Abstract
The choice between meiosis and alternative developmental pathways in budding yeast depends on the expression and activity of transcriptional activator Ime1. The transcription of IME1 is repressed in the presence of glucose, and a low basal level of IME1 RNA is observed in vegetative cultures with acetate as the sole carbon source. IREu, a 32-bp element in the IME1 promoter, exhibits upstream activation sequence activity depending on Msn2 and -4 and the presence of acetate. We show that in the presence of glucose IREu functions as a negative element and that Sok2 mediates this repression activity. We show that Sok2 associates with Msn2. Sok2 functions as a general repressor whose availability and activity depend on glucose. The activity of Sok2 as a repressor depends on phosphorylation of T598 by protein kinase A (PKA). Relief of repression of Sok2 depends on both the N-terminal domain of Sok2 and Ime1. In the absence of glucose and the presence of Ime1 Sok2 is converted to a weak activator. Overexpression of Sok2 or mild expression of Sok2 with its N-terminal domain deleted leads to a decrease in sporulation. Previously it was reported that overexpression of Sok2 suppresses the growth defect resulting from a temperature-sensitive PKA; thus Sok2 has a positive role in mitosis. We show that Candida albicans Efg1, a homolog of Sok2, complements sok2Δ in repressing IREu. Our results demonstrate that Sok2, a positive regulator of mitosis, and Efg1, a positive regulator of filamentation, function as negative regulators of meiosis. We suggest that cells use the same regulators with opposing effects to ensure that meiosis will be an alternative to mitosis.
In the budding yeast Saccharomyces cerevisiae the choice between meiosis-sporulation and alternative developmental pathways such as the mitotic cell cycle, filamentous growth, and G1 arrest depends on the expression and activity of master regulator Ime1. Cells with IME1 deleted arrest in meiosis at G1, prior to any meiotic event, i.e., transcription of meiosis-specific genes, premeiotic DNA replication, meiotic recombination, and nuclear divisions (16, 43). IME1 encodes a transcriptional activator (24, 42) that is recruited to the promoters of early meiosis-specific genes by interacting with sequence-specific DNA-binding protein Ume6 (33).
The environmental signals that determine the decision to exit mitosis and embark upon meiosis regulate both the transcription and the activity of Ime1 (16, 33). Cells grown in vegetative media with glucose as the sole carbon source (SD) have undetectable levels of transcripts of IME1. A low but detectable basal level is present in vegetative media with acetate as the sole carbon source (SA). Upon nitrogen depletion in the presence of acetate (SPM) the level of IME1 mRNA is transiently increased in MATa/MATα diploids but not in cells carrying only one of the two mating type alleles (16). This regulated transcription results from a combinatorial effect of multiple elements present in the remarkably large 5′ region (over 2,100 bp long) of IME1 (35). The nitrogen signal is transmitted through a single upstream repression sequence (URS) element, whereas the glucose signal is transmitted to at least three distinct upstream activation sequence (UAS) elements in the promoter of IME1 (35; G. Shenhar and Y. Kassir, unpublished data).
The cyclic AMP (cAMP)-dependent protein kinase (PKA) signal pathway plays a pivotal role in the decision between mitosis and meiosis (26). Mutations that result in high PKA activity, such as mutation of constitutively active RAS2val19 and deletion of BCY1 (the regulatory subunit of PKA) result in sporulation deficiency. On the other hand, mutations that result in no PKA activity, such as temperature-sensitive mutations in the gene for adenylate cyclase (CYR1) or RAS exchange factor (CDC25), cause cell cycle arrest and entry into meiosis in the presence of nitrogen (4, 26). The PKA signal pathway negatively regulates the transcription of IME1 (27). Biochemical evidence indicates that this pathway transmits a glucose signal (20, 46). We have shown, accordingly, that this signal pathway transmits the glucose signal to IREu, a 32-bp UAS element in the 5′ region of IME1 (35).
This paper further characterizes the mode by which the PKA pathway regulates the function of IREu. This element, by itself, exhibits low UAS activity in the presence of glucose and a 10-fold-increased activity in the absence of glucose and the presence of acetate (35). Two known targets of PKA, Msn2 and its homolog Msn4 (11, 25, 37, 41), bind to and promote the UAS activity of IREu (35). In this report we characterize the role of a third protein, Sok2, a putative DNA-binding protein (47). SOK2 was identified as a gene dosage suppressor for the temperature-conditional growth defect of a tpk1Δ tpk2-ts tpk3Δ strain (47). TPK1 to -3 encode the catalytic subunits of PKA (4). Suppression by Sok2 requires the presence of residual PKA activity (47). Sok2 is apparently a negative regulator of transcription: in its absence the levels of GAC1 and SSA3 are increased, whereas when overexpressed it causes a reduction in their expression (47). Moreover, Sok2 is also a negative regulator of filamentous growth (47). In this report we show that in the presence of glucose IREu functions as a negative element and that Sok2 mediates this activity. We show that Sok2 functions as a general repressor whose activity depends on glucose. The carbon source regulates the availability of Sok2, as well as its activity. The activity of Sok2 as a repressor depends on phosphorylation of T598 by PKA. Relief of repression of Sok2 in the absence of glucose and the presence of acetate as the sole carbon source depends on both the N-terminal domain of Sok2 and Ime1.
MATERIALS AND METHODS
Yeast strains.
The relevant genotypes of the strains used are described in Table 1. For Y1075 and Y1076, a one-step replacement of IME1 by ime1::hisG-URA3-hisG with a deletion from −1118 to +946 was accomplished following transformation of Y1064 and Y1065, respectively, with a 7.2-kb XhoI-SacII fragment from P1408. URA+ transformants were patched on 5-fluoro-orotic acid plates to select for derivatives which had recombined out the URA3 gene. For Y1077 and Y1080, the gal1-lacZ chimera was integrated at GAL1 by transformation of Y1064 and Y1075, respectively, with pRY171 (49) cut with XhoI. For Y1078 and Y1093, the CDC25 allele in Y1064 and Y1089 (isogenic to Y1065 but rim11::LEU2) was replaced by cdc25-2::URA3 following transformation with a SalI-PvuII fragment from P1902 (27). For Y1078a, Y1161, and Y1086, the IREu-his4-lacZ chimera was integrated at the leu2-3,112 allele by transformation of Y1078, Y1064, and Y1076, respectively, with YIp1994 digested with PpuMI. For Y1162 and Y1170, a one-step deletion protocol was used to replace the SOK2 alleles in Y1161 and Y1132, respectively, with a sok2Δ::TRP1 fragment (from plasmid P2145).
TABLE 1.
List of strains and relevant genotypes
| Strain | Relevant genotype | Remarks |
|---|---|---|
| Y1064 | MATaura3-52 leu2,3-112 trp1Δ his3::hisG ade2-1 met gal80::hisG gal4::hisG | |
| Y1065 | MATα ura3-52 trp1Δ leu2-3,112 his3::hisG ade2-R8 gal80::hisG gal4::hisG | |
| Y1075 | MATaime1::hisG | Isogenic to Y1064 |
| Y1076 | MATα ime1::hisG | Isogenic to Y1065 |
| Y1077 | MATagal80::hisG gal4::hisG gal1::gal1-lacZ-URA3 | Isogenic to Y1064 |
| Y1078 | MATacdc25-2::URA3 | Isogenic to Y1064 |
| Y1078a | MATaleu2,3-112::LEU2-IREu-his4-lacZ cdc25-2::URA3 | Isogenic to Y1064 |
| Y1080 | MATagal80::hisG gal4::hisG ime1::hisG gal1::gal1-lacZ-URA3 | Isogenic to Y1075 |
| Y1086 | MATα ime1::hisG leu2,3-112::LEU2-IREu-his4-lacZ | Isogenic to Y1076 |
| Y1093 | MATα rim11::LEU2 cdc25-2::URA3 | Isogenic to Y1065 |
| Y1132 | MATα msn2::HIS3 msn4::URA3 leu2,3-112::LEU2-IREu-his4-lacZ | Isogenic to Y1065 (35) |
| Y1161 | MATa leu2-3,112::LEU2-IREu-his4-lacZ | Isogenic to Y1064 |
| Y1162 | MATa sok2Δ::TRP1 leu2-3,112::LEU2-IREu-his4-lacZ | Isogenic to Y1161 |
| Y1170 | MATα msn2::HIS3 msn4::URA3 sok2Δ::TRP1 leu2,3-112::LEU2-IREu-his4-lacZ | Isogenic to Y1132 |
| Y422 | MATa/MATα ura3-52/ura3-52 trp1Δ/trp1Δ leu2-3,112/leu2-3,112 ade2-1/ade2-R8 his4-519/HIS4 his6-1/HIS6 can1/CAN1 | Isogenic to Y1064 × Y1065 |
Plasmids.
pAS2 carries pADH1-GAL4(1-147)-HA-ADHt on a 2μm TRP1 CYH2 vector (14). pBIST carries pGAL1-EFG1 in YEplac195 (45). YCp1376 carries ime1 (−1122 to +202)-lacZ on a URA3 ARS1 CEN4 vector (35). P1408 carries ime1 (−3762 to −1118)-hisG-URA3-hisG-ime1 (+946 to +2132) on Bluescript. YEp1784 carries sok2 with a deletion between +18 and +565 (verified by sequencing) on a 2μm TRP1 vector. In this deletion Sok2 is expressed from an internal ATG, leading to the formation of truncated Sok2 lacking the N-terminal 247 amino acids. This plasmid was constructed by replacing the URA3 marker in PMW61 (47) with TRP1, using in vivo recombination. YIp1994 carries ime1 (−1153 to −1122)-his4-lacZ (designated IREu-his4-lacZ) on a LEU2 vector (35). P2108 carries SOK2 (+1 to +2361) on pGEM T-easy cloning vector. This plasmid was constructed by inserting a 2.7-kb PCR fragment derived from oligonucleotides sok2+1 and sok2-2675B using genomic DNA as a template into pGEM-T-easy vector (Promega). P2145 carries sok2::TRP1 on Bluescript. This plasmid was constructed in two steps. First a 3.8-kb BamHI-HindIII fragment from pMW61 (47) was ligated to a Bluescript vector cut with the same enzymes. Then TRP1 on a 1.4-kb EcoRI fragment was inserted into the resulting plasmid cut with EcoRI. This insertion created a deletion of most of the coding region. YIp2218 carries UASGAL1-UASHIS4-his4-lacZ on a LEU2 vector (M. Cohen-Koren, personal communication). YIp2254 carries ime1 (−4401 to +201)-lacZ on a LEU2 vector. This plasmid was constructed in two steps. First a 2.6-kb NcoI-SacI fragment from YCp1376 and a 3.8-kb EcoRI-NcoI fragment from YCp214 (35) were ligated together to vector YIpLac128 (9) cut with SacI and EcoRI. Then a 6.5-kb SacI-EcoRI fragment from the resulting plasmid and a 1.5-kb SacI-PstI fragment from pMC1781 were ligated to YIpLac128 (9) cut with PstI and EcoRI. YIp2296 carries ime1 (−1153 to +202)-lacZ on a LEU2 vector. This plasmid was constructed in two steps. First a 0.8-kb PCR fragment derived from oligonucleotides ime1-IREu and ime1-348R was inserted into pGEM-T-easy vector (Promega). Then a 0.6-kb NcoI-SphI fragment from the resulting plasmid and a 6.5-kb NcoI-PpuMI fragment from YIp2254 were ligated to YIpLac128 (9) cut with PpuMI and SphI. YIp2297 carries ime1 (−1149 to +202)-lacZ on a LEU2 vector. This plasmid was constructed in two steps. First a 0.8-kb PCR fragment derived from oligonucleotides ime1-nIREu and ime1-348R was inserted into pGEM-T-easy vector (Promega). Then a 0.6-kb NcoI-SphI fragment from the resulting plasmid and a 6.5-kb NcoI-PpuMI fragment from YIp2254 were ligated to YIpLac128 (9) cut with PpuMI and SphI. YEp2314 carries pADH1-GAL4(1-147)-SOK2 on a TRP1 2μm vector. This plasmid was constructed by three-piece ligation between a 2.3-kb BamHI-NcoI fragment from P2108, a 2.8-kb NcoI-SacI fragment from pAS2, and YEpLac112 (9) cut with SacI and BamHI. YEp2342 carries pSOK2-3xHA-SOK2 on a 2μm TRP1 vector. This plasmid was constructed in several steps. A 120-bp PCR fragment derived from oligonucleotides HA-ATG and HA-RevNcoI using pMPY-3xHA (38) as a template was inserted into pGEM-T-easy vector (Promega) to create P2339. A 1.7-kb PCR fragment derived from oligonucleotides SOK2-1r and SOK2-1784 using genomic DNA as a template was inserted into pGEM-T-easy vector (Promega) to create P2172. In the second step we performed a three-piece ligation between a 120-bp SacII-NcoI fragment from P2339, a 1.7-kb SacII-SalI fragment from P2172, and vector YIpLac211 (9) cut with NcoI and SalI. In the third step we used three-piece ligation between a 1.9-kb SalI-NcoI fragment from the resulting plasmid, a 2.3-kb BamHI-NcoI fragment from P2108, and vector YIpLac128 (9) cut with BamHI and SalI. YIp2344 carries ime1 (−4401 to −1154 and −1122 to +202)-lacZ on a LEU2 vector. This plasmid was constructed in three steps. First a 0.4-kb XhoI fragment from YCp1975 (35) was inserted into YIpLac128 (9) cut with SalI. Then a 2.2-kb SphI-PpuMI fragment from the resulting plasmid and a 0.6-kb NcoI-SphI fragment from YIp2296 were ligated to a 6.7-kb PpuMI-NcoI fragment from YIp2254. Then a 3.5-kb NheI-SacI fragment from the resulting plasmid was inserted into YIp2254 cut with the same enzymes. P2372 carries 3xHA-SOK2 on Bluescript vector. This plasmid was constructed by inserting a 2.4-kb XbaI-XmaI 3xHA-SOK2 fragment from YEp2342 into Bluescript cut with the same enzymes. YEp2382 carries CMVp-tTA and 7xtetO-cyc1-3xHA-SOK2 on a 2μm URA3 vector. This plasmid was constructed by inserting a NotI-HindIII fragment from P2372 into pcm190 (8) cut with the same enzymes. YEp2420 carries pSOK2-HA-lacZ on a 2μm LEU2 vector. This plasmid was constructed by inserting a 1.8-kb BamHI-SalI fragment from YEp2342 into E366R (29) cut with the same enzymes. YEp2432 carries pSOK2-3xHA-SOK2 on a 2μm HIS3 vector. This plasmid was constructed by ligating a 4.2-kb SalI-SacI fragment from YEp2342 into pRS423 (40) cut with the same enzymes. YEp2452 carries pSOK2-3xHA-sok2T598A on a 2μm HIS3 vector. This plasmid was constructed by site-directed mutagenesis using YEp2432 and oligonucleotide SOK2T598A. YEp2486 carries pCDC28-3xHA-SOK2 on a 2μm URA3 vector. This plasmid was constructed by three-piece ligation between a 2.5-kb NotI-SalI 3xHA-SOK2 fragment from P2372, a 0.4-kb NotI-SacII fragment carrying pCDC28 from P2301 (M. Cohen-Koren, personal communication), and vector pRS426 (40) cut with SacII and SalI. YEp2520 carries pSOK2-3xHA-sok2T598A on a 2μm URA3 vector. This plasmid was constructed by inserting a 4.2-kb SacI-SalI fragment from YEp2452 into pRS426 (40) cut with the same enzymes. YEp2530 carries pADH1-gal4(1-147)-sok2T598A on a TRP1 2μm vector. This plasmid was constructed by three-piece ligation between a 3.6-kb SpeI-SacI fragment from YEp2314, a 1.6-kb SpeI-BamHI fragment from YE2452, and vector YEpLac112 (9) cut with BamHI and SacI. YEp2534 carries pCDC28-GST on a 2μm TRP1 vector. This plasmid was constructed by three-piece ligation between a 0.45-kb SphI-NdeI fragment carrying pCDC28 from P2301 (M. Cohen-Koren, personal communication), 0.7-kb NdeI-KpnI fragment carrying the glutathione S-transferase (GST) gene from pRD56 (I. Herskowitz, personal communication), and vector YEplac112 (9) cut with SphI and KpnI. YEp2536 carries pCDC28-GST-MSN2 on a 2μm TRP1 vector. This plasmid was constructed by three-piece ligation between a 1.1-kb SphI-BamHI fragment from YEp2534, a 2.1-kb BamHI-HindIII fragment from YEp1784, and vector YEplac112 (9) cut with SphI and HindIII. YEp2558 carries pCDC28-3xHA-sok2T598A on a 2μm URA3 vector. This plasmid was constructed by three-piece ligation between a 2.3-kb BamHI-SacI fragment from YEp2520, a 0.5-kb SacI-SphI fragment from YEp2486, and vector pRS426 (40) cut with BamHI and SphI. YEp2573 carries pSOK2-3xHA-sok2T598A on a 2μm HIS3 kanr vector. This plasmid was constructed by three-piece ligation between a loxp-kanr-loxp cassette on a 1.5-kb NotI-XhoI fragment from a derivative of pUG6 (12), P2148, a 4.2-kb XhoI-SacI fragment from YEp2452, and vector pRS423 (40) cut with NotI and SacI. YEp2588 carries MSN2 on a 2μm LEU2 vector. This plasmid was constructed by inserting a 2.4-kb SacI-HindIII fragment from YEp1636 (35) into YEpLac181 (9) cut with the same enzymes.
Oligonucleotides.
Oligonucleotides and their sequences were as follows: HA-ATG, 5′ GGCCGGTCTAGAATGTACCCATACGATGTTCCT; HA-revnco, 5′ CCATGGCAGCGTAATCTGGAACGTC; ime1-IREu, 5′ AGCGCCTTTGATCCTTCCCCTCGAAGACGAAAA; ime1-348R, 5′ GCCGGCGAGCTCCATAAAAGAGGAAAAGT; ime1-nIREu, 5′ GCTCTAGACGTCTTCGAGGGGAAGGA; sok2+1, 5′ GGGCATGCCATGGCTATGCCCATCGGTAACCCA; Sok2-NC+1, 5′ CCATGGATGCCCATCGGTAACCCA; SOK2-2676B, 5′ CCGCGGATCCAAGGAATTCATAGTT; SOK2-1R, 5′ GAGATCTAAAAGGACCTTACCAGGG; Sok2-1784, 5′ GCCGGTGGTAAATACGCG; SOK2T598A, 5′ CCTTGAAAAAATGCGCAATGCCTAAC.
Media and genetic techniques.
PSP2 (SA) (minimal acetate medium) and SPM (sporulation medium) have been described (17). Synthetic dextrose medium (SD) has been described (39). Both SD and SA are vegetative media. Meiosis was induced as follows. Cells were grown to 107 cells/ml in PSP2 supplemented with the required amino acids, washed once with water, and resuspended in SPM. Yeast transformation with lithium acetate was done as described previously (9). Standard methods for DNA cloning and transformation were used (36). Site-directed mutagenesis was done as described previously (22). Proteins were extracted from at least three independent transformants and assayed for β-galactosidase (β-Gal) activity as described previously (28, 32, 41). Results are given as Miller units.
Metabolic labeling with 32P.
Cells were pregrown overnight at 30°C in selective medium and transferred into fresh phosphate-depleted medium at an optical density at 600 nm (OD600) of about 0.1. When cell density reached an OD600 of 0.6, 10 ml was pelleted and resuspended in 1 ml of phosphate-depleted medium containing 1 mCi of [32P]orthophosphate (NEN). Following a 30-min incubation at 30°C, proteins were extracted and immunoprecipitated with antibodies directed against 12CA5 and run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS PAGE) gel essentially as described previously (19). Gel was exposed to X-ray film.
Antibodies.
Mouse monoclonal antibodies directed against the hemagglutinin (HA) epitope (12CA5) were purchased from Boehringer. Mouse monoclonal antibodies directed against GST(B-14) were purchased from Santa Cruz Biotechnology.
Preparation of yeast protein extracts and Western and coimmunoprecipitation analyses.
Protein extracts for Western analysis were prepared from trichloroacetic acid-treated cells as described previously (6). The Western analysis procedure was essentally as described previously (6, 7). Protein extraction for immunoprecipitation and the immunoprecipitation procedure were essentially as described previously (1).
RESULTS
IREu functions as a glucose-regulated URS and UAS element.
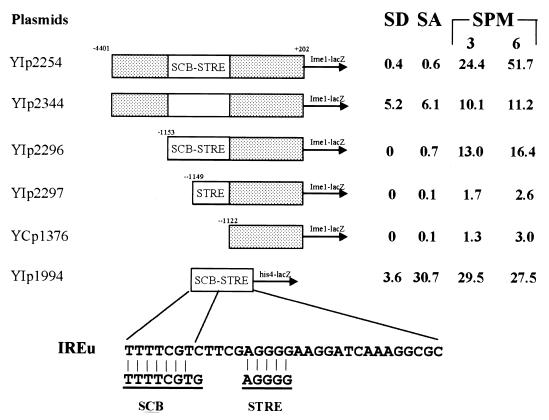
In a previous report we used two criteria to demonstrate that IREu exhibits UAS activity. First, we showed that the expression level of an ime1-lacZ chimera extending to IREu is reduced in comparison to that of a construct that includes IREu (35) (Fig. 1, compare YIp1376 to YIp2296). Second, we showed that IREu confers UAS activity to a silent his4-lacZ reporter (35) (Fig. 1, YIp1994). To strengthen the above conclusion and to show that within its natural context IREu functions as a UAS element, a nested deletion was constructed. Figure 1 shows that, as expected, nested deletion of IREu led to a 2.5- to 5-fold reduction in the expression of ime1-lacZ in SPM (Fig. 1, compare YIp2254 to YIp2344).
FIG. 1.
IREu serves as a URS element in SD and as a UAS element in SA. Strain Y422 carrying various lacZ plasmids either on a CEN vector (YCp plasmid) or integrated in the genomic LEU2 gene (YIp plasmid) was grown in PSP2 to 107 cells/ml. Cells were washed once in water and resuspended in SPM. Samples were taken to extract proteins and measure lacZ levels after 0 (SA), 3, and 6 h in SPM. In addition, proteins were extracted from 107 cells/ml grown in glucose-containing media (SD). The level of β-Gal is given in Miller units. The results are the averages of three or four independent transformants. Standard deviations were less than 10%. The sequence of IREu and its homology to the known SCB and STRE elements are given. Dotted boxes, sequences of IME1; open box, nested deletion; SCB-STRE box, IREu element.
The sequence of IREu (Fig. 1) reveals the presence of two known UAS elements, STRE (34) and an imperfect SCB (3) (seven out of eight residues are identical). Since both elements function as UAS elements (3, 34), it is possible that both contribute to the function of IREu. To determine if the putative SCB element is functional, we compared the level of expression of an ime1-lacZ chimera that extends to IREu (YIp2296) to those of one without IREu (YCp1376) and of one that extends to STRE (without SCB) (YIp2297). Figure 1 demonstrates that, without the SCB sequence (deletion of 4 bp from the 5′ end), the remaining STRE did not function as a UAS element. This result suggests that either the UAS activity is confined to the SCB sequence or that both sequences are required for the UAS activity of IREu. We favor the latter hypothesis, since we have shown that the two homologous transcriptional activators, Msn2 and Msn4, that bind STRE elements in stress genes (25, 37) bind IREu when synthesized in vitro (data not shown) and promote its UAS activity (35) (Fig. 2).
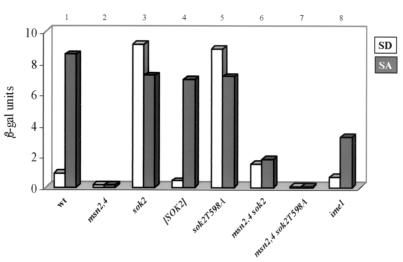
FIG. 2.
Positive and negative regulators of IREu. Expression of the IREu-his4-lacZ chimeric gene integrated at LEU2 by 107 cells/ml grown in either SD or SA was measured. The level of β-Gal is given in Miller units. The results are the averages of three or four independent transformants. Standard deviations were less than 10%. The following isogenic strains were used: Y1161 (wt; column 1), Y1132 (msn2Δ msn4Δ; column 2), Y1162 (sok2Δ; column 3), Y1170 (msn2Δ msn4Δ sok2Δ; column 6), Y1086 (ime1Δ; column 8), Y1162 carrying on a 2μm SOK2 plasmid (YEp2432; column 4) or sok2T598A (YEp2452; column 5) and Y1170 carrying on a 2μm sok2T598A plasmid (YEp2573; column 7).
The nested deletion of IREu reveals that, in addition to its positive role, IREu functions as a negative element. Deletion of only IREu from an ime1-lacZ construct led to a 10- to 13-fold increase in expression in vegetative media with either glucose (SD) or acetate (SA) as the sole carbon source (Fig. 1, compare YIp2254 to YIp2344). The presence of nitrogen repression element USC1 (35) in the ime1-lacZ construct hindered our ability to determine if this repression is due to the carbon or nitrogen source or both. However, since nitrogen has no effect on the expression of IREu-his4-lacZ (35) (Fig. 1, YIp1994), we conclude that it is the glucose that mediates the URS activity of IREu.
Sok2 is a negative regulator of IREu in the presence of glucose.
Previously we reported that the cAMP/PKA pathway transmits the glucose signal that represses the UAS activity of IREu in the presence of glucose (35). What might be the negative regulator that mediates this repression? A putative candidate is Sok2, a target of PKA that functions as a transcriptional repressor (47). We examined, therefore, the effect of Sok2 on the activity of IREu. Deletion of SOK2 had a dramatic effect: it led to a 10-fold increase in the expression of IREu-his4-lacZ in SD, but not in SA, media (Fig. 2, compare column 3 to column 1). Conversely, overexpression of Sok2 led to a twofold decrease in expression in SD (Fig. 2, column 4). These results suggest that Sok2 mediates the URS activity of IREu in glucose media.
Expression of Sok2 depends on the presence of glucose.
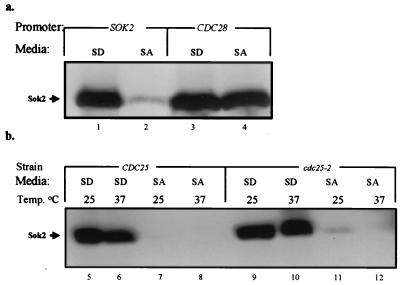
The availability of Sok2 and/or posttranslational modifications of Sok2 may be responsible for its carbon source-regulated repression activity. A pSOK2-3x-HA-SOK2 chimera on a 2μm plasmid was used to determine the level of expression of Sok2 in glucose and acetate media. Western analysis reveals that the steady-state level of Sok2 was dramatically reduced in acetate medium in comparison to that in glucose medium (Fig. 3a, compare lane 2 to lane 1). Two lines of evidence suggest that this effect results from transcriptional regulation. (i) The steady-state levels of Sok2 expressed from the constitutive CDC28 promoter in glucose and acetate media are identical (Fig. 3a, lanes 3 and 4). (ii) Fusion of the SOK2 promoter to lacZ led to 2.8 U of β-Gal in SD and only 0.3 U in SA. The lower level of Sok2 in the presence of acetate as the sole carbon source may explain why in this medium Sok2 does not function as a repressor.
FIG. 3.
Expression of Sok2 is reduced in acetate media. Proteins extracted from 107 cells/ml were subjected to immunoblot analysis using antibodies directed against HA. (a) Strain Y422 carrying on a 2μm plasmid either pSOK2-3xHA-SOK2 (YEp2432) (lanes 1 and 2) or pCDC28-3xHA-SOK2 (YEp2486) (lanes 3 and 4). Cells were grown in either SD (lanes 1 and 3) or SA (lanes 2 and 4). (b) Strains. Y1064 (wt) (lanes 5 to 8) and its isogenic Y1078 (cdc25-2) (lanes 9 to 12) carrying pSOK2-3xHA-Sok2 (YEp2432). Cells were grown in either SD (lanes 5, 6, 9, and 10) or SA (lanes 7, 8, 11, and 12) at 25°C (lanes 5, 7, 9, and 11) or shifted to 37°C for 4 h (lanes 6, 8, 10, and 12).
Sok2 functions as a general repressor.
Repression by Sok2 may be due to either the sequestering of a positive regulator (Msn2 or -4) or active repression. To determine how Sok2 represses transcription, we examined the ability of a Gal4 DNA-binding domain [Gal4(bd); amino acids 1 to 147]–Sok2 fusion protein to repress transcription of a UASGALI-UASHIS4-his4-lacZ reporter. Figure 4 shows that expression of Sok2 led to more than a twofold decrease in the expression of the reporter gene in SD, whereas in SA Sok2 did not repress transcription (compare columns 1 and 2). Similar results were obtained when we fused Sok2 to lexA and determined the ability of the fusion protein to repress transcription of lexOP-UASCYCI-lacZ (data not shown). In these systems the extent of repression by Sok2 was lower than the extent of repression on IREu (2-fold versus 10-fold). We assume that this difference reflects the use of different reporter genes as well as the expression of Sok2 from different promoters (pADHI versus pSOK2). We conclude, that Sok2 actively represses transcription, but only in the presence of glucose. Since in this assay, the Gal4(bd)-Sok2 chimeric protein was expressed from the ADH1 promoter, our results suggest that glucose regulates not only the availability of Sok2 but also its activity.
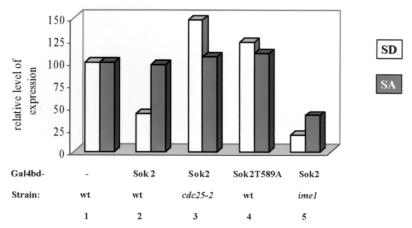
FIG. 4.
Sok2 is a transcriptional repressor whose activity depends on PKA. Proteins were extracted from cells grown in either SD or SA to 107 cells/ml. The level of β-Gal was measured, and relative levels are given. The results are the averages of three or four independent transformants. Standard deviations were less than 10%. Strains used are Y1064 (wt; columns 1, 2, and 4), Y1078 (cdc25-2; column 3), and Y1075 (ime1Δ; column 5) carrying UASGAL1-UASHIS4-his4-lacZ (YIp2218) integrated at LEU2. These strains were mated to wt (Y1065), cdc25-2 (Y1093), and ime1Δ (Y1076) strains, respectively. They also carried the following 2μm TRP1 plasmids: pADH1-gal4(bd) (pAS2 [14]) (column 1), pADH1-Gal4bd-SOK2 (YEp2314) (columns 2, 3, and 5), and pADH1-gal4(bd)-sok2T598A (YEp2530) (column 4).
Active PKA promotes repression by Sok2.
The cAMP-dependent PKA signal pathway transmits the glucose signal that inhibits the UAS activity of IREu (35). This result suggests that this pathway might affect both Sok2 and Msn2 and -4, the negative and positive regulators of IREu, respectively. It is possible, therefore, that the low level of Sok2 in acetate media is due to the low activity of PKA. In order to examine this hypothesis, we compared the steady-state levels of Sok2 in wild-type (wt) and cdc25-2 cells. CDC25 encodes the RAS-specific GDP/GTP exchange factor (reference 4 and references therein). Mutant cdc25-2 (temperature sensitive) produces a drastic decrease in the level of cAMP and consequently no activity of PKA (4). The steady-state levels of Sok2 in both wt and cdc25-2 cells at 25°C and following 4 h of incubation at 37°C were similar, namely, high levels in SD and low levels in SA (Fig. 3b, compare lanes 5 to 8 to lanes 9 to 12). We conclude from these results that PKA does not regulate the transcription of SOK2 or the accumulation of Sok2 protein.
It is possible that the cAMP signal pathway mediates the ability of Sok2 to repress transcription in the presence of glucose. We examined, therefore, the ability of Sok2 to repress transcription in the cdc25-2 mutant. At the permissive temperature cdc25-2 cells show essentially the same level of expression as wt cells (data not shown). Figure 4 shows that Gal4(bd)-Sok2 does not repress transcription of UASGALI-UASHIS4-his4-lacZ in cdc25-2 cells incubated for 4 h at the restrictive temperature (column 3). We conclude, therefore, that the activity of Sok2 as a repressor depends on a functional PKA.
The predicted amino acid sequence encoded by SOK2 shows a single PKA phosphorylation site, KKCT, amino acids 595 to 598. To determine whether phosphorylation of Sok2 by PKA is required for its ability to repress transcription, we mutated this threonine 598 residue to alanine. Diploid cells carrying only this sok2T598A allele on a 2μm plasmid had a phenotype similar to that of sok2Δ cells, namely, no repression of IREu-his4-lacZ; a high level of expression was observed in both SD and SA (Fig. 2, column 5). Moreover, Gal4(bd)-Sok2T598A did not repress transcription of the UASGALI-UASHIS4-his4-lacZ reporter gene (Fig. 4, column 4). Lack of repression was not due to lower levels of Sok2T598A (Western analysis; data not shown). We suggest that in the presence of glucose as the sole carbon source, PKA is highly active and phosphorylates T598. Phosphorylation of this residue is required for the ability of Sok2 to function as a repressor.
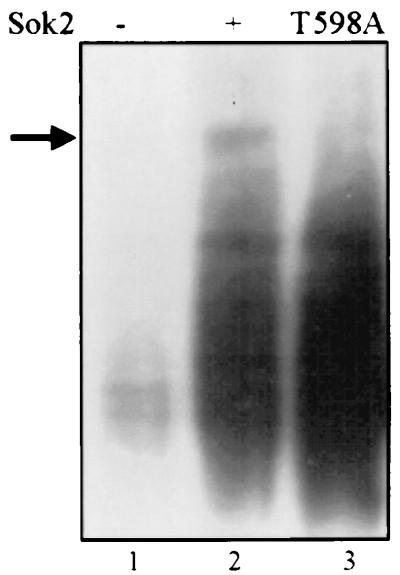
In the intact cell Sok2 is phosphorylated on T598.
In order to show whether Sok2 is a phosphoprotein, we metabolically labeled cultures with [32P]orthophosphate and analyzed the protein extracts by immunoprecipitation using antibodies directed against HA. Figure 5 shows that overexpression of 3xHA-Sok2 (from pCDC28) led to the detection of a specific band with the apparent molecular weight (MW) of Sok2 (compare lanes 1 and 2). This specific band was absent from cultures expressing the mutant 3xHA-Sok2T598A protein (Fig. 5, lane 3). These results support our suggestion that Sok2 is a phosphoprotein. Figure 5 shows a smear of nondiscrete radioactive bands in immunoprecipitate from cells expressing either HA-Sok2 or HA-Sok2T598A. Since the MWs of these bands are lower than the expected MW of Sok2, we assume that they represent phosphoproteins that are immunoprecipitated either nonspecifically by HA or by labeled proteins which associate with Sok2 in the immunoprecipitation. Our results suggest that phosphorylation of Sok2 is probably only on a single residue, threonine 598. We point out that, in order to detect the labeled Sok2 protein, gels were exposed for about 2 months. We assume that phosphorylation of only a single residue does not cause any apparent change in mobility and requires long exposure.
FIG. 5.
Sok2 is a phosphoprotein. Proteins were extracted from cells incubated with 32P for 30 min. Following immunoprecipitation with antibodies directed against HA, proteins were separated by SDS–10% PAGE and exposed to X-ray film. The strain used is Y422 carrying a vector (pRS426 [40]; lane 1; control), pCDC28-3x-HA-SOK2 (YEp2486; lane 2), or pCDC28-3xHA-sok2T598A (YEp2558; lane 3).
Ime1 is required to relieve repression activity of Sok2.
Positive regulators of IREu were identified as gene dosage suppressors that increase the expression of an IREu-cycl-lacZ chimeric gene (35; K. Robzyk, personal communication). Previously we reported the identification and characterization of MSN2 (35), and here we describe a second positive regulator, IME1 itself. Figure 2 (column 8) shows that Ime1 is required for the complete UAS activity of IREu in acetate media. Diploid cells with IME1 deleted show about threefold reduction in the expression of IREu-his4-lacZ, whereas expression of UASHIS4-his4-lacZ was not affected (data not shown). Note that deletion of both MSN2 and MSN4 had a more drastic effect, namely, no expression (35) (Fig. 2, column 2). Thus, both Ime1 and Msn2 and -4 are positive regulators of IREu. Ime1 is a transcriptional activator that does not bind DNA; rather it is recruited to promoters by association with a DNA-binding protein (33). We postulated, therefore, that Ime1 activates IREu by affecting the activity of either Msn2 and -4 or Sok2. We favor the latter since transcriptional activation of stress elements by Msn2 and -4 is independent of IME1. Activation by Msn2 and -4 occurs when Ime1 is absent, namely, in haploid cells grown in vegetative media (25, 37). We determined, therefore, if relief of Sok2 repression in SA depends on Ime1. We compared the ability of Sok2 to repress transcription of the heterologous UASGALI-UASHIS4-his4-lacZ in wt and ime1Δ diploids. In cells expressing only Gal4(bd) deletion of IME1 had no effect on the expression of the reporter (data not shown). Figure 4 (column 5) shows that in diploid cells with IME1 deleted, Gal4(bd)-Sok2 repressed transcription in both SD and SA, whereas in wt cells repression occurred only in SD. We conclude that Ime1 is required to relieve repression by Sok2.
In the presence of acetate Sok2 is converted into a weak activator.
Figure 4 shows that the level of expression of UASGALI-UASHIS4-his4-lacZ was increased in cdc25-2 cells expressing Gal4(bd)-Sok2 (column 3) as well as in wt cells expressing Gal4(bd)-Sok2T598A (column 4) in comparison to wt cells expressing only Gal4(bd) (column 1). Although this effect was mainly observed in SD, it suggests the possibility that in SA Sok2 is an activator rather than a repressor. We determined, therefore, the ability of Gal4(bd)-Sok2 to activate transcription of gal1-lacZ. In vegetative media with glucose as the sole carbon source (SD) similar levels of expression were observed in cells expressing either Gal4(bd) or Gal4(bd)-Sok2 (Table 2). However, in the presence of acetate, expression of either Gal4(bd)-Sok2 or Gal4(bd)-Sok2T598A led to a 50-fold increase in comparison to Gal4(bd) (Table 2). These results imply that, as suggested above, in acetate media Sok2 is converted into a weak activator. Moreover, this activity of Sok2 is independent of T598. However, in diploid cells with IME1 deleted, this phenomenon is not observed: the level of expression of gal1-lacZ is not increased in SA (Table 2). We conclude, therefore, that Ime1 is required to convert Sok2 to an activator. Table 2 also shows that in cells overexpressing Msn2 there was a fivefold increase in the ability of Gal4(bd)-Sok2 to activate transcription of gal1-lacZ. Since Msn2 is a transcriptional activator that enters the nucleus in the absence of glucose (11), it is not surprising that the increase in transcriptional activation of Sok2 took place only in SA. These results imply that Sok2 and Msn2 associate and that Msn2, similar to Ime1, might also be responsible for the conversion of Sok2 into a weak activator.
TABLE 2.
In acetate media, Sok2 functions as a weak transcriptional activator
| Expressed protein | Relevant genotypea | β-Gal activityb (Miller units) in:
|
|
|---|---|---|---|
| SD | SA | ||
| Gal4(bd) | wt | 0.01 ± 0.007 | 0.01 ± 0.001 |
| Gal4(bd)-Sok2 | wt | 0.02 ± 0.020 | 0.56 ± 0.106 |
| Gal4(bd)-Sok2T598A | wt | 0.03 ± 0.013 | 0.57 ± 0.035 |
| Gal4(bd)-Sok2 | wt + MSN2 | 0.04 ± 0.020 | 2.85 ± 0.150 |
| Gal4(bd)-Sok2 | imelΔ | 0.03 ± 0.005 | 0.02 ± 0.003 |
wt, Y153 × Y187 (14) carrying pADH1-gal4(bd) (pAS2 [14]), pADH1-gal4(bd)-SOK2 (YEp2314), and pADH1-gal4(bd)-sok2T598A (YEp2530); MSN2, strain carries MSN2 on a 2μm plasmid (YEp2588); ime1Δ, Y1080 × Y1076.
Results for wt strains are averages of results for Y153 × Y187, Y153 × Y187 carrying MSN2, and Y1077 × Y1076.
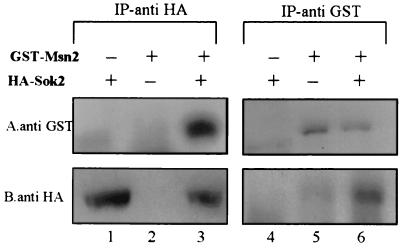
Sok2 associates with Msn2.
We used coimmunoprecipitation to determine if Sok2 associates with Msn2. For these experiments Sok2 was tagged with 3xHA and Msn2 was tagged with GST. Figure 6 shows that, in diploid cells expressing both proteins, GST-Msn2 was immunoprecipitated along with HA-Sok2 in an anti-HA immune complex (Fig. 6A, lane 3). However, in cells expressing either GST-Msn2 or HA-Sok2 alone, GST-Msn2 was not recovered (Fig. 6A, lanes 1 and 2). In the reciprocal experiment, HA-Sok2 was immunoprecipitated along with GST-Msn2 in an anti-GST immune complex (Fig. 6B, lane 6). In cells expressing either GST-Msn2 or HA-Sok2 alone, HA-Sok2 was not recovered (Fig. 6B, lanes 4 and 5). As a control, Fig. 6 shows that these proteins were recovered when the same antibody was used for both immunoprecipitation and detection. These results demonstrate that Sok2 and Msn2 interact.
FIG. 6.
Sok2 associates with Msn2. Coimmunoprecipitation of GST-Msn2 and HA-Sok2 is shown. Proteins were extracted from logarithmic cultures grown in SD. Anti-HA or anti-GST immune complexes were prepared from strain Y422 carrying plasmids YEp2382 (HA-Sok2) (lanes 1, 3, 4, and 6) and YEp2536 (GST-Msn2) (lanes 2, 3, 5, and 6). Proteins were separated by SDS–8% PAGE, and immunoblotting was done with anti-HA for the anti-GST immune complexes and with anti-GST for the anti-HA immune complexes. Following the stripping of bound antibodies, a second immunoblotting was performed using anti-GST and anti-HA, respectively. (A) probing with anti- GST; (B) probing with anti-HA. IP, immunoprecipitation.
Genetic evidence suggesting that Sok2 binds IREu.
As described above and as shown in Fig. 2, the expression of IREu was absolutely dependent on Msn2 and -4; in double-mutant msn2Δ msn4Δ cells, IREu-his4-lacZ was not expressed (Fig. 2, column 2). However, in sok2Δ strains this dependency was relieved, and in the triple-mutant sok2Δ msn2Δ msn4Δ strain, low but substantial levels of expression were observed (Fig. 2, compare columns 2 and 6). These results imply that Sok2 or a protein regulated by Sok2 or both bind IREu under all growth conditions and that only in the absence of both of these may an opportunist transcriptional activator bind to and activate IREu. To distinguish between these two possibilities, namely, whether Sok2 or another protein binds IREu, we determined the level of expression of IREu-his4-lacZ in cells carrying a mutant allele of SOK2 that express a protein lacking repression activity. Figure 2 shows that, in msn2Δ msn4Δ sok2Δ cells carrying sok2T598A on a multicopy plasmid, IREu-his4-lacZ was not expressed (compare column 7 to columns 2 and 6). These results indicate that Sok2 binds (directly or through a mediator) IREu.
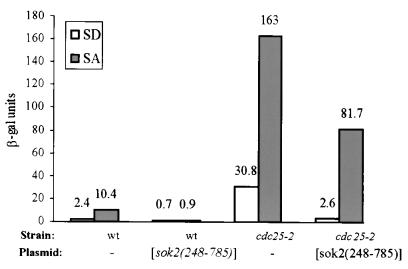
Relief of repression of Sok2 depends on its N terminus.
Overexpression of truncated Sok2 lacking the N-terminal 248 amino acids led to complete repression of IREu-his4-lacZ in both SD and SA (Fig. 7). Note that overexpression of the entire Sok2 protein had a mild effect, and only in SD (Fig. 2). We conclude, therefore, that the N-terminal domain is required to relieve repression. On the other hand, the repression activity of Sok2 is mediated by PKA through the C-terminal domain (amino acid T598). This result predicts that, in cells with low PKA activity, repression by Sok2(248-785) will be relieved. cdc25-2 cells carrying sok2(248-785) on a 2μm plasmid were shifted to the nonpermissive temperature for 4 h, proteins were extracted, and the level of expression of IREu-his4-lacZ was determined. Overexpression of Sok2(248-785) in wt cells led to an 11-fold decrease in the expression of IREu-his4-lacZ in SA medium, whereas in cdc25-2 cells only a 2-fold repression was observed (Fig. 7). The low repression activity of Sok2(248-785) in cdc25-2 cells grown in SA is probably due to residual PKA activity that remained from growth at the permissive temperature. These results suggest that separate domains of Sok2 are responsible for repression and its termination.
FIG. 7.
The N-terminal domain of Sok2 is required to relieve repression in SA. Shown is the expression of IREu-his4-lacZ chimeric gene integrated at LEU2. Cells were grown in SD or SA to 0.5 × 107 (wt) and 1 × 107 cells/ml (cdc25-2 strain) at 25°C and shifted to 37°C for 4 h. The level of β-Gal is given in Miller units. The results are the averages of three or four independent transformants. Standard deviations were less than 10%. The isogenic strains used were Y1161 (wt) and Y1087a (cdc25-2) carrying on a 2μm plasmid a vector (YEpLac112 [9]) or pSOK2-sok2(248-785) (YEp1784).
Sok2 is a negative regulator of meiosis.
The previous results suggest that the reduction in the activity of IREu in response to overexpression of Sok2 will affect the ability of diploid cells to enter and complete meiosis and sporulation. However, when SOK2 was present in the cell on a multicopy 2μm plasmid, the efficiency of sporulation was not affected (Table 3). Nevertheless, this result does not contradict the above prediction, since the expression of SOK2 was reduced in the presence of acetate as the sole carbon source. Thus, this condition did not promote overexpression of Sok2. Indeed, when the entire Sok2 was expressed from the CDC28 promoter, the level of sporulation was reduced to about 60% (Table 3). However no repression was observed when the sok2T598A allele was expressed (Table 3). Moreover, a twofold reduction in sporulation was observed in cells carrying sok2(248-785) on a 2μm plasmid (Table 3). These results show that Sok2 is a negative regulator of meiosis and that its repression activity depends on its steady-state levels, phosphorylation of T598, and its N-terminal domain.
TABLE 3.
Sok2 is a negative regulator of meiosisa
| Gene expressed | % Asci at:
|
|
|---|---|---|
| 24 h | 48 h | |
| None | 67 | 81 |
| pSOK2-SOK2 | 67 | 84 |
| pSOK2-sok2T598A | 66 | 79 |
| pSOK2-sok2(248–785) | 34 | 45 |
| pCDC28-SOK2 | 41 | 59 |
| pCDC28-sok2T598A | 63 | 80 |
Strain Y422 carried the genes on a 2μm plasmid. In each case, 400 cells were counted.
DISCUSSION
Regulated activity of IREu.
The carbon source plays a pivotal role in regulating the transcription of IME1: in a medium promoting vegetative growth with glucose as the sole carbon source IME1 is silent, whereas in the presence of acetate as the sole carbon source IME1 is transcribed (16). The carbon source signal is transmitted to at least three distinct elements in the unusually large upstream regulation region of this gene (35; G. Shenhar, unpublished data). In this report we focused on a single 32-bp element, IREu. We have shown previously that this element functions as a carbon source-regulated UAS element whose activity in SA is absolutely dependent on homologous transcriptional activators Msn2 and Msn4 (35). Here we show that an additional transcriptional activator, Ime1 itself, is required for complete UAS activity (Fig. 2). We have also shown that, in glucose media, IREu functions as a URS element (Fig. 1) and that this URS activity depends on Sok2 (Fig. 2).
How is this regulated activity accomplished? Two specific DNA-protein complexes, whose levels increase in the presence of acetate, are formed on IREu (35). By gel shift assay we have shown that one of these complexes, the lower-mobility one, depends on Msn2 and -4 (35). Furthermore, Msn2 binds IREu in a gel shift assay when in vitro transcribed and translated (data not shown). Since in vitro-synthesized Msn2 and -4 bind STRE sequences (25, 37) and since IREu contains such a sequence (Fig. 1) (35), we suggest that these proteins bind the STRE sequence in IREu (35). The sequence of IREu (Fig. 1) reveals the presence of an additional UAS element, an SCB-like sequence (Fig. 1). Within IREu this element is functional, since its deletion leads to complete loss of UAS activity (Fig. 1). The UAS activity of the SCB elements within the HO and CLN1,2 genes is regulated by the Swi4-Swi6 complex (18, 30). SOK2 encodes a DNA-binding protein that shows extensive homology to the DNA-binding domain of Swi4 (47), implying that Sok2 may bind to this SCB-like element in IREu. However, neither deletion nor overexpression of Sok2 had any effect on the DNA-protein complexes formed on IREu (data not shown). Furthermore, in vitro-transcribed and -translated Sok2 did not shift the position of IREu in a gel shift assay, even when mixed with in vitro-synthesized Msn2 or with yeast cell extract (data not shown). These results suggest that either Sok2 does not bind IREu or Sok2 binds IREu but the assay we used was not sensitive enough to detect this binding. The following genetic evidence supports the latter hypothesis. (i) In cells with both MSN2 and MSN4 deleted IREu was silent (Fig. 2). However, when in these cells SOK2 was also deleted, a 10-fold increase in β-Gal units was observed in both SD and SA (Fig. 2). These results suggest the possibility that either Sok2 or a protein regulated by it binds IREu and that in the absence of Sok2 an imposter transcription factor binds IREu, promoting its UAS activity. Since this imposter protein can activate transcription only in the physical absence of Sok2, in the triple-mutant msn2Δ msn4Δ sok2T598A strain, IREu-his4-lacZ was not expressed (Fig. 2), and our results point to Sok2 as the protein that binds IREu. (ii) Transcriptional activation of Sok2 in SA is increased in cells overexpressing Msn2, suggesting that these two proteins associate (Table 2). Indeed, by coimmunoprecipitation we show that these two proteins associate (Fig. 6). Since Msn2 binds the STRE element in IREu and since in the absence of the SCB element IREu is not active as a UAS (Fig. 1), we assume that a protein that binds SCB promotes the binding of Msn2. Since Sok2 interacts with Msn2, it might be the protein that promotes the binding of Msn2 to IREu.
Within the IREu element the SCB and STRE sequences are close (Fig. 1), suggesting the following two working hypotheses for the regulated activity of IREu (i) In the presence of glucose the presumed binding of Sok2 to the SCB element sequesters Msn2 and -4, leading to repression. In the presence of acetate Sok2 does not bind; thus repression is relieved and Msn2 and -4 bind the STRE sequence in IREu, promoting transcription (ii) Sok2 is required for the efficient binding of Msn2 and -4 to IREu; thus it binds IREu under all growth conditions. In the presence of glucose Sok2 represses transcription, whereas in the absence of glucose Sok2 does not interfere with the transcriptional activation function of Msn2 and -4. The following results support the second hypothesis. (i) Sok2 functions as a general repressor when tethered to heterologous promoters; thus, its activity as a repressor appears unrelated to the sequestering of activators. (ii) Sok2 associates with Msn2 (Fig. 6). (iii) Msn2 binds IREu under all growth conditions (35). (iv) In the absence of the SCB-like element, IREu exhibits no UAS activity (Fig. 1). We suggest that Sok2 and Msn2 or -4 form a heterodimer that binds IREu. Thus, without the SCB sequence, Sok2 cannot assist the binding of Msn2 or -4 to the STRE sequence.
The second hypothesis suggests that in the absence of Sok2, but in the presence of the SCB sequence, an imposter protein can promote the binding of Msn2 or -4 to STRE. Sok2 is highly homologous to the DNA-binding domains of Swi4, Phd1, and Mbp1; thus any of these proteins may substitute for Sok2 and promote the binding of Msn2 and -4. Indeed, deletion of SWI4 caused a three- to fourfold increase in the expression of IREu-cyc1-lacZ in SD (35; K. Robzyk, personal communication). However, the effect of Swi4 could not be observed for an IREu-his4-lacZ construct integrated into the genome (G. Shenhar, unpublished data), suggesting that Swi4 may be an imposter that can bind IREu under specific conditions.
The function of Sok2: repression versus activation.
In this report we show that Sok2 functions as a carbon source-regulated repressor. This repression depends on an active cAMP-dependent signal pathway. We suggest that PKA phosphorylates Sok2 on threonine 598, since this residue is in a consensus for PKA and since its mutation to alanine prevents the in vivo phosphorylation of Sok2 and prevents Sok2 from functioning as a repressor. Transcriptional repression, in general, is established by different modes that can roughly be divided into two groups: (i) specific repressors that sequester transcriptional activators and (ii) active repressors that are characterized by distinct repression and DNA-binding domains (for reviews see references 13 and 15). As discussed above, we assume that Sok2 represses transcription following its binding to IREu. The region in Sok2 that is homologous to the DNA-binding domain of Swi4 corresponds to amino acids 437 to 494, whereas repression depends on T598. We suggest, therefore, that separate domains in Sok2 are responsible for binding and repression. This implies that Sok2 functions as an active repressor. This conclusion is supported by the ability of Sok2 to repress transcription when tethered to heterologous reporters. Note that the C-terminal domain of Sok2 contains a high proportion of charged amino acids, a feature typical of several known repression domains (13). The mode by which Sok2 represses transcription is beyond the scope of this paper, and further work is required to elucidate it.
Several mechanisms explain why in acetate media Sok2 does not function as a repressor. The first is transcriptional regulation. The steady-state level of Sok2 is dramatically decreased in the presence of acetate as the sole carbon source (Fig. 3). The second is posttranslational modification of Sok2. In acetate media low activity of PKA results in reduced levels of phosphorylated Sok2. This conclusion is based on the observation that, in cdc25-2 cells or in cells carrying the sok2T598A allele, Sok2 did not repress transcription (Fig. 2 and 4). Accordingly, this Sok2T598A protein, unlike the wt Sok2, was not metabolically labeled with 32P (Fig. 5). The third is the carbon source-regulated availability of Ime1. In diploid cells with IME1 deleted derepression of UASGALI-UASHIS4-his4-lacZ in SA was incomplete (Fig. 4). Since IME1 is not transcribed in glucose media (16), it does not interfere with the ability of Sok2 to function as a repressor in this media. The fourth is the function of the N-terminal domain of Sok2. A truncated Sok2 protein lacking the N-terminal 248 amino acids functions as a repressor in both glucose and acetate media (Fig. 7). Thus, this region is required to cancel the repression activity of Sok2. We assume that the use of multiple levels of control ensures that Sok2 will function as a transcriptional repressor in vegetative cultures with glucose as the sole carbon source and will be converted into a weak activator only in the absence of glucose and the presence of Ime1 (Table 2). Thus, overexpression of Sok2 does not promote repression in acetate media (SA or SPM; Fig. 2 and Table 3), whereas its expression from a heterologous promoter or truncation of its N-terminal domain suffices for repression in the presence of acetate (Fig. 7 and Table 3).
The predicted amino acid sequence of Sok2 reveals that the N-terminal domain contains several glutamine-rich stretches. Glutamine domains play an important role in transcriptional activation and multimerization (5, 48). Four possible nonexclusive models for the function of this domain can be envisioned. (i) Association of Ime1 with the N-terminal domain of Sok2 converts it to an activator. (ii) The N-terminal domain might recruit a phosphatase required to dephosphorylate Sok2. (iii) This domain may be required for the association between Sok2 and Msn2 and -4. Thus, in its absence, Msn2 and -4 cannot efficiently bind IREu, and consequently IREu cannot function as a UAS element. (iv) The N-terminal domain recruits components of the transcription machinery such as the Swi-Snf chromatin-remodeling complex or the SAGA histone acetyltransferase complex. Since separate domains are required for promoting and relieving repression, we suggest that relief of repression is not just the opposite of repression but actually the promotion of transcriptional activation.
The choice between mitosis and meiosis.
In this report we show that Sok2 is a negative regulator of meiosis: overexpression of Sok2 leads to a decrease in the efficiency of spore formation (Table 3). This negative role results from its function as a repressor on a specific carbon source-regulated element in IME1, IREu (Fig. 2). Previously, Sok2 was identified as a positive regulator of mitosis: when present on a multicopy plasmid, it suppressed the temperature-sensitive phenotype of a tpk1Δ tpk2-ts tpk3Δ mutant (47). On the other hand, disruption of SOK2 exacerbates the phenotype of tpk2 mutants, leading to slower growth, and increases the resistance of ras2val19 mutants to elevated temperatures (47). Ward et al. (47) report that the consensus for the PKA phosphorylation site is within a region required for suppression of TPK. Thus, the same region required for repressing meiosis (Table 3) functions as a positive regulator of mitosis. An inverse effect is observed for Msn2 and -4. These proteins function as positive regulators of IME1 and meiosis (35) (Fig. 2) but as negative regulators of mitosis (41). Deletion of both MSN2 and MSN4 suppresses the lethality of a strain with the TPK1 to -3 genes deleted (41).
The above-summarized results lead us to suggest a model for how alternative developmental pathways are regulated by a single signal pathway (Fig. 8). Both Sok2 and Msn2 and -4 are targets of PKA (11, 41, 47). Phosphorylation of Sok2 promotes its function as repressor, whereas phosphorylation of Msn2 and -4 affects their ability to function as activators. These regulators have opposing effects on meiosis and mitosis: Sok2 is a positive regulator of mitosis and a negative regulator of meiosis, whereas Msn2 and -4 are negative regulators of mitosis and positive regulators of meiosis. The use of the same regulators but in reverse directions for different developmental pathways ensures that one pathway will be an alternative to the other one. When cells enter mitosis, meiosis will be repressed, whereas when cells enter meiosis, mitosis will be blocked. Thus, a single signal transduction pathway, cAMP-dependent PKA, suffices to control two alternative developmental pathways. A similar role for cAMP in determining a developmental switch was reported for the development of vertebrate neural crest cells. Low levels of cAMP promote the development of sympathoadrenal cells, whereas high-level cAMP activity has an antagonistic effect, namely, preventing this development (2).
FIG. 8.
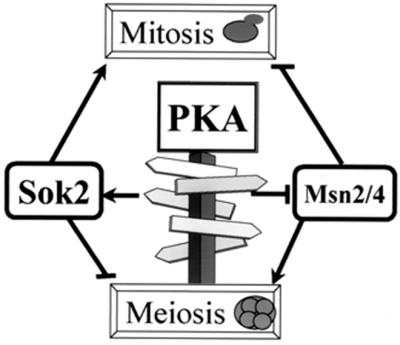
The role of PKA in determining the choice between mitosis and meiosis. Glucose increases the level of cAMP and consequently the activity of PKA. High activity of PKA leads to phosphorylation of several substrates, including Msn2 and -4 and Sok2. Msn2 and -4 are negative regulators of mitosis and positive regulators of meiosis. We suggest that nonphosphorylated Msn2 and -4 activate meiosis and repress mitosis, whereas PKA-phosphorylated Msn2 and -4 are neither inhibitors of mitosis nor activators of meiosis. The opposite relations are observed for Sok2, which is a negative regulator of meiosis and a positive regulator of mitosis. A PKA-phosphorylated Sok2 inhibits meiosis and promotes mitosis, whereas a nonphosphorylated Sok2 inhibits mitosis and promotes meiosis.
What is the role of PKA in controlling filamentous growth in relation to mitosis and meiosis? In S. cerevisiae, entry into invasive growth in haploids and pseudohyphae in diploids occurs in the presence of glucose and low levels of nitrogen. An intact cAMP-dependent PKA signal pathway promotes entry into mitosis or filamentous growth (21, 23, 31; S. Martin, M. Ansari, G. Shenhar, Y. Kassir, and H. Kuntzel, submitted for publication) but inhibits meiosis. Interestingly, the choice between meiosis and filamentous growth is accomplished by using the same regulators but in reverse directions. Msn2 and -4 are negative regulators of invasive growth (44) and, as discussed above, are positive regulators of meiosis. Overexpression of SOK2 homologs, the S. cerevisiae PHD1 and Candida albicans EFG1 genes, promotes filamentous growth (10, 45). Efg1 is a functional homolog of Sok2, as deduced from the observation that expression of Efg1 from the GAL1 promoter (pBIST) led to eightfold repression of IREu-his4-lacZ in SD (1.0 versus 9.2 β-Gal units) in cells with SOK2 (Y1162) deleted. Thus, a negative regulator of meiosis is homologous to positive regulators of filamentous growth. However, deletion of SOK2 rather than overexpression promotes filamentous growth. Thus, the effect of Sok2 on filamentous growth is not clear (for a discussion see reference 47), and further work is required to elucidate the relation between filamentous growth and meiosis.
ACKNOWLEDGMENTS
We thank B. Horwitz for critical reading of the manuscript and H. Kuntzel and G. Fink for helpful discussions. We thank S. Elledge, J. F. Ernst, G. Fink, S. Garrett, I. Herskowitz. A. Sugino, and A. Tzagoloff for kindly providing plasmids.
This work was supported by a grant from the Israel Science Foundation and the Ministry of Science and Culture, Niedersachsen, Germany.
REFERENCES
- 1.Ansari K, Martin S, Farkasovsky M, Ehbrecht I M, Kuntzel H. Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:30052–30058. doi: 10.1074/jbc.274.42.30052. [DOI] [PubMed] [Google Scholar]
- 2.Bilodeau M L, Boulineau T, Hullinger R L, Andrisani O M. Cyclic AMP signaling functions as a bimodal switch in sympathoadrenal cell development in cultured primary neural crest cells. Mol Cell Biol. 2000;20:3004–3014. doi: 10.1128/mcb.20.9.3004-3014.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis-and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 4.Broach J R. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991;7:28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- 5.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 6.Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 9.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 10.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 14.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 15.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 16.Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 17.Kassir Y, Simchen G. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:94–110. doi: 10.1016/0076-6879(91)94009-2. [DOI] [PubMed] [Google Scholar]
- 18.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 19.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraakman L, Lemaire K, Ma P, Teunissen A W R H, Donaton M C V, van Dijck P, Winderickx J, de Winde J H, Thevelin J M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 21.Kubler E, Mosch H U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandel S, Robzyk K, Kassir Y. IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev Genet. 1994;15:139–147. doi: 10.1002/dvg.1020150204. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto K, Uno I, Ishikawa T. Initiation of meiosis in yeast mutants defective in adenylate cyclase and cyclic AMP-dependent protein kinase. Cell. 1983;32:417–423. doi: 10.1016/0092-8674(83)90461-0. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura A, Treinin M, Mitsuzawa H, Kassir Y, Uno I, Simchen G. The adenylate cyclase/protein kinase cascade regulates entry into meiosis in Saccharomyces cerevisiae through the gene IME1. EMBO J. 1990;9:3225–3232. doi: 10.1002/j.1460-2075.1990.tb07521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 30.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SW14, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 31.Robertson L S, Fink G R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 33.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 35.Sagee S, Sherman A, Shenhar G, Robzyk K, Ben-Doy N, Simchen G, Kassir Y. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1985–1995. doi: 10.1128/mcb.18.4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 39.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith A, Ward M P, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith H E, Driscoll S E, Sia R A, Yuan H E, Mitchell A P. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics. 1993;133:775–784. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith H E, Mitchell A P. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2142–2152. doi: 10.1128/mcb.9.5.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanhill A, Schick N, Engelberg D. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol Cell Biol. 1999;19:7529–7538. doi: 10.1128/mcb.19.11.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. : [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Aelst L, Jans A W, Thevelein J M. Involvement of the CDC25 gene product in the signal transmission pathway of the glucose-induced RAS-mediated cAMP signal in the yeast Saccharomyces cerevisiae. J Gen Microbiol. 1991;137:341–349. doi: 10.1099/00221287-137-2-341. [DOI] [PubMed] [Google Scholar]
- 47.Ward M P, Gimeno C J, Fink G R, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkins R C, Lis J T. DNA distortion and multimerization: novel functions of the glutamine-rich domain of GAGA factor. J Mol Biol. 1999;285:515–525. doi: 10.1006/jmbi.1998.2356. [DOI] [PubMed] [Google Scholar]
- 49.Yocum R R, Hanley S, West R, Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]