Abstract
Nanoscale alterations in the cellular membrane transpire during cellular interactions with the extracellular environment through the endocytosis processes. Although the biological innuendos as well as alterations in cellular morphology during endocytosis are well-known, nanomechanical amendments in the cellular membrane are poorly understood. In this manuscript, atomic force microscope is employed to demonstrate the nanomechanical alterations in membrane dynamics during receptor mediated endocytosis of gold nanoparticles conjugated with either plectin-1 targeted peptide (PTP-GNP) or scrambled peptide (sPEP-GNP). Plectin-1 is aberrantly overexpressed at cell membrane of pancreatic cancer cells and is known to provide and maintain cellular mechanical integrity. During receptor mediated endocytosis of nanoparticles, we demonstrate temporal nanomechanical changes of cell membrane in both immortal pancreatic cancer Panc1 cells and patient derived primary pancreatic cancer cell, 4911. We further confirm the alterations of plectin-1 expression in Panc1 cell membrane during the receptor mediated endocytosis using classical streptavidin–biotin reaction and establish its association with nanomechanical alteration in membrane dynamics. Withdrawal of PTP-GNPs from the cell culture restores the plectin-1 expression at the membrane and reverses the mechanical properties of Panc1. We also show a distinctly opposite trend in nanomechanical behavior in cancer and endothelial cells when treated with sPEP-GNP and PTP-GNP, respectively, signifying receptor independent endocytosis process. This study illustrates the nanomechanical perspective of cell membrane in receptor mediated endocytosis of nanoparticles designed for organ specific drug delivery.
Keywords: gold nanoparticle, atomic force microscopy, endocytosis, plectin-1, membrane stiffness, pancreatic cancer
Graphical Abstract
INTRODUCTION
Nanoparticles are increasingly becoming a prime focus of various studies in cancer biology due to their potential therapeutic applications.1–4 Particularly, gold nanoparticles (GNPs), due to their strong binding affinity to thiol groups can be effectively surface modified with targeted peptides, antibodies, and anticancer drugs for effective therapeutic efficacy.5,6 Recently, nanoparticles have been recognized as superior candidates for organ specific drug delivery.1,7–9 Conventionally, a targeting agent for receptor protein including EGFR, iRGD and plectin-1, is conjugated on the surface of the nanoparticles, which is then internalized by receptor mediated endocytosis. These receptor proteins are often integral structural elements of cellular membrane and provide mechanical stability.10,11 During the endocytosis process, cell membrane undergoes dynamic morphology changes such as pit/pore formation, actin reorganization, and membrane ruffling often explored using atomic force microscope (AFM),12,13 which enables label free, high resolution surface analysis of live cells without compromising their structural integrity from fixation. Although various fluorescence imaging technologies and transmission electron microscopy have been widely used to visualize these structural dynamics, they involve extensive sample preparation and data acquisition.14 On the contrary, AFM provides a more quantitative approach to characterize real time dynamic amendments in membrane morphology and nanomechanics of live cells.1,15–17
High resolution atomic force microscope has been employed to establish a correlation between size of the pit and the amount of assembled clathrin along with actin dynamics during clathrin-mediated endocytosis process.12 Also, high resolution AFM morphology acquisition technique has been explored to study the ligand–receptor interaction using GNPs modified with transferrin (TF), which is uptaken by the TF receptors actively present on the surface of human nasopharyngeal carcinoma cells suggested by the pore formation.13 Previous studies have shown that surface membrane protein such as actin is responsible for cellular mechanical integrity.18,19 Its reorganization during various biological processes including endocytosis alters membrane stiffness levels.20 Using AFM, Zhang et al. monitored the dynamic alteration of membrane stiffness during clathrin-dependent receptor mediated endocytosis process in HeLa cells.20 AFM’s force–separation (F–S) curves have been used to track the effect of the size of GNPs in adhesion force during single GNP interaction with HeLa and Vero cell surface.21,22 Although these articles identified the size dependent nanoparticle interaction with cell membrane, the AFM study did not directly describe the endocytosis process. Moreover, these studies do not address the real-time dynamic nanomechanical changes occurring in cell membrane during receptor mediated endocytosis upon treatment with targeted gold nanoparticles.
Plectin-1, a high molecular weight protein (~500 kDa), that serves as key component in cytoskeleton network organization by linking intermediate filaments to microtubules and microfilaments, has been identified in pancreatic ductal adenocarcinoma (PDAC) pathogenesis.23,24 The most interesting observation is that plectin-1 is aberrantly overexpressed at cell membrane of PDAC cells,25 while being mostly localized in the cytoplasm and nucleus in other cells.26–28 Not surprisingly, it has been suggested as a useful biomarker for PDAC.25,29 A plectin-1-targeting peptide (PTP) with sequence KTLLPTP has been successfully explored by several groups to specifically target PDAC cells of both mouse and human origin, for imaging.1,29–34 In addition, a slightly modified sequence of the above peptide (KTLLPTPC) has been used to decorate the surface of polylactic-co-glycolic acid-polyethylene glycol (PLGA–PEG) nanoparticles to deliver therapeutic cargo to PDAC cells in a targeted manner.35 We, for the first time successfully demonstrated that redesigning PTP can govern highly stable PTP-GNP to execute highly targeted delivery of gemcitabine in PDAC tissue but not in the surrounding healthy pancreas tissue.7
Herein, we demonstrate dynamic alterations of nanomechanics of PDAC cell membrane during this receptor mediated endocytosis process. Using streptavidin and biotin reaction, we monitor the dynamic change in plectin-1 expression in the course of endocytosis process and examine nanomechanical attributes of cellular membrane using AFM in both Panc1 cells (immortal cell line) and 4911 (primary cell line is isolated from patient derived xenograft, PDX). We monitor time dependent correlation of nanomechanical alternation of cellular membrane with the available amount of plectin-1 at the cell surface. As a negative control, we choose human umbilical vein endothelial cells (HUVECs) in which, plectin-1 does not express on the cell membrane. Hence, PTP-GNP endocytosis in HUVEC cells does not involve receptor mediated endocytosis and the nanomechanical behavior of cell membrane during this internalization process is significantly different than receptor mediated endocytosis in PDAC cells. We also introduce scrambled peptide conjugated with GNP (sPEP-GNP) in PDAC cell culture. In this case, absence of plectin-1 binding peptide on GNP surface recruited receptor independent endocytosis. The dynamic alterations of cellular membrane nanomechanics follow the same trend as that of PTP-GNP interaction with HUVEC. Growth factor and cytokines available in the cell culture are often adsorbed on GNP surface during the incubation process and produce protein corona. This effectively restricts small targeting peptides to interact with cell surface receptors. To eliminate this unwanted hindrance we further shielded PTP-GNP surface with poly ethylene glycol (PEG) and extended the active plectin-1 mediated endocytosis of GNPs.
RESULTS
Plectin-1 is Overexpressed at the Surface of Pancreatic Cancer Cell Line.
Recently, plectin-1 has been identified as PDAC specific marker as they are aberrantly overexpressed at the surface of PDAC cell lines. In this study, we first validated expression of membrane bound plectin-1 expression in cell lines of different origin using classical streptavidin–biotin reaction as presented in Supporting Information (SI) Figure S1.
Endothelial (HUVEC) and normal pancreatic epithelial cells (HPDEC) express lower level of plectin-1 at their membrane, whereas it is highly expressed on the membrane of PDAC cells such as Panc1, AsPC-1, and MiaPaCa-2. Not surprisingly, primary pancreatic cancer cell lines isolated from PDX (4911 and 4535) also exhibited abundant expression of plectin-1 at their membrane (Figure 1). Among all the PDAC cells Panc1 presented the highest expression of plectin-1 (Figure 1).
Figure 1.
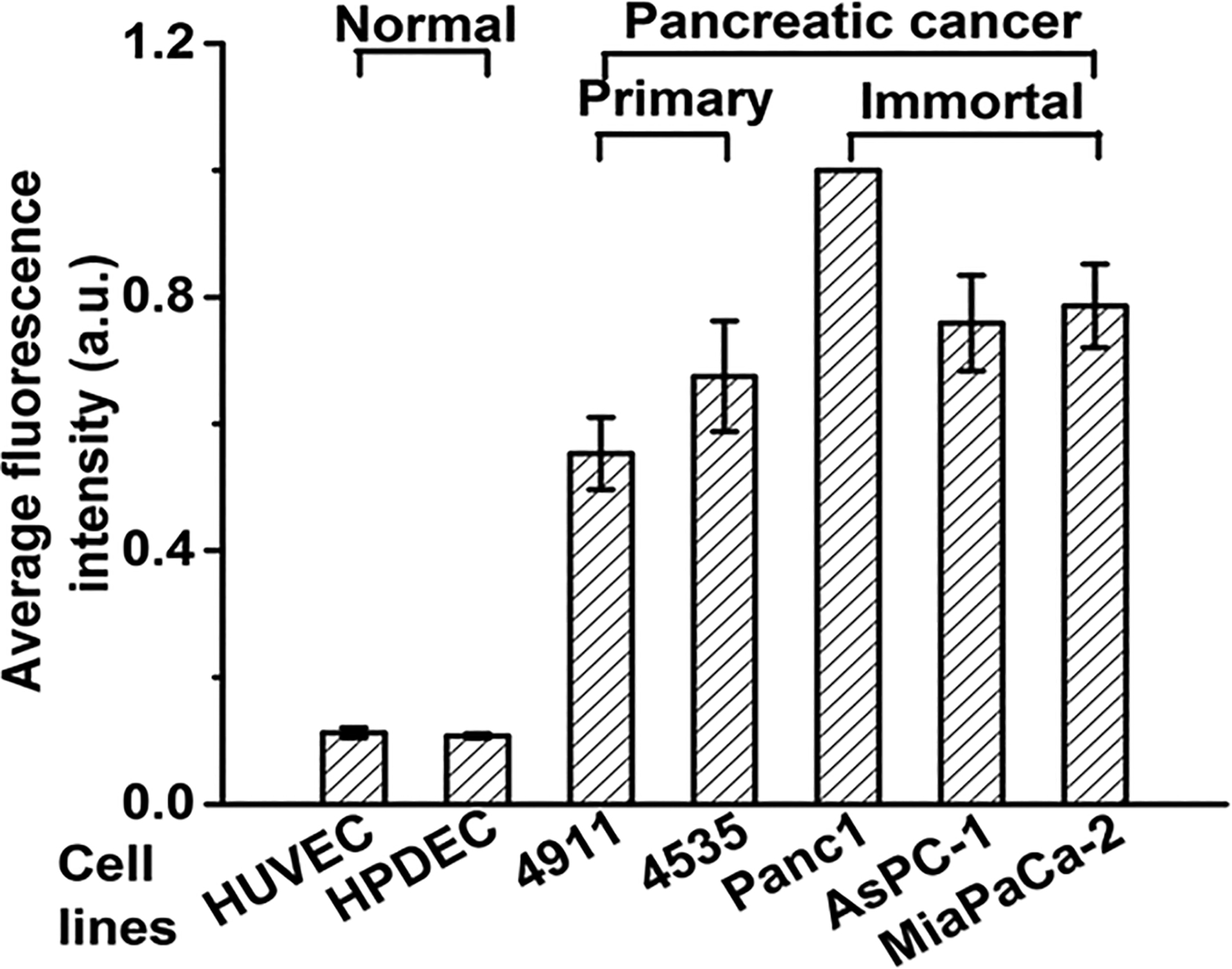
Surface plectin-1 expression in normal (endothelial and epithelial) and PDAC (primary and immortal) cell lines.
Dynamics of Membrane Nanomechanics upon PTP-GNP Treatment in Pancreatic Cancer Cell Lines.
The process of endocytosis involves dynamic alteration of the expression and orientation of structural proteins, which causes mechanical instability. We employed AFM to monitor the mechanical changes of cellular membrane during the PTP-GNP endocytosis. The principal of AFM relies on the interaction between the AFM tip and the cell membrane resulting in tip deflection which generates high resolution morphology as well as F–S curve (Figure 2A–C). This is further analyzed using appropriate mathematical models to obtain various characteristic nanomechanical properties. In this study, both Panc1 and 4911 cells were treated with PTP-GNP and we performed AFM to visualize the morphology of these cells and mechanical changes of cellular membrane. Figure 2B is a representative morphology of Panc1, whereas a representative F–S curve is shown in Figure 2C. PTP-GNP interacts with PDAC cells’ surface plectin-1 receptors (Figure 2D). Amidst continuous presence of PTP-GNP, we monitor the alteration in membrane dynamics in both Panc1 cells and 4911 cells for short time periods up to 30 min as well as longer time periods up to 6 h.
Figure 2.
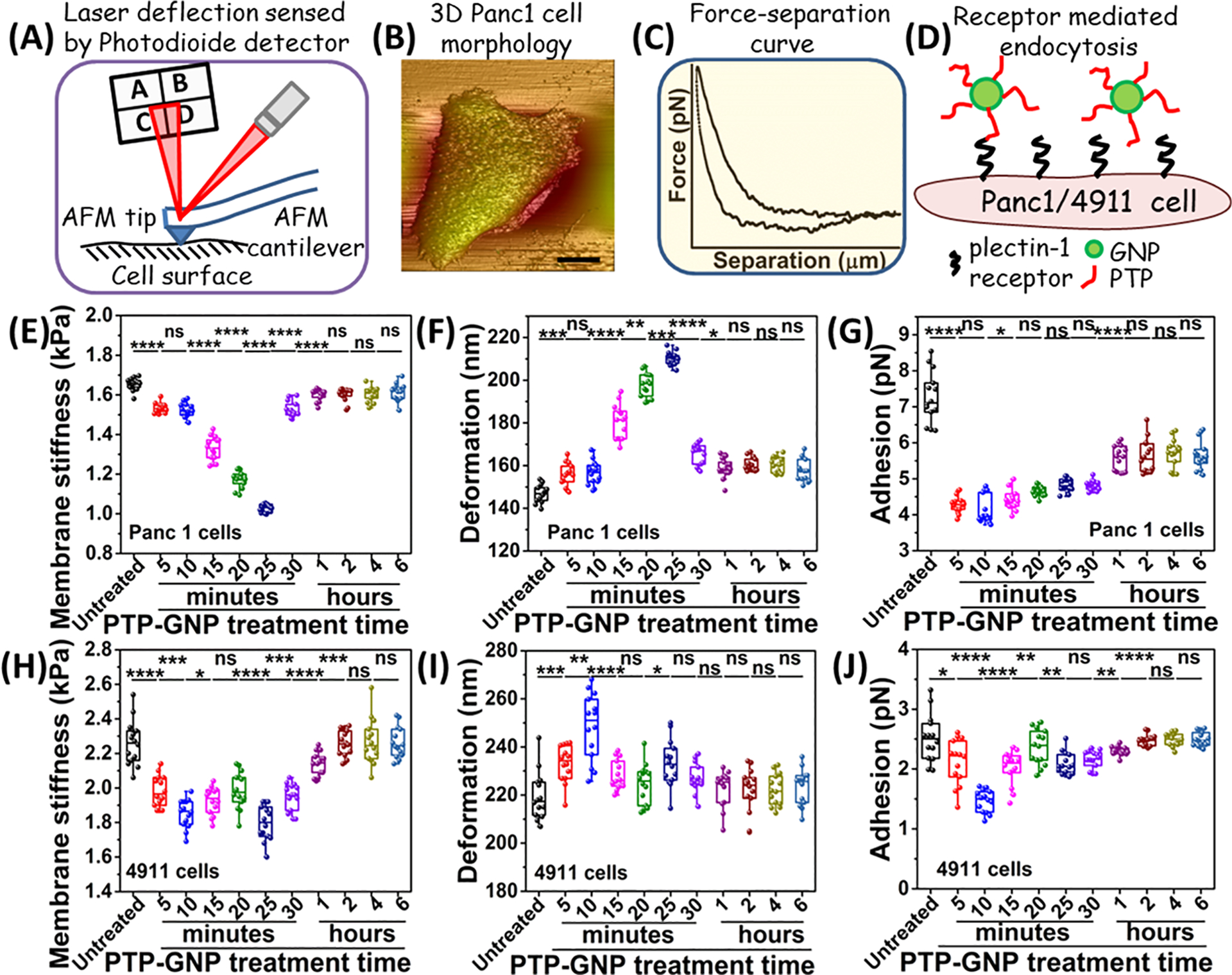
Dynamics of membrane nanomechanics upon PTP-GNP treatment in pancreatic cancer cell lines. (A) Schematic of AFM experimental setup. (B) Pictorial representation of Panc1 cell morphology. (C) A representative F–S curve. (D) Schematic representing a receptor mediated interaction between PTP-GNP and surface plectin-1 receptors. Alteration in Panc1 cells’ nanomechanical properties upon PTP-GNP treatment: (E) membrane stiffness, (F) deformation, and (G) adhesion. Alteration in 4911 cells’ nanomechanical properties upon PTP-GNP treatment: (H) membrane stiffness, (I) deformation, and (J) adhesion.
Membrane stiffness corresponding to the untreated Panc1 cells (1.65 ± 0.03 kPa) closely resembles our previously published data36 (Figure 2E). The cell membrane stiffness initially decreased systematically during the PTP-GNP treatment for first 25 min and reached to a minimum value of 1.02 ± 0.02 kPa (nearly 41% of the initial stiffness) followed by a quick recovery. We monitored no changes in stiffness for next 6 h. We also monitored adhesion for PTP-GNP treated Panc1 cells (Figure 2G). Untreated Panc1 cells exhibited adhesion value of 7.26 ± 0.7 pN. Upon PTP-GNP treatment, we observed ~50% decrease in adhesion value at fifth minute (3.52 ± 0.13 pN). Thereafter, the adhesion increased systematically until 25th minute with saturation observed at 1 h (Figure 2G). No further change in adhesion was observed up to 6 h. Compared to Panc1 cells, untreated 4911 cells exhibited stiffer membrane with stiffness value of 2.26 ± 0.13 kPa (Figure 2H). 4911 cells subjected to PTP-GNP treatment initially displayed a decreasing trend in membrane stiffness up to 10 min (1.85 ± 0.08 kPa) followed by a slight increase in membrane stiffness at 15th (1.92 ± 0.07 kPa) and 20th minute (1.98 ± 0.1 kPa). At 25th minute, the membrane stiffness dropped significantly (1.79 ± 0.1 kPa) compared to 20th minute, thereafter, began its recovery (Figure 2H). The recovery duration of membrane stiffness was longer in 4911 cells (t1 ~ 120 min) compared to Panc1 cells indicating prolonged dynamic PTP-GNP uptake process by 4911 cell membrane. These cells exhibited ~22% maximum decrease in membrane stiffness. We further monitored the adhesion parameter over 6 h duration for PTP-GNP treated 4911 cells (Figure 2J). Untreated 4911 cells exhibited lower adhesion value (2.26 ± 0.13 pN) compared to untreated Panc1 cells. Upon PTP-GNP treatment, we observed maximum decrease of 42% in adhesion parameter at 10th minute (1.46 ± 0.18 pN) following which, it began recovery. We observed a complete recovery of adhesion parameter at 2 h and remained more or less similar up to 6 h (Figure 2J). Deformation is another nanomechanical characteristic solely depended on interaction between AFM tip and cell membrane. We also quantified deformation of cellular membrane of both 4911 and Panc1 cells, during the PTP-GNP internalization at short and long time duration. Deformation values for both Panc1 cells and 4911 cells displayed a complementary behavior to the membrane stiffness (Figure 2F and 2I). Untreated Panc1 cells exhibited lower membrane deformation (146.77 ± 4.15 nm) compared to 25th minute of post PTP-GNP treatment (210.19 ± 3.13 nm). The 4911 cells also displayed similar trend in deformation during PTP-GNP treatments, where membrane deformation increased from 219.52 ± 9.97 nm to 233.19 ± 9.73 nm after 25 min of treatments. Deformation appeared to be a corollary of membrane stiffness. Together, these results indicate that the alterations in membrane dynamics (cells becoming softer and decreased adhesion) in Panc1 cells are a result of PTP-GNP treatment during the endocytosis process.
Nanomechanics of Cell Membrane for Receptor Independent Uptake of Gold Nanoparticles.
To understand the mechanical changes of cell membrane in receptor independent endocytosis we have employed (i) gold nanoparticles conjugated with scrambled peptide (sPEP-GNP) in PDAC cell culture and (ii) PTP-GNP in HUVEC culture; HUVEC has minimal membrane expression of plectin-1 (as shown in schematic from Figure 3A,B, respectively). We evaluated the response of membrane stiffness and deformation in the presence of gold nanoparticles for short time period up to 30 min in HUVECs (Figure 3D,F) as well as for long time period up to 6 h in PDAC culture (Figure 3C,E).
Figure 3.
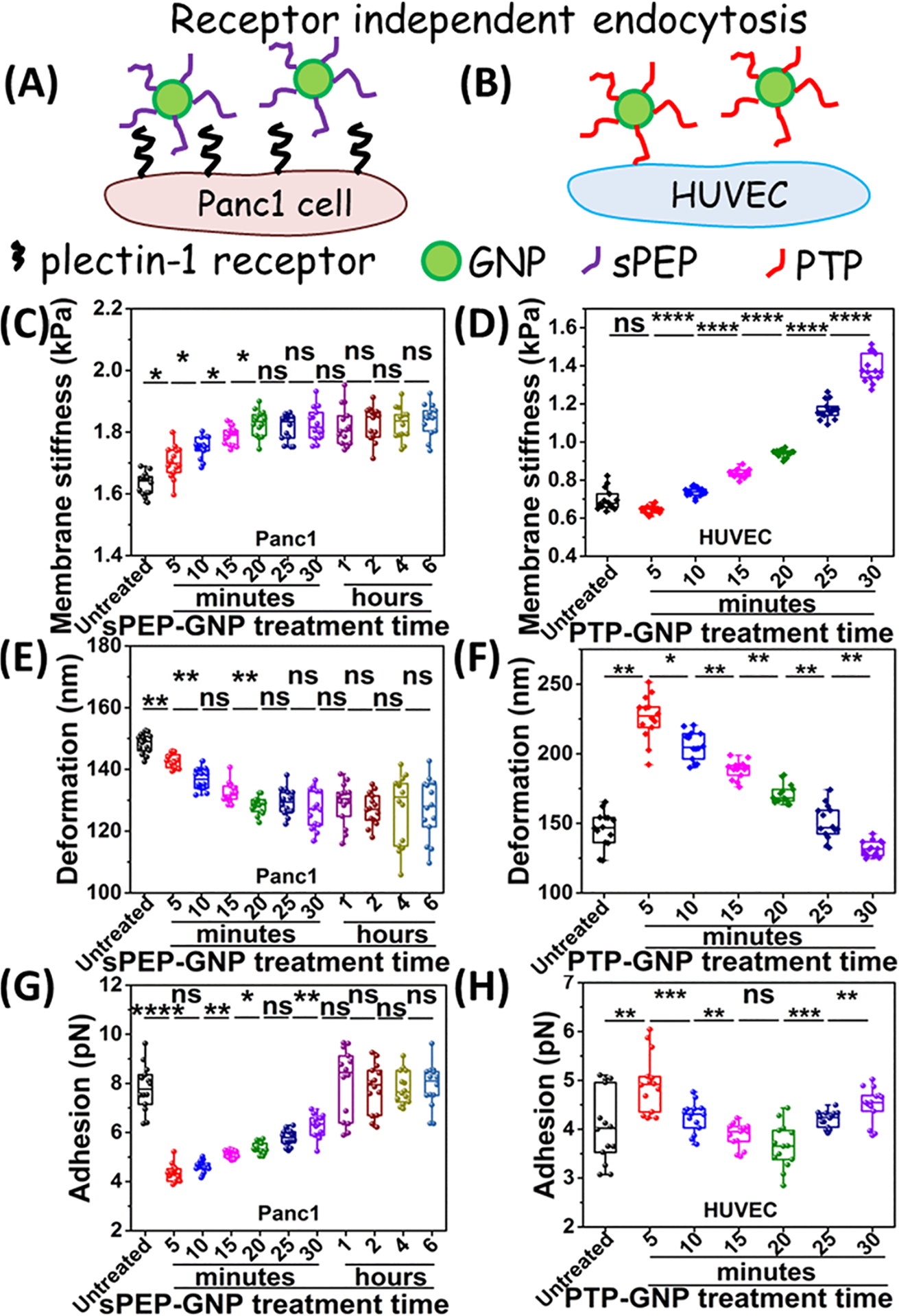
Dynamics of membrane nanomechanics during receptor independent uptake of GNPs. Schematics representing receptor independent uptake in (A) Panc1 treated with sPEP-GNP and (B) HUVECs treated with PTP-GNP. Alteration in membrane stiffness in (C) Panc1 cells upon sPEP-GNP treatment. (D) HUVEC cells upon PTP-GNP treatment. Alteration in deformation in (E) Panc1 cells upon sPEP-GNP treatment. (F) HUVEC cells upon PTP-GNP treatment. Alteration in adhesion in (G) Panc1 cells upon sPEP-GNP treatment. (H) HUVEC cells upon PTP-GNP treatment.
Untreated Panc1 cells exhibited similar stiffness value of 1.63 ± 0.03 kPa to those in Figure 2E and our previous study.36 Upon treatment with sPEP-GNP, we observed an increasing trend in membrane stiffness indicating the overall cell surface become harder up to 20th minute with maximum stiffness of 1.82 ± 0.04 kPa thereby, maintaining the membrane stiffness up to 6 h (Figure 3C). Such a trend was exactly opposite to the Panc1 cells treated with PTP-GNP in which, the cells appeared to become softer initially. In terms of change in membrane stiffness, we observed an increase of 11.1% stiffness value. Deformation on the other hand, showed a complementary trend compared to the membrane stiffness and was observed to be 128.32 ± 2.77 nm at 20th minute (corresponding to stiffest cells), significantly lower than untreated softer cells with deformation of 148.36 ± 3.17 nm (Figure 3E). Further, we monitored dynamic alterations in membrane nanomechanics in HUVECs for 30 min by treating them with PTP-GNP. Untreated HUVECs with stiffness value of 0.64 ± 0.01 kPa (Figure 3D) and deformation 225.87 ± 17.32 nm (Figure 3F) appeared softer than untreated PDAC cells. HUVECs exhibited an increasing trend in membrane stiffness as observed up to 30 min. We observed an increase of ~114% in the membrane stiffness value at the end of 30 min in PTP-GNP treated HUVECs. Such an increasing trend was similar to that observed in sPEP-GNP treated Panc1 cells. Deformation on the other hand, decreases to 131.6 ± 5.38 nm at 30th minute (Figure 3F). We also evaluated dynamic adhesion response in both PDAC and HUVEC with sPEP-GNP and PTP-GNP treatment, respectively (Figure 3G,H). Untreated Panc1 displayed an adhesion value of 7.8 ± 0.93 pN. Upon PDAC treated with sPEP-GNP, we observed an immediate ~45% decrease in its adhesion parameter corresponding to fifth minute (4.34 ± 0.35 pN). We observed a systematic recovery in adhesion parameter after the initial decrease with complete recovery at 1 h, which sustained up to 6 h (Figure 3G). Untreated HUVECs on the other hand, exhibited lower adhesion (4.05 ± 0.73 pN) compared to untreated PDAC cells (Figure 3H). HUVECs when treated with PTP-GNP displayed an initial increase in adhesion parameter at fifth minute (4.93 ± 0.59 pN) followed by a systematic drop until 20th minute (3.65 ± 0.44 pN). Following which, the adhesion value increased up to 30th minute (4.49 ± 0.36 pN) (Figure 3H). Together, these results indicate that during receptor independent endocytosis, the membrane of Panc1 cells and HUVEC become stiffer and the adhesion is recovered in contrast to receptor mediated endocytosis where membrane stiffness decreases and adhesion is partially recovered.
Dynamic Recovery of Panc1 Cell Membrane Stiffness upon Exposure to Various PTP-GNP Incubation Times.
To further ascertain the dynamic variation in membrane stiffness and deformation during the course of PTP-GNP internalization in plectin-1 expressing cells, we altered the incubation time of PTP-GNP and estimated mechanical relaxation using AFM. After specific incubation time, we replaced the media consisting of PTP-GNP with fresh DMEM media and observed that the stiffness and adhesion of the membrane initiated recovery process instantaneously over a course of 30 min window (Figure 4A,B, respectively).
Figure 4.
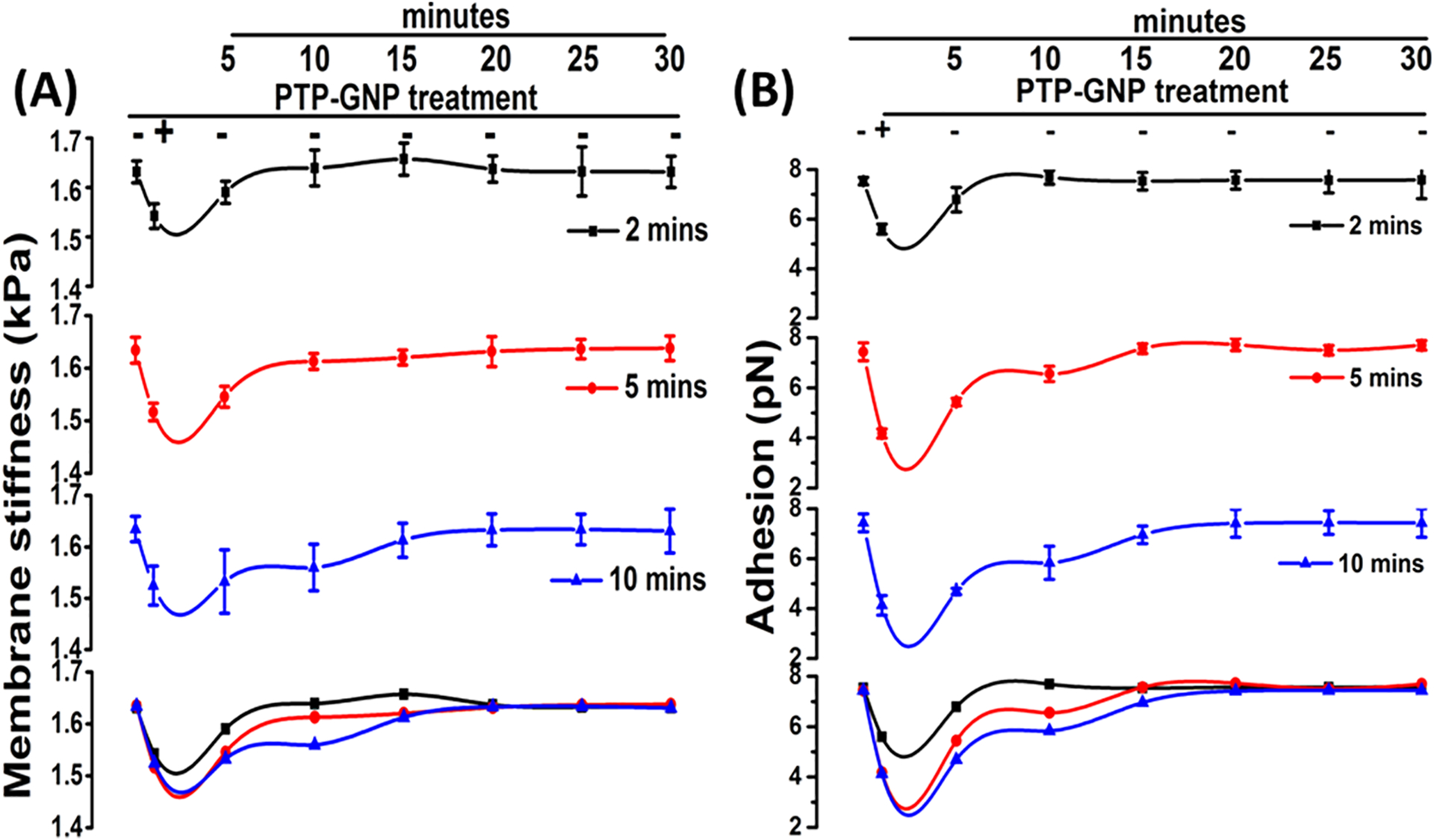
Dynamic recovery of Panc1 cell membrane nanomechanical properties upon exposure to PTP-GNP for various time durations. (A) Membrane stiffness. (B) Adhesion.
More importantly, we observed that longer incubation times leads to delayed recovery of membrane stiffness. Upon 2 min of incubation, Panc1 cell membrane recovered stiffness at 10th minute (1.63 ± 0.03 kPa compared to untreated cells with stiffness of 1.63 ± 0.02), while both 5 and 10 min of incubation successfully recovered membrane stiffness at 15th minute (1.62 ± 0.01 kPa compared to untreated cells with stiffness of 1.63 ± 0.02 kPa and 1.62 ± 0.03 kPa compared to untreated cells with stiffness of 1.63 ± 0.02 kPa, respectively, for 5 and 10 min incubation times). It was important to note that the rate of recovery was slower for 10 min incubation time as evident from merged panel (Figure 4A). We observed a similar trend in adhesion recovery compared to membrane stiffness and that longer incubation times lead to delayed recovery. For 2 min of incubation, membrane recovered its adhesion at 10th minute (7.19 ± 0.28 pN) compared to untreated Panc1 (7.03 ± 0.15 pN). Incubation for 5 min led to membrane adhesion recovery at 15th minute (7.06 ± 0.19 pN) compared to untreated Panc1 (6.93 ± 0.36), whereas 10 min of incubation led to complete recovery at 20th minute (6.91 ± 0.56 pN) compared to untreated Panc1 (6.97 ± 0.4 pN). Figure 4B (merged panel) also shows the comparative rate of recovery in which, it is clear that progressive increase in incubation times lead to slower recovery. We also evaluated the membrane deformation corresponding to various incubation times and noticed similar recovery of these properties as shown in SI Figure S2. We observed a quicker recovery in membrane deformation in the presence of 2 min of PTP-GNP incubation with a complete recovery of deformation levels at fifth minute (146.52 ± 3.06 nm compared to untreated cells with deformation of 146.15 ± 1.87 nm). The recovery of deformation levels for 5 and 10 min of incubation were more or less similar and a complete recovery was observed at the 10th minute post replacing the PTP-GNP containing media with fresh complete DMEM (SI Figure S2, merged panel). Together, these results confirm that AFM is highly capable of monitoring the dynamic response of Panc1 cell mechanics during endocytosis process and this is reversible.
Dynamic Response of Plectin-1 Receptors on Panc1 Cell Surface in Response to Endocytosis of PTP-GNP.
Plectin-1 surface protein is known to provide and maintain structural integrity in Panc1 cell.24 Plectin-1 dependent endocytosis process can involve plectin-1 internalization causing a disruption of structural integrity and, hence, the dynamic alteration in membrane nanomechanical properties. To test our hypothesis that plectin-1 expression is responsible for dynamic alteration of membrane mechanics, we initially treated Panc1 cells with PTP-GNP for 2–30 min and then 1–6 h with time durations maintained same as the AFM experiments. After corresponding time point treatments, the 96-well plate comprising of treated Panc1 cells was immediately placed on ice to freeze the cellular activities (specifically receptor internalization). Streptavidin–biotin based immunoassay was then employed to recognize the plectin-1 proteins at the membrane (SI Figure S1). The average fluorescence intensity for untreated Panc1 cells is highest due to the presence of large number of free plectin-1 receptors on cell membrane surface. With time, the number of free receptors on the Panc1 cell surface decreased due to the continuous receptor mediated endocytosis process, which is evident from average fluorescence intensity (Figure 5A).
Figure 5.
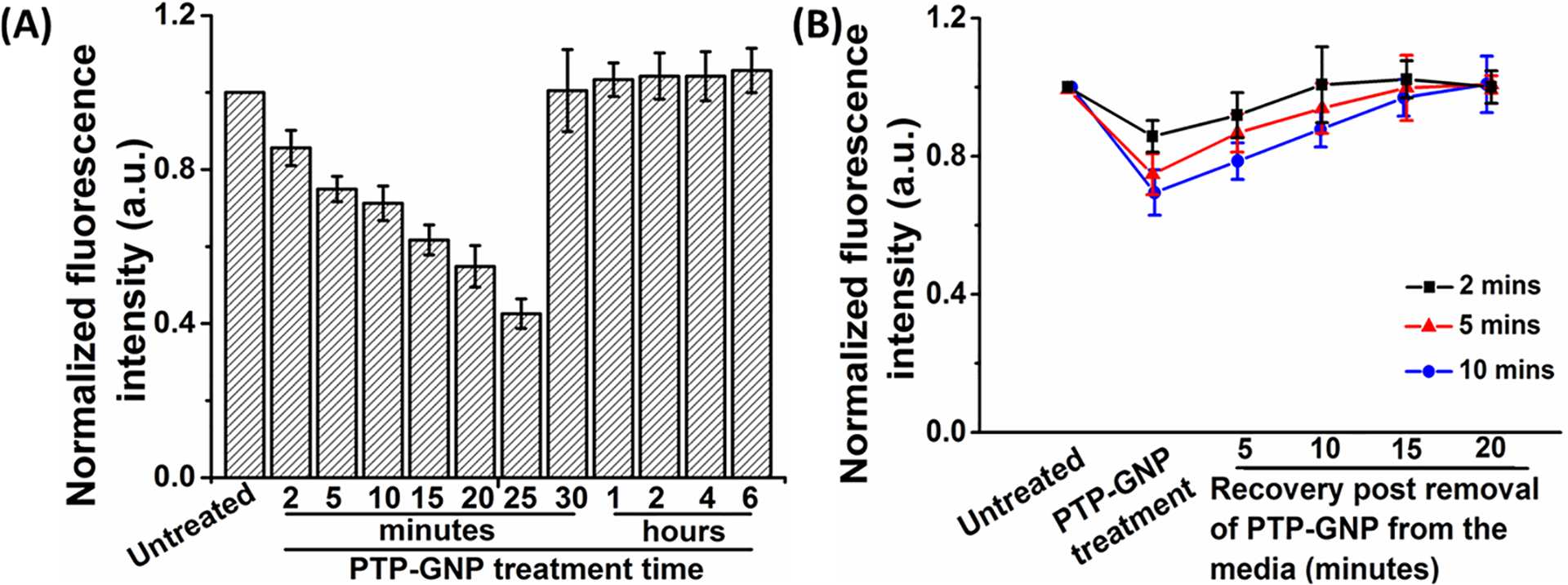
Dynamic response of Plectin-1 receptors on Panc1 cell membrane in response to endocytosis of PTP-GNP. (A) Dynamic alteration of plectin-1 expression on Panc1 cell membrane for various time points post PTP-GNP treatment. (B) Recovery of plectin-1 receptors on Panc1 cell membrane at various time points upon dynamic PTP-GNP treatment.
The decreasing trend in the average fluorescence intensity up to 25th minute closely follows the nanomechanical trend observed in Figure 2E. Thereafter, the average fluorescence intensity levels revert back to the original value indicating the recycling of plectin-1 receptors to the Panc1 cell membrane surface. At 30th minute and thereafter up to 6 h, the average fluorescence intensity remains more or less constant indicating that there is no active region on PTP-GNP to bind to the free plectin-1 receptors available on the Panc1 cell membrane surface. Maximum change in average fluorescence intensity was observed at 25th minute with ~48% decrease corresponding to a ~41% decrease in membrane stiffness confirming that alteration in membrane dynamics in Panc1 cells is dependent on plectin-1 receptors at corresponding time points. We further studied the recovery of plectin-1 receptors with withdrawal of GNP from cell culture. Panc1 cells were incubated with PTP-GNP for 2, 5, and 10 min. Following which, the PTP-GNP containing media was rinsed and replaced with fresh DMEM media void of PTP-GNP. Post media change after 2, 5, and 10 min of incubation, recovery of plectin-1 expression was monitored for 20 min in the interval of 5 min at which point, the 96-well plate was immediately placed on ice. At this time, the cells were treated with biotin complex followed by GFP-streptavidin molecules. From Figure 5B, it is evident that the average fluorescence intensity corresponding to the PTP-GNP incubation time closely follows that observed in Figure 5A. Post treatment with fresh media, plectin-1 receptors for 2 min incubation are observed to recover at the earliest with complete recovery after 10 min. Five minutes and 10 min PTP-GNP incubation times show complete recovery of plectin-1 receptors at 15th minute with the rate of recovery for 5 min incubation time much faster than the 10 min incubation time (Figure 5B). Such a trend was also observed in nanomechanical studies evident from Figure 3. Above results suggest a correlation between membrane nanomechanics in Panc1 cell and plectin-1 receptors.
DISCUSSION
There have been a plethora of studies conducted to understand the biological behavior of cells when treated with GNPs and the various formulations such as their selective uptake by cells, their exocytosis pattern as well as toxicity effects.3,37–39 So far, AFM has been extensively used to study morphological alterations such as pit formation and membrane ruffling on cell surface in the presence of GNP using AFM.25,40 While the biochemics behind the receptor mediated endocytosis is well studied, very little is known about the biomechanics of cell surface involved in receptor dependent endocytosis of GNPs. Using AFM tool and employing nanoindentation technique using optimal experimental parameters, here we have addressed the real time dynamic alteration in nanomechanics of live cells in the presence of various formulations of GNPs. Foremost, we evaluated surface plectin-1 expression in normal endothelial and epithelial cell lines (HUVEC and HPDEC, respectively) as well as primary PDX isolated cells (4911 and 4535) and immortal (Panc1, AsPC-1 and MiaPaCa-2) pancreatic cancer cell lines. Pancreatic cancer cell lines expressed significantly more surface plectin-1 expression compared to normal cell lines. Further, by using PTP-GNP to target plectin-1 surface receptor protein in Panc1 and 4911 cells (receptor mediated endocytosis), we observed that both cells became softer initially, thereafter recovering their membrane stiffness during the course of endocytosis process. On the contrary, the dynamic alteration of membrane stiffness followed opposite trend on Panc1 during interaction of scrambled peptide conjugated to GNP. Such a trend also resonated on HUVEC’s interaction with PTP-GNP. Deformation on the other hand, exhibited a complementary behavior to the stiffness. Membrane dynamics during receptor mediated endocytosis in Panc1 cells were further confirmed by varying the incubation times of PTP-GNP treatment and we observed that longer incubation times led to delayed recovery of membrane stiffness. Using classical streptavidin–biotin binding complex reaction, we observed a systematic decrease in expression of surface plectin-1 receptors post treatment of PTP-GNP in Panc1 cells and the trend was similar to the membrane nanomechanics observed with the AFM. Further, plectin-1 receptors recovery during the dynamic receptor mediated endocytosis for various incubation times agreed with AFM membrane recovery experiment. Overall, these results confirmed a direct correlation between membrane dynamics using AFM and surface plectin-1 receptors using streptavidin–biotin reaction during receptor mediated endocytosis in PTP-GNP treated Panc1 cells.
Endocytosis process is classified into phagocytosis, micro-pinocytosis, pinocytosis, clathrin independent, and receptor mediated endocytosis, which mostly involves clathrin and/or caveloae.3,41,42 Nanoparticle–ligand binding with the cell membrane receptors decreases local Gibbs free energy, which induces the membrane to wrap around the nanoparticle to form a closed vesicle structure.43 Recent studies have shown that physical attributes such as size, shape, surface chemistry, and charge of modified nanoparticles govern cellular uptake.15,44–49 Along with physical properties of nanoparticles, their nanomechanical behavior such as elasticity plays a significant role in endocytosis process.50–54 Soft spherical nanoparticles are found to be less efficiently wrapped compared to stiff nanoparticles. Experimental results supporting the simulations have been demonstrated in which, PEG-based hydrogel nanoparticles of uniform size and ranging elastic moduli (0.255–3000 kPa) were used. Softer nanoparticles exhibited reduced cellular uptake in immune cells (J774 macrophages), endothelial cells (bEnd.3), and cancer cells (4T1).50 Above well-established studies are informative and describe the physical attributes of nanoparticles governing various uptake mechanisms but fails to quantitatively address nanomechanical changes in real-time cell membrane mechanics during the endocytosis process.
As the endocytosis process is a dynamic event, it becomes pivotal to study the alterations in membrane nanomechanics with time. AFM solely relies on interaction between tip and the cell surface and as a result becomes an appropriate tool to quantify the dynamic changes in nanomechanical properties at cell surface with nanoscale spatial resolution. AFM generates a force–separation (F–S) curve based on the tip–sample interaction from which, several nanomechanical properties such as stiffness, deformation and adhesion can be extracted.36,55 In this study, we observed changes in stiffness and deformation for short time (up to 30 min) as well as long time (up to 6 h). Variable maximum changes in membrane stiffness in 4911 and Panc1 cell lines could be due to two varying factors: rate of internalization of PTP-GNP bound to plectin-1 receptors and continuous recycling of the receptors. Deformation exhibited an opposite trend to that of membrane stiffness. For equal applied force, stiffer cell membrane offered more resistance to the AFM tip that is indenting into the cell surface whereas, for softer cell membrane more deformation was observed for corresponding time points. A similar observation in dynamic alteration in membrane stiffness during clathrin-dependent receptor mediated endocytosis process was seen in serum-starved HeLa cells with endothelial growth factor (EGF, a receptor endocytosis stimulator).20 The authors observed an immediate spike in membrane stiffness upon EGF addition with peak stiffness observed within 60 s thereby, maintaining it more or less constant over next 10 min and ultimately returning back to baseline after 15 min. Wnt3a, another receptor endocytosis stimulator also showed similar trend in membrane stiffness, however, the rate of increase in membrane stiffness was 5 times slower and the process lasted 2 times longer than EGF indicating the dynamicity of receptor mediated endocytosis on nanomechanical alteration of membrane stiffness.20 This study, however did not involve a nanoparticle-cell membrane interaction and moreover, was performed in serum starved HeLa cells. Researchers often explore the adhesion nanomechanical property, which is the pull up force experienced by the tip from the cell surface membrane as it tries to retract at the end of tip–sample interaction.56,57 Adhesion parameter depends upon the tip geometry, sample type and their interactions. In this study, variable change in adhesion levels was observed for both receptor dependent and receptor independent endocytosis mechanisms (Figures 2G,J and 3G,H). These alterations emphasize on the dynamic nature of nanoparticle uptake mechanisms. During receptor dependent uptake, membrane adhesion fails to recover completely compared to receptor independent uptake (complete recovery). Although for PDAC cells, adhesion parameter can be used to distinguish type of uptake (receptor dependent v/s receptor independent); in depth studies need to be conducted to deduce the exact mechanism responsible for this variable behavior. Care was taken in analyzing the F–S curves such that the indentation was less than 10% of the overall height of the cell, within the acceptable depth in the field of AFM to avoid causing permanent damage to the cell as well as neglect the effect of substrate stiffness.58–60
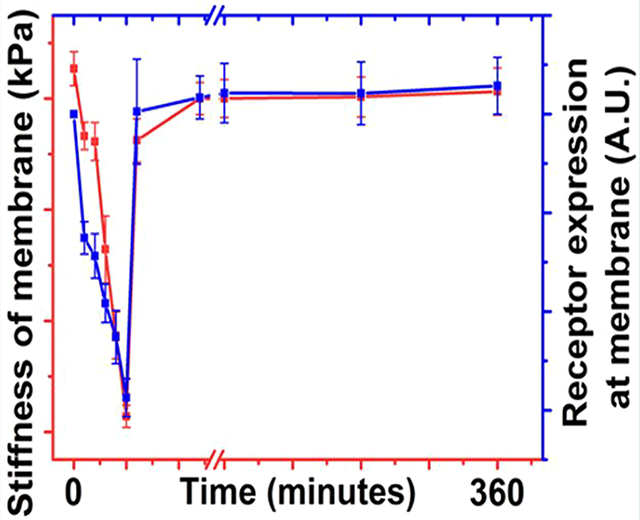
Plectin-1 is responsible for cellular structural integrity. PTP-GNP binds to the plectin-1 receptor on the surface of Panc1 cells and internalizes causing an overall reduced membrane surface plectin-1 expression. This was confirmed by streptavidin–biotin complex at various time points as seen from Figure 5A. AFM studies demonstrated reduction in membrane stiffness of Panc1 cells when treated with PTP-GNP. Comparative analysis of alteration in membrane stiffness and surface plectin-1 expressions at respective time instants confirmed that dynamic alteration in membrane nanomechanics was due to the change in surface plectin-1 expression and the trends in both nanomechanics and surface plectin-1 expression at various time points closely followed each other as seen from Figure 6.
Figure 6.
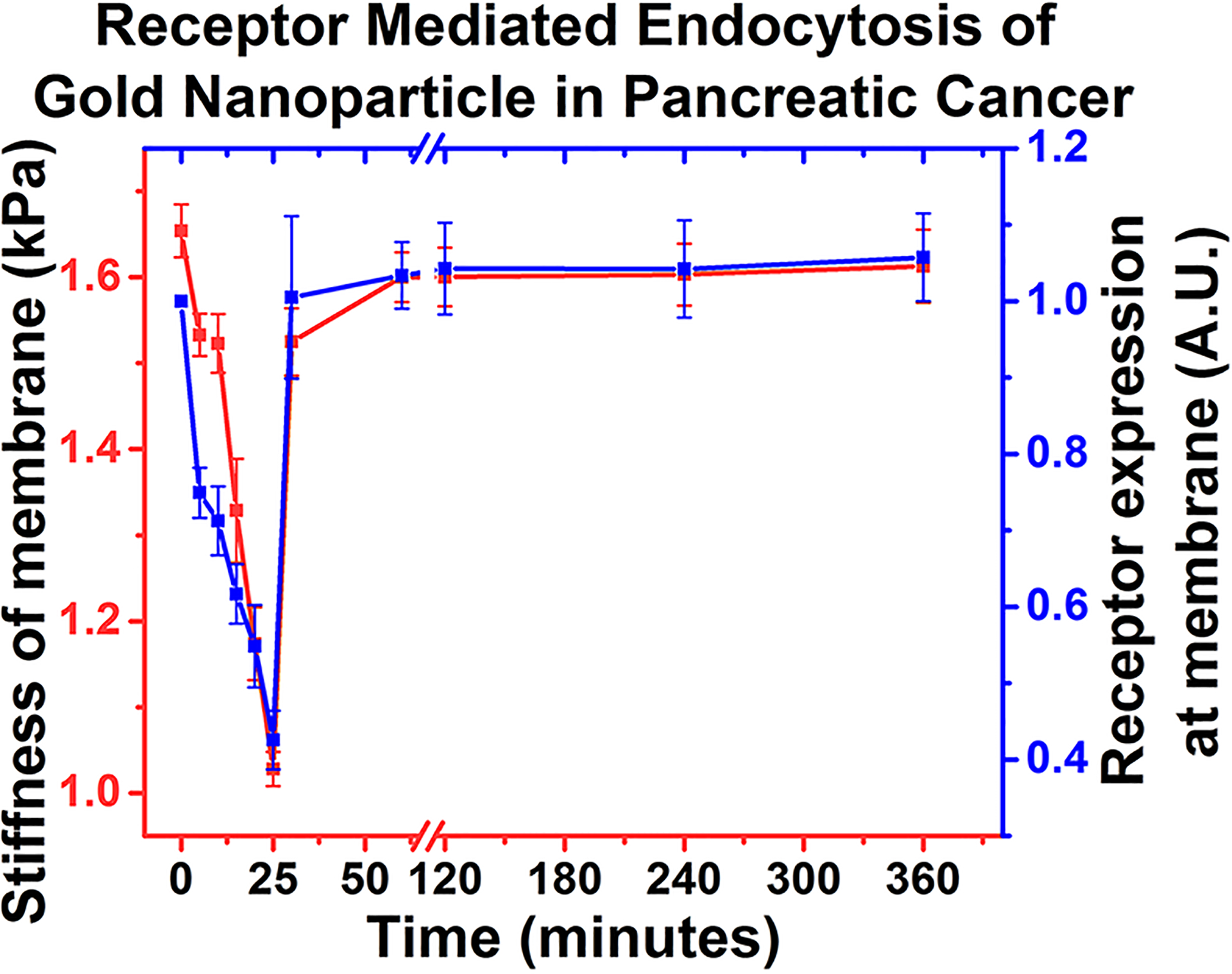
Receptor mediated endocytosis of gold nanoparticle in pancreatic cancer.
Longer PTP-GNP incubation times resulted in longer recovery time of surface stiffness and plectin-1 receptors to get back to normal levels suggesting that with longer incubation time, more plectin-1 receptors at the membrane surface were internalized. In case of PTP-GNP, saturation in cell membrane stiffness and surface plectin-1 expression levels was observed after a certain period of time as seen from Figure 2E and Figure 5A. Such behavior was also confirmed by measuring surface plectin-1 expression after pretreating Panc1 cells by PTP-GNP for 30 min followed by fresh PTP-GNP treatment (SI Figure S3A). This does not bode well with the dynamic and continuous nature of endocytosis process. These observations led to the theory of protein corona formation around nanoparticle, which is well-known.61–63 A possible reason could be that the PTP conjugation to the GNP does not cover the entire available surface area and various growth hormones and proteins present in serum containing medium can bind to free surface of GNP forming protein corona. To test this theory, Panc1 cells were treated with PEGylated PTP-GNP such that the available free surface was saturated with PEGs. Upon Panc1 treatment with PEGylated PTP-GNP, the surface plectin-1 expression was observed to be reduced over a period of over 8 h, a significant improvement from the results seen in SI Figure S3B. This confirmed the theory of protein corona formation and the prolonged reduced surface plectin-1 expressions demonstrated continuous plectin-1 receptor mediated endocytosis process driven by constant internalization and recycling of plectin-1 receptors at the surface of Panc1 cells.
CONCLUSION
Nanoindentation using AFM clearly recognizes the dynamic behavior of nanoparticle endocytosis both qualitatively and quantitatively in live cells. During cellular interaction of PTP-GNP, PTP interacts with plectin-1 available on PDAC cells and recruits receptor mediated endocytosis. Receptor mediated endocytosis of targeted GNPs causes transient and reversible alteration in plectin-1 expressions at the cell membrane of the PDAC cells. This results into structural modification, which ultimately leads to dynamic alteration of mechanical properties of cellular membrane. Hence, a direct correlation between the surface plectin-1 expression and membrane stiffness was clearly seen for receptor mediated endocytosis process. Additionally, we have shown that AFM has a strong potential to differentiate membrane nanomechanics in receptor mediated versus receptor independent endocytosis.
EXPERIMENTAL SECTION
Regents.
Gold(III) chloride trihydrate, sodium chloride and sodium hydroxide were purchased from Sigma. The plectin-1 targeting peptide with additional modification KTLLPTPYC and scrambled peptide KGHSGLMYC were purchased from Biomatik and Peptide2.0, respectively (>95% purity). mPEG3-SH was purchased from PurePEG. Endothelial basal medium, RPMI, Dulbecco’s Modified Eagle’s Medium (DMEM), Keratinocyte-SFM media, Bovine pituitary hormone and epidermal growth factor (EGF) was purchased from Life technologies. DMEM/F12 and Leibovitz (L-15) medium for AFM experiments along with PBS (1×, pH 7.4) was purchased from Thermofisher. Additionally, plectin-1 polyclonal primary antibody (catalog no. PA5–79829) and Streptavidin, Alexa Fluor 488 conjugate was also purchased from Thermofisher. Biotinylation kit was purchased from Abcam. Fetal Bovine Serum was obtained from Atlanta Biologicals. Collagen type 1 was purchased from Corning. Penicillin Streptomycin was purchased from Gibco.
Cell Lines Used in This Study with Purchasing Information and Validation.
Immortalized pancreatic cancer cell lines such as Panc1 (catalog no. CRL-1469), AsPC-1 (catalog no. CRL-1682) and MIA PaCa-2 (catalog no. CRL-1420) were purchased from American Type Culture Collection (ATCC) and used without further validations. Primary patient derived pancreatic cancer cell lines 4535 and 4911 were obtained from Mayo Clinic Pancreatic cancer Spore and used previously.64 Human umbilical vein endothelial cell line (HUVECs, catalog no. CC-2519) was purchased from Lonza. Human pancreatic ductal endothelial cells (HPDECs) were obtained from Dr. Peter Storz at Mayo Clinic Jacksonville, FL campus (a generous gift)
PTP-GNP Synthesis.
Gold nanoparticles were synthesized using plectin-1 targeted peptide or scrambled peptide assisted template following our previously published literature.36 Briefly, Gold(III) chloride trihydrate and peptide were dissolved in Milli-Q water in 10:1 molar ratio and stirred for 2 min at 37 °C. Following which, 1 M NaOH was added dropwise until the overall pH of the solution reached 12. Following a continuous stirring for 16 h, GNPs were isolated by ultracentrifugation at 38K RPM using Beckmann Optima L-80 XP ultracentrifuge (purchased from Beckman Coulter Inc., Brea, CA). Loose pellet of GNP at the bottom of the centrifuge tube was carefully isolated from the supernatant. Ultimately, Milli-Q water was added to the pellet to make the final volume the same as reaction volume and yield GNP stock solution. For experiments, however, the stock solution was further diluted 10 times to make a working stock solution. Both the original stock and working stock were stored at 4 °C until further use. Using Shimadzu UV–vis Spectrometer we confirmed the surface resonance peak of GNP. In our previous work, we have performed a comprehensive characterization of GNP prepared by plectin-1 targeted peptide or scrambled peptide via dynamic light scattering (DLS) as well as computational modeling. We observed a hydrodynamic size of ~5 nm using DLS for both PTP-GNP and sPEP-GNP. Further, using computational modeling simulation, we were able to decipher that around 53 peptides were available at GNP surface.7
Cell Culture.
Immortalized cell lines Panc1 and MiaPaCa-2 were cultured in complete DMEM media; whereas AsPC-1 was cultured in RPMI media both supplemented with 10% FBS and 1% penicillin streptomycin. HUVECs were grown on plates/wells coated with 30 μg/mL collagen type 1 solution and cultured in endothelial basal medium supplemented with 12 μg/mL bovine brain extract, 1 μg/mL hydrocortisone and 1 μg/mL GA-1000. HPDEC cells were cultured in Keratinocyte-SFM media and supplemented with bovine pituitary hormone and EGF. Additionally, primary patient derived pancreatic cancer cell lines 4911 and 4535 were cultured in DMEM/F12 culture media supplemented with 10% FBS and 1% penicillin streptomycin. Panc1 cells were arrested in S phase by treating them with 20 μg/mL aphidicolin for 12 h prior to the AFM experiment and their arrested phase was maintained during the course of the experiment as per our previous study.36 For experiments involving streptavidin–biotin, respective cell line was passaged and cultured into a 96 well plate in their respective medium for 1 day before each experiment at 37 °C in a humidified 5% CO2 atmosphere. Cells were cultured at concentration such that during the time of experiment all the cell lines were at ~8000 cells/well. For all AFM experiments, Panc1 and 4911 cells were cultured at 60–70% confluency in a 60 mm dish in their respective culture medium.
Cell Surface Plectin-1 Labeling Using Streptavidin–Niotin Conjugation.
Streptavidin–biotin reaction is commonly used to label the surface protein and other biomolecules containing primary amines.65 Streptavidin–biotin bonds are one of the strongest bonds with dissosiative constant (Ka) = 10−14 . Expression of surface plectin-1 protein in normal as well as PDAC cell lines was monitored via streptavidin–biotin complex. Briefly, plectin-1 antibody was prepared in 3% BSA solution and stored at 4 °C for immediate use. Biotinylation kit comprising of 1 μL of biotin (type A) modifier reagent was added to 10 μL of antibody to be labeled and mixed gently by withdrawing and redispensing the mixture. Further, the antibody mixture was added to lyophilized material (type A) conjugation mix vial. The antibody mixture and conjugation mix was then gently mixed with pipet. The mixture was allowed to sit at room temperature for 15 min. Following which, 1 μL of biotin (type A) quencher reagent was added to the above mixture and rested for 5 min at room temperature. The biotin conjugated antibody mix was stored at 4 °C for short-term use and at −20 °C for long-term use. For experimental purpose, working biotinylated antibody solution was prepared by mixing the conjugation mixture with 3% BSA in 1:1000 ratios. To execute streptavidin–biotin reaction, cells were washed with 1× PBS three times followed by biotinylated antibody treatment (100 μL/well) for 30 min. The solution was then aspirated, and the cells were rinsed with 1X PBS three times to get rid of any unbound biotinylated antibody conjugation. Cells were then treated with streptavidin conjugated fluorophore (purchased from Sigma, St. Louis, MO) bearing concentration of 5 μg/mL for 30 min in dark and at room temperature. After which, cells were washed with 1× PBS and 100 μL of 1× PBS was then added to each well. The cells were immediately monitored for plectin-1 determined by the fluorophore expression by employing SpectraMax i3X (purchased from Molecular Devices, San Jose, CA).
Experimental Design with Atomic Force Microscopy (AFM).
For all AFM experiments, Dimension Fastscan with ScanAsyst and ICON head (purchased from Bruker Corp, Santa Barbara, CA) was employed. To study the cellular nanomechanical properties, we used MLCT-Bio E probe consisting of triangular tip geometry. The probe characteristics such as nominal spring constant (k) and the tip radius values were 0.1 N/m and 20 nm, respectively with the 38 kHz resonant frequency made it suitable for soft cell samples. Before experiment, the laser was aligned on the tip to accurately sense deflections in the cantilever responsible for force–separation curve (F–S). Further, the tip was calibrated in fluid surrounding to account for changes in probe parameters due to hydrodynamic resistance by ramping on a hard surface (plain 60 mm culture dish containing Milli-Q water). The calibrated values for the probe were k = 0.112 N/m with deflection sensitivity = 40.3 nm/V and peak force tapping amplitude sensitivity of 773.4 nm/V at 1 kHz tapping frequency. Also, the peak force amplitude was set to 300 nm for accurate tip–sample interaction. Since, AFM is a surface mapping tool and it is well established that cell surface is highly heterogeneous due to the presence of actin fibers, microtubules and lipid layer,66,67 we confined our study to a 500 × 50 nm2 region over the nuclear membrane region to obtain homogeneity in experimental values; a technique well established from our previous work.36 Post various treatments, cells were identified using an optical microscope attached to the ICON head. Nanoindentation technique involves AFM tip approaching the cell surface followed by indenting into the cell surface at the user specified force levels and finally, retracting from the cell surface. The motion comprising of approach, indentation and retract yields an F–S curve. This technique solely relies on deflection in the cantilever as a result of various forces participating during the tip–sample interaction.68 Ramping parameters involved in nanoindentation technique were optimized as per our previous studies.36 For this study, ramp size was 4 μm, ramp rate was 1 Hz with 2048 samples/ramp and applied trigger threshold was 50 pN. For each treatment and each dynamic time point, atleast 14 cells were used with each cell indented no less than 10 times over a 500 × 50 nm2 region yielding at the minimum 140 F–S curve. All experiments were performed at 37 °C, maintained using a heated stage monitored closely by a temperature controller (Lake Shore Cryotronics, Inc., Westerville, OH).
Data Analysis of AFM Experiments.
Each individual ramp resulted in an F–S curve from which, several nanomechanical properties such as stiffness, deformation, and adhesion were extracted. However, these F–S curves had to be initially preprocessed followed by analyzing them using Bruker’s Nanoscope analysis v1.9 software. Briefly, each F–S curve was subjected to boxcar filter to reduce the overall noise followed by baseline correction. As the indentation in the cell membrane was comparable to the tip dimension, Hertzian contact model69 was employed by fitting the retrace curve to yield cell membrane stiffness.
Where, F = applied force, E = Young’s modulus (stiffness parameter), ϑ = Poisson’s ratio (material property, for cell = 0.3),70 R = tip radius, δ = indentation of tip into the cell surface.
Other nanomechanical properties such as deformation and adhesion were also obtained from the retrace curve for an acquired F–S curve.
Statistical Significance.
Origin Pro Lab software was used to calculate statistical significance and also to plot the data. First, each data set was first examined for normal distribution criteria and then One-way ANOVA test was applied to measure the significance. Statistical differences were determined to be significant at P < 0.05.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported partly by National Institutes of Health grants CA78383, CA150190, NHLBI (#HL140411) and Florida Department of Health (Cancer Research Chair Fund, Florida #3J) to DM and Mayo Clinic Pancreatic Cancer SPORE Career Enhancement Award, Eagles fifth District Cancer Telethon-Cancer Research Fund and Jay and Deanie Stein Career Development Award for Cancer Research at Mayo Clinic Jacksonville to SB.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsabm.0c01443.
PEGylation of PTP-GNP synthesis, results on effect of PTP-GNP protein corona formation on receptor dependent endocytosis process, and supplementary figures describing streptavidin–biotin reaction schematic, dynamic recovery of Panc1 cell membrane’s deformation upon exposure to PTP-GNP for various incubation durations and effect of PTP-GNP protein corona formation on receptor mediated endocytosis process(PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsabm.0c01443
The authors declare no competing financial interest.
REFERENCES
- (1).Petros RA; DeSimone JM Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discovery 2010, 9, 615–627. [DOI] [PubMed] [Google Scholar]
- (2).Zhang L; Gu F; Chan J; Wang A; Langer R; Farokhzad O Nanoparticles in medicine: therapeutic applications and developments. Clin. Pharmacol. Ther 2008, 83, 761–769. [DOI] [PubMed] [Google Scholar]
- (3).Nazir S; Hussain T; Ayub A; Rashid U; MacRobert AJ Nanomaterials in combating cancer: therapeutic applications and developments. Nanomedicine 2014, 10, 19–34. [DOI] [PubMed] [Google Scholar]
- (4).Bertrand N; Wu J; Xu X; Kamaly N; Farokhzad OC Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Delivery Rev 2014, 66, 2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ravindran A; Chandran P; Khan SS Biofunctionalized silver nanoparticles: advances and prospects. Colloids Surf., B 2013, 105, 342–352. [DOI] [PubMed] [Google Scholar]
- (6).Bhattacharyya S; Kudgus RA; Bhattacharya R; Mukherjee P Inorganic nanoparticles in cancer therapy. Pharm. Res 2011, 28, 237–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pal K; Al-Suraih F; Gonzalez-Rodriguez R; Dutta SK; Wang E; Kwak HS; Caulfield TR; Coffer JL; Bhattacharya S Multifaceted peptide assisted one-pot synthesis of gold nanoparticles for plectin-1 targeted gemcitabine delivery in pancreatic cancer. Nanoscale 2017, 9, 15622–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).De Jong WH; Borm PJ Drug delivery and nanoparticles: applications and hazards. Int. J. Nanomed 2008, 3, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Patra CR; Bhattacharya R; Mukhopadhyay D; Mukherjee P Fabrication of gold nanoparticles for targeted therapy in pancreatic cancer. Adv. Drug Delivery Rev 2010, 62, 346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kumar A; Zhang X; Liang X-J Gold nanoparticles: emerging paradigm for targeted drug delivery system. Biotechnol. Adv 2013, 31, 593–606. [DOI] [PubMed] [Google Scholar]
- (11).Liu R; Li X; Xiao W; Lam KS Tumor-targeting peptides from combinatorial libraries. Adv. Drug Delivery Rev 2017, 110, 13–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yoshida A; Sakai N; Uekusa Y; Imaoka Y; Itagaki Y; Suzuki Y; Yoshimura SH Morphological changes of plasma membrane and protein assembly during clathrin-mediated endocytosis. PLoS Biol. 2018, 16, No. e2004786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yang P-H; Sun X; Chiu J-F; Sun H; He Q-Y Transferrin-mediated gold nanoparticle cellular uptake. Bioconjugate Chem. 2005, 16, 494–496. [DOI] [PubMed] [Google Scholar]
- (14).Yoshida Y; Meerbeek BV; Snauwaert J; Hellemans L; Lambrechts P; Vanherle G; Wakasa K; Pashley DH A novel approach to AFM characterization of adhesive tooth–biomaterial interfaces. J. Biomed. Mater. Res 1999, 47, 85–90. [DOI] [PubMed] [Google Scholar]
- (15).Cartagena-Rivera AX; Wang W-H; Geahlen RL; Raman A Fast, multi-frequency, and quantitative nanomechanical mapping of live cells using the atomic force microscope. Sci. Rep 2015, 5, 11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Müller DJ; Dufrêne YF Force nanoscopy of living cells. Curr. Biol 2011, 21, R212–R216. [DOI] [PubMed] [Google Scholar]
- (17).Goksu EI; Vanegas JM; Blanchette CD; Lin W-C; Longo ML AFM for structure and dynamics of biomembranes. Biochim. Biophys. Acta, Biomembr 2009, 1788, 254–266. [DOI] [PubMed] [Google Scholar]
- (18).Lecuit T; Lenne P-F Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol 2007, 8, 633–644. [DOI] [PubMed] [Google Scholar]
- (19).Blanchoin L; Boujemaa-Paterski R; Sykes C; Plastino J Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev 2014, 94, 235–263. [DOI] [PubMed] [Google Scholar]
- (20).Zhang X; Ren J; Wang J; Li S; Zou Q; Gao N Receptor-mediated endocytosis generates nanomechanical force reflective of ligand identity and cellular property. J. Cell. Physiol 2018, 233, 5908–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shan Y; Ma S; Nie L; Shang X; Hao X; Tang Z; Wang H Size-dependent endocytosis of single gold nanoparticles. Chem. Commun 2011, 47, 8091–8093. [DOI] [PubMed] [Google Scholar]
- (22).Ding B; Tian Y; Pan Y; Shan Y; Cai M; Xu H; Sun Y; Wang H Recording the dynamic endocytosis of single gold nanoparticles by AFM-based force tracing. Nanoscale 2015, 7, 7545–7549. [DOI] [PubMed] [Google Scholar]
- (23).Reipert S; Steinböck F; Fischer I; Bittner RE; Zeöld A; Wiche G Association of mitochondria with plectin and desmin intermediate filaments in striated muscle. Exp. Cell Res 1999, 252, 479–491. [DOI] [PubMed] [Google Scholar]
- (24).Shin SJ; Smith JA; Rezniczek GA; Pan S; Chen R; Brentnall TA; Wiche G; Kelly KA Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 19414–19419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bausch D; Thomas S; Mino-Kenudson M; Fernández-Del CC; Bauer TW; Williams M; Warshaw AL; Thayer SP; Kelly KA Plectin-1 as a novel biomarker for pancreatic cancer. Clin. Cancer Res 2011, 17, 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Fuchs P; Zörer M; Rezniczek GA; Spazierer D; Oehler S; Castañón MJ; Hauptmann R; Wiche G Unusual 5′ transcript complexity of plectin isoforms: novel tissue-specific exons modulate actin binding activity. Hum. Mol. Genet 1999, 8, 2461–2472. [DOI] [PubMed] [Google Scholar]
- (27).Abrahamsberg C; Fuchs P; Osmanagic-Myers S; Fischer I; Propst F; Elbe-Bürger A; Wiche G Targeted ablation of plectin isoform 1 uncovers role of cytolinker proteins in leukocyte recruitment. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 18449–18454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Shaiken TE; Opekun AR Dissecting the cell to nucleus, perinucleus and cytosol. Sci. Rep 2014, 4, 4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kelly KA; Bardeesy N; Anbazhagan R; Gurumurthy S; Berger J; Alencar H; DePinho RA; Mahmood U; Weissleder R Targeted nanoparticles for imaging incipient pancreatic ductal adenocarcinoma. PLoS medicine 2008, 5, No. e.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang Q; Yan H; Jin Y; Wang Z; Huang W; Qiu J; Kang F; Wang K; Zhao X; Tian J A novel plectin/integrin-targeted bispecific molecular probe for magnetic resonance/near-infrared imaging of pancreatic cancer. Biomaterials 2018, 183, 173–184. [DOI] [PubMed] [Google Scholar]
- (31).Dong W; Cai Z; Pang J; Wang J; Tang N; Zhang W; Wang F; Xie Z; Lin F; Chang X Radiotherapy Enhancement for Human Pancreatic Carcinoma Using a Peptide-Gold Nanoparticle Hybrid. J. Biomed. Nanotechnol 2020, 16, 352–363. [DOI] [PubMed] [Google Scholar]
- (32).Kelly K, Weissleder R, Bardeesy N 2019, Google Patents.
- (33).Leung K 111In-Tetrameric Plectin-1 Targeting Peptide (4 (βAKTLLPTP-GGS (PEG5000)) KKK-111In-DOTA-βA-NH2) 2004, 1–4. [PubMed] [Google Scholar]
- (34).Konkalmatt PR; Deng D; Thomas S; Wu MT; Logsdon CD; French BA; Kelly KA Plectin-1 targeted AAV vector for the molecular imaging of pancreatic cancer. Front. Oncol 2013, 3, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sanna V; Nurra S; Pala N; Marceddu S; Pathania D; Neamati N; Sechi M Targeted nanoparticles for the delivery of novel bioactive molecules to pancreatic cancer cells. J. Med. Chem 2016, 59, 5209–5220. [DOI] [PubMed] [Google Scholar]
- (36).Kulkarni T; Angom RS; Das P; Bhattacharya S; Mukhopadhyay D Nanomechanical insights: Amyloid beta oligomer-induced senescent brain endothelial cells. Biochim. Biophys. Acta, Biomembr 2019, 1861, 183061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sau S; Agarwalla P; Mukherjee S; Bag I; Sreedhar B; Pal-Bhadra M; Patra CR; Banerjee R Cancer cell-selective promoter recognition accompanies antitumor effect by glucocorticoid receptor-targeted gold nanoparticle. Nanoscale 2014, 6, 6745–6754. [DOI] [PubMed] [Google Scholar]
- (38).Alkilany AM; Lohse SE; Murphy CJ The gold standard: gold nanoparticle libraries to understand the nano–bio interface. Acc. Chem. Res 2013, 46, 650–661. [DOI] [PubMed] [Google Scholar]
- (39).Walkey CD; Olsen JB; Song F; Liu R; Guo H; Olsen DWH; Cohen Y; Emili A; Chan WC Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 2014, 8, 2439–2455. [DOI] [PubMed] [Google Scholar]
- (40).Robinson MS; Watts C; Zerial M Membrane dynamics in endocytosis. Cell 1996, 84, 13–21. [DOI] [PubMed] [Google Scholar]
- (41).Kirkham M; Fujita A; Chadda R; Nixon SJ; Kurzchalia TV; Sharma DK; Pagano RE; Hancock JF; Mayor S; Parton RG Ultrastructural identification of uncoated caveolin-independent early endocytic vehicles. J. Cell Biol 2005, 168, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mayor S; Parton RG; Donaldson JG Clathrin-independent pathways of endocytosis. Cold Spring Harbor Perspect. Biol 2014, 6, No. a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Albanese A; Tang PS; Chan WC The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng 2012, 14, 1–16. [DOI] [PubMed] [Google Scholar]
- (44).Hillaireau H; Couvreur P Nanocarriers’ entry into the cell: relevance to drug delivery. Cell. Mol. Life Sci 2009, 66, 2873–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Panariti A; Miserocchi G; Rivolta I The effect of nanoparticle uptake on cellular behavior: disrupting or enabling functions? Nanotechnol., Sci. Appl 2012, 5, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Rejman J; Oberle V; Zuhorn IS; Hoekstra D Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J 2004, 377, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Safavi-Sohi R; Maghari S; Raoufi M; Jalali SA; Hajipour MJ; Ghassempour A; Mahmoudi M Bypassing protein corona issue on active targeting: zwitterionic coatings dictate specific interactions of targeting moieties and cell receptors. ACS Appl. Mater. Interfaces 2016, 8, 22808–22818. [DOI] [PubMed] [Google Scholar]
- (48).Verma A; Stellacci F Effect of surface properties on nanoparticle–cell interactions. Small 2010, 6, 12–21. [DOI] [PubMed] [Google Scholar]
- (49).Cho EC; Xie J; Wurm PA; Xia Y Understanding the role of surface charges in cellular adsorption versus internalization by selectively removing gold nanoparticles on the cell surface with a I2/KI etchant. Nano Lett. 2009, 9, 1080–1084. [DOI] [PubMed] [Google Scholar]
- (50).Anselmo AC; Zhang M; Kumar S; Vogus DR; Menegatti S; Helgeson ME; Mitragotri S Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano 2015, 9, 3169–3177. [DOI] [PubMed] [Google Scholar]
- (51).Yi X; Gao H Kinetics of receptor-mediated endocytosis of elastic nanoparticles. Nanoscale 2017, 9, 454–463. [DOI] [PubMed] [Google Scholar]
- (52).Guo P; Liu D; Subramanyam K; Wang B; Yang J; Huang J; Auguste DT; Moses MA Nanoparticle elasticity directs tumor uptake. Nat. Commun 2018, 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hui Y; Yi X; Wibowo D; Yang G; Middelberg AP; Gao H; Zhao C-X Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Science Advances 2020, 6, No. eaaz4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Myerson JW; Braender B; Mcpherson O; Glassman PM; Kiseleva RY; Shuvaev VV; Marcos-Contreras O; Grady ME; Lee HS; Greineder CF Flexible nanoparticles reach sterically obscured endothelial targets inaccessible to rigid nanoparticles. Adv. Mater 2018, 30, 1802373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kurland NE; Drira Z; Yadavalli VK Measurement of nanomechanical properties of biomolecules using atomic force microscopy. Micron 2012, 43, 116–128. [DOI] [PubMed] [Google Scholar]
- (56).Puech P-H; Poole K; Knebel D; Muller DJ A new technical approach to quantify cell–cell adhesion forces by AFM. Ultramicroscopy 2006, 106, 637–644. [DOI] [PubMed] [Google Scholar]
- (57).Siamantouras E; Hills CE; Liu K-K; Squires PE Examining Cell-Cell Interactions in the Kidney Using AFM Single-Cell Force Spectroscopy. In Diabetic Nephropathy; Springer, 2020; pp 189–201. [DOI] [PubMed] [Google Scholar]
- (58).Mahaffy R; Shih C-K; MacKintosh F; Käs J Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys. Rev. Lett 2000, 85, 880. [DOI] [PubMed] [Google Scholar]
- (59).Moeendarbary E; Valon L; Fritzsche M; Harris AR; Moulding DA; Thrasher AJ; Stride E; Mahadevan L; Charras GT The cytoplasm of living cells behaves as a poroelastic material. Nat. Mater 2013, 12, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Chen J Nanobiomechanics of living cells: a review. Interface Focus 2014, 4, 20130055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Del Pino P; Pelaz B; Zhang Q; Maffre P; Nienhaus GU; Parak WJ Protein corona formation around nanoparticles–from the past to the future. Mater. Horiz 2014, 1, 301–313. [Google Scholar]
- (62).Lundqvist M; Stigler J; Cedervall T; Berggård T; Flanagan MB; Lynch I; Elia G; Dawson K The evolution of the protein corona around nanoparticles: a test study. ACS Nano 2011, 5, 7503–7509. [DOI] [PubMed] [Google Scholar]
- (63).Lundqvist M Nanoparticles: tracking protein corona over time. Nat. Nanotechnol 2013, 8, 701–702. [DOI] [PubMed] [Google Scholar]
- (64).Ding L; Madamsetty VS; Kiers S; Alekhina O; Ugolkov A; Dube J; Zhang Y; Zhang J-S; Wang E; Dutta SK Glycogen Synthase Kinase-3 Inhibition Sensitizes Pancreatic Cancer Cells to Chemotherapy by Abrogating the TopBP1/ATR-Mediated DNA Damage Response. Clin. Cancer Res 2019, 25, 6452–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Hirsch JD; Eslamizar L; Filanoski BJ; Malekzadeh N; Haugland RP; Beechem JM; Haugland RP Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem 2002, 308, 343–357. [DOI] [PubMed] [Google Scholar]
- (66).Cho KA; Ryu SJ; Oh YS; Park JH; Lee JW; Kim H-P; Kim KT; Jang IS; Park SC Morphological adjustment of senescent cells by modulating caveolin-1 status. J. Biol. Chem 2004, 279, 42270–42278. [DOI] [PubMed] [Google Scholar]
- (67).Lingwood D; Kaiser H-J; Levental I; Simons K Portland Press Ltd, 2009.
- (68).Carvalho FA; Santos NC Atomic force microscopy-based force spectroscopy—biological and biomedical applications. IUBMB Life 2012, 64, 465–472. [DOI] [PubMed] [Google Scholar]
- (69).Hertz H (1882), MacMillan, New York. [Google Scholar]
- (70).Ebenstein DM; Coughlin D; Chapman J; Li C; Pruitt LA Nanomechanical properties of calcification, fibrous tissue, and hematoma from atherosclerotic plaques. J. Biomed. Mater. Res., Part A 2009, 91, 1028–1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


