, [3H] L-alanine, and [3H] L-glutamine uptake, with a minimal effect on the transport of [3H] L-isoleucine, [3H] L-histidine, and [3H] L-tryptophan (Fig. 4C and SI Appendix, Table S2). This result mapped the influence of a region far from the substrate-binding site on substrate specificity.
Fig. 4.
Tyrosine 93 in TM2 regulates substrate specificity. (A) Contacts between the TM helices TM2, TM6, and TM10 hold the conformation of TM6. Color codes for hLAT2 helices and residues are as used in Fig. 1. Oxygen atoms are shown in red and nitrogen atoms in blue. (B) hLAT2 sequence alignment of the motif GALCYAEL with that of human LATs. (C) Transport of 10-µM radiolabeled (3H) glycine, L-alanine, L-valine, L-isoleucine, L-glutamine, L-histidine, and L-tryptophan in HeLa cells by human wild-type LAT2 (white bars) and hLAT2 mutants T402A (black bars) and Y93A (gray bars). Data are expressed as mean ± SEM from at least three independent experiments run in quadruplicate.
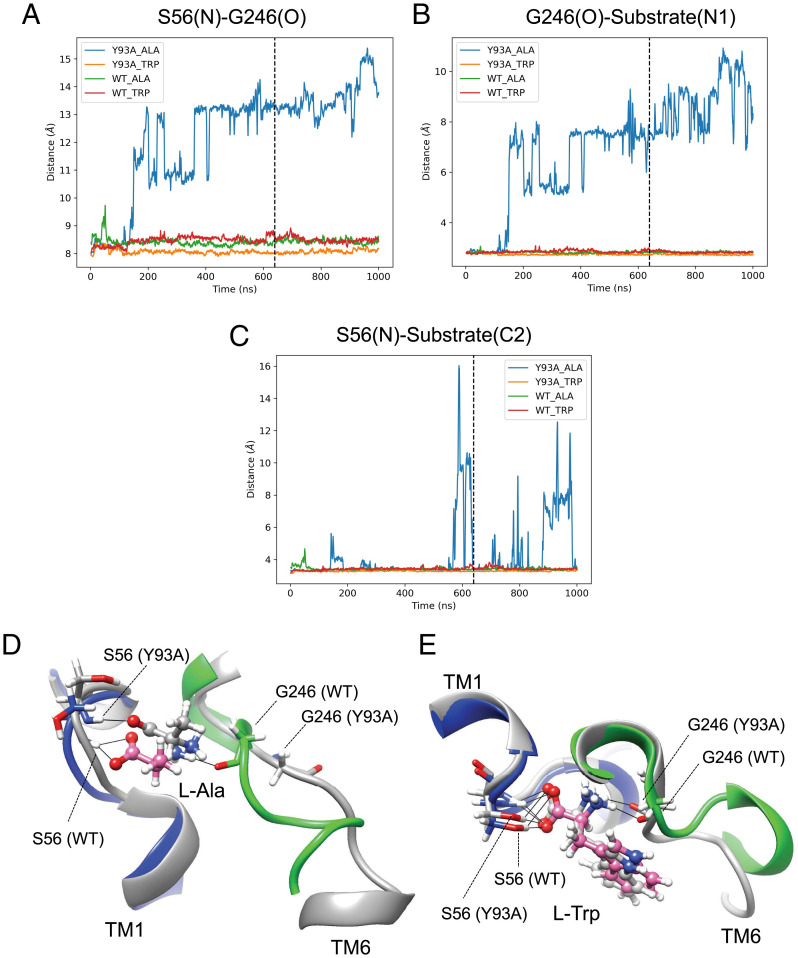
We then performed MD analysis of wild-type hLAT2 and the Y93A mutant to explore the molecular basis of the altered substrate selectivity. In the presence of bound L-alanine, MD showed that the π-stacking interactions Tyr-93 (TM2)–Trp-248 (TM6) and Tyr-93–Tyr-399 (TM10) are stable, whereas in the Y93A mutant Trp-248 occupies the space of the absent side chain of Tyr-93, thereby facilitating the displacement of Gly-246 (SI Appendix, Fig. S12). Displacement of Gly-246 in the unwound segment of TM6 widens the distance between the backbone atoms that bind the α-amino carboxyl moiety of L-alanine by ∼4 to 5 Å (Fig. 5A). As a result, first, the amino group of L-alanine detaches from Gly-246 in TM6 (Fig. 5B), and second, the connection between the carboxylate group of L-alanine with TM1 is lost (Fig. 5 C and D), thus destabilizing the wild-type binding pose of L-alanine, and consequently, reducing the transport activity for this amino acid (Fig. 4C and SI Appendix, Table S2). Interestingly, the Y93A mutation did not affect the external Km but decreased dramatically the Vmax of [3H] L-alanine and [3H] L-glutamine uptake (SI Appendix, Fig. S10 and Table S3). This suggests that the proper interaction of L-alanine, and eventually L-glutamine, with both unwound segments of TM1 and TM6 is a requirement to trigger transport. Whether the Y93A mutation compromises the transit from outward-facing to the occluded state or from this state to the inward-facing state remains to be established.
Fig. 5.
MD analysis of wild-type hLAT2/CD98hc and mutant Y93A bound to L-alanine or L-tryptophan. Evolution of the N atom of Ser-56–O atom of Gly-246 (A), N1 atom of the substrate–O atom of Gly-246 (B), and N atom of Ser-56–C2 atom of the substrate (C) distances for hLAT2/CD98hc wild type (WT) and Y93A mutant. The lines in A–C indicates the time (640 ns) corresponding to the snapshots shown in D and E. Snapshots of L-alanine (D) and L-tryptophan (E) bound to wild type and Y93A mutant. Cartoons and C atoms are shown in blue (TM1) and green (TM6) in wild-type hLAT2 and in gray in Y93A mutant. Substrate C atoms are shown in pink and gray in wild-type hLAT2 and Y93A mutant, respectively. O and N atoms in substrates and residues are shown in red and blue, respectively. Black lines connect atoms located at H-bond distance.
In sharp contrast, L-tryptophan, because of its larger side chain, establishes hydrophobic stacking with Gly-246 and Gly-247 at the unwound region of TM6, acquiring a wild-type pose and preventing the migration of Gly-246 (Fig. 5E and SI Appendix, Fig. S12). As expected, the Y93A mutation did not affect Km and had a minimal effect on Vmax of [3H] L-tryptophan influx (SI Appendix, Fig. S10 and Table S3). This observation would thus explain the activity of the Y93A mutant for large amino acids, particularly L-tryptophan (Fig. 4C and SI Appendix, Table S2).
Discussion
The recent structures of several members of the HAT family of transporters reveal that they have a very similar architecture. Intriguingly, each HAT has evolved to specialize in the transport of a subset of substrates, and this specialization drives the significant differences in the physiological functions of each member of the family. Here, we provide a significant advance in our understanding of the structural and molecular basis defining substrate specificity in the HAT family. Based on cryo-EM structure of hLAT2/CD98hc in inward-facing conformation, PELE analysis of amino acid substrate binding, and transport studies, we have identified residues within the substrate-binding site (Gly-246 and Asn-134) and in a scaffold of the unwound segment of TM6 (Tyr-93 in TM2) that are key for substrate specificity between the human transporters LAT2, LAT1, and Asc1.
The core of the substrate-binding site in all HATs is formed by unwound segments in TM1 and TM6, whose residues expose their amide backbone atoms, which serve as engaging platform for the α-amino carboxyl moiety of each substrate. Interestingly, TM6 is less conserved among human LATs than TM1 (25) (Fig. 3C). In particular, in hLAT2, Gly-246, which is a key residue interacting with the substrate α-amino group, is a serine residue in human Asc1 (Fig. 3C). LAT2 has a unique substrate selectivity profile, and it mediates the exchange of all neutral amino acids, including L-glutamine (26). Interestingly, our results showed that a serine residue at position 246 (as in hAsc1 or G246S hLAT2 mutant) resulted in a transporter with a substrate preference for small neutral amino acids. In contrast, a glycine residue in this position (as in hLAT2 or S246G hAsc1 mutant) shifted the transporter preference to large neutral amino acids. Small substrates such as glycine and L-alanine are not naturally well transported by hLAT1, but when the equivalent position to Gly-246 in hLAT2 is mutated in hLAT1 (G255A) (Fig. 3C), hLAT1 shows improved capacity to transport L-alanine and L-valine while reducing the transport of larger amino acids (19).
Therefore, Gly-246 in TM6 of hLAT2 and the equivalent position in other HATs of neutral amino acids are part of the mechanism that contributes to discriminating small from large substrates, and our PELE and transport kinetics experiments shed light on the molecular basis underlying the role of this glycine/serine residue. According to this model, the hydroxyl group of a serine residue in this position would stabilize the binding of the α-amino group of small amino acids, thus increasing their transport. This view is favored by the decreased external Km for L-alanine uptake in the hLAT2 G246S mutant. In contrast, this hydroxyl group would facilitate a shift of large substrates from the wild-type pose, favored by the interaction of their large side chains with other residues in the substrate vestibule and then decreasing their transport. Interestingly, L-glutamine and L-tryptophan uptake kinetics in hLAT2 G246S mutant point toward deficient catalytic steps of the transport cycle that reduce substrate translocation (L-glutamine and L-tryptophan) and increase external Km (L-glutamine). However, the transport cycle steps of L-glutamine and L-tryptophan uptake affected by G246S mutation are at present unknown. Interestingly, amino acid uptake experiments confirm, at least partially, the predictions based on the inward-open structure, which would suggest that interactions of the substrate with its binding site are very similar in the outward- and inward-facing conformations. However, another explanation might be that substrate–transporter interactions in the inward-open state are a more important determinant of transport rates than those in the outward-open state.
Structural analysis revealed an asparagine residue in hLAT2 TM3 (Asn-134) that pointed toward Gly-246 and that is not conserved in hLAT1 (S144). In fact, the N134S mutation greatly reduced the uptake of small neutral amino acids and L-glutamine in hLAT2 without affecting the transport of larger amino acids (Fig. 3D), suggesting that Asn-134 would be the main residue responsible for the differences in selectivity of small neutral amino acids and L-glutamine between hLAT2 and hLAT1 (26, 27). According to the PELE analysis, the N134S mutant would facilitate the binding of L-alanine out of the canonical substrate-binding mode and would constrain the poses of L-glutamine within the substrate-binding site, thus reducing transport. However, [3H] L-alanine and [3H] L-glutamine kinetic analysis as well as defective transport of glycine in hLAT2 N134S mutant cannot be fully explained in the scenario of the inward-facing conformation of hLAT2. In this regard, the bacterial alanine-glycine APC transporter AgcS contains a glutamine residue in TM3—the same TM region where Asn-134 is located—that presents H-bonding with the α-amino nitrogen atom of the substrate in the occluded conformation (38). This observation suggests that Asn-134 in hLAT2 might be important for the binding of small substrates in the occluded conformation during the transport cycle.
We found that the conformation of the substrate-binding site in hLAT2, and consequently substrate selectivity, is controlled by a region outside the binding site, which has a significant impact since several mutations in this region alter substrate preference and associate with some human diseases (10). The unwound segments of TM1 and TM6 are stabilized with multiple connections (Fig. 2 C and D). In particular, the unwound region of TM6 (Ser-242, Trp-248, and Asn-249) interacts with residues in TM2 (Tyr-93) and TM10 (Tyr-396 and Try 399). In contrast to TM1, these stabilizing interactions are not conserved among LATs and are possibly responsible for the differences observed in the conformation of the unwound region of TM6 in the structure of several LATs (19–25). Mutations Y396A and Y399A, predicted to destabilize the conformation of the unwound region in TM6 (Fig. 4A), almost completely abolished L-tryptophan and L-leucine uptake, thereby highlighting the relevance of these residues for the function of the transporter (23).
Interestingly, the Y93A mutation changed the substrate profile of hLAT2, eliminating the transport of small substrates (glycine, L-alanine, and, to a lesser extent, L-valine) and L-glutamine but sustaining that of large substrates (L-isoleucine, L-histidine, and L-tryptophan) (Fig. 4C). Indeed, the substrate selectivity profile for the Y93A mutant greatly resembles that of hLAT1 (27). MD analysis of wild-type hLAT2 and Y93A mutant in the presence of substrates (L-alanine and L-tryptophan) suggested a molecular mechanism that explains the changes in substrate selectivity in the Y93A mutant. In this regard, we propose that TM2 acts as a scaffold for the unwound region in TM6, maintaining the conformation of the binding site. Thus, significant modifications of residues in TM2 may alter the transport activity and/or selectivity profile of hLAT2. Similarly, in the bacterial APC amino acid transporter MhsT, interaction of the 235GMG237 motif (equivalent to 247GWN249 in hLAT2) in the unwound region of TM6 with the highly conserved glutamate residue 66 in TM2 (in an equivalent position to Tyr-93 in hLAT2) has recently been reported (39). Interestingly, the interaction between Glu-66 and the GMG motif changes depending on the size of the substrate bound, thereby regulating binding pocket volume and modulating transporter substrate selectivity (39).
Residue Tyr-93 in hLAT2 lies within a motif of TM2 that is conserved in human LATs 89G (A/S) L (C/S) (Y/F) A E (L/I)96 (Fig. 4B) and that is involved in connections of TM2 with TM6, TM7, and the TM10-11 loop. The fully conserved Ala-94 presents hydrophobic interactions with Leu-265 in TM7, and Glu-95 presents salt bridges with Arg-418, with the fully conserved Arg-410 and with Lys-421 within the TM10-11 loop. Interestingly, the A94T mutation associated with cataracts results in defective L-tryptophan transport, with no impact on the transport of L-alanine or L-tyrosine (10), while the R418C variant associated with ARHL reduces the transport activity of L-alanine and L-tyrosine by ∼50% (9). These results reinforce the idea that modifying interactions of the TM2 motif with other regions of the protein would affect the substrate selectivity profile of human LAT2 by reshaping the substrate-binding site.
Taken together, our findings provide structural basis that regulates substrate preference in HATs for neutral amino acids. Our results highlight the concept that substrate specificity in these transporters requires a variety of kinetic mechanisms to reach substrate specificity in a scenario of broad substrate range. Thus, a unique combination of residues in the binding site differentially regulates substrate interaction and/or translocation. Besides, particular differences in the conformation of the unwound TM6 region due to interaction with surrounding TMs are also key determinants of the substrate selectivity profile. Accordingly, we found that TM2 acts as a scaffold for TM6, regulating its conformation and its capacity to transport certain substrates. This scaffold region accumulates mutations associated with some human diseases, and our model suggests that these would cause disease by distorting the conformation of TM6, thus altering substrate selectivity.
Materials and Methods
Cell Lines.
HeLa cells were maintained at 37 °C in a humidified 5% CO2 environment in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 50 units/mL penicillin, 50 μg/mL streptomycin, and 2 mM L-glutamine. HEK293-6E cells (40) were cultured following the provider’s standard protocols in chemically defined F17 Freestyle medium (Invitrogen, Life Technologies) supplemented with 1 g/L pluronic F68 (Applichem), 4 mM L-glutamine, and 12.5 mg/L G418. HEK293-6E cells were grown in polycarbonate shake flasks with vented lids and with a capacity of 125 mL to 2 L (Triforest Plasticware) in a Brunswick S41il CO2 shaker with 25-mm orbital (Eppendorf, Merck KGaA) at 37 °C, 5% CO2, and 120 rpm (80 rpm for 3-L flasks) without exceeding 2 × 106 cells/mL during maintenance and 25% flask capacity.
Cryo-EM of hLAT2/CD98hc.
The specimen was vitrified using 3 μL freshly purified hLAT2/hCD98hc applied to glow-discharged Quantifoil R 0.6/1 Cu 300 mesh grids (Electron Microscopy Sciences). Grids were blotted for 2 s under 95% humidity and plunge frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM datasets were collected on a Titan Krios G2 electron microscope operating at 300 kV and equipped with a Gatan Imaging Filter and a K3 summit direct electron detector (Gatan) in counting mode at the Electron Bio-Imaging Centre (eBIC) and at the facilities of the University of Leicester. Parameters and statistics of cryo-EM data acquisition are shown in SI Appendix, Table S1.
Image Processing.
Movie-frames from K3 were aligned using MotionCor2 (41) with 35 patches per image, applying dose weighting. Contrast transfer function (CTF) parameters were determined using Gctf (42). Particles were selected using Topaz, after training the neural network using a manually selected subset (43). Subsequent image processing was performed using RELION 3.1 (31), cryoSPARC (44), and computational imaging system for transmission electron microscopy (cisTEM) (45). The initial data set was subjected to several rounds of reference-free 2D classification to obtain 319,613 particles that generated 2D averages with excellent signal to noise ratio for the TM helices and showing different views of hLAT2/hCD98hc (SI Appendix, Fig. S2). Selected particles were used to generate an ab initio model in cryoSPARC that was used for a first consensus refinement in RELION 3.1 (SI Appendix, Fig. S3). The particles were then classified into six subgroups using 3D classification in RELION 3.1. The quality of each subgroup was evaluated by analysis of the structural details in sections along the z-axis. The best group of particles was refined to a medium resolution density depicting all the TM helices in the hLAT2/hCD98hc complex. Subsequently, this 3D classification step was repeated six times, selecting the best class in each run and combining all the particles selected into one group (22). After removing all the duplicated particles, the final dataset comprised 176,132 particles, which were polished using RELION 3.1 and two rounds of CTF refinement in cisTEM. In this latter step, a mask was used, excluding the micelle and applying a low-pass filter outside the mask. Postprocessing and B-factor sharpening were performed using the protocols in RELION 3.1. Volumes were also postprocessed using SPoC (46), which helped to visualize the connectivity of the densities and the interpretation of the resulting volume, and it was used for representations in Fig. 1. Average resolutions of the structure were estimated as 3.9 Å using FSC using the gold-standard criterion and a cutoff of 0.143 in RELION (31) and 3.7 Å using ResMap (32) (SI Appendix, Fig. S2). Local resolution estimates using ResMap showed that most of LAT2 was resolved between 2.5 and 3.0 Å (SI Appendix, Fig. S2).
Model Building.
Modeling was started by preparing a model of hLAT2 based on the atomic structures of hLAT1 using Iterative-Threading ASSEmbly Refinement (I-TASSER) for automatic template matching and homology modeling (47). This hLAT2 homology model in complex with CD98hc from PDB 6IRS was fitted as a rigid body into our high-resolution hLAT2/CD98hc cryo-EM density. Subsequent manual adjustments in the atomic model were performed using Coot (48), and Phenix real space refinement was used as the final refinement step to improve the geometries of the model (49). N-terminal residues 1 to 40 for hLAT2 and 1 to 60 for CD98hc were not visible in our cryo-EM density and therefore not included in the atomic model. A digitonin molecule was added next to TM3, TM9, TM10, and TM12 rigid fitted in the extra density described in Fig. 1.
Mutagenesis and Transfection of Wild-Type hLAT2/CD98hc and Mutants.
HeLa cells were transiently transfected in a 24-well plate with 300 ng/well of the human Strep-TagII-LAT2 (N-terminally tagged) in pcDNA3.1+ (29) or pEGFP-C1 (Clontech) and 200 ng/well of the His-CD98hc (isoform f) (N-terminally tagged) in pcDNA4-His-MaxC (50) using Lipofectamine 2000 (Invitrogen). Single point mutations were introduced using the QuikChange mutagenesis kit (Stratagene). All mutations were verified by sequencing. Amino acid transport assays were carried out 24 h after transfection.
hLAT2/CD98hc Amino Acid Transport Assays in HeLa Cells.
Amino acid uptake measurements were performed on hLAT2/CD98hc and GFP/CD98hc-transfected HeLa cells. Uptake rates were measured as previously described (9). Briefly, replicate cultures were incubated with 10 μM cold L-amino acid (glycine, L-alanine, L-valine, L-isoleucine, L-glutamine, L-histidine, and L-tryptophan) and 1 μCi/mL [3H] L-amino acid at room temperature for 1 min in a sodium-free (137 mM choline chloride) transport buffer that also contained 5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, and 10 mM Hepes (pH 7.4). Transporter-mediated amino acid uptake was calculated by subtracting the uptake measured in GFP-transfected cells.
For kinetic studies, cells were incubated with 1 μCi/mL [3H] L-amino acid and varying concentrations of unlabeled L-amino acid (0 to 2,000 μM). One 24-well plate for each condition (wild-type, mutant, or GFP-transfected cells) and each substrate (L-alanine, L-glutamine, and L-tryptophan) was seeded with 4 × 104 HeLa cells per well. After 24 h, cells were transfected as stated above (Mutagenesis and Transfection of Wild-Type hLAT2/CD98hc and Mutants), and transport assays were performed 24 h after transfection. To improve amino acid uptake values reproducibility, transfection efficiency was determined by GFP-fluorescence analysis in a control 24-well plate and only experiments with >80% of transfected cells were used. Moreover, all 24 wells from a single 24-well plate were assayed at a time and for 1 min of transport, minimizing in that way point-by-point incubation variability. Cold substrates were prepared at 100 mM, aliquoted, and stored at −20 °C until use. Aliquots were thawed only once to reduce variability. Each replicate of the kinetic studies was performed simultaneously for wild-type and mutated versions of hLAT2 and the three substrates analyzed. The Michaelis–Menten and Eadie–Hofstee equations were then applied to calculate Km and Vmax values (SI Appendix, Table S3) using the GraphPad Prism software. Uptake was terminated by washing with an excess volume of chilled transport buffer. Data are expressed as the mean ± SEM of three experiments performed on different days and on different batches of cells.
PELE Enzyme-Substrate Interaction Modeling.
The cryo-EM structure of hLAT2 was prepared for PELE simulations with the Protein Preparation Wizard (PrepWizard) tool implemented in Schrödinger (51). Missing hydrogen atoms were added by the utility applyhtreat in the PrepWizard tool. PROPKA 3.0 was used to calculate the protonation state of titratable residues at pH 7.4 and, on the basis of the predicted pKa values, the hydrogen-bonding network was optimized. The resulting structure was subjected to a restrained minimization step with the all-atom optimized potentials for liquid simulations (OPLS-AA) force field (FF), keeping heavy atoms in place and optimizing only the positions of the hydrogen atoms.
The PELE software was used to map the enzyme-substrate interaction (52). PELE is a heuristic Monte Carlo (MC) procedure designed to map protein–ligand induced fit interactions and extensively used in drug design (53) and enzyme engineering (54). Each MC step involves a complex series of events, including ligand and protein (backbone) perturbation, side-chain sampling, and a minimization. Typically, tens to hundreds of thousands of MC steps are used to explore the substrate-binding (or migration) space, where we record structural parameters and the OPLS2005 FF enzyme-substrate interaction energy. Two sets of simulations were performed. First, from an initially docked structure obtained with the Glide software (55, 56), we ran a local PELE exploration for each substrate in the wild-type transporter. As expected, PELE retrieved the wild-type (also referred to as canonical) pose as the main minimum, which could not be obtained in the initial docking due to the closeness of the Apo form. The second set, performed for the wild type and the selected mutants started from the canonical pose, and the ligand was allowed to explore a larger space; the center of mass was allowed to move within an 8-Å window. For each enzyme-substrate system, simulations involved 128 computing cores running for 1,250 MC steps each, which involved on average ∼36 wall clock hours on the MareNostrum IV supercomputer at the Barcelona Supercomputing Center.
MD Simulations.
The structure of LAT2 in complex with L-alanine and L-tryptophan generated by PELE (Fig. 3A and SI Appendix, Fig. S8B) was prepared for MD simulations. The same structures were used to model the corresponding Y93A mutants in the holo form. To model the membrane in the system, hLAT2 coordinates were preoriented with respect to the membrane (parallel to the z-axis) by alignment with BasC (Protein Data Bank [PDB] 6F2W) in the Orientations of Proteins in Membranes (OPM) database (https://opm.phar.umich.edu/) (57). The protein was then embedded in a POPC lipid bilayer using the CHARMM-GUI Membrane Builder by the replacement method (58–61). Next, 192 lipid molecules were placed in the lipid bilayer (i.e., 100 and 92 lipids in the upper and bottom leaflet, respectively) with its center at z = 0. The system was then solvated using a TIP3 PM water layer of 225-Å thickness above and below the lipid bilayer. NaCl ions corresponding to 0.15 M (47 negative and 45 positive) were also added to the system using MC sampling. Finally, with the CHARMM-GUI Membrane Builder, we also generated the necessary scripts to perform minimization, equilibration, and production runs in AMBER, using the CHARMM36 FF (C36 FF), as explained below. The simulations were run for the four systems using the C36 FF for lipids and the CHARMM TIP3P water model, at constant temperature (300 K) and pressure (1 bar), under Periodic Boundary Conditions, and with Particle Mesh Ewald electrostatics. The simulation time step was set to 2 fs in conjunction with the SHAKE algorithm to constrain the covalent bonds involving hydrogen atoms. After standard Membrane Builder minimization (2.5 ps) and equilibration (375 ps in 6 steps), production simulation was run (1 µs for each trajectory).
Subcloning of hLAT2 and CD98hc proteins, transient transfection, and production of hLAT2/CD98hc heterodimer in HEK293-6E cells, hLAT2/CD98hc purification for cryo-EM, hLAT2/CD98hc purification for amino acid transport assays, reconstitution into proteoliposomes, and the amino acid transport assays in proteoliposomes are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
The datasets of LAT2/4F2hc used in this study were obtained at the UK National eBIC under Block Allocation Group (BAG) proposal EM20135 (Stop cancer—structural studies of macromolecular complexes involved in cancer by cryo-EM), funded by the Wellcome Trust, Medical Research Council (MRC), and Biotechnology and Biological Sciences Research Council (BBRSC), and at the Leicester Institute of Structural and Chemical Biology. We acknowledge the help of Diamond Light Source for access to eBIC and the help of Dr. Christos Savva at Leicester. We thank Nick Berrow (Protein Expression Facility at the Institute for Research in Biomedicine (IRB) Barcelona) for the construction of the vectors with the tagged versions of hLAT2 and hCD98hc used in this study. We thank Ismael Mingarro for his help with the calculation of ER membrane insertion of hLAT2 and V460E mutant. We also thank Rafael Fernandez Leiro at Centro Nacional de Investigaciones Oncológicas(CNIO) for discussions about image processing of the data. This work was funded by “la Caixa” Foundation, Health Research Grant 2020 (LCF/PR/HR20/52400017) to M.P. and O.L.; by the Spanish Ministry of Science, Innovation and Universities (MCIU/AEI) Grants SAF2015-64869-R-FEDER and RTI2018-094211-B-100-FEDER to M.P. and SAF2017-82632-P to O.L.; cofunded by the European Regional Development Fund (ERDF); the support of Catalan Government (Grant 2017 SGR 961) to M.P.; the support of the NIH Carlos III to CNIO; Grants Y2018/BIO4747 and P2018/NMT4443 from the Autonomous Region of Madrid and cofunded by the European Social Fund and the ERDF to O.L. C.F.R. is funded by BES-2015-071348 PhD fellowship by the MCIU/AEI. We gratefully acknowledge institutional funding from the Spanish State Research Agency of the Spanish Ministry of Science and Innovation—Programa Estatal de Fomento de la Investigación Científica y Técnica de Excelencia—Centres of Excellence “Severo Ochoa” CEX2019-000891-S and CEX2019-000913-S. IRB Barcelona is a member of the Centres de Recerca de Catalunya (CERCA) System of the Catalan Government. P.B. is supported by a Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) contract.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113573118/-/DCSupplemental.
Data Availability
The cryo-EM map and the structure of human LAT2/CD98hc apo are deposited in the PDB database and Electron Microscopy Data Bank (EMDB) database with accession codes PDB ID: 7B00 and EMD-11952, respectively. All other data are available in the main text and SI Appendix.
References
- 1.Bröer S., Palacín M., The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 436, 193–211 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Fotiadis D., Kanai Y., Palacín M., The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34, 139–158 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Bröer A., Rahimi F., Bröer S., Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 291, 13194–13205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhutia Y. D., Babu E., Ramachandran S., Ganapathy V., Amino acid transporters in cancer and their relevance to “glutamine addiction”: Novel targets for the design of a new class of anticancer drugs. Cancer Res. 75, 1782–1788 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Calonge M. J., et al. , Cystinuria caused by mutations in rBAT, a gene involved in the transport of cystine. Nat. Genet. 6, 420–425 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Feliubadaló L., et al. , International Cystinuria Consortium, Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat. Genet. 23, 52–57 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Kahya U., Köseer A. S., Dubrovska A., Amino acid transporters on the guard of cell genome and epigenome. Cancers (Basel) 13, 125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torrents D., et al. , Identification of SLC7A7, encoding y+LAT-1, as the lysinuric protein intolerance gene. Nat. Genet. 21, 293–296 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Espino Guarch M., et al. , Mutations in L-type amino acid transporter-2 support SLC7A8 as a novel gene involved in age-related hearing loss. eLife 7, e31511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knöpfel E. B., et al. , Dysfunctional LAT2 amino acid transporter is associated with cataract in mouse and humans. Front. Physiol. 10, 688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safory H., et al. , The alanine-serine-cysteine-1 (Asc-1) transporter controls glycine levels in the brain and is required for glycinergic inhibitory transmission. EMBO Rep. 16, 590–598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tărlungeanu D. C., et al. , Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell 167, 1481–1494.e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilches C., et al. , Cooperation of antiporter LAT2/CD98hc with uniporter TAT1 for renal reabsorption of neutral amino acids. J. Am. Soc. Nephrol. 29, 1624–1635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cascio L., et al. , Abnormalities in the genes that encode large amino acid transporters increase the risk of autism spectrum disorder. Mol. Genet. Genomic Med. 8, e1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng M., et al. , LAT2 regulates glutamine-dependent mTOR activation to promote glycolysis and chemoresistance in pancreatic cancer. J. Exp. Clin. Cancer Res. 37, 274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J. M., et al. , In vitro characterization of a small molecule inhibitor of the alanine serine cysteine transporter -1 (SLC7A10). J. Neurochem. 129, 275–283 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Sakimura K., Nakao K., Yoshikawa M., Suzuki M., Kimura H., A novel Na(+) -Independent alanine-serine-cysteine transporter 1 inhibitor inhibits both influx and efflux of D-Serine. J. Neurosci. Res. 94, 888–895 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Jersin R. A., et al. , Role of the neutral amino acid transporter SLC7A10 in adipocyte lipid storage, obesity, and insulin resistance. Diabetes 70, 680–695 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Lee Y., et al. , Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc. Nat. Struct. Mol. Biol. 26, 510–517 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wu D., et al. , Structural basis for amino acid exchange by a human heteromeric amino acid transporter. Proc. Natl. Acad. Sci. U.S.A. 117, 21281–21287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan R., et al. , Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT. Sci. Adv. 6, eaay6379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan R., Zhao X., Lei J., Zhou Q., Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 568, 127–130 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Yan R., Zhou J., Li Y., Lei J., Zhou Q., Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex. Cell Discov. 6, 82 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan R., et al. , Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter. Cell Discov. 7, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Errasti-Murugarren E., et al. , L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction. Nat. Commun. 10, 1807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pineda M., et al. , Identification of a membrane protein, LAT-2, that co-expresses with 4F2 heavy chain, an L-type amino acid transport activity with broad specificity for small and large zwitterionic amino acids. J. Biol. Chem. 274, 19738–19744 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Yanagida O., et al. , Human L-type amino acid transporter 1 (LAT1): Characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta 1514, 291–302 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Pineda M., et al. , The amino acid transporter asc-1 is not involved in cystinuria. Kidney Int. 66, 1453–1464 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Rosell A., et al. , Structural bases for the interaction and stabilization of the human amino acid transporter LAT2 with its ancillary protein 4F2hc. Proc. Natl. Acad. Sci. U.S.A. 111, 2966–2971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeckelmann J. M., Fotiadis D., Sub-nanometer cryo-EM density map of the human heterodimeric amino acid transporter 4F2hc-LAT2. Int. J. Mol. Sci. 21, 7094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, 42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosco J., et al. , ATP modulates SLC7A5 (LAT1) synergistically with cholesterol. Sci. Rep. 10, 16738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jungnickel K. E. J., Parker J. L., Newstead S., Structural basis for amino acid transport by the CAT family of SLC7 transporters. Nat. Commun. 9, 550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowalczyk L., et al. , Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc. Natl. Acad. Sci. U.S.A. 108, 3935–3940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deitmer J. W., Bröer A., Bröer S., Glutamine efflux from astrocytes is mediated by multiple pathways. J. Neurochem. 87, 127–135 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Hessa T., et al. , Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Ma J., et al. , Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS. Proc. Natl. Acad. Sci. U.S.A. 116, 2086–2090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Focht D., et al. , A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition. EMBO J. 40, e105164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loignon M., et al. , Stable high volumetric production of glycosylated human recombinant IFNalpha2b in HEK293 cells. BMC Biotechnol. 8, 65 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bepler T., Kelley K., Noble A. J., Berger B., Topaz-Denoise: General deep denoising models for cryoEM and cryoET. Nat. Commun. 11, 5208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beckers M., Sachse C., Permutation testing of Fourier shell correlation for resolution estimation of cryo-EM maps. J. Struct. Biol. 212, 107579 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43 (W1), W174-81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fort J., et al. , The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J. Biol. Chem. 282, 31444–31452 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., Sherman W., Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Lecina D., Gilabert J. F., Guallar V., Adaptive simulations, towards interactive protein-ligand modeling. Sci. Rep. 7, 8466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilabert J. F., Lecina D., Estrada J., Guallar V., “Monte Carlo techniques for drug design: The success case of PELE” in Biomolecular Simulations in Structure‐Based Drug Discovery, F. L. Gervasio and V. Spiwok, Eds. (Wiley, 2018), 87–103. [Google Scholar]
- 54.Acebes S.et al. , . Rational Enzyme Engineering Through Biophysical and Biochemical Modeling. ACS Catal. 6, 1624–1629 (2016). [Google Scholar]
- 55.Halgren T. A., et al. , Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Friesner R. A., et al. , Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Lomize A. L., Pogozheva I. D., Lomize M. A., Mosberg H. I., Positioning of proteins in membranes: A computational approach. Protein Sci. 15, 1318–1333 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jo S., Kim T., Im W., Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2, e880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jo S., Lim J. B., Klauda J. B., Im W., CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee J., et al. , CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu E. L., et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM map and the structure of human LAT2/CD98hc apo are deposited in the PDB database and Electron Microscopy Data Bank (EMDB) database with accession codes PDB ID: 7B00 and EMD-11952, respectively. All other data are available in the main text and SI Appendix.