Abstract
One strategy for population suppression seeks to use gene drive to spread genes that confer conditional lethality or sterility, providing a way of combining population modification with suppression. Stimuli of potential interest could be introduced by humans, such as an otherwise benign virus or chemical, or occur naturally on a seasonal basis, such as a change in temperature. Cleave and Rescue (ClvR) selfish genetic elements use Cas9 and guide RNAs (gRNAs) to disrupt endogenous versions of an essential gene while also including a Rescue version of the essential gene resistant to disruption. ClvR spreads by creating loss-of-function alleles of the essential gene that select against those lacking it, resulting in populations in which the Rescue provides the only source of essential gene function. As a consequence, if function of the Rescue, a kind of Trojan horse now omnipresent in a population, is condition dependent, so too will be the survival of that population. To test this idea, we created a ClvR in Drosophila in which Rescue activity of an essential gene, dribble, requires splicing of a temperature-sensitive intein (TS-ClvRdbe). This element spreads to transgene fixation at 23 °C, but when populations now dependent on Ts-ClvRdbe are shifted to 29 °C, death and sterility result in a rapid population crash. These results show that conditional population elimination can be achieved. A similar logic, in which Rescue activity is conditional, could also be used in homing-based drive and to bring about suppression and/or killing of specific individuals in response to other stimuli.
Keywords: gene drive, population suppression, selfish genetic element, Drosophila
Gene drive occurs when particular genetic elements are transmitted to viable, fertile progeny at rates greater than those of competing allelic variants or other parts of the genome (reviewed in ref. 1). There has long been interest in the idea that selfish genetic elements mediating gene drive could be used to spread a fitness cost into a population, thereby bringing about population suppression or elimination (2–5). Selfish elements known as homing endonuclease genes (HEGs), which encode a site-specific nuclease (synthetic versions use RNA-guided nucleases such as Cas9 to achieve site specificity), provide one approach to achieving this goal. A HEG is located at the same site in the genome as its target site, thereby disrupting it. When a HEG is present in heterozygotes, the wild-type (WT) allele, which contains an intact target site, is subject to cleavage. If this DNA break is repaired through homologous recombination (HR) using the HEG-bearing chromosome as the template, then the WT allele is converted to a HEG allele, thereby bringing about an increase in the HEG copy number (for a review, see ref. 1). The HEGs observed in nature use multiple methods, including inteins and self-splicing introns, that allow themselves to be copied into highly conserved sequences in essential genes without disrupting the function of these genes (6), thereby minimizing any associated fitness cost. In contrast, in 2003 (3) Austin Burt recognizes that if an engineered HEG inserts itself into a gene essential for viability or female fertility, population suppression can result as the HEG spreads and the frequency of loss-of-function (LOF) homozygotes (bisex or female specific) increases (7–11). Other approaches, some of which also utilize homing, seek to decrease population fitness by driving it to an all-male state by shredding the X chromosome during spermatogenesis (12–17). Population suppression through homing can fail when homing rates are low (7, 8) and/or repair of cleaved target sites in the essential gene results in the creation of uncleavable but functional (resistant) alleles (compare refs. 9, 10, and 18). Ecological factors such as inbreeding and spatial structure may also hinder suppression (19–24). These variables must be determined on a species- and locus-specific basis. Similar considerations apply to the use of Y-linked X shredders, which must also function when present on the highly heterochromatic Y chromosome.
An alternative approach to species-specific population suppression that utilizes gene drive but does not require homing or sex ratio distortion works in two steps. The first utilizes gene drive to spread through a population (population modification) one or more transgenes (Cargo) that confer conditional lethality in response to the presence of an environmental stimulus such as an otherwise benign chemical, infection with a virus, prokaryote or fungus, diapause, or a change in temperature (compare refs. 2, 4, 5, and 25). The second step, population suppression, follows upon introduction of the environmental stimulus. A central challenge with this approach, which has previously been envisioned as involving the activity of a dominantly acting Cargo transgene (compare refs. 2, 4, 5, and 25) is how to ensure the continued function of this (by definition) nonessential gene since LOF mutations that inactivate it will be strongly selected for. Here, we sidestep this problem through a two-part strategy. In the first, Cleave and Rescue–based gene drive (ClvR) is used to bring about population modification. Importantly, a necessary consequence of ClvR drive is that survival or fertility of all members of the modified population becomes dependent on the activity of a ClvR-encoded version of an essential gene (the Rescue). The potential for conditional population suppression is brought about by making the activity of the Rescue conditional. Here, we achieve this by embedding within the Rescue coding region an intein cassette that can only be correctly spliced out of the encoded protein (thereby achieving Rescue activity) at a low, permissive temperature. This strategy disfavors selection for escaper mutations that have lost the conditional feature of Rescue activity since point mutations, insertions, and deletions are most likely to result in loss of intein function, thereby preventing the creation of a functional Rescue protein. Similar considerations apply for related strategies in which the activity of the Rescue is made conditional through incorporation of an N-terminal conditional degron (Discussion). Regardless of the exact mechanism, the key points associated with this LOF-based strategy for conditional suppression are that survival of the population becomes dependent on a drive element–encoded version of an essential gene (the Rescue), the Rescue is engineered to be conditional, and most mutations (though not all; Discussion) in the sequences conferring conditionality will lead to self-elimination because they also disrupt Rescue function.
ClvR Selfish Genetic Elements as a Tool for Conditional Population Suppression.
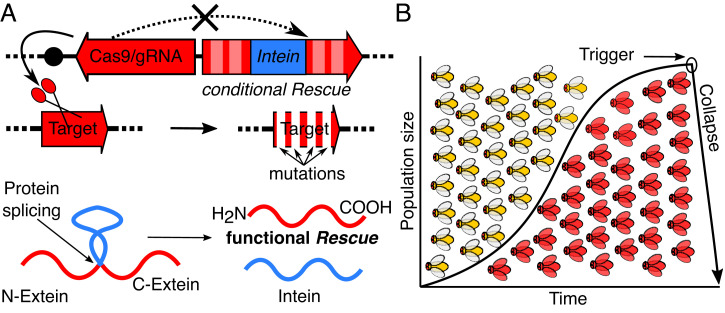
ClvR selfish genetic elements (26, 27) [also referred to as toxin antidote recessive embryo (TARE) in a related proof-of-principle implementation (28)] have two components (Fig. 1A). The first is a DNA sequence–modifying enzyme such as Cas9 and one or more guide RNAs (gRNAs). These constitute the Cleaver, are expressed in the germline, and act in trans to disrupt the endogenous version of an essential gene, creating potentially lethal LOF alleles in the germline and in the zygote due to maternal carryover of active Cas9/gRNA complexes. The second is a recoded version of the essential gene resistant to cleavage that acts in cis to guarantee the survival of those who carry it (the Rescue). The lethal LOF phenotype manifests itself in those who fail to inherit ClvR and have no other functional copies of the essential gene, while those who inherit ClvR and its associated Rescue survive. In this way, as with many other toxin-antidote–based selfish genetic elements found in nature (reviewed in ref. 29) and created de novo (30), ClvR gains a relative transmission advantage that can drive it to genotype (all individuals carry at least one ClvR allele at the locus) or allele (all alleles at the locus carry ClvR) fixation by causing the death of those who lack it (26–28, 31). Importantly, once a ClvR element has spread to genotype fixation (and unlike other selfish elements in nature), all endogenous WT alleles of the essential gene have been eliminated through cleavage and LOF allele creation. At this point, the only source of essential gene function comes from ClvR itself—a form of genetic addiction that creates a state of permanent genotype fixation. In consequence, if function of the Rescue, a kind of Trojan horse now omnipresent in a population, is condition dependent, so too will be the survival of that population.
Fig. 1.
TS-ClvR design and concept. (A) TS-ClvR drive element comprised of Cas9/gRNAs targeting an essential gene and a recoded Rescue (not recognized by Cas9/gRNAs), which includes a TS-intein within its coding region. After translation, the TS-intein can splice itself out to yield a functional Rescue protein. (B) Population suppression with a TS-ClvR. TS-ClvR–bearing flies (red) are released into a WT population (yellow). The TS-ClvR selfish element spreads into the population at the cost of WT. Once the TS-ClvR element has reached genotype fixation (all individuals in the population have at least one copy of TS-ClvR), all functional endogenous copy of the essential gene targeted by TS-ClvR will have been mutated to LOF. At this point, the conditional TS-Rescue within the ClvR element provides the only source of essential gene function in the population, making it subject to a collapse in response to a temperature shift.
Seasonal temperature is an environmental variable that could in principle be used to bring about conditional population suppression in rapidly reproducing species in which there are a number of generations each year. For more slowly reproducing species, in which drive to genotype fixation (and the co-occurring loss of endogenous essential gene function) requires a longer interval, other mechanisms of bringing about conditional lethality or sterility, presumably based on environmental triggers introduced by humans throughout the target population range, would be required (Discussion). Drosophila suzukii, an invasive species of Europe, Asia, and North and South America (32, 33), is one potential target for a temperature-based approach to conditional suppression. It has a number of generations per year and is often invasive in temperate climates that experience large seasonal temperature variations (34), providing opportunities for introducing a temperature-dependent population bottleneck as a method of suppression. As a proof-of-principle demonstration of this idea, we sought to create a version of ClvR in Drosophila melanogaster in which Rescue function is temperature sensitive (TS; TS-ClvR). We show that a TS-ClvR element can successfully spread a conditional Rescue into Drosophila populations at permissive temperatures. When populations now dependent on this transgene are shifted to nonpermissive temperatures, they rapidly become sterile and go extinct.
Results
Insertion of a TS-Intein into the Drosophila Essential Gene dribble (dbe) Results in Temperature-Sensitive LOF.
Traditional approaches to generation of dominant or recessive TS mutations in essential genes in metazoans are laborious as they involve random mutagenesis of whole genomes followed by large-scale screens at different temperatures for otherwise fit TS mutants. As an alternative, we sought to create TS versions of an essential gene by introducing a TS version of an intein into the protein coding sequences of Rescue transgenes within ClvRs previously shown to spread into WT populations (Fig. 1 and refs. 26 and 27). An intein is a protein-encoded autoprocessing domain able to excise itself from a polypeptide and rejoin the N- and C-terminal flanking sequences (exteins) to create a WT version of the encoded protein (35). Importantly, once an intein has been introduced into the coding sequence of an essential gene and that version provides the only source of essential gene function, loss of splicing activity through mutation cannot be selected for since the nonspliced version is nonfunctional.
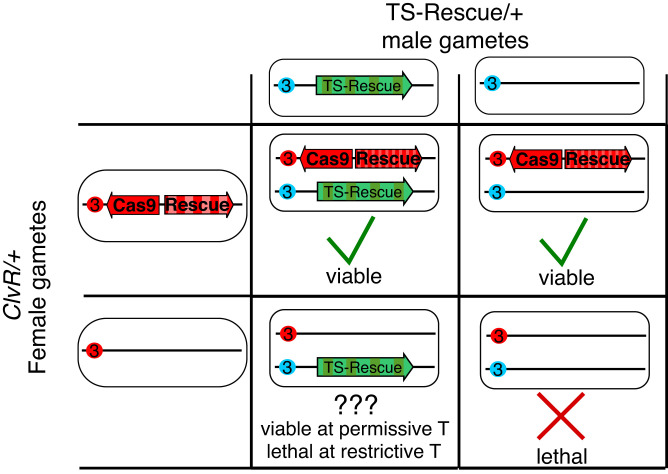
The Sce VMA intein, which is located within the Saccharomyces cerevisiae vacuolar membrane ATPase, is able to excise itself from a number of foreign proteins (36). TS versions of Sce VMA inteins have been isolated that allow spicing at a range of low but not higher temperatures [ranging from 18 to 30 °C (37, 38)]. A mechanistic requirement for successful intein splicing is that the C-terminal extein starts with a cysteine residue. Other less well characterized sequence contexts also regulate splicing efficiency (39–41). To determine if ClvR Rescue genes that contain the Sce VMA intein are functional, we generated six WT- and six TS-intein–bearing versions of Rescue transgenes for two previously described ClvR target genes that each include three cysteines in their coding regions [dribble (dbe), in ClvRdbe (27) and technical knockout (tko), in ClvRtko (26); SI Appendix, Fig. S1D]. We tested the ability of intein-bearing Rescue transgenes to provide essential gene function by examining progeny of a cross between females heterozygous for complete ClvRdbe or ClvRtko elements and males heterozygous for the corresponding WT-intein Rescue (Rescue-INTWT) or TS-intein Rescue (Rescue-INTTS) transgene (Fig. 2).
Fig. 2.
Crossing scheme to identify conditional Rescue transgenes. Maternally inherited (red centromere) and paternally inherited (blue centromere) third chromosomes are indicated.
When present in females, ClvRdbe and ClvRtko cleave and create LOF alleles of their target genes in the maternal germline and the zygote with a frequency of >99.9% (26, 27). Thus, in the absence of another source of Rescue activity, essentially all viable progeny should be ClvR bearing (in an outcross, the 50% that fail to inherit ClvR die because they lack a functional copy of the essential gene). In contrast, if the Rescue-INTWT or Rescue-INTTS in heterozygous males is active, ∼33% of viable progeny should be non–ClvR-bearing (as compared with 0.1% for a full element), and these should all carry the intein-bearing Rescue (Fig. 2). From crosses carried out at 23 and 27 °C, we identified one version of the dbe Rescue that retained function, in which the intein was inserted N-terminal to cysteine 2 of the dbe coding sequence (SI Appendix, Tables S1 and S2). The dbe Rescue transgene carrying the WT-intein was functional at 23 and 27 °C. The Rescue carrying the TS-intein was also functional at 23 °C but was largely (though not completely) nonfunctional at 27 °C (Fig. 3 and SI Appendix, Table S2). Flies carrying the dbe Rescue-INTTS construct were then used as a genetic background in which to create flies carrying a full ClvRdbe-INTTS (referred to as TS-ClvRdbe) drive element carrying the other components found in ClvRdbe (27). These include Cas9 expressed under the control of the germline regulatory sequences from the nanos gene, four gRNAs targeting the endogenous dbe locus expressed under the control of individual U6 promoters, and an OpIE-td-tomato marker gene (SI Appendix, Fig. S1 B and C).
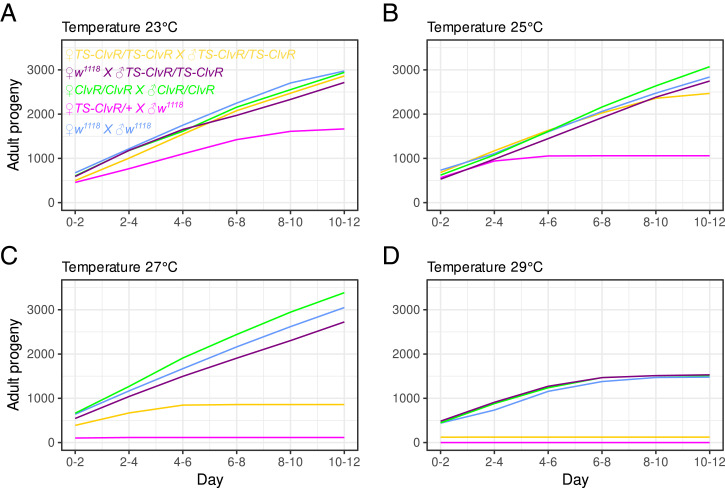
Fig. 3.
Cumulative adult fly output at different temperatures: (A) 23 °C, (B) 25 °C, (C) 27 °C, (D) 20 °C. Shown is the cumulative adult progeny output of four replicates in which five females were crossed to five males over 12 d. Crosses were heterozygous ♀TS-ClvRdbe/+ × ♂w1118 in magenta, homozygous ♀TS-ClvRdbe/TS-ClvRdbe × ♂TS-ClvRdbe/TS-ClvRdbe in yellow, ♀w1118 × ♂TS-ClvRdbe/TS-ClvRdbe in violet, ♀w1118 × ♂w1118 (control) in blue, and the original non-TS ♀ClvRdbe × ♂ClvRdbe (control) in green.
TS-ClvRdbe Efficiently Creates LOF Alleles at Permissive Temperatures.
A TS-ClvR must be able to efficiently create LOF alleles at all relevant environmental temperatures, and Cas9 activity has been shown to be temperature sensitive, with reduced activity at lower temperatures (42, 43). To test the ability of Cas9 to create dbe LOF alleles at temperatures permissive for intein splicing, we crossed heterozygous TS-ClvRdbe females to w1118 (WT) males at 22 °C and scored viable progeny for inheritance of the TS-ClvRdbe marker. If the TS-ClvRdbe Cas9/gRNAs successfully create dbe LOF alleles in the maternal germline and in the early embryo, viable progeny should be largely or exclusively TS-ClvRdbe bearing. ClvR was present in 93.8% of the offspring, a lower frequency than previously reported for the original ClvRdbe [>99% (27)], in which crosses were carried out at 26 °C. This is likely due to reduced Cas9 activity, since similar tests with the original ClvRdbe stock at 22 °C also resulted in a reduced drive inheritance of 95.9% (SI Appendix, Table S3).
To test whether escapers from the cross of female TS-ClvRdbe/+ to male w1118 had acquired functional resistance alleles (an uncleavable but functional version of the endogenous dbe gene) that could thwart drive, we carried out several tests (summarized in SI Appendix, Fig. S2). First, we backcrossed all 91 escaper males (the 93 females were not tested because they were not virgin) to heterozygous TS-ClvRdbe females and found an average drive inheritance rate in progeny of 86.1% (30 did not produce offspring), indicating that the dbe locus was still sensitive to LOF allele creation. After mating, DNA was extracted from all males that were still alive and the dbe target locus sequenced. Results of the sequencing analysis are summarized in SI Appendix, Data S1. In short, all of the sequenced flies retained WT target sites at the dbe locus. Mixed sequencing signals were observed at specific sites from some individuals, probably indicating cleavage of only one copy of the dbe locus. Second, as a way of getting a more general population level feel for the possible existence of undetected escapers or alleles at other loci that prevent LOF allele creation—and thus depress drive and population suppression—we carried out 12 drive experiments that each began with an individual escaper male of particular concern as the starting material. In brief, we selected 12 of the above 61 fertile escaper males that had a lower-than-average bias toward being TS-ClvRdbe bearing. To provide a representative sample, we selected at least one cross from each of the four original bottles (A to D). For each of these 12 crosses (of a non–TS-ClvRdbe-bearing escaper male to heterozygous +/TS-ClvRdbe–bearing females) we collected all male and female offspring (nonvirgin, non-ClvR, and ClvR bearing), moved them to 12 bottles, and carried out multigeneration drive experiments (see SI Appendix, Fig. S2). All populations attained TS-ClvRdbe genotype fixation within three to five generations. To determine if all WT dbe alleles had been rendered LOF once genotype fixation of TS-ClvRdbe was achieved, the 12 TS-ClvRdbe populations were transferred again and incubated at 29 °C. Each bottle (∼300 individuals) produced a few progeny, but none of these produced progeny in the next generation, arguing that all endogenous WT alleles of dbe had been eliminated. All together, these results show that while ClvR drive strength (as measured by LOF allele creation rate) is modestly reduced at low temperature due to reduced levels of Cas9/gRNA activity, this is not associated with the appearance of resistant alleles that prevent drive (SI Appendix, Data S1 and Fig. S2 for workflow).
TS-ClvRdbe Flies Suffer a Temperature-Dependent Loss of Reproductive Output.
In order to bring about condition-dependent population suppression following gene drive–based population modification, carriers must experience a high fitness cost under nonpermissive conditions. A major determinant of fitness is reproductive output. Dbe is a gene whose product is required in all proliferative cells (44). Thus, reproductive output is likely to be a sensitive indicator of dbe function and the effects of dosage at different temperatures. To explore these topics, we characterized the number of adult progeny produced by females having two, one, or no copies of TS-ClvRdbe. Because the only viable progeny of ClvR-bearing mothers are also ClvR bearing (with ClvR now providing the only zygotic source of essential gene function), reproductive output of these females is a function of both maternal and zygotic TS-ClvRdbe Rescue activity at different temperatures. We focused on females because population numbers are generally thought to be limited by female fertility, and adult sexual maturation and ongoing fertility requires cell proliferation and growth of somatic and germline cells. In contrast, young adult males already contain large numbers of mature sperm, which also have a long functional lifetime once deposited in the female reproductive tract (45). Consistent with this, when TS-ClvRdbe males, raised at 23 °C, were transferred to 29 °C as young adults, they showed an age- and temperature-dependent fertility loss similar to that of control w1118 males (SI Appendix, Tables S5 and S6). For each cross involving females, four replicate vials having five females and five males (derived from flies raised at 22 °C) were incubated at different temperatures ranging from 23 to 29 °C and transferred to fresh vials every 2 d. The cumulative adult fly output from these crosses over time is plotted in Fig. 3 (see also SI Appendix, Fig. S3 for individual plots). At the low temperature of 23 °C, crosses between homozygous WT (w1118) flies resulted in the production of progeny at a roughly constant rate, with only a modest drop off in production during days 10 to 12. The rate of offspring production over time was similar for crosses involving homozygous (non-TS) ClvRdbe males and females and for crosses between WT females and homozygous TS-ClvRdbe males, in which progeny carry one WT version of dbe and one TS version. In contrast, crosses between heterozygous TS-ClvRdbe females and WT males produced fewer absolute numbers of progeny. This is expected since the ∼50% of progeny that fail to inherit TS-ClvRdbe die due to lack of essential gene function. The rate of adult offspring production also decreased in the last two intervals (SI Appendix, Fig. S3), suggesting that in an otherwise LOF background, even at permissive temperatures, the combination of one maternal and zygotic copy of the dbe Rescue-INTTS results in some age-dependent loss of fecundity.
At higher temperatures (25 to 27 °C), the loss of reproductive output associated with maternal and zygotic dependence on the TS-ClvRdbe Rescue was more dramatic. At 29 °C, heterozygous TS-ClvRdbe females (the potentially viable progeny of which are heterozygous for TS-ClvRdbe) gave rise to no adult progeny, while homozygous TS-ClvRdbe females (the potentially viable progeny of which are homozygous for TS-ClvRdbe since their mothers were mated with homozygous TS-ClvRdb males) produced only a few viable progeny. Progeny production also ended somewhat prematurely at 29 °C for crosses in which the female parent was WT or ClvRdbe bearing. However, this appears to be a general temperature effect since the ability to produce progeny was lost at a similar rate for both sets of crosses. These results involving crosses of ClvRdbe/+ females to dbe Rescue-INTTS males at different temperatures, and data presented in SI Appendix, Tables S3 and S4 show that individuals carrying TS- ClvRdbe (the vast majority of which lack dbe function from the endogenous locus; SI Appendix, Table S3) are reproductively fit (as inferred by the number of TS-ClvRdbe–bearing adult progeny produced by TS-ClvRdbe–bearing females) at lower but not higher temperatures.
TS-ClvRdbe Spreads to Transgene-Bearing Genotype Fixation at a Permissive Temperature.
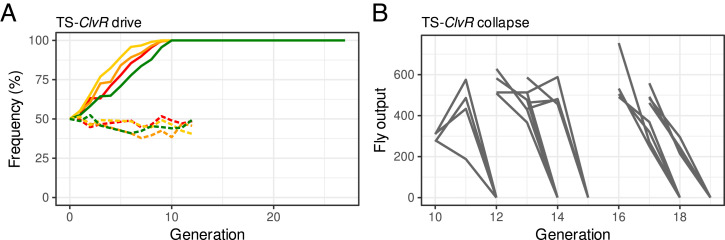
Population modification followed by suppression requires that drive into a WT population succeed at low, permissive temperatures. To test the ability of TS-ClvRdbe to achieve this end, we carried out a gene drive experiment at 22 °C. To seed the drive, we crossed heterozygous TS-ClvRdbe males (w1118; TS-ClvRdbe/+) to WT (w1118) females to create a starting TS-ClvRdbe allele frequency of 25% in four replicate populations. Mated females were allowed to lay eggs in a food bottle for 1 d and removed afterward. The drive experiments were kept in a temperature-controlled incubator at 22 °C. After ∼16 d, most progeny had developed into adults, which were then removed from the bottles, scored for the presence of the TS-ClvRdbe marker (td-tomato), and transferred to a fresh food bottle to repeat the cycle. Results of the drive experiment are shown in Fig. 4A. The TS-ClvRdbe construct reached genotype fixation between 9 and 10 generations in all four replicate drive populations, while a construct carrying only the dbe Rescue-INTTS but no Cas9/gRNAs did not increase in frequency. By generation 18, TS-ClvRdbe allele frequencies ranged from 93.2 to 97.6% (SI Appendix, Table S7).
Fig. 4.
Population modification at a permissive temperature followed by suppression at a restrictive temperature. (A) Shown are genotype frequencies of TS-ClvRdbe–bearing flies over discrete generations at 22 °C. TS-ClvRdbe is indicated with solid lines (replicates in different colors), dbe Rescue-INTTS controls with dashed lines. (B) Gray lines show individual population trajectories for all replicates when incubated at 29 °C. All populations produced some offspring when moved from 22 to 29 °C. These collapsed in the next generation due to complete sterility.
Populations in Which TS-ClvRdbe Is Ubiquitous Undergo a Population Collapse When Shifted to Elevated Temperature.
The goal of drive with a TS-ClvR is ultimately to bring about a population crash in response to an environmental temperature shift once LOF allele creation associated with population modification has rendered all members of the population dependent on the Rescue-INTTS. As a test of this hypothesis, we followed the fate of drive populations shifted to 29 °C at generations 10, 12, 13, 16, and 17. At each of these points, adults from the 22 °C drive population were allowed to lay eggs for 1 d at 22 °C in order to continue the drive and then moved to 29 °C to allow egg laying for a further 2 d. Adults were then removed and the fate of the 29 °C populations followed, as with the drive populations kept at 22 °C (SI Appendix, Table S8). Populations fixed for ClvRdbe (control) individuals produce many adult progeny over six generations when continuously housed at 29 °C (c.f. SI Appendix, Table S9). In contrast, populations of drive individuals—which at this point are heterozygous or homozygous for TS-ClvRdbe—give rise to only a few adult progeny per parent for one more generation (c.f. gray line leading from the number of generation 10 individuals transferred to 29 °C to the generation 11 adult progeny number). These latter adults were universally sterile, resulting in population extinction in the next generation (Fig. 4B).
Discussion
Our results show that gene drive can be used to spread a trait conferring conditional lethality into an insect population, resulting in a population crash when the restrictive condition is experienced, in this case a temperature shift. Additional Cargo genes, designed to bring about some other phenotype such as prevention of disease transmission prior to conditional population suppression could also be included in such gene drive elements. The implementation described herein used the ClvR gene drive mechanism, which concurrently renders LOF endogenous copies of an essential gene and replaces them with a TS version as spread occurs. A similar outcome (drive followed by condition-dependent suppression) could also be achieved using strategies in which a HEG homes into an essential gene locus, thereby disrupting its function, while also carrying a cleavage-resistant version of the essential gene as a rescuing transgene (46–50), that in this case is engineered to be temperature sensitive.
How do TS and related forms of conditional population suppression compare with other transgene-based population suppression approaches? Non-gene drive–based methods such as sterile male release and its variants offer one strategy (51–53), as does inundation with a dominant transgene that mediates killing in response to an environmental trigger, either as a solo entity or as a part of a multilocus self-limiting drive (54–57). These methods are attractive because they are mechanistically straightforward, but they will often have greater costs to implementation than drive-based approaches, since they are not self-sustaining (see refs. 1 and 57 for a more detailed discussion of these issues). As noted in the introduction, gene drive–based approaches that utilize HEGs and X-shredding provide, alone or in combination, a powerful way of bringing about population suppression or elimination in mosquitoes (11, 15, 17). They have a low threshold and can function in the presence of significant fitness costs. However, both have molecular and/or cell biological and genetic requirements that may only be met in some species: high rates of homing for HEGs; X/Y males, and the cell biological conditions that allow a transgene on the Y to bring about cleavage of the X that results in a loss of X-bearing sperm for X shredders. In addition, because drive in these systems results in the direct creation of genotypes that mediate suppression—inviable or sterile homozygotes for HEGs; a male bias for X shredders—other factors can in principle play roles during drive to limit the suppression achieved for specific introduction paradigms. These include population inbreeding, spatial structure, and selection for mutations in trans (at other loci) that suppress drive (19–24). The idea of modification followed by conditional suppression, particularly with drive elements such as ClvR that do not utilize homing to bring about their spread, provides an alternative paradigm, in which drive mediating population modification is separated in time from suppression. In consequence, in such systems (and in HEG-based systems that carry a TS-Rescue version of the essential gene disrupted by the HEG), there is likely to be minimal selection, during drive, for mutations in cis or trans that block the population suppression mechanism. This positive feature of conditional suppression notwithstanding, non-HEG and X-shredding drive mechanisms such as ClvR and various forms of underdominance will typically come with some kind of introduction threshold and spread more slowly than HEGs (in organisms with a high frequency of homing), particularly when rare (1). In addition, effective conditional suppression is contingent on being able to introduce the trigger at the right time and place. Thus, conditional suppression based on temperature sensitivity (TS-ClvR or engineered HEG with a TS-Rescue) requires a temperate climate and is only possible in species (often insects) that have many generations per year, sufficient to bring about drive to genotype fixation prior to onset of a restrictive temperature. Similar points apply to other seasonal variables such as humidity and daylight length. For organisms with a slower reproductive rate, conditional suppression following drive will require different stimuli, probably introduced throughout the target area by humans (though see later in this section for a discussion of strategies that utilize conditional killing following natural infection to eliminate only individuals that carry a specific human pathogen). The challenge with human-introduced triggers for population suppression is that they must be dispersed in such a way as to make timely contact with all individuals in the target population. In the case of a chemical trigger, this condition could apply in already managed environments (agriculture) or perhaps enclosed bodies of water. Viruses, bacteria, and fungi (see discussion below for an example of how a virus might act as a trigger) provide other potentially more self-sustaining and disseminating triggers for larger areas.
The conditional population suppression system described herein targets both males and females since dbe gene activity is required in both sexes. In such a system, complete suppression (population elimination) in response to a temperature shift is possible when ClvR-dependent drive results in transgene-bearing genotype fixation. This is because at this point, all endogenous alleles of the essential gene have been eliminated from the population and essential gene function derives only from the conditional Rescue. With such a system, the target environment may require some level of periodic repopulation with transgenic individuals as a result of incoming migration of WT. A modified system that would reduce this need, and work to maintain the transgene in the target environment in the face of incoming migration of WT, eliminates only females or female fertility under nonpermissive conditions (for modeling of a related system with these characteristics see ref. 58). ClvRs that bring about LOF and Rescue of two different genes, one that is needed for unconditional and sex-independent viability (mediating strong drive) and a second that is conditional and required for female viability or fertility (allowing for elimination of only females under nonpermissive conditions), could be used to achieve this goal. Drives that carry a version of the sex determination gene doublesex (dsx) with a TS-intein in the female-specific exon provide one possible implementation. ClvRs able to rescue the viability and fertility associated with LOF of two different essential genes at the same time have been created (26, 27), suggesting this approach is plausible. Finally, we note that the strategy for generating TS strains described here (replacement of a WT version of an endogenous gene with a TS version) could also be used as a method of engineering sex-specific sorting in inundative suppression strategies such as the sterile insect technique. Many existing SIT systems utilize strains in which TS sex sorting can be carried out (59). That said, these successes were hard won. TS strain creation involved large-scale mutagenesis and screening of many thousands of genotypes, followed by introgression of the TS trait into often complex and specific genetic backgrounds that must be otherwise fit. In many cases, the mutations that result in TS sensitivity remain unknown, limiting the ability to move the trait into related species (59). In cases in which the nature of the TS mutation is known (as for a number of TS mutations in D. melanogaster), the ability to create similar (hopefully also TS lethal) mutations in pest species using Cas9 provides one way to simplify this process (59), though the fitness costs associated with these changes can only be discovered empirically (60).
Success with any TS gene drive system in the wild will require knowledge of temperature fluctuations within a season in the region of interest, the life phases in which the target species is most susceptible (and resistant) to loss of essential gene function, and potentially further selections in rapidly reproducing organisms like yeast (37, 38) for TS-inteins best suited to the environmental temperature regimes involved. Also, because seasonal temperatures do not change in an all or none fashion, gradual shifts toward nonpermissive conditions will provide opportunities for selection to take place on sequences within the intein coding region that reduce or eliminate temperature sensitivity. The targeting of biosynthetic essential genes such as dbe, whose transient LOF is unlikely to result in an immediate fitness cost (as is seen for some other TS mutants that cause immediate paralysis; c.f. ref. 61) probably provides some level of environmental phenotypic buffering in this regard but would not eliminate selection. While next-generation ClvR elements can be cycled through a population, replacing old, failed elements with new ones (27), strategies that forestall the need for such cycles of modification for as long as possible would be useful. This can be achieved by building into the Rescue transgene mechanistic redundancy with respect to how temperature sensitivity is achieved, thereby necessitating multiple mutational hits for the Rescue to lose its TS characteristic. As an example, an N-terminal TS degron (the N-terminal location preventing the loss of degron activity through frameshift or stop codons) that promotes the degradation of a linked C-terminal protein at elevated temperature provides one such approach (62). Insertion of multiple copies of a common TS-intein at different positions provides another.
Regardless of the mechanisms by which redundancy in terms of conditionality are built into a drive system, it will be important to determine the frequency with which mutations in cis (within the TS-Rescue) and in trans (in other genes) arise that can lead to Rescue function that is no longer conditional (suppressor mutations) and if such mutations are present in wild populations. Similar considerations also apply for non-TS conditional systems. In the case of TS-ClvR, dominant suppressor mutations can be identified by mutagenizing males of the TS strain and crossing these to females also of the TS strain (in which all endogenous copies of the essential gene have been rendered LOF) at the permissive temperature. If progeny raised at the nonpermissive temperature survive and are fertile, a suppressor mutation has likely been created. Recessive cis- and transacting suppressors can be identified in similar, albeit more labor-intensive screens. Screens in yeast similar to those used to identify the original TS-intein (37, 38) can also be utilized to identify cis-acting suppressor (revertant) mutations. In addition, it will also be important to explore the behavior of TS ClvRs in populations that originate from diverse ecologies, particularly with respect to environmental temperature range, since these populations could have floating within them alleles of genes encoding proteins such as heat shock proteins/chaperones that may—as a byproduct of activities needed to promote normal protein folding at elevated temperatures—also function as suppressors of TS lethality by promoting the folding and/or stability of TS proteins.
Finally, we note that a similar logic to that presented here, in which Rescue activity is conditionally blocked, could be used to bring about species-specific suppression in response to other stimuli. Small molecules provide one example. These could block intein splicing activity (63), promote the degradation of a target protein (64), or decrease the stability of specific transcripts (65). Target genes that might be particularly amenable to such approaches, which will likely alter expression only transiently following application, include those encoding proteins whose loss results in rapid cell death, such as inhibitors of apoptosis (66). Virus infection provides a further opportunity for engineering conditional lethality. As an example, virus-encoded protease activity, required for viral polyprotein processing in many systems, serves as an “honest” and specific indicator of infection. If one or more viral protease target sites are engineered into the products of key host essential genes—and these versions replace WT counterparts during drive—cleavage at these sites in organisms that are virally infected could result in a lethal LOF phenotype. This could be used to directly suppress populations in response to introduction of a naturally occurring and otherwise benign virus. A similar strategy could also be used to selectively eliminate members of a disease vector population that are infected with a human, animal, or plant pathogenic virus in the context of a simple population modification scenario.
Materials and Methods
Synthesis of TS-Rescues for tko and dbe Target Genes.
All constructs in this work were assembled with Gibson cloning (67). Enzymes were from NEB and cloning and DNA extraction kits from Zymo. Inteins were gene synthesized as gblocks from IDT. We started from our previously cloned Rescue constructs (26, 27). The Rescue for tko was derived from the ortholog of Drosophila virilis, the one for dbe from D. suzukii. Both genes have three cysteines in their coding sequences. We used Gibson assembly to insert a WT-intein and a TS-intein [mutation D324G (37, 38)] after each of the cysteines for a total of 12 constructs. In addition, the plasmids had a dominant OpIE–green fluorescent protein (GFP) marker, an attP site, and homology arms to facilitate CRISPR-HR–mediated insertion into the fly genome at the 68E map position on chromosome 3.
The constructs were injected into w1118 flies along with a preloaded Cas9/gRNA RNP complex having a gRNA (both from IDT) targeting chromosome 3 at 68E (SI Appendix, Fig. S1A). Details were as described previously (27). All Gibson cloning primers and construct GenBank files are in SI Appendix, Data S1. Embryonic injections were carried out by Rainbow Transgenic Flies. Injected G0 flies were outcrossed to w1118 and screened for ubiquitous GFP expression.
Screening Crosses for Temperature-Dependent Rescue Activity.
To determine if any of the intein-bearing Rescues showed temperature-dependent Rescue activity, we set up crosses between heterozygous virgins that carry the original non-TS ClvR element and heterozygous males carrying the different Rescue-INT(TS or WT) versions (crossing scheme in Fig. 2). All crosses were set up in triplicates and incubated at 23 or at 27 °C. None of the intein-Rescues for tko were able to provide adequate gene function at either temperature (SI Appendix, Table S1). For dbe, the Rescue transgenes carrying the WT-intein inserted after cysteine 2 and 3 were able to rescue flies at both temperatures. Rescue transgenes containing the TS-intein inserted after cysteines 1 or 3 were not able to provide Rescue function at either temperature. In contrast, Rescue transgenes carrying the TS-intein inserted after cysteine 2 showed promising behavior, with most progeny dying at 27 °C but not at 23 °C (SI Appendix, Table S2, highlighted in red). We used these flies to build a fully functional TS-ClvR selfish element. Note: For the WT-intein inserted after cysteine 1 of dbe, we did not obtain transformants after a first round of injections. Since the TS-intein version of that construct did not show Rescue activity, this insertion position was not further pursued.
Synthesis of TS-ClvRdbe Flies.
Cas9 and a set of four gRNAs (each driven by a U6 promoter) that target endogenous alleles of dbe were integrated into the attP site within the TS-intein Rescue construct, as described previously (26, 27). The gRNA scaffolds were optimized as described previously by replacing the T base at position 4 with a G and extending the duplex by 5 bp (68, 69).
The construct was modified further using Gibson assembly to add in a new OpIE-td-tomato marker gene (the original plasmid had a 3xP3-GFP marker that would have been hard to screen for in the ubiquitous GFP background of the TS-Rescue–carrying flies) and was injected into flies carrying the TS-Rescue alongside a helper plasmid providing a source of PhiC31 integrase (Rainbow Transgenic Flies) (SI Appendix, Fig. S1B). Injected G0 flies were outcrossed to w1118 and screened for ubiquitous td-tomato expression. Positive transformants were balanced over TM3, Sb to subsequently generate a homozygous stock of TS-ClvRdbe flies carrying the TS-Rescue and Cas9/gRNAs (SI Appendix, Fig. S1C). Primers and construct GenBank files are in SI Appendix, Data S1.
Crosses to Determine Cleavage to LOF of TS-ClvRdbe.
We crossed homozygous TS-ClvRdbe and ClvRdbe (control) males to w1118 virgins to generate heterozygous offspring. Heterozygous TS-ClvRdbe (or ClvRdbe control) virgins were crossed to w1118 males, incubated at a permissive temperature of 22 °C, and the offspring was scored for the presence of the dominant TS-ClvRdbe marker. Results are shown in SI Appendix, Table S3.
Analysis of Escapers.
From the experiment to determine cleavage to LOF, we recovered 91 males that did not carry the TS-ClvRdbe marker. We also recovered 72 males that did not carry the ClvRdbe marker from the control crosses with the original ClvRdbe flies. All of them were crossed to heterozygous TS-ClvRdbe/+ (or ClvRdbe for the controls) females and incubated at 22 °C again. After they mated, we took the male out of each vial and extracted genomic DNA. We amplified an amplicon spanning all four cut sites within the endogenous dbe locus and sequenced it. The offspring of the crosses was again scored for the presence of the TS-ClvRdbe (or ClvRdbe) marker. Afterward, we selected 12 vials with low cleavage to LOF rates and transferred all the offspring to a food bottle to start a gene drive experiment as described below. In these gene drive experiments, we did not score marker frequencies. The drive experiment was continued until TS-ClvRdbe (or ClvRdbe controls) reached genotype fixation in all bottles. This took from three to five generations. Bottles with TS-ClvRdbe were subsequently transferred again and incubated at 29 °C to test if a population collapse could be induced. All results with a more detailed description are shown in SI Appendix, Data S1. The populations did crash, indicating that no functional endogenous alleles exist in these drive populations.
Crosses to Test for Temperature-Dependent Rescue Function of TS-ClvRdbe.
We set up crosses involving females and males (all reared at 22 °C) of the following genotypes: homozygous TS-ClvRdbe (10 vials), w1118 (control, 5 vials), and ClvRdbe (control, 5 vials). These were incubated at a potentially restrictive temperature of 29 °C. Offspring output of generations F1 and F2 are shown in SI Appendix, Table S4.
Crosses to Determine Fecundity of TS-ClvRdbe Flies over a Range of Temperatures.
We set up five different crosses (genotypes below). These included five females and five males (four replicates) that had been reared at 22 °C. After setting up the cross, the vials were incubated at 23, 25, 27, and 29 °C. Every 48 h, adults were transferred to a fresh food vial, and this was repeated five times. We scored the adult fly output in each of these vials. Results are shown in Fig. 3 and SI Appendix, Fig. S3. Crosses were as follows:
♀TS-ClvRdbe/+ × ♂w1118,
♀TS-ClvRdbe/TS-ClvRdbe × ♂TS-ClvRdbe/TS-ClvRdbe,
♀w1118 × ♂TS-ClvRdbe/TS-ClvRdbe,
♀ClvRdbe × ♂ClvRdbe (control), and
♀w1118 × ♂w1118 (control).
Gene Drive Experiment.
We seeded four replicate populations by crossing heterozygous TS-ClvRdbe/+ males (or Rescue-INTTS/+ that do not have Cas9/gRNAs as a control) to w1118 females (25% starting allele frequency). Flies were placed in food bottles, incubated at 22 °C, and allowed to lay eggs for 1 d. Afterward, they were removed from the bottles, and the eggs were allowed to develop into adults. After ∼16 to 17 d, a large number had eclosed as adults. These were anesthetized on a CO2-pad, scored for the dominant TS-ClvRdbe marker, and transferred to a fresh food bottle to repeat the cycle. Counts are in SI Appendix, Data S1.
TS-ClvRdbe and ClvRdbe (Control) Populations at 29 °C.
After the TS-ClvRdbe flies in the gene drive experiment reached genotype fixation (generation 10 and following), we first transferred them to a fresh food bottle to continue the gene drive experiment. After they laid eggs in that bottle for 1 d, we transferred them again to a fresh bottle. That second bottle was now incubated at 29 °C. Flies were given 2 d to lay eggs in that bottle before they were removed again. Eggs were allowed to develop into adults that were then scored and put in a fresh food bottle that was again kept at 29 °C. Flies were kept in that bottle for 1 wk prior to removal, so as to maximize the number of eggs laid. However, no progeny developed within these bottles. Results are shown in Fig. 4B (gray lines) and SI Appendix, Table S8.
As a control experiment, we used the previously characterized ClvRdbe stock, which carries a WT copy of the recoded Rescue (27). ClvRdbe flies were taken from a gene drive experiment [generation 44 (27)], transferred to a fresh food bottle, and incubated alongside the TS-ClvRdbe bottles at 29 °C. They were allowed to lay eggs for 2 d, after which adults were removed. After the eggs developed into adults, we determined the adult population number and transferred these individuals to a fresh food bottle to repeat the cycle. This was repeated for a total of six transfers with no obvious reduction in population size. Results are shown in SI Appendix, Table S9.
Supplementary Material
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This work was carried out with support to B.A.H. from the US Department of Agriculture, National Institute of Food and Agriculture (NIFA) specialty crop initiative under US Department of Agriculture NIFA Award No. 2012-51181-20086 and the California Institute of Technology (Caltech) Resnick Sustainability Institute. G.O. was supported by a Baxter Foundation Endowed Senior Postdoctoral Fellowship and the Caltech Resnick Sustainability Institute. T.I. was supported by NIH Training Grant No. 5T32GM007616-39.
Footnotes
This paper results from the NAS Colloquium of the National Academy of Sciences, “Life 2.0: The Promise and Challenge of a CRISPR Path to a Sustainable Planet,” held December 10–11, 2019, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. NAS colloquia began in 1991 and have been published in PNAS since 1995. The complete program and video recordings of presentations are available on the NAS website at http://www.nasonline.org/CRISPR. The collection of colloquium papers in PNAS can be found at https://www.pnas.org/cc/the-promise-and-challenge-of-a-crispr-path.
Competing interest statement: The authors have filed patent applications on ClvR and related technologies (US Application No. 15/970,728 and No. 16/673,823; provisional patent No. CIT-8511-P).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107413118/-/DCSupplemental.
Data Availability
A ll study data are included in the article and/or supporting information.
References
- 1.Hay B. A., Oberhofer G., Guo M., Engineering the composition and fate of wild populations with gene drive. Annu. Rev. Entomol. 66, 407–434 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Whitten M. J., Insect control by genetic manipulation of natural populations. Science 171, 682–684 (1971). [DOI] [PubMed] [Google Scholar]
- 3.Burt A., Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaChance L. E., Knipling E. F., Control of insect populations through genetic manipulations. Ann. Entomol. Soc. Am. 55, 515–520 (1962). [Google Scholar]
- 5.Schliekelman P., Gould F., Pest control by the introduction of a conditional lethal trait on multiple loci: Potential, limitations, and optimal strategies. J. Econ. Entomol. 93, 1543–1565 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Stoddard B. L., Homing endonucleases from mobile group I introns: Discovery to genome engineering. Mob. DNA 5, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deredec A., Burt A., Godfray H. C. J., The population genetics of using homing endonuclease genes in vector and pest management. Genetics 179, 2013–2026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godfray H. C. J., North A., Burt A., How driving endonuclease genes can be used to combat pests and disease vectors. BMC Biol. 15, 81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond A., et al. , A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond A. M., et al. , The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13, e1007039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyrou K., et al. , A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galizi R., et al. , A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5, 3977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galizi R., et al. , A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci. Rep. 6, 31139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fasulo B., et al. , A fly model establishes distinct mechanisms for synthetic CRISPR/Cas9 sex distorters. PLoS Genet. 16, e1008647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simoni A., et al. , A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol. 38, 1054–1060 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meccariello A., et al. , Engineered sex ratio distortion by X-shredding in the global agricultural pest Ceratitis capitata. BMC Biol. 19, 78 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond A., et al. , Gene-drive suppression of mosquito populations in large cages as a bridge between lab and field. Nat. Commun. 12, 4589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.KaramiNejadRanjbar M., et al. , Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc. Natl. Acad. Sci. U.S.A. 115, 6189–6194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull J. J., Lethal gene drive selects inbreeding. Evol. Med. Public Health 2017, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bull J. J., Remien C. H., Gomulkiewicz R., Krone S. M., Spatial structure undermines parasite suppression by gene drive cargo. PeerJ 7, e7921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhole S., Lloyd A. L., Gould F., Gene drive dynamics in natural populations: The importance of density dependence, space, and sex. Annu. Rev. Ecol. Evol. Syst. 51, 505–531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomulkiewicz R., Thies M. L., Bull J. J., Evading resistance to gene drives. Genetics 217, iyaa040 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champer J., Kim I. K., Champer S. E., Clark A. G., Messer P. W., Suppression gene drive in continuous space can result in unstable persistence of both drive and wild-type alleles. Mol. Ecol. 30, 1086–1101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook F., Bull J. J., Gomulkiewicz R., Gene drive escape from resistance depends on mechanism and ecology. bioRxiv [Preprint] (2021). (Accessed 5 September 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould F., Schliekelman P., Population genetics of autocidal control and strain replacement. Annu. Rev. Entomol. 49, 193–217 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Oberhofer G., Ivy T., Hay B. A., Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. U.S.A. 116, 6250–6259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberhofer G., Ivy T., Hay B. A., Gene drive and resilience through renewal with next generation Cleave and Rescue selfish genetic elements. Proc. Natl. Acad. Sci. U.S.A. 117, 9013–9021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Champer J., et al. , A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burga A., Ben-David E., Kruglyak L., Toxin-antidote elements across the tree of life. Annu. Rev. Genet. 54, 387–415 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Chen C.-H., et al. , A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316, 597–600 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Champer J., Kim I. K., Champer S. E., Clark A. G., Messer P. W., Performance analysis of novel toxin-antidote CRISPR gene drive systems. BMC Biol. 18, 27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ørsted I. V., Ørsted M., Species distribution models of the Spotted Wing Drosophila (Drosophila suzukii, Diptera: Drosophilidae) in its native and invasive range reveal an ecological niche shift. J. Appl. Ecol. 56, 423–435 (2019). [Google Scholar]
- 33.Reyes J. A., Lira-Noriega A., Current and future global potential distribution of the fruit fly Drosophila suzukii (Diptera: Drosophilidae). Can. Entomol. 152, 587–599 (2020). [Google Scholar]
- 34.Little C. M., Chapman T. W., Hillier N. K., Plasticity is key to success of Drosophila suzukii (Diptera: Drosophilidae) invasion. J. Insect Sci. 20, 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills K. V., Johnson M. A., Perler F. B., Protein splicing: How inteins escape from precursor proteins. J. Biol. Chem. 289, 14498–14505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kane P. M., et al. , Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science 250, 651–657 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Zeidler M. P., et al. , Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 22, 871–876 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Tan G., Chen M., Foote C., Tan C., Temperature-sensitive mutations made easy: Generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges. Genetics 183, 13–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong S., Williams K. S., Wotkowicz C., Xu M. Q., Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J. Biol. Chem. 273, 10567–10577 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Hirata R., Anraku Y., Mutations at the putative junction sites of the yeast VMA1 protein, the catalytic subunit of the vacuolar membrane H(+)-ATPase, inhibit its processing by protein splicing. Biochem. Biophys. Res. Commun. 188, 40–47 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Cooper A. A., Stevens T. H., Protein splicing: Excision of intervening sequences at the protein level. BioEssays 15, 667–674 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Xiang G., Zhang X., An C., Cheng C., Wang H., Temperature effect on CRISPR-Cas9 mediated genome editing. J. Genet. Genomics 44, 199–205 (2017). [DOI] [PubMed] [Google Scholar]
- 43.LeBlanc C., et al. , Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 93, 377–386 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Chan H. Y., Brogna S., O’Kane C. J., Dribble, the Drosophila KRR1p homologue, is involved in rRNA processing. Mol. Biol. Cell 12, 1409–1419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnakenberg S. L., Siegal M. L., Bloch Qazi M. C., Oh, the places they’ll go: Female sperm storage and sperm precedence in Drosophila melanogaster. Spermatogenesis 2, 224–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M., Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Champer J., et al. , A CRISPR homing gene drive targeting a haplolethal gene removes resistance alleles and successfully spreads through a cage population. Proc. Natl. Acad. Sci. U.S.A. 117, 24377–24383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adolfi A., et al. , Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat. Commun. 11, 5553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandul N. P., Liu J., Bennett J. B., Marshall J. M., Akbari O. S., A confinable home-and-rescue gene drive for population modification. eLife 10, e65939 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham T. B., et al. , Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLoS Genet. 15, e1008440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kandul N. P., et al. , Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun. 10, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das S. R., Maselko M., Upadhyay A., Smanski M. J., Genetic engineering of sex chromosomes for batch cultivation of non-transgenic, sex-sorted males. PLoS Genet. 16, e1009180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobson S. L., When more is less: Mosquito population suppression using sterile, incompatible and genetically modified male mosquitoes. J. Med. Entomol. 58, 1980–1986 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Gould F., Huang Y., Legros M., Lloyd A. L., A killer-rescue system for self-limiting gene drive of anti-pathogen constructs. Proc. Biol. Sci. 275, 2823–2829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noble C., et al. , Daisy-chain gene drives for the alteration of local populations. Proc. Natl. Acad. Sci. U.S.A. 116, 8275–8282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webster S. H., Vella M. R., Scott M. J., Development and testing of a novel killer-rescue self-limiting gene drive system in Drosophila melanogaster. Proc. Biol. Sci. 287, 20192994 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oberhofer G., Ivy T., Hay B. A., Split versions of Cleave and Rescue selfish genetic elements for measured self limiting gene drive. PLoS Genet. 17, e1009385 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akbari O. S., et al. , Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth. Biol. 3, 915–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen T. N. M., Choo A., Baxter S. W., Lessons from Drosophila: Engineering genetic sexing strains with temperature-sensitive lethality for sterile insect technique applications. Insects 12, 243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choo A., et al. , Precise single base substitution in the shibire gene by CRISPR/Cas9-mediated homology directed repair in Bactrocera tryoni. BMC Genet. 21, 127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Bliek A. M., Meyerowitz E. M., Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature 351, 411–414 (1991). [DOI] [PubMed] [Google Scholar]
- 62.Faden F., et al. , Phenotypes on demand via switchable target protein degradation in multicellular organisms. Nat. Commun. 7, 12202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z., et al. , Small-molecule inhibitors for the Prp8 intein as antifungal agents. Proc. Natl. Acad. Sci. U.S.A. 118, e2008815118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verma R., Mohl D., Deshaies R. J., Harnessing the power of proteolysis for targeted protein inactivation. Mol. Cell 77, 446–460 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Umuhire Juru A., Cai Z., Jan A., Hargrove A. E., Template-guided selection of RNA ligands using imine-based dynamic combinatorial chemistry. Chem. Commun. (Camb.) 56, 3555–3558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S. L., Hawkins C. J., Yoo S. J., Müller H. A., Hay B. A., The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98, 453–463 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Dang Y., et al. , Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oberhofer G., Ivy T., Hay B. A., Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. U.S.A. 115, E9343–E9352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A ll study data are included in the article and/or supporting information.