Abstract
The emergence of drug resistance against the known hookworm drugs namely albendazole and mebendazole and their reduced efficacies necessitate the need for new drugs. Chemically diverse natural products present plausible templates to augment hookworm drug discovery. The present work utilized pharmacoinformatics techniques to predict African natural compounds ZINC95486082, ZINC95486052 and euphohelionon as potential inhibitory molecules of the hookworm Necator americanus β tubulin gene. A library of 3390 compounds was screened against a homology-modelled structure of β tubulin. The docking results obtained from AutoDock Vina was validated with an acceptable area under the curve (AUC) of 0.714 computed from the receiver operating characteristic (ROC) curve. The three selected compounds had favourable binding affinities and were predicted to form no interactions with the resistance-associated mutations Phe167, Glu198 and Phe200. The compounds were predicted as anthelmintics using a Bayesian-based technique and were pharmacologically profiled to be druglike. Further molecular dynamics simulations and MM-PBSA calculations showed the compounds as promising anthelmintic drug leads. Novel critical residues comprising Leu246, Asn247 and Asn256 were also predicted for binding. Euphohelionon was selected as a template for the de novo fragment-based design of five compounds labelled A1, A2, A3, A4 and A5; with four of them having SAscore values below 6, denoting easy synthesis. All the five de novo molecules docked firmly in the binding pocket of the β tubulin with no binding interactions with the three known resistance mutation residues. Binding energies of −8.2, −7.6, −7.3, −7.2 and −6.8 kcal/mol were obtained for A1, A2, A3, A4 and A5, respectively. The identified compounds can serve as treasure troves from which future potent anthelmintics can be designed. The current study strives to assuage the hookworm disease burden, especially making available molecules with the potential to circumvent the chemoresistance.
Keywords: Necator americanus, Anthelmintics, Natural products, Pharmacoinformatics, Molecular modelling, Drug discovery
1. Introduction
Hookworm infection remains a significant health burden globally. The disease is caused by the hookworm, an intestinal parasitic worm, that infects over 600 million persons especially in resource-limited countries and results in 135,000 deaths annually [1,2]. The hookworm infection belongs to a larger group of diseases called Neglected Tropical Diseases (NTDs) known to cause debilitating effects. Some NTDs are caused by parasitic worms including schistosomes, filarial and guinea worms [3]. Areas largely affected by hookworm include countries in sub-Saharan Africa, South Asia, Latin America, the Caribbean, the Middle East, and North Africa [4]. The human hookworm infection is primarily caused by Ancylostoma duodenale and Necator americanus [5] with the latter accounting for more than 85% of all hookworm infections [5,6]. The infection is known to affect growth as well as limit memory and cognition in children [7].
The β (beta) tubulin gene of the hookworm is critical for intracellular processes, cellular division, overall mobility, entry and migration of the parasite in the host cell. It is a subunit of the microtubule which, plays a crucial role in cell division and maintenance of the cytoskeleton [8]. It binds to two molecules of guanosine-5- triphosphate (GTP), at the positive end of microtubules [9]. Beta tubulin has so far been extensively exploited as a crucial target for anthelmintic and as a target for several other compounds [8]. Treatment using benzimidazoles including albendazole and mebendazole selectively target the beta-tubulin gene isotype, disrupting the polymerization of microtubules. Benzimidazoles confer their anthelminthic effect on susceptible nematodes by binding to their beta-tubulin gene resulting in the prevention of microtubule polymerization causing the destabilization of the intracellular processes and cellular division within the parasite and an overall immobility effect [10]. Community-based treatment methods normally employ a mass drug administration (MDA) strategy involving a combination regimen of albendazole, Ivermectin (IVM), and praziquantel (PZQ) [11–14]. This control strategy is, however, considered a high-risk approach in both humans and animals since it rather induces resistance [11]. For instance, there are reported cases of drug resistance in livestock in parts of Mali, Australia, South Africa and Paraguay [11]. Drawing lessons from this, it is therefore recommended a careful use of human anthelminthic in a way that will allow the prolonged use of these drugs and at the same time manage the problem of drug resistance and treatment failures.
Genetic mutations in the beta-tubulin gene are among the factors that have been reported to confer resistance in several parasitic nematodes including local strains of N. americanus [15,16]. These mutations occur at codons 167, 198 and 200 result in amino acid changes [15,17,18]. The changes lead to the substitution of phenylalanine with tyrosine at codons 167 and 200, and glutamate with alanine at amino acid position 198 [17,19,20]. Therefore, the binding and stabilization by alternative small molecule inhibitors with these residues Glu198, Phe167 and Phe200 at the active site of the beta-tubulin molecule is highly undesirable since it can lead to a decrease in binding affinity [21]. There is the need for molecules with different modes of binding.
Natural products (NPs) are structurally and chemically diverse compared to synthetic libraries [22]. Studies have shown that more than 50% of NPs pass the Lipinski rule of five for drug-likeness [23]. This makes natural products more absorbable than their synthetic counterparts do. Recent studies have reported a group of Dichapetalin compounds notably Dichapetalin A with promising anthelminthic activity against N. americanus [24].
In silico strategies for drug discovery presents an advantage in terms of time and cost for identifying novel chemotherapeutic agents by exploring a compendium of small compounds retrieved from freely available public databases. These methods have been used to identify some promising therapeutics against diseases [25,26]. Some of these databases include the African natural product database (AfroDB) and the North African Natural Product database (NANPDB) which contains a diverse and highly potent chemical landscape of natural compounds that could be explored for potential anthelminthic leads [27,28].
We sought to identify potential novel anthelminthic leads by performing virtual screening of natural products of African origin against a modelled 3D structure of beta-tubulin receptor of N. americanus. Also, we aimed at generating novel molecules using de novo fragment-based drug design. We characterized the mechanisms of binding for the resistance-associated mutations within the active site. In addition, pharmacological profiling, and relevant biological activity predictions were performed to determine the therapeutic utility of the potential lead molecules.
2. Methodology
A series of computational applications and techniques were employed in this study (Supplementary Table 1).
2.1. Template selection and homology modelling of N. americanus beta-tubulin gene
A search in Protein Data Bank (PDB; http://www.rcsb.org/) revealed no solved tertiary structure of N. americanus beta tubulins, hence the primary sequence of the protein (Gene ID: NECAME_11014, Accession number: W2T758) was retrieved from UniProt [29]. For template identification, Aguayo-Ortiz et al. employed the Iterative Threading ASSEmbly Refinement (I-TASSER) software tool to predict a suitable template based on a binding site containing residues associated with mutations [30]. Using a similar approach, our protein sequence comprising of 449 amino acids was submitted for template and binding site identification via the Iterative Threading ASSEmbly Refinement (I-TASSER) server [31], which predicted four plausible templates. The D chain of the subunit of the multimeric structure of tubulin tyrosine ligase (T2R-TTL) [PDB ID: 5C8Y], was selected for further studies due to the presence of amino acid residues associated with resistance as well as a high sequence identity to the template. The crystallographic structure of T2R-TTL (PDB ID: 5C8Y, resolution: 2.59 Å) was downloaded from PDB and used as a template in homology modelling of the beta-tubulin receptor using MODELLER v 9.17 [32]. The align2d function in MODELLER v9.17 was used to align the sequence of the target with the template and five candidate 3-D models generated using MODELLER v 9.17. The best model was then chosen based on the lowest value of the Discrete Optimized Potential Energy (DOPE) and high GA341 scores [32]. The selected best model and the template were structurally aligned using SuperPose [33].
2.2. Model refinement and assessment
The selected best model was refined to fix steric clashes and bumps using the WHAT IF server [34]. This is due to models having undesired bond lengths, bond angles, torsion angles and contacts. The refined structure was then energy minimized using GROMOS43B1 force field in Swiss-PdbViewer v 4.10 [35] to correct local bond and angle geometry and to relax the close contacts in the geometric chain. The WHAT IF program implements WHAT_CHECK to ascertain and fix steric clashes based on the overlap of two non-bonding atoms of distance cutoff set at 0.4 Å. The minimized model was visualized with PyMOL v 1.74 [36] and further optimized using molecular dynamics simulations. This was performed to evaluate the overall stability, folding, and obtain insight into the conformational changes as well as the dynamics of the refined model. The molecular dynamics (MD) simulation of the structure was performed with the Linux version of GROMACS v 5.1.4 [37] software package by utilizing the GROMOS 96_43a1 force field and the simple point charge (SPC) water model by passing the “-water spce” command. The modelled structure was first immersed in a periodic water box of cubic shape (1 nm thick). After solvating the receptor, the net charge on the protein was +8e. Genion command in GROMACS was used to add eight chloride (Cl-) counter ions to neutralize the net charge on the protein. Electrostatic energy was computed via the particle mesh Ewald method with a computational load of 0.19. The cutoff distance for the calculation of the Coulomb and van der Waals interaction was 1.0 Å with the cutoff scheme being Verlet for minimization of 50000 steps. The system was subjected to a two-step ensemble process (NVT and NPT) at a temperature of 300 K and pressure of 1 bar (P) for 2 ps. Linear Constraint Solver (LINCS) constraints were performed for all bonds, with position restraint on the protein and allowing only the water molecules to move to equilibrate with the protein structure. The final production run was performed for the minimization of 5 ns under the same equilibration conditions of 300 K and 1 bar. The results were analyzed using GROMACS v 5.1.4 and GRACE v 5.1.25 [38] which uses the command xmgrace in a Linux terminal. The last frame of the minimized protein in gro format was saved as a pdb format using Visual Molecular Dynamics (VMD) v1.9.3 [39]. The minimized model was then validated by generating a Ramachandran plot [40] using the PROCHECK v 3.5.4 [41]. Other programs such as ERRAT, VERIFY3D and Qmean [42,43] were also used to corroborate the PROCHECK results.
2.3. Prediction and analysis of binding site
The potential binding site of the beta-tubulin model containing the amino acid residues of interest was predicted using the Computed Atlas of Surface Topography of proteins (CASTp) [44] and further visualized in PyMOL [36].
2.4. Protein preparation for docking
AutoDockTools v 4.2.6 [45] was used in the preparation of the protein model which involved the assignment of Gasteiger partial charges [46]. All existing water or solvent molecules were removed to eliminate the influence of solvent interactions in the protein-ligand docking. The receptor file saved in Protein Data Bank partial charge and atom type (pdbqt) format was used as an input receptor file for AutoDock Vina.
2.5. Selection and preparation of ligands
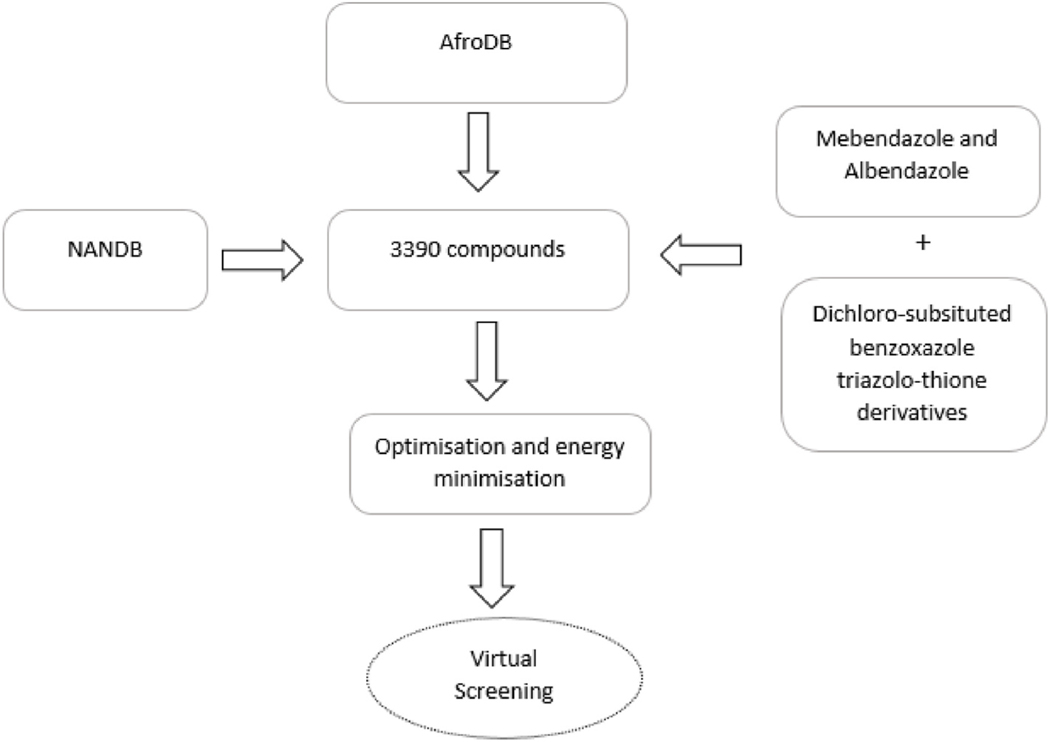
An integrated natural product library (Fig. 1) comprising 885 compounds from the African Natural Product Database (AfroDb) [28], a subset of ZINC [47] and 2500 compounds from the North African Natural Product Database (NANPDB) [27] databases were retrieved in Structural Data Format (sdf). The two libraries contain structurally diverse natural products of African origin [27,28]. Additionally included in the screening dataset were two known anthelmintics namely albendazole and mebendazole [48–50] as well as three potent dichloro-substituted benzoxazole-triazolo-thione derivatives namely 6, 8-dichloro [1,2,4]triazolo[3,4-b] [1,3]benzoxazole-3(2h)-thione (PubChem ID 53327690); 5,7-dichloro-1,3-Benzoxazole-2-thiol (PubChem IDs 723308); and 6,8-dichloro-2-{[(4nitrophenyl)amino]methyl} [1,2, 4]triazolo[3,4b] [1,3]benzoxazole-3(2h)-thione (PubChem ID 53327692) [51]. The structures of the controls are provided in the Supplementary Figure 1. The sdf files of the five compounds were retrieved via PubChem, a freely available chemical structure database [52]. The total set of 3390 compounds were further optimized and the energy was minimized using Open Babel 2.3.1 within the Pyrx 0.8 interface [53] with MMFF94 force field energy minimization. All ligand files were then converted to compatible pdbqt files using AutoDock Tools.
Fig. 1.
A flow chart for the preparation and selection of ligands for virtual screening. The library consisted of African natural products and five known hookworm beta-tubulin inhibitors.
2.6. Virtual screening
AutoDock Vina [54] tool in PyRx v 0.8 [55] was used to virtually screen a library of 3390 compounds against the binding pocket of the predicted model. The grid box with dimensions (22.5 Å × 22.5 Å x 22.5 Å) and centre (−18.35 Å, −8.23 Å, −22.48 Å) was set around the predicted active site residues of the receptor. A default exhaustiveness of 8 was used. After completion of the virtual screening process, the top hits, with the lowest binding energies (kcal/mol) relative to the known inhibitors, were selected for further visualization in PyMOL [36]. The top 30 hits with reasonably good binding energies and binding poses were selected for further evaluation.
2.7. Docking protocol validation
Five molecules comprising albendazole and mebendazole, as well as three potent dichloro-substituted benzoxazole-triazolo-thione derivatives with PubChem CIDs [52] 53327690, 723308 and 53327692 were used as actives in the validation of docking protocol. Albendazole and mebendazole are broad-spectrum anthelmintic drugs targeting beta-tubulin [48,50]. Moreover, the choice of including the three dichloro-substituted benzoxazole-triazolo-thione derivatives was based on their reported efficacious inhibitory activities against helminth beta tubulins [51]. The SMILES of the five molecules were used to obtain a dataset of decoys via the Directory of Useful Decoys- Enhanced (DUD-E) [56]. Fifty decoys were obtained for each ligand, then each set of decoys has similar physicochemical properties to the parent ligand but is dissimilar in topology [57]. Furthermore, the dataset of 255 compounds comprising of 250 decoys and 5 ligands were screened against the predicted active site of the modelled beta tubulin receptor using AutoDock Vina. The docking result containing the respective binding energies of the ligands and their decoys was used to compute the Area Under the Curve (AUC) of the Receiver Operating Curve (ROC) via easyROC v 1.3.1 [58]. The variables were set to “nonparametric” as the method for curve fitting, “DeLong (1988) [+]”as the method for standard error estimation and confidence interval, and a “Type I” error of 0.05.
2.8. Protein-ligand interaction profiling
Ligplot+ [59] was used to study the 2D protein-ligand interaction which includes the hydrogen bonding and the hydrophobic contacts. The best poses of the hits were saved in a ‘.pdb’ file format. The protein-ligand complex for these poses was visualized using PyMOL. The complexes were then loaded into the Ligplot+ workspace to generate the 2D schematic intermolecular interactions between the protein and the ligands.
2.9. Exploring the anthelmintic activity of the hits
Prediction of Activity Spectra for Substances (PASS) [60] which a is Bayesian-based machine learning technique was used to predict the anthelmintic activity of the top 30 hits. Anthelmintics are drugs for the treatment of helminth infections, including hookworm infection. A DrugBank and literature search for anthelmintic activity was also done to support the PASS predictions based on the structural information of the hits.
2.10. Drug likeness and pharmacological profiling
The SMILES of the hits were used to obtain the pharmacological profiles and physicochemical properties of the PASS predicted anthelmintic hits via SwissADME [61]. Parameters considered include molecular weight, number of hydrogen bond acceptors and donors for evaluating drug-likeness and gastrointestinal absorption, P-gp substrates, and cytochrome P450 inhibition for evaluating the pharmacokinetics.
2.11. Toxicity prediction analysis
The toxicity profiles of the PASS anthelmintic predicted compounds were evaluated and analyzed using the OSIRIS property explorer in DataWarrior v 4.5.2 [62]. OSIRIS explorer uses a precomputed set of known toxic compounds and fragments to predict relevant properties comprising tumorigenicity, mutagenicity reproductive effect and irritancy [62]. ADVERPred [63] was also used to predict the nephrotoxicity and hepatotoxicity of the compounds.
2.12. Molecular dynamics of protein-ligand complexes and MM-PBSA analyses
The MD simulations of the protein-ligand complex were performed in GROMACS v 2018 [64]. The protein topology of the modelled beta-tubulin was generated using the GROMOS 96_43a1 force field whereas the ligand topologies were generated via PRODRG [65], with defined settings (Chirality: Yes, Charges: Full, and EM: No). A protein-ligand complex was then constructed. The complex was solvated with SPC water in a cubic box of size 1.0 nm and neutralized with Na and Cl ions. Energy minimization of the complex was conducted for 50,000 steps using the steepest descent algorithm. The ligands were restrained before the NVT and NPT ensemble simulations. Equilibration simulation was run for 100 ps for each ensemble. A production MD of 100 ns simulation was performed on the complexes. The g_mmpbsa [66] was used to compute the free binding energies of the complexes over the 100 ns simulation using the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method. R programming package was used to generate graphs for the MM-PBSA simulation.
2.13. De novo design of inhibitors
A potential lead complex was used as input for the de novo design of novel ligands via e-LEA3D [67]. A binding site radius of 10 Å and a weight in a final score of 1 were selected in setting up the docking function. The coordinates of the active site as used in molecular docking studies was considered. The maximum number of conformers, number of generations and population size were set at 1, 30 and 30, respectively.
3. Results and discussion
3.1. Template identification, homology modelling of proteins, molecular dynamic simulations and validation
Aguayo-Ortiz et al. used the crystal structure of Ovis aries beta-tubulin as a template to generate the homology model of the beta-tubulin of the nematode Trichinella spiralis and to predict a binding site that contained conserved residues [30]. Similarly, using I-TASSER [31,68], the D chain of the crystal structure of tubulin tyrosine ligase (T2R-TTL) with PDB ID 5C8Y was chosen as the best template for homology modelling based on the presence of conserved amino acid residues Phe167, Glu198 and Phe200, within the binding site. A sequence alignment with the template also showed that the beta-tubulin is homologous to the subunit of the T2R-TTL with 88% sequence identity and 94.3% sequence similarity (Supplementary Figure 2). In addition, the results of sequence alignment revealed that the predicted active site contained residues Phe167 and Glu198. Model 5 was selected as the best structural model of the beta-tubulin of N. americanus (UniProt ID W2T75) based on the lowest discrete optimized potential energy (DOPE) score of −53055.30859 obtained (Table 1). The DOPE score and GA341 are methods used for evaluating the accuracy and reliability of modelled protein structures. DOPE is an atomic distance-dependent statistical method that is useful in evaluating the energies of generated models [69]. Models with the lowest DOPE score and highest GA341 score are considered reasonably better. The resulting protein model is a monomer, folded into a β domain that consists of 11-stranded β-sheets, and 11 α-helices (Fig. 2A and B). Structural alignment of the 3D model and template using SuperPose [33] gave a root mean square deviation (RMSD) of 1.32 Å. The RMSD value obtained indicates an acceptable difference between the protein model and the template [33]. In addition, the Modeller objective function (molpdf) was used to measure the sum of all the restrains.
Table 1.
Comparison of DOPE, GA341 and molpdf scores of the five models predicted using MODELLER with the lowest discrete optimized protein energy (DOPE) score corresponded to Model 5.
| Models | Molpdf score | DOPE score | GA341 score |
|---|---|---|---|
|
| |||
| Model 1 | 2642.68408 | −52703.71875. | 1.00 |
| Model 2 | 2614.13916 | −52903.56250. | 1.00 |
| Model 3 | 2478.50610 | −52848.83594. | 1.00 |
| Model 4 | 2537.23853 | −52964.20312. | 1.00 |
| Model 5 | 2550.17090 | −53055.30859. | 1.00 |
Fig. 2.
(A) 3D model of the beta-tubulin of N. americanus (UniProt ID, W2T75). The helices are shown in red, beta sheets in yellow and loops in green. (B) Structural alignment between the protein model and template generated using SuperPose and visualized in PyMOL. The model is shown in purple and the template in yellow. The superposition of the two structures had an acceptable RMSD of 1.32 Å in SuperPose. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The overall structure of the model is similar to the template protein as expected (Fig. 2B). Additionally, the structural alignment revealed conserved key amino acid residues at positions 167 and 198 but only a residue difference at position 200 where phenylalanine (aromatic non- polar residue) is present in the model structure but a tyrosine (aromatic polar residue) in the template; these two residues differ by only a hydroxyl group. Further, there were binding site residue differences at positions 165 and 166, where two serine residues (polar amino acid) were present in the model while asparagine and threonine (also polar amino acids) occupied those respective positions in the template. In addition, there were amino acid differences at positions 317 and 370, where methionine and valine (hydrophobic amino acid residues) were present in the model and isoleucine (also hydrophobic amino acid) at the respective positions in the template.
3.2. Model refinement and assessment
Protein model refinement and energy minimization were performed on our best model using Swiss-PdbViewer [70] and GROMACS [64,71], respectively. Analysis of the GROMACS generated trajectories of the refined protein model indicated that the RMSD increased from the beginning but after a period of 0.5 ns, it remained relatively stable for the rest of the duration of the simulation (Fig. 3). This suggests that the model had very low RMSD for the backbone with less Root Mean Square (RMS) fluctuations and flexibility, and thus indicating that the model was well-conditioned.
Fig. 3.
RMSD plot of the molecular dynamics simulation using GROMACS. A plot of RMSD in nanometres (nm) against time in nanoseconds (ns). The RMSD increased from 0 ns to 0.5 ns and levelled off with slight fluctuations towards the end of 1 ns.
The quality of the optimized structure was finally evaluated by generating a Ramachandran plot using PROCHECK [41]. Ramachandran plots highlight the most favoured, allowed, generously allowed and disallowed regions of the modelled protein structure. Ideally, a model of reasonably high quality should have at least 90% residues in the core regions [40]. The Ramachandran plot for the predicted model revealed that 92.3% of residues were within the most favourable region, whilst 7.7% were in the allowed region (Fig. 4), supportive that the predicted model was of reasonably high quality. In addition, the overall quality factor predicted by the ERRAT for the model was 89.327 (Supplementary Figure 3) which corroborates the quality of the protein model structure. ERRAT [72] provides the overall quality factor for non-bonded atomic interactions and the generally accepted value should be greater than 50 for a high-quality model. When the model was further validated using VERIFY 3D [42], 88.77% of the residues were predicted as having an average 3D-1D score greater than 0.2.
Fig. 4.
Ramachandran plot of beta-tubulin model from N. americanus obtained by PROCHECK: 92.3% residues in favourable regions (A, B, L); 7.7% residues in the additional allowed region (a, b, l, p); 0.0% residues in generously allowed regions (-a,-b,-p,-l); and 0% residues in disallowed regions.
3.3. Prediction and analysis of binding site
The residues within the binding site of tubulins generally have a high sequence and structural conservation among helminth tubulins which are different from other families of tubulins. The predicted binding pocket included all residues whose mutation are linked to anthelminthic resistance (Supplementary Figure 4) and had a computed area of 401.920 Å2 and volume of 164.250 Å3. Forty-three (43) residues formed the putative binding pocket and these included conserved residues Phe167, Glu198 and Phe200 (Fig. 5, Supplementary Figure 4).
Fig. 5.
Predicted binding cavity of the modelled structure via CASTp. The predicted binding pocket is depicted as red. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Virtual screening
Molecular docking is a useful computational technique in informing the selection of lead compounds [73]. After screening an integrated library of 3390 against the active site, the top hits were selected for further analysis based on their binding energies. The docking poses of the top hits were further visualized using PyMOL. Hits that were not docked firmly in the active site pocket were ferreted out. The binding energies and docking poses were used as criteria to select 30 hits for further evaluation. These compounds had lower binding energies than the five known inhibitors. Table 2 shows the docking results of the top 30 hits ranked according to their binding energies. S,5Z,8Z,11Z,13E, 17Z)-15-hydroxy-1-(2,4,6- trihydroxyphenyl)-15- methylicosa-5,8,11, 13,17- pentaen-1-one had the lowest binding energy of −8.7 kcal/mol, suggesting a plausible strong molecular interaction. The binding energies of the top 30 hits ranged from −8.7 kcal/mol to −7.7 kcal/mol. Additionally, the screening results of the known hookworm beta-tubulin inhibitors namely albendazole and mebendazole and three other dichloro substituted benzoxazole-triazolo-thione derivatives were included (Table 2). Mebendazole had the lowest binding energy of −7.0 kcal/mol amongst the known inhibitors followed by the compound of PubChem ID 53327692 with a binding energy of −6.4 kcal/mol (Table 2). The docking results showed the top 30 hits having relatively lower binding energies than the known inhibitors.
Table 2.
Docking results for the top 30 hits and 5 beta-tubulin inhibitors. The table shows the IDs or common names, IUPAC names and their binding energies (kcal/mol) of the hits. The compounds are arranged in increasing order of binding energy. The lower the binding energy, the better the binding affinity. The IUPAC names were generated in MarvinSketch.
| No. | ZINC ID/Common name/PubChem ID | IUPAC Name | Binding affinity (kcal/ mol) |
|---|---|---|---|
|
| |||
| S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6- trihydroxyphenyl)-15- methylicosa-5,8,11,13,17- pentaen-1-one | (5Z,8Z,11Z,13E,15S,17Z)-15-hydroxy-15-methyl-1-(2,4,6-trihydroxyphenyl)icosa-5,8,11,13,17- pentaen-1-one | −8.7 | |
| ZINC14760755 | 3-{[(2E)-3,7-dimethylocta-2,6-dien-1-yl]oxy}−1,8-dihydroxy-6-methyl-9,10-dihydroanthracen-9- one | −8.6 | |
| ZINC95485927 | (3S,4aR,6aR,6bS,8aR,12aS,14aR,14bR)- 4,4,6a,6b,8a,11,11,14b-octamethyl- 1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,14,14a,14b- icosahydropicen-3-yl (2E)-3-(4-hydroxy- 3- methoxyphenyl)prop-2-enoate |
−8.5 | |
| ZINC95486082 | (2S)-2-[2,2-dimethyl-8-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H-1-benzopyran-6-yl]-7-hydroxy- 3,4-dihydro-2H-1-benzopyran-4-one | −8.5 | |
| ZINC95486263 | 2-{4-[5-(5,7-dihydroxy-4-oxo-4H-chromen-2-yl)-2-hydroxyphenoxy]-3-hydroxyphenyl}−5,7- dihydroxy-4H-chromen-4-one | −8.5 | |
| Campesterol | (1R,3aS,3bR,9aS,9bS,11aR)-1-[(2R,5R)-5,6-dimethylheptan-2-yl]-9a,11a-dimethyl-1H,2H,3H,3aH,3bH,4H,5H,5aH,6H,9H,9bH,10H,11H-cyclopenta[a]phenanthren-7-ol | −8.4 | |
| ZINC14780716 | (2E)-1-[2,4-dihydroxy-5-(3-methylbut-2-en-1-yl)phenyl]-3-[4-hydroxy-3-(3-methylbut-2-en-1-yl) phenyl]prop-2-en-1-one | −8.3 | |
| ZINC95485922 | 2-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-1,3,5,8-tetrahydroxy-4-(3-methylbut-2-en-1-yl)-9H- xanthen-9-one | −8.3 | |
| ZINC95486052 | (2S)-2-[2,2-dimethyl-8-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H-1-benzopyran-6-yl]-5,7- dihydroxy-3,4-dihydro-2H-1-benzopyran-4-on | −8.3 | |
| ZINC95485928 | 7-[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]-6,8,12-trihydroxy-2,2-dimethyl-2,5-dihydro-1,10- dioxatetraphen-5-one | −8.2 | |
| Orthidine_A | N-[(2S,3S)-7-[(1E)-2-carbamimidamidoethenyl]-3-(3,4-dihydroxyphenyl)-2,3-dihydro-1,4- benzodioxin-2-yl]guanidin | −8.2 | |
| Robustaflavone | 6-[5-(5,7-dihydroxy-4-oxo-4H-chromen-2-yl)-2-hydroxyphenyl]-5,7-dihydroxy-2-(4- hydroxyphenyl)-4H-chromen-4-one | −8.2 | |
| Tetrahydrorobustaflavone | 6-[5-(5,7-dihydroxy-4-oxo-4H-chromen-2-yl)-2-hydroxyphenyl]-5,7-dihydroxy-2-(4- hydroxyphenyl)-4H-chromen-4-one | −8.2 | |
| ZINC13480348 | (2R)-7-{[(2E)-3,7-dimethylocta-2,6-dien-1-yl]oxy}- 5,10-dihydroxy-2-methyl-4-oxo-1,2,3,4- tetrahydroanthracen-2-yl acetate | −8.1 | |
| Siphonellinol_C | (3R,5aR,6R,7S,9aR)-6-{2-[(5S,6S)-5-hydroxy-6-[(2E)-4-hydroxy-4-methylpent-2-en-1-yl]-2,6- dimethylcyclohex-1-en-1-yl]ethyl}−2,2,5a,7-tetramethyl-hexahydro-3H-1-benzoxepine-3,7-diol | −8.1 | |
| ZINC28462577 | 2-{4-[5-(5,7-dihydroxy-4-oxo-4H-chromen-2-yl)-2-hydroxyphenoxy]phenyl}−5,7-dihydroxy-4H- chromen-4-one | −8.0 | |
| ZINC95486072 | (1R,4S,5R,8R,13R,14S,17S,18R)-4,5,9,9,13,20,20-heptamethyl-24 oxahexacyclo [15.5.2.01,18.04,17.05,14.08,13]tetracosane-10,22-dione |
−8.0 | |
| ZINC95486073 | (1R,4S,5R,8R,13R,14S,17S,18R)-4,5,9,9,13,20,20-heptamethyl-24 oxahexacyclo [15.5.2.01,18.04,17.05,14.08,13]tetracosan-22-one |
−8.0 | |
| ZINC95486081 | (2S)-7-hydroxy-2-[(2R,3S)-2-hydroxy-3-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H-1-benzopyran-6- yl]-3,4-dihydro-2H-1-benzopyran-4-one | −7.9 | |
| 6,10-dimethyl-9-methylene-2-(4-methyl-1,2- dioxabicyclo [2.2.2] oct-5-en-l-yl) undec-5-ene | 1-[(5E)-6,10-dimethyl-9-methylideneundec-5-en-2-yl]-4-methyl-2,3-dioxabicyclo[2.2.2]oct-5-ene | −7.9 | |
| Spinescen | (5S)-3-[(13S)-13-hydroxy-13-[(2S,2′S,5S,5′R)-5’-[(1S)-1,5,6-trihydroxyundecyl]-[2,2′-bioxolan]- 5-yl]tridecyl]-5-methyl-5H-furan-2-one | −7.9 | |
| Euphohelionon | (1R,3S,4S,5S,8R,10R,12R,13R,14S,15S)-8,15-bis(benzoyloxy)-3,7,11,11,14-pentamethyl-16- oxatetracyclo[11.2.1.01,5.010,12]hexadec-6-en-4-yl benzoate | −7.9 | |
| Anchinopeptolide_A | (2S)-2-{[(2S,3R,4S)-3-(2-carbamimidamidoethyl)-4-(3-carbamimidamidopropyl)-2,4-dihydroxy-1-[(1S)-1-{[(1E)-2-(4-hydroxyphenyl)ethenyl]carbamoyl}ethyl]-5-oxopyrrolidin-2-yl] formamido}-N-[(1E)-2-(4-hydroxyphenyl)ethenyl]propanamide | −7.9 | |
| ZINC33833639 | (4aS,6aS,6bR,8aR,10S,12aR,12bS,14bS)-10-hydroxy-4a-(hydroxymethyl)-2,2,6a,6b,9,9,12a- heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,12b,13,14b-icosahydropicen-5-one | −7.8 | |
| ZINC95485992 | (2E)-1-{2,4-dihydroxy-5-[(3S)-3-hydroxy-4-methylpent-4-en-1-yl]phenyl}−3-{4-hydroxy-3-[(3E)- 3-methylpent-3-en-1-yl]phenyl}prop-2-en-1-one | −7.8 | |
| ZINC95486074 | (1R,4S,5R,8R,10S,13R,14S,17S,18R,22S)-4,5,9,9,13,20,20-heptamethyl-24-oxahexacyclo [15.5.2.01,18.04,17.05,14.08,13]tetracosane-10,22-diol |
−7.8 | |
| ZINC95486075 | (3S,4aR,6aR,6bS,8R,8aS,12aS,14aS,14bR)-8a-(hydroxymethyl)-4,4,6a,6b,11,11,14b- heptamethyl-1,2,3,4,4a,5,6,6a,6b,7,8,8a,9,10,11,12,12a,14,14a,14b-icosahydropicene-3,8-diol | −7.8 | |
| ZINC13365959 | 3-[(1S)-1-(1H-indol-6-yl)-3-methylbut-2-en-1-yl]-6-(3-methylbut-2-en-1-yl)-1H-indole | −7.7 | |
| ZINC13485435 | (1R,10S)-6,13-bis(3-methylbut-2-en-1-yl)-8,17-dioxatetracyclo[8.7.0.02,7.011,16]heptadeca 2,4,6,11(16),12,14-hexaene-5,14-diol |
−7.7 | |
| ZINC15120680 | 2-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-1,3,5,8-tetrahydroxy-9H-xanthen-9-one | −7.7 | |
| Mebendazole | methyl N-(6-benzoyl-1H-1,3-benzodiazol-2- yl)carbamate | −7.0 | |
| PubChem CID 53327692 | 6,8-dichloro-2-{[(4nitrophenyl)amino]methyl} [1,2,4]triazolo[3,4b] [1,3]benzoxazole-3(2h)- thione | −6.4 | |
| Albendazole | methyl N-[6-(propylsulfanyl)-1H-1,3-benzodiazol-2- yl]carbamate | −5.6 | |
| PubChem CID 53327690 | 6,8-dichloro [1,2,4]triazolo[3,4-b] [1,3]benzoxazole-3(2h)-thione | −5.6 | |
| PubChem CID 723308 | 5,7-Dichloro-1,3-Benzoxazole-2-Thiol | −4.8 | |
3.5. Docking protocol validation
The docking protocol of AutoDock Vina [54] was validated using the ROC curve analysis. Five hookworm’s beta-tubulin inhibitory ligands comprising albendazole, mebendazole, and PubChem IDs 53327690, 723308 and 53327692 with their respective decoys were screened against the beta-tubulin model to generate the ROC curve. The ROC curve evaluates a docking model’s ability to discriminate between actives and decoys [74]. An area under the curve (AUC) value of 1 is considered a perfect classification, and below 0.5 is poor discrimination [75,76]. Accordingly, an AUC value of 0.7–0.8 is considered acceptable and an AUC value of 0.8–0.9 is interpreted as excellent. Moreover, an AUC value of above 0.9 is considered outstanding [75,77]. By screening 5 actives and 250 decoys against hookworm beta-tubulin via AutoDock Vina, an AUC value of 0.714 was computed from the ROC curve (Fig. 6). The computed AUC value is considered acceptable [75].
Fig. 6.
A ROC curve generated after screening 255 compounds consisting of five inhibitors and 250 decoys against the 3D model of the beta-tubulin of N. americanus. An acceptasble AUC of 0.714 was obtained.
To support the ROC evaluation of the AutoDock Vina, previously reported studies had employed AutoDock Vina to successfully screen compounds against helminth beta-tubulin receptors [51,78] The predicted compounds were experimented and confirmed as potent helminth inhibitors, suggesting that AutoDock Vina is an effective tool for deriving potential inhibitors for beta tubulins of N. americanus.
3.6. Characterization of the mechanism of binding
Hydrogen bonding and hydrophobic interactions are critical in the stabilization of a ligand in the binding pocket of a receptor [79]. The protein-ligand interactions depend on the structure and functional groups of the ligand [79]. LigPlot+ [59] was used to generate the 2-D schematic representation of the protein-ligand interactions. A total of 25 out of 30 hits formed hydrogen bonds of varying lengths with residues of the active site. This suggests stabilization of the ligands in the binding pockets. Orthidine A formed the highest number of five hydrogen bonds with the residues (Fig. 7A, Table 3), denoting the high stability of Orthidine A in the binding pocket [80]. ZINC2842577 and S, 5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6-trihydroxyphenyl)-15- methylicosa-5,8,11,13,17-pentaen-1-one formed four hydrogen bonds (Fig. 7B and C, Table 3). Moreover, three hydrogen bonding were observed anchinopeptolide A, ZINC14760755, and ZINC95486263 protein-ligand complexes (Fig. 7D, E and F, Table 3). The shortest hydrogen bond length of 2.64 Å was observed with robustaflavone. The shorter the hydrogen bond length, the stronger the bond [81]. Euphohelionon, anchinopeptolide A, ZINC95486082, and ZINC95486052 were the only compounds that did not interact with Phe167, Glu198 and Phe200 (Table 3). Euphohelionon formed one hydrogen bond of length 3.19 Å with Asn256 and hydrophobic intermolecular contacts with 10 residues namely Leu253, Ala248, Val255, Lys252, Ala352, Ala314, Lys350, Leu246, Gln245, and Thr351 (Table 3). ZINC95486082 formed eleven hydrophobic contacts and two hydrogen bonds (Table 3). ZINC95486052 interacted with the beta tubulin via hydrophobic interactions with residues namely Asn247, Thr312, Ala315, Val313, Leu253, Met257, Asn256, Lys350, Asn247, Lys252, Met316, Leu246, and Asn348. Table 3 shows the protein-ligand interactions of some 13 selected hits.
Fig. 7.
Binding mode evaluation of (A) Orthidine A, (B) S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6-trihydroxyphenyl)-15-methylicosa-5,8,11,13,17-pentaen-1-one, (C) ZINC28462577, (D) anchinopeptolide A, (E) ZINC14760755, and (F) ZINC95486263 protein-ligand complexes. The figure shows the ligand represented as sticks docked inside the binding pockets with a surface representation. The Ligplot+ representations of the protein-ligand complex interactions are also shown. The hydrophobic contacts are shown as red spoke arcs whereas the hydrogen bonds are represented as short dotted green lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Protein-ligand interactions of some selected hits generated with Ligplot+. The table shows the binding energies, interacting residues in active binding site number of hydrogen bonds, with their bond length and the number of hydrophobic contacts.
| ZINC ID/Compound Name | Binding energy (kcal/mol) | Interacting residues in the active site | Number of hydrogen bonds [amino acid residue (Å)] | Number of hydrophobic interactions | Total number of bonds |
|---|---|---|---|---|---|
|
| |||||
| Orthidine A | −8.2 | Asn256, Thr351, Asn348, Asn247, Ala248, Leu246, Lys350, Met257, Ala314, Ile345, Val313, Thr312 | 5 [Asn256: (3.02)] [Thr351: (3.13; 2.94)] [Asn348: (3.22)] [Asn247: (3.12)] |
7 | 12 |
| S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1- (2,4,6-trihydroxyphenyl)-15-methylicosa-5,8,11,13,17-pentaen-1-one | −8.7 | Thr237, Gln134, Val236, Phe167, Phe200, Glu198, Ser165, Leu253, Asn256, Thr312, Asn348, Val313, Lys350, Met257, Leu246, Ala314, Cys239, Ala352, Met316, Ala248, Leu240, Val236 | 4 [Val236: (2.87)] [Glu134: (2.70)] [Glu198: (2.99)] [Asn256: (3.21)] |
20 | 24 |
| ZINC28462577 | −8.0 | Lys350, Asn256, Gln134, Glu198, Val236, Leu240, Leu250, Thr237, Met257, Leu246, Ala248, Asn247, Leu253, Met316, Ala314, Ala315, Val368, Cys239 | 4 [Glu198: (3.05)] [Gln134: (2.75)] [Asn256: (3.31)] [Lys350: (3.32)] |
14 | 18 |
| Anchinopeptolide A | −7.9 | Val255, Lys252, Ala248, Leu253, Asn247, Ala314, Met316, Lys350, Leu246, Ala352, Gln245, Asn256, Thr351, Ala315 | 3 [Thr351: (3.10)] [Asn247: (3.17)] [Asn256: (3.15)] |
10 | 13 |
| ZINC14760755 | −8.6 | Ala248, Ala314, Met316, Met257, Leu246, Val368, Thr351, Ser165, Gln134, Phe200, Thr237, Leu240, Glu198, Cys239, Leu253, Lys350, Ala315 | 3 [Lys350: (2.77)] [Ala315: (3.01; 3.16)] |
14 | 17 |
| ZINC95486263 | −8.5 | Glu198, Gln134, Asn247, Ser165, Leu250, Met257, Leu240, Thr237, Leu253, Val236, Met316, Ala314, Val236, Val368, Cys239, Ala315, Leu246, Ala248, Lys252 | 3 [Glu198: (3.10)] [Gln134: (2.89)] [Asn247: (3.25)] |
16 | 19 |
| ZINC95485922 | −8.3 | Leu250, Val368, Phe200, Ser165, Glu198, Val236, Cys239, Asn247, Ala248, Leu246, Asn256, Lys350, Val313, Thr312, Ala314, Leu253, Met257, Asn348, Met257 | 3 [Asn247: (2.86)] [Lys350: (3.13)] [Asn256: (2.97)] |
15 | 18 |
| ZINC95486082 | −8.5 | Asn247, Ala315, Ala314, Cys239, Leu246, Met316, Leu253, Lys350, Asn256, Thr312, Val313 | 2 [Asn247: (3.32 Å; 3.07)] | 9 | 11 |
| robustaflavone | −8.2 | Ala352, Leu246, Val368, Ala315, Ala314, Cys239, Met316, Val236, Met257, Glu198, Phe167, Ser165, Gln134, Leu250, Phe200, Leu253, Asn256, Lys350 |
2 [Asn256: (3.21)] [Gln134: (2.64)] |
16 | 18 |
| euphohelionon | −7.9 | Leu253, Ala248, Val255, Lys252, Ala352, Asn256, Ala314, Lys350, Leu246, Gln245, Thr351 | 1 [Asn256: (3.19)] | 10 | 11 |
| ZINC95486052 | −8.3 | Asn247, Thr312, Ala315, Val313, Leu253, Met257, Asn256, Lys350, Asn247, Lys252, Met316, Leu246, Asn348 | 0 | 13 | 13 |
| 6,10-dimethyl-9-methylene-2-(4-methyl- 1,2-dioxabicyclo [2.2.2] oct-5-en-l-yl) undec-5-ene | −7.9 | Glu198, Leu250, Val236, Phe200, Ser165, Met257, Val368, Cys239, Met316, Ala315, Ala314, Lys350 Thr237, Leu253, Gln134, Leu240 | 0 | 16 | 16 |
3.7. The anthelmintic activities of the hits
The anthelmintic related biological activity was predicted and reinforced using structural similarity evaluation with known anthelmintics. The main goal was to predict scaffolds that could be optimized for the development of anthelmintics inhibitors. PASS [60] was used to predict the pharmacological activities of the top 30 hits. The prediction was based on the structural-activity relationship between the compound of interest and a training set of over 26000 compounds with known biological activities [60]. The relevant biological activity considered in this study was the propensity of the hits to be anthelminthic. PASS was used in previous studies to predict the anthelmintic activity of novel compounds [82,83]. Experimental results from these studies mostly corroborated the PASS predictions, indicating that PASS is a plausible Bayesian-based technique for predicting the anthelmintic activity of a compound. For a given compound, PASS predicts Probable activity (Pa) and Probable inactivity (Pi), with both ranging between 0.000 and 1.000 for a predicted activity. A total of 19 compounds out of the 30 hits were predicted to be anthelmintic, with Pa > Pi (Table 4) which denote their likely anthelmintic potential [83]. The compound 6,10-dimethyl-9-methylene-2-(4-methyl-1,2-dioxabicyclo[2.2.2]oct-5-en-l-yl)undec-5-ene obtained from NANPDB had the highest Pa of 0.759 and Pi of 0.003 and with such Pa > Pi, there is a high propensity for it to be a potent anthelmintic warranting further pharmacological evaluation [82,83]. Furthermore, a Drugbank similarity search [84] of 6,10-dimethyl-9-methylene-2-(4-methyl-1,2-dioxabicyclo[2.2.2]oct-5-en-l-yl) undec-5-ene was done to support the PASS predictions. The hit had a DrugBank similarity score of 0.511 to terpenin-4-ol, an isomer of terpineol. Terpine-4-ol has been reported to be an anthelmintic with potent activity against the eggs and larvae of helminth Haemonchus contortus [85,86]. The DrugBank [84] performs structure similar searches by using locally developed SMILES string comparison method to identify related structures. The similarity scoring ranges from 0–1, with 1 being the exact compound.
Table 4.
PASS anthelmintic predictions of the 19 natural product hits from AfroDB and NANPDB. The table shows the ZINC IDs or names of the compounds, their probability of activity (Pa) and the probability of anthelmintic inactivity (Pi).
| No. | ZINC ID/Compound name | Pa | Pi | Pa>Pi |
|---|---|---|---|---|
|
| ||||
| 6,10-dimethyl-9-methylene-2-(4-methyl-1,2- dioxabicyclo[2.2.2]oct-5-en-l-yl)undec-5-ene | 0.759 | 0.003 | Yes | |
| ZINC15120680 | 0.722 | 0.003 | Yes | |
| ZINC95485928 | 0.687 | 0.004 | Yes | |
| ZINC95485922 | 0.627 | 0.004 | Yes | |
| ZINC14760755 | 0.58 | 0.004 | Yes | |
| ZINC14780716 | 0.482 | 0.02 | Yes | |
| ZINC13480348 | 0.474 | 0.008 | Yes | |
| Robustaflavone | 0.464 | 0.024 | Yes | |
| Tetrahydrorobustaflavone | 0.464 | 0.024 | Yes | |
| ZINC95486263 | 0.461 | 0.024 | Yes | |
| ZINC28462577 | 0.457 | 0.025 | Yes | |
| S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6- trihydroxyphenyl)-15-methylicosa-5,8,11,13,17-pentaen-1-one | 0.39 | 0.018 | Yes | |
| Euphohelionon | 0.378 | 0.02 | Yes | |
| ZINC95486052 | 0.376 | 0.02 | Yes | |
| ZINC95485992 | 0.367 | 0.058 | Yes | |
| ZINC95486082 | 0.316 | 0.032 | Yes | |
| Spinescen | 0.291 | 0.118 | Yes | |
| ZINC13485435 | 0.24 | 0.172 | Yes | |
| ZINC95486081 | 0.207 | 0.084 | Yes | |
ZINC15120680 had the second highest Pa of 0.722 and Pi of 0.003. It also had a DrugBank similarity score of 0.599, 0.572 and 0.571 to Kaempherol, Quercetin and Genistein, respectively. Kaempherol, quercetin and genistein are naturally occurring products with multiple pharmacological actions, including anthelmintic activity [87]. Kaempherol and quercetin for instance were found to contribute to the anthelmintic property of 10 East African plants [88]. Moreover, these two natural products were reported to cause paralysis and death of known helminths Taenia solium (tape worm) and Pheritima posthuma (earthworm) [89]. Phytochemical analysis of the root tuber peels revealed rich genistein contents [87,90]. Genistein appears to have an eclectic mechanism of action against helminth parasites [91]. For instance, genistein was found to alter the carbohydrate metabolism of cestode parasites [87,92]. It also disrupts the spines and the tegumental surface of trematodes, leading to total paralysis or death. Moreover, genistein and its derivatives interfere with the microtriches, vesiculations and nuclear pyknosis of cestodes E. multicularis and E. granulosus [87,93].
ZINC95486052 and ZINC95486082 were structurally similar to naringenin with DrugBank similarity scores of 0.768 and 0.818, respectively. Naringenin was reported to have a synergistic effect on the synthesis and shedding of cuticle larval stages of helminth H. contortus [94]. A substructure probe via PubChem [52] revealed a common 2-phenylchromen-4-one substructure in ZINC15120680 and its similar DrugBank compounds (kaempherol, quercetin and genistein). ZINC95486052, ZINC95486082, ZINC28462577, ZINC95485922, ZINC14760755, robustaflavone, tetrahydrorobustaflavone and ZINC95485928 all had a 2-phenylchromen-4-one substructure. This substructure is native to the flavonoid family of natural products. Studies have also attributed the high anthelmintic property in flavonoids to the presence of reactive 2-phenylchromen-4-one structure [89].
ZINC95485928 was predicted to possess anthelmintic activity with Pa of 0.687 and Pi of 0.004. It had a DrugBank structural similarity score of 0.500 to Diosmetin. Diosmetin extracts from plant species D. insularis was found to be efficacious against gastrointestinal nematodes in goats [95]. The authors further suggested that flavones, a family of natural products could probably be responsible for anthelmintic activities [95]. Furthermore, the NANPDB hit Euphohelionon, which is a constituent of the Euphorbia helioscopia L. plant [96] was predicted to have an anthelminthic activity with Pa of 0.378 and Pi of 0.02. A study of the methanolic extracts of E. helioscopia, the parent plant of euphohelionon, revealed significant larvicidal and ovicidal effects against H. contortus worms, both in vitro and in vivo [97]. Table 4 summarizes the PASS anthelmintic predictions of the 19 hit compounds from AfroDB [28] and NANPDB [27].
3.8. Drug likeness and pharmacological profiling
Lipinski’s Rule of Five (RO5) [98] was used to evaluate the drug-likeness of the 19 predicted anthelmintic hits. The rule has been applied in analyzing African natural product libraries [28,99,100]. For a molecule to be qualified as “drug-like” and orally active, it must not violate more than one of the following criteria: a molecular weight ≤500 Da; logarithm of n-octanol/water partition ≤5, which determines the lipophilicity; hydrogen bond donors ≤5; and hydrogen bond acceptors ≤10. Table 5a provides the physicochemical properties and evaluation of the drug-likeness of the 19 predicted anthelmintic hits of which 12 did not violate any of the RO5 criteria. Furthermore, only three hits violated one of the RO5s. In total, 15 out of 19 hits were predicted to be druglike via SwissADME [61]. The remaining four hits with the least drug-likeness were tetrahydrorobustaflavone, robustaflavone, ZINC13480348 and euphohelionon.
Table 5a.
Physicochemical properties of the 19 predicted anthelmintic compounds comprising MW = Molecular weight, HBD=Number of hydrogen bond donors, HBA=Number of hydrogen bond acceptors, LOGP = Coefficient of partition, and the drug-likeness based on the number of Lipinski violations.
| No. | Compound ID | MW (g/mol) | HBA | HBD | LOGP | VIO-LATIONS |
|---|---|---|---|---|---|---|
|
| ||||||
| 6,10-dimethyl-9-methylene-2-(4-methyl-1,2-dioxabicyclo[2.2.2]oct-5-en-l-yl)undec-5-ene | 318.49 | 2 | 0 | 4.76 | 1 | |
| ZINC15120680 | 396.43 | 6 | 4 | 1.98 | 0 | |
| ZINC95485928 | 462.53 | 6 | 3 | 2.93 | 0 | |
| ZINC95485922 | 464.55 | 6 | 4 | 2.93 | 0 | |
| ZINC14760755 | 392.49 | 4 | 2 | 3.38 | 0 | |
| ZINC14780716 | 392.49 | 4 | 3 | 3.7 | 0 | |
| ZINC13480348 | 452.54 | 6 | 2 | 2.92 | 0 | |
| Robustaflavone | 538.46 | 10 | 6 | 0.25 | 2 | |
| Tetrahydrorobustaflavone | 538.46 | 10 | 6 | 0.25 | 2 | |
| ZINC95486263 | 554.46 | 11 | 6 | 0.02 | 3 | |
| ZINC28462577 | 538.46 | 10 | 5 | 0.52 | 1 | |
| S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6-trihydroxyphenyl)-15-methylicosa-5,8,11,13,17-pentaen-1-one | 440.57 | 5 | 4 | 3.35 | 0 | |
| Euphohelionon | 648.78 | 7 | 0 | 6.05 | 2 | |
| ZINC95486052 | 408.49 | 5 | 2 | 2.9 | 0 | |
| ZINC95485992 | 436.54 | 5 | 4 | 3.28 | 0 | |
| ZINC95486082 | 392.49 | 4 | 1 | 3.46 | 0 | |
| Spinescen | 638.92 | 8 | 4 | 3.02 | 1 | |
| ZINC13485435 | 392.49 | 4 | 2 | 3.73 | 0 | |
| ZINC95486081 | 380.43 | 5 | 2 | 2.62 | 0 | |
Pharmacological profiling studies are important components of the drug development pipeline. The pharmacokinetic properties considered for this study included human intestinal absorption, Permeability Glycoprotein (P-gp) binding, blood-brain barrier and cytochrome P450 (CYPs450) inhibition [101]. A total of 10 hits were predicted to have high intestinal absorption. Permeability glycoprotein (P-gp) is an ATP-dependent efflux pump that moves substances from the intracellular space to the extracellular space [102]. P-gp has a broad substrate specificity [103] and the excretion of drugs back into the gut lumen by P-gp pharmacokinetically reduces the potency of some drugs which are P-gp substrates [104]. Moreover, studies have implicated P-gp as a contributing factor in helminth drug resistance [105]. Only three of the hits were predicted to be P-gp substrates. Another important pharmacokinetic parameter is the Cytochromes P450 (CYPs) inhibition. Cytochromes P450 (CYPs), which are major enzymes involved in drug metabolism belong to a protein super family containing heme [106]. Inhibition of CYPs may lead to possible drug-drug interactions [107]. The CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4 inhibition of the compounds were predicted via SwissADME (Table 5b). Toxicity predictions were also performed on the compounds due to their potential contribution to drug attrition [108]. The mutagenicity, tumorigenicity, irritancy and reproductive effect were predicted using Osiris Property Explorer (Table 5c). Furthermore, the nephrotoxicity and hepatotoxicity were predicted via ADVERPred, which utilized Structure−Activity Relationships (SARS) models built using the PASS [63]. The probability of activity (Pa) and probability of inactivity (Pi) for these parameters for the 19 compounds have been provided in Table 5c.
Table 5b.
Prediction of the pharmacokinetics of the 19 predicted anthelmintic compounds showing gastrointestinal absorption (GI), Cytochrome P450 (CYP450) enzymes inhibition and permeability glycoprotein (P-gp) substratess
| ZINC ID/Compound Name | GI Absorption | P-gp substrate | CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 |
|---|---|---|---|---|---|---|---|
|
| |||||||
| 6,10-dimethyl-9-methylene-2-(4-methyl-1,2dioxabicyclo[2.2.2]oct-5-en-l-yl)undec-5-ene | High | No | Yes | Yes | Yes | Yes | No |
| ZINC15120680 | High | No | Yes | No | Yes | No | No |
| ZINC95485928 | Low | No | No | Yes | No | No | No |
| ZINC95485922 | Low | No | No | No | No | No | No |
| ZINC14760755 | High | No | No | Yes | No | No | No |
| ZINC14780716 | High | No | Yes | No | Yes | No | Yes |
| ZINC13480348 | High | No | No | No | Yes | No | Yes |
| Robustaflavone | Low | No | No | No | No | No | No |
| tetrahydrorobustaflavone | Low | No | No | No | No | No | No |
| ZINC95486263 | Low | No | No | No | Yes | No | No |
| ZINC28462577 | Low | No | No | No | Yes | No | No |
| ZINC15120680 | Low | No | Yes | No | Yes | No | No |
| S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6-trihydroxyphenyl)-15-methylicosa- 5,8,11,13,17-pentaen-1-one | Low | No | No | No | Yes | No | Yes |
| Euphohelionon | High | No | No | No | No | Yes | No |
| ZINC95486052 | High | No | No | Yes | Yes | No | Yes |
| ZINC95485992 | High | Yes | No | No | Yes | No | Yes |
| ZINC95486082 | Low | No | No | Yes | Yes | No | Yes |
| Spinescen | High | Yes | No | No | No | No | Yes |
| ZINC13485435 | High | Yes | No | Yes | Yes | No | Yes |
Table 5c.
Toxicity predictions of the 19 predicted anthelmintic compounds using DataWarrior and ADVERPred. The mutagenicity, tumorigenicity, irritancy, reproductive effect, hepatotoxicity and nephrotoxicity were considered. ADVERPred predicts the probability of activity (Pa) and probability of inactivity (Pi).
| ZINC ID/Compound Name | Mutagenic | Tumorigenic | Reproductive Effective | Irritant | Hepatotoxicity | Nephrotoxicity | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Pa | Pi | Pa | Pi | |||||
|
| ||||||||
| 6,10-dimethyl-9-methylene-2-(4-methyl-1,2-dioxabicyclo[2.2.2]oct-5-en-l- yl)undec-5-ene | None | High | None | None | None | None | None | None |
| ZINC15120680 | None | None | None | None | None | None | None | None |
| ZINC95485928 | High | None | None | None | None | None | None | None |
| ZINC95485922 | None | None | None | None | None | None | None | None |
| ZINC14760755 | None | None | None | None | None | None | None | None |
| ZINC14780716 | None | None | None | None | None | None | None | None |
| ZINC13480348 | High | None | None | High | None | None | None | None |
| Robustaflavone | None | None | None | None | 0.522 | 0.184 | None | None |
| tetrahydrorobustaflavone | None | None | None | None | 0.522 | 0.184 | None | None |
| ZINC95486263 | None | None | None | None | 0.711 | 0.096 | None | None |
| ZINC28462577 | None | None | High | None | 0.722 | 0.092 | None | None |
| ZINC15120680 | None | None | None | None | None | None | None | None |
| S,5Z,8Z,11Z,13E,17Z)-15-hydroxy-1-(2,4,6-trihydroxyphenyl)-15-methylicosa-5,8,11,13,17-pentaen-1-one | None | High | None | High | None | None | None | None |
| Euphohelionon | None | None | None | None | None | None | 0.244 | 0.232 |
| ZINC95486052 | None | None | None | High | None | None | None | None |
| ZINC95485992 | None | None | None | None | None | None | None | None |
| ZINC95486082 | None | None | None | None | None | None | None | None |
| Spinescen | None | None | High | None | 0.384 | 0.271 | 0.369 | 0.103 |
| ZINC13485435 | None | None | None | None | None | None | None | None |
3.9. Molecular dynamics simulation of selected compounds
Three African natural compounds namely ZINC95486082, ZINC95486052, and euphohelionon were selected for downstream MD simulation. These compounds had low binding energies, relatively good pharmacological profiles, predicted anthelmintic propensity and most importantly did not interact with the mutation-associated residues. Additionally, two known inhibitors comprising PubChem ID 53327692 and mebendazole were selected as controls. Both compounds had relatively better binding energies amongst the five inhibitors in this study. A 100 ns MD simulation was performed to expound on the structural stability and conformational changes when situated in diverse and dynamic physiological conditions [109]. The parameters studied included the root mean square deviation (RMSD), the radius of gyration (Rg), and the root mean square fluctuation (RMSF). The RMSD evaluates the deviation of the protein-ligand complex during the simulation from the initial protein backbone atomic coordinates and is used as a plausible measure of a protein’s stability [110]. An RMSD plot over simulation time revealed the backbones of the five complexes stabilized after 25 ns (Fig. 8A). The ZINC95486082-beta-tubulin complex rose from 0 nm to 0.5 nm during the start of the simulation. It then peaked around 23 ns, and later averaged around 0.75 nm over the remaining simulation time. Furthermore, the ZINC95486052-beta-tubulin complex maintained a steady RMSD of approximately 0.64 nm after the initial 20 ns of simulation. Additionally, the RMSD of Euphohelionon-beta-tubulin complex rose steadily from 0 nm and peaked around 1.0 nm, after which it began to decline. It then maintained an average RMSD of approximately 0.77 nm. The mebendazole-beta tubulin complex had the highest deviation of 1.0 nm from its protein backbone. The PubChem CID 53327692-beta tubulin complex also maintained an RMSD of approximately 0.64 nm.
Fig. 8.
Graphical representations of Rg, RMSD, and RMSF of hookworm beta tubulin-ligand complexes over a 100 ns MD simulation. (A) Graph of backbone RMSD of the unbound protein and the respective complexes versus time in picoseconds. (B) The radius of gyration graph of the unbound protein and the other complexes versus time in picoseconds (C) Graph of RMSF of the unbound protein and the other complexes versus the number of residues. In all the three graphs (A, B and C), black, red, green, blue, yellow and purple plots represent the unbound protein, Mebendazole, PubChem ID 53327692 (Dichloro), ZINC9486052, ZINC9486082 and euphohelionon beta-tubulin complexes, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The compactness of the complexes was evaluated using the Rg [111]. The Rg of all five complexes decreased over the first 20 ns. The trend then stabilized over the remaining simulation time (Fig. 8B). ZINC95486082–beta tubulin, ZINC95486052–beta-tubulin, Euphohelionon–beta-tubulin, Mebendazole-beta tubulin and PubChem ID 53327692–beta-tubulin complexes had average Rg of approximately 2.1 nm, 2.13 nm, 2.12 nm, 2.14 nm, and 2.13 nm, respectively. A stably folded and compact protein maintains a reasonable steady radius of gyration throughout the simulation [112]. As indicated in Fig. 7B, the trends appeared steady indicating the protein remained stably folded after binding.
Finally, the stability of the individual residues was assessed using their RMSF plots [109]. All the complexes possessed similar residue fluctuations within the same regions, with little deviation from the unbound protein core (Fig. 8C). Sizeable residue fluctuations were observed in the regions from residue index 420–449 which might be the region with the highest flexibility.
3.10. Evaluation of potential leads using the MM-PBSA calculations
The binding free energies of the complexes were estimated using MM-PBSA calculations. The calculations address some limitations of current scoring functions [113]. Contributing terms to the binding free energy include the electrostatic, polar, non-polar, and van der Waals energies. The binding free energies were computed in terms of average energies and standard deviations. Mebendazole had a relatively low average binding free energy of −150.810 kJ/mol (Table 6), thus affirming its binding energy against hookworm beta-tubulin. In addition, PubChem ID 53327692 had binding energy of −122.239 kJ/mol. The energy contributions per individual residues to the binding was also evaluated, whereby residues contributing >5.0 kJ/mol or < −5.0 kJ/mol were considered critical [114]. The amino acid residue, Glu198 contributed 34.3977 kJ/mol and 30.2668 kJ/mol to ligand binding of mebendazole and PubChem ID 53327692, respectively (Supplementary Figure 5).
Table 6.
The MM-PBSA predicted scores for the ZINC95486052, ZINC95486082, euphohelionon, and PubChem ID 53327692 complexes. The table shows the average binding energies and their contributing terms in average ± standard deviations in kJ/mol.
| Name | Electrostatic (kJ/mol) | Van Der Waals (kJ/mol) | Non-polar (kJ/mol) | Polar (kJ/mol) | Binding energy (kJ/mol) |
|---|---|---|---|---|---|
|
| |||||
| ZINC95486052 | 341.594 ± 79.830 | −195.548 ± 39.296 | −17.911 ± 3.429 | 179.040 ± 88.856 | 307.175 ± 46.596 |
| ZINC95486082 | 247.491 ± 55.392 | −165.756 ± 71.527 | −15.864 7.108 | 270.694 ± 93.924 | 336.564 ± 70.495 |
| Euphohelionon | −68.456 ± 35.677 | −186.699 ± 64.469 | −15.841 ± 5.685 | 147.377 ± 47.152 | −123.620 ± 70.683 |
| Mebendazole | −81.725 ± 11.668 | −217.842 ± 11.900 | −17.814 ± 0.850 | 166.572 ± 15.425 | −150.810 ± 14.230 |
| PubChem ID 53327692 | −76.394 ± 16.633 | −246.863 ± 10.661 | −20.181 ± 0.860 | 221.199 ± 29.753 | −122.239 ± 21.460 |
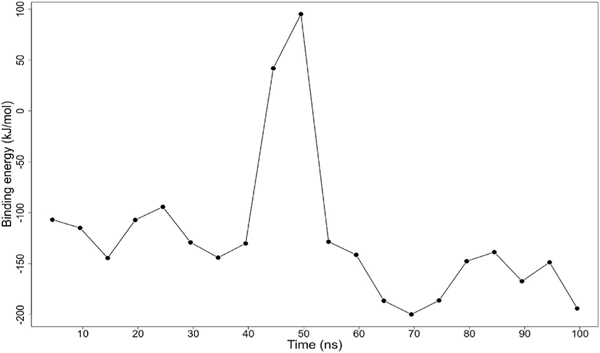
Euphohelionon had the lowest binding free energy of −123.620 kJ/mol amongst the three potential compounds (Table 6). Its binding energy plot over the simulation time showed initial binding energy of 106 kJ/mol. It then averaged within the range of −150 kJ/mol and −90 kJ/mol, further peaking at 50 ns. A final binding energy of −194 kJ/mol was obtained at 100 ns (Fig. 9). The van der Waal, electrostatic and polar energies contributed favourably to the free binding energy (Table 6). An energy decomposition plot revealed Leu246, Leu253 and Asp327 contributed energies beyond the ± 5.0 kJ/mol threshold (Fig. 10; Supplementary Table 2). The observed high energies corroborate the aforementioned assertion that Leu246 and Leu253 are critical for ligand binding and stabilization. Besides, the resistance mutation associated residues Phe167, Glu198 and Phe200 contributed energies of −0.0220 kJ/mol, 2.4911 kJ/mol and −0.0742 kJ/mol to ligand binding, respectively (Fig. 10; Supplementary Table 2). This fell short of the threshold required for critical residues, suggesting that perhaps these are not the foremost ligand-binding residues.
Fig. 9.
Binding energy plot of euphohelionon complex. The graph shows binding energy (kJ/mol) versus time over 100 ns simulation.
Fig. 10.
The per-residue decomposition of the binding energy plot of the euphohelionon-beta-tubulin complex. The per-residue binding energies of the likely mutated residues (Phe167, Glu198, and Phe200) are coloured red. The three residues contributed minimal energies to the complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
ZINC95486052 was predicted by AutoDock Vina to have binding energies of −8.5 kcal/mol. However, MM-PBSA calculations showed it had high binding free energy of 307.175 kJ/mol. This was predominantly due to high electrostatic and polar energies of 341.594 kJ/mol and 179.040 kJ/mol, respectively. Also, ZINC95486082 had high binding free energy of 336.564 kJ/mol. Molecules with high binding free energies may have limited lead likeness [114], hence a decreased propensity for further exploitation [115]. A binding energy decomposition per residue evaluation revealed Glu198 contributed favourably (>5.0 kJ/mol) to ligand binding of both compounds. Leu256 was observed to contribute favourable binding energies (< −5.0 kJ/mol) in both complexes.
3.11. Selection of parent molecule for de novo ligand design
The parent compound was selected based on its binding energy, protein-ligand interactions, predicted anthelmintic activity and free binding energies. After virtual screening an integrated library of 3385 compounds of African origin against the modelled beta-tubulin receptor, 30 hit compounds were selected based on their binding energies and how they are firmly docked in the binding pockets. All the selected hits had lower binding energy than the five known inhibitors. Compounds that form strong biomolecular interactions with resistance-associated mutation residues could lose interactions, leading to loss of affinity [21]. Four compounds namely anchinopeptolide A, euphohelionon, ZINC95486082 and ZINC 95486052 formed no molecular interactions with the resistance mutation associated residues (Table 3). However, anchinopeptolide A violated three rules of the RO5s (Table 5a). More so, it was not predicted as anthelmintic, hence warranted no further downstream exploration. ZINC95486082 and ZINC95486052 had binding of −8.5 kcal/mol and −8.3 kcal/mol, respectively. They were both predicted anthelmintics with Pa>0.3 and Pa>Pi (Table 4). Substructure searches revealed a common 2-phenylchromen-4-one substructure for ZINC95486082 and ZINC95486052. The 2-phenylchromen-4-one substructure was reported to be responsible for the high anthelmintic properties in flavonoids [89]. Both ZINC95486052 and ZINC95486082 were structurally similar to naringenin, a known anthelmintic with similarity scores of 0.768 and 0.818, respectively. However, evaluation of free binding energies of the two compounds via MM-PBSA calculations revealed high binding energies of 307.175 and 336.564 kJ/mol, respectively (Table 6), hence possibly decreasing their likelihood to be a lead compound [114].
Euphohelionon had binding energy of −7.9 kcal/mol and formed one hydrogen bond with critical residue Asn256 and nine hydrophobic contacts. It was predicted as anthelmintic with a Pa of 0.378 and Pi of 0.02. When the Pa>0.3 and Pa>Pi for a particular biological activity, the compound warrants further experimental validation. Furthermore, euphohelionon is sourced from the E. helioscopia a local African plant, which has reported anthelmintic activities. Additionally, euphohelionon had a generally good pharmacological profile (Table 5). Further evaluation of the binding energies via MM-PBSA calculations revealed a good binding free energy of −123.620 kJ/mol (−29.546 kcal/mol). The energy contribution of the individual residues also revealed that three critical residues namely Leu246, Leu253 and Asp327, contributed energies beyond the ± 5.0 kJ/mol to the euphohelionon-beta-tubulin binding. Furthermore, per energy decomposition of the residues revealed it would likely not interact with the known resistance-associated mutation residues. Considering these factors, euphohelionon was selected as a parent compound for de novo design of potential inhibitor molecules of hookworm beta-tubulin. The step-by-step stages in the selection of a parent compound have been illustrated in Fig. 11.
Fig. 11.
The schema shows the step-by-step approaches involved in the filtering and selection of compounds. Firstly, 3390 compounds comprising 3385 African natural products and 5 known beta-tubulin inhibitors were screened against the modelled beta-tubulin of the hookworm. Thirty compounds with low binding energies were selected as top hits for downstream analysis. The physicochemical and pharmacological properties, protein-ligand interactions and biological activity predictions of the 30 compounds were assessed in silico. Three natural product compounds were selected as leads based on their good pharmacological profiles, mechanism of binding, and predicted anthelmintic activity.
3.12. De novo fragment-based design of novel potential anthelmintics
The e-LEA3D server [67] was used to generate a total of 50 new molecules using euphohelionon as a parent compound. Duplicates and error generating molecules were eliminated. A total of 15 molecules with good physicochemical properties were used in molecular docking studies. The molecules with binding energies < −6.0 kcal/mol were selected for downstream analysis. Five novel molecules with identifications A1, A2, A3, A4 and A5 had good binding energies relative to the known inhibitors. Binding energies of −8.2 kcal/mol, −7.6 kcal/mol, −7.3 kcal/mol, −7.2 kcal/mol and −6.8 kcal/mol were obtained for A1, A2, A3, A4, and A5, respectively (Table 7). Only A1 had better binding energy than the parent compound, euphohelionon. Insights into the mechanism of binding revealed similar interactions akin to the euphohelionon-beta-tubulin complex. A1 formed a total of thirteen hydrophobic contacts with binding site residues including Leu246, Lys350 and Asn256. In addition, A2 formed two hydrogen bonds and six hydrophobic contacts (Fig. 12, Table 7). A3 formed nine hydrophobic contacts as well as three hydrogen bonds of varying lengths with Ala314. Furthermore, A4 formed two hydrogen bonds with Asn256 and Asn247. A5 formed a total of 11 hydrophobic contacts. Most importantly, none of the molecules interacted with the resistance mutation related residues (Table 7).
Table 7.
A summary of the protein-ligand interactions of the five novel compounds generated with Ligplot+. The table shows the binding energy, the synthetic accessibility score, the interacting residues in the active site, and the respective intermolecular bonds.
| Compound | Binding Energy (kcal/mol) | Synthetic accessibility Score (SAscore) | Interacting Residues in the active site | Number of hydrogen bonds [amino acid residues (Ǻ)] | Number of hydrophobic bonds |
|---|---|---|---|---|---|
|
| |||||
| A1 | −8.2 | 4.51 | Ala315, Met316, Leu246, Leu253, Thr351, Lys350, Cys239, Ala314, Asn256, Ala248, Asn247, Val25, Lys252 |
0 | 13 |
| A2 | −7.6 | 4.94 | Val249, Asn347, Lys350, Asn256, Leu253, Ala352, Leu246, Thr351 | 2 [Thr351: 2.88; 3.10] | 6 |
| A3 | −7.3 | 6.04 | Asn247, Ala352, Ala248, Leu246, Lys252, Leu253, Asn256, Thr351, Ala314, Ala315, Lys350 | 3 [Ala315: 3.22; 2.92] [Lys350: 2.97] |
|
| A4 | −7.2 | 3.10 | Lys252, Ala248, Leu253, Leu246, Lys350, Thr251, Asn247, Asn256 | 2 [Asn247: 2.14] [Asn256: 2.90] |
|
| A5 | −6.8 | 3.22 | Val368, Ala315, Ala352, Ala314, Met316, Lys350, Thr351, Leu246, Leu253, Val236, Met257 | 0 | 11 |
Fig. 12.
Ligplot+ representation of the protein-ligand interaction of the beta tubulin-A2 complex. The ligand forms six hydrophobic contacts (represented as red spook arcs) and two hydrogen bonding (represented as green dotted lines) with residues in the active site. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The novelty, physicochemical and pharmacokinetic profiles of the five compounds were assessed via SwissADME [61]. A search using the SMILES of the five compounds uncovered that all the molecules had no exact structure in the database, asserting their uniqueness. More so, all the molecules complied with R05s [98] except A1 which violated it. A1 violated two R05s comprising MW = 538.57 g/mol and logP = 4.35. Synthetic accessibility score (SAscore) is a useful parameter for estimating the ease of synthesizing a molecule. SAscore ranges from 1 (easy to synthesize) to 10 (difficult to synthesize). Molecules with SAscore <6.0 have lower propensity to synthesize [116,117]. Estimated SAscore values of 4.51, 4.94, 6.04, 3.1, and 3.22 were obtained for A1, A2, A3, A4 and A5, respectively. All the molecules were found to be within the acceptable threshold, except A3. Pharmacokinetic predictions also revealed all the molecules except A3 had high intestinal absorption (Supplementary Table 3). Undesirable pharmacological profiles of a compound can be improved through chemical modifications. Amongst the hits, A5 had no predicted toxicity (Supplementary Table 3). Additionally, anthelmintic activity predictions via PASS revealed A5 had a PA of 0.448 and Pi of 0.028. When Pa>Pi and Pa>0.3, there is a high propensity for it to be a potent anthelmintic warranting further pharmacological evaluation [82,83]. Also, drug off-target activity as a result of drugs having different biological activities other than intended [118, 119] cannot be discounted. The two-dimensional structures of the five de novo molecules have been provided (Table 8).
Table 8.
The names, generated IUPAC names via ChemAxon and two-dimensional chemical structures of the novel potential lead compounds generated using euphohelionon as a template via e-LEA3D [67].
| Name | Generated IUPAC Name | Generated Chemical structure |
|---|---|---|
|
| ||
| A1 | (2R)-11-[4-(9H-carbazol-4-yl)phenyl]-2-fluoro-6-hydroxy-7-phenyl-4-oxa-1-azatricyclo[7.3.1.05, 13] trideca-5(13),6,8,11-tetraen-10-one |

|
| A2 | 7-[(2R,3R,4R)-2-(9H-carbazol-4-yl)-4-methyloxan-3-yl]-6-hydroperoxy-4-oxa-1-azatricyclo[7.3.1.05, 13] trideca-5(13),6,8,11-tetraen-10-one |

|
| A3 | (2S)-2-[(2R)-11-[(2S,3aR,6aR)-1-(9H-carbazol-4-yl)-octahydrocyclopenta[b]pyrrol-2-yl]-6-methyl-10-oxo-7-phenyl-4-oxa-1-azatricyclo[7.3.1.05, 13]trideca-5(13),6,8,11-tetraen-2-yl]-2-amino-N-hydroxyacetamide |

|
| A4 | 7-(9H-carbazol-4-yl)-6-hydroxy-4-oxa-1-azatricyclo[7.3.1.05, 13]trideca-5(13),6,8,11-tetraen-10-one |

|
| A5 | (2S,3R)-2-(4-methylphenyl)-3-phenyloxane |

|
4. Contribution to the field
Computational techniques have been proven to be a faster and cheaper means of exploring novel drugs. This study supplements current efforts by identifying three African natural compounds with good pharmacological profiles and most importantly predicted to have no interactions with the resistance-associated residues. The three-dimensional structure of the hookworm beta-tubulin was modelled with high confidence. Moreover, the de novo fragment-based design was employed to identify five unique compounds with the potential of circumventing the mutation associated residues. Additionally, a novel mechanism of binding was elucidated which could aid in the design of future potent hookworm beta-tubulin inhibitors.
5. Conclusion
This study identified three molecules with anthelminthic potential by screening a library composed of 3385 African natural products against a modelled 3D structure of the beta-tubulin gene product of Necator americanus. The AutoDock Vina molecular docking protocol was validated using the ROC with an acceptable AUC of 0.714. The three molecules, ZINC95486082, ZINC95486052 and euphohelionon, were predicted to have distinct mechanisms of binding relative to those of known drugs. These compounds did not interact with the benzimidazole resistance-related residues Phe167, Glu198 and Phe200 in N. Americanus beta-tubulin. A sub-structure analysis indicated that ZINC95486082 and ZINC95486052 contained 2-phenylchromen-4-one, a known anthelmintic moiety. Euphohelionon was selected as a parent compound for de novo ligand design because of its predicted anthelmintic properties, molecular interactions, and low binding energy consolidated with MM-PBSA calculations. Also, five novel molecules were generated using the fragment-based drug design and four can be synthesized easily. They also had favourable binding energies when compared to known inhibitors, and exhibited no contacts with any of the resistance mutation residues. These compounds can subsequently be synthesized and experimentally characterized for the development of much-needed novel anthelmintics.
Supplementary Material
Acknowledgements
The high-performance computing platform (Zuputo) used for the modelling and simulations was generously provided by the West African Centre for Cell Biology of Infectious Pathogens, University of Ghana.
Funding
This study was supported by a National Institutes of Health grant to MDW (ID #: 1R01AI099623 and ID# U19AI129916). The funder had no role in the design of the study, data collection and analysis, and in the preparation of the manuscript and the decision to publish.
List of Abbreviations
- ADMET
Absorption, Distribution, Metabolism, Excretion and Toxicity
- BBB
Blood-Brain Barrier
- DOPE
Discrete optimized potential energy
- FDA
Food and Drugs Authority
- H-bond
Hydrogen bond
- I-TASSER
Iterative Threading ASSEmbly Refinement
- IVM
Ivermectin
- MDA
Mass Drug Administration
- MD
Molecular Dynamics
- NP
Natural Product
- NPs
Natural Products
- PDB
Protein Data Bank
- SDF
Structure Data File
- SMILES
Simplified Molecular Input Line Entry System
- RMSD
Root Mean Square Deviation
- MMPBSA
Molecular Mechanics Poisson–Boltzmann Surface Area
Footnotes
Ethics approval and consent to participate
Not Applicable.
Consent to publish
Not Applicable.
Availability of data and materials
Accession numbers and their databases have been mentioned in the manuscript. Any other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100734.
References
- [1].Hotez PJ. One world health: Neglected tropical diseases in a flat world. PLoS Neglected Trop Dis 2009;3(4). 10.1371/journal.pntd.0000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ngui R, Ching LS, Kai TT, Roslan MA, Lim YAL. Molecular identification of human hookworm infections in economically disadvantaged communities in Peninsular Malaysia,. Am J Trop Med Hyg May 2012;86(5):837–42. 10.4269/ajtmh.2012.11-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aagaard-Hansen J, Lise Chaignat C. Neglected tropical diseases: equity and social determinants. Equity, Soc. Determ. Public Heal. Program. 2019:135–57 [Online]. Available: https://www.who.int/neglected_diseases/Social_determinants_NTD.pdf. [Google Scholar]
- [4].Pullan RL, Smith JL, Jasrasaria R, Brooker SJ. Global numbers of infection and disease burden of soil-transmitted helminth infections in 2010. Parasites Vectors 2014;7:37. 10.1186/1756-3305-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Loukas A, et al. Vaccinomics for the major blood-feeding helminths of humans. OMICS Sep. 2011;15(9):567–77. 10.1089/omi.2010.0150. [DOI] [PubMed] [Google Scholar]
- [6].Hotez P, Brooker S, Bethony JM, Bottazzi ME, Loukas A, Xiao S. Hookworm infection. N Engl J Med 2004;351(8):799–807. 10.1056/NEJMra032492. [DOI] [PubMed] [Google Scholar]
- [7].Hotez PJ, Bethony J, Bottazzi ME, Brooker S, Buss P. “Hookworm: ‘The great infection of mankind. PLoS Med 2005;2(3). 10.1371/journal.pmed.0020067. 0187–0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fennell B, et al. Microtubules as antiparasitic drug targets. Expet Opin Drug Discov May 2008;3(5):501–18. 10.1517/17460441.3.5.501. [DOI] [PubMed] [Google Scholar]
- [9].Horio T, Murata T. The role of dynamic instability in microtubule organization. Front Plant Sci 2014;5:511. 10.3389/fpls.2014.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kwa MS, Veenstra JG, Van Dijk M, Roos MH. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J Mol Biol March. 1995;246(4):500–10. 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- [11].Geerts S, Gryseels B. Drug resistance in human helminths: current situation and lessons from livestock. Clin Microbiol Rev April. 2000;13(2):207–22. 10.1128/cmr.13.2.207-222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Humphries D, et al. Effectiveness of albendazole for hookworm varies widely by community and correlates with nutritional factors: a cross-sectional study of school-age children in Ghana. Am J Trop Med Hyg Feb. 2017;96(2):347–54. 10.4269/ajtmh.16-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Warren KS. An integrated system for the control of the major human helminthparasites. Acta Leiden 1990;59(1–2):433–42. [PubMed] [Google Scholar]
- [14].V Brown G, Warren KS. For the new millennium: control of helminth diseases throughout the world. Med J Aust October. 1993;159(7):461–3. 10.5694/j.1326-5377.1993.tb137967.x. [DOI] [PubMed] [Google Scholar]
- [15].Lubega GW, Prichard RK. Specific interaction of benzimidazole anthelmintics with tubulin: high-affinity binding and benzimidazole resistance in Haemonchuscontortus. Mol Biochem Parasitol Jan. 1990;38(2):221–32. [DOI] [PubMed] [Google Scholar]
- [16].Schwenkenbecher JM, Albonico M, Bickle Q, Kaplan RM. Characterization of beta-tubulin genes in hookworms and investigation of resistance-associated mutations using real-time PCR. Mol Biochem Parasitol December. 2007;156(2):167–74. 10.1016/j.molbiopara.2007.07.019. [DOI] [PubMed] [Google Scholar]
- [17].Rashwan N, Bourguinat C, Keller K, Gunawardena NK, de Silva N, Prichard R. Isothermal diagnostic assays for monitoring single nucleotide polymorphisms in necator americanus associated with benzimidazole drug resistance. PLoS Neglected Trop Dis December. 2016;10(12):e0005113. 10.1371/journal.pntd.0005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McSorley HJ, Loukas A. The immunology of human hookworm infections. Parasite Immunol August. 2010;32(8):549–59. 10.1111/j.1365-3024.2010.01224.x. [DOI] [PubMed] [Google Scholar]
- [19].Diawara A, et al. Association between response to albendazole treatment and beta-tubulin genotype frequencies in soil-transmitted helminths. PLoS Neglected Trop Dis 2013;7(5):e2247. 10.1371/journal.pntd.0002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Albonico M, Wright V, Bickle Q. Molecular analysis of the beta-tubulin gene of human hookworms as a basis for possible benzimidazole resistance on Pemba Island. Mol Biochem Parasitol April. 2004;134(2):281–4. 10.1016/j.molbiopara.2003.12.008. [DOI] [PubMed] [Google Scholar]
- [21].Vasavi CS, Ramasamy T, Munusami P. Exploring the drug resistance mechanism of active site, non-active site mutations and their cooperative effects in CRF01_AE HIV-1 protease: molecular dynamics simulations and free energy calculations. J Biomol Struct Dyn July. 2018:1–43. 10.1080/07391102.2018.1492459. [DOI] [PubMed] [Google Scholar]
- [22].Butler MS. The role of natural product chemistry in drug discovery. J Nat Prod December. 2004;67(12):2141–53. 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- [23].Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod March. 2012;75(3):311–35. 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chama MA, et al. Isolation, characterization, and anthelminthic activities of a novel dichapetalin and other constituents of Dichapetalum filicaule. Pharm Biol July. 2016;54(7):1179–88. 10.3109/13880209.2015.1059861. [DOI] [PubMed] [Google Scholar]
- [25].Asiedu SO, Kwofie SK, Broni E, Wilson MD. Computational identification of potential anti-inflammatory natural compounds targeting the p38 mitogen- activated protein kinase (MAPK): implications for COVID-19-induced cytokine storm. Biomolecules April. 2021;11(5). 10.3390/biom11050653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Broni E, Kwofie SK, Asiedu SO, Miller WA, Wilson MD. A molecular modeling approach to identify potential antileishmanial compounds against the cell division cycle (cdc)-2-Related kinase 12 (CRK12) receptor of leishmania donovani. Biomolecules March. 2021;11(3):458. 10.3390/biom11030458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ntie-Kang F, et al. NANPDB: a resource for natural products from Northern african sources. J Nat Prod July. 2017;80(7). 10.1021/acs.jnatprod.7b00283. 2067–2076. [DOI] [PubMed] [Google Scholar]
- [28].Ntie-Kang F, et al. AfroDb: a select highly potent and diverse natural product library from african medicinal plants. PloS One 2013;8(10):e78085. 10.1371/journal.pone.0078085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].UniProt “. A hub for protein information. Nucleic Acids Res January. 2015;43: D204–12. 10.1093/nar/gku989. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aguayo-Ortiz R, et al. Towards the identification of the binding site of benzimidazoles to β-tubulin of Trichinella spiralis: insights from computational and experimental data. J Mol Graph Model April. 2013;41:12–9. 10.1016/j.jmgm.2013.01.007. [DOI] [PubMed] [Google Scholar]
- [31].Roy A, Kucukural A, and Zhang Y, “I-TASSER: a unified platform for automated protein structure and function prediction,” Nat Protoc, vol. 5, no. 4, pp. 725–738, April. 2010, doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eswar N, et al. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinform. Oct. 2006;5. 10.1002/0471250953. bi0506s15. Unit-5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Maiti R, Van Domselaar GH, Zhang H, Wishart DS, SuperPose “. A simple server for sophisticated structural superposition. Nucleic Acids Res 2004;32:590–4. 10.1093/nar/gkh477. WEB SERVER ISS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vriend G. What IF: a molecular modeling and drug design program. J Mol Graph 1990;8(1):52–6. 10.1016/0263-7855(90)80070-v. Mar. [DOI] [PubMed] [Google Scholar]
- [35].Kaplan W, Littlejohn TG. Swiss-PDB viewer (deep view). Briefings Bioinf May 2001;2(2):195–7. [DOI] [PubMed] [Google Scholar]
- [36].DeLano WL. “PyMOL: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40:82–92. 10.1038/s41598-017-03842-2. [DOI] [Google Scholar]
- [37].Berendsen HJC, van der Spoel D, van Drunen R. “GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun September. 1995; 91(1):43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- [38].Vaught A. “Graphing with gnuplot and xmgr: two graphing packages available under Linux,” Linux J [Online]. Available: http://dl.acm.org/citation.cfm?id=326327.326334;Aug.1996. 1996,28. [Google Scholar]
- [39].Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph February. 1996;14(1):27–8. 33–38. [DOI] [PubMed] [Google Scholar]
- [40].Ho BK, Brasseur R. The Ramachandran plots of glycine and pre-proline. BMC Struct Biol August. 2005;5:14. 10.1186/1472-6807-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Laskowski RA, MacArthur MW, Moss DS, and Thornton JM, “PROCHECK: a program to check the stereochemical quality of protein structures,” J Appl Crystallogr, vol. 26, no. 2, pp. 283–291, April. 1993, doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- [42].Eisenberg D, Lüthy R, Bowie JU. “VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 1997;277:396–404. 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- [43].Kalman M, Ben-Tal N. Quality assessment of protein model-structures using evolutionary conservation. Bioinformatics May 2010;26(10):1299–307. 10.1093/bioinformatics/btq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res July. 2018;46(W1):W363–7. 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem December. 2009;30(16):2785–91. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gasteiger J. and Marsili M, “Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges,” Tetrahedron, vol. 36, no. 22, pp. 3219–3228, January. 1980, doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- [47].Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J Chem Inf Model 2005;45(1):177–82. 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dayan AD. Albendazole, mebendazole and praziquantel. Review of non-clinical toxicity and pharmacokinetics. Acta Trop 2003;86(2–3):141–59. 10.1016/S0001-706X(03)00031-7. [DOI] [PubMed] [Google Scholar]
- [49].Venkatesan P. Albendazole. J. Antimicrob. Chemother. 1998;41(2):145–7. 10.1093/jac/41.2.145. [DOI] [PubMed] [Google Scholar]
- [50].Islam N, Chowdhury NA. Mebendazole and pyrantel pamoate as broad-spectrum anthelmintics. Southeast Asian J Trop Med Publ Health Mar. 1976;1:81–4. [PubMed] [Google Scholar]
- [51].V Satyendra R, Vishnumurthy KA, Vagdevi HM, Rajesh KP, Manjunatha H, Shruthi A. Synthesis, in vitro antioxidant, anthelmintic and molecular docking studies of novel dichloro substituted benzoxazole-triazolo-thione derivatives. Eur J Med Chem 2011;46(7):3078–84. 10.1016/j.ejmech.2011.03.017. [DOI] [PubMed] [Google Scholar]
- [52].Kim S, et al. PubChem substance and compound databases, vol. 44; 2016. p. 1202–13. 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminf October. 2011;3:33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Trott O, Olson AJ, “AutoDock Vina. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem January. 2010;31(2):455–61. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dallakyan S, Olson Arthur J. Small molecule library screening by docking with PyRx. Methods Mol Biol 2015;1263:243–50. 10.1111/j.1747-0285.2007.00471.x. [DOI] [PubMed] [Google Scholar]
- [56].Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem July. 2012;55(14):6582–94. 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem 2006;49(23):6789–801. 10.1021/jm0608356. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Goksuluk D, Korkmaz S, Zararsiz G, Karaagaoglu AE. easyROC: an interactive web-tool for ROC curve analysis using R language environment. R J. 2016;8: 213–30. [Google Scholar]
- [59].Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model October. 2011;51(10):2778–86. 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- [60].Parasuraman S. Prediction of activity spectra for substances. J Pharmacol Pharmacother 2011;2(1):52–3. 10.4103/0976-500X.77119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717. 10.1038/srep42717. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sander T, Freyss J, von Korff M, Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model February. 2015; 55(2):460–73. 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- [63].Ivanov SM, Lagunin AA, V Rudik A, Filimonov DA, V Poroikov V. ADVERPred- Web service for prediction of adverse effects of drugs. J Chem Inf Model 2018;58 (1):8–11. 10.1021/acs.jcim.7b00568. 1. [DOI] [PubMed] [Google Scholar]
- [64].Abraham MJ, et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software 2015;1(2): 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- [65].Schüttelkopf AW, van Aalten DMF. “PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr Sect D Biol Crystallogr August. 2004;60(8):1355–63. 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- [66].Kumari R, Kumar R, Lynn A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model July. 2014;54(7):1951–62. 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- [67].Douguet D. “e-LEA3D : a computational-aided drug design web server, vol. 38; 2010. p. 615–21. 10.1093/nar/gkq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf 2008;9: 1–8. 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Shen M-Y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci November. 2006;15(11):2507–24. 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis January. 1997;18(15):2714–23. 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- [71].Berendsen HJC, Drunen R va. “GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun September. 1995;91(1–3):43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- [72].Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci September. 1993;2(9):1511–9. 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Morris GM, Lim-Wilby M. Molecular docking. Methods Mol Biol 2008;443: 365–82. 10.1007/978-1-59745-177-2_19. [DOI] [PubMed] [Google Scholar]
- [74].Jain AN. Bias, reporting, and sharing: computational evaluations of docking methods. J Comput Aided Mol Des 2008;22(3–4):201–12. 10.1007/s10822-007-9151-x. [DOI] [PubMed] [Google Scholar]
- [75].Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment,. J Thorac Oncol 2010;5(9):1315–6. 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- [76].Florkowski CM. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin Biochem Rev August. 2008;29(Suppl 1):S83–7 [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/18852864. [PMC free article] [PubMed] [Google Scholar]
- [77].Hevener KE, et al. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model February. 2009;49(2):444–60. 10.1021/ci800293n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sravani T, Paarakh PM, Shruthi SD. In silico and in vitro anthelmintic activity of β-sitosterol isolated from rhizomes of Hedychium spicatum Buch.-Ham. Indian J. Nat. Prod. Resour. 2014;5(3):258–61. [Google Scholar]
- [79].Du X, et al. Insights into protein-ligand interactions: mechanisms, models, and methods. Int J Mol Sci January. 2016;17(2):144. 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fu Y, Zhao J, Chen Z. Insights into the molecular mechanisms of protein-ligand interactions by molecular docking and molecular dynamics simulation: a case of oligopeptide binding protein. Comput. Math. Methods Med. 2018;2018:1–12. 10.1155/2018/3502514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Remer LC, Jensen JH. Toward a general theory of hydrogen Bonding: the short, strong hydrogen bond [HOH···OH]-. J Phys Chem A October. 2000;104(40):9266–75. 10.1021/jp002726n. [DOI] [Google Scholar]
- [82].Jamkhande PG, Barde SR. Evaluation of anthelmintic activity and in silico PASS assisted prediction of Cordia dichotoma (Forst.) root extract. Ancient Sci Life 2014;34(1):39–43. 10.4103/0257-7941.150779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Garg K, Bansal Y, Bansal G, Goel RK. Design, synthesis, and PASS-assisted evaluation of novel 2-substituted benzimidazole derivatives as potent anthelmintics. 2014. p. 2690–7. 10.1007/s00044-013-0856-1. [DOI] [PubMed] [Google Scholar]
- [84].Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res Jan. 2018;46(D1):D1074–82. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Grando TH, et al. In vitro activity of essential oils of free and nanostructured Melaleuca alternifolia and of terpinen-4-ol on eggs and larvae of Haemonchus contortus. J Helminthol May 2016;90(3):377–82. 10.1017/S0022149X15000401. [DOI] [PubMed] [Google Scholar]
- [86].Oliveira CB, et al. In vitro activity of essential oils of free and nanostructured Melaleuca alternifolia and of terpinen-4-ol on eggs and larvae of Haemonchus contortus. J Helminthol 2015;90(3):377–82. . [DOI] [PubMed] [Google Scholar]
- [87].Tandon V, Das B. “Genistein : is the multifarious botanical a natural anthelmintic too? J Parasit Dis 2018. 10.1007/s12639-018-0984-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mengistu G, Hoste H, Karonen M, Salminen J-P, Hendriks WH, Pellikaan WF. The in vitro anthelmintic properties of browse plant species against Haemonchus contortus is determined by the polyphenol content and composition. Vet Parasitol April. 2017;237:110–6. 10.1016/j.vetpar.2016.12.020. [DOI] [PubMed] [Google Scholar]
- [89].Lasisi AA, Kareem SO. Evaluation of anthelmintic activity of the stem bark extract and chemical constituents of Bridelia ferruginae ( Benth ) Euphorbiaceae. Afr J Plant Sci 2011;5(August):469–74. 10.5897/AJPS.9000108. [DOI] [Google Scholar]
- [90].Pal P, Tandon V. Anthelmintic efficacy of flemingia vestita (fabaceae): genistein- induced alterations in the esterase activity in the cestode,raillietina echinobothrida. J Biosci 1998;23(1):25–31. 10.1007/BF02728520. [DOI] [Google Scholar]
- [91].Das B, Tandon V, Saha N. Anthelmintic efficacy of Flemingia vestita (Fabaceae): alteration in the activities of some glycolytic enzymes in the cestode, Raillietina echinobothrida. Parasitol Res Jul. 2004;93(4):253–61. 10.1007/s00436-004-1122-8. [DOI] [PubMed] [Google Scholar]
- [92].Tandon V, Das B, Saha N. Anthelmintic efficacy of Flemingia vestita (Fabaceae): effect of genistein on glycogen metabolism in the cestode, Raillietina echinobothrida. Parasitol Int June. 2003;52(2):179–83. [DOI] [PubMed] [Google Scholar]
- [93].Naguleswaran A, et al. “In vitro metacestodicidal activities of genistein and other isoflavones against Echinococcus multilocularisand Echinococcus granulosus. Antimicrob Agents Chemother November. 2006;50(11):3770–8. 10.1128/AAC.00578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Klongsiriwet C, Quijada J, Williams AR, Mueller-Harvey I, Williamson EM, Hoste H. Synergistic inhibition of Haemonchus contortus exsheathment by flavonoid monomers and condensed tannins. Int. J. Parasitol. Drugs Drug Resist. 2015;5(3):127–34. 10.1016/j.ijpddr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Oliveira Santos F, et al. In vitro anthelmintic and cytotoxicity activities the Digitaria insularis (Poaceae). Vet Parasitol Aug. 2017;245:48–54. 10.1016/j.vetpar.2017.08.007. [DOI] [PubMed] [Google Scholar]
- [96].Mohamed A, et al. Euphorbia helioscopia: chemical constituents and biological activities. Int J Phytopharm January. 2012;3:78–90. [Google Scholar]
- [97].Lone BA, Chishti MZ, Ahmad F, Tak H, Bandh SA. Veterinary Parasitology in vitro and in vivo anthelmintic activity of Euphorbia helioscopia L. Vet Parasitol 2012; 189(2–4):317–21. 10.1016/j.vetpar.2012.04.023. [DOI] [PubMed] [Google Scholar]
- [98].Lipinski CA. “Lead- and drug-like compounds: the rule-of-five revolution,” Drug Discov. Today Technol Dec. 2004;1(4):337–41. 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- [99].Onguéné PA, et al. The potential of anti-malarial compounds derived from African medicinal plants, part III: an in silico evaluation of drug metabolism and pharmacokinetics profiling. Org. Med. Chem. Lett. September. 2014;4(1):1–9. 10.1186/s13588-014-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ntie-Kang F, et al. ConMedNP “. A natural product library from Central African medicinal plants for drug discovery. RSC Adv 2014;4(1):409–19. 10.1039/c3ra43754j. [DOI] [Google Scholar]
- [101].Doogue MP, Polasek TM. The ABCD of clinical pharmacokinetics. Ther. Adv. Drug Saf. 2013;4(1):5–7. 10.1177/2042098612469335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Li M, et al. Pungent ginger components modulate human cytochrome P450 enzymes in vitro. Nat. Publ. Gr. 2013;34(9):1237–42. 10.1038/aps.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ogihara T, et al. What kinds of substrates show P-glycoprotein-dependent intestinal absorption? Comparison of verapamil with vinblastine. Drug Metabol Pharmacokinet June. 2006;21(3):238–44. 10.2133/dmpk.21.238. [DOI] [PubMed] [Google Scholar]
- [104].Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin Pharmacokinet 2003;42(1):59–98. 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- [105].Kerboeuf D, Blackhall W, Kaminsky R, von Samson-Himmelstjerna G. P- glycoprotein in helminths: function and perspectives for anthelmintic treatment and reversal of resistance. Int J Antimicrob Agents September. 2003;22(3):332–46. 10.1016/s0924-8579(03)00221-8. [DOI] [PubMed] [Google Scholar]
- [106].K Vuppala P, Janagam DR, Balabathula P. Importance of ADME and bioanalysis in the drug discovery. J. Bioequiv. Availab. June. 2013;5(4). 10.4172/jbb.10000e31. [DOI] [Google Scholar]
- [107].Lynch T, Neff AP. The effect of cytochrome P450 metabolism on drug response,interactions, and adverse effects. Am Fam Physician August. 2007;76(3):391–6. [PubMed] [Google Scholar]
- [108].Li AP. Accurate prediction of human drug toxicity: a major challenge in drug development. Chem Biol Interact 2004;150(1):3–7. 10.1016/j.cbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- [109].Mazumder M, et al. Investigations on binding pattern of kinase inhibitors with PPAR γ: molecular docking, molecular dynamic simulations, and free energy calculation studies. PPAR Res 2017;2017. 10.1155/2017/6397836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Musyoka TM, Kanzi AM, Lobb KA, Bishop ÖTastan. Structure based docking and molecular dynamic studies of plasmodial cysteine proteases against a South African natural compound and its analogs. Sci Rep 2016;6:1–12. 10.1038/srep23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Falsafi-Zadeh S, Karimi Z, Galehdari H. VMD DisRg: new User-Friendly Implement for calculation distance and radius of gyration in VMD program. Bioinformation 2012;8(7):341–3. 10.6026/97320630008341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lobanov MI, Bogatyreva NS, V Galzitskaia O. Radius of gyration is an indicator of compactness of protein structure. Mol. Biol. (Mosk). 2008;42(4):701–6. 10.1134/S0026893308040195. [DOI] [PubMed] [Google Scholar]
- [113].Wang C, Greene D, Xiao L, Qi R, Luo R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2018;4:87. 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kwofie SK, et al. Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput Biol Med 2019;113: 103414. 10.1016/j.compbiomed.2019.103414. [DOI] [PubMed] [Google Scholar]
- [115].Broni E, Kwofie SK, Asiedu SO, Miller WA, Wilson MD. “A molecular modeling approach to identify potential antileishmanial compounds against the cell division cycle (Cdc)-2-related kinase 12 (crk12) receptor of leishmania donovani. Biomolecules March. 2021;11(3):1–32. 10.3390/biom11030458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ertl P, Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions 2009;11: 1–11. 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Islam A, Plillay T. Identification of promising anti-DNA Gyrase antibacterial compounds using de novo design, molecular docking and molecular dynamics studies. J Biomol Struct Dyn 2019. 10.1080/07391102.2019.1617785. vol. 0, no. 0, p. 000. [DOI] [PubMed] [Google Scholar]
- [118].Rudmann DG. On-target and off-target-based toxicologic effects. Toxicol Pathol February. 2013;41(2):310–4. 10.1177/0192623312464311. [DOI] [PubMed] [Google Scholar]
- [119].Lim H, Poleksic A, Xie L. Exploring landscape of drug-target-pathway-side effect associations. AMIA Jt. Summits Transl. Sci. Proc. AMIA Jt. Summits Transl. Sci May 2018;2017:132–41 [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/29888057. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.