Abstract
Chronic inflammation is a hallmark of atherosclerosis and results from an imbalance between proinflammatory and proresolving signaling. The human GPR32 receptor, together with the ALX/FPR2 receptor, transduces biological actions of several proresolving mediators that stimulate resolution of inflammation. However, since no murine homologs of the human GPR32 receptor exist, comprehensive in vivo studies are lacking. Using human atherosclerotic lesions from carotid endarterectomies and creating a transgenic mouse model expressing human GPR32 on a Fpr2×ApoE double-KO background (hGPR32myc×Fpr2–/–×Apoe–/–), we investigated the role of GPR32 in atherosclerosis and self-limiting acute inflammation. GPR32 mRNA was reduced in human atherosclerotic lesions and correlated with the immune cell markers ARG1, NOS2, and FOXP3. Atherosclerotic lesions, necrotic core, and aortic inflammation were reduced in hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice as compared with Fpr2–/–×Apoe–/– nontransgenic littermates. In a zymosan-induced peritonitis model, the hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice had reduced inflammation at 4 hours and enhanced proresolving macrophage responses at 24 hours compared with nontransgenic littermates. The GPR32 agonist aspirin-triggered resolvin D1 (AT-RvD1) regulated leukocyte responses, including enhancing macrophage phagocytosis and intracellular signaling in hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice, but not in Fpr2–/–×Apoe–/– nontransgenic littermates. Together, these results provide evidence that GPR32 regulates resolution of inflammation and is atheroprotective in vivo.
Keywords: Cardiology, Inflammation
Keywords: Atherosclerosis, G protein–coupled receptors
Introduction
Resolution of acute inflammation is an active, coordinated response aimed at terminating the inflammatory response and restoring tissue homeostasis (1). Resolution of inflammation is regulated by a superfamily of lipid mediators called specialized proresolving mediators (SPMs) that consist of several structurally distinct families biosynthesized from polyunsaturated fatty acids, such as the resolvins (Rv) and lipoxins (LX) (1, 2). SPMs exert potent bioactions in both antiinflammation and proresolution, namely counterregulating neutrophil recruitment and production of proinflammatory mediators while also enhancing clearance of apoptotic neutrophils (i.e., efferocytosis), microbes, and debris (1). Without an effective and timely resolution response, the initial inflammatory insult can lead to chronic inflammation (2), a pathology associated with many widely occurring human diseases, including cardiovascular diseases.
Atherosclerosis is characterized by persistent, chronic vascular inflammation (3), indicative of a defective endogenous inflammation-resolution response (3–5). Indeed, impaired efferocytosis, SPM biosynthesis, and imbalance between SPMs and proinflammatory mediators have recently emerged as key features of nonresolving inflammation in atherosclerosis (3, 6). Specifically, reduced levels of SPMs, including resolvin D1 (RvD1), in blood, saliva, and locally in plaques, correlate with atheroprogression and plaque severity (6–8). Furthermore, RvD1 actions potently limit vascular inflammation, regulate vascular homeostasis (9, 10), and promote plaque stability in murine models of atherosclerosis (6). Therefore, targeting the RvD1 pathway may represent a potential strategy for stimulating resolution in atherosclerotic inflammation.
The bioactions of SPMs are mediated by their cognate GPCRs (1, 10). Two specific GPCRs for RvD1 have been identified in humans, namely ALX/FPR2 and GPR32 (11), which mediate the biological actions of RvD1 in a context-dependent manner (10). GPR32 and ALX/FPR2 also bind several other SPMs in addition to RvD1, including RvD3, RvD5, and LXA4 (1, 10). Since mice lack the murine homologue for GPR32, the proresolving actions observed by these ligands in experimental murine models are mainly attributed to Fpr2, the murine homolog for ALX/FPR2 (12). Accordingly, little is currently known about GPR32 signaling and its function in vivo, and thus the role of GPR32 in the pathology of disease, such as atherosclerosis, remains to be determined. The aim of this study was to determine the role of human GPR32 in vivo during inflammation resolution and atherosclerosis.
Results
GPR32 is reduced in human atherosclerotic lesions.
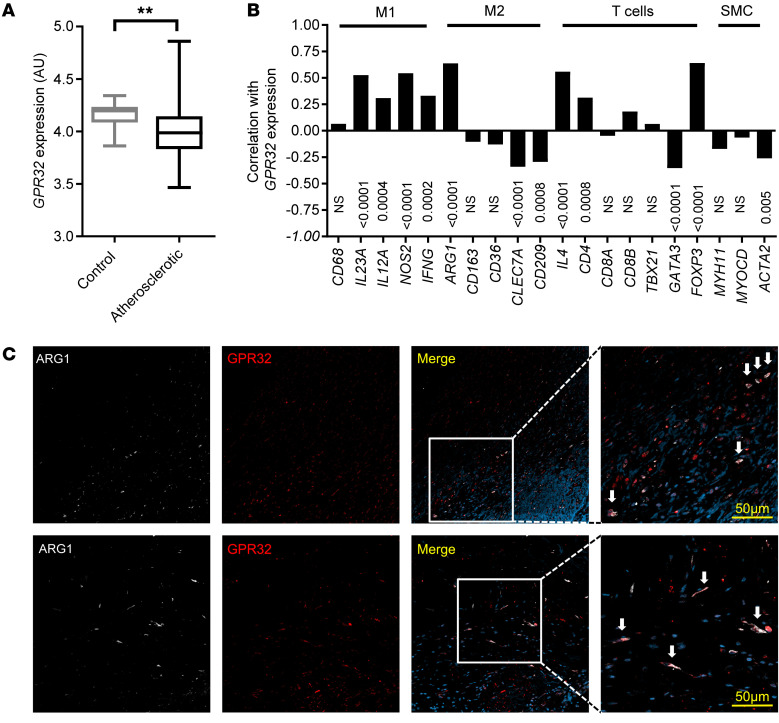
GPR32 mRNA levels were significantly lower in human carotid atherosclerotic lesions compared with those in control arteries (Figure 1A). Further transcriptomic characterization of the atherosclerotic lesions revealed that GPR32 mRNA expression significantly correlated with several immune cell markers. The strongest correlations (r > 0.5) were observed with the M1 macrophage markers IL23A (r = 0.53) and NOS2 (also termed iNOS; r = 0.54), the M2 macrophage marker arginase 1 (ARG1) (r = 0.63), the Treg marker FOXP3 (r = 0.64), and IL4 (r = 0.56) (Figure 1B). Immunofluorescent GPR32 staining revealed that GPR32 expression in the atherosclerotic lesions was predominantly colocalized with ARG1 (Figure 1C) and the general macrophage marker CD68 (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI142883DS1), but was not limited to this leukocyte population, since colocalization with CD3+CD8– lesional lymphocytes was also detected (Supplemental Figure 1, A and B). Structural cells of the vascular wall were positive for GPR32, although detected to colocalize to a lower degree with smooth muscle actin (SMA) for vascular smooth muscle cells (VSMC) and vWF for endothelial cells in comparison with leukocyte populations (Supplemental Figure 1, A and B). Consistent with the strong association (Figure 1B) and colocalization (Figure 1C) between GPR32 and the M2 macrophage marker ARG1, GPR32 protein surface expression was significantly higher on M2- compared with M1-differentiated human macrophages (Supplemental Figure 1C). An inflammatory challenge with LPS (100 ng/mL) reduced the macrophage GPR32 surface expression to a significantly larger degree on M2 than M1 macrophages (Supplemental Figure 1D).
Figure 1. Human GPR32 expression in nonatherosclerotic and atherosclerotic arteries.
(A) GPR32 mRNA expression in human nonatherosclerotic control arteries (n = 10) and atherosclerotic plaques from carotid arteries (n = 127). Results are expressed as median AUs with minimum to maximum bars. **P < 0.01 versus control arteries using unpaired Student’s t test with Welch’s correction. (B) Correlation between GPR32 mRNA levels and immune cell markers using Pearson’s r correlation coefficient and (C) representative immunofluorescence staining from human atherosclerotic plaques (n = 4). GPR32 (red) colocalized with ARG1 (white) in the atherosclerotic plaques, indicated by white arrows in the magnified areas. Original magnification, ×20. Scale bars: 50 μm (zoomed images).
Generation and characterization of hGPR32mycTg×Fpr2–/–×Apoe–/– mice.
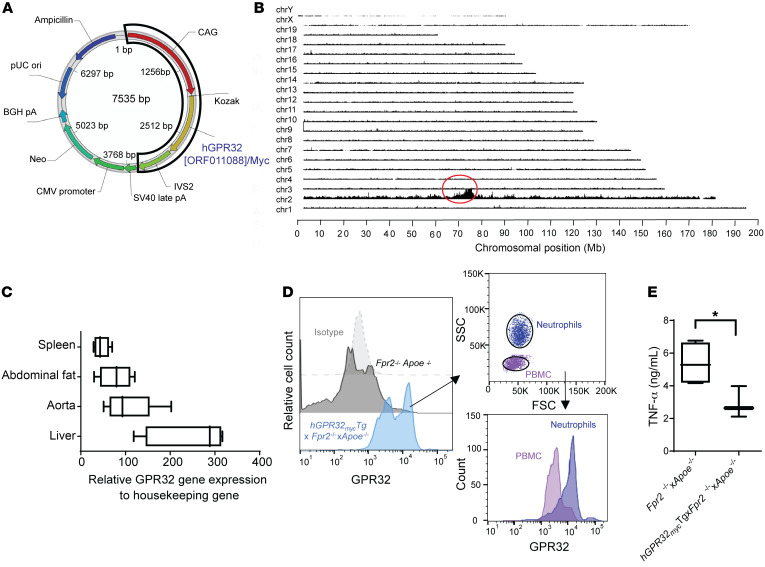
In order to specifically investigate the role of GPR32 signaling in murine atherosclerosis in vivo, we generated a transgenic mouse strain by introducing the human GPR32 Tg into the previously generated hyperlipidemic Apoe-KO mice lacking the Fpr2 receptor (Fpr2–/–×Apoe–/– mice; detailed in Methods) (13, 14). The design of the Tg included the synthetic CAG promotor for strong ubiquitous expression in mammalian cells and a myc-tagged human GPR32 (hGPR32myc) for additional identification of the Tg (Figure 2A). The Tg insertion was identified on chromosome 2, with only 1 Tg-Tg fusion site and an estimated copy number greater than 10 (Figure 2B). The hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice were viable, healthy, and fertile, but had significantly lower body weight (BW) compared with the Fpr2–/–×Apoe–/– nontransgenic littermates (Supplemental Figure 2A). The underlying cause for this BW difference remains to be determined and should be acknowledged as a potential limitation of this model. MRI did not detect any significant differences between the groups in gross tissue morphology in terms of visceral or subcutaneous adipose tissue disposition (Supplemental Figure 2, B and C). Expression of genes related to metabolism was not significantly different in liver or abdominal fat tissue derived from hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic compared with nontransgenic Fpr2–/–×Apoe–/– mice (Supplemental Figure 2, D and E). Fasting glucose, glucose tolerance (Supplemental Figure 2F), blood leukocyte numbers, and differential cell counts (Supplemental Table 1) were not significantly different between the groups. Interestingly, the hGPR32mycTg×Fpr2–/–×Apoe–/– mice had dark brown fur and were visually distinguishable from the black Fpr2–/–×Apoe–/– littermates, as confirmed by genotyping (Supplemental Figure 2G) and in line with the insertions of the Tg at chromosome 2 that carries the agouti gene that controls the coat color of mice.
Figure 2. Generation of a new mouse strain expressing a human GPR32 Tg on an Apoe–/–×Fpr2–/– double-KO background.
(A) Schematics of the vector construction containing the hGPR32(ORF011088)/Myc Tg. (B) TLA sequence coverage across the mouse genome from an hGPR32mycTg×Fpr2–/–×Apoe–/– mouse showing the different chromosomes on the y axis and the chromosomal position on the x axis. The red circle indicates the Tg integration site. (C) Human GPR32 mRNA expression in different mouse tissues (n = 5). (D) Flow cytometry histogram (representative of n = 3) showing GPR32 expression in naive whole blood from Fpr2–/–×Apoe–/– (dark gray) and hGPR32mycTg×Fpr2–/–×Apoe–/– (light blue) mice (left) and differential expression of hGPR32myc in PBMC (purple) and neutrophil (dark blue) cell subsets (right). Isotype control cells are shown in light gray. (E) TNF-α levels in supernatants of LPS-stimulated (100 ng/mL, 24 hours) BMDMs from Fpr2–/–×Apoe–/– (n = 4, white box) and hGPR32mycTg×Fpr2–/–×Apoe–/– (n = 3, black box) mice. Results are expressed as median with minimum to maximum bars. *P < 0.05 between genotypes using Student’s unpaired t test.
hGPR32myc Tg mRNA expression was readily detected in several organs, including the aorta, spleen, liver, and abdominal fat (Figure 2C). hGPR32myc expression was not detected in the nontransgenic littermates. hGPR32myc Tg protein expression was detected by flow cytometry in naive whole-blood leukocytes, which identified higher expression on neutrophils compared with PBMCs (Figure 2D).
The inserted hGPR32myc Tg was functional, as demonstrated by significantly lower TNF-α levels released from LPS-stimulated bone marrow–derived macrophages (BMDMs) isolated from hGPR32mycTg×Fpr2–/–×Apoe–/– compared with Fpr2–/–×Apoe–/– mice (Figure 2E). Since GPR32 is a ligand-activated receptor (11) and since the endogenous ligands are produced by murine BMDMs (15), we next measured the known SPM GPR32 ligands in these incubations by targeted lipidomics using liquid chromatography–tandem mass spectrometry (LC-MS/MS). The GPR32 ligands RvD1, RvD3, LXA4, and RvD5 were all detected in LPS-stimulated BMDMs from both genotypes. No significant differences were observed between BMDMs derived from hGPR32mycTg×Fpr2–/–×Apoe–/– compared with BMDMs from Fpr2–/–×Apoe–/– mice for RvD1 (2.47 ± 2.47 vs. 10.1 ± 3.2 pg/mL, P = 0.132), RvD3 (523.1 ± 54.3 vs. 455.7 ± 63.1 pg/mL, P = 0.463), LXA4 (8.4 ± 0.75 vs. 7.0 ± 1.3 pg/mL, P = 0.403), and RvD5 (4.37 ± 2.47 vs. 2.17 ± 2.17 pg/mL, P = 0.514; n = 3). Hence, the observed reduction in TNF-α levels and the presence of GPR32 ligands in LPS-stimulated hGPR32mycTg×Fpr2–/–×Apoe–/– BMDMs indicates that hGPR32 is functionally expressed in these mice, with implications for counterregulating inflammation.
hGPR32mycTg×Fpr2–/–×Apoe–/– mice have reduced atherosclerotic plaque burden in vivo.
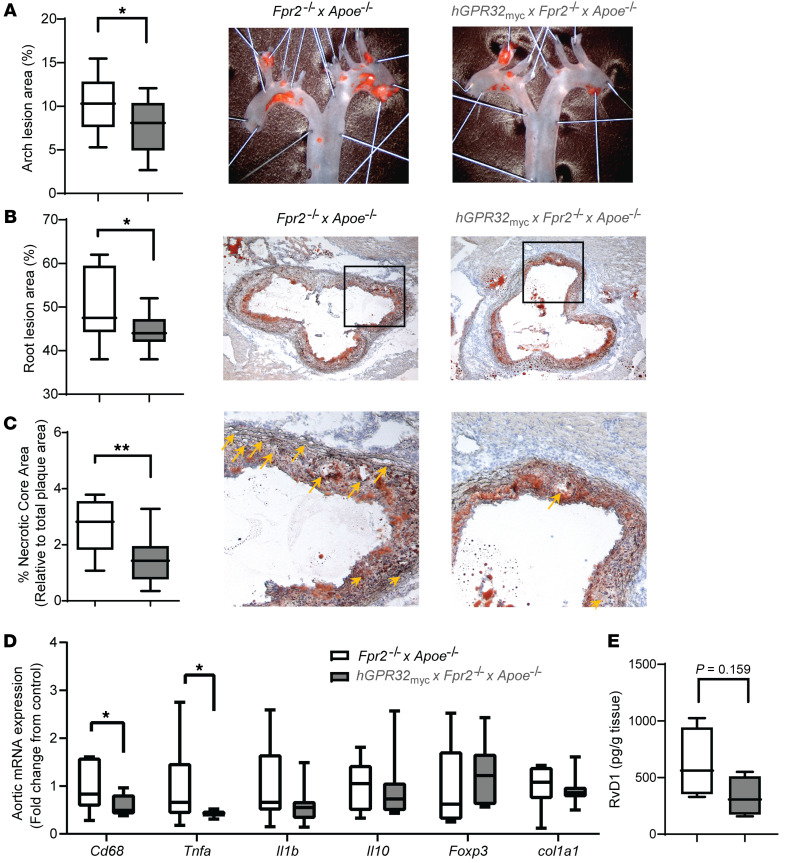
To investigate the role of GPR32 in atherosclerosis, we compared atherosclerotic lesions in female hGPR32myc transgenic mice and their Fpr2–/–×Apoe–/– nontransgenic littermates at 24 weeks of age after 12 weeks on a high-fat diet (HFD). The hGPR32mycTg×Fpr2–/–×Apoe–/– mice had a significantly reduced atherosclerotic lesion burden in both the aortic arch (Figure 3A) and the aortic root (Figure 3B) compared with Fpr2–/–×Apoe–/– mice. This was concomitant with a significantly smaller relative necrotic core area size in lesions from the hGPR32mycTg×Fpr2–/–×Apoe–/– compared with nontransgenic Fpr2–/–×Apoe–/– mice (Figure 3C). No significant changes were observed between the 2 genotypes regarding blood lipids (Supplemental Figure 3A), leukocyte number, and differential count (Supplemental Table 2) or blood chemistry, including glucose levels (Supplemental Table 3),
Figure 3. Reduced atherosclerotic plaque burden and inflammation in aortas from hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice in vivo.
Atherosclerotic lesion burden in aortic arches (A) and aortic roots (B) from Fpr2–/-×Apoe–/– (open; n = 10) and hGPR32mycTg×Fpr2–/–×Apoe–/– (gray; n = 10) mice after 12 weeks HFD. Representative micrographs are shown for (A) en face staining of aortic arches and (B) aortic root for each genotype. Original magnification, ×10 (A); ×5 (B). (C) Relative necrotic core area expressed as percentage of the total plaque area. Representative micrographs for each genotype are shown, and the magnified areas with necrotic cores highlighted by yellow arrows shown in C are indicated by a black box in the aortic roots in B. Original magnification, ×20. (D) Fold change in aortic mRNA levels in hGPR32mycTg×Fpr2–/–×Apoe–/– (n = 7) relative to Fpr2–/–×Apoe–/– mice (n = 9). (E) Levels of RvD1 in aortic lesions, expressed as pg/g tissue (n = 4). Results are expressed as median with minimum to maximum bars. *P < 0.05; **P < 0.01 between genotypes using Student’s unpaired t test.
To identify potential mechanisms underlying the atheroprotective actions of GPR32, we assessed atherosclerotic plaque composition and gene expression. Abdominal aortas derived from hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice compared with nontransgenic Fpr2–/–×Apoe–/– mice exhibited significantly lower mRNA levels for the macrophage marker Cd68 (Figure 3D), accompanied by a trend for lower CD68 immunostainings in the aortic root lesions (P = 0.07; Supplemental Figure 3B). No significant differences were observed in stainings for collagen content, α-SMA, or CD3+ cells (Supplemental Figure 3C). The proinflammatory cytokine Tnfa was significantly reduced in hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic compared with Fpr2–/–×Apoe–/– mice (Figure 3D). A similar trend was observed for Il1b (P = 0.153), whereas the antiinflammatory cytokine Il10, collagen 1a (col1a1), and the Treg-associated transcription factor Foxp3 were not differentially expressed between the 2 genotypes (Figure 3D). The GPR32 ligand RvD1 was detected using targeted LC-MS/MS analysis of aortic lesions from both hGPR32mycTg×Fpr2–/–×Apoe–/– and Fpr2–/–×Apoe–/– mice (Figure 3E).
Similar results for atherosclerosis were observed when the experimental protocol was prolonged, and mice were fed HFD for 16 weeks. hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice had a 30% lower (P = 0.058) atherosclerotic lesion burden compared with Fpr2–/–×Apoe–/– littermates (Supplemental Figure 4A). This was concomitant with significantly fewer GR-1+ (Ly6G/Ly6C+) cells in plaques from hGPR32mycTg×Fpr2–/–×Apoe–/– mice (Supplemental Figure 4B), indicating a reduced aortic infiltration of neutrophils (Ly6G+) and monocytes (Ly6C+).
Despite the lower BW of hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice (Supplemental Figure 4C), there was no significant correlation between lesion size and BW in either genotype (Supplemental Figure 4D). However, a trend toward a negative correlation (r = –0.5111; P = 0.08) between lesion size and hGPR32myc expression was observed in hGPR32mycTg×Fpr2–/–×Apoe–/– mice (Supplemental Figure 4E). Taken together, these findings point to a role for GPR32 signaling in limiting aortic inflammation, with implications for atheroprotection in vivo.
Inflammation-resolution responses in hGPR32mycTg×Fpr2–/–×Apoe–/– mice during zymosan-induced peritonitis.
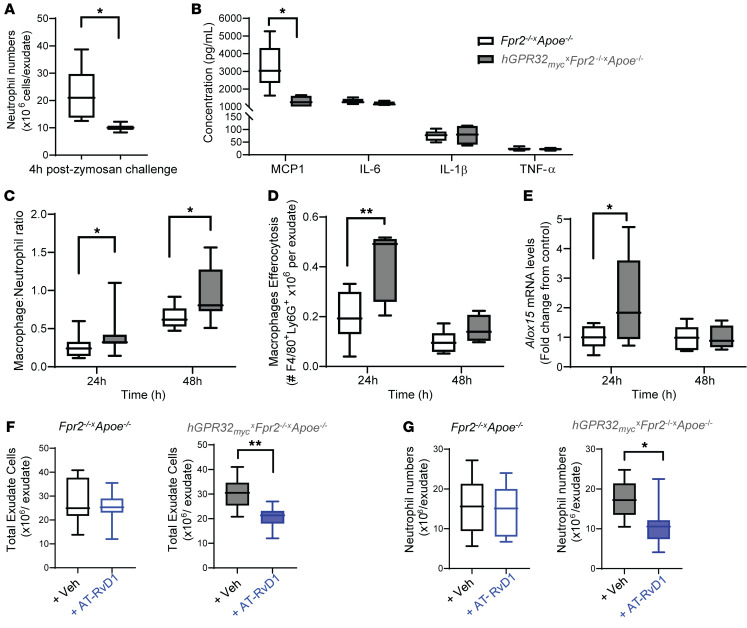
The well-established murine model of acute, self-limited inflammation (1, 16) initiated by i.p. injection of zymosan A (1 mg/mouse) induced a temporal leukocytic infiltration into the peritoneal cavity. In mice sacrificed at 4 hours after zymosan challenge, representing the initiation phase, flow cytometry revealed that neutrophil numbers (detected by Ly6G+ events) were approximately 50% lower in hGPR32mycTg×Fpr2–/–×Apoe–/– mice compared with Fpr2–/–×Apoe–/– mice (Figure 4A). Exudate levels of the proinflammatory chemokine CCL2 were also significantly lower in hGPR32mycTg×Fpr2–/–×Apoe–/– mice, whereas no significant differences were observed for exudate levels of IL-6, IL-1β, or TNF-α (Figure 4B).
Figure 4. Regulation of inflammation-resolution responses in hGPR32mycTg×Fpr2–/–×Apoe–/– mice during acute peritonitis.
Fpr2–/–×Apoe–/– (white box) and hGPR32mycTg×Fpr2–/–×Apoe–/– (gray box) mice were subjected to a zymosan challenge (1 mg/mouse, i.p.), and peritoneal exudates were collected at the indicated time points. (A) Exudate neutrophil numbers (Ly6G+ cells) and (B) cytokine levels after 4 hours (n = 7, white box;n = 3, gray box). (C) Exudate macrophage/neutrophil ratio after 24 hours (n = 17, white box; n =11, gray box) and 48 hours (n = 8, white box; n =7, gray box), respectively.(D) Efferocytosis assessed as Ly6G+ macrophages and (E) Alox15 mRNA expression in exudate cells after 24 hours (n = 11, white box; n =5, gray box) and 48 hours (n = 6, white box; n =4, gray box), respectively. *P < 0.05; **P < 0.01 between genotypes. (F) Total exudate cells and (G) neutrophils in Fpr2–/–×Apoe–/– (open) and hGPR32mycTg×Fpr2–/–×Apoe–/– (filled) mice injected with vehicle (black outline) or AT-RvD1 (100 ng/mouse; blue outline) immediately prior to zymosan challenge (vehicle: n = 10 for Fpr2–/–×Apoe–/– and n = 7 for hGPR32mycTg×Fpr2–/–×Apoe–/–; AT-RvD1, n = 8 for both genotypes). *P < 0.05; **P < 0.01 vs. vehicle. Results are expressed as median with minimum to maximum bars. Unpaired Student’s t test was used to compare 2 groups, and 2-way ANOVA was used for comparing multiple groups.
Since GPR32 mediates signals that promote resolution of inflammation (2, 10), we next examined the exudates at 24 and 48 hours after zymosan challenge, representing the resolution phase.
While there was a significantly lower percentage of neutrophils (65.09 ± 2.38 vs. 55.69 ± 2.2 × 106 neutrophils/exudate, P = 0.016 at 24 hours and 43.94 ± 1.90 vs. 33.5 ± 3.77 × 106 neutrophils/exudate, P = 0.028 at 48 hours) in exudates from hGPR32mycTg×Fpr2–/–×Apoe–/– mice compared with Fpr2–/–×Apoe–/– mice, no differences were observed in neutrophil numbers at either 24 hours (16.1 ± 1.1 vs. 16.2 ± 1.3 × 106 neutrophils/exudate, P = 0.949; n = 17, 11) or 48 hours (8.8 ± 1.3 vs. 6.9 ± 1.2 × 106 neutrophils/exudate, P = 0.296, n = 8, 6. A significantly higher macrophage-to-neutrophil ratio was observed in exudates from hGPR32mycTg×Fpr2–/–×Apoe–/– compared with Fpr2–/–×Apoe–/– mice (Figure 4C), which was mainly driven by a higher number of macrophages (3.99 × 106 vs. 6.01 × 106 macrophages/exudate P = 0.073 at 24 hours, 5.46 × 106 vs 5.85 × 106 macrophages/exudate P = 0.296). This was concomitant with a significantly higher number of macrophages containing engulfed apoptotic neutrophils in hGPR32mycTg×Fpr2–/–×Apoe–/– exudates, indicative of enhanced efferocytosis (Figure 4D), a key process in resolution of inflammation, at 24 hours (1). Furthermore, hGPR32mycTg×Fpr2–/–×Apoe–/– mice had significantly higher Alox15 mRNA expression in their exudate leukocytes compared with Fpr2–/–×Apoe–/– mice (Figure 4E). In addition, significantly lower mRNA levels of the inflammatory cytokines Il1b (0.96 ± 0.08 vs. 0.40 ± 0.08 after 24 hours, P = 0.003 and 1.13 ± 0.13 vs. 0.50 ± 0.09 after 48 hours, P = 0.003) and Tnfa (1.06 ± 0.13 vs. 0.64 ± 0.06 after 48 hours, P = 0.01) were observed in exudates from hGPR32mycTg×Fpr2–/–×Apoe–/– mice compared with Fpr2–/–×Apoe–/– mice. Finally, we confirmed in a separate experimental series that the macrophage-to-neutrophil ratio observed in exudates from hGPR32mycTg×Fpr2–/–×Apoe–/– mice at 24 hours was significantly higher when compared with both regular Apoe–/– mice and Fpr2–/–×Apoe–/– mice generated independently of the transgenic breeding (Supplemental Figure 5). Taken together, these results indicate that the hGPR32myc Tg confers an intrinsic antiinflammatory and proresolving response in vivo following an acute inflammatory challenge.
AT-RvD1 regulates leukocyte responses in hGPR32mycTg×Fpr2–/–×Apoe–/– mice. RvD1 and its aspirin-triggered epimer AT-RvD1 regulate leukocyte responses and accelerate resolution of inflammation in animal models in vivo (1, 16), actions in mice that have currently only been attributed to its signaling via Fpr2 (14, 16). Here, we administered AT-RvD1 to hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice to assess the impact of ligand-mediated actions of the hGPR32myc Tg on leukocyte responses in the absence of Fpr2 in vivo. AT-RvD1 (100 ng, i.p.) injected into mice immediately prior to zymosan challenge resulted in significantly lower numbers of total exudate cells (Figure 4F) as well as neutrophils (Figure 4G) when compared with those in vehicle-treated hGPR32mycTg×Fpr2–/–×Apoe–/– transgenic mice. This action was lost in nontransgenic Fpr2–/–×Apoe–/– mice lacking the hGPR32myc Tg (Figure 4, F and G), which suggests that AT-RvD1 regulates leukocyte responses during inflammation resolution by activating hGPR32myc in vivo.
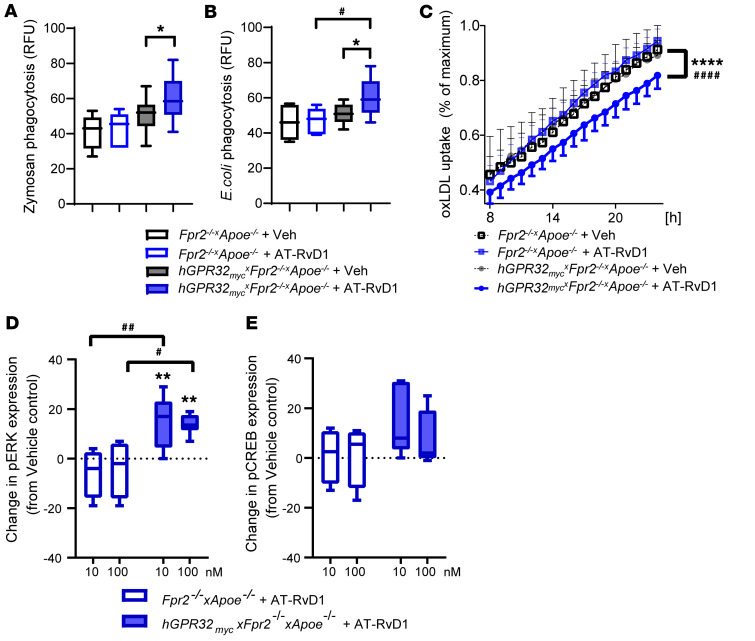
AT-RvD1 regulates macrophage phagocytosis and oxLDL uptake in hGPR32mycTg×Fpr2–/–×Apoe–/– mice.
Administration of AT-RvD1 (100 nM) in vitro significantly enhanced phagocytosis of zymosan (Figure 5A) and E. coli (Figure 5B) in naive peritoneal macrophages derived from hGPR32mycTg×Fpr2–/–×Apoe–/– mice, whereas AT-RvD1 did not significantly alter phagocytosis in nontransgenic Fpr2–/–×Apoe–/– mice (Figure 5, A and B). On the other hand, AT-RvD1 significantly reduced oxidized low-density lipoprotein (oxLDL) uptake in hGPR32mycTg×Fpr2–/–×Apoe–/– macrophages, whereas no AT-RvD1–induced effects were observed in Fpr2–/–×Apoe–/– macrophages (Figure 5C). Taken together, these results indicate that AT-RvD1 mediates direct proresolving and atheroprotective actions in macrophages via GPR32 in hGPR32mycTg×Fpr2–/–×Apoe–/– mice.
Figure 5. AT-RvD1 alters leukocyte responses and regulates phosphorylation of ERK1/2 in hGPR32mycTg×Fpr2–/–×Apoe–/– but not Fpr2–/–×Apoe–/– mice.
Naive resident peritoneal macrophages were isolated and seeded in a 96-well plate (0.1 × 106 cells/well). Uptake of pH-rodo labeled (A) zymosan (n = 6) or (B) E. coli (n = 10) by peritoneal macrophages from Fpr2–/–×Apoe–/– (open boxes) and hGPR32mycTg×Fpr2–/–×Apoe–/– (filled boxes) that were pretreated with vehicle (black outline) or AT-RvD1 (100 nM, 15 minutes, 37°C; blue outline) followed by 60 minutes of incubation with the phagocytic stimuli. Results are expressed as relative fluorescence units (RFU). (C) Peritoneal macrophages were preincubated with AT-RvD1 (100 nM, 15 minutes, 37°C) followed by the addition of FITC-labeled oxLDL (10 μg/ml) to assess phagocytosis of oxLDL. Images were recorded every 1 hour for a total of 24 hours. Data are presented as percentages of maximum phagocytosis (after 24 hours). Results are expressed as mean ± SEM. Changes in (D) pERK and (E) pCREB activation in peritoneal macrophages treated with AT-RvD1 (10 or 100 nM; 5 minutes, 37°C) from vehicle control in Fpr2–/–×Apoe–/– (open box, n = 4) and hGPR32mycTg×Fpr2–/–×Apoe–/– (filled box, n = 6) mice. Results are expressed as median with minimum to maximum bars. *P < 0.05; **P < 0.01; ****P < 0.0001 vs. vehicle-treated hGPR32mycTg×Fpr2–/–×Apoe–/– macrophages. #P < 0.05; ##P < 0.01; ####P < 0.0001 in AT-RvD1–treated Fpr2–/–×Apoe–/– vs. AT-RvD1–treated hGPR32mycTg×Fpr2–/–×Apoe–/– macrophages; 2-way ANOVA.
AT-RvD1-GPR32–dependent intracellular signaling in hGPR32mycTg×Fpr2–/–×Apoe–/– mice.
Naive mouse peritoneal macrophages stimulated with AT-RvD1 (10 and 100 nM) for 5 minutes and stained for flow cytometric analysis of intracellular phosphorylated targets revealed that AT-RvD1 significantly increased phosphorylation of pERK1/2 compared with vehicle-treated hGPR32mycTg×Fpr2–/–×Apoe–/– macrophages (Figure 5D). A similar trend was observed for phosphorylated cAMP response element binding protein (pCREB), which did not reach statistical significance (Figure 5E). In contrast, these phosphoproteins were unaltered after AT-RvD1 stimulation of macrophages collected from nontransgenic Fpr2–/–×Apoe–/– mice (Figure 5, D and E). These results indicate that hGPR32myc Tg regulates protein phosphorylation as intracellular signals in isolated macrophages from hGPR32mycTg×Fpr2–/–×Apoe–/– mice.
Discussion
The results from the present study point to a resolution of inflammation mediated through the RvD1 receptor GPR32 in atherosclerosis. First, we demonstrate that human atherosclerotic lesions exhibit decreased GPR32 mRNA levels compared with those in control arteries, with predominant expression in macrophages with a proresolving phenotype. Second, by generating a transgenic hyperlipidemic mouse strain that lacks the murine RvD1 receptor Fpr2 while expressing the human GPR32 receptor, we found that proresolving signaling through GPR32 was atheroprotective independently of Fpr2, through leukocyte regulated responses, decreased proinflammatory cytokines, reduced oxLDL uptake, and enhanced phagocytosis. Third, the murine model identified agonist-induced signaling by GPR32 ligands, which strengthened the translational implications of the findings for human disease. Taken together, these results point to stimulating GPR32 as a potential therapeutic strategy for reducing atherosclerotic cardiovascular diseases by means of stimulating resolution of inflammation.
In the present study, the expression levels of the RvD1 receptor GPR32 were significantly lower in atherosclerotic compared with healthy human arteries (P = 0.004). While this observation follows the expression pattern of the proresolving receptor ERV1/ChemR23 for RvE1 (17), the mRNA levels for the other RvD1 receptor, ALX/FPR2, are increased in human atherosclerotic lesions (18). The latter study and the present results hence implicate a dichotomous role for these 2 RvD1 receptors in atherosclerosis, which is further supported by the observation that GPR32 expression, but not ALX/FPR2 expression, is reduced on blood leukocytes from patients with chronic heart failure and associated with disease severity and unresponsiveness to the immunomodulatory actions of RvD1 (19). These observations that GPR32 affects systemic inflammatory responses, taken together with our findings that GPR32 expression is reduced in atherosclerosis and in LPS-stimulated M2 macrophages, raises the possibility of a deficit in proresolving signaling as a common characteristic associated with cardiovascular disease pathology.
Macrophages play a critical role in atherosclerosis, and depending on their phenotype, they can either induce plaque inflammation (e.g., M1) or transduce the resolution of inflammation and tissue repair (e.g., M2) (4). Resolution-phase macrophages (rM) share characteristics with both M1 and M2 macrophages, supporting the idea that the positive correlation of GPR32 with both iNOS and ARG1 (20) reflects GPR32 expression in rM macrophages. GPR32 has indeed been implicated as a key player in mediating the actions of RvD1 in human macrophage polarization to M2- and rM-like phenotypes (21), which was supported by the higher GPR32 expression in M2 macrophages in the present study. Atherosclerotic lesion GPR32 expression was not limited to macrophages, since correlation and colocalization with markers of lesional lymphocytes were also observed. Indeed, RvD1 signaling via GPR32 promotes de novo induction of Foxp3+ Tregs (19), a cell type that regulates excessive immune responses and reduces atherosclerosis via promoting resolution (22). Finally, GPR32 expression was detected in structural cells of the vascular wall similarly to that of other SPM receptors (5, 17) and the GPR32 expression previously reported in leukocytes, vascular cells, and adipocytes (10, 21, 23, 24). Furthermore, in the gene atlas of the human protein-encoding transcriptomes (available at BioGPS; http://biogps.org) the highest levels of GPR32 mRNA were found in liver, heart, CD34+ hematopoietic cells, CD33+ myeloid cells, cardiac myocytes, and SMC (25), further supporting the involvement of GPR32 in the immune and cardiovascular systems. Taken together, our findings in human atherosclerotic lesions support the idea that regulatory immune cell signaling may be dysregulated in human atherosclerosis, which is consistent with a failure in the resolution response.
The exploration of the causal role of GPR32 in atherosclerosis has been hampered by the lack of a murine homolog for this receptor, whereas the role of Fpr2 and their shared ligands, such as RvD1 and AT-LXA4, has been more extensively studied in experimental models of atherosclerosis (6, 13, 18). Hence, introducing a human GPR32 to a hyperlipidemic Apoe–/– mouse that lacks the endogenous murine RvD1 receptor Fpr2 enabled us to study the role of GPR32 signaling in atherosclerosis independently of Fpr2. Having established that the human GPR32 Tg was expressed and functional, mice were subjected to HFD to induce atherosclerosis. The observed antiatherogenic effects in the hGPR32mycTg×Fpr2–/–×Apoe–/– mice suggest that GPR32 signaling is protective in atherosclerosis in vivo and extend the previously reported atheroprotective effects transduced by ChemR23 and Fpr2 (13, 17). Since these protective effects were found to be independent of lipid levels, it suggests that GPR32 signaling in atherosclerosis involves modulating the inflammation-resolution pathways rather than altering lipid metabolism. Indeed, the reduced atherosclerosis burden was accompanied by lowering of aortic proinflammatory gene expression as well as reduced numbers of macrophages and neutrophils in the lesions. Additionally, our findings are in line with earlier findings demonstrating that transgenic overexpression of human proresolving receptors is protective in experimental murine models (26), indicating that receptor expression levels may be crucial for the regulation of the responses transduced. Hence, together with our findings, this implicates a role for GPR32 signaling in modulating atheroprogression.
In the present study, we establish the presence of GPR32 agonists produced locally in murine BMDM and atherosclerotic lesions in line with previous studies (6, 27). Reduced levels of SPMs, in particular, the GPR32 agonist RvD1, have been associated with atheroprogression (6), further supporting the notion that proresolving signaling is dysregulated in atherosclerosis. Akin to RvD1, its aspirin-triggered epimer AT-RvD1, which displays enhanced resistance to further enzymatic conversion (1), potently limits neutrophil infiltration and accelerates resolution in animal models (1, 16). However, the actions of AT-RvD1 and RvD1 in murine models in vivo have hitherto only been attributed to their signaling via Fpr2. The present study is the first, to our knowledge, to show that GPR32, independently of Fpr2, regulates neutrophil infiltration in aortic lesions and acute peritonitis. These results are consistent with earlier findings in humans showing that GPR32 signaling reduces human neutrophil chemotaxis in vitro (23). In that context, neutrophil influx into the aortic wall is associated with lesion formation and further augments recruitment of mononuclear phagocytes into the site (28). Furthermore, disruption of proresolution signaling is associated with enhanced neutrophilic infiltration into the vascular wall and aortic dilation in angiotensin II–infused hyperlipidemic mice (29). Taken together, these results suggest that atheroprotective signaling via GPR32 may involve limiting neutrophil recruitment during atherosclerotic development in vivo.
On the basis of those findings, we investigated the effects of AT-RvD1 and GPR32 on the resolving inflammatory exudate. It should be recognized that while this model has not been previously studied in hyperlipidemic mice, it was well suited to study the role of GPR32-mediated signaling on leukocyte responses during self-resolving inflammatory challenge in vivo in the absence of differences in cholesterol levels between groups and the almost 50% reduction in neutrophil numbers between the 24- and 48-hour time points, in line with an active self-limited inflammatory response. Introduction of the human GPR32 to these mice resulted in dampened inflammation at 4 hours and enhanced resolution responses at 24 hours, as demonstrated by enhanced efferocytosis, a fundamental process in resolution (1), and upregulation of Alox15, a key enzyme in SPM biosynthesis and an emerging M2 or rM macrophage marker. Because RvD1 and its more stable analogue, AT-RvD1, interact with GPR32 (16), we also tested to determine whether AT-RvD1 could regulate leukocyte responses in hGPR32mycTg×Fpr2–/–×Apoe–/– mice. Indeed, AT-RvD1 reduced total leukocyte and neutrophil numbers in the hGPR32mycTg×Fpr2–/–×Apoe–/– mice, but not the Fpr2–/–×Apoe–/– nontransgenic mice, indicating that AT-RvD1 mediates its proresolving actions via GPR32 in vivo. Furthermore, we also showed that AT-RvD1 enhances phagocytosis of zymosan and E. coli in macrophages isolated from hGPR32mycTg×Fpr2–/–×Apoe–/– mice, which is consistent with earlier findings with Rv and human macrophages (11, 21). On the other hand, AT-RvD1 reduced the uptake of oxLDL, supporting the notion that macrophage GPR32 signaling may mediate protective effects in atherosclerosis. These results extend our previous findings for RvE1 macrophage signaling through ChemR23 (17) and provide further evidence for direct atheroprotective actions mediated by Rv by means of stimulating resolution.
Having found that AT-RvD1 elicited proresolving actions in vivo and with peritoneal macrophages in hGPR32mycTg×Fpr2–/–×Apoe–/– mice, we investigated the downstream intracellular signaling pathways elicited by AT-RvD1 in these mice. AT-RvD1 activated phosphorylation of pERK1/2, with a similar trend for pCREB, in macrophages from hGPR32mycTg×Fpr2–/–×Apoe–/– mice, but not Fpr2–/–×Apoe–/– mice, consistent with intracellular signaling pathways observed for RvD2-GPR18 and MaR2-LGR6 in mouse peritoneal macrophages (30, 31). Furthermore, these results are also in line with recent findings in which individual SPMs, including RvD1, activate distinct intracellular signaling pathways in peripheral blood leukocytes in humans, with the most pronounced activation observed for pERK1/2 and pCREB in neutrophils and CD14+ monocytes (32). Hence, these results demonstrate that AT-RvD1 elicits protein phosphorylation as intracellular signals in isolated macrophages via GPR32, similarly to those observed for other SPMs and with human cells.
In summary, this study uncovered that GPR32 signaling attenuates atherosclerosis and mediates proresolving leukocyte responses in vivo in hyperlipidemic Apoe–/– mice lacking the receptor Fpr2. We extended previous observations on SPM receptor expression in human atherosclerotic lesions to now include dysregulated GPR32 expression in atherosclerotic compared with normal arteries. We provide evidence for the in vivo role of the RvD1 receptor GPR32 in atheroprotection by means of promoting proresolving leukocyte responses, enhancing phagocytosis, and reducing oxLDL uptake, with implications for atherosclerotic lesion size, necrotic core formation, and aortic inflammation. The effects of hGPR32myc in the regulation of immune responses and the amelioration of atherosclerosis progression in vivo, together with the dysregulated expression of GPR32 in human atherosclerosis, further shows that a defect in the GPR32 signaling pathway may underpin the impaired resolution mechanisms in atherosclerosis progression. Hence, stimulating GPR32 may represent a potential therapeutic strategy for reducing atherosclerotic cardiovascular diseases by means of stimulating resolution of inflammation.
Methods
Human studies
Human atherosclerotic plaques were obtained from the Biobank of Karolinska Carotid Endarterectomies (BiKE) project. The study included a total of 127 atherosclerotic plaque tissue samples from patients that underwent carotid endarterectomy (CEA) at Karolinska University Hospital. Disease-free, nonatherosclerotic, iliac arteries, referred to as normal arteries, were obtained from organ donors and used as controls. Isolated RNA from 127 patients and 10 controls was subjected to global mRNA analysis with Affymetrix HG-U133 plus 2.0 expression arrays as previously described (13, 33, 34). Data were normalized with signal space transformation-robust multichip analysis, yielding log2-transformed expression values (35, 36). Paraffin-embedded atherosclerotic sections were evaluated by immunofluorescence staining, as described below.
Immunofluorescence staining
For staining of human atherosclerotic tissues, sections were incubated overnight at 4°C with the following primary Abs: rabbit anti-human GPR32 N-terminal (catalog GTX108119, GeneTex), rabbit anti-human Arg1 (catalog NB100-59740; Bio-Techne), mouse anti-human SMA (catalog M0851, Agilent Dako), mouse anti-human vWF (catalog M0616, Agilent Dako), mouse anti-human CD8 (catalog NCL-CD8-295, Novocastra Leica Biosystems), mouse anti-human CD68 (catalog NCL-CD68, Novocastra Leica Biosystems), and goat anti-human CD3 (catalog PM110, Biocare Medical). Next, the sections were incubated for 1 hour at room temperature with secondary Abs conjugated with DyLight 488 or 594 (Invitrogen, Thermo Fisher), and nuclei were stained with DAPI (S3023, Agilent Dako).
Primer and vector design
The vector used to generate the hGPR32myc transgenic mice was designed using Vector Builder software, version 1.0 (VectorBuilder GmbH) and synthesized by Cyagen Biosciences. The ORF of human GPR32 (GenBank NM_001506), including a myc-reporter tag, was cloned in a mammalian expression vector (Figure 1A). In order to drive a strong ubiquitous expression of hGPR32myc in mammalian cells, the strong synthetic CAG promoter based on the chicken β-actin gene and a Kozak consensus sequence was included in the expression vector backbone. A synthetic intron IVS2 was added to increase stability and expression of the mRNA, while the simian virus 40 late polyadenylation signal (SV40 late pA) and bovine growth hormone polyadenylation signal (BGH pA) were added to enhance transcriptional termination. The neomycin resistance gene under the control of the CMV promoter allowed for selection and stable expression in mammalian cells. For amplification of the vector in E. coli, a bacterial pUC ori and ampicillin resistance were included. The transgenic insert was extracted from the vector backbone by digesting 1 μg of vector DNA simultaneously with 20 IU each of AscI (GG/CGCGCC), MluI (A/CGCGT), and XhoI (C/TCGAG) (all from New England Biolabs) at 37°C overnight. The 3833 bp long fragment containing the transgenic insert was separated on a 1% agarose gel, isolated and purified, and used for oocyte injection by the Karolinska Center for Transgene Technologies (KCTT) core facility at the Karolinska Institute, Department of Comparative Medicine.
Animals
Animals were bred and kept under standard housing conditions on a 12-hour light/12-hour dark cycle. Fpr2–/– mice were generated at the William Harvey Institute (London, United Kingdom), as previously described (14). Of note, the insertions of the gene cassette and GFP reporter in reverse orientation into intron 1 of the Fpr2 gene prevents the transcriptional read-through of the Fpr2 and Fpr3 genes (14, 37), both of which are murine homologues to the human ALX/FPR2 (12). Thus, these mice are also termed Fpr2/Fpr3-KO mice (37), but are referred to as Fpr2–/– herein. The Fpr2–/–×Apoe–/– mice were generated at Karolinska Institute as described (13).
Generation and characterization of hGPR32myc transgenic mice
Transgenic mice expressing the hGPR32myc Tg were generated by random transgenesis in Fpr2–/–×Apoe–/– mice with the help of the KCTT core facility at Karolinska Institute. The hGPR32myc Tg was injected into fertilized oocytes of superovulating Fpr2–/–×Apoe–/– females to generate the F1 generation of hyperlipidemic hGPR32myc transgenic mice that lack the Fpr2 receptor (hGPR32mycTg×Fpr2–/–×Apoe–/–). A transgenic male from the F1 generation was established as a potential founder and bred hemizygously with Fpr2–/–×Apoe–/– females. Targeted locus amplification (TLA) sequencing (Cergentis) identified the Tg integration site, possible genomic rearrangements, and estimation of Tg copy number. Genotyping was performed with DNA isolated from mouse ear biopsies. Two sets of primers were used, covering the entire Tg including the myc tag. For GPR32, the forward and reverse primers were 5′-CAGAGGCTGCAGTGACAGGCAA-3′ and 5′-GTCCGGAATTTCAGGTGCGCAGA-3′, respectively (519 bp product). For GPR32myc, the forward and reverse primers were 5′-ACTCTACATCACCTTTGTGTTCCTC-3′ and 5′-TCTTCTGAGATGAGTTTTTGTTCTTC-3′, respectively (747 bp product).
The transgenic mice were viable and fertile and showed no adverse health effects, breeding issues, or obvious behavioral or developmental defects after monitoring the transgenic mouse line for several generations. No gross morphological or tissue defects were observed upon histological examination by light microscopy. Peripheral blood analysis for red and white blood cells and plasma analysis for triglycerides, cholesterol, and glucose levels showed no differences between transgenic and nontransgenic littermates (Supplemental Figure 2, B and F, and Supplemental Table 1). Organs, blood, and bone marrow were collected from 8- to 12-week-old male and female mice to confirm the expression of the hGPR32myc Tg and characterization of the hGPR32mycTg×Fpr2–/–×Apoe–/– mice in comparison with their nontransgenic Fpr2–/–×Apoe–/– littermates. Blood was collected into EDTA-coated tubes (Sarstedt), and samples from spleen, liver, abdominal fat, and abdominal aorta were collected and snap-frozen for subsequent total RNA isolation using the RNeasy Lipid Tissue Kit (QIAGEN) as described below.
Genome TLA sequencing
Spleens from 8-week-old hGPR32mycTg×Fpr2–/–×Apoe–/– mice were collected and sent to Cergentis for TLA to determine the integration site of the Tg, to assess the presence of structural variants surrounding the Tg integration site, to assess the Tg sequence itself, and to estimate the Tg copy number. Two Tg-specific primer pairs were designed for the TLA as follows: set 1: forward, 5′-AATACTCTGAGTCCAAACCG-3′ (β-globin), reverse, 5′-TCGTTACAAATGCAAGCTAAA-3′ (Tg locations 3441 and 3172, respectively); set 2: forward, 5′-GATGACTGTCTTCCGTATGG-3′, reverse, 5′-GCATCCGGAAGAGTTCATC-3′ (Tg locations 1982 and 1886, respectively). The primer sets were used in individual TLA. PCR products were purified, library prepared using the Illumina NexteraXT protocol, and sequenced on an Illumina sequencer (Illumina). Reads were mapped using the Smith-Waterman algorithm alignment tool BWA-SW, which allows for partial mapping, optimally suited for the identification of break-spanning reads. The mouse mm10 genome version was used for mapping.
Murine BMDMs
BMDMs were obtained from hGPR32mycTg×Fpr2–/–×Apoe–/– mice by flushing mouse femurs and tibiae with RPMI 1640 medium (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS. The cells were subsequently centrifuged (300g, 10 minutes, 4°C), suspended in PBS+/+ (Gibco, Thermo Fisher Scientific), seeded in a 10 mL dish, and incubated for 60 minutes at 37°C. Adherent cells were cultured for 6 days in RPMI 1640 media supplemented with 10% FBS, 1% penicillin-streptomycin, and 2 mM l-glutamine (all Gibco, Thermo Fisher Scientific) and mouse rM-CSF (10 ng/mL; Preprotech Nordic). Mouse BMDMs were then plated onto 96-well plates (1 × 105 cells/well) and stimulated for 24 hours with 100 ng/ml LPS (serotype O111:B4, MilliporeSigma); cell supernatants were collected and assessed for TNF-α levels using ELISA (R&D Systems) or for select SPMs using targeted LC-MS/MS analysis as described below.
Experimental animal models
Atherosclerosis.
For the induction of atherosclerosis, 12-week-old female hGPR32mycTg×Fpr2–/–×Apoe–/– (hGPR32myc transgenic) mice and Fpr2–/–×Apoe–/– (nontransgenic) littermates were fed an HFD (Clinton/Cybulsky High Fat Rodent Diet With Regular Casein and 1.25% Added Cholesterol, D12108C, Research Diets Inc.) and water ad libitum for 12 or 16 weeks. At the end of each experiment, mice were sacrificed by asphyxiation in a CO2-enriched atmosphere, blood was collected via cardiac puncture into EDTA-coated tubes (Sarstedt), and the animals were perfused with 10 mL sterile RNAse-free PBS prior to harvesting of organs. Samples from spleen, liver, abdominal fat, and abdominal aorta were dissected and snap-frozen for RNA isolation. Aortic roots and arches were dissected and preserved for histological lesion analysis and en face lesion analysis, respectively.
Acute inflammation: peritonitis.
For self-resolving acute peritonitis, male hGPR32mycTg×Fpr2–/–×Apoe–/– mice and Fpr2–/–×Apoe–/– littermates were injected i.p. with 1 mg/mouse zymosan (MilliporeSigma), and peritoneal exudates were collected at 4 hours (acute phase) or 24 and 48 hours (resolution phase) by peritoneal lavage with 5 mL DPBS–/– (Gibco, Thermo Fisher Scientific). In some experiments, mice received vehicle (0.1% EtOH in PBS) or 100 ng/mouse AT-RvD1 (Cayman Chemicals; i.p.) immediately prior to zymosan injection and peritoneal exudates were collected after 24 hours. Exudate leukocyte numbers were determined using Trypan blue staining, which was analyzed using an automated cell counter (TC20, Bio-Rad). Differential counts and efferocytosis were assessed using flow cytometry (FACSVerse, BD Biosciences) as outlined below. Cytokine and chemokine levels were assessed in cell-free supernatants using ELISA, and RNA was isolated from pelleted peritoneal cells.
Blood and plasma analysis
Blood counts were analyzed using an automated hematology analyzer (Scil Vet abc hematology analyzer). Cholesterol and triglycerides were measured in EDTA-collected plasma using kits from Randox following the manufacturer’s instructions
Evaluation of atherosclerosis
The atherosclerotic lesion burden in the aortic arch was determined by en face analysis. In brief, dissected aortic arches were fixed in 4% neutral buffered formalin, cut longitudinally, pinned, and stained with 5% (w/v) Sudan IV solution (Sigma-Aldrich). Images were acquired using a Leica Microsystems color video camera. ImageJ (NIH) was used to quantify the total area of the aortic arch and the area of the atherosclerotic lesions, respectively, on the micrographs in a blinded fashion. Aortic roots were embedded in OCT (Histolab Products AB) and frozen at –80°C. Serial 10 μm thick cryosections were collected starting from the aortic valve cusps for a total distance of 800 μm and fixed in either formalin or acetone.
Formalin-fixed sections that were collected every 100 μm from the aortic valve cusps were stained with Oil Red O (Sigma-Aldrich), and images were captured with a light microscope equipped with a DC300 camera (Leica). Aortic root plaque burden was determined in a blinded fashion by planimetry of digital micrographs of 5 consecutive sections using ImageJ and expressed as lesion surface area relative to aortic root surface areas. Furthermore, the area of necrotic cores, as defined as unstained zones devoid of cells or extracellular matrix, was measured by planimetry of digital micrographs at a distance of 600 μm from the aortic valve cusps in a blinded fashion using ImageJ. The relative necrotic core area was first calculated for each individual plaque, and then the average of all plaques within the aortic root of 1 animal was formed. Lesion composition was analyzed using immunohistochemistry in lesion-matched sections (detailed in Supplemental Methods).
Real-time quantitative PCR
Samples from spleen, liver, abdominal fat, abdominal aortas, and peritoneal exudate cells were collected and snap-frozen for total RNA isolation on a QIAcube automated liquid handling system using QIAzol and QIAgen RNEasy Mini Kits (QIAgen). RNA concentration was spectrophotometrically determined using a Nanodrop 1000 (Thermo Fisher Scientific) and quality assessed by using a 2100 bioanalyzer (Agilent Technologies) prior to reverse transcription to cDNA using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific). Real-time quantitative PCR (RT-qPCR) was performed on a 7900HT Fast Real-Time PCR system (Thermo Fisher Scientific) using TaqMan Assay-On-Demand primers and probes (Supplemental Table 4; Thermo Fisher Scientific). Results were expressed as fold changes from control using the ΔCT method by comparing the threshold cycle (CT) for the gene of interest with that obtained using hypoxanthine phosphoribosyl-transferase (Hprt) and TATA-binding protein (Tbp) as housekeeping genes.
Flow cytometry
GPR32 expression was assessed in whole blood from hGPR32mycTg×Fpr2–/–×Apoe–/– (hGPR32myc transgenic) mice and Fpr2–/–×Apoe–/– (nontransgenic) mice. Briefly, red blood cells were lysed using Fix/Lyse Solution (Thermo Fisher Scientific) for 15 minutes at room temperature and washed twice with Flow Cytometry Staining Buffer (Thermo Fisher Scientific). Cells were then incubated with Fc block for 10 minutes, followed by addition of 1 μg rabbit anti-human GPR32 N-terminal (GTX108119, GeneTex) or rabbit IgG isotype control (GTX35035, GeneTex) for 30 minutes at room temperature. Cells were washed and labeled with an Alexa Fluor 647 secondary goat anti-rabbit Ab (Invitrogen) for 30 minutes and protected from light. Cells were then washed and staining assessed using BD Fortessa (BD Biosciences) and FlowJo Software (Tree Star).
To analyze the cellular composition of exudates, 0.5 × 106 peritoneal exudate cells were washed in FACS staining buffer (PBS containing 5% FBS), incubated with Fc block for 10 minutes at 4°C, and labeled with the following fluorescently conjugated rat anti-mouse Abs: F4/80-PE (catalog 12-4801-82, eBioscience), CD11b-APC (catalog RM2805, Invitrogen), and Ly6G–Pacific blue (catalog 127612, BioLegend).
For the assessment of macrophage efferocytosis of apoptotic neutrophils in vivo, inflammatory peritoneal exudate cells were fixed in 4% paraformaldehyde and labeled for extracellular F4/80-PE for 30 minutes at room temperature. The cells were then permeabilized (Cytoperm; BD Bioscience) and labeled for intracellular neutrophils using Ly6G–Pacific blue for 30 minutes at room temperature (38). All Abs were used at a 1:100 dilution. Cells were washed and staining was assessed using the BD FACSVerse flow cytometer (BD) and the FlowJo software package (Tree Star).
Peritoneal macrophage phagocytosis
Peritoneal macrophages from naive hGPR32mycTg×Fpr2–/–×Apoe–/– and Fpr2–/–×Apoe–/– mice were collected in 5 ml of DPBS–/– (Gibco, Thermo Fisher Scientific). Cells were counted and seeded (1 × 105 cells/well) in 96-well, flat, clear-bottom, black-walled plates (Corning) and allowed to adhere for 2 hours at 37°C. Nonadherent cells were washed and cells allowed to recover for 24 hours in 10% FBS RPMI media (as described for BMDMs above). Macrophages were preincubated with either vehicle (0.01% EtOH) or 100 nM AT-RvD1 (Cayman Chemicals) in DPBS+/+ (Gibco, Thermo Fisher Scientific) for 15 minutes at 37°C, followed by the addition of pH-rodo fluorescently labeled zymosan (1:10; macrophage;zymosan) or E. coli (1:50; macrophage: E. coli) (both from Invitrogen, Thermo Fisher Scientific). Phagocytosis was assessed after 1 hour using an Infinite M1000 Microplate Reader and Magellan software, version 6.6 (Tecan). For the assessment of oxLDL phagocytosis, cells were incubated with 10 μg/mL FITC-labeled oxLDL in 2% FBS RPMI 1640 medium (Thermo Fisher Scientific) in an IncuCyte ZOOM live-cell imaging and analysis platform (Essen BioScience) for 24 hours. Four images per well from 3 replicates were taken every 2 hours for a total of 24 hours using a ×20 objective lens and analyzed using IncuCyte basic software following the manufacturer’s instructions.
Cell signaling
To assess intracellular signaling via the transgenic GPR32myc receptor, peritoneal cells from hGPR32mycTg×Fpr2–/–×Apoe–/– and Fpr2–/–×Apoe–/– mice were incubated with 10 and 100 nM AT-RvD1, respectively, for 5 minutes at 37°C. Incubation was stopped by addition of 1.6% of paraformaldehyde for 10 minutes at room temperature. Cells were permeabilized in 80% ice-cold methanol and stored at –20°C. After permeabilization, cells were washed twice with Flow Cytometry Staining Buffer (eBioscience, Thermo Fisher Scientific) and then stained with anti-mouse/human PE-pERK1/2 (T202/Y204) (clone MILAN8R) (eBioscience, Thermo Fisher Scientific) and APC-pCREB (S133) (clone 4D11) (Invitrogen, Thermo Fisher Scientific) or their respective isotype controls. Levels of pERK1/2 and pCREB were then assessed using flow cytometry on a BD Fortessa Flow Cytometer (BD Bioscience) and the FlowJo software package (Tree Star).
ELISA
Supernatants of LPS-stimulated BMDMs were collected and frozen at –80°C until analysis. Samples were diluted 1:5 in dilution buffer, and TNF-α concentration was quantified using the Mouse TNF-α DuoSet ELISA Kit (R&D Systems) according to the manufacturer′s protocol. Exudate samples from the peritonitis model were quantified by ELISA MAX Mouse MCP-1/IL-6 ELISA Kits (both from BioLegend) and the Mouse IL-1β DuoSet ELISA Kit (R&D Systems) according to the manufacturer’s protocol.
Solid-phase extraction and LC-MS/MS analysis
BMDM macrophage incubations were stopped by the addition of 2 volumes ice-cold methanol, and 250 μL ice-cold MeOH was added to snap-frozen aortic samples prior to manual homogenization. Deuterated internal standards for d4-LTB4, d5-LXA4 (640 pg), and d5-RvD2 (1200 pg) representing each chromatographic region of the targeted SPM were added to facilitate quantification. Following protein precipitation by the added MeOH, samples were extracted by solid-phase extraction on C18 columns (Biotage) and subjected to LC-MS/MS analysis. The system consisted of a QTrap 5500 (AB Sciex) equipped with Shimadzu LC-20AD HPLC and a Shimadzu SIL-20AC autoinjector (Shimadzu Corp.). SPMs were eluted in increasing concentrations of organic solvent, starting with methanol/water (50/50, vol/vol) from 0 to 2 minutes, then ethanol/water (80/20, vol/vol) from 2 to 14.5 minutes, and finally methanol/water (98/2, vol/vol) from 14.6 minutes to 25 minutes (0.5 mL/min flow rate). To monitor and quantify levels of targeted lipid mediators, a multiple reaction monitoring (MRM) coupled with information-dependent acquisition (IDA) and an enhanced product ion scan (EPI) method were developed with signature ion pairs, Q1 (parent ion) and Q3 (characteristic daughter ion) for each molecule. Data acquisition was performed in negative ionization mode, and identification was conducted using published criteria (39) with a minimum of 6 diagnostic ions. Linear calibration curve for each compound was obtained with r2 values ranging from 0.990 to 0.999, and detection limit was approximately 0.1 pg in this system.
Statistics
Correlations between GPR32 and immune marker mRNA levels in human atherosclerotic plaques were performed using Pearson’s correlation with FDR adjusted P values. Experimental data are expressed as median value with minimum to maximum bars. Statistically significant differences were determined by 2-tailed, nonpaired Student′s t test for comparisons between 2 groups or 2-way ANOVA comparing 2 or more groups, followed by Šidák’s post hoc test. P < 0.05 was considered significant. Analyses were performed using GraphPad Prism, version 8 (GraphPad Software).
Study approval
Human material was obtained from the BiKE project, which used written, informed consent in accordance with the Declaration of Helsinki. The use of material for the present study was approved by the Karolinska Institute ethics committee. All animal experiments were performed in accordance with institutional and national guidelines for the care and use of laboratory animals and were approved by the Ethical Committee of Northern Stockholm.
Author contributions
ST, HA, GK, MC, SCP, GA, VM, LT, ALG, RB, and ASC conducted experiments. ST, HA, VM, GK, ALG, and LT acquired and analyzed data. ST and MB designed the transgenic mouse strain. HA, ST, and MB conceived the project, managed all studies, and wrote the manuscript. All authors edited the manuscript. HA, MB, and SCP revised the manuscript with input from all coauthors. Order of first authors is alphabetical.
Supplementary Material
Acknowledgments
The authors thank Jessica Lundgren, Selameyhune Assefa, André Ilveus, and Anna-Lena Gustafsson for valuable support in the animal facility; Stephan Teglund and Erik Nilsson (KCTT) for generation of the transgenic mouse; Anneli Olsson for valuable experimental help in the lab; and Stephen Malin for supportive discussions. The authors also thank Peter Damberg for MRI measurements and analysis; Kristina Edvardsson for mouse handling support (Karolinska Experimental Research and Imaging Centre [KERIC]); and Fernando Santos, Elba Ochoa Bernad, and Rafael Gracia (Solutex) for LC-MS/MS measurements. The work was supported by the Swedish Research Council (no. 2019-01486), the Swedish Heart and Lung Foundation (grants 2018057, 120180572, and 20170311 to MB; 20184251 to ST; and 20190625 and 20190196 to HA), the King Gustaf V and Queen Victoria Freemason Foundation, and Region Stockholm (no. 20170365). ST was supported by a Deutsche Forschungsgemeinschaft fellowship (no. MU3857/1-1). HA was also supported by a Horizon 2020 Marie Skłodowska-Curie fellowship (no. 656817) and the Foundation for Geriatric Diseases at Karolinska Institute. SCP was supported by the Professor Nanna Svartz Foundation. DFJK and RB were supported by the Swedish Heart-Lung Foundation (grants 20180571, 20180572, and 20170311 to MB) and the Novo Nordisk Foundation (no. NNF15CC0018346 and 0064142).
Version 1. 10/26/2021
In-Press Preview
Version 2. 12/15/2021
Electronic publication
Funding Statement
ST
HA
SCP
DFJK, RB
DFJK, RB
MB
MB
MB
MB
HA
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(24):e142883.https://doi.org/10.1172/JCI142883.
See the related Commentary at Proresolving receptor tames inflammation in atherosclerosis.
Contributor Information
Hildur Arnardottir, Email: hildur.arnardottir@ki.se.
Silke Thul, Email: silke.thul84@gmail.com.
Sven-Christian Pawelzik, Email: svenchristianpawelzik@gmail.com.
Glykeria Karadimou, Email: glykeria.karadimou@ki.se.
Gonzalo Artiach, Email: gonzalo.artiach.castanon@gmail.com.
Alessandro L. Gallina, Email: alessandro.gallina@ki.se.
Victoria Mysdotter, Email: victoria.mysdotter@stud.ki.se.
Miguel Carracedo, Email: miguelcarracedoortiz@gmail.com.
Laura Tarnawski, Email: laura.tarnawski@ki.se.
April S. Caravaca, Email: april.caravaca@ki.se.
Roland Baumgartner, Email: roland.baumgartner@ki.se.
Daniel F.J. Ketelhuth, Email: daniel.ketelhuth@ki.se.
Peder S. Olofsson, Email: peder.olofsson@ki.se.
Gabrielle Paulsson-Berne, Email: Gabrielle.Berne@ki.se.
Göran K. Hansson, Email: goran.hansson@ki.se.
Magnus Bäck, Email: magnus.back@ki.se.
References
- 1.Serhan CN, Levy BD. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J Clin Invest. 2018;128(7):2657–2669. doi: 10.1172/JCI97943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugimoto MA, et al. Mediators of the resolution of the inflammatory response. Trends Immunol. 2019;40(3):212–227. doi: 10.1016/j.it.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Kasikara C, et al. The role of non-resolving inflammation in atherosclerosis. J Clin Invest. 2018;128(7):2713–2723. doi: 10.1172/JCI97950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Back M, et al. Inflammation and its resolution in atherosclerosis: mediators and therapeutic opportunities. Nat Rev Cardiol. 2019;16(7):389–406. doi: 10.1038/s41569-019-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carracedo M, et al. The resolution of inflammation through omega-3 fatty acids in atherosclerosis, intimal hyperplasia, and vascular calcification. Semin Immunopathol. 2019;41(6):757–766. doi: 10.1007/s00281-019-00767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fredman G, et al. An imbalance between specialized pro-resolving lipid mediators and pro-inflammatory leukotrienes promotes instability of atherosclerotic plaques. Nat Commun. 2016;7:12859. doi: 10.1038/ncomms12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazan HA, et al. Circulating inflammation-resolving lipid mediators RvD1 and DHA are decreased in patients with acutely symptomatic carotid disease. Prostaglandins Leukot Essent Fatty Acids. 2017;125:43–47. doi: 10.1016/j.plefa.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thul S, et al. Low salivary resolvin D1 to leukotriene B4 ratio predicts carotid intima media thickness: a novel biomarker of non-resolving vascular inflammation. Eur J Prev Cardiol. 2017;24(9):903–906. doi: 10.1177/2047487317694464. [DOI] [PubMed] [Google Scholar]
- 9.Merched AJ, et al. Atherosclerosis: evidence for impairment of resolution of vascular inflammation governed by specific lipid mediators. FASEB J. 2008;22(10):3595–3606. doi: 10.1096/fj.08-112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirault J, Back M. Lipoxin and resolvin receptors transducing the resolution of inflammation in cardiovascular disease. Front Pharmacol. 2018;9:1273. doi: 10.3389/fphar.2018.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnamoorthy S, et al. Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc Natl Acad Sci U S A. 2010;107(4):1660–1665. doi: 10.1073/pnas.0907342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Back M, et al. Update on leukotriene, lipoxin and oxoeicosanoid receptors: IUPHAR Review 7. Br J Pharmacol. 2014;171(15):3551–3574. doi: 10.1111/bph.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petri MH, et al. Aspirin-triggered lipoxin A4 inhibits atherosclerosis progression in apolipoprotein E–/– mice. Br J Pharmacol. 2017;174(22):4043–4054. doi: 10.1111/bph.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufton N, et al. Anti-inflammatory role of the murine formyl-peptide receptor 2: ligand-specific effects on leukocyte responses and experimental inflammation. J Immunol. 2010;184(5):2611–2619. doi: 10.4049/jimmunol.0903526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, et al. NLRP3 Inflammasome deficiency protects against microbial sepsis via increased lipoxin B4 synthesis. Am J Respir Crit Care Med. 2017;196(6):713–726. doi: 10.1164/rccm.201604-0892OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamoorthy S, et al. Resolvin D1 receptor stereoselectivity and regulation of inflammation and proresolving microRNAs. Am J Pathol. 2012;180(5):2018–2027. doi: 10.1016/j.ajpath.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laguna-Fernandez A, et al. ERV1/ChemR23 signaling protects against atherosclerosis by modifying oxidized low-density lipoprotein uptake and phagocytosis in macrophages. Circulation. 2018;138(16):1693–1705. doi: 10.1161/CIRCULATIONAHA.117.032801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petri MH, et al. The role of the FPR2/ALX receptor in atherosclerosis development and plaque stability. Cardiovasc Res. 2015;105(1):65–74. doi: 10.1093/cvr/cvu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiurchiu V, et al. Resolution of inflammation is altered in chronic heart failure and entails a dysfunctional responsiveness of T lymphocytes. FASEB J. 2019;33(1):909–916. doi: 10.1096/fj.201801017R. [DOI] [PubMed] [Google Scholar]
- 20.Bystrom J, et al. Resolution-phase macrophages possess a unique inflammatory phenotype that is controlled by cAMP. Blood. 2008;112(10):4117–4127. doi: 10.1182/blood-2007-12-129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid M, et al. Resolvin D1 polarizes primary human macrophages toward a proresolution phenotype through GPR32. J Immunol. 2016;196(8):3429–3437. doi: 10.4049/jimmunol.1501701. [DOI] [PubMed] [Google Scholar]
- 22.Sharma M, et al. Regulatory T cells license macrophage pro-resolving functions during atherosclerosis regression. Circ Res. 2020;127(3):335–353. doi: 10.1161/CIRCRESAHA.119.316461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norling LV, et al. Resolvin D1 limits polymorphonuclear leukocyte recruitment to inflammatory loci: receptor-dependent actions. Arterioscler Thromb Vasc Biol. 2012;32(8):1970–1978. doi: 10.1161/ATVBAHA.112.249508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claria J, et al. Resolvin D1 and resolvin D2 govern local inflammatory tone in obese fat. J Immunol. 2012;189(5):2597–2605. doi: 10.4049/jimmunol.1201272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devchand PR, et al. Human ALX receptor regulates neutrophil recruitment in transgenic mice: roles in inflammation and host defense. FASEB J. 2003;17(6):652–659. doi: 10.1096/fj.02-0770com. [DOI] [PubMed] [Google Scholar]
- 27.Viola JR, et al. Resolving lipid mediators maresin 1 and resolvin D2 prevent atheroprogression in mice. Circ Res. 2016;119(9):1030–1038. doi: 10.1161/CIRCRESAHA.116.309492. [DOI] [PubMed] [Google Scholar]
- 28.Drechsler M, et al. Hyperlipidemia-triggered neutrophilia promotes early atherosclerosis. Circulation. 2010;122(18):1837–1845. doi: 10.1161/CIRCULATIONAHA.110.961714. [DOI] [PubMed] [Google Scholar]
- 29.Petri MH, et al. Resolution of inflammation through the lipoxin and ALX/FPR2 receptor pathway protects against abdominal aortic aneurysms. JACC Basic Transl Sci. 2018;3(6):719–727. doi: 10.1016/j.jacbts.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang N, et al. Novel resolvin D2 receptor axis in infectious inflammation. J Immunol. 2017;198(2):842–851. doi: 10.4049/jimmunol.1601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang N, et al. Maresin 1 activates LGR6 receptor promoting phagocyte immunoresolvent functions. J Clin Invest. 2019;129(12):5294–5311. doi: 10.1172/JCI129448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norris PC, et al. A cluster of immunoresolvents links coagulation to innate host defense in human blood. Sci Signal. 2017;10(490):eaan1471. doi: 10.1126/scisignal.aan1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folkersen L, et al. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ Cardiovasc Genet. 2010;3(4):365–373. doi: 10.1161/CIRCGENETICS.110.948935. [DOI] [PubMed] [Google Scholar]
- 34.Gistera A, et al. Transforming growth factor-β signaling in T cells promotes stabilization of atherosclerotic plaques through an interleukin-17-dependent pathway. Sci Transl Med. 2013;5(196):196ra100. doi: 10.1126/scitranslmed.3006133. [DOI] [PubMed] [Google Scholar]
- 35.Sarajlic P, et al. Artificial intelligence models reveal sex-specific gene expression in aortic valve calcification. JACC Basic Transl Sci. 2021;6(5):403–412. doi: 10.1016/j.jacbts.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artiach G, et al. Omega-3 polyunsaturated fatty acids decrease aortic valve disease through the resolvin E1 and ChemR23 axis. Circulation. 2020;142(8):776–789. doi: 10.1161/CIRCULATIONAHA.119.041868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brancaleone V, et al. A vasculo-protective circuit centered on lipoxin A4 and aspirin-triggered 15-epi-lipoxin A4 operative in murine microcirculation. Blood. 2013;122(4):608–617. doi: 10.1182/blood-2013-04-496661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnardottir HH, et al. Aging delays resolution of acute inflammation in mice: reprogramming the host response with novel nano-proresolving medicines. J Immunol. 2014;193(8):4235–4244. doi: 10.4049/jimmunol.1401313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colas RA, et al. Identification and signature profiles for pro-resolving and inflammatory lipid mediators in human tissue. Am J Physiol Cell Physiol. 2014;307(1):C39–C54. doi: 10.1152/ajpcell.00024.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.