Abstract
Sequence specific recognition and functional inhibition of biomedically relevant double-helical RNAs is highly desirable, but remains a formidable problem. The present study demonstrates that electroporation of a triplex-forming peptide nucleic acid (PNA), modified with 2-aminopyridine (M) nucleobases, inhibited maturation of endogenous microRNA-197 in SH-SY5Y cells, while having little effect on maturation of microRNA-155 or −27a. In vitro RNA binding and Dicer inhibition assays suggested that the observed biological activity was most likely due to a sequence-specific PNA-RNA triplex formation that inhibited the activity of endonucleases responsible for microRNA maturation. The present study is the first example of modulation of activity of endogenous non-coding RNA using M-modified triplex-forming PNA.
Graphical Abstract
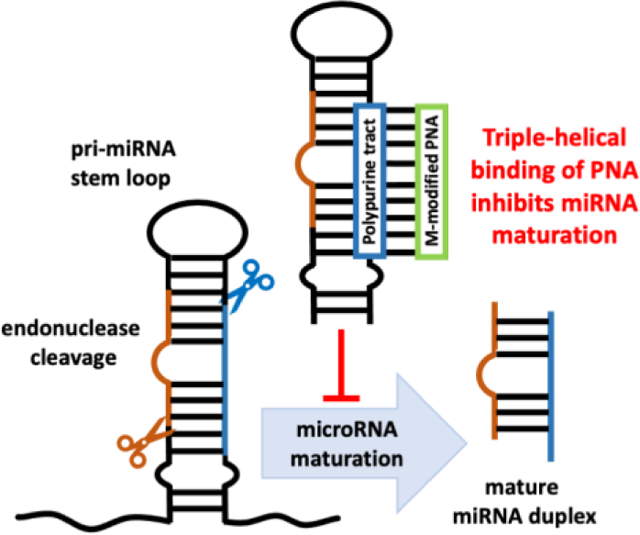
Peptide nucleic acids (PNAs) are DNA mimics that bind single- and double-stranded DNA and RNA with high affinity and sequence selectivity.1–4 When binding to polypurine tracts in double-stranded DNA (dsDNA), PNA can form either a 1:1 PNA-DNA triple-helix or a 2:1 PNA-DNA strand-invasion triplex that displaces the pyrimidine rich strand of DNA as a P-loop.2, 5–7 Binding of PNA to double-stranded RNA (dsRNA) remained unexplored until 2010, when Rozners and co-workers8 showed that PNA formed unusually strong and sequence selective triple helix with dsRNA through the major groove Hoogsteen hydrogen-bonding (Figure 1). Substitution of cytosine (pKa ~4.5) with the more basic 2-aminopyridine (M, pKa ~6.7) enabled strong binding of PNAs at physiologically relevant conditions, including pH 7.4.9 Interestingly, binding of the M-modified PNAs to dsRNA was at least an order of magnitude stronger than to the same sequence of dsDNA.9–12 These results suggested a hypothesis that M-modified triplex-forming PNAs might sequence-specifically recognize and functionally control dsRNA in biological systems. In this communication, we confirm this hypothesis by showing that triple-helical binding of M-modified PNA was able to inhibit the maturation of an endogenous microRNA (miRNA or miR), miR-197 in SH-SY5Y cells.
Figure 1.
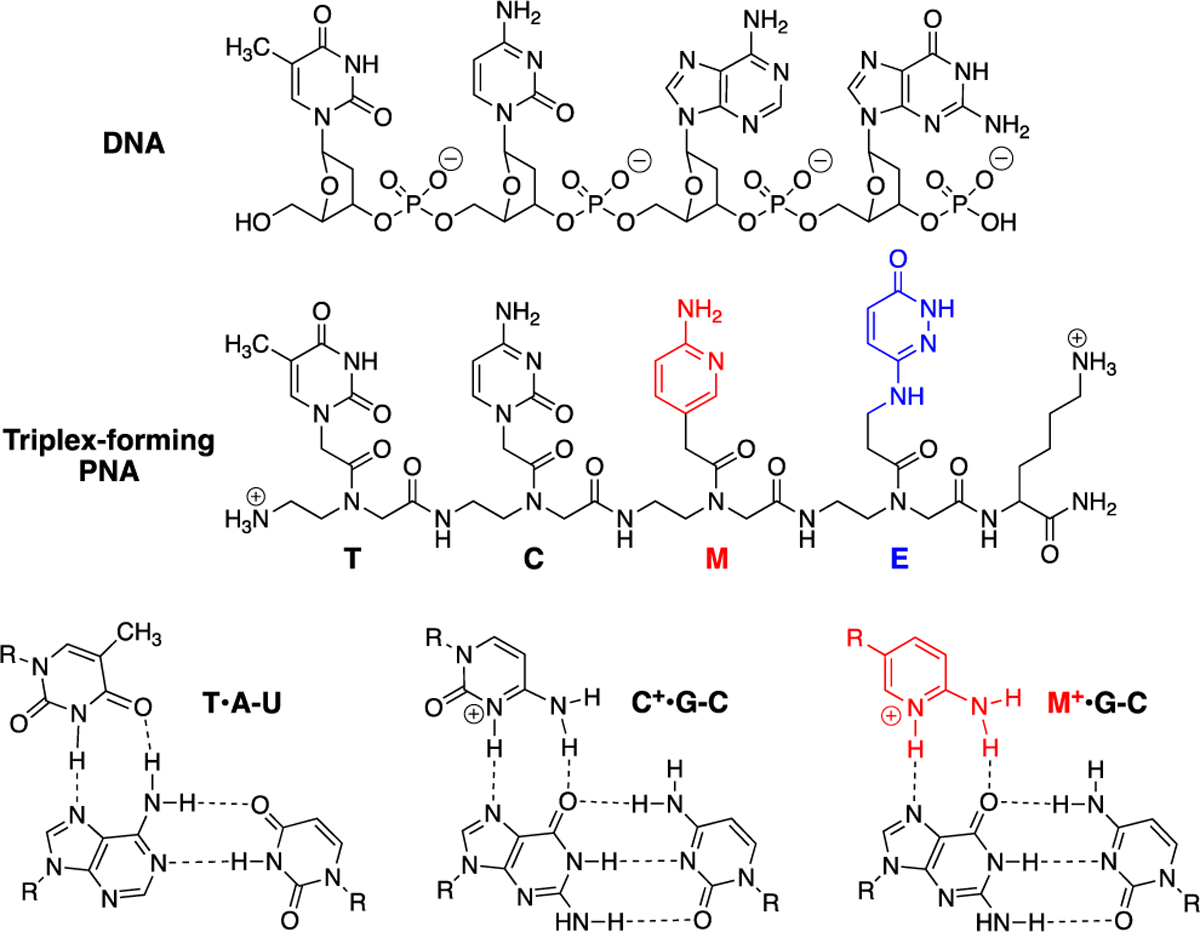
Structures of DNA, triplex-forming PNA, and Hoogsteen hydrogen-bonded base triplets.
In an earlier study,13 we used ~1200 nucleotides long reporter mRNAs encoding Renilla luciferase (RLuc) as a model system to study PNA-dsRNA triplex in vitro and in cells. The mRNAs had an artificially designed hairpin in the 5′-UTR that contained a purine rich target site for a triplex-forming 9-mer PNA (nine nucleotides long). The 9-mer PNA suppressed in vitro translation of RLuc from the matched mRNA but not from the mutant mRNA that had a single mismatch in the PNA binding site.13 After electroporation in MCF7 cells, the 9-mer PNA reduced RLuc translation from a plasmid containing the matched mRNA (but not mutant) by ~60%.13 These encouraging preliminary results suggested that PNAs had an underappreciated capability to recognize and control the function of structured RNAs in cells and prompted us to explore if PNA-dsRNA triple helix formation may be also able to modulate an endogenous biological process. We chose biogenesis of miRNAs as the test system for our PNAs because the biology of these small double-stranded regulatory RNAs is relatively well understood.
MiRNAs are small non-coding RNAs that influence all developmental processes and progress of diseases, especially cancer, by regulating expression of most mRNAs.14 Canonical miRNAs are transcribed as long primary miRNAs (pri-miRNAs) and processed in nucleus by Microprocessor, a heterotrimeric protein complex consisting of one molecule of the Drosha endonuclease and two molecules of DGCR8 protein, which liberates ~60 nucleotides long hairpin precursor miRNA (pre-miRNA). Pre-miRNA is exported to the cytoplasm by Exportin 5 and the hairpin loop is cleaved off by Dicer endonuclease to liberate the mature miRNA, which is loaded in Argonaute protein followed by expulsion of the passenger strand and retention of the guide strand, to form the RNA-induced silencing complex (RISC). The biogenesis of miRNAs is tightly controlled at all of these steps15, 16 and has been explored as a new target for traditional small molecule17, 18 and oligonucleotide drug development.19, 20
At the outset of our study, we hypothesized that triple-helical binding of PNA1 to the polypurine tract (underlined italic in Figure 2A) of miR-197 precursors (either pri-miRNA or pre-miRNA) would prevent their recognition and processing by Drosha and Dicer, the endonucleases responsible for miRNA maturation. We chose miR-197, a putative onco-miRNA, because it has a short polypurine tract (eight purines interrupted by one U) suitable for triplex formation. Downregulation of miR-197 induces apoptosis in cancer cells.21 miR-197 is also implicated in inflammatory skin disorders and autism spectrum disorders through modulation of cellular migration and differentiation.22, 23 The PNA design having four lysine residues (one at amino and three at carboxyl end) was motivated by our previous studies10, 11 showing that lysine conjugation improved cellular uptake of PNAs and increased their affinity for target RNA without decreasing sequence specificity. For recognition of U interrupting the polypurine tract of miR-197 hairpins, we used 3-oxo-2,3-dihydropyridazine (E nucleobase, Figure 1) originally developed by Nielsen and co-workers24 for E•T-A triplets in DNA. We25 and others26 found that E forms stable and selective E•U-A triplets in RNA; however, the exact hydrogen bonding scheme has not been determined.27
Figure 2.
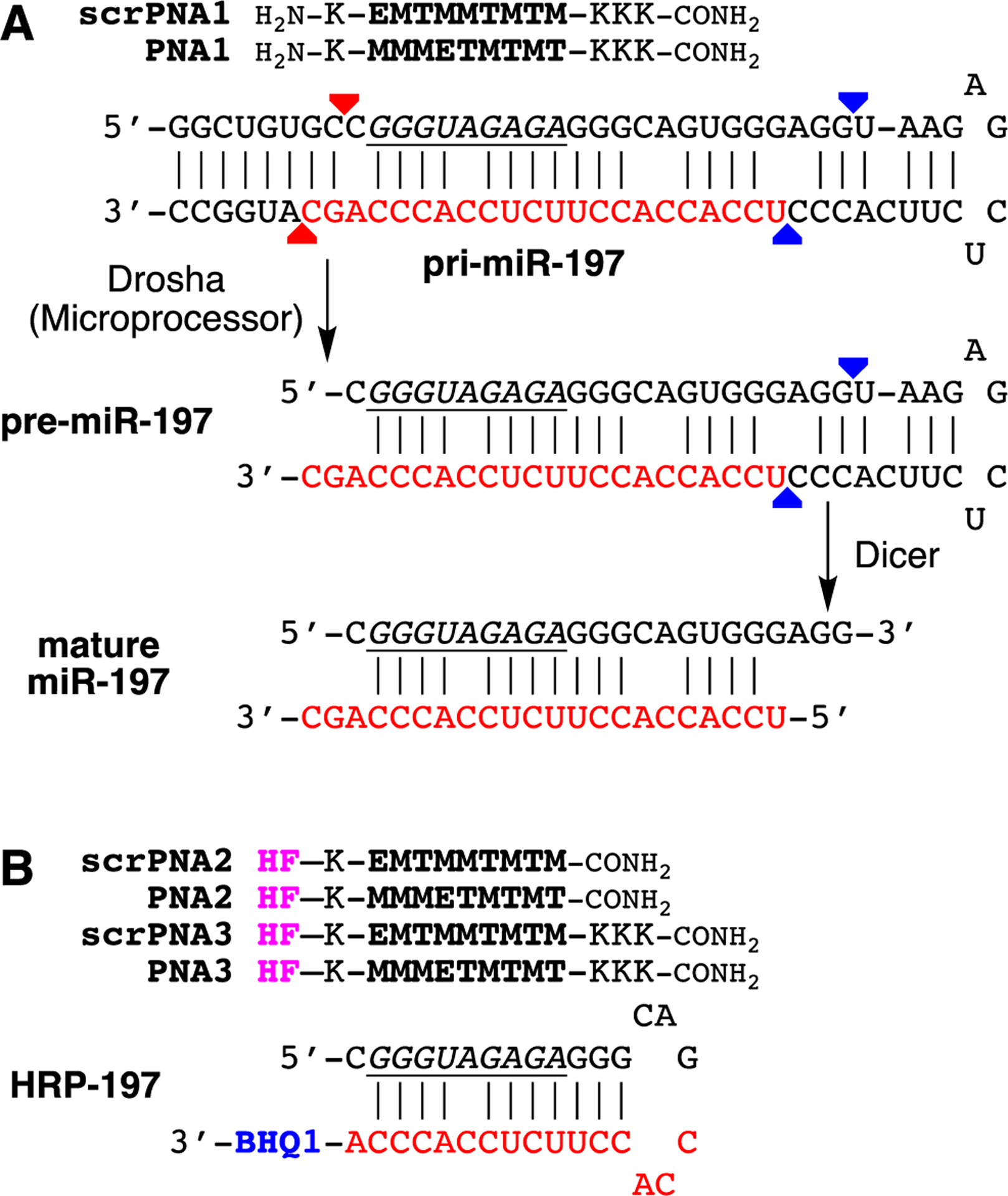
(A) Sequences of triplex-forming PNAs and pri-miR-197 hairpin. The mature miRNA is indicated in red, the Dicer and Drosha cleavage sites are shown with blue and red arrows, respectively, and the PNA binding site is highlighted in underlined italic. (B) Sequences of fluorescently labeled triplex-forming PNAs and HRP-197 modelling the purine rich PNA binding site of pri-miR-197 used in in vitro binding assay. Abbreviations: scr, scrambled PNA sequence; HF, HiLyte Fluor 488 dye; BHQ1, Black Hole Quencher 1.
We started testing our hypothesis with in vitro confirmation that PNA formed a sequence specific triple helix with the polypurine tract of pri-miR-197 hairpin. The target site in pri-miR-197 hairpin has one A-C mismatched base pair unlike the fully matched dsRNA, which has been customarily used as PNA target.11 However, our previous studies9, 10 have suggested that M-modified triplex-forming PNAs were able to recognize non-canonical and mismatched base pairs in dsRNA hairpins related to pri-miR-197. To confirm this expectation, we used PNA2 labeled with HiLyte Fluor 488 dye28 (HF in Figure 2B) and HRP-197 modified with black hole quencher 1 (BHQ1) following the methodology developed in our previous studies.13, 29 The hairpin structure of HRP-197 models the PNA binding site (underlined italic in Figure 2) of pri-miR-197. Titration of PNA2 with HRP-197 resulted in a gradual decrease of fluorescence intensity (Figure 3) that was fit to bimolecular binding model giving a value of equilibrium association constant at 37 °C (KA 37) at 1.66 ± 0.48 × 109 M−1. Titration of the PNA having a scrambled sequence, scrPNA2 with HRP-197 gave ~25-fold weaker binding with KA 37 value at 0.06 ± 0.01 × 109 M−1. Similar results were obtained with PNA3 (KA 37 = 2.76 ± 0.38 × 109 M−1) and scrPNA3 (KA 37 = 0.14 ± 0.02 × 109 M−1) that had three additional lysine residues attached to the carboxy end. These results were consistent with our previous studies9–11 and confirmed that cationic triplex-forming PNAs had high affinity and sequence selectivity for binding dsRNA even when the target site contains a mismatch base pair.
Figure 3.
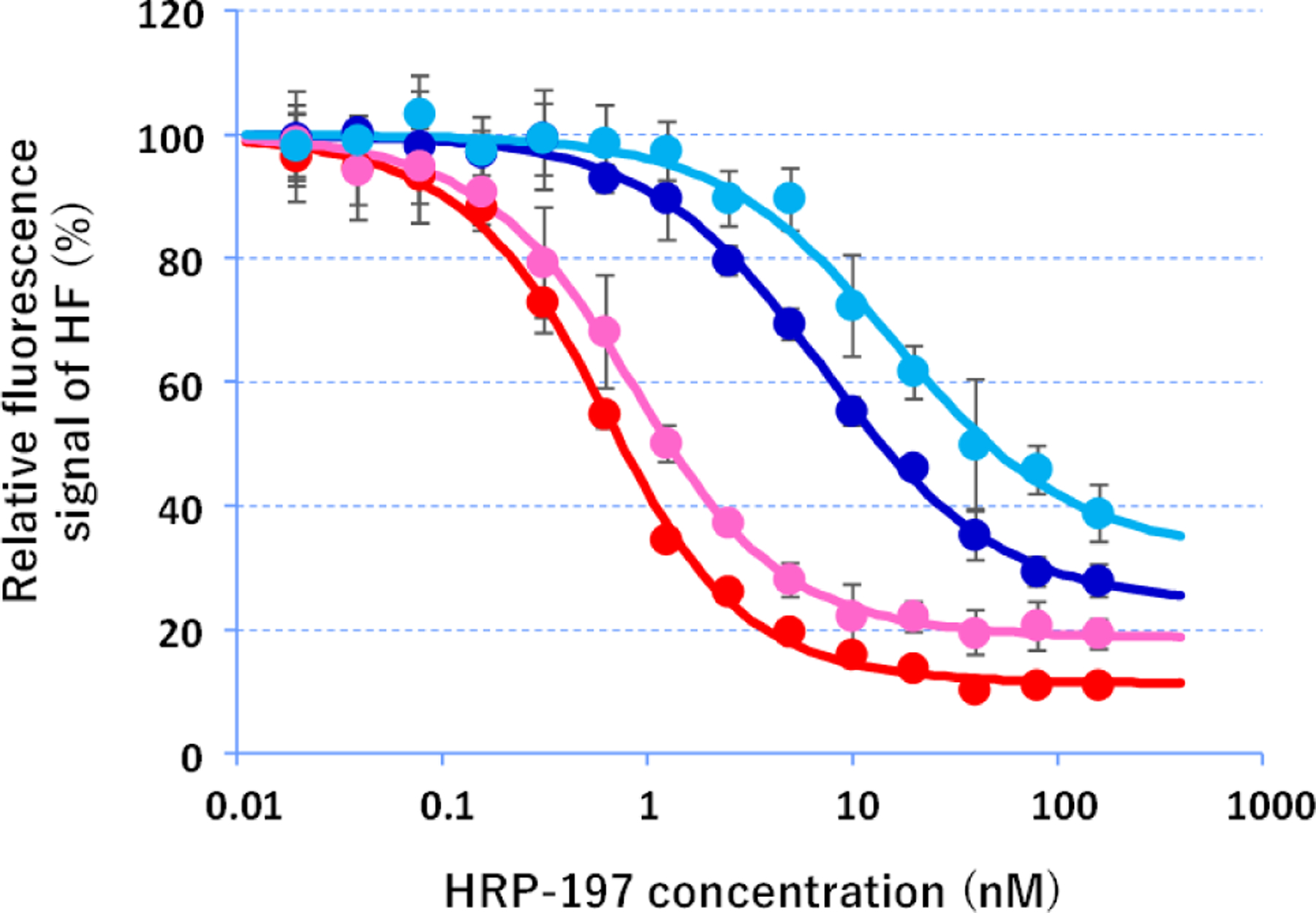
In vitro binding of PNAs to HRP197. Fluorescently-labeled PNA2 (pink), scrPNA2 (light blue), PNA3 (red), or scrPNA3 (dark blue) at 500 pM were mixed with various concentrations of BHQ1-modified HRP-197 in a buffer containing 30 mM HEPES-KOH (pH 7), 100 mM KCl, 10 ng/μL tRNA, and 0.01% CHAPS. Fluorescence signals were measured at 37 °C using 490 nm excitation and 530 nm emission. Values are means ± standard deviation of triplicate samples. Lines indicate theoretical signal change obtained by fitting the experimental data.
Next, we tested if the triple helix formation could inhibit activity of Dicer enzyme that processes miRNA precursors. A part of pri-miR-197 hairpin (Figure 2A), containing a sequence of the stem loop of hsa-mir-197 registered in miRBase,30, 31 was labeled with Cy3 dye at its 3′ end (250 nM) and treated with human recombinant Turbo Dicer enzyme for 3 h at 37 °C. Fluorescence imaging of Cy3 signals after denaturing PAGE and subsequent SYBR Gold staining of the gel showed increasing cleavage of the pri-miR-197 hairpin dependent on the enzyme concentration (Figure S7). Based on this result, Cy3-labeled pri-miR-197 hairpin was treated with 0.1 unit/μL of the enzyme in the presence or absence of 500 nM PNA, and inhibition of the enzymatic reaction was evaluated after gel electrophoresis (Figure 4). Matched PNA1 (but not the scrambled scrPNA1) efficiently inhibited Dicer cleavage of the pri-miR-197. These results confirmed that PNA binding was able to interfere with cleavage of pri-miR hairpins by Dicer.
Figure 4.
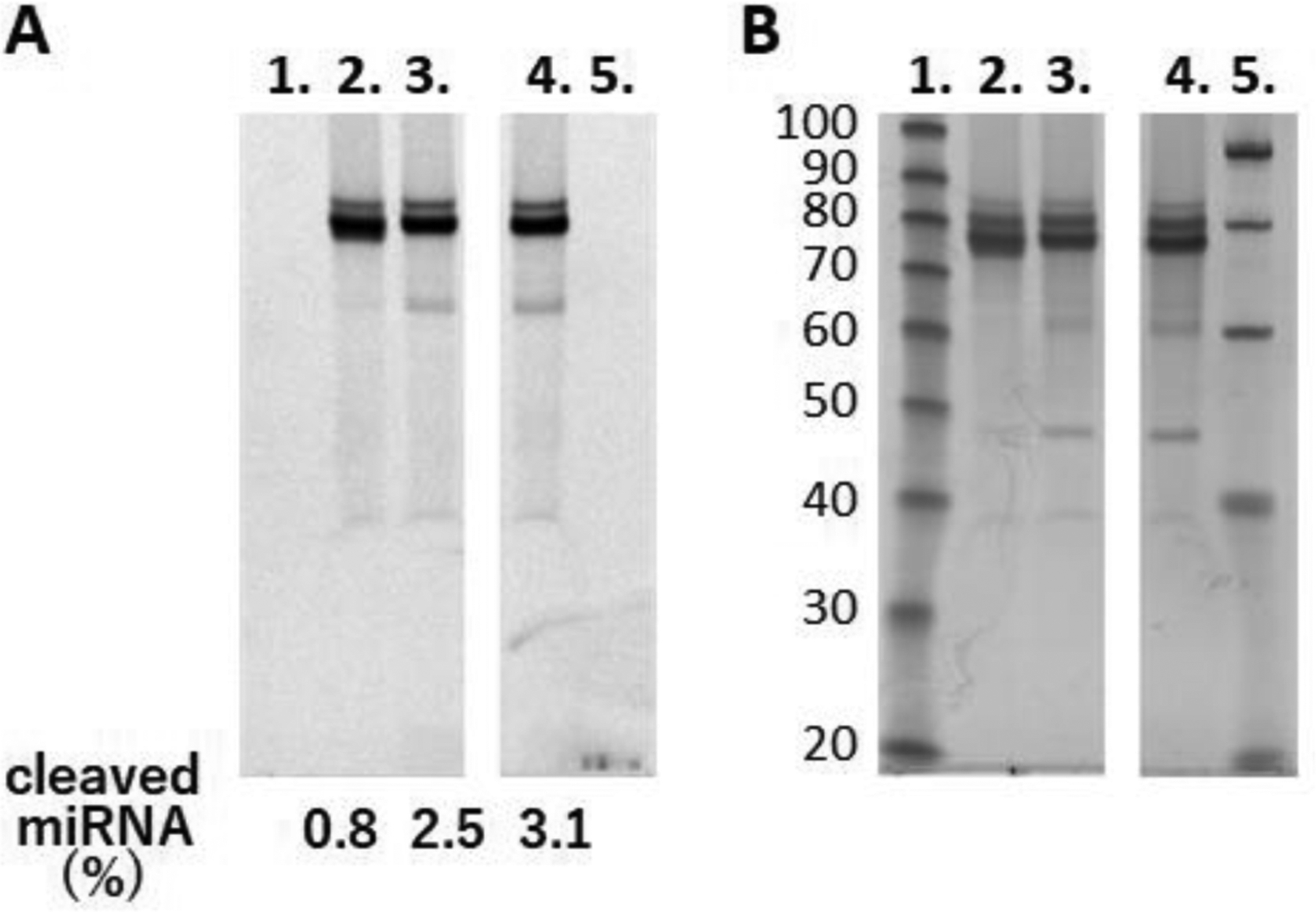
Inhibition of Dicer-mediated cleavage of pri-miR-197 by PNA. Products of Dicer cleavage reaction were separated by denaturing PAGE and imaged by (A) Cy3 fluorescence and (B) SYBR Gold staining. Samples are 10 base pair ladder (lane 1), reaction mixtures with PNA1 (lane 2), with scrPNA1 (lane 3), and without PNA (lane 4), and 20 base pair ladder (lane 5). Ratio values of the cleaved product calculated from fluorescence signals of Cy3 are shown under the gel image.
To confirm our hypothesis that sequence specific triplex formation can inhibit maturation of endogenous miRNA, we electroporated PNA1 and scrPNA1 into SH-SY5Y cells, a human neuroblastoma cell line known to express sufficient levels of miR-197.32 After 72 h-incubation, the level of mature miR-197 was evaluated using qRT-PCR. In addition, we selected two miRNAs, miR-155 and miR-27a, which compared to miR-197 are expressed in SH-SY5Y cells at relatively high and low levels, respectively, as controls to monitor nonspecific variations in mature miRNA levels (Figures S8a to S8c). Multiplex reverse transcription (RT) was performed using RT-primers for respective miRNAs to reduce handling deviations of the RT samples.33 We obtained linear standard curves for all miRNA targets by using the same dilution series of the RT product (Figure S8d). The results in Figure 5 confirmed that PNA1 significantly reduced levels of mature miR-197 in a sequence specific manner. As expected, PNA1 did not reduce the levels of either mature miR-155 (Figure 5A) or mature miR-27a (Figure 5B). When the expression level of miR-197 was normalized for that of miR-155, PNA1 inhibited mature miR-197 to ~30% compared to cells not treated with PNA (dark red rhombs in Figure 5A). Normalization for miR-27a gave a similar ~30% inhibition of miR-197 (Figure S9). The control scrPNA1 did not affect the expression level of miR-197 normalized for miR-155, although the standard deviations were larger than for PNA1 or PNA(−).
Figure 5.
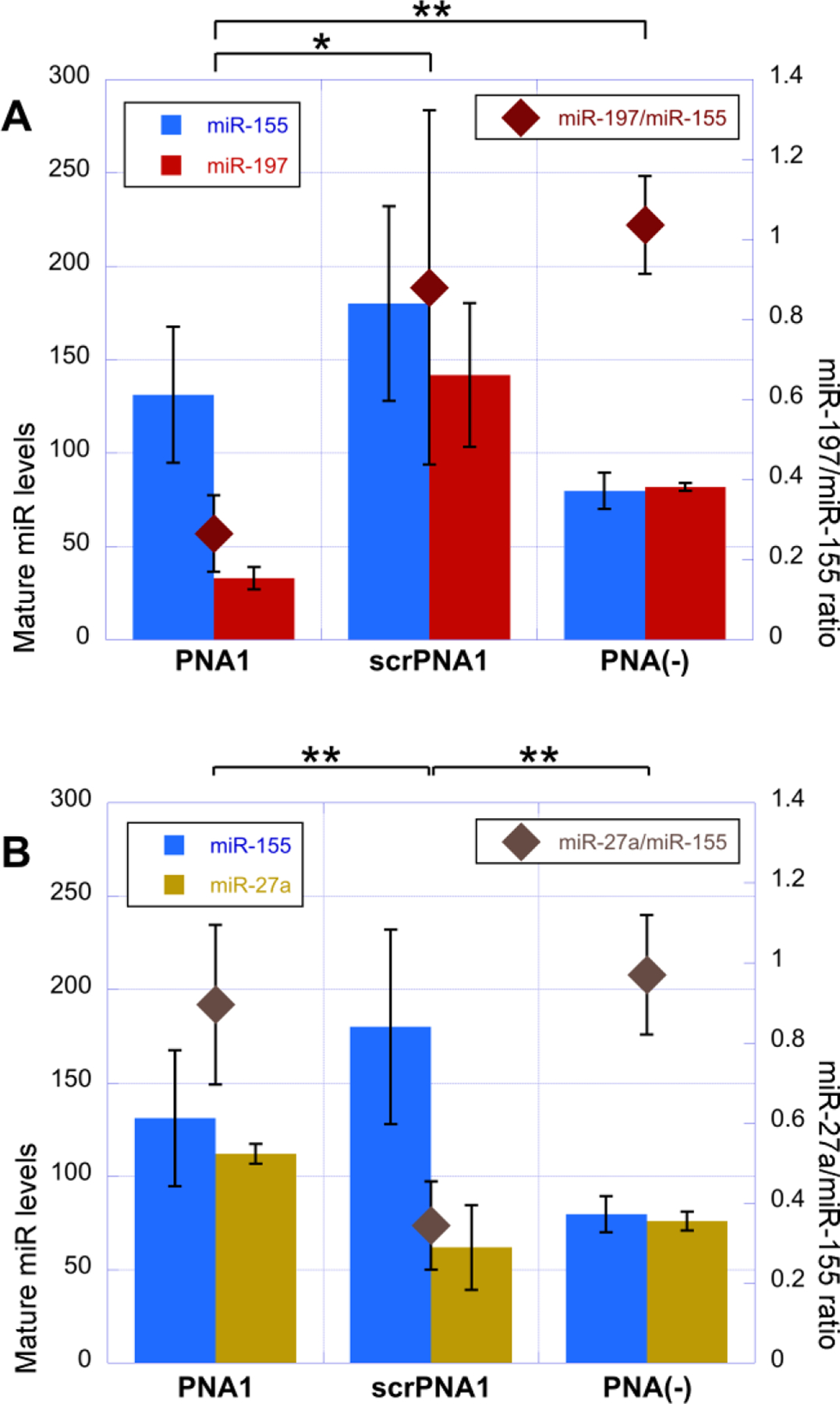
Levels of mature miRNA in cells after 72 hours treated with PNAs. (A) Levels of miR-155 (blue) and miR-197 (red) are represented on left axis, and their relative values (dark red) are plotted on right axis. (B) Levels of miR-155 (blue) and miR-27a (dark yellow) are represented on left axis, and their relative values (brown) are plotted on right axis. Values are means ± standard deviations of experiments replicated four times. Asterisk and double asterisks indicate two-tailed P values of less than 0.1 and 0.02, respectively, as calculated using Student’s t-test.
Somewhat unexpectedly, scrPNA1 selectively decreased the relative level of miR-27a (Figures 5B and S9). Closer inspection of pri-miR-27a hairpin revealed that scrPNA1 had a partial sequence match of seven nucleobases with pri-miR-27a hairpin (Figure S10) that could explain the apparent inhibition of miR-27a maturation. The larger errors correlated with decreased cell proliferation after electroporation of scrPNA1 (Figure S11), most likely due to apoptosis, which is a reported result of inhibition of miR-27a.34, 35 A bioinformatics survey of H. Sapiens miRNAs in the miRBase30, 31 revealed that the nine nucleobase target of PNA1 was unique in miR-197. However, a search for partial 7-mer complementarity (as with the pri-miR-27a hairpin) found additional four matches for PNA1 and a total of ten matches for scrPNA1 (Table S8). These results illustrate the importance of careful sequence design that minimizes off-target sites for triplex-forming PNAs.
In a related project that explores detection of post-transcriptional adenosine to inosine RNA editing, we observed that the fluorophore labeled PNA2 also reduced the levels of mature miR-197 in SH-SY5Y and human cervix epithelioid carcinoma (HeLa) cell lines (Figure S12). Collectively, the qRT-PCR results confirmed our hypothesis that triplex-forming PNA is able to inhibit maturation of endogenous pri-miRNA hairpins in live cells.
Overall, we noted that all non-targeting PNAs somewhat enhanced expression of miRNAs (c.f., PNA1 and srPNA1 with PNA(−) in Figure 5). We do not have a compelling explanation for this phenomenon, but it is conceivable that PNAs had some subtle non-specific stimulatory effect on complex protein-RNA interactions that regulate miRNA maturation.15, 16
While the PNAs have untapped potential as therapeutic compounds, we envision that the immediate applications of the triplex-forming PNAs may be as broadly useful tools to study structure and function of various non-coding RNAs. In this study, we used qRT-PCR to confirm that triplex-forming PNA efficiently inhibited maturation of endogenous miR-197 in SH-SY5Y cells. This finding is important as the first example of modulation of activity of endogenous non-coding RNA using M-modified triplex-forming PNA. Taken together with our previous studies,9–13 the current results confirm that M-modified triplex-forming PNAs are uniquely suited for sequence-specific recognition of dsRNA in biological systems.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Tatsuya Ohyama for the bioinformatics survey of potential PNA targets.
Funding Sources
This work was supported by U.S. National Science Foundation (CHE-1708761 to E.R.), U.S. National Institutes of Health (R35 GM130207 to E.R.), Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant No. 17H06351, 18KK0164, and 19K05723), Japan, The Hirao Taro Foundation of KONAN GAKUEN for Academic Research, and Chubei Itoh Foundation.
Footnotes
Supporting Information Available. This material is available free of charge via the Internet. General experimental procedures, synthesis, purification, and LC-MS characterization of PNA oligomers, and details of qRT-PCR procedures.
REFERENCES
- (1).Buchardt O, Egholm M, Berg RH, and Nielsen PE (1993) Peptide nucleic acids and their potential applications in biotechnology, Trends Biotechnol. 11, 384–386. [DOI] [PubMed] [Google Scholar]
- (2).Nielsen PE, Egholm M, Berg RH, and Buchardt O (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide, Science 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- (3).Egholm M, Buchardt O, Nielsen PE, and Berg RH (1992) Peptide nucleic acids (PNA). Oligonucleotide analogs with an achiral peptide backbone, J. Am. Chem. Soc 114, 1895–1897. [Google Scholar]
- (4).Egholm M, Nielsen PE, Buchardt O, and Berg RH (1992) Recognition of guanine and adenine in DNA by cytosine and thymine containing peptide nucleic acids (PNA), J. Am. Chem. Soc 114, 9677–9678. [Google Scholar]
- (5).Wittung P, Nielsen P, and Norden B (1997) Extended DNA-recognition repertoire of peptide nucleic acid (PNA): PNA-dsDNA triplex formed with cytosine-rich homopyrimidine PNA, Biochemistry 36, 7973–7979. [DOI] [PubMed] [Google Scholar]
- (6).Bentin T, Hansen GI, and Nielsen PE (2006) Structural diversity of target-specific homopyrimidine peptide nucleic acid-dsDNA complexes, Nucleic Acids Res. 34, 5790–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hansen ME, Bentin T, and Nielsen PE (2009) High-affinity triplex targeting of double stranded DNA using chemically modified peptide nucleic acid oligomers, Nucleic Acids Res. 37, 4498–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Li M, Zengeya T, and Rozners E (2010) Short Peptide Nucleic Acids Bind Strongly to Homopurine Tract of Double Helical RNA at pH 5.5, J. Am. Chem. Soc 132, 8676–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zengeya T, Gupta P, and Rozners E (2012) Triple Helical Recognition of RNA Using 2-Aminopyridine-Modified PNA at Physiologically Relevant Conditions, Angew. Chem., Int. Ed 51, 12593–12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Muse O, Zengeya T, Mwaura J, Hnedzko D, McGee DW, Grewer CT, and Rozners E (2013) Sequence Selective Recognition of Double-Stranded RNA at Physiologically Relevant Conditions Using PNA-Peptide Conjugates, ACS Chem. Biol 8, 1683–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hnedzko D, McGee DW, Karamitas YA, and Rozners E (2017) Sequence-selective recognition of double-stranded RNA and enhanced cellular uptake of cationic nucleobase and backbone-modified peptide nucleic acids, RNA 23, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ryan CA, and Rozners E (2020) Impact of Chirality and Position of Lysine Conjugation in Triplex-Forming Peptide Nucleic Acids, ACS Omega 5, 28722–28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Endoh T, Hnedzko D, Rozners E, and Sugimoto N (2016) Nucleobase-Modified PNA Suppresses Translation by Forming a Triple Helix with a Hairpin Structure in mRNA In Vitro and in Cells, Angew. Chem., Int. Ed 55, 899–903. [DOI] [PubMed] [Google Scholar]
- (14).Bartel DP (2018) Metazoan microRNAs, Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ha M, and Kim VN (2014) Regulation of microRNA biogenesis, Nat. Rev. Mol. Cell Biol 15, 509–524. [DOI] [PubMed] [Google Scholar]
- (16).Treiber T, Treiber N, and Meister G (2019) Regulation of microRNA biogenesis and its crosstalk with other cellular pathways, Nat. Rev. Mol. Cell Biol 20, 5–20. [DOI] [PubMed] [Google Scholar]
- (17).Disney MD (2019) Targeting RNA with Small Molecules To Capture Opportunities at the Intersection of Chemistry, Biology, and Medicine, J. Am. Chem. Soc 141, 6776–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Li Y, and Disney MD (2018) Precise Small Molecule Degradation of a Noncoding RNA Identifies Cellular Binding Sites and Modulates an Oncogenic Phenotype, ACS Chem. Biol 13, 3065–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li Z, and Rana TM (2014) Therapeutic targeting of microRNAs: current status and future challenges, Nat. Rev. Drug Discovery 13, 622–638. [DOI] [PubMed] [Google Scholar]
- (20).Cha W, Fan R, Miao Y, Zhou Y, Qin C, Shan X, Wan X, and Cui T (2018) MicroRNAs as novel endogenous targets for regulation and therapeutic treatments, MedChemComm 9, 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, and De Maria R (2014) Antitumor effect of miR-197 targeting in p53 wild-type lung cancer, Cell Death Differ. 21, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lerman G, Sharon M, Leibowitz-Amit R, Sidi Y, and Avni D (2014) The crosstalk between IL-22 signaling and miR-197 in human keratinocytes, PloS one 9, e107467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang YM, Zheng YF, Yang SY, Yang ZM, Zhang LN, He YQ, Gong XH, Liu D, Finnell RH, Qiu ZL, Du YS, and Wang HY (2019) MicroRNA-197 controls ADAM10 expression to mediate MeCP2’s role in the differentiation of neuronal progenitors, Cell Death Differ. 26, 1863–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Eldrup AB, Dahl O, and Nielsen PE (1997) A Novel Peptide Nucleic Acid Monomer for Recognition of Thymine in Triple-Helix Structures, J. Am. Chem. Soc 119, 11116–11117. [Google Scholar]
- (25).Gupta P, Zengeya T, and Rozners E (2011) Triple helical recognition of pyrimidine inversions in polypurine tracts of RNA by nucleobase-modified PNA, Chem. Commun 47, 11125–11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kim KT, Chang D, and Winssinger N (2018) Double-Stranded RNA-Specific Templated Reaction with Triplex Forming PNA, Helv. Chim. Acta 101, e1700295. [Google Scholar]
- (27).Ong AAL, Toh D-FK, Patil KM, Meng Z, Yuan Z, Krishna MS, Devi G, Haruehanroengra P, Lu Y, Xia K, Okamura K, Sheng J, and Chen G (2019) General recognition of U-G, U-A, and C-G pairs by double-stranded RNA-binding PNAs incorporated with an artificial nucleobase, Biochemistry 58, 1319–1331. [DOI] [PubMed] [Google Scholar]
- (28).Hnedzko D, McGee DW, and Rozners E (2016) Synthesis and properties of peptide nucleic acid labeled at the N-terminus with HiLyte Fluor 488 fluorescent dye, Bioorg. Med. Chem 24, 4199–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Endoh T, Annoni C, Hnedzko D, Rozners E, and Sugimoto N (2016) Triplex-forming PNA modified with unnatural nucleobases: the role of protonation entropy in RNA binding, Phys. Chem. Chem. Phys 18, 32002–32006. [DOI] [PubMed] [Google Scholar]
- (30).Griffiths‐Jones S (2004) The microRNA Registry, Nucleic Acids Res. 32, D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, and Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature, Nucleic Acids Res. 34, D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Flores O, Kennedy EM, Skalsky RL, and Cullen BR (2014) Differential RISC association of endogenous human microRNAs predicts their inhibitory potential, Nucleic Acids Res. 42, 4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Le Carré J, Lamon S, and Léger B (2014) Validation of a multiplex reverse transcription and pre-amplification method using TaqMan® MicroRNA assays, Front. Genet 5, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C, Dorsey SG, and Faden AI (2014) Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins, J. Neurosci 34, 10055–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Su C, Huang D-P, Liu J-W, Liu W-Y, and Cao Y-O (2019) miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1, Oncol. Lett 18, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


