Significance
Metagenome sequencing provides a useful repository to extract evolutionary information and assist protein structure predictions. The sequence-search process, however, becomes increasingly prohibitive due to the huge library size. We hypothesize that there exist inherent evolutionary linkages between microbial niches and protein families that can be used to construct precise multiple sequence alignments (MSAs). To examine the hypothesis, we built a model library of four major biomes containing 4.25 billion sequences. Large-scale protein folding experiments revealed that MSAs collected from individually linked microbiomes can generate more accurate contact and structure models than those from all microbiome sequences but use significantly fewer computing resources. These results demonstrate the potential to solve the metagenome-search problem using a microbiome targeted approach.
Keywords: microbiome, protein structure prediction, deep learning, multiple sequence alignments, protein homologous families
Abstract
Information derived from metagenome sequences through deep-learning techniques has significantly improved the accuracy of template free protein structure modeling. However, most of the deep learning–based modeling studies are based on blind sequence database searches and suffer from low efficiency in computational resource utilization and model construction, especially when the sequence library becomes prohibitively large. We proposed a MetaSource model built on 4.25 billion microbiome sequences from four major biomes (Gut, Lake, Soil, and Fermentor) to decode the inherent linkage of microbial niches with protein homologous families. Large-scale protein family folding experiments on 8,700 unknown Pfam families showed that a microbiome targeted approach with multiple sequence alignment constructed from individual MetaSource biomes requires more than threefold less computer memory and CPU (central processing unit) time but generates contact-map and three-dimensional structure models with a significantly higher accuracy, compared with that using combined metagenome datasets. These results demonstrate an avenue to bridge the gap between the rapidly increasing metagenome databases and the limited computing resources for efficient genome-wide database mining, which provides a useful bluebook to guide future microbiome sequence database and modeling development for high-accuracy protein structure and function prediction.
Given the rapid explosion of protein sequences, computer-based approaches play an increasingly important role in protein structure determination and structure-based function annotations (1, 2). Two types of strategies have been widely considered for protein three-dimensional (3D) structure prediction (2): the first is template-based modeling, which constructs structural models using solved structures as templates, where its success requests for the availability of homologous templates in the Protein Data Bank (PDB); the second is template free modeling (FM) approach (or ab initio modeling), which dedicates to model the “Hard” proteins that do not have close homologous structures in the PDB. Due to the lack of reliable physics-based force fields, the most efficient FM methods, including Rosetta (3), QUARK (4), and I-TASSER (Iterative Threading ASSEmbly Refinement) (5), and most recently AlphaFold (6) and trRosetta (7), rely on a prior spatial restraints derived, usually through deep neural-network learning (8, 9), from the coevolution information based on multiple sequence alignments (MSAs) of homologous sequences (10). Hence, to model 3D structure of the “Hard” proteins, a sufficient number of homologous sequences is critical to ensure the accuracy of deep machine-learning models and the quality of subsequent 3D structure constructions (11).
Considerable effort was recently paid to the utilization of metagenome sequence data to enhance the MSA and FM model constructions. For example, Ovchinnikov et al. used the Integrated Microbial Genomes database to generate contact-map predictions and create high-confidence models for 614 Pfam protein families that lack homologous structures in the PDB (12). Using UniRef20 (13), Michel et al. combined contact-map prediction with the CNS (Crystallography & NMR System) folding method (14) to model protein structure for 558 Pfam families of unknown structure with an estimated 90% specificity. Most recently, Wang et al. examined the usefulness of the Tara Oceans microbial genomes and found that the microbiome genomes can provide additional help on high-quality MSA construction and protein structure and function modeling (15). This result demonstrated a significant role of the microbiome sequences, which represent one of the largest reservoirs of microbial species on this planet, in FM structural folding and structure-based function annotations.
Despite the success of metagenome-assisted 3D structure modeling, there are still thousands of Pfam families whose structure cannot be appropriately modeled with a satisfactory confidence. One critical reason is that despite the rapid accumulation of sequences, the current sequence databases are far from complete, and very few homologous sequences are available for many of the FM targets. On the other hand, the metagenome sequence databases have become extremely large (e.g., the Joint Genome Institute database contains more than 60 billion microbial genes and keeps increasing with at least 20,000 new sequences added per day) (16, 17), which makes a thorough and balanced database search increasingly slow and difficult. In a recent study, Zhang et al. showed that using current data mining tools, the quality of MSAs from metagenome library is not always proportional to the effective number of homologous sequences (Neff, reference SI Appendix, Eq. S1), partly due to the complexity of the sequence family relations and the bias of sequence database searches (10). The recent CASP experiments also witnessed various examples where the folding simulations for FM targets are negatively impacted by the contact/distance predictions due to the biased MSAs from the large metagenome datasets despite the high Neff value (18, 19). Therefore, a balanced sequence mining with accurate MSA construction is of critical importance to help improve the efficiency of sequence database searching and the subsequent 3D structure modeling.
In this work, we hypothesize that there exists an inherent evolutionary linkage between microbial niches (biome) and protein families, where a targeted approach built on linked biome families can be used to improve both efficiency and accuracy of MSA construction and protein structure predictions. To examine the hypothesis, we collected a model library of 4.25 billion microbiome sequences from the EBI metagenomic database (MGnify database) (20) that cover four major biomes (Gut, Lake, Soil, and Fermentor). The “marginal effect” analyses showed profoundly different effects of specific biomes on supplementing homologous sequences for different Pfam families. A machine-learning model named MetaSource is then developed to predict the source biome of target proteins, which can significantly improve the contact-map and 3D structure models accuracy with using more than threefold lower computer memory and CPU time. These results have validated the important biome-sequence–Pfam associations, which can lead a way toward better efficiency and effectiveness of the microbiome-based targeted approach to protein structure and function predictions.
Results and Discussions
Biome-Specific Microbiome Samples Contain Billions of Different Functional Genes from Thousands of Genera.
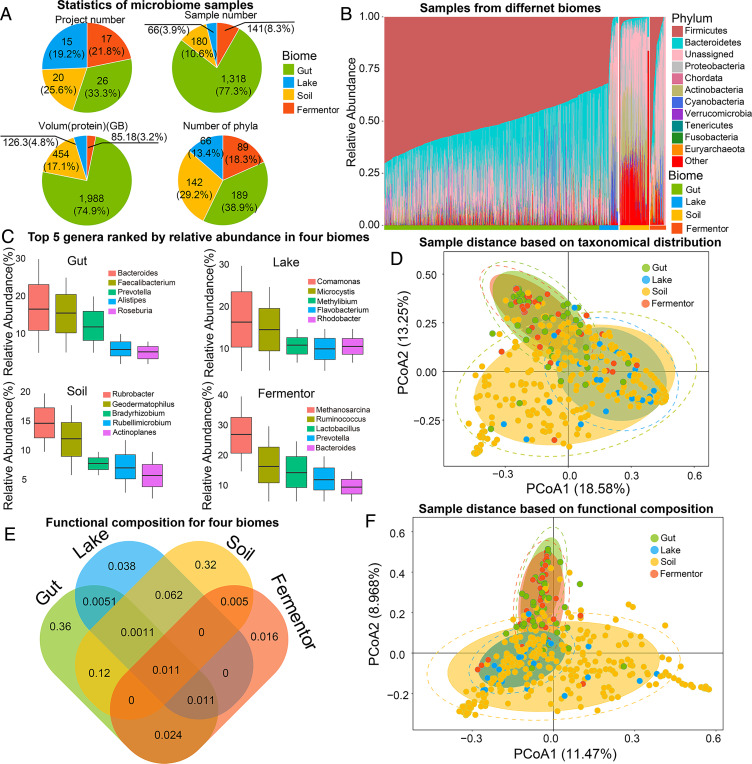
The 1,705 microbiome samples were collected from four typical microbial niches (biomes) (Gut, Lake, Soil, and Fermentor, Fig. 1A). Processed by the EBI pipeline version 4.1, a total of 4.25 billion protein sequences (functional genes) were predicted from these biomes, where a biome-specific taxonomic profile can be observed in Fig. 1B. The taxonomical profile illustrates that each biome is enriched for a specific set of abundant phyla, followed with a large number of low-abundant phyla, which represents a common distribution in microbial community (21–23).
Fig. 1.
Taxonomic and functional profiling for different microbiome samples. (A) The basic statistics of microbiome samples collected from the four biomes. (B) Species distribution on phylum level for samples in four biomes. The species distribution is categorized by their biomes and labeled with different colors. For all the samples, the top 10 phyla ranked by the average counts among all samples are illustrated. “Unassigned” means the species cannot be identified by a known phylum. “Other” represents the combination of the rest of the phyla. (C) Top-five genera ranked by relative abundances for four biomes. (D) PCoA result based on taxonomic profile on genus level for samples from the four biomes. Samples from the same biome are labeled with the same color. The CIs of samples in the same biome are marked in circles. (E) The shared and specific functional distribution for four biomes. The number labeled in the figure means the number (in billion) of specific or sheared sequences annotated by GO database. (F) PCoA result based on functional distribution for samples from the four biomes based on GO annotation. Samples from the same biome are labeled with the same color. The CIs of samples in the same biome are marked in circles.
Among the 1,705 microbiome samples, 169 phyla were identified, covering the common members in the kingdom of Bacteria and Archaea. With further classification on the genus level, 8,721 genera were identified, and different top-five genera ranked by relative abundance in four biomes also illustrate a biome-specific taxonomic profile (Fig. 1C). These results indicate that the biomes host different microbiome cohorts, and further investigation revealed the correlation between microbial communities’ taxonomic profile and their living biome: in the Gut biome, for example, Firmicutes (average relative abundance: 0.41 ± 0.28) and Bacteroidetes (average relative abundance: 0.26 ± 0.14) were the dominant phyla. Members in phylum Firmicutes were involved in energy resorption associated with reduced low-grade inflammation in obesity (24). Bacteroidetes play an important role in the development of immune dysregulation and systemic disease (25, 26). In Lake and Soil biome, phylum Proteobacteria is the dominant phylum (average relative abundance: 0.23 ± 0.18 and 0.35 ± 0.16, respectively), which takes part in nitrogen fixation and oxidation of iron, sulfur, and methane (27). In the Fermentor biome, phylum Firmicutes is the dominating phylum (average relative abundance: 0.46 ± 0.36), in which most members play the role of anaerobic fermentation (28), the main function of most Fermentor.
To illustrate the divergence among the biomes, statistical tests were performed based on the species distribution: the Wilcox test (nonparametric statistical test, single-tail test) for each pair of four biomes indicate a statistical difference among four biomes (SI Appendix, Table S1). Furthermore, the Principal Coordinates Analysis (PCoA) indicates a biome-specific taxonomic profile for 1,705 microbiome samples (Fig. 1D): samples collected from the same biome could cluster into one group (reflected by a concentrated confidence circle). Moreover, samples from the Lake biome were closer to those of the Soil biome, while those of the Gut biome and Fermentor biome were closer. This phenomenon could be attributed to the similar environments between Gut and Fermentor (oxygen-limited environment), as well as between Lake and Soil environment (open-air environment).
Among the 4.25 billion protein sequences obtained from these four biomes, we observed the biome-specific functional profiles. In total, 1.25 billion proteins could be annotated by Gene Ontology (GO) database. Similar to taxonomic profile, these four biomes host different functional annotations (Fig. 1E): 0.36 billion (68.4%) annotations were only detected in Gut biome, 0.038 (29.9%) billion annotations only in Lake biome, 0.32 (62.7%) billion annotations only in Soil biome, and 0.016 billion (24.2%) of gene annotations only in Fermentor biome. The PCoA result based on functional profiles presents clear differences among these four biomes (Fig. 1F). Again, samples from Gut and Fermentor biomes were closer, while samples from Lake and Soil were closer, similar with the PCoA result based on taxonomic profiles. These results illustrated a statistically biome-specific taxonomical profile and functional composition and the feasibility of using the biome as a label to search for the protein with the specific function.
Metagenome-Sourced Proteins Assisted Successful Structure Modeling for Thousands of Protein Families without Homologous Templates.
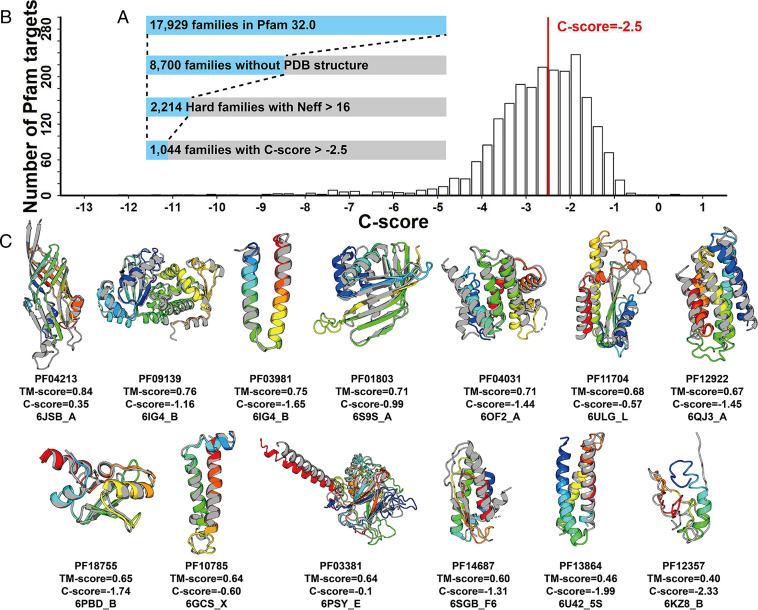
Recent studies have shown that metagenome sequences can help improve the performance of protein structure prediction (12, 15), especially for the Pfam families without solved structures. We selected 2,214 Pfam families with the Neff of MSA >16 (= 24) from 8,700 Pfam families that have no member with solved structures. These Pfam families were all categorized as Hard targets by LOMETS (LOcal MEta-Threading Server) because no homologous templates could be detected from the PDB by threading (29). Here, the Neff = 16 was selected as cutoff, because we found most of the monomer targets with Neff > 16 are foldable by contact-assisted I-TASSER (C-I-TASSER) from the 168 benchmark targets (SI Appendix, Fig. S1). Starting with microbiome sequences, we extended the deep-learning C-I-TASSER (19) method to predict structure models for the 2,214 unsolved Pfam families. Based on the benchmark results showing that models with a confidence score (C-score) ≥−2.5 usually have a correct fold (see Eq. 2 and SI Appendix, Fig. S2), 47% (1,044/2,214) of the Pfam families were found to be foldable by C-I-TASSER (Fig. 2A). In Fig. 2B, we presented the C-score histogram distribution of the C-I-TASSER models on the 2,214 unknown Pfam families. Considering false discovery rate (FDR) obtained from the benchmark tests (see Materials and Methods), there should be around 971 [= 1,044 × (1 – 6.96%)] Pfam families with high-confidence models.
Fig. 2.
Structural modeling results for unknown Pfam Hard families. (A) Number of Pfam families at each stage of the analysis, where each set is a subset of the previous set. (B) The C-score distribution of the Pfam Hard families with Neff >16. (C) Structural models on 13 newly solved Pfam families with C-score >−2.5. In each case, the C-I-TASSER model is shown in rainbow color, and the solved experimental structure of a member from the same Pfam family is shown in gray.
Here, the benefit for the inclusion of the microbiome sequences is obvious, as the average number of homologous sequences (1,646) in the MSAs collected from the metagenome database is 3.6-fold higher than that in the original Pfam MSA (459) built on the UniProt genome database, where 2,166 families have the number increased and 48 have the number reduced (SI Appendix, Fig. S3A). As a result, the average Neff of the metagenome MSAs (75.9) is 196% higher than that of the Pfam MSAs (25.6) (SI Appendix, Fig. S3C). Especially, although the average sequence similarity to the query of the metagenome MSA (46.7%) is 11% higher than the original Pfam MSAs (42.2) (SI Appendix, Fig. S3B), the average Meff value, which is a measure of the diversity of MSAs by HHblits (30), is 97% higher than that of the Pfam MSA (Meff = 3.9) (SI Appendix, Fig. S3D). Such a comprehensive MSA from microbiome, which covers more diverse and homologous sequences, will help the deep-learning algorithms to better derive coevolution information involved in residue pairs and therefore result in more accurate contact maps to assist the C-I-TASSER structure modeling. A more detailed comparison of MSAs collected from original Pfam families and using microbiome metagenome is supplied in SI Appendix, Text S2.
The C-I-TASSER modeling was performed on the Pfam database version 32.0 released in September 2018. The new Pfam version 33.0 reported 28 new families with solved structures for at least one member among the 2,214 modeled Pfam families, which provides an opportunity to assess the performance of the prediction. Since only one member from each Pfam family was modeled by C-I-TASSER, the modeled member may be different from the member with solved structure. For these cases, we superposed the solved structure to the C-I-TASSER model using TM-align (Template-modeling-align) (31) and calculated the TM-score (Template-modeling-score) between the C-I-TASSER model and the experimental structure. The comparison between the C-I-TASSER models and the solved experimental structures is listed in SI Appendix, Table S2. Although all the families are nonhomologous to the PDB structures, 50% of the C-I-TASSER models have been correctly folded with TM-scores >0.5. This result is roughly consistent with the estimation that 47% of the 2,214 Pfam families are foldable by C-I-TASSER. Fig. 2C presents the 13 Pfam families which have a C-score >−2.5. While most of the targets have a correct fold, there are two cases (PF3864 and PF12357) whose TM-scores are below 0.5. For PF3864, C-I-TASSER predicted it as a three-helix bundle but the solve structure covers only two helices. Therefore, TM-score is 0.46 when normalized with full-length query sequence. If we normalized the TM-score by the length of the solved structure as what is done in CASP, the TM-score will be increased to 0.55. For PF12357, the solved structure shows that the major components of the structure are coils. Although the deep-learning predicted contact maps have a random-like pattern, the simulation of C-I-TASSER showed some convergency, resulting in a C-score = −2.33 marginally above the cutoff, which gives the false-positive model with a TM-score = 0.4.
As a deep-learning guided structure folding method, the performance of C-I-TASSER generally improves with the increase of homologous sequences. However, we observed a number of cases which C-I-TASSER fail to fold even with a relatively high Neff (SI Appendix, Fig. S1). SI Appendix, Fig. S4A presents the structures of the 12 targets whose Neff >16 but with TM-score <0.5. Most of these targets are from one part of protein complexes and contain flexible regions, that is, a helix or strand that is largely far away from the core region. The coevaluation across interacting protein chains could results in false contact predictions. As shown in SI Appendix, Table S3, half of the targets have incorrect contact maps with the top-L/5 long-range contact accuracy below 0.2. Meanwhile, the hydrophobic energy terms in the C-I-TASSER force field tend to fold the targets into a compact structure when isolated monomer proteins are modeled. SI Appendix, Fig. S4B presents a typical example ivli_A2, which comes from a triple-chain complex. Although C-I-TASSER generated reasonable short- and medium-range contact maps, the long-range contacts are completely wrong (SI Appendix, Fig. S4C). These false-positive long-range contacts, together with the generic hydrophobic force field of C-I-TASSER folded the protein into a compact structure while the native structure is far more extended due to the interaction with other component chains. An extended version of C-I-TASSER incorporating protein complex interactions in both contact prediction and folding simulations will be essential to address this issue.
The C-I-TASSER models, MSAs for all 2,214 Pfam families with unsolved structures, and the benchmark dataset for C-I-TASSER modeling are downloadable at https://github.com/HUST-NingKang-Lab/MetaSource/releases.
Enrichment of Homologous Sequences from Different Biomes.
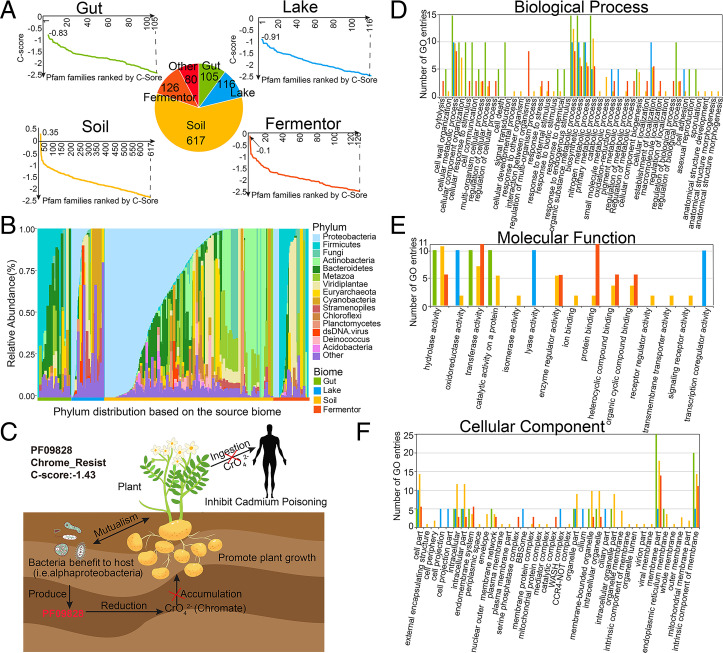
For the 1,044 Pfam families foldable by C-I-TASSER, an enrichment of homologous sequences from a specific biome can be observed, that is, 964 Pfam families (964/1,044, 92.3%) could be identified with a single biome whose Neff value is larger than the other three biomes, including 105 families for Gut, 116 families for Lake, 617 families for Soil, and 126 for Fermentor (Fig. 3A). For the remaining 80 Pfam families, two or more biomes contributed equally, which may be caused by the limited number of metagenome sequences (average 8.3 ± 3.1 metagenome sequences) aligned. We observed that sequences from the Soil biome could assist in folding more Pfam families than other biomes, that is, 39.6% sequences in the Soil biome could be aligned to Pfam families, while only 33.1% for the Gut biome, 30.8% for the Lake biome, and 24.3% for the Fermentor biome. These results are understandable as the metagenome in the Soil biome has been shown to have the highest species richness and most functional genes among these four biomes (32). However, it is worth mentioning that though microbiome sequences from Soil biome could supplement more Pfam than sequences from other biomes, this is not a winner-take-all situation: other biomes still work better than Soil biome for specific Pfam families.
Fig. 3.
The taxonomic and functional properties of the Pfam families foldable by C-I-TASSER. (A) C-score distribution for Pfam families after replenishing by metagenome sequences. The vertical axis represents the C-score. For each panel, horizontal axis represents the Pfam families (31). (B) The relative abundance of species distribution for Pfam families which were foldable by C-I-TASSER. The species distribution is divided into four biomes and labeled with different colors. Calculated by the average count among all samples, the top 10 phyla are illustrated and ranked. “Other” represents the combination of the rest of the phyla. (C) Proteins in PF09828 are involved in the reduction of chromate accumulation and are essential for chromate resistance. Bacteria that hosts in plant produce the proteins identified as PF09828 to reduce the accumulation of chromate, resulting in the fast growth of the plant and preventing the transmission of cadmium to humans through the food chain leads to cadmium poisoning. For all the Pfam families which were foldable by C-I-TASSER, after aligning the Pfam species to the Interpro database, their protein functions were annotated by GO annotations and classified by three top annotations: Biological Process (D), Molecular Function (E), and Cellular Component (F).
To assess the utilization efficiency (UE) of metagenome sequences in Pfam structure modeling, we define , where is the number of sequences from the metagenome datasets that are homologous to the ith Pfam family, and is the total number of metagenome sequences considered. In Table 1 (column 7), we list the UE values for different metagenome datasets on the Pfam families that are foldable by C-I-TASSER. It is shown that the utilization efficiencies of the three single biomes (Lake, Soil, and Fermentor with UE = 0.19, 0.49, and 0.94, respectively) are considerably higher than that from the combined dataset (0.15), although Gut’s UE are relatively low (0.04). If we count the number of Pfam families assisted by specific biomes, Soil and Fermentor assisted 907 and 2,000 families foldable per TeraByte (TB) sequences, respectively, which are 2 to 5 times higher than that of the combined dataset, where the latter is comparable to those in previous metagenome structure modeling works (12, 15). These results suggest that targeted MSA collections from specific microbial biomes could improve the utilization efficiency of metagenome sequences compared with the approach that simply combines all available sequence datasets.
Table 1.
Summary of utilization efficiency of metagenome sequences
| Dataset | V* | Np† | Nf‡ | Nf/V§ | Nf/Np¶ | UE# | P value (Integrated Microbial Genome/Tera/combined)ǁ |
| Integrated Microbial Genome | 1.41 | 2.25 | 614 | 435.5 | 272.9 | 0.10 | NA/NA/NA |
| Tera Oceans | 0.15 | 0.20 | 68 | 453.3 | 348.7 | 0.30 | NA/NA/NA |
| Combined | 2.4 | 4.3 | 1,044 | 435.0 | 245.7 | 0.15 | 4E-19/E-26/NA |
| Gut | 1.4 | 2.1 | 105 | 75.1 | 50 | 0.04 | 5E-20/9E-18/6E-23 |
| Lake | 0.3 | 0.7 | 116 | 386.7 | 178.5 | 0.19 | 4E-22/3E-17/8E-22 |
| Soil | 0.68 | 1.4 | 617 | 907.4 | 440.7 | 0.49 | 9E-20/3E-19/5E-20 |
| Fermentor | 0.06 | 0.1 | 126 | 2,000.0 | 1,326.3 | 0.94 | 9E-21/5E-19/8E-20 |
For predicted Pfam families with unsolved structures, the statistic results for metagenome sequence utilization efficiency were calculated for results based on combined dataset (metagenomes from all of the four biomes), four single biomes, compared with datasets from previous studies.
*V: Volume size of protein sequence datasets (in TB).
†Np: No. of protein sequences (in billions).
‡Nf: No. of foldable Pfam families with C-score >−2.5.
§Nf/V: No. of foldable Pfam families/Volume size of dataset.
¶Nf/M: No. of foldable Pfam families/Number of sequences (per billion sequence).
#UE: Utilization efficiency of metagenome sequences in Pfam structure modeling (per 1,000 metagenome sequences).
ǁP value: P value calculated on the UE values relative to “Integrated Microbial Genome,” “Tera Oceans,” and “Combined data,” respectively.
To decipher the important role of the solved Pfam families in their living environment, taxonomic profile and functional composition analyses were applied for each of the 964 Pfam families with single corresponding biome (Fig. 3). The taxonomic profile for the 964 Pfam families illustrates specificity of contributions of microbial biome’s sequences to Pfam structure modeling (Fig. 3B). Overall, similar to microbiome samples (Fig. 1B), the heterogeneous species distribution reflects a biome-specific enrichment pattern for the 964 Pfam families. Moreover, the dominating species in specific Pfam families are often the dominating species in assisted microbiome samples for MSA constructions. For example, In Pfam families labeled with Gut biome (Figs. 1B and 3B), phylum Firmicutes and Bacteroidetes (both belonging to Gut) were the dominate phyla in Pfam families (0.41 ± 0.28 and 0.26 ± 0.14, respectively) and corresponding source biome (0.48 ± 0.31 and 0.31 ± 0.15, respectively), which indicates that this biome-specific enrichment pattern was influenced by the species composition of the microbiome samples.
In addition to structure modeling, the functional composition for the 964 Pfam families provides a useful insight into this biome-specific enrichment pattern. Based on the GO annotation, for example, 368 Pfam families were aligned to GO level-3 Biological Process (286), Molecular Function (90), and Cellular Component (189) (Fig. 3 D–F). By analyzing the functional annotations for these Pfam families, the biome-specific enrichment pattern could also be detected, reflected by the fact that many function annotations were only detected in a single biome, including 129 (45.1%) for Biological Process, 69 (76.7%) for Molecular Function, and 109 (57.7%) for Cellular Component (SI Appendix, Table S4).
Further functional analysis based on the biological process annotations reveals their important roles in helping the host species to adapt to their environment (Fig. 3D). In Gut and Fermentor biomes, for example, the main functions are associated with anaerobic energy metabolism (52.7% and 68.7% annotations for Gut and Fermentor, respectively). Enrichment of these functions could help their host to efficiently utilize the carbon sources to live in the oxygen-free environment and produce metabolites to interact with their host (33, 34). In the Lake biome, the main functions are associated with bacteria-specific cell motility (60.3%) to help their host adapt to the flowing water environment (35). Moreover, in the Soil biome, the functional roles of Pfam families with a C-score ≥−2.5 were connected to such processes as nitrogen fixation (28.8%) and oxidation of iron (20.3%), sulfur (16.8%), and methane (10.3%) to take part in the soil chemical element cycle or adapt to the iron-enriched environment (36–38).
The aforementioned association of protein families with biomes are rooted in the intricate but potentially important properties of the protein structures and functions: to adapt their biomes, functional genes from microbial species have to evolve so that the species could gain the advantage over other species in that specific niche, thus certain functional genes (or protein families) would be highly likely to be enriched in a specific niche, though not exclusive to be present in such a niche. A typical example for supplementing homologous sequence for Pfam family 3D structure and function prediction is on a previously unsolved Pfam family PF09828, which contained 713 homologous sequences in the family and 98.3% sequences (701/713) of them are identified as bacteria (Fig. 3C). After the sequences from the four biomes were included in the MSA construction search, the number of homologous sequences in the MSA for this family increased from 713 to 5,582 (Soil: 5,348, Lake: 151, Gut: 4, Fermentor: 79), resulting in a relatively high-accuracy contact-map and 3D structure prediction (C-score = −1.43). Interestingly, 526 sequences of the bacteria-sourced sequences in Pfam sequences (73.8% = 526/713) are classified into phylum Proteobacteria, the dominant phyla in the Soil biome that counts for 93.0% of the homologous sequences supplemented in the MSA (Fig. 3B). Further functional analysis reveals its role in the Soil biome: bacteria that hosts in plant produce the proteins identified as PF09828 to reduce the accumulation of chromate in plant (39). The reduction of chromate in plant could promote the growth of the plant and prevent the transmission of cadmium to humans through the food chain, which leads to cadmium poisoning (40). In SI Appendix, Text S3 and Table S5, we list 10 other examples to showcase the biome-sequence–Pfam relationships. Taken together, these data illustrate potential correlations between the composition of the Pfam families and the source biomes used to supplement the MSAs for structure and functional modeling.
Marginal Effect Analyses Reveal Biome-Sequence–Pfam Relationship.
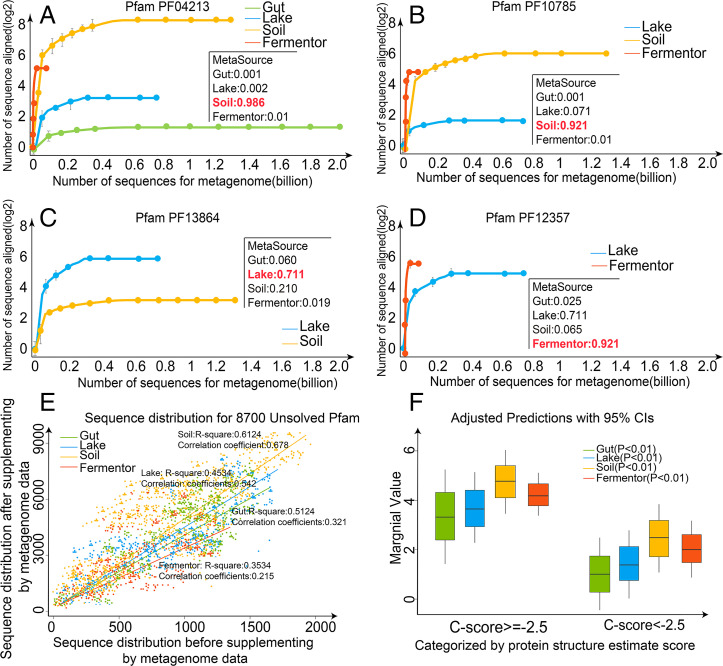
The results of the last section have strongly indicated that the protein sequences from different biomes have profoundly different effects on supplementing homologous sequences of different Pfam families. To quantitatively examine the effect, we define the marginal effect of ith biome on jth Pfam family by , where is the number of homologous sequences for the jth family when searching the query through ith biome dataset, and is the number of homologous sequences in the jth family from the Pfam database. In Dataset S1, we list the marginal effects of the four biomes on all the 8,700 unknown Pfam families; the data shows that the contributions of different biomes to a specific Pfam can be drastically different, as reflected by their ME values. In Fig. 4 A–D, we present the contribution of biomes on the MSA collections for four examples from PF04213, PF10785, PF13864, and PF12357, where the microbiome samples were randomized for the MSA collections at different sequence numbers. For different Pfam families, the sequence homology pools are dominated by different biome datasets, suggesting again a strong link between biome and Pfam in regard to homologous sequence supplement.
Fig. 4.
Evaluation of marginal effect for Pfam families. Collected from the four biomes, the homology sequences distribution of Pfam family (A) PF04213, (B) PF10785, (C) PF13864, and (D) PF12357 are illustrated, where the source biome of these Pfams was estimated by MetaSource. (E) The sequence distribution of metagenome data from the four biomes for all 8,700 Pfam families with unsolved structures. After the sequences from four biomes were aligned to 8,700 Pfam families with unsolved structures, respectively, the marginal effect is estimated by comparison of the number of Pfam family’s homologous sequences before and after the use of the metagenome sequences. (F) Marginal effect categorized by protein structure estimate scores.
To examine the overall trend of marginal effect, we plot in Fig. 4E the versus values for all 8,700 Pfam families from four microbial biomes. For each biome, a linear regression was established based on the marginal value distribution of the 8,700 Pfam families. The correlation coefficients of simulated curve for four biomes are 0.678, 0.542, 0.321, and 0.215 for Soil, Lake, Gut, and Fermentor, respectively, suggesting that the metagenome sequences are estimated to process a statistically positive effect to supplement the homologous sequences for Pfam families. On average, the marginal effect value is 5.28 ± 3.25, 3.85 ± 2.96, 3.48 ± 3.11, and 4.12 ± 1.65 for Soil, Lake, Gut, and Fermentor, respectively. This rank of average marginal values for four biomes is largely consistent with the rank of species richness for the four biomes (SI Appendix, Fig. S5). Although the Soil biome has the highest overall marginal effect value, there are several hundreds of the Pfams families which have their highest marginal value from other three biomes, suggesting again the importance of biome-specific metagenome sequence selection to maximize the efficiency of MSA collection.
In Fig. 4F, we split Pfam families into two groups based on C-I-TASSER folding results. It was shown that the ME value for the families with C-score −2.5 is much higher than that with C-score <−2.5 (5.27 ± 3.44 versus 1.28 ± 0.85 with a P value = 3.86e-26 in Student’s t test). Therefore, marginal effect value is also strongly correlated with the ability of a biome-specific metagenome sequence to assist the 3D structure assembly simulation through supplementing more homologous sequences.
MetaSource Prediction Model for Effective Homologous Sequence Supplements.
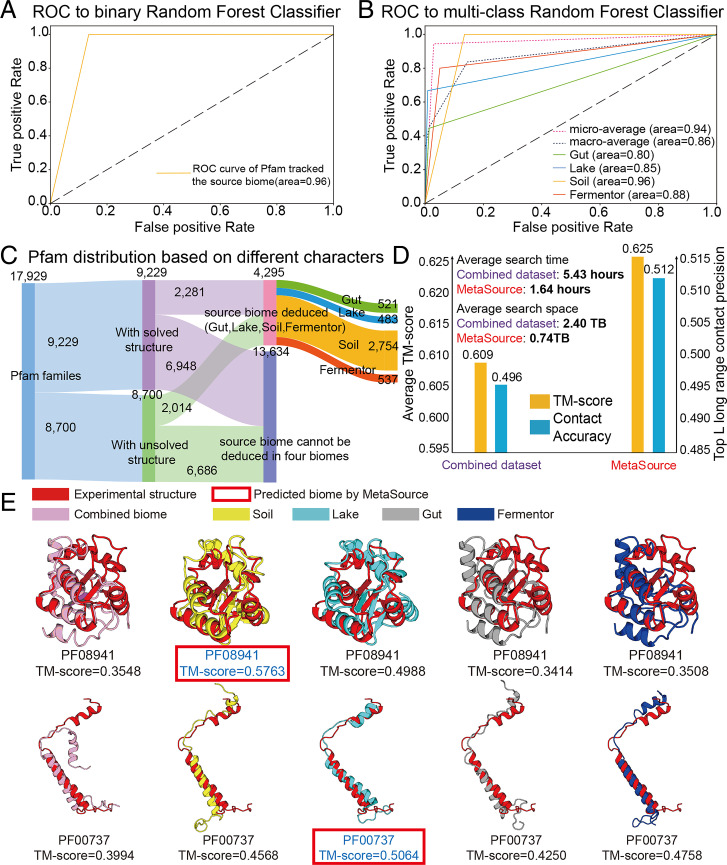
Previous analysis has revealed a profoundly different effect of specific biomes on supplementing homologous sequences for different Pfam families. Here, we proposed the MetaSource prediction model to identify one or a set of biomes, which can better supplement homologous sequence collections for specific Pfam families. First, to determine whether the source biome of the query Pfam family is one of the four biomes, a binary-classification model was constructed by using the 964 Pfam families labeled with a single biome as the training dataset and 7,736 (= 8,700 − 964) Pfam families with unsolved structures as the testing dataset. As shown in Fig. 5A, MetaSource achieves an area under curve (AUC) of 0.96 under 0.001 permutation P value on the binary-classification test. Second, to predict the most probable source biome out of the four biomes for a Pfam family, the multiclass random forest algorithm was chosen to construct this model. In this context, a biome that could supplement the largest number of homologous sequences was considered as the “correct” biome. The 964 Pfam families labeled with a single biome were used, with 20 cross-validation iterations (Fig. 5B), showing a strong predictive power of MetaSource for the Pfam families, with a microaverage AUC of 0.94, under 0.001 permutation P value. The top 20 important features used in MetaSource were supplied in SI Appendix, Fig. S6.
Fig. 5.
The source biomes predicted by MetaSource for Pfam families. (A) The receiver operating characteristic (ROC) analysis of binary-classification MetaSource model. This model was constructed to determine whether the source biome of the query Pfam family is one of the four biomes. (B) The ROC analysis of multiple-classification MetaSource model. This model was constructed to predict the source biome for Pfam families. To evaluate the overall prediction accuracy, the microaverage (obtained by aggregating the contributions of all classes to compute the average metric) and macroaverage value (calculated by the metric independently for each class and taking the average) were applied. (C) The Pfam classification result for all the Pfam families based on the prediction result of MetaSource model. (D) Average TM-score, accuracy of top-L contacts, and average MSA search time for the combined and MetaSource predicted biome datasets. (E) Case studies of modeling Pfam (PF08941 and PF00737) with MSA from different biomes. The model with the highest TM-score is shown in blue font. The model labeled with red frame is the source biome predicted by MetaSource.
To further examine the practical usefulness of the Metagenome database and MetaSource model in 3D structure modeling, we incorporated the 204 Pfam families with known structure into our analysis as the validation dataset (Fig. 5C). First of all, C-I-TASSER using the MSA from genome database (Uniclust30 + Uniref90, step 2 results of DeepMSA, SI Appendix, Fig. S7) can generate a model with TM-score = 0.583, which is 2.5% higher than a C-I-TASSER model that solely uses the Uniclust30 genome database (step 1 of DeepMSA) with P value = 2.1E-4 by one-tailed paired Student’s t test (SI Appendix, Fig. S8). Uniclust30 and Uniref90 database are built from the same UniprotKB database, which means no potentially new sequences should exist in Uniref90 over Uniclust30. Thus, this increase of modeling performance is due to the iteratively homolog refinement searching by DeepMSA. After adding metagenome database in the step 3 of DeepMSA, the TM-score of C-I-TASSER models increases to 0.609, which is 4.5% higher than only using genome databases with P value = 3.8E-11. This result demonstrates again the usefulness of the metagenome database in 3D structure prediction by extending the MSA coverage and diversity. Overall, by combining the DeepMSA tool with metagenome database, TM-score of the C-I-TASER model increased by 7% compared with the simple one-step HHblit MSA collection approach.
Furthermore, given the rapid increase of metagenome databases, selecting a subset of related homologous sequences would be helpful for improving both the speed and accuracy of MSA collection and protein 3D structure modeling. As listed in SI Appendix, Table S6, MetaSource was able to predict the biomes which resulted in the highest Neff (or the highest TM-score) with an accuracy of 79.9% (or 80.2%) (permutation P value: 0.001) on the 204 solved Pfam families. In Fig. 5D, we further compare the average contact accuracy and TM-score of the C-I-TASSER models when using MSAs collected from the combined dataset and the dataset chosen by MetaSource. It was shown that, although the volume of the sequence database is much smaller (0.74 TB/per target and 2.40 TB/per target for MetaSource and Combined datasets, respectively), using the targeted dataset from MetaSource results in a higher contact accuracy (0.512) and TM-score (0.625) than that of the combined dataset (0.496 and 0.609), which corresponds to a P value = 2.0E-5 for contact and a P value = 6.3E-6 for TM-score in Student’s t test. As shown in SI Appendix, Fig. S9A, 70% of the targets have the TM-score increased by biomes chosen by MetaSource. Accordingly, the speed of MSA search based on MetaSource (1.65 h/per target) is also faster than that from the combined dataset (5.44 h/per target). The result may be understandable because sequences from the “wrong” source biome can produce “noise” to the MSA collection and deep learning–based contact prediction, where the identification of correct source biomes could help depress such noises. Here, it is noted that although the overall TM-score increase (2.6% = (0.625-0.609)/0.609) is slightly lower than that due to the introduction of metagenome (4.5% = (0.609-0.583)/0.583), the MetaSource approach has additional impact on improving the speed of MSA construction, which is important in the long run to the field with the rapid increase of the metagenome databases.
In Fig. 5E, we present two Pfam examples from PF08941 and PF00737 with known structure, for which MetaSource predicted Soil and Lake as the best source biome, respectively. In both cases, only the models with the MetaSource predicted biomes could create a model with a TM-score above 0.5. We also noticed that, although the MSA from the combined biome contains more sequences than single biome, the structure models are clearly worse than the MSA from some single biome (Soil or Lake), probably due to the noise contribution from irrelevant metagenome sequences. Taxonomic profile analyses also showed that PF08941 and PF00737 are mainly composed with proteins from phylum Proteobacteria and Cyanobacteria, which dominate in Soil and Lake biomes, respectively (27, 41).
Nevertheless, there are overall 29 out of the 204 test cases (14%) where C-I-TASSER failed to generate correct fold with a TM-score >0.5. As listed in SI Appendix, Table S7, most of the failed cases are due to the following: 1) the target is part of a protein complex as discussed in SI Appendix, Fig. S4; or 2) the number of effective sequences, Neff, is too low for both combined and MetaSource MSAs. There are also a few cases which failed due to bad tail orientation and sparse MSA at local structural regions. Despite the failures, the TM-score of the models using MetaSource is higher than that using combined sequences for most of the cases, suggesting that the failure was not due to the use of MetaSource. Overall, although the Neff of MetaSource (= 41.7) is lower than the combined MSA (51.2), the average TM-score of MetaSource (= 0.348) is higher than the latter (0.329), demonstrating the efficiency of MetaSource to selecting correct biomes even for the difficult cases. By checking the entire testing set, we identified one case (PF07072) for which the C-I-TASSER model using MetaSource MSA is obviously worse than that using combined MSA (TM-score = 0.527 versus 0.574); this failure was due to the incorrect biome prediction by MetaSource that predicted the biome “soil” as the targeted biome while the biome with the highest Neff and TM-score is “fermentor” (SI Appendix, Table S6). Since PF07072 consists of a high portion of the sequences that cannot be classified to any known phylum (29.8% = 290/972), improving the accuracy of MetaSource for such families will be important to address this issue.
In addition to C-I-TASSER, we also examined the usefulness of the MetaSource for other recently developed deep learning–based protein modeling methods, including DMPfold (42), trRosetta (7), and AlphaFold2 (43). As summarized in SI Appendix, Fig. S9 B–D, the direct use of MetaSource selected biomes has resulted in an TM-score increase for 65, 73, and 86% of models by DMPfold, trRosetta, and AlphaFold2, respectively, where the P values for the average TM-score changes are 8.1E-4, 7.3E-7, and 5.7E-7, respectively, showing that the overall improvement is statistically significant. These results demonstrated the generality of the concept for utilizing the intrinsic links of protein and biome sequences to enhance the sequence-based ab initio prediction structure prediction.
Conclusions
As a grand reservoir of novel genes and proteins, microbial communities contain a large number of uncultured species that are unique for adapting their living environments. Nowadays, the metagenome sequencing technology has been advanced enough to sequence microbial communities in many of the known biomes on Earth, while more complete gene catalogs of microbial communities have been obtained from some biomes than others due to the accessibility of the species as well as their functional genes. While these microbiome sequences have been shown useful for boosting the accuracy and capacity of deep learning–based protein structure and function predictions, the model training and metagenome search were largely blind and fall short in efficiency in source tracking the most relevant biome datasets for specific protein targets. For designing a more effective targeted approach, deeper insights should be obtained to link microbiome biomes with protein family homologous sequences.
In this study, we utilized a model library of 2.4 TB microbiome sequencing data, representing 4.25 billion microbiome sequences from four major biomes (Gut, Lake, Soil, and Fermentor) to investigate the usefulness of metagenome sequences from specific biomes for protein structure prediction of individual Pfam families. First, the inclusion of these microbiome sequences has boosted MSAs with credible multiplicity for 2,214 out of 8,700 Pfam families with unsolved structures. By applying C-I-TASSER ab initio structure folding pipeline, highly reliable folds were constructed for 1,044 Hard Pfam families, which account for 12% of all unknown Pfam families.
To further examine the association between the metagenome sequences and Pfam families, we quantified the marginal effect of metagenome sequences on Pfam families, where the data shows that metagenome sequences from different biomes have drastically discriminable power to different Pfam families. Accordingly, instead of searching through all the metagenome sequences to find the homology sequences, a simple machine-learning model, MetaSource, was constructed for source tracking the most relevant biome datasets for specific Pfam family structure modeling. The utilization of the MetaSource predicted biomes have resulted in 3.2-fold reduction in the database size and 3.3-fold increase in MSA construction speed but with 3.2% of contact-map accuracy and 2.6% of TM-score increase in the C-I-TASSER final models. This result is particularly encouraging in this postgenomic era when the number of genome and metagenome sequences increases exponentially, and the speed and memory requests become a major bottleneck for sequence mining and MSA collection through large-scale sequence database searching (10). These findings could be used as a useful bluebook to guide the modeling of protein structure and function based on the deeper insights into the biome–protein association.
In addition to “biome,” other biological labels may also be used by the targeted modeling approach. As an example, we have examined the use of phylum label to train a “PhylaSource” model to guide the homology sequence search for the targeted MSA construction. Since not all metagenome sequencing data are currently labeled with phylum, we downloaded all the prokaryotic and viral genomes from the National Center for Biotechnology Information (NCBI) that contain phylum label and then trained PhylaSource on 964 Pfam families with known structures (see details in SI Appendix, Text S4). As shown in SI Appendix, Fig. S10 A and B, the use of the PhylaSource model did result in a slightly (but statistically significantly) higher contact accuracy compared with the use of the combined datasets (0.488 versus 0.476 in top-L long-range contact with P value = 1.5E-5) and C-I-TASSER TM-score (0.617 versus 0.615 with P value = 2.3E-5), despite the threefold reduction in the size of searched sequence data (236 GB versus 736 GB by PhylaSource and combined, respectively). However, the overall accuracy is not as good as that by MetaSource (Fig. 5D), probably due to the fact that the sequence dataset with phylum label available (7.18E5) is significantly lower than the microbiome data used by MetaSource (4.25E9). Nevertheless, this result demonstrated again the generality of the targeted approach built on the inherent linkage of protein families and ecological and evolutional species groups.
Furthermore, we examined the usefulness of modeling sequence distance (e.g., E-value) to guide the homology sequence search. For this, we trained a model named “EvalueSource” to predict what E-value cutoffs should be used for HHblits or hmmer at the step 3 of the DeepMSA search when using a Pfam family as input (see details in SI Appendix, Text S5). As shown in SI Appendix, Fig. S10D, using the predicted E-value cutoff could also result in a slightly higher contact accuracy (0.508 versus 0.496) and TM-score (0.613 versus 0.609), compared with that using the default E-value in DeepMSA, but the difference is not statistically (or marginally) significant with a P value = 0.062 and 0.055 for contact and TM-score, respectively. Meanwhile, the average TM-score is significantly lower than that of MetaSource (0.613 versus 0.625) with a P value = 3.5E-5. Since EvalueSource used the same combined sequence database, it does not result in any search space and time reductions as MetaSource or PhylaSource does. In this context, predicting generic sequence distances such E-value does not seem to be able to generate similar effects as targeted models built on the ecological group labels for improving both accuracy and speed of protein structure prediction procedures. This is probably due to the fluctuation of sequence distances among different protein families, while the inherent linkage between protein families and the ecological species groups could not be captured by the generic sequence distances such as E-value cutoffs.
Finally, we should emphasize that this study only considers four microbiome biomes (Gut, Lake, Soil, and Fermentor) with C-I-TASSER structure modeling method as a proof of concept. Much more metagenome datasets, including other ecological indexes, could be straightforwardly incorporated into this model. Moreover, with the rapid progress of the field, C-I-TASSER considering only contact-map restraints may no longer represents the state of the art of protein structure prediction. We have made brief tests on some of more advanced methods, including DMPfold (42), trRosetta (7), and AlphaFold2 (43), which demonstrated similar enrichments on the performance of structure modeling. Thus, with the rapid accumulation of metagenome sequence databases and the method developments involving more thorough sequence-based restraints armed with more advanced deep-learning methods, we expect that the targeted metagenome selection approach should have more sensitive and pronounced impacts on the efficiency and effectiveness of the protein structure prediction and structure-based protein function annotations.
Materials and Methods
Microbial Community Cohorts Collected from Four Biomes.
We collected metagenome data from the EBI database (https://www.ebi.ac.uk/metagenomics/). We then referred to the EBI database; the microbial niches were annotated in a hierarchical classification tree, named as biome (44). Hence, to cover all the typical biomes, samples under three top-layer biomes were screened: “Engineered” biome (the affiliate biome “Fermentor” was selected as a representative biome), “Environmental” biome (the affiliate biome “Soil” and “Lake” were selected as representative biomes), and “Host-associated” biome (“Gut” biome as representative biome). The samples from Gut biome were collected from human gut covering different countries (SI Appendix, Fig. S11) and animal (mice, pigs, cattle, etc.) intestines.
Since the EBI data has been processed by a different processing pipeline, we reanalyzed the 1,705 metagenomes uniformly using pipeline version 4.1. If the data are processed by the pipeline older than version 4.1, the raw reads were downloaded and performed by the SeqPrep (version 1.2) and Trimmomatic (version 0.35) for quality control. The proteins were then predicted by FragGeneScan (version 1.20) and Prodigal (version 2.6.3). Finally, a total of 4.25 billion protein sequences were collected from the 1,705 high-quality samples. Moreover, the taxonomic profiles were predicted by MAPseq (version 1.2.2), and the functional composition was calculated by InterProScan based on GO annotation (version 5.25-64.0).
Taxonomic and Functional Analysis for Pfam Families.
To decipher the association between microbial communities and Pfam families, the taxonomic and functional distributions for the Pfam families were analyzed. Since the original species for every homology sequence of a Pfam family could be tracked, all the species information (species classification and count number for each species) was obtained from the Pfam database and used as the taxonomic profile. Moreover, the InterPro annotation and their associated GO terms for each family were used as the functional annotation, which was stored in Pfam database. All these data are available at the file transfer protocol (FTP) site, under the release version 32.0 (ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam32.0/).
Pfam Family Dataset Construction for 3D Structure Modeling.
Pfam (version 32) is a database that contains 17,929 protein families, each represented by a hidden Markov model (HMM). Typically, each Pfam entry is comprised of a seed alignment, which forms the basis to build a profile HMM using HMMER (45). The profile HMM is then queried against a sequence database, and all matches scoring above the curated threshold are aligned back to the profile HMM to generate the full alignment. Pfam includes 9,229 protein families that have at least one member with experimentally determined structures. For the remaining 8,700 families, there is no structural information available for any member, where a breakdown of the Pfam families in this study is shown in SI Appendix, Fig. S12.
From the 9,229 known Pfam families, 372 have been randomly selected as a benchmark dataset to investigate the performance of C-I-TASSER and MetaSource. The families are Hard targets as defined by LOMETS (29) since there is no homologous template with a sequence identity <30% in the PDB library. To generate a representative sequence, we searched each of the Pfam families against the SCOPe database, where the best hit with the PDB ID appearing in the Pfam structure member list will be selected. Finally, 168 Pfam families are used for benchmarking C-I-TASSER and the remaining 204 families for testing MetaSource.
Out of the 8,700 unknown families, we first removed the entries with less than 50 amino acids of sequence length, resulting in a set of 8,266 Pfam families. We select one representative sequence for C-I-TASSER modeling for each given Pfam family. To do so, we first ran “HMMsearch” to search the family against the UniRef100 database, with the sequences hit ranked by their E-values. For the best hit with the lowest E-value, we run DeepMSA (10) to build MSA, where 2,251 Pfam families with Neff score >16 (see Neff definition in SI Appendix, Eq. S1) and defined as “Hard” targets by LOMETS are selected for modeling. Finally, the 1,044 Pfam families with model having C-score −2.5 from C-I-TASSER are selected for training MetaSource model (SI Appendix, Fig. S12).
Procedures of the MSA Collection.
To predict the structure and function of the 8,700 unknown Pfam families, the metagenome database of a combination of the four biomes (Gut, Lake, Soil, and Fermentor) were attempted to supplement the Pfam homologous sequence using a three-step procedure outline by DeepMSA (10) (SI Appendix, Fig. S7). In step 1, HHblits (30) from HH-suite is used to search the query sequence against UniClust30 (46) to generate the first-level MSA. In step 2, the Jackhmmer from the HMMER (45) package is used to search the query sequence against UniRef90 (47) to extract full-length sequences (hits), and HHblits is used to convert the full-length sequences into a custom HHblits format database. Starting from the first-level MSA, HHblits is again applied to search this custom database to generate the second level MSA. In step 3, the second level MSA is converted by hmmbuild from the HMMER package into an HMM, and the HMM is then searched against the metagenome sequence database (the combination of the four biomes) by HMMsearch from the HMMER package to extract full-length hits. Similar to step 2, hits from HMMsearch are built into a custom HHblits database. The second level MSA is used to jump-start an HHblits search against this custom HHblits database to get the third level MSA. For each MSA, a Neff score is computed by SI Appendix, Eq. S1, where the families with Neff score ≥16 are selected as “effective Pfam families.” Finally, the homologous sequences in the MSA are collected for further protein structure modeling.
The same MSA construction pipeline is also used for MSA generation for 168 C-I-TASSER benchmark dataset and 204 MetaSource testing dataset. Additionally, for MetaSource testing dataset, we also generated four sets of MSAs which used four biomes as database individually instead of the combined metagenome in DeepMSA step 3. Those five sets MSAs (from four biomes individually and the combined one) of MetaSource testing dataset are used for building five sets of C-I-TASSER models to check the correctness of MetaSource.
Contact-Assisted Structure Prediction by C-I-TASSER.
Based on the collected MSAs, residue–residue contact maps are predicted using five deep-learning and coevolution based predictors, including TripletRes (9), ResTriplet (48), NeBcon (49), ResPRE (50), and ResPLM (19). The consensus contacts are collected from top-L contacts from the five predictors, respectively. These contacts are implemented in the C-I-TASSER simulation through the following potential:
| [1] |
where is the distance between residue pair i and j; is the contact prediction C-score for this residue pair; D is a protein length-dependent parameter to change the gradient of the well, which ranges from 14 to 18 Å (SI Appendix, Fig. S13).
Starting from the representative sequence of a Pfam family, homologous templates are detected from the PDB library by LOMETS metathreading server (29), which consists of 11 individual threading programs, CEthreader (51), CNFsearch (52), FFAS3D (53), HHpred (54), HHsearch (54), MUSTER (55), Neff-MUSTER (5), PPAS (56), PROSPECT2 (57), SP3 (58), and SparksX (59). The consensus contact and distance restraints are collected from the LOMETS template alignments, which are combined with the sequence-based contact potential as Eq. 1 to guide the I-TASSER structural assembly simulations (5). Finally, the decoy conformations from the C-I-TASSER simulation trajectories are clustered by SPICKER (60), where the largest cluster is further refined at atomic level by FG-MD (fragment-guided molecular dynamics) (61) and returned as the final model.
C-I-TASSER Model Quality Estimation.
To estimate the model quality, we run C-I-TASSER on 168 Pfam proteins that have known structures in PDB, where all 168 proteins were “Hard” targets according to LOMETS classification (56). The data in SI Appendix, Fig. S2 demonstrate a strong correlation between the TM-score of the C-I-TASSER models and the C-score of the folding simulations, which has a Pearson correlation coefficient (PCC = 0.801). Here, the C-score is defined by:
| [2] |
where and are the highest Z-score of the templates by the i-th LOMETS2 threading program and the corresponding Z-score cutoff for distinguishing between good and bad templates. These Z-score–related parameters describe the significance of the LOMETS threading alignments. is the satisfaction rate of top-L long-range contacts in the final model, that is, , where is the number of the top-L predicted contacts with residue separation >24, (or 0) if the i-th contact is satisfied (or not satisfied) in the final C-I-TASSER model. measures the degree of structure convergence in the C-I-TASSER simulation and is calculated by , where is the number of decoys in the SPICKER cluster, is the total number of structure decoys generated in C-I-TASSER simulation, and is the average rmsd of the structure decoys to the cluster centroid. Weight parameters () are decided by maximizing the PCC. If we select a C-score cutoff of −2.5, the Matthews correlation coefficient on the benchmark dataset reached a maximum of 0.614 and an FDR of only 6.96% (SI Appendix, Fig. S4).
MetaSource Model Construction and Evaluation for Predicting the Source Biome of Pfam Families.
To identify the source biome that has the largest number of homologous sequences for a given Pfam family, we construct a machine-learning model named MetaSource (https://github.com/HUST-NingKang-Lab/MetaSource). As depicted in SI Appendix, Fig. S14, the pipeline consists of four consecutive steps:
-
1.
Investigation of the biome-sequence–Pfam association: The sequences collected from four biomes (Gut, Lake, Soil, and Fermentor) were used for supplementing the homologous sequences of Pfam families (SI Appendix, Fig. S14A). Furthermore, by comparing the homologous sequence number before and after supplementing the metagenome sequence for Pfam families, the marginal effect analysis was applied to perform a quantitative assessment on the biome-sequence–Pfam association (SI Appendix, Fig. S14C).
-
2.
Training dataset construction using Pfam families with unsolved structure (SI Appendix, Fig. S14D): The Pfam families foldable by C-I-TASSER (e.g., C-score −2.5) are used as the training dataset for the prediction model since the biome genomes have a stronger contribution to the MSA construction for these Pfam families (SI Appendix, Fig. S14B). For the Pfam families foldable by C-I-TASSER, the biome with highest Neff was used as the data label after supplementing the homology sequences from four biomes, respectively. The taxonomic profile on genus level for Pfam families was used as the feature for the training set. To reduce the complexity of the data, the genera with an average relative abundance less than 0.001 were filtered out. Furthermore, to select the features with a significant difference in the distribution of multiple groups, the Kruscal–Wilcox test was performed with a P value over 0.05 and q-value over 0.05 calculated by the Bonferroni method.
-
3.
Constructing the MetaSource model (SI Appendix, Fig. S14E): Given our relatively small dataset, we sought to identify a model that would tend toward low variance, and the random forest algorithm was applied. First, to predict whether the source biome could be one of the four common biomes (Gut, Lake, Soil, Fermentor), a binary random forest algorithm was applied. The positive dataset was 964 Pfam families foldable by C-I-TASSER, and the negative dataset was 7,736 (= 8,700 − 964) Pfam families with unsolved data. Then, to predict the single biome that could effectively supplement homologous sequences for the specific Pfam family, a multilabel random forest classifier was applied using 964 Pfam families foldable by C-I-TASSER as training data. To find the best combination of model parameters, grid search was applied to exhaustively search over all parameter values. Then, the model was trained by 20 cross-validation iterations, and in each iteration, the model was trained on three-fourths of the dataset. Finally, the capacity to predict the source biome that was left out was assessed.
-
4.
Validating the MetaSource using the Pfam families with solved structure (SI Appendix, Fig. S14F): To validate the performance of MetaSource prediction model, the Pfam families with solved structure were used to evaluate the performance of MetaSource on prediction of source biome. For each Pfam family with known structure, we built MSA after querying the homology sequences from Gut, Lake, Soil, Fermenter, and combined four biomes, respectively. Those five MSAs were passed to C-I-TASSER to build five structures individually and then were compared with experimental structure. If the biome with highest Neff is consistent with the prediction source biome using MetaSource, the prediction will be considered correct. All the four steps are implemented by Python, using the scikit-learn package (version 0.22).
Supplementary Material
Acknowledgments
This work is supported in part by the National Institute of General Medical Sciences (grants GM136422, S10OD026825), the National Institute of Allergy and Infectious Diseases (Grant AI134678), the NSF (grants IIS1901191, DBI2030790, MTM2025426), National Natural Science Foundation of China Grants (32071465, 31871334, and 31671374), and Ministry of Science and Technology's National Key Research and Development Program Grant (No. 2018YFC0910502).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110828118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Baker D., Sali A., Protein structure prediction and structural genomics. Science 294, 93–96 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 18, 342–348 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simons K. T., Kooperberg C., Huang E., Baker D., Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268, 209–225 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Xu D., Zhang Y., Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J., et al. , The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senior A. W., et al. , Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Yang J., et al. , Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U.S.A. 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S., Sun S., Li Z., Zhang R., Xu J., Accurate de novo prediction of protein contact map by ultra-deep learning model. PLOS Comput. Biol. 13, e1005324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., et al. , Deducing high-accuracy protein contact-maps from a triplet of coevolutionary matrices through deep residual convolutional networks. PLoS Comput. Biol. 17, e1008865 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C., Zheng W., Mortuza S. M., Li Y., Zhang Y., DeepMSA: Constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins. Bioinformatics 36, 2105–2112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha R., et al. , Assessing the accuracy of contact predictions in CASP13. Proteins 87, 1058–1068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ovchinnikov S., et al. , Protein structure determination using metagenome sequence data. Science 355, 294–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium, UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brünger A. T., et al. , Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., et al. , Fueling ab initio folding with marine metagenomics enables structure and function predictions of new protein families. Genome Biol. 20, 229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen I. A., et al. , The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayers E. W., et al. , GenBank. Nucleic Acids Res. 48, D84–D86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abriata L. A., Tamò G. E., Dal Peraro M., A further leap of improvement in tertiary structure prediction in CASP13 prompts new routes for future assessments. Proteins 87, 1100–1112 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Zheng W., et al. , Deep-learning contact-map guided protein structure prediction in CASP13. Proteins 87, 1149–1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell A. L., et al. , MGnify: The microbiome analysis resource in 2020. Nucleic Acids Res. 48, D570–D578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd-Price J., et al. , IBDMDB Investigators, Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunagawa S., et al. , Tara Oceans Coordinators, Ocean plankton. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Thompson L. R., et al. , Earth Microbiome Project Consortium, A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramanan D., et al. , Helminth infection promotes colonization resistance via type 2 immunity. Science 352, 608–612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckburg P. B., et al. , Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson G. P., et al. , Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole P., Ramachandran V., Terpolilli J., Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Detman A., et al. , Cell factories converting lactate and acetate to butyrate: Clostridium butyricum and microbial communities from dark fermentation bioreactors. Microb. Cell Fact. 18, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng W., et al. , LOMETS2: Improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins. Nucleic Acids Res. 47, W429–W436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remmert M., Biegert A., Hauser A., Söding J., HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Skolnick J., TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flemming H. C., Wuertz S., Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Fenn K., et al. , Quinones are growth factors for the human gut microbiota. Microbiome 5, 161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito T., et al. , Genetic and biochemical analysis of anaerobic respiration in Bacteroides fragilis and its importance in vivo. MBio 11, e03238-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hentchel K. L., et al. , Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater. ISME J. 13, 523–536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber K. A., Achenbach L. A., Coates J. D., Microorganisms pumping iron: Anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Fierer N., Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Jansson J. K., Hofmockel K. S., Soil microbiomes and climate change. Nat. Rev. Microbiol. 18, 35–46 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Sturm G., et al. , Chromate resistance mechanisms in Leucobacter chromiiresistens. Appl. Environ. Microbiol. 84, e02208-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., et al. , Characteristics and in situ remediation effects of heavy metal immobilizing bacteria on cadmium and nickel co-contaminated soil. Ecotoxicol. Environ. Saf. 192, 110294 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Cabello-Yeves P. J., Rodriguez-Valera F., Marine-freshwater prokaryotic transitions require extensive changes in the predicted proteome. Microbiome 7, 117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greener J. G., Kandathil S. M., Jones D. T., Deep learning extends de novo protein modelling coverage of genomes using iteratively predicted structural constraints. Nat. Commun. 10, 3977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell A. L., et al. , EBI Metagenomics in 2017: Enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 46, D726–D735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eddy S. R., Profile hidden Markov models. Bioinformatics 14, 755–763 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Mirdita M., et al. , Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 45, D170–D176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H.; UniProt Consortium, UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y., Zhang C., Bell E. W., Yu D. J., Zhang Y., Ensembling multiple raw coevolutionary features with deep residual neural networks for contact-map prediction in CASP13. Proteins 87, 1082–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He B., Mortuza S. M., Wang Y., Shen H. B., Zhang Y., NeBcon: Protein contact map prediction using neural network training coupled with naïve Bayes classifiers. Bioinformatics 33, 2296–2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Hu J., Zhang C., Yu D.-J., Zhang Y., ResPRE: High-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics 35, 4647–4655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng W., et al. , Detecting distant-homology protein structures by aligning deep neural-network based contact maps. PLOS Comput. Biol. 15, e1007411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., Tagami A., Dobele G., Lindström M. E., Sevastyanova O., The impact of lignin structural diversity on performance of cellulose nanofiber (CNF)-starch composite films. Polymers (Basel) 11, E538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu D., Jaroszewski L., Li Z., Godzik A., FFAS-3D: Improving fold recognition by including optimized structural features and template re-ranking. Bioinformatics 30, 660–667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Söding J., Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Wu S., Zhang Y., MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins 72, 547–556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu S., Zhang Y., LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D., Xu D., Guo J. T., Ellrott K., Xu Y., PROSPECT II: Protein structure prediction program for genome-scale applications. Protein Eng. 16, 641–650 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Hughes C. S., et al. , Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y., Faraggi E., Zhao H., Zhou Y., Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics 27, 2076–2082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Skolnick J., SPICKER: A clustering approach to identify near-native protein folds. J. Comput. Chem. 25, 865–871 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Zhang J., Liang Y., Zhang Y., Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.