Abstract
The loss of functional β cell mass contributes to development and progression of type 2 diabetes (T2D). However, the molecular mechanisms differentiating islet dysfunction in T2D from nondiabetic states remain elusive. In this issue of the JCI, Son et al. applied reverse engineering to obtain the activity of gene expression regulatory proteins from single-cell RNA sequencing data of nondiabetic and T2D human islets. The authors identify unique patterns of regulatory protein activities associated with T2D. Furthermore, BACH2 emerged as a potential transcription factor that drives activation of T2D-associated regulatory proteins in human islets.
Pancreatic β cell failure in type 2 diabetes
Type 2 diabetes (T2D) is a chronic and progressive disease that is increasing worldwide (1). Although both insulin resistance and impaired insulin secretion contribute to the development of T2D, the loss of functional β cell mass plays a central role in the progression of the disease (1, 2). T2D risk loci in genome-wide association studies (GWAS) are often related to β cell development and function (3). Longitudinal studies have shown that a decline in insulin secretion determines the conversion from normoglycemia to glucose intolerance to T2D, and the long-term dependence on insulin treatment in T2D (4). Thus, the preservation and restoration of healthy β cells are logical therapeutic goals, yet remain insufficiently addressed by current therapies. The shortcoming of current T2D treatment reflects our limited understanding of the pathology of β cell failure in human T2D (1). For more than 700 T2D risk loci identified by GWAS, each locus has a relatively small effect and some are at noncoding regions without known functions (3). Human T2D islets show activation of multiple stress pathways, including endoplasmic reticulum stress, oxidative stress, and inflammation (1, 5). However, the contribution of each stress pathway to islet failure and the interplay between genetic susceptibility and stress pathways remain ill defined.
Filling a knowledge gap
Single-cell RNA sequencing (scRNA-Seq) of human islet cells has been tapped to fill a knowledge gap in the pathogenesis of β cell failure in T2D (6). However, a consensus for a T2D-specific gene expression signature is unsettled because lists of differentially expressed genes in separate scRNA-Seq data show little agreement (6). In this issue of the JCI, Son, Ding, et al. utilized 6,137 sets of scRNA-Seq data from human pancreatic islet cells. Human islet cells from four donors without diabetes and six donors with T2D were reverse engineered to determine activities of regulatory proteins in each cell (Figure 1) (7). Regulatory proteins included 1,813 transcription factors (TFs), 969 co-TFs, and 3,370 signal-transducing proteins whose activity directly regulates gene expression level. Based on the assumption that an activity of a given regulatory protein (a master regulator protein, MR) determines the expression level of its target genes, transcriptome data were converted into protein activity through two algorithms. Both algorithms were previously validated in cancer cells and others (8, 9). The first, Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNe), analyzes a large set of gene expression data (over 100 required) to identify clusters of genes (regulons) whose expression changes together through direct interaction (8). Once regulons are identified by ARACNe, scRNA-Seq data of each cell are interrogated to assign the activity of approximately 6,000 regulatory proteins by virtual inference of protein activity by enriched regulon analysis (VIPER) (10). VIPER calculates the activity of each regulatory protein by combining activation/repression levels of its target genes; activity will receive a high score if multiple positively regulated genes are highly upregulated and multiple negatively regulated genes are strongly repressed. The current study used metaVIPER, a version of VIPER capable of assigning MR to a regulon incorporating MR and target gene information previously obtained in a wide range of cells (9).
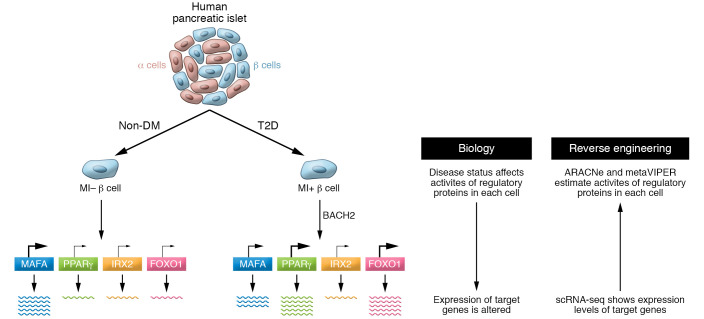
Figure 1. Reverse engineering of single-cell RNA sequencing data profiles regulatory protein activity in human islet cells.
The activity of regulatory proteins in each human islet cell changes, depending on factors such as cell identity (α vs. β cells) and disease status (nondiabetic [non-DM] vs. T2D). Son, Ding, et al. (7) used reverse engineering to extrapolate the activity of each regulatory protein from the expression level of target genes. Gene expression profiles were obtained by scRNA-Seq analysis, and regulatory protein activity was predicted using two algorithms, ARACNe and metaVIPER. Metabolically inflexible (MI) β cells showed transcription factor (TF) and co-TF activity changes, as exemplified in this model by increases in PPARγ and FOXO1 activities in T2D islets. Notably, BACH2 was implicated as one driver of T2D regulatory protein activities.
The activities of MAFA (β cell TF) and IRX2 (α cell TF) assigned to each islet cell showed high correlation with INS (insulin) and GCG (glucagon) mRNA expression, respectively, validating the performance of ARACNe and metaVIPER in processing human islet scRNA-Seq data.
What are the advantages of converting scRNA-Seq data to activity of regulatory proteins? A major limitation of scRNA-Seq is the substantial proportion of genes that remain unassessed (gene dropout) due to low sequencing depth (6). The metaVIPER algorithm can assess activity for regulatory proteins whose expression levels are too low for detection by scRNA-Seq (7). Moreover, the data obtained indicate activity that is more biologically relevant than the expression level of a regulatory protein. Since the activity of each regulatory protein is estimated from approximately 50 target genes, the activity data have high fidelity (9). Importantly, Son, Ding, et al.’s profiling of regulatory protein activities provided a landscape of nondiabetic and T2D human islets, leading to a number of exciting findings (7). First, the authors identified human α and β cell clusters enriched in T2D islets and defined a pattern of regulatory protein activity representative of T2D islets. Second, they found BACH2 as a regulatory protein that drives T2D-islet protein activity signatures. Third, they showed that a small subset of regulatory proteins, such as BACH2, drives T2D disease phenotypes and can be targeted to reverse the process. Indeed, suppression of BACH2 improved β cell function in a mouse T2D model and human T2D islets (7).
Son, Ding, et al. grouped human islet cells into 10 clusters consisting of four β cell– and six α or α cell–like clusters based on profiles of protein activities. The activities of the following key islet regulatory proteins were evaluated to characterize clusters: metabolic inflexibility (MI) and stress drivers (PPARγ, PPARα, FOXO1, etc.), endocrine progenitor markers (RFX6 and RFX7), and stem-like-cell TFs (NANOG, MYCL, and POU5F1) (11, 12) (Figure 1). Interestingly, a major factor that governed clustering was MI. Nondiabetic islets were enriched with metabolically flexible, high-INS-expressing β cells (MI–1) and metabolically inflexible, weak-GCG-expressing α cells (MI+5). In comparison, T2D islets were dominated by metabolically inflexible β cells (MI+2) that also showed high activity of stress response regulatory proteins. For α cells, T2D islets were enriched with metabolically flexible cells that showed progenitor/stem-cell-like features (MI–5), a cluster that may reflect β to α cell transition. With T2D-associated signature in hand, 15 proteins were chosen for their high activities in T2D over nondiabetic islet cells (MI+2 vs. MI–1 for β cells and MI–5 vs. MI+5 for α cells). Single-cell gain-of-function sequencing (scGOF-Seq) of nondiabetic islets overexpressing 15 candidate regulators yielded three promising candidates (BACH2, CUX2, and AFF3) that change protein activities from nondiabetic to a T2D-like profile. When these candidate proteins were downregulated in T2D islets, BACH2 changed the regulatory protein profile from T2D-like to nondiabetic (as determined by Perturb-Seq; ref. 13). BACH2, a member of the BACH family of basic leucine zipper TFs, is highly expressed in lymphocytes and some neuronal cells (14). BACH2 polymorphisms are associated with autoimmune diseases, including type 1 diabetes, which is attributed to a role of BACH2 in the regulation of T lymphocyte function (15). However, BACH2 is also involved in β cell development and stress responses. BACH2 protects β cells from apoptosis under cytokine exposure (16). Altered expression of BACH2 has been seen in dedifferentiated mouse β cells (17). More relevant to T2D, BACH2 and its target genes showed signs of activation in an assay for transposase-accessible chromatin using sequencing (ATAC-Seq) of human T2D islets (18). In the current study, the immunoreactivity of BACH2 was markedly increased in both α and β cells of human pancreas from T2D donors. An increase in Bach2 mRNA levels was also noted in islets isolated from diabetic (db/db) mice. Compound 8, a pharmacologic inhibitor of BACH1 and BACH2, improved glucose homeostasis of db/db mice after 14-day treatment and glucose-stimulated insulin secretion in human T2D islets, indicating that BACH2 mediates β cell dysfunction in T2D.
A leap forward and more work needed
As is often the case for groundbreaking discoveries, Son, Ding, et al. (7) raise issues for future investigation. T2D is a highly heterogeneous disease, with each case at different stages of progression and with different combinations of genetic and environmental stressors. It will be important to test whether the T2D regulatory protein activity profile in the current study is reproducible in other cohorts of T2D scRNA-Seq data. Identification of multiple clusters for β and α cells in the current study agree with heterogeneity of β and α cells defined by transcriptomes in scRNA-Seq (6). Do subclasses of α and β cells defined by protein activity profile correlate with subclasses defined by gene expression? Another limitation is that the candidate regulatory proteins were validated in a limited number of donors. Further studies expanding the number of donors are needed to strengthen the results, determine hierarchy among candidate regulators, and identify additional regulators. The function of BACH2 in the development of β cell failure in T2D and the long-term durability of BACH2 inhibition to reverse β cell failure require further study. While the current study indicates that suppression of BACH2 activity improves β cell function, it has also been reported that BACH2 suppression in β cells exacerbates β cell apoptosis under cytokine exposure (16).
Despite its limitations, the study by Son, Ding, et al. (7) is a substantial leap forward in the use of transcriptome data to understand molecular pathways linking β cell demise to T2D. Camunas-Solar et al. performed scRNA-Seq in human islet cells that were also evaluated by patch clamp to combine gene expression with electrophysiology (19). Bosi et al. integrated three scRNA-Seq data sets and proposed defective insulin secretion, sterol regulatory element–binding protein (SREBP) signaling, and oxidative stress as candidate pathways representing T2D pathogenesis (20), and Wigger et al. obtained a serum lipidome, islet bulk transcriptome, and islet proteome from each donor to allow correlation between the three omics (21). By integrating information obtained from multiple approaches, we will gain a better understanding of the molecular pathways controlling β cell dysfunction in T2D and how these may be targeted for treatment.
Acknowledgments
YI is supported by the NIH (R01-DK090490), Department of Veteran Affairs (I01 BX005107), and Fraternal Orders of Eagles Diabetes Research Center at the University of Iowa.
Version 1. 12/15/2021
Electronic publication
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(24):e154482. https://doi.org/10.1172/JCI154482.
See the related article at BACH2 inhibition reverses β cell failure in type 2 diabetes models.
References
- 1.Halban PA, et al. β-Cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. J Clin Endocrinol Metab. 2014;99(6):1983–1992. doi: 10.1210/jc.2014-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso L, et al. TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 2021;37(2):109807. doi: 10.1016/j.celrep.2021.109807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahn SE. The importance of the beta-cell in the pathogenesis of type 2 diabetes mellitus. Am J Med. 2000;108(suppl 6a):2S–8S. doi: 10.1016/s0002-9343(00)00336-3. [DOI] [PubMed] [Google Scholar]
- 5.Imai Y, et al. Islet inflammation: a unifying target for diabetes treatment? Trends Endocrinol Metab. 2013;24(7):351–360. doi: 10.1016/j.tem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YJ, Kaestner KH. Single-cell RNA-seq of the pancreatic islets--a promise not yet fulfilled? Cell Metab. 2019;29(3):539–544. doi: 10.1016/j.cmet.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Son J, et al. BACH2 inhibition reverses β cell failure in type 2 diabetes model. J Clin Invest. 2021;131(24):e153876. doi: 10.1172/JCI153876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 9.Ding H, et al. Quantitative assessment of protein activity in orphan tissues and single cells using the metaVIPER algorithm. Nat Commun. 2018;9(1):1471. doi: 10.1038/s41467-018-03843-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez MJ, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48(8):838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim-Muller JY, et al. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metab. 2014;20(4):593–602. doi: 10.1016/j.cmet.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benner C, et al. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixit A, et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167(7):1853–1866. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richer MJ, et al. T cell fates zipped up: how the Bach2 basic leucine zipper transcriptional repressor directs T cell differentiation and function. J Immunol. 2016;197(4):1009–1015. doi: 10.4049/jimmunol.1600847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson CC, et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat Genet. 2021;53(7):962–971. doi: 10.1038/s41588-021-00880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marroqui L, et al. BACH2, a candidate risk gene for type 1 diabetes, regulates apoptosis in pancreatic β-cells via JNK1 modulation and crosstalk with the candidate gene PTPN2. Diabetes. 2014;63(7):2516–2527. doi: 10.2337/db13-1443. [DOI] [PubMed] [Google Scholar]
- 17.Kim-Muller JY, et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat Commun. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bysani M, et al. ATAC-seq reveals alterations in open chromatin in pancreatic islets from subjects with type 2 diabetes. Sci Rep. 2019;9(1):7785. doi: 10.1038/s41598-019-44076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camunas-Soler J, et al. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metab. 2020;31(5):1017–1031. doi: 10.1016/j.cmet.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosi E, et al. Integration of single-cell datasets reveals novel transcriptomic signatures of β-cells in human type 2 diabetes. NAR Genom Bioinform. 2020;2(4):lqaa097. doi: 10.1093/nargab/lqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wigger L, et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab. 2021;3(7):1017–1031. doi: 10.1038/s42255-021-00420-9. [DOI] [PubMed] [Google Scholar]