Significance
For exocytosis, a secretory granule recruits three proteins associated with the plasma membrane, syntaxin, SNAP-25, and Munc18. All three form clusters. In artificial lipid bilayers, nonstoichiometric factors such as bilayer composition, thickness, and curvature cause syntaxin to cluster even when it contains little more than its transmembrane domain. We have also observed such clusters in live cells. Where single granules had docked at the plasma membrane, they recruited complexes of syntaxin and Munc18-1 into densely packed clusters. The clusters may impede fusion but are well suited to keep granules anchored at the plasma membrane.
Keywords: exocytosis, syntaxin, Munc18, protein clustering
Abstract
Syntaxin helps in catalyzing membrane fusion during exocytosis. It also forms clusters in the plasma membrane, where both its transmembrane and SNARE domains are thought to homo-oligomerize. To study syntaxin clustering in live PC12 cells, we labeled granules with neuropeptide-Y-mCherry and syntaxin clusters with syntaxin-1a green fluorescent protein (GFP). Abundant clusters appeared under total internal reflection (TIRF) illumination, and some of them associated with granules (“on-granule clusters”). Syntaxin-1a-GFP or its mutants were expressed at low levels and competed with an excess of endogenous syntaxin for inclusion into clusters. On-granule inclusion was diminished by mutations known to inhibit binding to Munc18-1 in vitro. Knock-down of Munc18-1 revealed Munc18-dependent and -independent on-granule clustering. Clustering was inhibited by mutations expected to break salt bridges between syntaxin’s Hb and SNARE domains and was rescued by additional mutations expected to restore them. Most likely, syntaxin is in a closed conformation when it clusters on granules, and its SNARE and Hb domains approach to within atomic distances. Pairwise replacements of Munc18-contacting residues with alanines had only modest effects, except that the pair R114A/I115A essentially abolished on-granule clustering. In summary, an on-granule cluster arises from the specific interaction between a granule and a dense cluster of syntaxin-Munc18-1 complexes. Off-granule clusters, by contrast, were resistant to even the strongest mutations we tried and required neither Munc18-1 nor the presence of a SNARE domain. They may well form through the nonstoichiometric interactions with membrane lipids that others have observed in cell-free systems.
Four proteins catalyze membrane fusion when a secretory granule or a synaptic vesicle undergoes exocytosis. The first, synaptobrevin (Syb), resides in the granule or vesicle membrane, while the three others are either rooted in the plasma membrane (syntaxin) or are associated with it (Munc18-1 and Synaptosomal Associated Protein 25kD (SNAP-25). Syb, syntaxin, and SNAP-25 are collectively called SNAREs and form a ternary complex that bridges the membranes where they are about to fuse (1). Before fusing, secretory granules must dock at the plasma membrane and recruit from the latter an “acceptor complex” for Syb that includes syntaxin, SNAP-25, and probably Munc18-1 (2–4). Syntaxin readily forms complexes with both Munc18-1 (5, 6) and SNAP-25 (7). Diverse other proteins may also be found at granule sites (8).
Interestingly, syntaxin, SNAP-25, and Munc18-1 all form clusters in the plasma membrane, as first seen in membrane sheets from sonicated PC12 cells immunostained for syntaxin (9–11). The clusters measure 68 to 80 nm in diameter in superresolution images (12, 13) and harbor 75 syntaxin molecules each (13). In membrane sheets, the clustering of syntaxin requires its transmembrane and SNARE domains but apparently no other proteins (14) and no granules (10). Syntaxin coclusters with phosphatidylinositol 4,5-bisphosphate (PIP2), and about a quarter of docked granules assembled a Syntaxin/PIP2 cocluster (15). In live cells, such coclusters were not seen (16). Clusters also form in artificial lipid bilayers containing both cholesterol and PIP2 (17). Syntaxin forms large aggregates in giant unilamellar liposomes (18) and smaller ones in stacked lipid bilayers (12). It clustered in lipid bilayers even when its entire cytosolic domain was removed except for a “juxta-membrane” stump 15 residues long. Multiple factors may encourage syntaxin to cluster such as hydrophobic mismatch (19) and membrane curvature (20). However, three are thought to predominate (21): 1) In the hydrocarbon core of the lipid bilayer, syntaxin transmembrane domains cluster when they must compete with cholesterol for phospholipid molecules (17, 22); 2) In the polar layer of the bilayer's internal leaflet, the positively charged juxta-membrane domain of syntaxin coclusters with negatively charged PIP2 or phosphatidylinositol 3,4,5 trisphosphate (12, 23); and finally 3) in the cytosol, syntaxin SNARE domains are thought to homo-oligomerize (21). None of the three factors are stoichiometric. Moreover, because syntaxin must be in its open conformation for its SNARE domain to interact, that conformation is assumed to predominate in clusters (10, 13, 14, 20, 24, 25).
Where granules populate the plasma membrane sparsely enough to be distinguished individually, the recruitment of syntaxin, SNAP-25, and Munc18-1 to them can be observed in live cells and at the level of individual granules (8, 26, 27). All three proteins are then seen to form clusters at granule sites. In movies, syntaxin clusters formed while granules docked at the plasma membrane and dispersed again when granules left it (26). Although syntaxin readily formed a new cluster where a granule had freshly docked, granules rarely or never docked at the abundant preexisting clusters that were not associated with a granule [“off-granule” clusters (26, 28)]. This suggests that granules recruit syntaxin and not vice versa. Like off-granule clusters in membrane sheets, on-granule clusters in live cells harbor about 50 syntaxin molecules in PC12 cells (27) and are of subdiffraction size (26). Unlike off-granule clusters, however, on-granule clusters exclude syntaxin mutants that either carry the point mutation I233A (28) or lack their N-terminal domain (29). This suggests that granules carry a component that specifically recognizes a conformation or domain of syntaxin either by itself or in combination with another molecule. The protein may be Munc18-1, (29) which like syntaxin, forms clusters at granule sites (11, 26).
Here, we explored syntaxin clusters in live cells at the level of single granules and single clusters. We find that on-granule clusters harbor syntaxin in its closed conformation and in a complex with Munc18. They coexist with off-granule clusters that do not require Munc18-1 and can form with syntaxin in an open conformation. In live cells, the nonstoichiometric interactions emphasized in previous work account for off-granule clusters, while a specific interaction between granules, syntaxin, and Munc18-1 results in on-granule clusters.
Results
Recruitment of Syntaxin to Granule Sites.
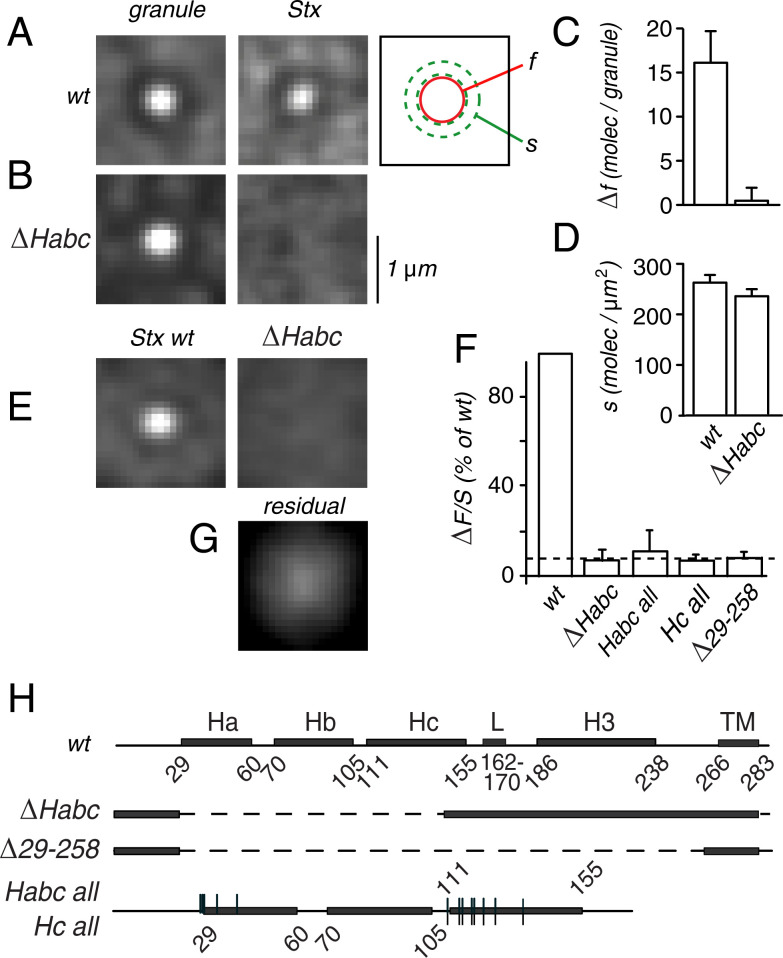
We imaged individual granules and their association with syntaxin-1a (Stx-1a), a neuronal isoform of syntaxin. PC12 cells were cotransfected with the red fluorescent granule marker neuropeptide-Y-mCherry (NPY-mCh) and with green fluorescent Stx-1a (Stx-GFP). Neither the mCherry within the granule nor the GFP on the external side of Stx-1a are likely to disturb interactions in the cytosol, because both are separated from it by a membrane. A crippled cytomegalovirus (CMV) promoter ensured that Stx-GFP was expressed in only small amounts to avoid out-competing endogenous Stx-1a. In images of single cells, both red and green fluorescence appear punctate, and the dots represent single granules (red) or single Stx-1a clusters [SI Appendix, Fig. S1A (28)]. We located granules in the red image and then excised small square images centered on them. Squares of the same size and locations were also excised from the green image, and the resulting image pairs averaged for all granules in a cell (Fig. 1A). Each of the paired images shows a central spot representing either the granule (Left) or its associated Stx-1a cluster (Middle). Selected to be solitary, the granules in our analysis were surrounded by a granule-free zone that was visible as a faint dark ring in averaged images. The Stx-GFP fluorescence was measured in a small central circle, f, and in a surrounding annulus, s, coincident with the ring (Fig. 1A, Right and SI Appendix, Fig. S1B). The value of s is taken to represent the local surface density of Stx-GFP molecules in the absence of a granule. Its average was 261 molecules/μm2 in this cell (Fig. 1D), about one-half the surface density of endogenous syntaxin-1A [540/μm2 (27)]. Evidently, Stx-GFP competed with an excess of unlabeled protein. The difference Δf = f – s is taken to represent the number of Stx-GFP molecules associated with the granule (Fig. 1C). Its high variability arises in part because one-half of the granules have no cluster (28), as was confirmed by analysis of histograms (SI Appendix, Fig. S1C). Ultimately, values of s and Δf were averaged for each cell, and their ratio, ΔF/S, was taken as proportional to the affinity of Stx-1a for a granule-associated but unidentified binding site.
Fig. 1.
Recruitment of syntaxin to granule sites. (A) Two images were taken of a cell (SI Appendix, Fig. S1A), one in the red and another in the green fluorescence channel. Left, regions centered on 82 individual granules were excised from the red channel and the images therein averaged. Middle, average from identical locations in the green channel. Right, regions used for measuring the local surface fluorescence (s) and the fluorescence of the granule site (f). (B) As in A but from a cell expressing the mutant ΔHabc (34 granules). A and B printed with the black level set well above the background; one contrast setting was used on the Left and another in the Middle. (C) Average granule-related fluorescence in the syntaxin images used in A and B measured as the difference Δf = f – s. (D) Values of s in the two cells. (E) Left, Stx-1a images as in A (Middle) averaged over 33 cells expressing Stx-GFP (coverslip 0161). Right, as in B (Right) but from 31 cells expressing ΔHabc (coverslip 0163). Both coverslips from the same culture. Together, mutant and wild-type measurements constitute one experiment and provide the ratio mutant/wild type. (F) Other mutants with strong effects. From two to four experiments each, including 57 to 88 cells per mutant and an equal or greater number for wild-type Stx-GFP. Dashed line, average of ratios from all four mutants (0.080 ± 0.016, n = 11). (G) Residual signal. Images as in B (Right) were collected from all qualifying cells expressing the mutants and then printed at fivefold-higher contrast. (H) domain structure of Stx-1a showing the helical segments Ha, Hb, Hc, the linker region L, the SNARE domain H3, and the transmembrane domain TM. The first and last residues of each from ref. 5. The mutant ΔHabc lacked residues 28 through 145 (2) and mutant Δ29-258 lacked residues 29 through 258 (13). In mutant Hc all, the contacts with Munc18-1 were replaced with alanines (S110, R114, I115, T118, Q119, T122, K126, and N135). Their locations were marked by upward vertical lines. In Habc all, the residues in the Ha domains were replaced as well (R28, F29, M30, F34, and R41, downward vertical lines). The scale in B applies to all images.
Fig. 1B is from a cell transfected with a mutant, ΔHabc. Lacking Stx-1a’s Ha, Hb, and Hc domains [residues 28 to 145 (2)], it fails to bind to Munc18-1 in vitro (6). No cluster was seen even though the cell was chosen to have a surface density of ΔHabc similar to that of wild-type Stx-GFP in Fig. 1A. Fig. 1 C and D compare Δf and s in the two cells. The values for Δf differed significantly (P < 0.0001, n = 82 granules for wild type, 34 for ΔHabc), those for s did not. ΔHabc apparently did not enter on-granule clusters, although endogenous syntaxin probably still did.
Because expression levels of Stx-GFP and its mutants varied strongly from cell to cell, we selected cells with S values in a restricted range corresponding to Stx-GFP surface densities of 147 to 588 molecules/μm2. Moreover, since ΔF varies linearly with S at moderate expression (28), we could correct for expression differences by using the ratio ΔF/S. Fig. 1E averages results from multiple cells as in Fig. 1A (Middle) and Fig. 1B (Right). Again, ΔHabc failed to enter granule clusters. Whatever molecules recruit Stx-1a to a granule evidently do not bind Stx-1a without its Habc domain.
Careful examination showed a small residual signal even with ΔHabc and three other mutants selected from a broader scan of Munc18-1 interaction sites (Fig. 1 F and H). Their signals were equally dim even though the mutants are expected to differ strongly in severity (Fig. 1F). For instance, outright removal of the Habc domain (ΔHabc) was no more effective than exchanging for alanines either 13 (Habc all) or 8 of its Munc18-interacting residues (Hc all). Even near-complete removal of the cytosolic domain diminished ΔF/S no further (Δ29 -258). Evidently, residues 29 through 258 do not participate in the residual signal. On average, it accounted for 8.0 ± 1.6% of wild type (dashed in Fig. 1F). Green fluorescence of immature mCherry cannot explain it (SI Appendix, Supplementary Text, Fluorescence Bleedthrough). When images with all four mutants in Fig. 1F were collected, averaged, and printed at higher contrast (Fig. 1G), a spot remained in the center, but it was broader than in Fig. 1E (Left). Its fluorescence apparently arose at some distance from granules.
Clustered Syntaxin Is Bound to Munc18-1.
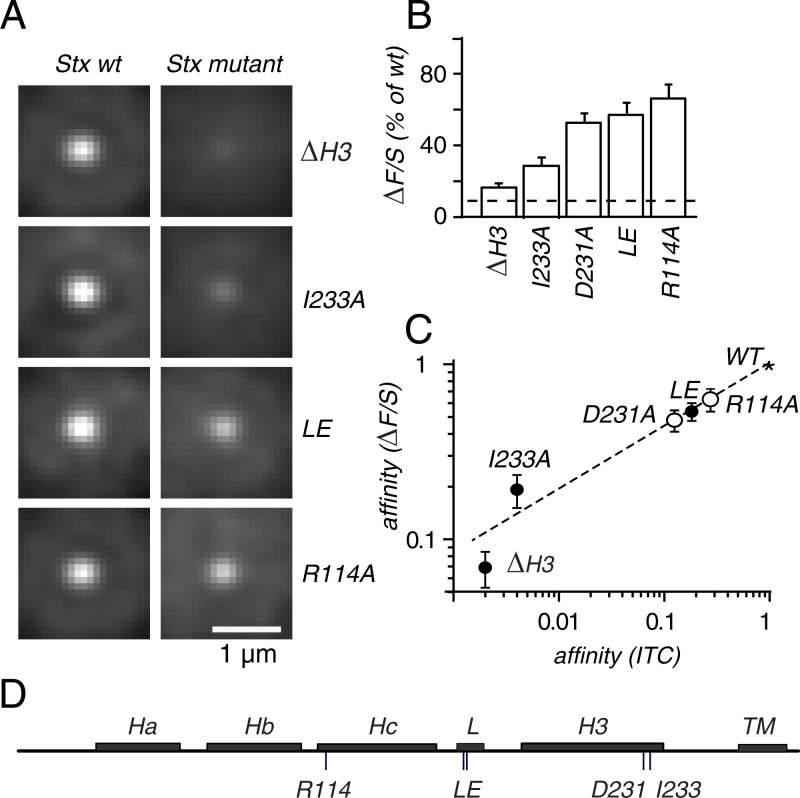
The mutants Δ29-258 and ΔHabc in Fig. 1F abolish the binding of Stx-1a to Munc18-1 in vitro (6), and Fig. 2 tests whether other mutants that inhibit binding similarly reduce recruitment. Outright removal of the SNARE domain (ΔH3) and the point mutation I233A both strongly diminished recruitment. In the linker domain, the effect of replacing L165 and E166 with alanines (LE) was more modest, but it was stronger than in our previous work (26). We consider the present result from a 10-fold larger number of cells more reliable. Finally, two Munc18-1 mutations, K46E and E59K, had previously been found to reduce the binding of Munc18-1 to Stx-1a in vitro (30). K46 contacts Stx-1a D231, and E59 forms a salt bridge with Stx-1a R114 (5). Interestingly, the Stx-1a mutations D231A and R114A measurably reduced ΔF/S.
Fig. 2.
Mutations inhibiting in vitro binding also inhibit clustering. (A), as in Fig. 1E but with mutants as indicated. ΔH3 lacks most of the SNARE domain [residues 203 through 254 (2)], whereas the other mutations are alanine substitutions and their locations are marked as short vertical lines in D. (B) As in Fig. 1F, ΔF/S values were less than wild type throughout (P < 0.02). In addition, the value of ΔH3 was higher than the residual signal (dashed line from Fig. 1, P < 0.01). Values for LE, D231A, and R114A were statistically indistinguishable. (C) The residual signal was subtracted from the data in B. The remainder was converted into affinities as described in SI Appendix, Fig. S2. The affinities on the abscissa were calculated from dissociation constants in the literature, as follows. Dots, Stx-1a mutants in table 1 of ref. 6; from Left to Right, they were Syx-1a1-179, Syx-1aI233A, and Syx-1aLE. Circles, Munc18-1 mutants K46E, E59K in table 1 of ref. 30. Isothermal titration calorimetry throughout. All values as a fraction of wild type (asterisk).
Our results in vivo may be compared to results in vitro obtained by others (Fig. 2C). First, we subtracted the residual signal from ΔF/S values and then converted the remainder into affinities (SI Appendix, Fig. S2). Next, affinities in vitro were calculated from the data in refs. 6 and 30 and compared with those determined from ΔF/S. Mutants Δ29-258 and ΔHabc were omitted as their signals were undetectable in vitro and in vivo. For the remaining mutants, the effects on ΔF/S in vivo seemed correlated with those on binding in-vitro. The finding is consistent with the idea that the Stx-1/Munch18-1 complex forms similarly both in clusters and in vitro. Effects in vitro were larger than in vivo. Most strikingly, I233A diminished the recruitment of Stx-1a by 7-fold and the binding of Munc18-1 more than 100-fold. Other proteins almost certainly contribute to the binding of Stx-1a to granules and are expected to add their binding energies (Discussion).
Munc18-Dependent and -Independent Clustering.
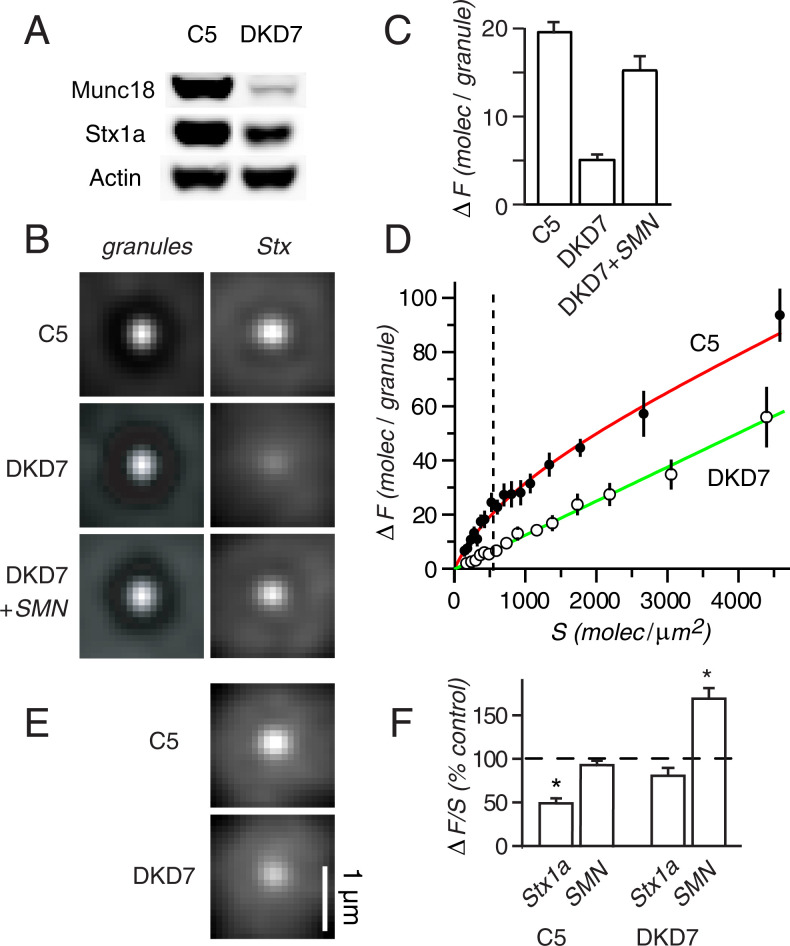
If Stx-1a must bind to Munc18-1 in order to cluster, then diminishing Munc18-1 levels should diminish Stx-GFP recruitment. To test this point, we compared DKD7, a PC12 cell line expressing Munc18-1 and Munc18-2 at low levels, to a control cell line, C5, that harbored Munc18-1 at wild-type levels (31). As in the earlier work, levels of Munc18-1 and Stx-1a were strongly reduced in DKD7 cells (Fig. 3A). The number of granules visible in TIRF was diminished modestly and their probability of exocytosis strongly (SI Appendix, Table S1).
Fig. 3.
Munc18-dependent and -independent clustering. (A) Levels of Munc18-1 , Stx-1a, and actin in C5 and DKD7 cells probed with antibodies indicated. (B) The granules and Stx-GFP images in C5 (Upper) and DKD7 cells (Below). Stx-GFP expression levels calculated from S values (in molecules/μm2) as follows. C5: 83 cells from seven cultures; S = 415 ± 10. DKD: 73 cells in seven cultures; S = 418 ± 10. DKD7+SMN: 65 cells in four cultures; S = 420 ± 9. All syntaxin images at the same contrast; granule images on auto-scale. (C) ΔF values for the cells in B. In DKD7 cells, the Stx-GFP plasmid contained the full CMV promoter. Dashed vertical line, endogenous Stx-1a level. (D), from measurements as in C but with a wider range of Stx-GFP expression levels. C5, 373 cells in eight cultures; DKD7, 280 cells in eight cultures. The cells were sorted according to their S values and then grouped in packets of 20. For each group, the mean ΔF was plotted against the mean S value. Results for C5 were fitted with ΔF = Bmax* (S / (S + k) + A * S, (red curve). The best fit was provided with Bmax = 37 ± 5 molec/granule, k = 899 ± 200 molec/μm2, and A =0.012 ± 0.004 μm2/granule. Green, linear component drawn separately (ΔF = 0.012 S). (E), as in B but at higher expression levels. They were, in molec/μm2, S = 3540 in C5 cells and S = 3550 in DKD7 cells. (F), as in Fig. 2 but cells in addition were transfected with unlabeled SMN or Stx-1a). Empty plasmid served as control. Asterisks differ with P < 0.01.
Cells were selected to express Stx-GFP at a surface density similar to that of endogenous Stx-1a in wild-type cells. Relative to C5, recruitment in DKD7 cells was strongly diminished but not abolished (Fig. 3 B and C). When calculated as in Fig. 1F, ΔF/S in DKD7 was 0.30 ± 0.05 times as large as in C5 but still significantly larger than the residual signal in Fig. 1 (P < 0.003, seven cultures). Recruitment was rescued in DKD7 cells by transfecting them with Munc18-1-SMN, an unlabeled Munc18-1 mutation that was resistant to the Munc18-1 knockdown plasmid (31). Next, clusters were examined over a wider range of Stx-GFP expression levels (Fig. 3D). In C5 cells, a plot of ΔF against S suggested the sum of two components, one saturating and the other linear (red curve). In DKD7 cells, only a linear component was present. It was indistinguishable from that in C5 cells, as seen when the linear component of C5 cells is drawn separately (green in Fig. 3D). We suggest that a portion of the Stx-1a recruitment to granules absolutely requires Munc18-1, and that it predominates at the low Stx-1a level found endogenously in wild-type cells (dashed vertical line). At strong Stx-GFP overexpression, Stx-1a forms on-granule clusters even without Munc18-1 (Fig. 3E).
Fig. 3F compares the effects of overexpressing unlabeled SMN or unlabeled Stx-1a. In C5 cells, SMN surprisingly failed to enhance recruitment; suggesting that Munc18-1 is not limiting at wild-type levels. The overexpression of unlabeled Stx-1a diminished recruitment, presumably because the added Stx-1a joined endogenous syntaxin in competing for granules. In DKD7 cells, by contrast, overexpressing unlabeled Stx-1a had no significant effect. Evidently, the lessened Stx-GFP recruitment in such cells was not for lack of endogenous Stx-1a.
Syntaxin Clusters in a Closed Conformation.
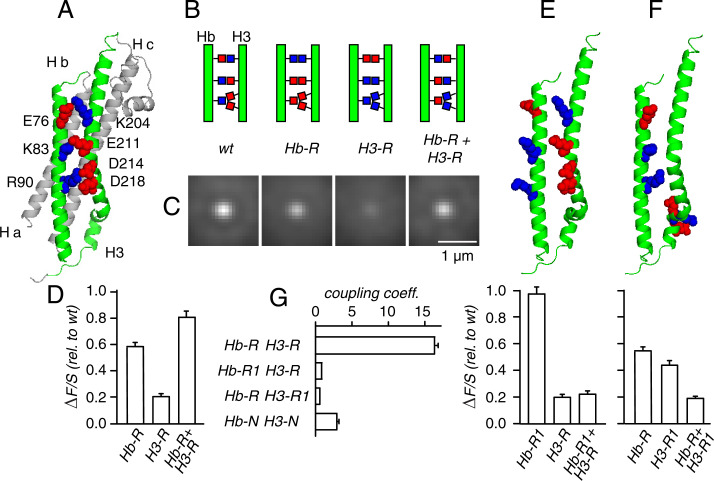
When Stx-1a binds to Munc18-1 in vitro, it is in a “closed conformation” [Fig. 4A (5)] wherein the Ha, Hb, and Hc domains cover the H3 (or SNARE) domain and leave no room for the proteins SNAP-25 and Syb that are required during membrane fusion. However, any clustering by homotypic aggregation of syntaxin SNARE domains would require open syntaxin (13, 14). To test for open Stx-1a, we took advantage of the three salt bridges that connect the Hb and H3 domains in closed Stx-1a. At each, an acidic (red) opposes a basic residue (blue in Fig. 4 A and B). To disrupt the bridges, we made a triple mutation reversing the charges on Hb (called Hb-R) and a quadruple mutation reversing the charges on H3 (called H3-R). Fig. 4B shows the mutated residues schematically, and Fig. 4 C and D demonstrate the effects of the two sets of mutations. On their own, both strongly diminished ΔF/S, consistent with a close proximity between Hb and SNARE domains. However, combining them (Hb-R + H3-R) largely rescued ΔF/S. The rescue was specific to the residues mutated. Charged residues close to those mutated in Hb-R lie on the outside surface of the molecule and are highlighted in Fig, 4E, Upper. Reversal of their charges (mutant Hb-R1) had no measurable effect on its own, and Hb-R1 failed to rescue ΔF/S when combined with H3-R (Fig. 4E, Lower). In H3, we found three more charged residues close to those mutated in H3-R. They are D231, R232, and E234 and are highlighted in Fig. 4F (Upper). Their charge reversal (mutant H3-R1) strongly diminished Stx-1a recruitment, but combining Hb-R with H3-R1 not only failed to rescue ΔF/S but diminished it further (Fig. 4F, Lower).
Fig. 4.
Complementing mutations rescue clustering. (A) The structure of Stx-1a (5) with Hb and H3 domains in green and the remainder in gray. Basic (blue) and acidic residues (red) form three salt bridges between Hb and H3. (B) Cartoon drawings of Hb and H3 show the bridging residues in wild type (wt) and in three mutants as follows. Hb-R, triple mutant reversing charges in the Hb domain (E76K/K83E/R90E), H3-R, quadruple mutant reversing charges in the SNARE domain (K204D/E211K/D214K/D218K), and Hb-R+ H3-R combining all seven replacements. (C) Clusters formed by wt and mutants as in Fig. 1E. Each experiment from one culture. Cells were transfected with either wt, Hb-R, H3-R or with both Hb-R and H3-R. Each image from between 24 to 39 cells containing about 25 acceptable granules/cell. (D) Six experiments as in C. (E) Upper as in A but highlighting charged Hb residues facing away from the molecule. Lower, effects of a triple mutant with reversed charges (set Hb-R1, E74K/K84E/K92E), of the set Hb-R and of the combination Hb-R1+ H3-R. From triplicate experiments except that the data for H3-R are from D. (F) Upper as in A but highlighting additional charged residues in H3. Lower, effects of a triple mutant reversing the highlighted charges (set H3-R1, D231K/R232E/E234K), of set Hb-R and of the combination Hb-R + H3-R1. From triplicate experiment except that the data for Hb-R are from D . (G) The coupling coefficients Ω of the indicated mutant pairs were calculated as in refs. 32 and 33 and then averaged. The SEs were 0.03 for (Hb-R1+ H3-R) and 0.22 for (Hb-R + H3-R1). For neutral replacements, see SI Appendix, Fig. S3. The affinities were calculated as in Fig. 2C. PC12-TM cells throughout.
We also replaced the charged residues in Fig. 4B with neutral polar amino acids (SI Appendix, Fig. S3). On their own, the two sets of mutants Hb-N and H3-N were less effective than the corresponding charge reversals. Nonetheless, the combined mutation Hb-N + H3-N partially rescued ΔF/S from the effect of H3-N (P < 0.04, n = 3). Evidently, only part of the rescue in Fig. 4 C and D is electrostatic.
To quantify how strongly two sets of mutations interact, we follow ref. 32 and define a coupling coefficient, Ω:
where a1, a2, and a3 indicate affinities relative to wild type. The affinities a1 and a2 apply when the mutations are applied singly and a3 applies when they are combined. If the effects of two mutations are independent, the change in affinity for the double mutant is the product of the changes for the single mutants, and Ω equals 1. Other values for Ω indicate coupling. Fig. 4G shows strong coupling between the mutants Hb-R and H3-R (Ω = 16.2 ± 1.4, n = 6) but not between Hb-R1 and H3-R or between Hb-R and H3-R1. Evidently, Hb and H3 are very near each other, and Stx-1a is closed. With neutral substitutions, Ω = 2.9 ± 0.6 for the pair Hb-N + H3-N.
An energy of interaction can be calculated as ΔΔGint = RT ln Ω, where R is the gas constant, and T is the absolute temperature (32, 33). ΔΔGint is 1.6 ± 0.05 kcal/mole for the pair Hb-R + H3-R and 0.6 ± 0.1 kcal/mole for Hb-N + H3-N. The two values suggest that two-thirds of the interaction energy is electrostatic.
The Interface with Munc18.
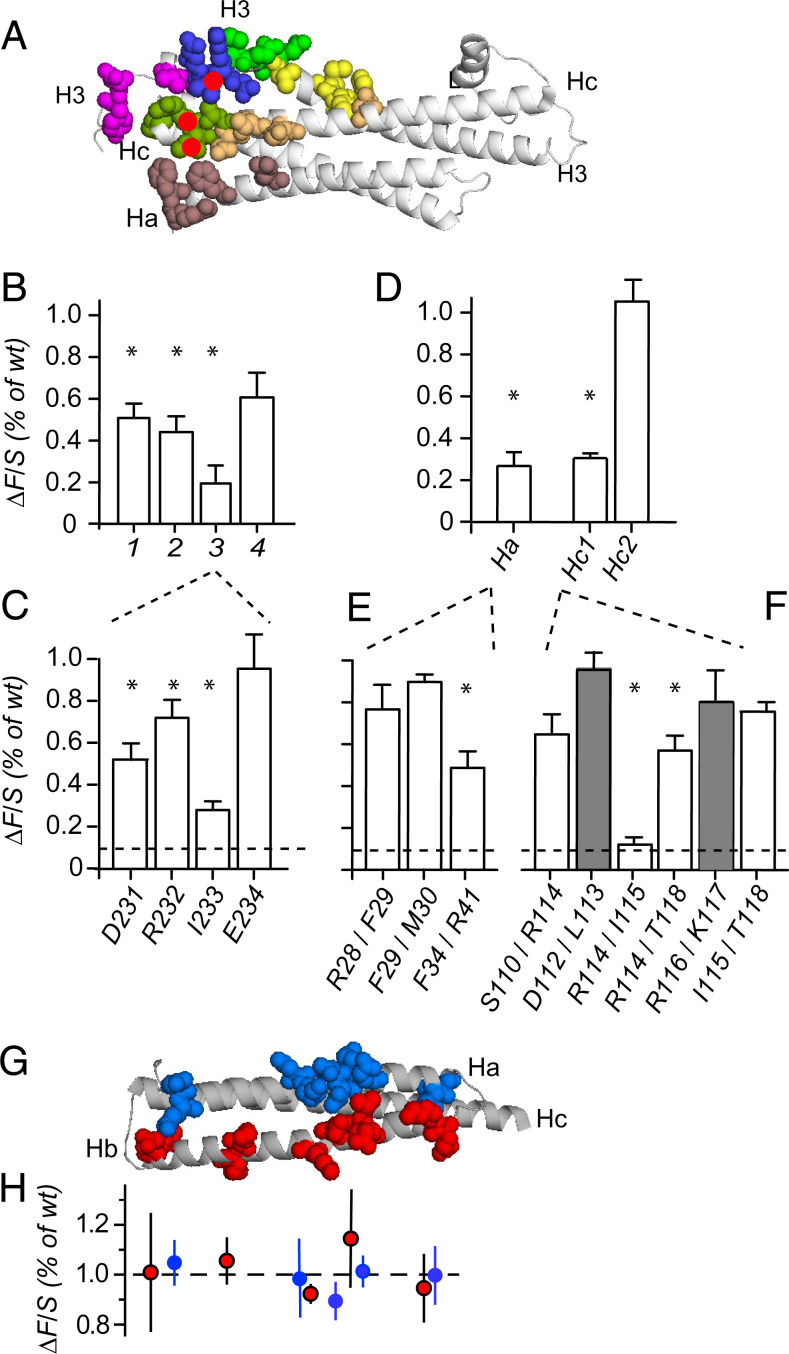
To learn which syntaxin residues are most important for recruitment, we replaced with alanines all residues that contact Munc18-1 and are not already alanines. Fig. 5A highlights them in different colors. First, residues in the H3 domain were replaced four at a time (Fig. 5B). Those in sets 1 (yellow) and 2 (green) contact domain 3 of Munc18, whereas those in sets 3 (blue) and 4 (magenta) contact Munc18-1 domain 1. Set 3 was the strongest, and Mutant 4 had little or no effect. When the mutations in 3 were applied individually, I233A was the strongest (Fig. 5C).
Fig. 5.
Most alanine substitutions diminish syntaxin recruitment only modestly. (A) Stx-1a (gray) with residues contacting Munc18-1 (5) in color; the SNARE domain (H3), Ha, and Hc are marked. Munc18-1 and Stx-1a’s N-terminal peptide removed for clarity. Red dots mark residues R114, I115, and I233 (Discussion). (B) The SNARE domain mutants are as follows. Set 1 (H213, F216, M217, and V223; yellow), set 2 (E224, S225, E228, and M229; green), set 3 (D231, R232, I233, and E234; blue), and set 4 (N236, V237, D242, and Y243; magenta). (C) Effects of individual residues in set 3. Among them, I233A had the largest effect (P < 0.02, n = 4). (D) Ha domain (R28, F29, M30, F34, and R41; brown) and two Hc domain mutants: Hc1 (S110, R114, I115, and T118; olive) and Hc2 (Q119, T122, K126, and N135; orange). (E and F) Among the pairwise replacements in Ha and Hc1, R114A/I115A had the largest efffect (P < 0.05, n = 3). Shaded residues do not contact Munc18-1 and are statistically no different from wild type. Two to four cultures and 55 to 85 cells per mutant. ΔF/S was determined as in Fig. 2 A and B. The asterisks in B through F denote statistically significant differences from wild type (P < 0.05). (G) Residues 27 through 157 of Stx-1a are in gray. Residues in colors were replaced with alanines. From Left to Right, they are D31/E32, D45/K46, E49/N50, E52/E53, and I61/L62 in the Ha domain (blue) and E103/E104, K92/K94, D81/K84, E77/L78, and E69/K70 in the Hb domain (red). (H) Effects of replacements in Ha (blue dots) and Hb (red dots). There are 52 to 94 cells for each mutant; the experiments were in duplicate or triplicate.
In the Ha domain, five residues contact domain 1 of Munc18-1 (brown in Fig. 5A). Replacing them all strongly diminished ΔF/S (Fig. 5D). Among pairwise mutations, only F34A/R41A significantly diminished recruitment (Fig. 5E).
Since replacing all nonalanine residues in the Hc domain abolished ΔF/S (Fig. 1F), residues were replaced four at a time (Fig. 5D). Mutant Hc2 (orange in Fig. 5A) was ineffective even though three residues contact Munc-18 domain 1 and one (N135) contacts domain 3. The large effect of Hc1 (olive in Fig. 5A) was explored in pairwise mutations in Fig. 5F. The effects were surprisingly modest for all pairs but one. Mutating nearby residues not in contact Munc18-1 (D112A/L113A and R116A/K117A, shaded in Fig. 5F) had no effect. However, R114A/I115A essentially abolished recruitment with ΔF/S statistically no larger than the residual signal. On their own, R114A and I115A caused modest effects (R114A, 0.68 ± 0.01, P < 0.02; I115A, 0.69 ± 0.01, P < 0.002; n = 4 in each). Apparently, the contribution of Hc to clustering strongly depends on a collaboration between R114 and I115.
As a control, we made pairwise replacements of Ha and Hb domain residues that point to the outside of the Stx-1a molecule and contact no other residue in the Munc18-Stx-1a complex (Fig. 5G). Most of the 10 pairs were acidic or basic. Their replacements had no measurable effect with ΔF/S = 102 ± 2% of wild type in n = 10 pairs of residues (Fig. 5H). Whatever recognition takes place between Stx-1a and granules, the residues mutated in Fig. 5G apparently play no part in it.
Off-Granule Syntaxin Clusters.
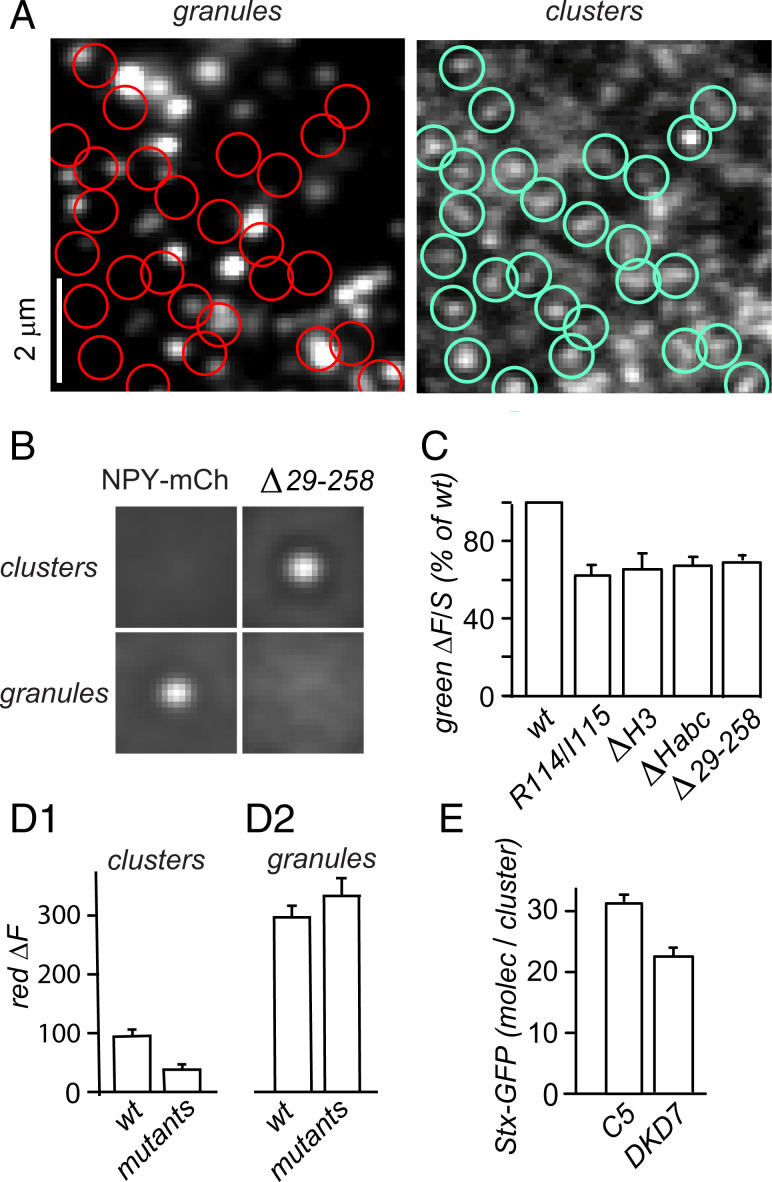
Aside from the on-granule clusters analyzed so far, cells also carry Stx-1a clusters that do not colocalize with granules (9, 10, 28, 29). Stx-1a mutants that are excluded from granule sites are expected to label such “off-granule” clusters in isolation. For example, the mutant Δ29-258 formed abundant clusters (Fig. 6B). Using the algorithm used earlier to locate granules in the red image, qualifying clusters were identified in the green channel instead (green circles, Right) and their locations copied into the NPY-mCh image (red circles, Left). Few or no red circles colocalized with a granule. As expected, averaged image pairs centered on Stx-1a clusters (“clusters” in Fig. 6B) showed a prominent spot in the Stx-GFP image but not in the NPY-mCh image. When image pairs were centered instead on granules (“granules” in Fig. 6B), the spot appeared in the NPY-mCh and not in the Stx-GFP channel. Evidently, our algorithm misses granules when aimed at clusters of Δ29-258, and such clusters lie off granule.
Fig. 6.
Mutants excluded from granule sites form off-granule clusters. (A) Granules (NPY-mCh) and Stx-1a clusters in a cell expressing the mutant Δ29-258. The clusters were located in the green image (blue circles) and then copied into the granule image (red). (B) Averages of image pairs centered on either Δ29-258 clusters or granules. The clusters are shown at one contrast and the granules at another. (C) Clusters of wild-type Stx-GFP and its mutants were located on the cells used in Figs. 1, 2, and 5. Their brightness was less in mutants than in the wild type (P < 0.02, triplicate experiments). Mean wild-type ΔF/S was 0.313 ± 0.011 (13 cultures). (D) Red fluorescence associated with either clusters (D1) or granules (D2). The clusters were detected as in A; their red fluorescence, if any, resulted from granules that happened to lie nearby. The granules were detected as in Fig. 1. The ΔF signals were read off NPY-mCh images in camera units. Cells expressed either Stx-GFP or one of the four mutants. Results with the mutants were averaged. In D1 but not D2 the red fluorescence was brighter when cells expressed Stx-GFP than when they expressed its mutants (P < 0.001, n = 13). (E) The clusters detected in DKD7 were dimmer than in C5 cells (P < 0.005, 7 cultures). Average S values in molecules/μm2 were 450 ± 10 in 90 C5 cells and 413 ± 8 in 92 DKD7 cells. With clusters directly detected in the green channel, C5 cells carried twice more Stx-GFP molecules than were associated with granules in Fig. 3C, presumably because only about one-half the granules carry a cluster [SI Appendix, Fig. S1C (28)]. This was confirmed by detecting clusters as in A–C and then selecting those that colocalized with a granule. Among 83 C5 cells, 29 harbored at least eight qualifying granules each. The granules were found to carry ΔF = 39 ± 4 molecules, nearly twice as many as the granules included in Fig. 3C (21 ± 2 molecules).
Fig. 6C compares clusters identified in the green channel. Some were formed by Stx-1a and others by Δ29-258 or the three other mutants that were excluded from granule sites in Figs. 1, 2, and 5. As in Fig. 6A (Right), off-granule clusters were abundant. They were also uniformly 30 to 40% dimmer than with wild-type Stx-GFP. Clusters with Stx-GFP, of course, form both on and off granule, and we attribute their greater average brightness to on-granule clusters. To confirm that some Stx-GFP clusters indeed lie on granule in our assay, we identified clusters as in A (Right) and measured the red fluorescence associated with them. Red fluorescence was brighter with wild type than with mutants (Fig. 6 D, 1, clusters) as expected from a presence of on-granule clusters. At the granule sites themselves (Fig. 6 D, 2, granules), the red fluorescence was vastly stronger, and there was no difference between wild type and mutants.
Since the mutants Δ29-258, ΔHabc and ΔH3 do not significantly bind to Munc18-1 in vitro (6), it was no surprise that Stx-GFP readily formed off-granule clusters in DKD7 cells. Just as the mutants in Fig. 6C formed clusters that were 30 to 40% dimmer than with Stx-GFP, so the Stx-GFP in DKD7 cells formed clusters that were 33% dimmer than in C5 cells (Fig. 6E). Moreover, Munc18-1 deprivation inhibited cluster formation clearly less effectively off (Fig. 6E) than on granule (Fig. 3C). Fig. 6 is consistent with off-granule clusters requiring no Munc18.
Although the mutations R114A/I115A, ΔH3, ΔHabc, and Δ29-258 strongly differed from one another, they formed clusters of near-equal brightness (Fig. 6C). It follows that they inhabited their clusters in equal numbers. Apparently, none of the residues 29 through 258 matter for how many syntaxin molecules are recruited. Moreover, the two mutants harboring an H3 domain (ΔHabc and R114A/I115A) were as bright as the two others that did not (ΔH3 and Δ29-258). Off-granule clustering therefore needs no H3 domain. On granule, R114A/I115A failed to form clusters despite its intact H3 domain (Fig. 5F), hence an H3 domain does not guarantee clustering. On-granule clustering does require an H3 domain (Fig. 2) but most likely for binding Munc18-1 (5, 6) and not for homo-oligomerization.
A recent study used superresolution microscopy to test whether off-granule clusters vary in size (34). Clusters of syntaxin mutant TMR (equal to Δ29-258) tended to be larger in diameter than those harboring a partial H3 domain, but the difference was small. We suggest that if such variations exist between our mutants, they were similarly small.
Discussion
We have imaged Stx clusters at granule sites in live cells sparingly transfected with Stx-GFP. Through mutations, we have sought to determine what portions of Stx-1a are required for its recruitment into clusters. For off-granule clusters, such requirements are minimal. Residues 257 through 288 (mutant SxTMH in ref. 12) readily formed clusters in PIP2-containing artificial lipid bilayers, even though this greatly truncated protein contained little more than the transmembrane region and the polybasic “juxta-membrane” domain (residues 260 through 265). The similar mutant Δ29-258 (Syx-TMR in refs. 13 and 34) also formed clusters in PC12 cells and did so both on membrane sheets (14) and in live cells (13). Off-granule, such clusters were no dimmer than those formed by the full-length Stx-1a mutant R114A/I115A (Fig. 6C). For off-granule clusters, therefore, nearly the entire cytoplasmic domain is dispensable, and so is Munc18-1 (Fig. 6D).
Stx-1a clustering on granules has strikingly different requirements. Residues 257 to 288 are insufficient (Fig. 1F), and the juxta-membrane domain is unnecessary (29). But other, more subtle mutations have large effects (I233A in Fig. 2, R114A/I115A in Fig. 5, and A111S/I115S in ref. 29). They suggest specific interactions between Stx-1a and a binding partner. Four findings point to Munc18-1 as that partner: 1) Munc18-1 is known to cluster at granule sites and/or bind to granules (11, 26, 35). 2) Munc18-1 is required for on-granule clusters, at least at physiological Stx-1a levels (Fig. 3). 3) Mutations that abolish Stx-1a binding to Munc18-1 in vitro also abolish its inclusion into on-granule clusters [Δ29-258 and ΔHabc (29)]. 4) Binding and inclusion into clusters are diminished in proportion (Fig. 2).
Our results suggest a high-affinity interaction between granules, Munc18-1 and Stx-1a. As long as both Stx-1a and Munc18-1 were present endogenously, they jointly out-competed Δ29-258, ΔHabc, and ΔΗ3 and excluded these mutants from on-granule clusters. To find which site on Stx-1a interacts most strongly with Munc18, we replaced pairs of Munc18-1–contacting residues with alanines. The effects were generally modest, and strong effects were observed in only two regions of the molecule. In the SNARE domain, mutant I233A was already known to diminish binding in vitro (6). In the Hc domain, R114A/I115A reduced recruitment to undetectable levels (Fig. 5), as did A111S/I115S (29). R114 makes an ionic bond with E59 of Munc18, while I115 contacts T56 in Munc18-1 and contributes to the “hydrophobic pocket” found in ref. 5. The two regions are separated by 117-120 residues in the Stx-1a peptide chain but are adjacent to each other in the closed form of Stx-1a. Red dots mark these three residues in Fig. 5A, and we suggest that they define Stx-1a’s binding site for Munc18. Stx-1 did bind to granules even without Munc18-1 but at reduced affinity (Fig. 3 B, C, and D). Binding without Munc18-1 is reminiscent of previous work in which chromaffin granules docked to the plasma membrane even without Munc18-1 (36).
Two earlier studies had used fluorescence recovery after photobleaching (13) and correlation analysis of immunostained fixed images (14) as proxies for syntaxin clustering. Unlike in Fig. 1 and ref. 27, the mutant ΔHabc was fully functional even though it cannot form a Stx-1a/Munc18-1 complex. The two studies were not directly based on the analysis of clusters, and no distinction was made between on- and off-granule syntaxin. They may have sampled Stx-1 assemblies different from the clusters investigated here.
An open Stx-1a conformation is generally assumed for clusters in membrane sheets. However, a strong interaction between two specific sets of mutants showed proximity between Hb and SNARE domains and hence a closed conformation. Others have estimated the distances between mutated residues by double-mutant analysis and by calculating the energy of interaction brought about by the double mutations (37). In an exhaustive study, double mutants were made in a bacterial enzyme and its protein inhibitor. Their energies of interaction, ΔΔGint, were determined and the distances between the partners in each pair measured by X-ray crystallography. ΔΔGint = 0.4 kcal/mole was observed for two residues separated by 0.71 nm, and no distances > 8 nm led to measurable coupling (33). Double mutants seem uniquely suited to detect close proximity. To compare our result with ref. 33, we assume that each copy of closed Stx-1a in a cluster can bind to Munc18-1 and hence to a granule. For the pair Hb-R and H3-R in Fig. 4, ΔΔGint was 1.6 ± 0.05 kcal/mole. The value presumably should be divided by three, as it represents the sum of energies brought about by the three residues mutated in Hb-R and the four in H3-R. The result (0.53 kcal/mole) is consistent with the idea that Hb and SNARE domains approach to within atomic distances where the three salt bridges form.
Ultimately, Syb must bind to Stx-1a before fusion can occur. Soluble Syb was found to hasten the washout of Munc18-1 from plasma membrane sheets as if it competed with that protein for Stx-1a. The result implies that Syb had access to Stx-1a, perhaps in a “half-open” conformation of Stx-1a (11). If Stx-1a in the Syb accessible pool was in fact clustered, then the opening of Stx-1a molecules must have been intermittent. Syb can bind to Munc18-1 in vitro, although it is displaced from that protein by the Habc domain of Stx-1a (38). Vsp33 (the Munc18-1 homolog used in vacuole fusion) can simultaneously bind the SNARE domains of both Vam3 (a syntaxin homolog) and Nyv1 (an R-SNARE–like Syb), at least if both SNAREs lack their N-terminal domains and are thus open (39). It is unclear how known structures of Munc18-1 would stably accommodate both Syb and closed Stx-1a. However, even fast fusion needs only three trans-SNARE complexes per granule (40–42). It is probable that most of the 50 Stx-1a/Munc18-1 complexes in a cluster do not participate in fusion and remain closed. Fig. 4 does not rule out a minority presence of open Stx-1a.
SNAP-25 is included in the acceptor complex for Syb. It coclusters with Stx-1a in fixed membrane sheets, possibly as part of the ternary SNARE complexes, and joins Syb and Munc18-1 in 100-nm clusters in fixed cultured neurons (43). In live endocrine cells, SNAP-25 forms clusters at granule sites (26) and may be present there as abundantly as Stx-1a (27). We do not know how, or whether, such SNAP-25 molecules also interact with Stx-1a/Munc18-1 complexes.
Presumably, the Stx-1a/Munc18-1 complex binds specifically to components on the granule membrane. An obvious partner would be Syb, but since on-granule Stx-1a clusters readily form also in cells deprived of Syb (28), Stx-1a/Munc18-1 complexes need another partner. In pancreatic beta cells, that partner may be granuphilin (44) [also called Slp-4 (45)]. Granuphilin binds to granules via Rab27a (46, 47), immunoprecipitates Stx-1a and Munc18-1 (35, 48, 49), and both proteins in turn precipitate granuphilin. The LE mutation of Stx-1a fails to do so (48, 49); therefore, inasmuch as LE promotes an open conformation (50, 51), Stx-1a is closed while in contact with granuphilin. Finally, the deletion of granuphilin in β cells prevents the docking of granules at the plasma membrane (52, 53). It is most likely that a Rab27/granuphilin complex on the granule mediates docking by binding to a Stx-1a/Munc18-1 complex at the plasma membrane (35, 52). In chromaffin and PC12 cells, melanophilin is apparently used for a similar purpose (35, 54). Following refs. 35 and 52, we suggest that docking ensues when a cluster of Stx-1a/Munc18-1 engages Rab27/granuphilin or Rab27/melanophilin. In so doing, Stx-1a/Munc18-1 anchors the granule to the plasma membrane. Such anchoring may be the main purpose of syntaxin clusters. Aside from docking, of course, clusters may supply the small group of Stx-1a/Munc18-1 complexes that ultimately unfurl their Habc domains, bind Syb and SNAP-25, and cause fusion.
Surprisingly, both granuphilin and melanophilin inhibit exocytosis (46, 52, 53). However, whereas the association with a granuphilin cluster approximately halves a granule’s chance of fusion (53), the association with an Stx-1a cluster increases it (26, 28). Apparently, Rab27/granuphilin and Stx-1a/Munc18-1 have both synergistic and opposing functions in exocytosis. To an endocrine cell, a partial inhibition of fusion may be an acceptable price for secure docking.
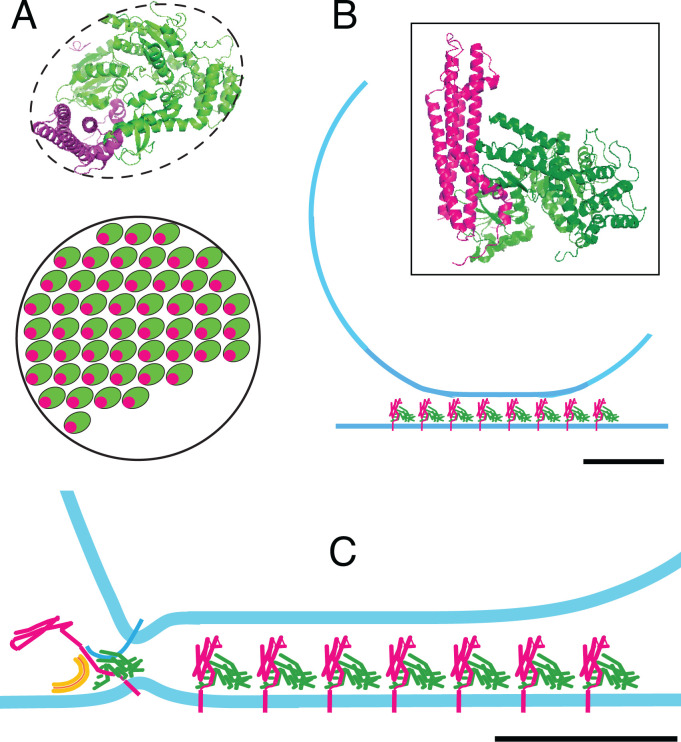
Finally, we wondered how many Stx-1a/Munc18-1 complexes can fit into a cluster. We view the size of the cluster as limited by the contact area between granule and plasma membrane. Fig. 7A (Upper) shows the Stx-1a/Munc18-1 complex (5) with the Stx-1a helices oriented perpendicular to the drawing (or the plasma membrane). Fig. 7A (Lower) is a cartoon of a single syntaxin cluster measuring 75 nm in diameter as in immunostained membrane sheets (12, 13). In a live, untransfected PC12 cell, the cluster harbors 50 Stx-1a molecules (27) as well as a similar number of Munc18-1 molecules, at least in beta cells (26). With each Stx-1a complexed to a Munc18, the complexes fill most of the cluster. Their distribution there is unknown, but they would cover 80% of the cluster’s area if tightly packed and leave room for other molecules only in small quantities.
Fig. 7.
Protein crowding in an on-granule cluster. (A) Upper, single Stx-1a/Munc18-1 complex (5) with Stx-1a helices oriented at right angle to the page. Lower, Stx-1a cluster in a PC12 cell. Its assumed diameter of 75 nm (12, 13) is an overestimate because it ignores the width of the point–spread function in a stimulated emission and depletion microscope (13). The cluster harbors 50 complexes (27) that occupy 80% of the circle, as is apparent if complexes are drawn tightly packed. (B) The granule (diameter typically 120 nm) resting on a cluster of complexes in the plasma membrane. Membranes are light blue. Inset, single complex with the Stx-1a helices aligned parallel to the page. Stx-1a is magenta or red, and Munc18-1 is green. (C) is similar to B but with a nascent fusion site added for illustration. The leftmost Stx-1a/Munc18-1 complex has opened and engaged a Syb molecule in the granule membrane (dark blue) as well as a SNAP-25 molecule (orange). Scale bars are 2.5 nm for single complexes (Upper in A and Inset in B) and 25 nm elsewhere.
Fig. 7B shows a side view of a granule resting on a bed of Stx-1a/Munc18-1 complexes. A single complex is shown in the inset. The complex alone would require an 8-nm distance between granule and plasma membrane. More space is required for additional proteins that granules carry to their docking site (not shown), including synaptotagmin (36), Rab27, Rab3 (26), and the granuphilin/rabphilin-like proteins (8, 53, 54). Not all will be present simultaneously. For example, the large protein CAPS/Munc13 (55) may be present only while it helps to open closed Stx-1a (56). We expect that crowding tends to make the interior of a cluster less hospitable for fusion. Perhaps ternary SNARE complexes operate instead at the periphery of the contact between fusing membranes as was found for the fusion of yeast vacuoles (57). In a speculative drawing (Fig. 7C), Stx-1a and Munc18-1 have started preparing for fusion. The N-terminal domain of Stx-1a has unfurled, and Syb has bonded with Munc18-1, perhaps in a groove between two antiparallel α-helices of Munc18-1 (39). A SNAP-25 molecule has been recruited as well.
Materials and Methods
Cells.
PC12-GR5 cells were used unless indicated otherwise and grown at 37 °C, 10%CO2 in high-glucose Dulbecco's modified essential medium containing 7% newborn calf serum, 7% horse serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin as in ref. 28. They were plated on poly-L-lysine–coated coverslips and transfected with plasmids (0.5 μg NPY-mCh and 1.0 μg Stx-1aGFP or its mutants) using lipofectamine 2000 following manufacturer instructions. Alternatively, the Neon transfection kit was used (Thermo-Fisher). Plasmids (2 μg NPY-mCh and 2 μg Stx1a-mGFP) were mixed into 110 μl Neon’s solution R, and 5 × 105 cells were suspended in the mixture. Up to 10-fold more Stx-1a–mGFP DNA or its mutants were sometimes used to increase expression levels. The cell suspension was loaded into a 110-μl tip, subjected to one 30-ms–long pulse of 1,040 V in the Neon electroporator, diluted with 0.5 ml growth medium, loaded on a coverslip, and placed in an incubator to allow cells to adhere. Finally, 2-ml growth medium was gently added to each well and the plate returned to the incubator. C5 and DKD7 cells were gifts from Dr. S. Sugita (Division of Fundamental Neurobiology, University Health Network, Toronto, ON M5T 2S8, Canada) and grown as described (31). PC12-TM cells were a gift from T. F. Martin (Department of Biochemistry, University of Wisconsin, Madison, WI) and grown as described (55). Coverslips were double coated with poly-L-lysine and rat tail collagen I. Transfection proceeded as with GR5 cells except with 1 × 106 cells and a “solution R” containing 8 μg NPY-mCh and 15 to 20 μg Stx-1a–mGFP or its mutants. Cells were imaged about 24 h after electroporation.
Plasmids.
NPY-mCherry was as described (28). In the Stx-1a–GFP plasmid, a (GGS)4 linker connected the C terminal of Stx-1a with the N terminal of monomeric enhanced GFP, and a crippled CMV promoter diminished expression. The construct served as the basis for all other mutants. Most are described in the Figure legends. Those in Figs. 1 A–E, 2, and 5 G and H were generated in-house by standard methods except that Habc all and Hc all in Figs. 1F and 5 B–F were made by Bio Basic Inc. SMN was generated by mutagenesis PCR from Munc18-1 complementary DNA, a kind gift from Dr. T.C. Südhof, by making the following nucleotide replacements: 249T/c, 252C/g, 256T/a, 257C/g, and 258T/c. Constructs in Fig. 4 were from ProNovus Bioscience LLC. In DKD7 cells where it was difficult to obtain sufficient Stx-GFP expression we either used the Stx-GFP plasmid with the full CMV promoter (Fig. 3 B, C, and E) or isolated suitable cells with a fluorescence-activated cell sorter (Fig. 3F). All plasmids were verified by sequencing.
Antibodies.
Primary antibodies detected Munc18-1 (ab3451r diluted 1:1,000) from Abcam, Stx-1 (sc-12736m diluted 1:200), and Actin (sc-1616g diluted 1:200), both from Santa Cruz Biotechnology. They were labeled with secondary antibodies conjugated with infrared dyes, as follows: Donkey anti-mouse IgR (Rockland 610-732-124, 1:10000) and Donkey anti-rabbit IgR (Rockland 611-732-127, 1:10000) conjugated with Dye 800, and donkey anti-goat IgR (Rockland 605-730-125, 1:5,000) conjugated with Dye 700DX. Signals were detected and quantified with the Odyssey Infrared Imaging System (Li-Cor Biosciences).
Imaging and Analysis.
The imaging equipment has been described [(28), SI Appendix, Supplementary Text, TIRF]. Cells were first viewed under yellow light (568 nm) that excited the red fluorescence of the granule marker, NPY-mCherry. A cell with distinct granules was chosen, brought into focus, and then imaged during a 0.1-s exposure to both 568- and 488-nm light. If the cell had sufficient green fluorescence, the image pair was stored for analysis. Images pairs for 20 to 50 cells were taken on each coverslip and collected into a stack. Throughout, we measured the average fluorescence in small regions of interest. In each image, a cell was outlined in the green channel along with a region that carried no cell and served as background. All other analyses were automated (SI Appendix, Supplementary Text, Analysis) using the program Metamorph (Universal Imaging Co.).
Stx-GFP expression varied widely among cells. Therefore, we generally (except in Fig. 3) selected cells with S values corresponding to Stx-GFP surface densities between 147 and 588 molecules/μm2. We excluded cells with less than eight acceptable granules and cells in which the SE for Δf was >10% of their S value. Also, the Stx-1a recruitment itself varied among wild-type cultures. For example, ΔF/S varied from 0.121 to 0.204 in the experiments of Fig. 2 (mean 0.158 + 0.005, 23 cultures). In all experiments therefore, we determined ΔF/S in wild-type and mutant cells from the same cultures. In Fig. 3, C5 and DKD7 cells were plated on the same day, harvested, and finally transfected on the next day. Cells were imaged 24 h after transfection. The ratio of ΔF/S values for mutant and wild-type or for DKD7 and C5 cells was the final result in each experiment. We discarded experiments in which the wild-type culture (or C5 cells) had an average ΔF/S < 0.100. Values are given ±standard error (SE), except for duplicate experiments where the standard deviation (SD) is given. Significance was tested with Student’s t test.
Contrast in Printed Images.
To visually compare images from different coverslips, we corrected them for differences in Stx-1a expression levels. First, the Stx-1a images of all qualifying granules in a wild-type cell were averaged (e.g., Fig. 1 A or B, Middle). The resulting images in turn were averaged over all cells on the coverslip (e.g., Fig. 1E). In this “coverslip average,” each cell is weighted equally.The background readings associated with each cell were averaged as well, the result was subtracted from the coverslip average, and the S value was measured. All pixel values were then multiplied by a constant (1,120 units) and divided by S, thus canceling out differences in Stx-1a–GFP expression levels. The coverslip with cells expressing mutant Stx-GFP was processed identically. The two resulting coverslip averages were printed at the same contrast. Because images of clusters have low contrast, the black level was set at about 1,000 units below S, and the white level at the brightest pixel in the wild-type image.
Supplementary Material
Acknowledgments
We thank Dr. S. Sugita for the C5 and DKD7 PC12 cell lines, Dr. T.H.F. Martin for his PC12 cells, and both for advice on how to maintain them. Heidi Owen helped with manuscript preparation, and the following provided thoughtful comments on the manuscript: Drs. Reinhard Jahn, D. Fasshauer, J. Taraska, W. Zagotta, and M. Lindau. This work was supported by NIH Grant MH60600 and the Vollum Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025748118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Rathore S. S., et al. , Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc. Natl. Acad. Sci. U.S.A. 107, 22399–22406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pobbati A. V., Stein A., Fasshauer D., N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313, 673–676 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Li F., Tiwari N., Rothman J. E., Pincet F., Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. Proc. Natl. Acad. Sci. U.S.A. 113, 10536–10541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misura K. M., Scheller R. H., Weis W. I., Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404, 355–362 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D., Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 27, 923–933 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misura K. M., Gonzalez L. C. Jr., May A. P., Scheller R. H., Weis W. I., Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a. J. Biol. Chem. 276, 41301–41309 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Trexler A. J., Sochacki K. A., Taraska J. W., Imaging the recruitment and loss of proteins and lipids at single sites of calcium-triggered exocytosis. Mol. Biol. Cell 27, 2423–2434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang T., et al. , SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20, 2202–2213 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang T., Margittai M., Hölzler H., Jahn R., SNAREs in native plasma membranes are active and readily form core complexes with endogenous and exogenous SNAREs. J. Cell Biol. 158, 751–760 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zilly F. E., Sørensen J. B., Jahn R., Lang T., Munc18-bound syntaxin readily forms SNARE complexes with synaptobrevin in native plasma membranes. PLoS Biol. 4, e330 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honigmann A., et al. , Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 20, 679–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieber J. J., et al. , Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317, 1072–1076 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Sieber J. J., Willig K. I., Heintzmann R., Hell S. W., Lang T., The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys. J. 90, 2843–2851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoyagi K., et al. , The activation of exocytotic sites by the formation of phosphatidylinositol 4,5-bisphosphate microdomains at syntaxin clusters. J. Biol. Chem. 280, 17346–17352 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Omar-Hmeadi M., Gandasi N. R., Barg S., PtdIns(4,5)P2 is not required for secretory granule docking. Traffic 19, 436–445 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Murray D. H., Tamm L. K., Clustering of syntaxin-1A in model membranes is modulated by phosphatidylinositol 4,5-bisphosphate and cholesterol. Biochemistry 48, 4617–4625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Bogaart G., et al. , Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milovanovic D., et al. , Hydrophobic mismatch sorts SNARE proteins into distinct membrane domains. Nat. Commun. 6, 5984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Destainville N., Schmidt T. H., Lang T., Where biology meets physics—A converging view on membrane microdomain dynamics. Curr. Top. Membr. 77, 27–65 (2016). [DOI] [PubMed] [Google Scholar]
- 21.van den Bogaart G., Lang T., Jahn R., Microdomains of SNARE proteins in the plasma membrane. Curr. Top. Membr. 72, 193–230 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Murray D. H., Tamm L. K., Molecular mechanism of cholesterol- and polyphosphoinositide-mediated syntaxin clustering. Biochemistry 50, 9014–9022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khuong T. M., et al. , Synaptic PI(3,4,5)P3 is required for syntaxin1A clustering and neurotransmitter release. Neuron 77, 1097–1108 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Jahn R., Scheller R. H., SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ullrich A., et al. , Dynamical organization of syntaxin-1A at the presynaptic active zone. PLOS Comput. Biol. 11, e1004407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandasi N. R., Barg S., Contact-induced clustering of syntaxin and munc18 docks secretory granules at the exocytosis site. Nat. Commun. 5, 3914 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Knowles M. K., et al. , Single secretory granules of live cells recruit syntaxin-1 and synaptosomal associated protein 25 (SNAP-25) in large copy numbers. Proc. Natl. Acad. Sci. U.S.A. 107, 20810–20815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barg S., Knowles M. K., Chen X., Midorikawa M., Almers W., Syntaxin clusters assemble reversibly at sites of secretory granules in live cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20804–20809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin P., et al. , Syntaxin clusters at secretory granules in a munc18-bound conformation. Mol. Biol. Cell 29, 2700–2708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han G. A., et al. , Munc18-1 domain-1 controls vesicle docking and secretion by interacting with syntaxin-1 and chaperoning it to the plasma membrane. Mol. Biol. Cell 22, 4134–4149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han L., et al. , Rescue of Munc18-1 and -2 double knockdown reveals the essential functions of interaction between Munc18 and closed syntaxin in PC12 cells. Mol. Biol. Cell 20, 4962–4975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranganathan R., Lewis J. H., MacKinnon R., Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron 16, 131–139 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Schreiber G., Fersht A. R., Energetics of protein-protein interactions: Analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248, 478–486 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Merklinger E., et al. , The packing density of a supramolecular membrane protein cluster is controlled by cytoplasmic interactions. eLife 6, 679–688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuboi T., Fukuda M., The Slp4-a linker domain controls exocytosis through interaction with Munc18-1.syntaxin-1a complex. Mol. Biol. Cell 17, 2101–2112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Wit H., et al. , Synaptotagmin-1 docks secretory vesicles to syntaxin-1/SNAP-25 acceptor complexes. Cell 138, 935–946 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Carter P. J., Winter G., Wilkinson A. J., Fersht A. R., The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell 38, 835–840 (1984). [DOI] [PubMed] [Google Scholar]
- 38.Xu Y., Su L., Rizo J., Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry 49, 1568–1576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker R. W., et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua Y., Scheller R. H., Three SNARE complexes cooperate to mediate membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 98, 8065–8070 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohrmann R., de Wit H., Verhage M., Neher E., Sørensen J. B., Fast vesicle fusion in living cells requires at least three SNARE complexes. Science 330, 502–505 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Shi L., et al. , SNARE proteins: One to fuse and three to keep the nascent fusion pore open. Science 335, 1355–1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertsinidis A., et al. , Ultrahigh-resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ. Proc. Natl. Acad. Sci. U.S.A. 110, E2812–E2820 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Takeuchi T., Yokota H., Izumi T., Novel rabphilin-3-like protein associates with insulin-containing granules in pancreatic beta cells. J. Biol. Chem. 274, 28542–28548 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Fukuda M., Mikoshiba K., Synaptotagmin-like protein 1-3: A novel family of C-terminal-type tandem C2 proteins. Biochem. Biophys. Res. Commun. 281, 1226–1233 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Tomas A., Meda P., Regazzi R., Pessin J. E., Halban P. A., Munc 18-1 and granuphilin collaborate during insulin granule exocytosis. Traffic 9, 813–832 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Yi Z., et al. , The Rab27a/granuphilin complex regulates the exocytosis of insulin-containing dense-core granules. Mol. Cell. Biol. 22, 1858–1867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torii S., Takeuchi T., Nagamatsu S., Izumi T., Rab27 effector granuphilin promotes the plasma membrane targeting of insulin granules via interaction with syntaxin 1a. J. Biol. Chem. 279, 22532–22538 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Fukuda M., Slp4-a/granuphilin-a inhibits dense-core vesicle exocytosis through interaction with the GDP-bound form of Rab27A in PC12 cells. J. Biol. Chem. 278, 15390–15396 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Dulubova I., et al. , A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 18, 4372–4382 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greitzer-Antes D., et al. , Tracking Ca2+-dependent and Ca2+-independent conformational transitions in syntaxin 1A during exocytosis in neuroendocrine cells. J. Cell Sci. 126, 2914–2923 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Gomi H., Mizutani S., Kasai K., Itohara S., Izumi T., Granuphilin molecularly docks insulin granules to the fusion machinery. J. Cell Biol. 171, 99–109 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuno K., Fujita T., Gomi H., Izumi T., Granuphilin exclusively mediates functional granule docking to the plasma membrane. Sci. Rep. 6, 23909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desnos C., et al. , Rab27A and its effector MyRIP link secretory granules to F-actin and control their motion towards release sites. J. Cell Biol. 163, 559–570 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabachinski G., Kielar-Grevstad D. M., Zhang X., James D. J., Martin T. F., Resident CAPS on dense-core vesicles docks and primes vesicles for fusion. Mol. Biol. Cell 27, 654–668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai Y., et al. , Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95, 591–607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Seeley E. S., Wickner W., Merz A. J., Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108, 357–369 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.