Significance
Activated protein C (aPC) is a promising therapeutic and exhibits multiple pharmacological benefits in preclinical studies, including in sepsis. aPC activates protease-activated receptor-1 (PAR1) and signals via β-arrestin-2 to promote endothelial barrier stabilization and anti-inflammatory and anti-apoptotic activities. The present study demonstrates that different aPC/PAR1 cytoprotective responses are uniquely regulated by discrete β-arrestin-2–mediated signaling pathways that are modulated by coreceptors. Thus, β-arrestin-2 functions as a central driver of distinct, aPC/PAR1-induced cytoprotective signaling pathways to control barrier protection and anti-apoptotic activities.
Keywords: biased signaling, cytoprotection, GPCR, endothelial dysfunction
Abstract
Endothelial dysfunction is associated with vascular disease and results in disruption of endothelial barrier function and increased sensitivity to apoptosis. Currently, there are limited treatments for improving endothelial dysfunction. Activated protein C (aPC), a promising therapeutic, signals via protease-activated receptor-1 (PAR1) and mediates several cytoprotective responses, including endothelial barrier stabilization and anti-apoptotic responses. We showed that aPC-activated PAR1 signals preferentially via β-arrestin-2 (β-arr2) and dishevelled-2 (Dvl2) scaffolds rather than G proteins to promote Rac1 activation and barrier protection. However, the signaling pathways utilized by aPC/PAR1 to mediate anti-apoptotic activities are not known. aPC/PAR1 cytoprotective responses also require coreceptors; however, it is not clear how coreceptors impact different aPC/PAR1 signaling pathways to drive distinct cytoprotective responses. Here, we define a β-arr2–mediated sphingosine kinase-1 (SphK1)-sphingosine-1-phosphate receptor-1 (S1PR1)-Akt signaling axis that confers aPC/PAR1-mediated protection against cell death. Using human cultured endothelial cells, we found that endogenous PAR1 and S1PR1 coexist in caveolin-1 (Cav1)–rich microdomains and that S1PR1 coassociation with Cav1 is increased by aPC activation of PAR1. Our study further shows that aPC stimulates β-arr2–dependent SphK1 activation independent of Dvl2 and is required for transactivation of S1PR1-Akt signaling and protection against cell death. While aPC/PAR1-induced, extracellular signal–regulated kinase 1/2 (ERK1/2) activation is also dependent on β-arr2, neither SphK1 nor S1PR1 are integrated into the ERK1/2 pathway. Finally, aPC activation of PAR1-β-arr2–mediated protection against apoptosis is dependent on Cav1, the principal structural protein of endothelial caveolae. These studies reveal that different aPC/PAR1 cytoprotective responses are mediated by discrete, β-arr2–driven signaling pathways in caveolae.
Endothelial dysfunction, a hallmark of inflammation, is associated with the pathogenesis of vascular diseases and results in endothelial barrier disruption and increased sensitivity to apoptosis (1, 2). There are limited treatment options for improving endothelial dysfunction, which is prevalent in diseases such as sepsis, a condition with high morbidity and mortality (3, 4). Activated protein C (aPC) is a promising therapeutic that exhibits multiple pharmacological benefits in preclinical studies, including sepsis (5–7). In endothelial cells, protease-activated receptor-1 (PAR1), a G protein–coupled receptor (GPCR), is the central mediator of aPC cytoprotective responses, including endothelial barrier stabilization, anti-inflammatory, and anti-apoptotic activities (6). The signaling pathways by which aPC/PAR1 elicits different cytoprotective responses are poorly defined.
aPC-dependent endothelial cytoprotection requires compartmentalization of PAR1 and the aPC coreceptor, endothelial protein C receptor (EPCR), in caveolin-1 (Cav1)–rich microdomains (8, 9). aPC activates PAR1 through the proteolytic cleavage of the receptor’s N-terminal arginine (R)-46 residue, which is distinct from the thrombin canonical cleavage site at (R)-41 (10). Several studies indicate that aPC/PAR1 requires β-arrestin-2 (β-arr2) to promote cytoprotection (11–13). We showed that aPC-activated PAR1 signals via β-arr2 and dishevelled-2 (Dvl2) scaffolds, and not heterotrimeric G proteins, to induce Rac1 activation and endothelial barrier protection (11). β-arr2 and Dvl2 are also required for aPC-mediated inhibition of cytokine-induced immune cell recruitment, an anti-inflammatory response (12). In addition, aPC/PAR1 stimulates Akt signaling and protects against endothelial cell death induced by tumor necrosis factor-alpha (TNF-α) and staurosporine (14, 15). However, the role of β-arr2 and Dvl2 scaffolds in mediating aPC/PAR1 anti-apoptotic responses is not known.
The interaction of GPCRs with coreceptors can alter the active conformation of receptors, β-arrestin recruitment, and biased signaling (16) and is relevant to aPC/PAR1-driven endothelial cytoprotective signaling. aPC-activated PAR1 cooperates with PAR3 and sphingosine-1-phosphate receptor-1 (S1PR1) to promote cytoprotection (17–19). aPC cleaves PAR3 at a noncanonical N-terminal (R)-41 site to promote endothelial barrier protection in vitro and in vivo (19). In contrast to PAR3, aPC signals indirectly to S1PR1 to enhance basal endothelial barrier stabilization and to protect against barrier disruption (17, 18). However, the mechanism by which aPC/PAR1 transactivates S1PR1 and the role of S1PR1 in other aPC-mediated cytoprotective responses, such as cell survival, is not known.
In this study, we assessed whether S1PR1 and the β-arr2 and Dvl2 scaffolds function as universal mediators of aPC/PAR1 cytoprotection by examining their function in anti-apoptotic responses. Using a combined pharmacological inhibitor and small interfering (si)RNA knockdown approach in human cultured endothelial cells, we define a β-arr2-sphingosine kinase-1 (SphK1)-S1PR1-Akt signaling axis that confers aPC/PAR1-mediated protection against cell death. Our studies further demonstrate that aPC-stimulated activation of SphK1 is dependent on β-arr2 and not Dvl2, whereas neither SphK1 nor S1PR1 are required for aPC-β-arr2–induced, extracellular signal–regulated kinase 1/2 (ERK1/2) signaling. This study reveals that different aPC/PAR1 cytoprotective responses are mediated by discrete β-arr2–driven signaling pathways modulated by coreceptors localized in caveolae.
Results
PAR1 and S1PR1 Colocalize in Caveolae.
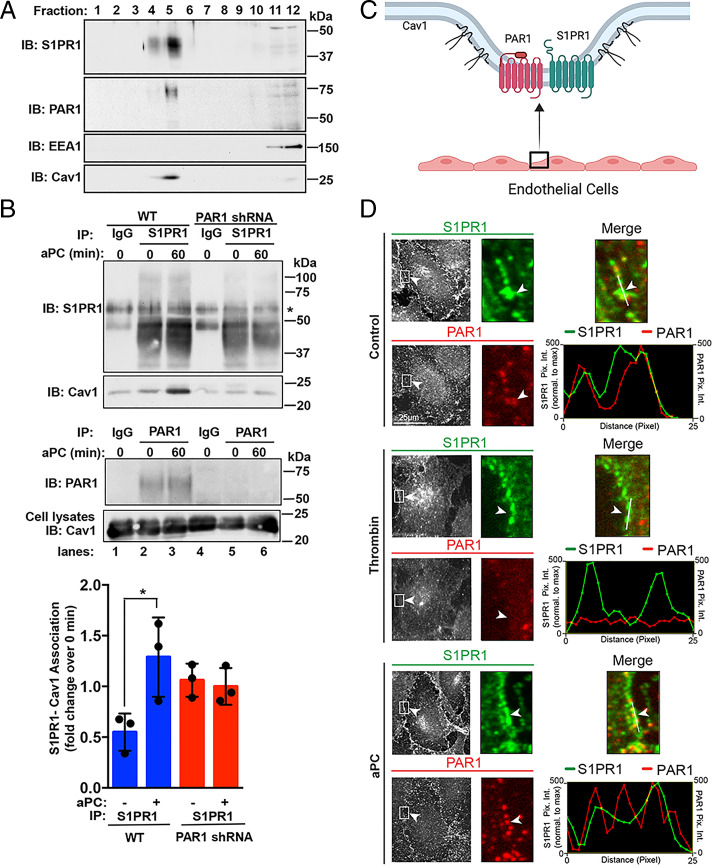
Enrichment of GPCRs in caveolae augments cell signaling efficiency and specificity (20). Cav1 is a structural protein essential for caveolae formation and modulates the activity of signaling molecules (21). PAR1 and EPCR have been shown to localize to caveolae, which is required for aPC-stimulated endothelial barrier protection (8, 9). This prompted us to investigate whether S1PR1 localized to caveolae in endothelial cells. The localization of endogenous S1PR1 and PAR1 in Cav1-enriched fractions was examined in human umbilical vein endothelial cell (HUVEC)–derived EA.hy926 cells using sucrose gradient fractionation. PAR1 segregated into Cav1-enriched fractions, as we reported previously (Fig. 1A) (11). S1PR1 was also detected in Cav1-enriched fractions (Fig. 1A), suggesting that both PAR1 and S1PR1 reside in caveolae in human cultured endothelial cells. Next, we examined coassociation of S1PR1 with Cav1 by immunoprecipitation. Endothelial EA.hy926 cells were stimulated with aPC, S1PR1 was immunoprecipitated, and Cav1 was detected by immunoblotting. aPC induced a significant increase in Cav1 interaction with S1PR1, compared to unstimulated cells (Fig. 1B, lanes 2 and 3). However, in PAR1-deficient cells, aPC failed to enhance S1PR1-Cav1 interaction over control cells (Fig. 1B, lanes 5 and 6). These data suggest that PAR1 and S1PR1 are both localized in caveolae (Fig. 1C) and that PAR1 is required for aPC-induced S1PR1 coassociation with Cav1.
Fig. 1.
PAR1 and S1PR1 coexist in caveolae and colocalize at the plasma membrane. (A) EA.hy926 cell lysates and subjected to sucrose gradient fractionation. Fractions were immunoblotted (IB) for S1PR1, PAR1, EEA1, and Cav1. (B) EA.hy926 cells wild-type (WT) and PAR1 shRNA-expressing cells were treated with 20 nM aPC, immunoprecipitated, and immunoblotted as shown. Data (mean ± SD, n = 3) were analyzed by t test (*P < 0.01). (C) Schematic of endothelial cell PAR1 and S1PR1 colocalization in caveolae. (D) EA.hy926 cells were treated with 10 nM α-thrombin or aPC for 60 min or untreated (Control) and immunostained for endogenous PAR1 (red) and S1PR1 (green), and colocalization was assessed by immunofluorescence confocal microscopy. (Insets) Magnifications of the cell periphery (white arrowhead) boxed area. Line scan analysis was performed in ImageJ to assess PAR1 and S1PR1 colocalization and relative to S1PR1. (Scale bar, 25 µm.)
We next examined PAR1 and S1PR1 colocalization in endothelial cells using immunofluorescence confocal microscopy. PAR1 and S1PR1 localized at the plasma membrane in unstimulated control cells and remained at the cell surface after aPC treatment, suggesting that neither PAR1 nor S1PR1 are internalized (Fig. 1D). This observation is consistent with previous studies showing aPC stimulation fails to promote PAR1 internalization in endothelial cells (9, 22). Unlike aPC, thrombin promoted PAR1 internalization resulting in diminished PAR1-S1PR1 colocalization (Fig. 1D). Thus, PAR1 and S1PR1 coexist in caveolae and remain at the cell surface after prolonged aPC stimulation (Fig. 1D). While S1PR1 has been implicated in aPC-mediated endothelial barrier stabilization (17, 18), the role of S1PR1 in aPC/PAR1-driven anti-apoptotic responses in endothelial cells is not known and was examined next.
S1PR1 Mediates aPC-PAR1 Anti-apoptotic Activity.
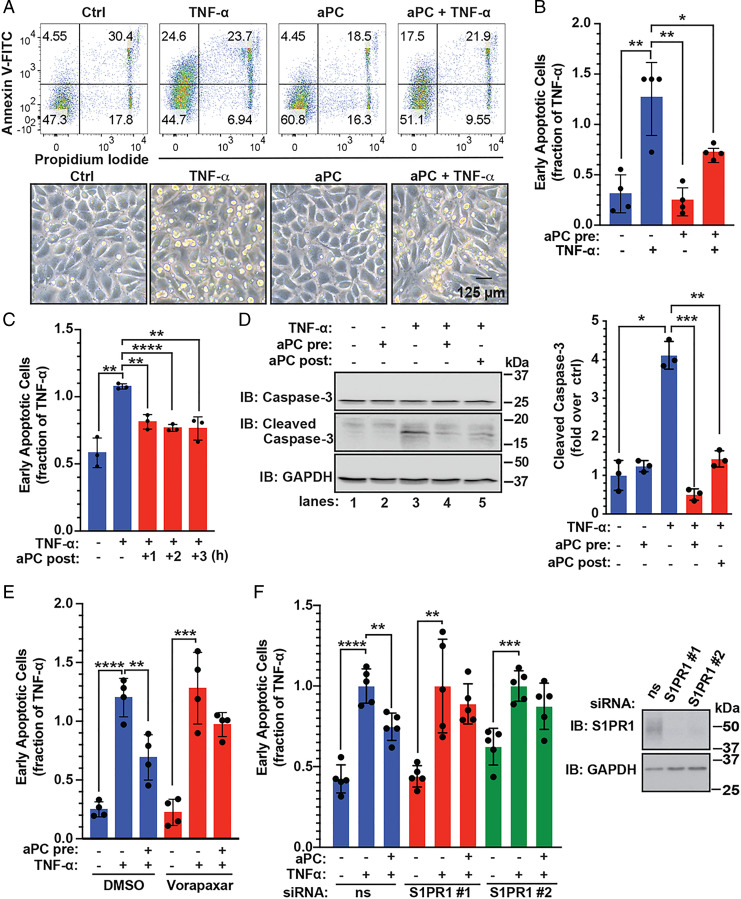
To assess the role of S1PR1 in aPC/PAR1-driven anti-apoptotic responses, aPC-mediated protection against TNF-α–induced cell death was examined. Endothelial cells incubated with TNF-α exhibited a significant increase in cell death as detected by Annexin V-FITC staining and flow cytometry, compared to untreated control cells (Fig. 2 A and B). In contrast, pretreatment with aPC for 4 h resulted in a significant reduction in TNF-α–induced cell death (Fig. 2 A and B). Phase–contrast images of TNF-α–treated endothelial cells revealed cell shrinkage and rounding indicative of cell death, which was not detected in control cells and reduced in aPC-treated cells (Fig. 2 A, Lower). Next, we determined if aPC treatment was sufficient to reverse TNF-α–initiated cell death by treating endothelial cells with TNF-α before aPC exposure. Similar to aPC pretreatment, posttreatment with aPC for 2 or 3 h caused a significant reduction in TNF-α–induced cell death (Fig. 2C). To further assess the effect of aPC treatment on the TNF-α–induced apoptosis, we examined cleavage of caspase-3, a key effector of cellular apoptosis. TNF-α induced a significant increase in caspase-3 cleavage (Fig. 2D, lanes 1 through 3), which was reversed by pre- and posttreatment with aPC (Fig. 2D, lanes 3 through 5). These results indicate that aPC confers protection against TNF-α–induced apoptosis.
Fig. 2.
PAR1 and S1PR1 mediate aPC anti-apoptotic activity in endothelial cells. (A) EA.hy926 cells were pretreated with 20 nM aPC for 4 h and then treated with 10 ng/mL TNF-α for 20 h or left untreated, control (Ctrl). (Top) Cell death was determined by flow cytometry using Annexin V-FITC and PI. (Bottom) Phase–contrast images of endothelial cells. (Scale bar, 125 µM.) (B, Top Right) The data (mean ± SD, n = 4) of the early apoptotic response were analyzed by t test. (C) EA.hy926 cells were treated with TNF-α for 20 h and then aPC was added for 1, 2, or 3 h. The data (mean ± SD, n = 3) were analyzed by t test. (D) EA.hy926 cells were pretreated with aPC for 3 h and followed by TNF-α for 20 h, or aPC was added 3 h post–TNF-α. Cleaved caspase-3 and GAPDH were detected by immunoblotting (IB). Data (mean ± SD, n = 3) was analyzed by t test. (E) EA.hy926 cells were pretreated with 10 µM vorapaxar for 1 h, stimulated with aPC for 4 h, then treated with TNF-α for 20 h, and apoptosis determined. Data (mean ± SD, n = 4) were analyzed by two-way ANOVA. (F) EA.hy926 cells transfected with nonspecific (ns) or two different, S1PR1-specific siRNAs were stimulated with aPC for 4 h and treated with TNF-α for 20 h. S1PR1 and GAPDH were detected by immunoblotting. Data (mean ± SD, n = 5) were analyzed by two-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Next, the function of PAR1 and S1PR1 in aPC-induced anti-apoptotic activity was determined. The PAR1 selective antagonist vorapaxar effectively blocked aPC-mediated protection against TNF-α–induced cell death (Fig. 2E), suggesting a critical role for PAR1. We used siRNA to assess S1PR1 function in aPC/PAR1-mediated anti-apoptotic responses. Depletion of S1PR1 was confirmed by immunoblot (Fig. 2F). As expected, TNF-α–induced cell death was significantly reduced by aPC in nonspecific siRNA control cells (Fig. 2F). However, aPC failed to protect against TNF-α–induced apoptosis in S1PR1-depleted cells (Fig. 2F). Thus, S1PR1 is an important mediator of aPC/PAR1-induced anti-apoptotic activities in endothelial cells; however, the mechanism by which S1PR1 facilitates aPC/PAR1-promoted cell survival is not known.
Akt-Mediated aPC Anti-apoptotic Activity Occurs via an S1PR1-Dependent Pathway.
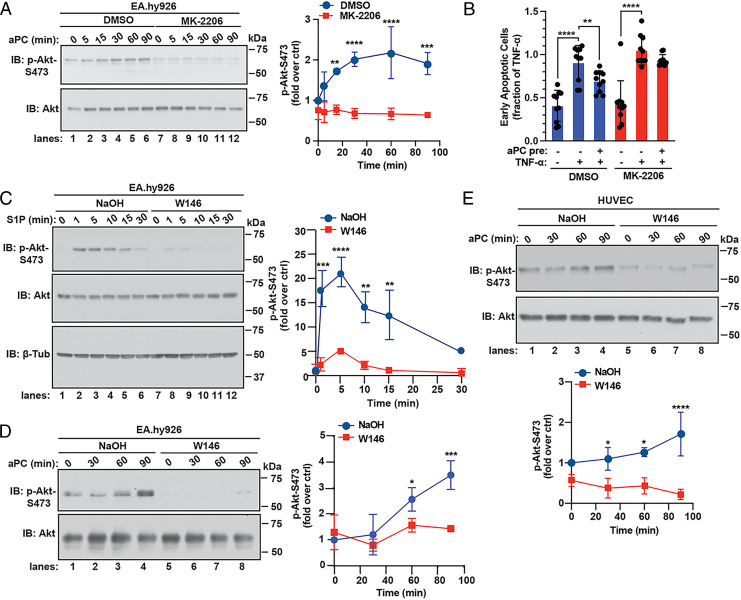
Akt1 is the major isoform that contributes to endothelial cell function, including cell survival (23). To determine the role of Akt in aPC-mediated anti-apoptotic responses, we used the Akt allosteric inhibitor MK-2206 that targets the closed, inactive conformation preventing phosphorylation and activation (24, 25). In endothelial cells, aPC stimulated a significant increase in Akt S473 phosphorylation that remained elevated for 90 min (Fig. 3A, lanes 1 through 6). Pretreatment with MK-2206 resulted in significant reduction of aPC-induced Akt S473 phosphorylation (Fig. 3A, lanes 7 through 12), confirming that MK-2206 effectively inhibits aPC-induced Akt activation. MK-2206 was then used to assess Akt function in aPC/PAR1 anti-apoptotic responses. In control cells, aPC-mediated significant protection against TNF-α–induced cell death (Fig. 3B). However, aPC failed to protect against apoptosis in cells pretreated with MK-2206 (Fig. 3B), indicating that Akt activity is required for aPC-induced prosurvival effects in endothelial cells.
Fig. 3.
Akt mediates aPC anti-apoptotic activity through an S1PR1-dependent pathway. (A) EA.hy926 cells were pretreated with 1 µM MK-2206 or DMSO vehicle for 1 h, stimulated with 20 nM aPC for various times, and Akt and phospho-Akt-S473 were detected by immunoblotting (IB). Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. (B) EA.hy926 cells were pretreated 1 μM MK-2206 or DMSO for 1 h, followed by aPC for 4 h, then treated with 10 ng/mL TNF-α for 20 h, and apoptosis measured. Data (mean ± SD, n = 10) were analyzed by two-way ANOVA. (C) EA.hy926 cells were pretreated with 10 µM W146 or NaOH vehicle for 30 min and stimulated with 1 µM S1P for various times. Akt, phospho-Akt-S473, and β-tubulin were detected by immunoblotting. Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. (D) EA.hy926 cells were pretreated with W146 or NaOH as described above, stimulated with aPC for various times, and Akt and phospho-Akt-S473 were detected by immunoblotting. Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. (E) HUVECs were pretreated with W146 or NaOH, stimulated with aPC, and data analyzed as described in D. ctrl, control. *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
The function of S1PR1 in aPC-induced, Akt-dependent cell survival was next examined using W146, an S1PR1 selective antagonist. Activation of S1PR1 with sphingosine-1-phosphate (S1P) caused a significant rapid increase in Akt S473 phosphorylation (Fig. 3C, lanes 1 through 6), which was abolished in W146-treated cells (Fig. 3C, lanes 7 through 12). In control cells, aPC induced significant increases in Akt phosphorylation at later times (Fig. 3D, lanes 1 through 4) that was virtually ablated in cells pretreated with the S1PR1 antagonist W146 (Fig. 3D, lanes 5 through 8). A similar inhibitory effect of W146 on aPC-induced Akt phosphorylation was observed in primary HUVECs (Fig. 3E). However, inhibition of S1PR1 with W146 did not perturb aPC/PAR1-stimulated ERK1/2 phosphorylation in either EA.hy926 cells or HUVECs (SI Appendix, Fig. S1), suggesting that S1PR1 antagonism specifically blocks Akt activation. These findings suggest that S1PR1 is required for aPC induction of Akt-driven cell survival signaling and not ERK1/2 signaling. However, the mechanisms by which aPC/PAR1 transactivates S1PR1-Akt signaling is not known.
aPC-Stimulated SphK1 Activity Is Required for Akt Activation.
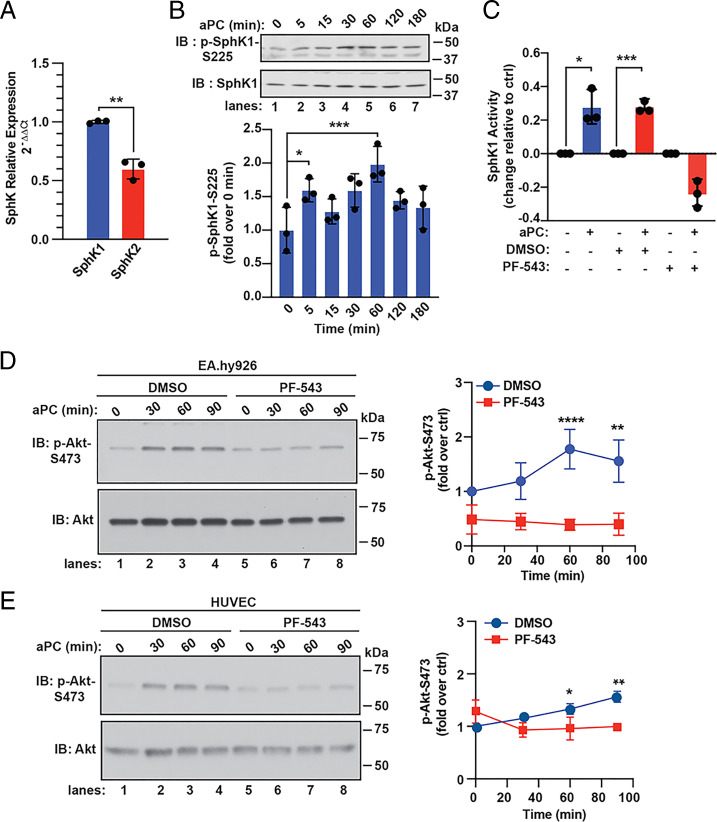
SphK catalyzes the phosphorylation of sphingosine to form S1P, the natural ligand for S1PR1 activation (26). SphK exists as two isoforms with SphK1 exhibiting a higher abundance in endothelial EA.hy926 cells compared to SphK2 (Fig. 4A). SphK1 phosphorylation at serine (S)-225 is highly correlated with activation (27). In aPC-treated endothelial cells, a significant increase in SphK1 S225 phosphorylation was detected using anti–phospho-SphK1 antibodies (Fig. 4B). To directly test if aPC stimulates SphK1 activity, we used a luminescence assay optimized to measure SphK1 activity in endothelial cells (SI Appendix, Fig. S2A). aPC induced a robust increase in SphK1 activity following 15 min of stimulation (Fig. 4C), which was markedly reduced by the SphK1 selective inhibitor PF-543 (28), compared to control cells (Fig. 4C). Thus, aPC/PAR1 stimulates SphK1 S225 phosphorylation and increases SphK1 activity.
Fig. 4.
aPC activates SphK1 and mediates aPC-induced Akt activity. (A) SphK1 and SphK2 expression was determined by qPCR. Data (mean ± SD, n = 3) were analyzed by t test. (B) EA.hy926 cells were stimulated with 20 nM aPC for various times. SphK1 and phospho-SphK1 were detected by immunoblotting (IB). Data (mean ± SD, n = 3) were analyzed by t test. (C) EA.hy926 cells were pretreated with 100 nM PF-543 or DMSO vehicle for 30 min, stimulated with aPC for 15 min, and SphK1 activity measured. Data (mean ± SD, n = 3) was analyzed by t test. (D) EA.hy926 cells were treated with PF-543 or DMSO as described in C, stimulated with aPC for various times, and Akt and phospho-Akt-S473 were detected by immunoblotting. Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. (E) HUVECs were pretreated with PF-543 or DMSO, stimulated with aPC, and analyzed as described in D. *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
To determine whether SphK1 activity is linked to aPC/PAR1-S1PR1–dependent Akt activation, endothelial cells were treated with PF-543. In control cells, aPC stimulated a significant increase in Akt phosphorylation (Fig. 4D, lanes 1 through 4), whereas aPC failed to induce Akt signaling in PF-543–pretreated cells (Fig. 4D, lanes 5 through 8). PF-543 similarly blocked aPC-induced Akt phosphorylation in HUVECs (Fig. 4E, lanes 1 through 4 versus 5 through 8). In contrast to Akt, aPC-induced ERK1/2 phosphorylation was not altered by the PF-543 inhibitor in either EA.hy926 or HUVECs compared to control cells (SI Appendix, Fig. S2 B and C). These results suggest that aPC-stimulated SphK1 activity is specifically linked to transactivation of the S1PR1-Akt signaling axis and not to the ERK1/2 signaling pathway.
β-arr2 Initiates aPC-Induced, SphK1-Dependent S1PR1-Akt Prosurvival Signaling.
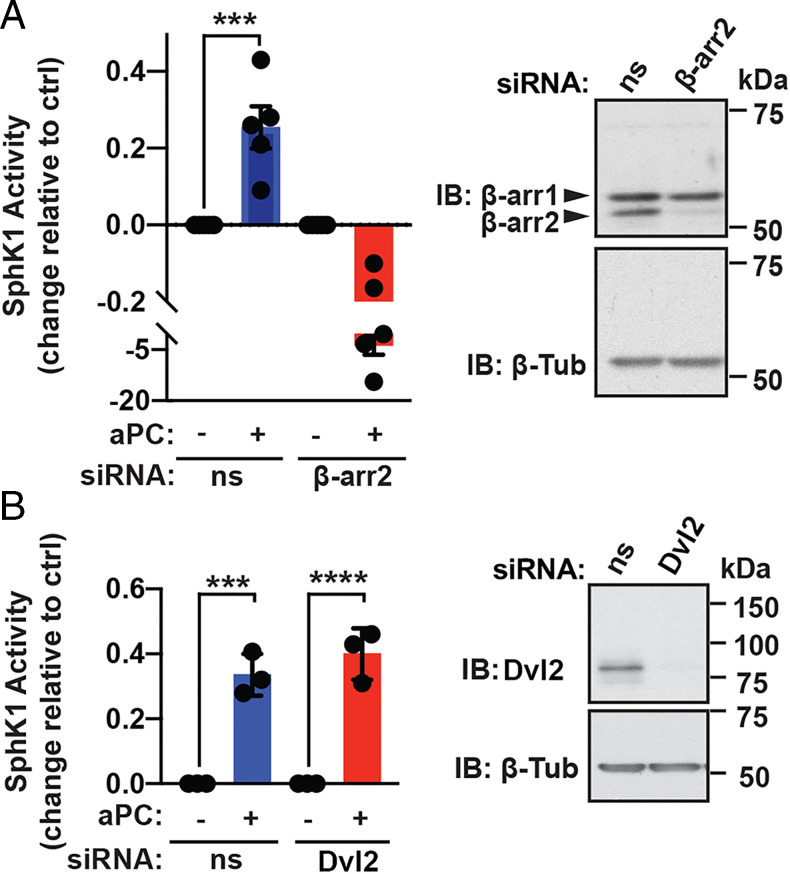
β-arr2 and Dvl2 function as scaffolds and facilitate aPC/PAR1-induced, Rac1-mediated endothelial barrier protection and protection against immune cell recruitment (11, 12). However, it is not known if β-arr2 and Dvl2 are similarly required for aPC-induced SphK1 activation and were examined using siRNA-targeted depletion. Depletion of β-arr2 and Dvl2 by siRNA was confirmed by immunoblot (Fig. 5 A and B). aPC stimulated a significant increase in SphK1 activity in endothelial cells transfected with nonspecific siRNA (Fig. 5A), whereas aPC-induced SphK1 activity was markedly reduced upon β-arr2 depletion (Fig. 5A). In contrast, aPC-mediated activation of SphK1 was not altered in Dvl2-depleted endothelial cells compared to nonspecific, siRNA-transfected cells (Fig. 5B), suggesting that β-arr2 functions as a key effector of aPC/PAR1-induced SphK1 activation. These findings further suggest that aPC stimulates divergent β-arr2–dependent cytoprotective signaling pathways.
Fig. 5.
β-arr2 and not Dvl-2 mediates aPC-induced SphK1 activity. (A) EA.hy926 cells were transfected with nonspecific (ns) siRNA or β-arr2 siRNA, stimulated with 20 nM aPC for 15 min, and SphK1 activity measured. Data (mean ± SD, n = 5) was analyzed by t test. β-arr2 and β-tubulin were detected by immunoblotting (IB). (B) EA.hy926 cells were transfected with ns siRNA or Dvl-2 siRNA and SphK1 activity determined. Dvl-2 and β-tubulin were detected by immunoblotting. Data (mean ± SD, n = 5) was analyzed by t test. ***P < 0.001; ****P < 0.0001.
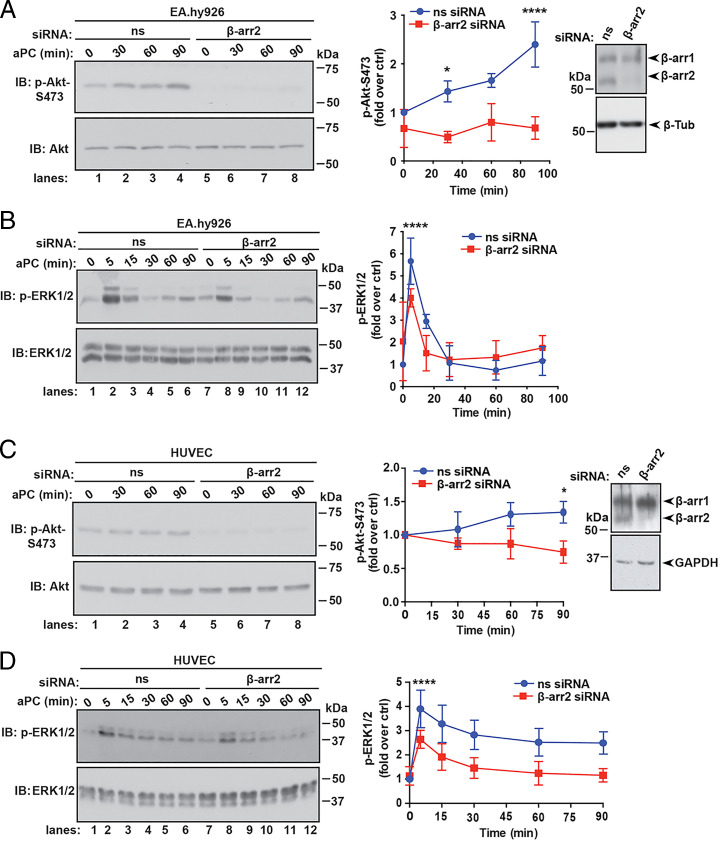
Next, we determined whether aPC/PAR1 transactivation of S1PR1-mediated Akt signaling is dependent on β-arr2 using siRNA-targeted depletion. β-arr2 knockdown was confirmed by immunoblotting (Fig. 6 A and C). aPC stimulated a significant increase in Akt phosphorylation in nonspecific, siRNA-transfected EA.hy926 cells (Fig. 6A, lanes 1 through 4), which was markedly decreased in β-arr2–deficient cells (Fig. 6A, lanes 5 through 8). Interestingly, the loss of β-arr2 expression also resulted in a significant decrease in aPC-induced ERK1/2 phosphorylation (Fig. 6B), indicating that aPC/PAR1-dependent β-arr2 function is required for ERK1/2 signaling, as we previously reported (11). In HUVECs, aPC-promoted Akt phosphorylation was also inhibited in β-arr2–deficient cells compared to control cells (Fig. 6C, lanes 1 through 4 versus 5 through 8). A similar effect of β-arr2 knockdown on aPC-induced ERK1/2 phosphorylation was observed in HUVECs (Fig. 6D, lanes 1 through 6 versus 7 through 12). These findings suggest that β-arr2 functions as master regulator of two distinct aPC/PAR1 signaling pathways, Akt and ERK1/2, that are differently regulated by downstream effectors.
Fig. 6.
β-arr2 drives aPC anti-apoptotic responses. (A) EA.hy926 cells transfected with nonspecific (ns) or β-arr2 siRNA were stimulated with 20 nM aPC for various times. Akt, phospho-Akt-S473, β-arr2, and β-tubulin were detected by immunoblotting (IB). Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. (B) EA.hy926 cells transfected with ns and β-arr2 siRNA were treated with aPC for various times and immunoblotted for ERK1/2 and phospho-ERK1/2. Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. (C) HUVECs were transfected, stimulated with aPC as described in A, and immunoblotted for Akt, phospho-Akt, β-arr2, and GAPDH. (D) HUVEC were transfected, stimulated with aPC as described in B, and immunoblotted for ERK1/2 and phospho-ERK1/2. Data (mean ± SD, n = 3) was analyzed by two-way ANOVA. *P < 0.05; ****P < 0.0001.
β-arr2 and Cav1 Mediate aPC/PAR1-Induced Cell Survival.
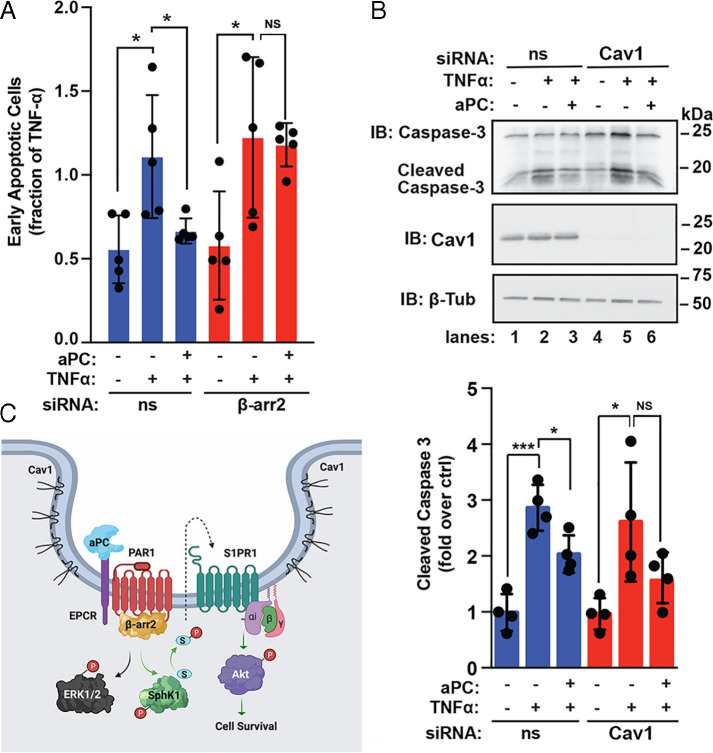
Next, we assessed the function of β-arr2 in aPC-mediated prosurvival in endothelial cells depleted of β-arr2 by examining aPC-mediated protection against TNF-α–induced cell death using Annexin V-FITC staining and flow cytometry. aPC significantly reduced TNF-α–induced cell death in nonspecific, siRNA-transfected cells (Fig. 7A); however, the capacity of aPC to protect against TNF-α–induced cell death was lost in β-arr2–deficient cells (Fig. 7A). These findings are consistent with a critical role for β-arr2 in aPC-mediated cell survival. We next examined the function of Cav1, a key structural protein essential for caveolae formation (29), in aPC/PAR1-induced protection against apoptosis. In these assays, we used siRNA-targeted depletion of Cav1 and assessment of caspase-3 cleavage, a central mediator of programmed cell death. In nonspecific, siRNA-transfected cells, incubation with TNF-α caused a marked increase in caspase-3 cleavage that was significantly decreased in aPC-treated cells (Fig. 7B). However, in endothelial cells depleted of Cav1, aPC failed to significantly alter TNF-α–induced caspase-3 cleavage (Fig. 7B). Taken together, our studies reveal a function for β-arr2 as a key regulator of aPC/PAR1-induced anti-apoptotic responses mediated by a distinct SphK1-SPR1-Akt–mediated prosurvival signaling pathway that does not integrate into the β-arr2–mediated ERK1/2 signaling (Fig. 7C).
Fig. 7.
aPC-induced anti-apoptotic responses requires β-arr2 and Cav1. (A) EA.hy926 cells transfected with ns or β-arr2 siRNA were stimulated with 20 nM aPC for 4 h, treated with 20 ng/mL TNF-α for 20 h, and apoptosis determined. Data (mean ± SD, n = 5) were analyzed by two-way ANOVA (*P < 0.05). NS, not significant. (B) EA.hy926 cells transfected with ns or Cav1 siRNA were pretreated with aPC and then stimulated with TNF-α, as described above. Cell lysates were immunoblotted (IB) using caspase-3, Cav1, or β-tubulin antibodies. Data (±SD, n = 3) were analyzed by t test (*P < 0.05; ***P < 0.001). (C) Model of aPC/PAR1 transactivation of S1PR1 via β-arr2–mediated SphK1 activation. Activation of PAR1 by aPC bound to its coreceptor EPCR cleaves and activates PAR1, which promotes β-arr2–dependent SphK1 activation and transactivation of S1PR1, resulting in Akt activation and cell survival. SphK1 catalyzes phosphorylation of sphingosine to form S1P, a ligand for S1PR1. The aPC-PAR1-β-arr2–driven SphK1-S1PR1-Akt signaling axis is distinct from the β-arr2–mediated ERK1/2 signaling pathway.
Discussion
Endothelial dysfunction results in barrier disruption and increased sensitivity to apoptosis. In this study, we delineate a GPCR-β-arr2–driven signaling pathway that regulates endothelial cellular resistance to apoptosis. Both PAR1 and S1PR1 reside in Cav1-rich microdomains and are required for aPC protection against cell death. We demonstrate that aPC/PAR1-promoted anti-apoptotic activity is mediated by Akt signaling induced by transactivation of the S1PR1 coreceptor. Moreover, aPC/PAR1-stimulated transactivation of S1PR1-Akt signaling is mediated by β-arr2–dependent SphK1 activation and occurs independent of Dvl2. We further show that β-arr2 functions as the central mediator of the aPC/PAR1-induced SphK1-S1PR1-Akt prosurvival signaling pathway, which is distinct from the β-arr2–induced ERK1/2 signaling pathway. Together, these studies reveal that different aPC/PAR1-induced cytoprotective responses are mediated by discrete β-arr2–driven signaling pathways that are uniquely modulated by coreceptors.
A major finding of our study is that aPC stimulates unique, β-arr2–driven signaling pathways. We previously showed that aPC-activated PAR1 signals preferentially through β-arr2 and not Gi proteins to confer protection against thrombin-induced barrier disruption (11). We also demonstrated that aPC/PAR1 stimulates β-arr2–dependent polymerization of Dvl2 and Rac-1 activation, which facilitates endothelial barrier protection. A recent study showed that aPC occupancy of EPCR in endothelial cells promotes β-arr2– and Dvl2-mediated inhibition of cytokine-induced monocyte recruitment, an anti-inflammatory response (12). These studies suggest that β-arr2 and Dvl2 scaffolds might function as universal effectors of different aPC/PAR1 cytoprotective responses. In this study, we report that β-arr2, but not Dvl2, is essential for aPC-stimulated SphK1 activity, which is required for transactivation of S1PR1-induced Akt activity. In addition, aPC/PAR1-activated SphK1-S1PR1 signaling does not intersect with β-arr2–mediated ERK1/2 signaling, consistent with discrete, β-arr2–regulated signal transduction pathways. aPC cytoprotective responses are well documented in other cell types, such as neurons, podocytes, and immune cells (30–32), but whether these responses are also regulated by discrete, β-arr2–mediated signaling pathways is not known.
Caveolae organize cellular signal transduction by bringing effectors in close proximity to receptors through binding to Cav1 (21). A unique feature of aPC/PAR1 cytoprotective signaling is the localization of key effectors, including PAR1, EPCR, and β-arr2 in caveolae, a subtype of lipid rafts (8, 11). Previous studies reported that caveolae are required for aPC/EPCR/PAR1 complex formation and β-arr2–mediated cytoprotection (8, 9, 11, 33). Here, we now show that S1PR1 also resides in caveolae together with PAR1. In addition, aPC increased S1PR1 binding to Cav1 through a PAR1-dependent mechanism. However, it is not known if S1PR1 binds directly to Cav1 or other components, such as EPCR or PAR1, after recruitment to caveolae. Importantly, aPC-PAR1–induced anti-apoptotic responses mediated by SphK1-S1PR1-Akt signaling also require Cav1, indicating that caveolae facilitate aPC/PAR1 anti-apoptotic activities.
aPC cleaves and activates a small population of PAR1 compartmentalized in caveolae and stabilizes an active conformation that preferentially binds to β-arr2 rather than Gi proteins (9, 11). Unlike activated GPCR rapid amplification of G protein signaling, catalyzed by exchange of GDP for GTP, aPC/PAR1 activation of β-arr2–dependent signaling is stoichiometric, protracted, and dependent on localization of critical effectors in caveolae (9, 11). The localization of aPC/PAR1 in caveolae in close proximity to other GPCRs, such as S1PR1, has enabled aPC/PAR1 to diversify signal transduction triggering different cytoprotective responses. We speculate that distinct cell types will exhibit different aPC/PAR1 cytoprotective responses depending on the cellular wiring, coreceptor expression, and cellular function. While aPC/PAR1 transactivation of S1PR1 is complex, the β-arr2–driven pathway likely evolved to allow aPC/PAR1 to couple indirectly to Gi-Akt signaling through a neighboring GPCR, such as S1PR1, since aPC-activated PAR1 does not couple directly to Gi (11). S1PR1 is not the only coreceptor that is required for aPC/PAR1-induced cytoprotective activities, and future studies are needed to understand the specific functions of other coreceptors, such as PAR3 (19), in enabling molecular effectors to convey signal transduction for different cytoprotective responses. The global phosphoproteome of aPC signaling in endothelial cells was recently reported and provides a valuable resource for identifying effectors of aPC cytoprotective signaling (34).
GPCR signaling is diverse, complex, and occurs at the plasma membrane and from endosomes to orchestrate effective inflammatory responses (35). Similarly, aPC/EPCR/PAR1 cytoprotective signaling is complex, occurs in caveolae, and modulated by different coreceptors in distinct cell types to yield different cytoprotective responses. Despite progress in understanding how aPC/PAR1 signaling is propagated in endothelial cells, the mechanisms responsible for turning off aPC signaling are not known for either PAR1 or coreceptors, including transactivated S1PR1. While S1P activation of S1PR1 is rapidly desensitized and internalized by the classic, GRK2-mediated phosphorylation and β-arrestin–dependent internalization through clathrin-coated pits (36–38), it is not known how aPC/PAR1-transactivated S1PR1 signaling is regulated. Interestingly, in endothelial cells lacking β-arr2, aPC/PAR1-transactivated, S1PR1-dependent Akt signaling is virtually abolished (Fig. 6), consistent with a role for β-arr2 in turning on rather than turning off signaling. If, in fact, regulation of transactivated S1PR1 signaling required β-arr2, we would expect enhanced Akt signaling, which is not observed. It is possible that β-arr1 functions predominantly in desensitization of transactivated S1PR1 signaling, but this has not been tested. Moreover, aPC/PAR1 signaling has a slow onset and is prolonged, suggesting that classic desensitization mechanisms may function differently in this particular cellular context. In summary, this study demonstrates that distinct aPC/PAR1 cytoprotective responses are driven by discrete β-arr2–mediating signaling pathways that are specifically modulated by different coreceptors in endothelial cells.
Materials and Methods
Cell Culture.
EA.hy926 cells (ATCC, #CRL-2922) were grown at 37 °C, 8% CO2 in 10% fetal bovine serum (FBS) in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, #10–013-CV and #10437–028) supplemented with fresh 20% preconditioned media every 2 d and used up to passage 8. Pooled primary HUVECs (Lonza, #C2519A) were grown at 37 °C, 5% CO2 in endothelial cell growth medium-2 (Lonza, #CC-3162), media was changed every 2 d and used up to passage 6. EA.hy926 and HUVECs were grown for 4 to 5 d until confluence and then incubated overnight in 0.4% FBS-DMEM. Cells were then washed and serum starved in DMEM containing 10 mM Hepes, 1 mM CaCl2, and 1 mg/mL bovine serum albumin (BSA) for 1 h prior to agonist, antagonist, and inhibitor treatments, as described in Agonists, Inhibitor, and Antagonist Treatments.
Antibodies.
The following antibodies were used in this study: anti–β-arr2 A2CT (a generous gift from Robert Lefkowitz, Duke University, Durham, NC), anti-Dvl2 (CST, #3216), anti-S1PR1 (Santa Cruz Biotechnology, #sc-25489), Cav1 (CST, #610060), anti-GAPDH (GeneTex, #GTX627408), anti–β-tubulin (CST, #86298S), anti–caspase-3 (CST, #9662S), anti-cleaved caspase-3 (CST, #9661), anti–phospho-SphK1-S225 (ECM Biosciences, #SP1641), anti-SphK1 (ECM Biosciences, #SP1621), anti–phospho-Akt-S473 (CST, #4060), anti-Akt (CST, #9272S), anti–phospho-ERK1/2 (CST, #9106L), anti-ERK1/2 (CST, #9102L), anti-PAR1 WEDE antibody (Beckman Coulter, #IM2584), and EEA1 (BD Biosciences, #610457) antibodies. Secondary antibodies were anti-mouse or anti-rabbit horseradish peroxidase–conjugated antibodies (Bio-Rad, #170–6516 and #170–6515) and fluorescent anti-mouse Alexa-488 (Invitrogen, #A-11001) and anti-rabbit Alexa-594 (Invitrogen, #A-11012) antibodies.
Agonists, Inhibitor, and Antagonist Treatments.
Cells were stimulated with aPC (Hematologic Technologies, #HCAPC-0080), TNF-α (PeproTech, #300–01A), S1P (Tocris, #1370), or α-thrombin (Enzyme Research Laboratories, #HT 1002a). Serum-starved cells were pretreated at 37 °C with 100 nM PF-543 (Tocris, #5754), or 10 µM W146 (Tocris, #3602) for 30 min or with 1 µM MK-2206 (Selleck, #S1078) or 10 µM Vorapaxar (Axon Medchem, #1755) for 1 h.
Transfections with siRNAs.
Cells were seeded at 1.4 × 105 cells per well in a 12-well plate, grown overnight, and transfected with siRNA using the TransIT-X2 System (Mirus, #MIR 600) according to the manufacturer’s instructions and experiments conducted 72 h posttransfection. The following siRNAs were used: 50 nM β-arr2 siRNA (Dharmacon) 5′-GGACCGCAAAGTGTTTGTG-3′, 12.5 nM Dvl2 siRNA #2 (Qiagen, #SI00063441) 5′-CACGCTAAACATGGAGAAGTA-3′, 25 nM of S1PR1 #1 (Qiagen, #SI00376201) 5′-ATGATCGATCATCTATAGCAA-3′, 25nM S1PR1 #2 (Qiagen, #SI00376208) 5′-TAGCATTGTCAAGCTCCTAAA-3′ siRNAs or AllStars Negative Control nonspecific siRNA (Qiagen, #1027281), and 50 nM CAV1 siRNA 5′-CCCACTCTTTGAAGCTGTTGGGAAA-3′ (Invitrogen, #CAV1HSS141467).
Cell Death Assays and Flow Cytometry.
EA.hy926 cells were seeded at 1.4 × 105 cells per well in a 12-well plate. Serum-starved cells were pretreated with 20 nM aPC for 4 h and then incubated with 10 ng/mL TNF-α for 20 h or posttreated with 20 nM aPC for either 1, 2, or 3 h. Cells were harvested using Cellstripper (Corning, #25–056-CL) and washed with cold phosphate-buffered saline (PBS). Cells were resuspended in 40 μL 1× Annexin V–binding buffer (Biolegend, #422201) plus 2 μL Annexin V-FITC (BioLegend, #640906), incubated at room temperature for 15 min, and protected from light. Cells were washed with 160 μL of 1× Annexin V–binding buffer, followed by a 5 min centrifugation at 550 × g. Pelleted cells were resuspended in 200 μL 1× Annexin V–binding buffer plus 2 μL of 100 μg/mL propidium iodide (PI) (Sigma-Aldrich, #P4170). Data acquisition was performed on a BD FACS Canto II Flow cytometer (BD Biosciences) on a log scale with 30,000 singlet gate events collected per sample. Data compensation and analysis were performed with FlowJo version 10 software (Tree Star). The gating strategy was as follows: Annexin V– and PI-negative events were backgated to forward scatter-area (FSC-A)/side scatter-area (SSC-A) to determine cell debris. A “not gate” was made based on cell debris in FSC-A/SSC-A. Doublet discrimination was performed using FSC-A versus forward scatter-height (FSC-H) and SSC-A versus side scatter-height (SSC-H). The resulting gated cells were analyzed for Annexin and PI staining and reported as percent of singlets.
Caspase-3 Cleavage Assay.
EA.hy926 cells were seeded at 6.2 × 105 cells per well in a 6-well plate. Serum-starved cells were pretreated with 20 nM aPC for 4 h followed by treatment with 20 ng/mL TNF-α for 20 h. Cells were washed with PBS and lysed in radioimmunoprecipitation assay buffer (50 mM Tris HCl, pH 8.0,150 nM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors (1 mM PMSF, 2 µg/mL aprotinin, 10 µg/mL leupeptin, 1µg/mL pepstatin, and 1 µg/mL trypsin protease inhibitor). Cells were sonicated at 10% amplitude for 10 s and clarified by centrifugation at 20,817 × g for 15 min. Cell lysates were immunoblotted using caspase-3, cleaved caspase-3, GAPDH, and β-tubulin antibodies and quantified by densitometry analysis using ImageJ software.
Signaling Assays.
EA.hy926 cells were seeded in a 24-well plate at 1.4 × 105 cells per well. HUVECs were seeded at 1.77 × 105 cells per well in a 24-well plate. Serum-starved cells were treated with either 20 nM aPC or 1 µM S1P for various times and then lysed in 2× Laemmli sample buffer (LSB) containing 200 mM dithiothreitol (DTT). Equivalent amounts of cell lysates were immunoblotted with phospho-SphK1-S225, SphK1, phospho-Akt-S473, Akt, phospho-ERK1/2, and ERK1/2 antibodies. Immunoblots were quantified as described in Caspase-3 Cleavage Assay.
SphK1 Activity Assay.
EA.hy926 cells were seeded in 6-well plates at 6 × 105 cells per well. Serum-starved cells were stimulated with 20 nM aPC for 15 min, washed with cold PBS, and SphK1 activity luminescence assay performed according to the manufacturer instructions (Echelon, #K-3500). In brief, cells were resuspended in reaction buffer with 1 mM DTT and sonicated for 10 s at 10% amplitude. Cell lysate protein was quantified using the bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, #23221 and #23224) and normalized to 1.5 mg/mL protein for each sample. Then, 400 mM sphingosine solution and 10 µL of each sample were aliquoted into each well of a 96-well plate. The reaction was initiated with the addition of 20 µM ATP to each sample, incubated for 30 min, and followed by the addition of K-LUMa ATP detector per well for 10 min to stop the reaction. Luminescence was determined using the Tristar LB 941 Plate Reader (Berthold Technologies). A reduction in luminescence compared to control indicates ATP depletion or consumption and used as an assessment of SphK1 activity. To generate positive or negative values for increased or reduced SphK1 activity, respectively, background luminescence was subtracted from the raw luminescence units for each sample. Using an ATP standard curve (SI Appendix, Fig. S2A), the concentration of ATP after the 30-min reaction was determined then subtracted from the starting ATP concentration of 20 µM. This yielded the concentration of ATP consumed in 30 min. The difference from control values was plotted for each sample, with three or more replicates for each experiment.
Immunofluorescence Confocal Microscopy.
EA.hy926 cells were seeded on coverslips in 12-well plate at a density of 1.4 × 105 cells per well. Serum-starved cells were stimulated with 20 nM aPC for 1 h or 10 nM α-thrombin for 1 h, washed with cold PBS, and incubated with PBS for 10 min. Endogenous PAR1 was labeled with anti-PAR1 WEDE antibody at 1:500 for 1 h on ice, cells were treated with or without agonists, fixed for 5 min with 4% paraformaldehyde, and permeabilized with 0.1% Triton-X 100. The detection of S1PR1 was determined using anti-S1PR1 antibody diluted at 1:100 in 0.03% BSA, 0.01% Triton-X 100, and 0.01% normal goat serum overnight at 4 °C. Secondary fluorescent antibodies anti-mouse Alexa-488 and anti-rabbit Alexa-594 diluted at 1:750 in 0.03% BSA, 0.01% Triton-X 100, and 0.01% normal goat serum were incubated at room temperature for 1 h. Slides were mounted using ProLong Gold Antifade Mountant (Invitrogen, #P10144). Confocal images were acquired sequentially using the same settings with an Olympus IX81 spinning-disk microscope equipped with a CoolSNAP HQ2 CCD camera (Andor) and 63× Plan Apo objective (1.4 NA) with appropriate excitation–emission filters. Line scan analysis was performed using Image J software (NIH).
Immunoprecipitation Assays.
EA.hy926 wild-type cells and EA.hy926 cells stably expressing PAR1-specific short hairpin (sh)RNA pSilencer Retro (9) were seeded at 4.95 × 106 cells per 10-cm dishes and grown overnight. Cells were then serum starved overnight, treated with 20 nM aPC, and lysed in Triton-X 100 lysis buffer (50 mM Tris⋅HCl, pH 7.4, 100 mM NaCl, 10 mM NaF, 1% Triton-X 100 supplemented with protease inhibitors). Cell lysates were homogenized, clarified by centrifugation, and protein concentrations determined by BCA. Equivalent amounts of lysates were subjected to immunoprecipitations using the anti-S1PR1 and anti-PAR1 WEDE antibodies. Immunoprecipitates were resuspended in 2× LSB containing 200 mM DTT and eluents immunoblotted using S1PR1, Cav1, and PAR1 antibodies and developed by chemiluminescence.
Sucrose Fractionation.
EA.hy926 cells were seeded at 4.95 × 106 cells per 10-cm dish. Cells were washed with cold PBS, lysed in sodium carbonate buffer (150 mM sodium carbonate, pH 11, 1 mM EDTA, supplemented with protease inhibitors) with a dounce homogenizer, passed through 18-G needle 10×, and sonicated on ice at 10% amplitude. Cell lysates were mixed with equal volume of 80% sucrose in MES-buffered saline (25 mM MES pH 6.5, 150 NaCl, and 2 mM EDTA) supplemented with 300 mM sodium carbonate for a total of 1.6 mL in a 12-mL ultracentrifuge tube (Beckman Coulter, #343778). Around ∼6 mL 35% MES-buffered saline supplemented with 150 mM sodium carbonate was added to the top of the tube gently without perturbing solution on the bottom and 4 mL 5% sucrose in MES-buffered saline supplemented with 150 mM sodium carbonate on top of the 35% MES-buffered saline solution. Samples were placed in SW41 rotor and ultracentrifuged for 18 to 20 h at 4 °C at 229,884 × g. The 1 mL fractions were collected sequentially, and samples were immunoblotted using S1PR1, EEA1, anti-PAR1, and Cav1 antibodies.
qRT-PCR.
EA.hy926 cells seeded at 3.2 × 105 cells per well of a 6-well plate were grown to confluency, and RNA was extracted using Direct-zol RNA Miniprep Plus Kit (#R2072, Zymo Research) and used to generate complementary DNA (cDNA). RNA was quantified and cDNA synthesized from 1 μg RNA using SuperScript IV VILO Master Mix with ezDNase enzyme kit (#111766050, Thermo Fisher Scientific). qRT-PCR was performed with TaqMan Fast Advanced Master Mix (#4444964, Thermo Fisher Scientific) and TaqMan Gene Expression Probes SphK1 (#Hs00184211_m1), SphK2 (#Hs01016542_g1), and 18S (#Hs03003631_g1) using a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). SphK1 and SphK2 messenger RNA transcript levels were normalized to 18S expression and analyzed using the comparative threshold cycle method. The number of cycles until threshold (Ct) was determined for each target. To normalize for variation, the Ct value for 18S was subtracted from the Ct value for each target, and the differences in expression relative to SphK1 were then determined using the 2-ΔΔCt method. Control reactions without cDNA for each probe were conducted in every assay to ensure specificity of the reactions.
Models.
The cartoons in Figs. 1 and 7 were created with BioRender.com.
Data Analysis.
Data were analyzed with Prism 9.0 statistical software and Microsoft Excel. Statistical analysis methods are indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank members of the J.T. Laboratory for advice and guidance. We also thank Dr. Antonio De Maio (University of California San Diego) for assistance with flow cytometry and Cara R. Rada and Hilda Mejia-Peña for assisting with HUVEC cell culture. This work was supported by NIH/National Institute of General Medical Sciences (NIGMS) Grants R01 GM116597 and R35 GM127121 (J.T.), NIH/NIGMS Grant R25 GM083275 (M.C.-A. and L.J.C.), NIH/National Heart, Lung, Blood Institute Grant T32 HL007444 (C.A.B.), NIH/NIGMS Grant K12 GM068524 (O.M.-I. and D.N.), NIH/National Institute of Child Health and Human Development Grant P50 HD12303 (M.A.L.), and the University of California President’s Postdoctoral Fellowship award (O.M.-I. and D.N.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106623118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Prober J. S., Sessa W. C., Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Charreau B., Signaling of endothelial cytoprotection in transplantation. Hum. Immunol. 73, 1245–1252 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Dombrovskiy V. Y., Martin A. A., Sunderram J., Paz H. L., Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: A trend analysis from 1993 to 2003. Crit. Care Med. 35, 1244–1250 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Goldenberg N. M., Steinberg B. E., Slutsky A. S., Lee W. L., Broken barriers: A new take on sepsis pathogenesis. Sci. Transl. Med. 3, 88ps25 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Kerschen E. J., et al. , Endotoxemia and sepsis mortality reduction by non-anticoagulant activated protein C. J. Exp. Med. 204, 2439–2448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin J. H., Zlokovic B. V., Mosnier L. O., Activated protein C: Biased for translation. Blood 125, 2898–2907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin J. H., Zlokovic B. V., Mosnier L. O., Activated protein C, protease activated receptor 1, and neuroprotection. Blood 132, 159–169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae J.-S., Yang L., Rezaie A. R., Receptors of the protein C activation and activated protein C signaling pathways are colocalized in lipid rafts of endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 104, 2867–2872 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo A., Soh U. J. K., Paing M. M., Arora P., Trejo J., Caveolae are required for protease-selective signaling by protease-activated receptor-1. Proc. Natl. Acad. Sci. U.S.A. 106, 6393–6397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosnier L. O., Sinha R. K., Burnier L., Bouwens E. A., Griffin J. H., Biased agonism of protease-activated receptor 1 by activated protein C caused by noncanonical cleavage at Arg46. Blood 120, 5237–5246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soh U. J. K., Trejo J., Activated protein C promotes protease-activated receptor-1 cytoprotective signaling through β-arrestin and dishevelled-2 scaffolds. Proc. Natl. Acad. Sci. U.S.A. 108, E1372–E1380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy R. V., Ardeshirylajimi A., Dinarvand P., Yang L., Rezaie A. R., Occupancy of human EPCR by protein C induces β-arrestin-2 biased PAR1 signaling by both APC and thrombin. Blood 128, 1884–1893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanki H., et al. , β-arrestin-2 in PAR-1-biased signaling has a crucial role in endothelial function via PDGF-β in stroke. Cell Death Dis. 10, 100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosnier L. O., Griffin J. H., Inhibition of staurosporine-induced apoptosis of endothelial cells by activated protein C requires protease-activated receptor-1 and endothelial cell protein C receptor. Biochem. J. 373, 65–70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Ceunynck K., et al. , PAR1 agonists stimulate APC-like endothelial cytoprotection and confer resistance to thromboinflammatory injury. Proc. Natl. Acad. Sci. U.S.A. 115, E982–E991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen A., et al. , Functionally distinct and selectiviely phosphorylated GPCR subpopulations co-exist in a single cell. Nat. Commun. 9, 1050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feistritzer C., Riewald M., Endothelial barrier protection by activated protein C through PAR1-dependent sphingosine 1-phosphate receptor-1 crossactivation. Blood 105, 3178–3184 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Finigan J. H., et al. , Activated protein C mediates novel lung endothelial barrier enhancement: Role of sphingosine 1-phosphate receptor transactivation. J. Biol. Chem. 280, 17286–17293 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Burnier L., Mosnier L. O., Novel mechanisms for activated protein C cytoprotective activities involving noncanonical activation of protease-activated receptor 3. Blood 122, 807–816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrom R. S., Insel P. A., The evolving role of lipid rafts and caveolae in G protein-coupled receptor signaling: Implications for molecular pharmacology. Br. J. Pharmacol. 143, 235–245 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fridolfsson H. N., Patel H. H., Caveolin and caveolae in age associated cardiovascular disease. J. Geriatr. Cardiol. 10, 66–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuepbach R. A., Feistritzer C., Brass L. F., Riewald M., Activated protein C-cleaved protease activated receptor-1 is retained on the endothelial cell surface even in the presence of thrombin. Blood 111, 2667–2673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., et al. , Akt1 regulates pathological angiogenesis, vascular maturation and permeability in vivo. Nat. Med. 11, 1188–1196 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnett S. F., et al. , Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 385, 399–408 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirai H., et al. , MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Ther. 9, 1956–1967 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Siow D., Wattenberg B., The compartmentalization and translocation of the sphingosine kinases: Mechanisms and functions in cell signaling and sphingolipid metabolism. Crit. Rev. Biochem. Mol. Biol. 46, 365–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitson S. M., et al. , Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 22, 5491–5500 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnute M. E., et al. , Modulation of cellular S1P levels with a novel, potent and specific inhibitor of sphingosine kinase-1. Biochem. J. 444, 79–88 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto T., Kogo H., Nomura R., Une T., Isoforms of caveolin-1 and caveolar structure. J. Cell Sci. 113, 3509–3517 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Guo H., et al. , Activated protein C prevents neuronal apoptosis via protease activated receptors 1 and 3. Neuron 41, 563–572 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Madhusudhan T., et al. , Cytoprotective signaling by activated protein C requires protease-activated receptor-3 in podocytes. Blood 119, 874–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue M., et al. , Activated protein C targets immune cells and rheumatoid synovial fibroblasts to prevent inflammatory arthritis in mice. Rheumatology (Oxford) 58, 1850–1860 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Bae J.-S., Yang L., Rezaie A. R., Lipid raft localization regulates the cleavage specificity of protease activated receptor 1 in endothelial cells. J. Thromb. Haemost. 6, 954–961 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Lin Y., et al. , Phosphoproteomic analysis of protease-activated receptor-1 biased signaling reveals unique modulators of endothelial barrier function. Proc. Natl. Acad. Sci. U.S.A. 117, 5039–5048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birch C. A., Molinar-Inglis O., Trejo J., Subcellular hot spots of GPCR signaling promote vascular inflammation. Curr. Opin. Endocr. Metab. Res. 16, 37–42 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watterson K. R., et al. , Dual regulation of EDG1/S1P(1) receptor phosphorylation and internalization by protein kinase C and G-protein-coupled receptor kinase 2. J. Biol. Chem. 277, 5767–5777 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Reeves P. M., Kang Y.-L., Kirchhausen T., Endocytosis of ligand-activated sphingosine 1-phosphate receptor 1 mediated by the clathrin-pathway. Traffic 17, 40–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnon T. I., et al. , GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science 333, 1898–1903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.