Abstract
Although severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines have shown efficacy against SARS-CoV-2, it is unknown if coronavirus vaccines can also protect against other coronaviruses that may infect humans in the future. Here, we show that coronavirus vaccines elicited cross-protective immune responses against heterologous coronaviruses. In particular, we show that a severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) vaccine developed in 2004 and known to protect against SARS-CoV-1 conferred robust heterologous protection against SARS-CoV-2 in mice. Similarly, prior coronavirus infections conferred heterologous protection against distinct coronaviruses. Cross-reactive immunity was also reported in patients with coronavirus disease 2019 (COVID-19) and in individuals who received SARS-CoV-2 vaccines, and transfer of plasma from these individuals into mice improved protection against coronavirus challenges. These findings provide the first demonstration to our knowledge that coronavirus vaccines (and prior coronavirus infections) can confer broad protection against heterologous coronaviruses and establish a rationale for universal coronavirus vaccines.
Keywords: COVID-19, Vaccines
Keywords: Adaptive immunity, Cellular immune response
Introduction
Coronaviruses have garnered attention for their potential to cause pandemics. In less than 20 years, there have been outbreaks from 3 coronaviruses with pandemic potential: severe acute respiratory syndrome 1 coronavirus (SARS-CoV-1), Middle East respiratory syndrome coronavirus (MERS), and, recently, severe acute respiratory syndrome 2 coronavirus (SARS-CoV-2). Various vaccines have shown efficacy in preventing coronavirus disease 2019 (COVID-19), but whether these vaccines protect against other coronaviruses remains unknown. It is also unclear whether prior coronavirus infections confer protection against other coronaviruses. Knowing whether coronavirus vaccines offer broad protection against different coronaviruses is crucial for vaccine development, because it would suggest that coronavirus vaccines can protect individuals, even if they are not completely matched to a specific coronavirus antigen. Moreover, knowing whether prior coronavirus infections provide cross-protection against other coronaviruses could help explain differences in COVID-19 susceptibility among humans.
In this study, we evaluated cross-reactive and cross-protective immunity elicited by coronavirus vaccines and coronavirus infections. Our studies show that coronavirus vaccines and coronavirus infections confer protection against heterologous coronaviruses.
Results
SARS-CoV-2 vaccines induce cross-reactive antibody responses against other coronaviruses in humans.
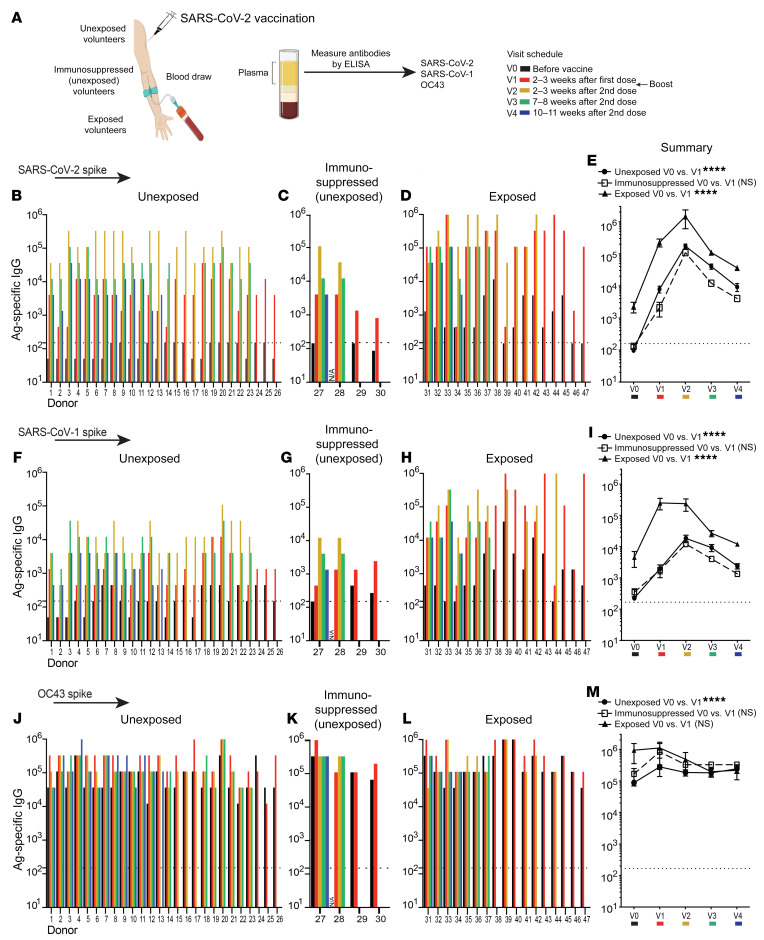
We first measured antibody responses following vaccination of humans with SARS-CoV-2 vaccines (Pfizer-BioNTek, Moderna, and Johnson & Johnson). We obtained plasma samples from human volunteers before vaccination and at several time points after vaccination (Figure 1A). These vaccinated individuals were identified as being unexposed to SARS-CoV-2 (asymptomatic and serologically negative); unexposed to SARS-CoV-2 and on immunosuppressive drugs (asymptomatic and serologically negative); or previously exposed to SARS-CoV-2 (symptomatic and RT-PCR+, prior to vaccination). Consistent with the RT-PCR results, we observed nucleocapsid-specific antibodies in most exposed individuals, but not in the unexposed individuals (Supplemental Figures 1, A–D; supplemental material available online with this article; https://doi.org/10.1172/JCI151969DS1). As expected, vaccination of humans with SARS-CoV-2 vaccines resulted in an increase in SARS-CoV-2 spike–specific antibodies (Figure 1, B–E). Consistent with prior reports (1, 2), the vaccine prime induced a more substantial increase in SARS-CoV-2–specific antibodies in individuals who were previously exposed to SARS-CoV-2 (Figure 1, D and E). Importantly, the SARS-CoV-2 vaccines also induced an increase in SARS-CoV-1 spike–specific antibodies, and previously exposed individuals showed more pronounced antibody responses relative to those of unexposed individuals (Figure 1, F–I).
Figure 1. Cross-reactive antibody responses following SARS-CoV-2 vaccination.
Antibody responses after SARS-CoV-2 vaccination. (A) Participants 1–17, 24–29, and 31–47 received the Pfizer/BioNTek vaccine; participants 18–23 received the Moderna vaccine; and participant 30 received the Johnson & Johnson vaccine. Participants were determined to be unexposed (participants 1–26) prior to vaccination on the basis of a negative serology test for SARS-CoV-2 spike and nucleocapsid proteins before vaccination (0–7 days prior to vaccination). Participants 27–30 were unexposed, under immunosuppressive regimens, and did not interrupt their treatments at the time of vaccination (treatments for participant 27: azathioprine and prednisone; participant 28: anti–IL-6 monoclonal antibody; participant 29: prednisone; and participant 30: methotrexate). Exposed participants 31–47 tested positive for SARS-CoV-2 by RT-PCR prior to vaccination. SARS-CoV-2 spike–specific antibody responses after vaccination in (B) unexposed, (C) unexposed immunosuppressed, and (D) exposed participants. (E) Summary of SARS-CoV-2 spike antibody responses. SARS-CoV-1 spike–specific antibody responses after vaccination in (F) unexposed, (G) unexposed immunosuppressed, and (H) exposed participants. (I) Summary of SARS-CoV-1 spike antibody responses. OC43 spike–specific antibody responses after vaccination in (J) unexposed, (K) unexposed immunosuppressed, and (L) exposed participants. (M) Summary of OC43 spike antibody responses. The y axis indicates the endpoint titer (the highest plasma dilution at which the absorbance was greater than 2 times that of the negative controls: human pre-2019 plasma; see Methods). Data shown are from an ongoing longitudinal study, in which participants were vaccinated on different dates, hence the heterogeneity in the available time points after infection. Antibody responses were evaluated by ELISA. Dashed lines represent the LOD. In panels E, I, and M, the indicated P values compare V0 and V1 from each group by paired Wilcoxon test. ****P < 0.0001, by paired Wilcoxon test (P > 0.05, NS). All participants except participant 28 (lack of V0 data) were included in the analysis. Error bars indicate the SEM.
We then quantified antibody responses against the spike protein of OC43, which is an endemic coronavirus that causes common colds in humans. All patients had high levels of preexisting antibody titers against OC43, but SARS-CoV-2 vaccination increased antibody titers against this endemic coronavirus in most unexposed (including immunosuppressed) participants (22 of 29, 76%) (Figure 1, J–M), consistent with earlier studies (3). Prior to vaccination, antibody responses to OC43 tended to be higher in individuals who were previously exposed to SARS-CoV-2 (Figure 1M).
We also evaluated bystander antibody levels before and after vaccination to determine whether SARS-CoV-2 vaccination increased noncoronavirus-specific immune responses. We found that antibodies against the influenza virus HA protein were not increased following SARS-CoV-2 vaccination, demonstrating that the increase in post-vaccination antibodies was specific to coronaviruses (Supplemental Figure 1, E–H). Taken together, these data show that SARS-CoV-2 vaccination elicits cross-reactive antibodies against other coronaviruses besides SARS-CoV-2.
Patients with COVID-19 show cross-reactive antibody responses against other coronaviruses.
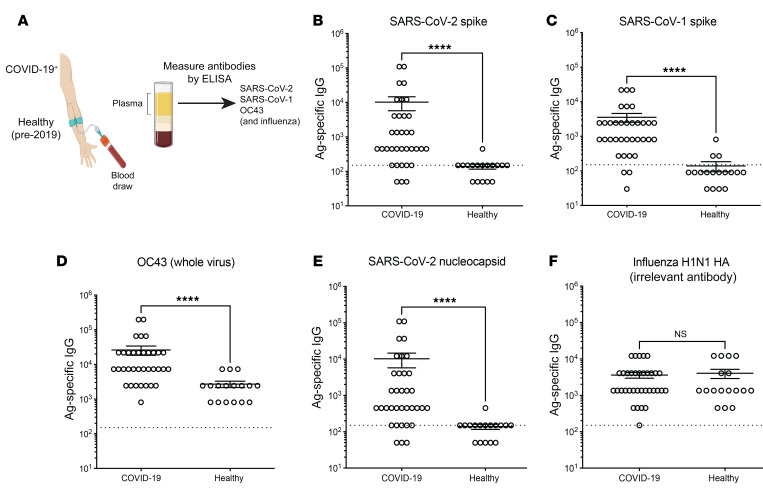
We next assessed whether cross-reactive antibodies could also be detected during a natural SARS-CoV-2 infection. We compared antibody responses in plasma from RT-PCR+, symptomatic patients with mild to severe COVID-19 as well as in healthy control plasma harvested before 2019 (Figure 2A. As expected (4), individuals with COVID-19 had higher levels of SARS-CoV-2 spike–specific antibodies (Figure 2B), as well as SARS-CoV-1 spike–specific (Figure 2C) and OC43-specific (Figure 2D) antibodies, relative to control individuals. We also measured antibody levels against the SARS-CoV-2 nucleocapsid protein for these 2 groups and found them to be significantly higher in patients with COVID-19 (Figure 2E). We did not observe any increase in influenza-specific antibodies in the COVID-19 cohort (Figure 2F). These data demonstrate that patients with COVID-19 develop cross-reactive antibody responses that recognize other coronaviruses.
Figure 2. Cross-reactive antibody responses following SARS-CoV-2 infection in humans.
Antibody responses following SARS-CoV-2 infection. (A) Participants in the COVID-19 group had a positive RT-PCR test accompanied by mild to severe symptoms. Serum samples (35 COVID-19 and 17 healthy controls) were collected once from week 3 to week 45 following symptom onset for the COVID-19 cohort. The healthy control cohort refers to human plasma collected prior to 2019. (B) SARS-CoV-2 spike–specific antibody responses. (C) SARS-CoV-1 spike–specific antibody responses. (D) OC43-specific antibody responses. OC43-infected cell lysate was used as a coating antigen. (E) SARS-CoV-2 nucleocapsid–specific antibody responses. (F) Influenza virus H1N1 HA–specific antibodies. Antibody responses were evaluated by ELISA. Dashed lines represent the LOD. ****P < 0.0001, by nonparametric Mann-Whitney U test. Error bars indicate the SEM.
Characterization of cross-reactive antibody responses with multiple SARS-CoV-2 vaccine modalities.
Our experiments above showed that SARS-CoV-2 vaccines induced antibody responses against heterologous coronaviruses in humans. Most of the vaccinated volunteers received mRNA vaccines, and we then assessed whether this effect was generalizable to other vaccine platforms. We primed C57BL/6 mice intramuscularly with various SARS-CoV-2 vaccines similar to the approved vaccines or experimental vaccines that have been used around the world during the COVID-19 pandemic, including adenovirus-based, vesicular stomatitis virus–based (VSV-based), mRNA-based, receptor-binding domain (RBD) protein–based, spike protein–based, and inactivated virus–based vaccines. We boosted mice homologously at approximately 3 weeks to recapitulate the regimen in most human trials, and we evaluated antibody responses 2 weeks after the boost.
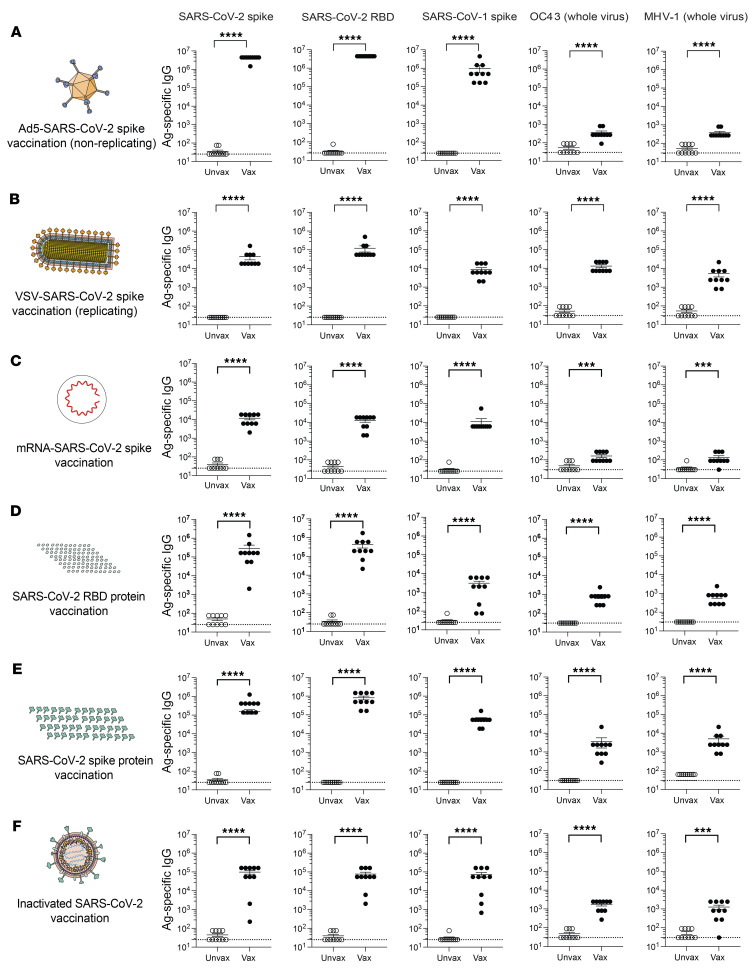
Consistent with our data in humans, vaccination of mice with an adenovirus vector expressing SARS-CoV-2 spike (Ad5-SARS-CoV-2 spike) resulted in potent antibody responses against SARS-CoV-2 and SARS-CoV-1 and a more modest, but statistically significant, increase in antibody responses against more distant coronaviruses, including OC43 and mouse hepatitis virus 1 (MHV-1) (Figure 3A). We found that cross-reactive antibody responses were also elicited by VSV-based, mRNA-based, RBD protein–based, spike protein–based, and inactivated virus–based vaccines (Figure 3, B–F). We also performed control experiments to measure cross-reactive antibody levels in mice that received sham vaccines lacking coronavirus spike transgenes. Vaccination with sham vectors did not elicit SARS-CoV-1– or SARS-CoV-2–specific antibodies (Supplemental Figure 2). Altogether, these data showed that multiple SARS-CoV-2 vaccine platforms were able to elicit cross-reactive antibody responses that recognized other coronaviruses.
Figure 3. Cross-reactive antibody responses following SARS-CoV-2 vaccination in mice.
(A) Antibody responses after Ad5-SARS-CoV-2 spike vaccination. (B) Antibody responses after VSV-SARS-CoV-2 spike vaccination. (C) Antibody responses after mRNA-SARS-CoV-2 spike vaccination. (D) Antibody responses after SARS-CoV-2 RBD vaccination. (E) Antibody responses after SARS-CoV-2 “whole” spike vaccination. (F) Antibody responses after inactivated SARS-CoV-2 vaccination. Mice were primed intramuscularly and boosted after 3 weeks (see Methods for vaccine dosing information). Antibody responses were evaluated by ELISA at week 2 after the boost. Experiments were conducted using wild-type C57BL/6 mice, except for VSV-SARS-CoV-2 spike vaccination, in which k18-hACE2 (C57BL/6) mice were used. Dashed lines represent the LOD. Data are from 2 independent experiments with 5 mice/group. Data from all experiments are shown. ***P < 0.001 and **** P < 0.0001, by Mann-Whitney U Test. Error bars indicate the SEM.
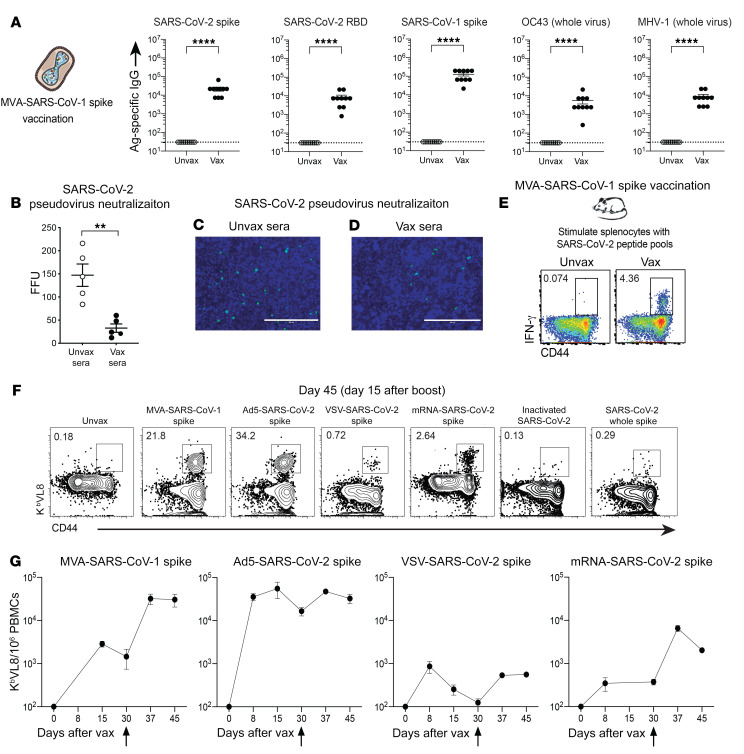
We then interrogated whether a vaccine against a different SARS coronavirus spike protein could also induce cross-reactive antibodies. Similarly, cross-reactive antibodies were observed with an experimental SARS-CoV-1 spike vaccine developed in 2004, based on modified vaccinia Ankara (MVA-SARS-1 spike), which was previously shown to protect mice and macaques against a SARS-CoV-1 challenge (refs. 5, 6 and Figure 4A). Interestingly, sera from MVA-SARS-CoV-1–vaccinated mice partially neutralized SARS-CoV-2 pseudovirus in vitro (Figure 4, B–D). These data show that immunization with a SARS-CoV-1 vaccine also elicited cross-reactive neutralizing antibodies against SARS-CoV-2 and other coronaviruses.
Figure 4. SARS-CoV-1 vaccination induces cross-reactive antibodies and T cells.
(A) Antibody responses after MVA-SARS-CoV-1 spike vaccination. (B) SARS-CoV-2 pseudovirus neutralization assay. SARS-CoV-2 pseudoviruses (200 FFU) were incubated with mouse sera diluted 1:4 prior to addition onto a HEK293-hACE2 cell monolayer. (C and D) Representative microscopic images of SARS-CoV-2 pseudovirus neutralization using sera from (C) unvaccinated mice (Unvax sera) or (D) SARS-CoV-1–vaccinated (Vax sera) mice. Scale bars: 400 μm. (E) Representative FACS plots showing cross-reactive SARS-CoV-2–specific CD8+ T cells in SARS-CoV-1–vaccinated mice. Cross-reactive CD8+ T cells were detected by intracellular cytokine staining after a 5-hour stimulation with SARS-CoV-2 spike overlapping peptide pools, in a 37°C, 5% CO2 incubator. Cells were gated on live CD8+ lymphocytes. Data are from splenocytes on post-boost day 15. (F) Representative FACS plots showing cross-reactive (VNFNFNGL-specific) CD8+ T cells in mice vaccinated with a SARS-CoV-1 vaccine and in mice vaccinated with various other SARS-CoV-2 vaccines. Cells were gated on live CD8+ lymphocytes. Data are from PBMCs on post-boost day 15. (G) Summary of CD8+ T cell responses among vaccine platforms. All mice were primed and boosted intramuscularly (see Methods for vaccine dosing information). Vertical arrows in G indicate the time of the boost. Experiments were done using wild-type C57BL/6 mice, except for VSV-SARS-CoV-2 spike vaccination, in which k18-hACE2 (C57BL/6) mice were used. In A, data are from 2 independent experiments with 5 mice/group; data from all experiments are shown, and dashed lines represent the LOD. In B, data are from 1 experiment with 5 mice/group. In E and F, representative results of experiments performed twice with 5 mice/group are shown. Panel G shows a summary of the 2 experiments combined. **P < 0.01 and ****P < 0.0001, by Mann-Whitney U test. Error bars indicate the SEM.
Following a viral infection, viral control is facilitated by CD8+ T cells. To measure cross-reactive CD8+ T cell responses, we harvested splenocytes from mice that received the SARS-CoV-1 vaccine and stimulated these cells with SARS-CoV-2 spike peptides (Supplemental Table 1) for 5 hours, followed by intracellular cytokine staining (ICS) to detect cross-reactive (SARS-CoV-2 spike–specific) CD8+ T cells. Interestingly, the SARS-CoV-1 vaccine elicited SARS-CoV-2–specific CD8+ T cell responses (Figure 4E), suggesting the presence of conserved CD8+ T cell epitopes in SARS-CoV-1 and SARS-CoV-2. To identify cross-reactive CD8+ T cell epitopes, we performed spike sequence alignment (Supplemental Figure 3) followed by epitope mapping. We identified 2 highly conserved epitopes in the spike protein, in particular the VVLSFELL and VNFNFNGL epitopes, which are highly conserved among other SARS-like coronaviruses (Supplemental Figure 4A). These 2 epitopes were identified in a prior study in SARS-CoV-2–infected mice (7). The VNFNFNGL CD8+ T cell response has also been reported to be elicited after SARS-CoV-1 infection in C57BL/6 mice (8), and we show that it was also immunodominant after SARS-CoV-2 vaccination (Supplemental Figure 4A). Using MHC class I epitope prediction algorithms (see Methods), we found that both VVLSFELL and VNFNFNGL were predicted to bind the mouse MHC-I Kb.
We reasoned that Kb VNFNFNGL tetramers could be used to track cross-reactive CD8+ T cells following SARS-CoV-1 or SARS-CoV-2 vaccination across multiple vaccine platforms. The spike protein vaccine and the inactivated virus vaccine did not generate robust Kb VNFNFNGL (Kb VL8) CD8+ T cell responses (Figure 4F), probably because CD8+ T cell priming is dependent on intracellular protein expression. However, we observed robust Kb VL8 CD8+ T cell responses above the limit of detection (LOD) after vaccination with viral vectors or mRNA (Figure 4F). Among the different vaccines, adenovirus-based, MVA-based, and mRNA-based vaccines generated the strongest Kb VL8 CD8+ T cell response (Figure 4, F and G).
We then performed single-cell T cell receptor–sequencing (scTCR-Seq) analyses to determine whether the cross-reactive Kb VL8 response exhibits a biased TCR usage. We showed at RNA and protein levels that most of the Kb VL8 response contained a TCR composed of V7α/Vβ11 (Supplemental Figure 4, B–E). We are currently using this scTCR-Seq information to develop a TCR-transgenic mouse that could be used to study cross-reactive CD8+ T cells among different sarbecovirus infections. Altogether, our data showed that a SARS-CoV-1 vaccine also generated antibody and T cell responses that recognized other coronaviruses. In particular, these data suggested that an old SARS-CoV-1 vaccine could protect against SARS-CoV-2.
A SARS-CoV-1 vaccine protects against a SARS-CoV-2 challenge.
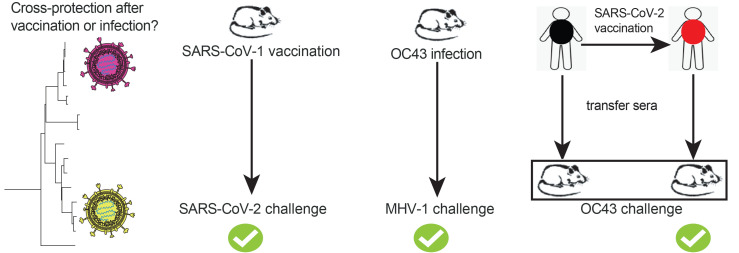
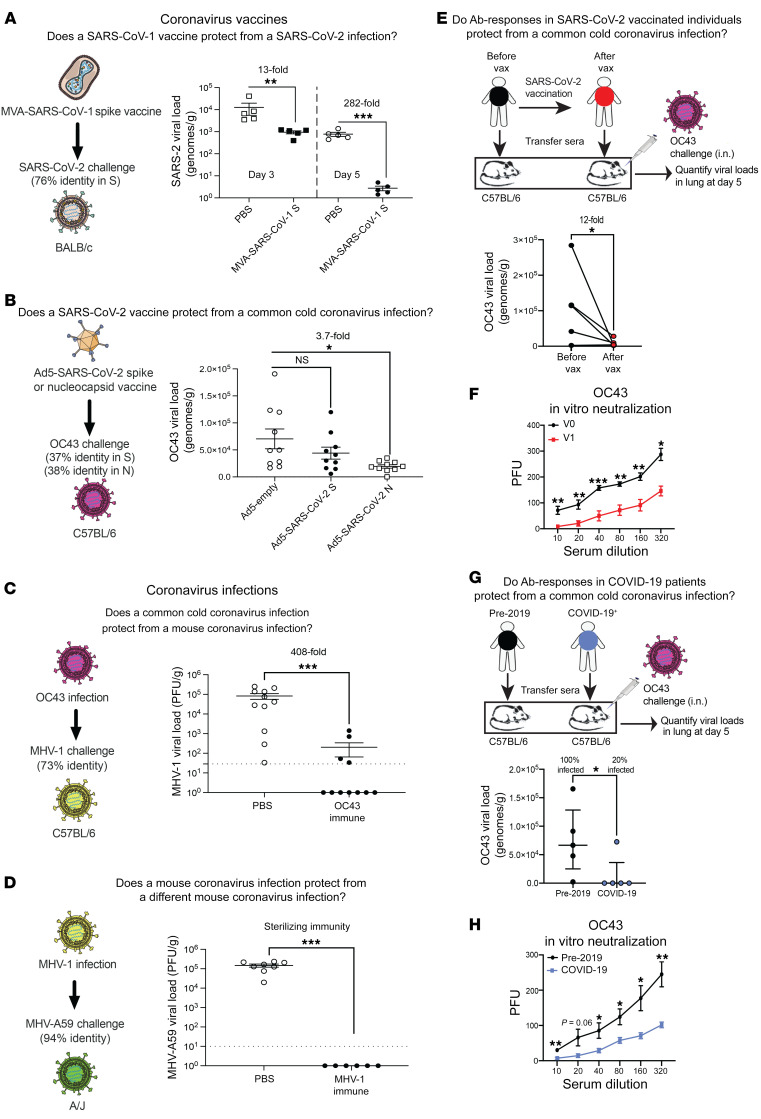
There are concerns about emerging SARS-CoV-2 variants and the possibility that they could escape vaccine-elicited protection (9). Furthermore, it is possible that SARS-CoV-1 may spill over again into the human population. Thus, a critical question is whether SARS-CoV-2 vaccines could also protect against SARS-CoV-1, as well as other bat coronaviruses. To answer this simple question, we performed challenge experiments to evaluate whether coronavirus vaccines could protect against heterologous coronaviruses. SARS-CoV-1 is a select agent, so we were not able to challenge SARS-CoV-2–vaccinated animals with SARS-CoV-1 in our BL3 facilities. Instead, we evaluated whether an old SARS-CoV-1 vaccine developed in 2004 could protect against a SARS-CoV-2 challenge. We immunized mice with a SARS-CoV-1 spike vaccine developed by Bernard Moss (MVA-SARS-1 spike; ref. 5), and then challenged mice intranasally with SARS-CoV-2. On post-challenge day 5, we harvested lungs and measured viral loads by RT-PCR. Strikingly, this SARS-CoV-1 vaccine conferred a 282-fold decrease in viral loads following a SARS-CoV-2 challenge (Figure 5A). We also observed improved control of SARS-CoV-2 at an earlier time point (day 3; Figure 5A). These data demonstrate that a sarbecovirus vaccine with a large antigenic mismatch (only 76% identity) could still confer robust protection following a heterologous sarbecovirus challenge.
Figure 5. Cross-protective immunity following coronavirus vaccination or coronavirus infection.
(A) Viral loads after SARS-CoV-2 (MA10) challenge in SARS-CoV-1–vaccinated mice. LOD = 0.007 genomes/g. (B) Viral loads after OC43 challenge in SARS-CoV-2–vaccinated mice. LOD = 27 genomes/g. (C) Viral loads after MHV-1 challenge in OC43-immune mice. (D) Viral loads after MHV-A59 challenge in MHV-1–immune mice. In A–D, mice were intramuscularly primed and boosted after 3 weeks (see Methods). Mice were challenged intranasally 2 weeks after the boost. (E) Viral loads after OC43 challenge in mice that received 50 μL human plasma (before or after vaccination). Plasma was adoptively transferred into naive mice, and on day 1 after transfer, mice were challenged intranasally with OC43. OC43 IgG titers before vaccination (V0) ranged from 12,150 to 109,350, and after vaccination (V1), they ranged from 328,050 to 984,150. (F) OC43 PRNT in plasma before and after vaccination. (G) Viral loads after OC43 challenge in mice that received 50 μL human plasma (pre-2019 vs. COVID-19). Plasma was adoptively transferred into naive mice, and on day 1 after transfer, mice were challenged intranasally with OC43. OC43 IgG titers were 4050 for all pre-2019 samples and 63,450 for all COVID-19 samples. (H) OC43 PRNT in plasma from pre-2019 donors and patients with COVID-19. Lung viral loads were quantified by RT-PCR (A, B, E, and G) or plaque assays (C, D, F, and H). Data are from day 5 after challenge unless otherwise indicated. Dashed lines represent the LOD. A–D data are from 2 independent experiments with 3–5 mice/group. Data in E–H are from 1 experiment with 5 mice/group. Data from all experiments are shown. In A–D, F, and H, error bars indicate the SEM. Panels E and G show paired values. Values below the LOD in log scales represent zero values. *P < 0.05, **P < 0.01, and ***P < 0.001, by nonparametric Mann-Whitney U test (A–D, F, and H) and paired t test (E and G).
Humans are constantly exposed to endemic coronaviruses, including the embecovirus OC43, and our next question was whether SARS-CoV-2 vaccines protect against this endemic coronavirus. To answer this question, we immunized mice with an Ad5 vector expressing either SARS-CoV-2 spike or nucleocapsid, and then challenged these mice intranasally with OC43. On post-challenge day 5, we harvested lungs and measured viral loads by RT-PCR. The SARS-CoV-2 nucleocapsid vaccine conferred a 3.7-fold viral load decrease relative to control viral loads following this common cold coronavirus challenge (Figure 5B). We observed no significant heterologous protection with the spike-based vaccine (Figure 5B). These data suggest that the degree of cross-protection is affected by the genetic similarity between the vaccine antigen and the challenge antigen. In other words, a sarbecovirus vaccine conferred robust protection against a related sarbecovirus challenge (Figure 5A), but only slight (or negligible) protection against an embecovirus challenge (Figure 5B).
Prior coronavirus infections confer protection against future coronavirus infections.
Similar to our data on patients with COVID-19, we found that coronavirus infections in mice also induced cross-reactive antibody responses. In particular, a common cold coronavirus (OC43) infection elicited cross-reactive antibodies against SARS-CoV-2, SARS-CoV-1, and MHV-1 (Supplemental Figure 5A). Cross-reactive antibodies were also generated after an MHV-1 infection (Supplemental Figure 5B). We thus tested whether mice that had prior coronavirus infections were better protected following heterologous coronavirus challenges. In our first model, we challenged OC43-immune mice with MHV-1. Note that OC43 and MHV-1 are 2 embecoviruses that are more genetically distant, relative to SARS-CoV-1 and SARS-CoV-2. Interestingly, OC43-immune mice exhibited a 408-fold viral load reduction following a heterologous MHV-1 challenge (Figure 5C).
In our second model, we challenged MHV-1–immune mice with MHV-A59. Although these 2 coronaviruses have similar names, they are genetically distinct. Note that MHV-1 and MHV-A59 are more genetically distant than SARS-CoV-2 and RaTG13. Interestingly, MHV-1–immune mice exhibited sterilizing immune protection against a heterologous MHV-A59 challenge (Figure 5D). These data demonstrate that prior coronavirus infections can confer protection against subsequent infections with related coronaviruses. Moreover, the degree of heterologous protection appeared to be influenced by the genetic similarity between the initial coronavirus infection and the subsequent coronavirus infection (Figure 5, C and D).
Mechanism: antibodies are sufficient for cross-protection.
Measuring cross-protection in humans is difficult, because most people are already seropositive for endemic coronaviruses. In addition, susceptibility to coronavirus infection can be influenced by many variables, including the immune history of the host, comorbidities, age, and socioeconomic status, rendering it difficult to determine whether SARS-CoV-2 vaccination or SARS-CoV-2 infection protect against other coronaviruses. Therefore, we developed a reductionist animal model that allowed us to better discern heterologous immune protection by vaccine-elicited antibodies, using plasma from humans who received SARS-CoV-2 vaccines. In our first experiment, we obtained longitudinal plasma from human volunteers, before and after receiving SARS-CoV-2 vaccines. We then transferred these donor-matched human plasma into naive C57BL/6 mice via intraperitoneal injection, and on the following day, we challenged these mice with the common cold coronavirus OC43. Mice that received post-vaccination human plasma exhibited a 12-fold lower OC43 viral load relative to mice that received pre-vaccination human plasma (Figure 5E). To explore the mechanism of immune protection, we performed plaque reduction neutralization titer (PRNT) assays using these donor-matched plasma samples (before and after vaccination). Human plasma harvested 2–3 weeks after SARS-CoV-2 vaccination showed more robust in vitro OC43 neutralization relative to matched pre-vaccination plasma (Figure 5F). These data show that SARS-CoV-2 vaccination in humans elicited humoral responses that conferred protection against a different coronavirus.
In our second experiment, we obtained plasma from patients with COVID-19 versus plasma from individuals before the 2019 pandemic. We transferred these human plasma samples into naive C57BL/6 mice, and on the following day, we challenged these mice with OC43. Plasma from COVID-19 patients induced sterilizing immunity to OC43 in 80% of the mice, whereas all of the mice that received pre-2019 human plasma showed detectable viral loads (Figure 5G). Human plasma from patients with COVID-19 also showed more robust in vitro OC43 neutralization by PRNT assay relative to that observed in pre-2019 plasma (Figure 5H). These data showed that antibody responses elicited by SARS-CoV-2 infection in humans conferred protection against an endemic coronavirus. Altogether, these data demonstrate that immunity elicited by SARS-CoV-2 vaccination or SARS-CoV-2 infection could provide cross-protection against common cold coronavirus infections.
Discussion
Several SARS-CoV-2 vaccines have been used in humans, but it is unknown if these vaccines could also protect against other viruses, including pandemic or endemic coronaviruses. In this study, we show that SARS-CoV-2 vaccination in humans elicits cross-reactive antibodies against SARS-CoV-1 and the common cold coronavirus OC43. Our subsequent studies in mice demonstrated that a SARS-CoV-1 vaccine protected against a SARS-CoV-2 challenge and that prior coronavirus infections can protect against subsequent infections with other coronaviruses.
Coronavirus vaccines have been previously shown to elicit cross-reactive antibodies (10–12), but until now, it has been unclear whether these antibodies cross-protect in vivo. We believe our data bring clarity to the question of cross-protection and suggest that cross-protection is proportional to the level of genetic conservation. For example, vaccination with a SARS-CoV-1 spike vaccine conferred robust protection against a SARS-CoV-2 challenge (which was 76% antigen-matched), but cross-protection was more limited when the challenge virus was more distant (Figure 5, A and B). Similarly, we observed a positive correlation between genetic similarity and cross-protection in the context of coronavirus infections. For example, OC43-immune mice showed partial protection against an MHV-1 challenge (73% antigenically matched). However, MHV-1-immune mice showed sterilizing protection against an MHV-A59 challenge (94% antigenically matched). Furthermore, our challenge data in Figure 5, A–D, suggest that it is more likely for a vaccine to confer cross-protection within (but not across) subgenera.
Immune cross-reactivity in the context of coronavirus vaccination or coronavirus infection is based on genetic conservation. Interestingly, vaccination with “whole” spike protein induced higher levels of cross-reactive antibodies relative to vaccination with RBD protein (Figure 3, D and E). This is likely due to the higher number of conserved epitopes in the “whole” spike protein, relative to RBD alone. Our data also suggest that the level of OC43-MHV cross-reactivity may vary between viral vector platforms, as replicating viral vectors (Figure 3B and Figure 4A) tended to generate higher levels of these cross-reactive responses compared with nonreplicating Ad5 vectors (Figure 3A).
Additionally, our human plasma transfer experiments suggest that SARS-CoV-2 spike-based vaccination confers partial protection against OC43 (Figure 5, E and F). However, SARS-CoV-2 spike-based vaccination in mice did not confer significant protection against OC43 (Figure 5B). This difference may be explained by the high levels of OC43-specific antibody in human plasma. It is possible that preexisting humoral immunity to OC43 improves the maturation of OC43-specific antibody in humans, upon SARS-CoV-2 vaccination.
Prior studies have suggested that recent endemic coronavirus infections in humans are associated with less severe COVID-19 (13). However, other studies have shown contradicting results (10). Such discrepancy can be explained by the heterogeneous “immune histories” of humans, including the fact that humans are frequently reexposed with endemic coronaviruses. In addition, age, sex, and preexisting conditions in humans can significantly influence COVID-19 susceptibility, making it difficult to quantify the antiviral effect of cross-reactive immune responses elicited by prior coronavirus infections. Our plasma transfer studies elucidate this issue of heterologous protection, as we transferred donor-matched plasma from humans (before and after vaccination) into naive, sex-matched, genetically identical recipient mice that had no preexisting immunity to any coronavirus (Figure 5, E and F).
A limitation of our study is that we only evaluated heterologous immune protection at an early point after vaccination or after infection, and it is possible that cross-protection declines over time. Future studies will determine the durability of cross-protection and whether cross-reactive antibodies are produced by plasma cells or short-lived plasmablasts. There is a critical point that is worth discussing further, as it could facilitate vaccine preparedness for future pandemics: we show that a single coronavirus vaccine based on the original SARS-CoV-1 could confer robust heterologous protection against SARS-CoV-2 and that the vaccine antigen did not need to fully match the viral challenge. There are ongoing discussions about how to better prepare for future coronavirus pandemics, and our data suggest that it would be reasonable to archive a stockpile of vaccine candidates based on known sequenced coronaviruses. Upon the start of an outbreak, the most “antigenically matched” vaccine from the preexisting catalog could be immediately tested and thus save vaccine manufacturing time. Even if vaccine protection is only partial with an antigenically mismatched vaccine, this approach may slow down viral transmission and mitigate clinical outcomes until more antigenically matched vaccines are developed. Overall, these findings provide a framework for the rational design of pancoronavirus vaccines and may help vaccine preparedness for future pandemics.
Methods
Human participants
Participants who met the following criteria were included in the study: age 18 or older, a SARS-CoV-2 infection and/or a scheduled COVID-19 vaccination, and the ability and willingness to provide informed consent. Candidates who were under the age of 18 or were unwilling or unable to provide informed consent were excluded from the study. Enrollment started on August 2020 and is expected to be completed by August 2022. The study population consisted of residents across the Chicago area. Adults of different ages, races, and ethnicities were included in the study. Participants were deidentified by assigning them a 4-letter study code to be used for the duration of the study. Participants who were considered to be exposed before vaccination had a positive RT-PCR test for SARS-CoV-2 any time prior to vaccination. Blood was collected by phlebotomy using BD Vacutainer 10 mL tubes containing sodium heparin. Anticoagulated blood was added to LeucoSep tubes (Greiner Bio), and plasma was separated by density gradient centrifugation. To protect the individual’s identity, all samples were labeled with their assigned 4-letter study code and stored in the principal investigator’s laboratory freezers.
Mice, vaccinations, infections, and challenges
Six- to 8-week-old C57BL/6, BALB/c, A/J mice were used. For VSV-SARS-2 spike vaccinations, k18-hACE2 (on C57BL/6 background) mice were used. All mice were purchased from The Jackson Laboratory (approximately half of the mice were males and half were females) and housed at Northwestern University’s Center for Comparative Medicine (CCM) or the UIC. Mice were immunized intramuscularly (50 μL per quadriceps) with Ad5-SARS-CoV-2 spike (109 PFU); VSV expressing SARS-CoV-2 spike protein (VSV-SARS-CoV-2 spike; 107 PFU); mRNA-based vaccine encoding SARS-CoV-2 spike protein (mRNA-SARS-CoV-2 spike; 5 μg); SARS-CoV-2 “whole-spike” protein (SARS-CoV-2 spike; 100 μg with 1:5 Adju-Phos, InvivoGen); SARS-CoV-2 RBD protein (100 μg with 1:5 Adju-Phos); γ-irradiated SARS-CoV-2 (inactivated SARS-CoV-2; 2.5 × 105 PFU); and MVA expressing SARS-CoV-1 spike protein (MVA-SARS-CoV-1 spike; 107 PFU). The vaccine doses were chosen empirically on the basis of prior studies by us and others (5, 14–22).
We obtained Ad5-SARS-CoV-2 spike from the University of Iowa Viral Vector Core (VVC-U-7643, Iowa City, Iowa, USA); VSV-SARS-CoV-2 spike from Sean Whelan (Washington University in St. Louis, St. Louis, Missouri, USA); and MVA-SARS-CoV-1 spike from the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, (NR-623, originally developed by Bernard Moss at the NIH (5). We obtained Ad5-SARS-CoV-2 nucleocapsid from David Masopust (University of Minnesota, Minneapolis, Minnesota, USA); this vector has been used in prior studies (15, 23).
We synthesized mRNA vaccines encoding for the codon-optimized SARS-CoV-2 spike protein from the strain USA-WA1/2020. Constructs were purchased from Integrated DNA Technologies (IDT) and contained a T7 promoter site for in vitro transcription of mRNA. The sequences of the 5′- and 3′-UTRs were identical to those in a Dengue virus mRNA vaccine documented in a previous publication (19). mRNA was synthesized from linearized DNA with T7 In Vitro Transcription Kits, following the manufacturer’s protocol. RNA was generated with pseudouridine in place of uridine with the Incognito mRNA Synthesis Kit (CellScript, catalog C-ICTY110510). A 5′ cap-1 structure and a 3′ poly-A tail were enzymatically added. mRNA was encapsulated into lipid nanoparticles using the NanoAssemblr Benchtop system (Precision NanoSystems). mRNA was dissolved in Formulation Buffer (catalog NWW0043, Precision NanoSystems) and run through a laminar flow cartridge with GenVoy ILM (catalog NWW0041, Precision NanoSystems) encapsulation lipids at a flow ratio of 3:1 (RNA: GenVoy-ILM), with a total flow rate of 12 mL/min, to produce mRNA–lipid nanoparticles (mRNA-LNPs). These mRNA-LNPs were characterized for encapsulation efficiency and mRNA concentration via the RiboGreen assay using the Quant-iT RiboGreen RNA Assay Kit (catalog R11490, Invitrogen, Thermo Fisher Scientific).
SARS-CoV-2 spike and RBD proteins used for vaccinations were produced by Sergii Pshenychnyi at Northwestern’s University Recombinant Protein Production Core using the following plasmids produced under HHSN272201400008C and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), NIH: vector pCAGGS containing the SARS-related coronavirus 2; Wuhan-Hu-1 spike glycoprotein gene (soluble, stabilized); NR-52394 and RBD; and NR-52309. Protein vaccines were administered with 1:5 Adju-Phos (InvivoGen).
Inactivated SARS-CoV-2 was obtained from BEI Resources, NIAID, NIH (SARS-related coronavirus 2, isolate USA-WA1/2020, γ-irradiated, NR-52287). MHV-1 was purchased from the American Type Culture Collection (ATCC, VR-261), and OC43 was received from BEI Resources, NIAID, NIH (NR-52725). MHV-A59 was a gift from Susan Weiss (University of Pennsylvania, Philadelphia, Pennsylvania, USA).
For OC43 and MHV challenges, mice were infected intranasally (25 μL/nostril) with OC43 (2 × 106 PFU) or mouse hepatitis virus (MHV-1/MHV-A59; 106 PFU).
For SARS-CoV-2 challenges, mouse-adapted SARS-CoV-2 (MA10) was provided by Ralph Baric (University of North Carolina, Chapel Hill, North Carolina, USA) (24). SARS-CoV-2 (MA10) was propagated and titered on Vero-E6 cells (ATCC, CRL1586). BALB/c mice were anesthetized with isoflurane and challenged via intranasal inoculation with 8 × 103 foci-forming units (FFU SARS-CoV-2 (MA10). Lungs were isolated from mice 5 days after infection and homogenized in PBS. RNA was extracted from lung homogenate using a Zymo Research Quick-RNA 96 Kit (R1052). Viral genomes were quantified via quantitative RT-PCR with the N1 Primer/Probe Kit from Integrated DNA Technologies (IDT, catalog 10006713).
Protein-specific ELISA (SARS-CoV-2 spike, RBD, nucleocapsid; SARS-CoV-1 spike; OC43 spike)
Antigen-specific total antibody titers were measured by ELISA as described previously (16, 25). Briefly, 96-well, flat-bottomed MaxiSorp plates (Thermo Fisher Scientific) were coated with 1 μg/mL of the respective protein for 48 hours at 4°C. Plates were washed 3 times with wash buffer (PBS plus 0.05% Tween 20). Blocking was performed with blocking solution (200 μL PBS plus 0.05% Tween 20 plus 2% BSA) for 4 hours at room temperature. Six microliters of sera (plasma for human ELISAs) was added to 144 μL blocking solution in the first column of the plate, 1:3 serial dilutions were performed until row 12 for each sample, and plates were incubated for 60 minutes at room temperature. Plates were washed 3 times with wash buffer followed by addition of HRP-conjugated goat anti–mouse IgG secondary antibody (SouthernBiotech; diluted in blocking solution 1:5000) at 100 μL/well and incubated for 60 minutes at room temperature. For the ELISAs with human plasma samples, goat anti–human IgG (H+L) conjugated to HRP at 1:1000 (Jackson ImmunoResearch) was used. After washing the plates 3 times with wash buffer, 100 μL/well SureBlue Substrate (SeraCare) was added for 1 minute. The reaction was stopped using 100 μL/well KPL TMB Stop Solution (SeraCare). Absorbance was measured at 450 nm using a Spectramax Plus 384 (Molecular Devices). In all ELISA plots, the y axis indicates the endpoint titer (the sera or plasma dilution at which absorbance was greater than 2 times the average for the negative controls human pre-2019 plasma or mouse-naive sera). SARS-CoV-2 spike and RBD proteins used for ELISAs were produced by Sergii Pshenychnyi and Irina Shepotinovskaya at the Northwestern Recombinant Protein Production Core using the following plasmids produced under HHSN272201400008C and obtained from BEI Resources, NIAID, NIH: vector pCAGGS containing the SARS-related coronavirus 2; Wuhan-Hu-1 spike glycoprotein gene (soluble, stabilized); NR-52394 and RBD; and NR-52309. SARS-CoV-2 nucleocapsid protein was obtained through BEI Resources, NIAID, NIH (NR-53797). SARS-CoV-1 spike protein was also obtained through BEI Resources, NIAID, NIH (NR-722). OC43 spike protein was purchased from Sino Biological (40607-V08B).
Virus-specific ELISAs (OC43; MHV-1)
Virus-specific ELISAs were performed as described earlier (16, 25). In brief, 96-well, flat-bottomed MaxiSorp plates (Thermo Fisher Scientific) were coated with 100 μL/well of the respective viral lysate (OC43-, or MHV-1–infected cell lysates) diluted 1:10 in PBS for 48 hours at room temperature. Plates were washed 3 times with wash buffer (PBS plus 0.5% Tween 20) followed by blocking with blocking solution (200 μL/well PBS plus 0.2% Tween 20 plus 10% FCS) for 2 hours at room temperature. Five microliters of sera (plasma for human ELISAs) was added to 145 μL blocking solution in the first column of the plate, and 1:3 serial dilutions were performed until row 12 for each sample followed by incubation at room temperature for 90 minutes. Plates were washed 3 times with wash buffer, followed by addition of 100 μL/well HRP-conjugated goat anti–mouse IgG (SouthernBiotech), diluted 1:5000 in blocking solution. Plates were incubated for 90 minutes at room temperature. Goat anti–human IgG (H+L) conjugated to HRP (1:1000; Jackson ImmunoResearch) was used when ELISA was performed with human samples. After washing the plates 3 times with wash buffer, 100 μL/well SureBlue Substrate (SeraCare) was added for 8 minutes. The reaction was stopped using 100 μL/well KPL TMB Stop Solution (SeraCare). Absorbance was measured at 450 nm using a Spectramax Plus 384 (Molecular Devices).
Virus propagation
OC43 was propagated in a 80%–90% confluent monolayer of HCT-8 cells (ATCC, CCL-244) in T175 flasks at a MOI of 0.01, diluted in 5 mL RPMI supplemented with 2% FBS, 1% penicillin/streptomycin, and 1% l-glutamine. Infected cells were incubated at 33°C for 2 hours in a humidified 5% CO2 incubator. After incubation, the flasks were supplemented with 20 mL 2% RPMI and incubated for 5 days at 33°C in a CO2 incubator. MHV-A59 and MHV-1 were expanded in 17CL-1 cells (gift from Susan Weiss, University of Pennsylvania, Philadelphia, Pennsylvania, USA) following a previously published protocol (26).
OC43 and MHV quantification by plaque assay
For MHV quantification, 106 cells/well L2 cells (gift from Susan Weiss, University of Pennsylvania) were seeded onto 6-well plates in 10% DMEM (10% FBS, 1% penicillin/streptomycin, and l-glutamine). After 2 days, when cells reached approximately 100% confluence, the media were removed. Ten-fold serial dilutions of viral stock or homogenized lung were prepared in 1% DMEM (1% FBS, 1% penicillin/streptomycin, and l-glutamine), added to the wells, and incubated at 37°C for 1 hour, with gentle rocking of the plates every 10 minutes. After incubation, 3.5 mL 1% agarose diluted 1:1 with 20% 2X-199 media (2X-199 media supplemented with 20% FBS, 1% penicillin/streptomycin, and l-glutamine) was overlaid onto the monolayer, and the plates were incubated at 37°C in 5% CO2 for 2 days. On day 2, the agar overlay was gently removed, and the monolayer was stained with 1% crystal violet for 15 minutes. After staining, the crystal violet was aspirated, plates were washed once with 2 mL water per well, and then dried to visualize plaques. Quantification of OC43 stocks for challenge studies was similar to the quantification of MHV-A59 (26), except that 5 mL agar overlay was added on an infected monolayer of L2 cells and incubated at 33°C in a CO2 incubator for 5–6 days. The monolayer was stained with 1% crystal violet, and plaques were quantified by manual counting. For viral load quantification in lung, tissue was collected in round-bottomed 14 mL tubes (Falcon) containing 2 mL 1% FBS DMEM. Tissues were ruptured using a Tissue Ruptor Homogenizer (QIAGEN). Homogenized tissues were clarified using a 100 μm strainer (USA Scientific Inc.) to remove debris, and clarified tissue lysates were used for the plaque assay.
Quantification of OC43 by RT-PCR
Lungs were isolated from mice and homogenized in 1% FBS DMEM. RNA was extracted from lung homogenate using a PureLink Viral RNA/DNA Mini Kit (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions. OC43 viral loads in lungs were determined using 1-step quantitative real-time RT-PCR. RT-PCR was performed using OC43-nucleocapsid–specific TaqMan primers and a probe labeled with a 5′-FAM reporter dye and a 3′-BHQ quencher (IDT) and an AgPath-ID One-Step RT-PCR kit (AgPath AM1005, Applied Biosystems) on an ABI QuantStudio 3 platform (Thermo Fisher Scientific). Each sample was tested in duplicate in 25 μL reactions containing 12.5 μL of a 2× RT-PCR buffer, 1 μL 25× RT-PCR enzyme mix provided with the AgPath kit, 0.5 μL (450 nM) forward primer, 0.5 μL (450 nM) reverse primer, 0.5 μL (100 nM) probe, and 10 μL RNA. In parallel, each sample was also tested for the β-actin gene as an internal control to verify RNA extraction quality using mouse β-actin–specific TaqMan primers and probe labeled with 5′-FAM and 3′-BHQ (IDT). Thermal cycling involved reverse transcription at 45°C for 10 minutes and denaturation at 95°C for 15 minutes, followed by 45 cycles of amplification (15 seconds at 95°C and 1 minute at 60°C.) To avoid cross-contamination, single-use aliquots were prepared for all reagents including primers, probes, buffers, and enzymes.
Quantification of SARS-CoV-2 by RT-PCR
Lungs were isolated from mice and homogenized in PBS. RNA was extracted from lung homogenate using a Zymo Research Quick-RNA 96 Kit (R1052). Viral genomes were quantified via RT-PCR with the TaqMan RNA-to-Ct One-Step Kit (Thermo Fisher Scientific, catalog 4392653) and primer/probe sets with the following sequences: forward, 5′-GACCCCAAAATCAGCGAAAT-3′, reverse, 5′-TCTGGTTACTGCCAGTTGAATCTG-3′, probe, 5′-ACCCCGCATTACGTTTGGTGGACC-3′ (IDT, catalog 10006713). A SARS CoV-2 copy number control was obtained from BEI Resources, NIAID, NIH (NR-52358) and used to quantify SARS-CoV-2 genomes.
Reagents, flow cytometry, and equipment
Dead cells were gated out using LIVE/DEAD fixable dead cell stain (Invitrogen, Thermo Fisher Scientific). The SARS-CoV-2 spike overlapping peptide pools obtained from BEI Resources, NIAID, NIH (NR-52402) were used for intracellular cytokine staining. Biotinylated MHC class I monomers (Kb VL8) were obtained from the NIH tetramer facility at Emory University (Atlanta, Georgia, USA). Cells were stained with fluorescence-labeled antibodies against CD44 (IM7 on Pacific blue, BioLegend, catalog 103020); CD8α (53-6.7 on PerCP-Cy5.5, BD Pharmingen, catalog 551162); IFN-γ (XMG1.2 on APC, BD Pharmingen, catalog 554413); and Vβ11 (RR3-15 on FITC, BioLegend, catalog 125905). Fluorescence-labeled antibodies were purchased from BD Pharmingen, except for anti-CD44, which was from BioLegend. Flow cytometric samples were acquired with a BD FACSCanto II or a BD LSR II and analyzed using FlowJo software (Tree Star).
SARS-CoV-2 pseudovirus neutralization assays
A SARS-CoV-2 pseudovirus was generated by transfection of HEK-293T cells (ATCC, CRL-1573) with a pCAGGS vector expressing the SARS-CoV-2 spike glycoprotein (BEI Resources, NIAID, NIH: NR-52310). Twenty-four hours later, transfected cells were infected with VSVΔG*G-GFP at a MOI of 0.5. After 24 hours, GFP foci were visualized, and the supernatant was harvested and passed through a 0.45 μM filter. This SARS-CoV-2 pseudovirus was concentrated using an Amicon Ultra-15 filter (UFC910024, MilliporeSigma) and then stored at –80°C. Titers were measured by infecting HEK-293T-hACE2 cells (BEI Resources, NIAID, NIH, NR-52511) and counting GFP foci under a fluorescence microscope after 24 hours.
The SARS-CoV-2 pseudovirus neutralization assay was performed by mixing serial dilutions of MVA-SARS-CoV-1 immune mouse sera (or naive sera) with 200 FFU SARS-CoV-2 pseudovirus in a 96-well plate and incubated for 2 hours. After incubation, 100 μL of the sera-virus mixture was transferred to a 96-well half-area plate containing HEK-293T-hACE2 cells. The next day, GFP foci were counted in each well under a fluorescence microscope.
MHC class I binding predictions
The MHC class I binding predictions were made on May 17, 2021, using the Immune Epitope Database (IEDB) analysis resource tool NetMHCpan, version 4.1 (http://tools.iedb.org/mhci/) (27).
scTCR-Seq data acquisition and analysis
C57BL/6 mice were immunized intramuscularly with 109 PFU Ad5-SARS-2 spike, and on day 28, splenic CD8+ T cells were MACS sorted using negative selection (STEMCELL Technologies). Purified CD8+ T cells were stained with Kb VL8, LIVE/DEAD stain, and flow cytometry antibodies against CD8 and CD44. Live, CD8+, CD44+, and Kb VL8+ cells were FACS sorted to approximately 99% purity on a FACS Aria Cytometer (BD Biosciences) and delivered to the Northwestern University NU-Seq core for scTCR-Seq using the Chromium NextGem 5′ v2 kit (10X Genomics). Once the library was sequenced, the output file in BCL format was converted to fastq files and aligned to the mouse genome in order to generate a matrix file using the Cell Ranger pipeline. These upstream quality control (QC) steps were performed by Ching Man Wai and Matthew Schipma at the Northwestern University NUSeq Core (Evanston, Illinois, USA). TCR analyses were performed using the scRepertoire package (28). Only cells expressing both TCRα and TCRβ chains were selected. For cells with more than 2 TCR chains, only the top 2 expressed chains were used. scTCR-Seq accession data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE173567; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173567).
Adoptive plasma transfers
C57BL/6 mice received 50 μL heat-inactivated human plasma from different human donors (before vaccination, after vaccination, before 2019, or SARS-CoV-2 convalescent). Each mouse received plasma from 1 different human donor. On the next day, mice were infected intranasally with 5 × 107 PFU OC43. Lungs were harvested on day 5 after infection and ruptured using a Tissue Ruptor Homogenizer (QIAGEN). Viral loads were quantified by RT-PCR as described above.
Statistics
Statistical analyses were performed using a Mann-Whitney U test, a paired t test, a 1-way ANOVA with multiple comparisons, or a paired Wilcoxon test. Dashed lines in ELISA/plaque assay figures represent the LOD. Data were analyzed using GraphPad Prism (GraphPad Software).
Study approval
Human specimens.
All protocols used for participant recruitment, enrollment, blood collection, sample processing, and immunological assays with human samples were approved by the IRB of Northwestern University (STU00212583). All participants voluntarily enrolled in the study by signing an informed consent form after receiving detailed information about the clinical study.
Mouse studies.
All mouse experiments were performed with approval from the IACUCs of Northwestern University and the UIC (study approval nos. IS00003258 and 20-107). All mouse experiments with BL2 agents were performed with approval from the IACUC of Northwestern University. SARS-CoV-2 infections of mice were performed at the UIC following BL3 guidelines, with approval from the UIC’s IACUC.
Author contributions
TD, NP, and PPM designed and conducted all mouse BSL-2 experiments. SS helped with the MHV-1 challenge studies. MP expressed the nucleocapsid protein and helped to analyze viral sequences. TC conducted the scTCR-Seq analyses. LV performed the human blood draws. LV and IJK provided feedback on the human antibody studies. JR and JC made the mRNA vaccine and performed the SARS-CoV-2 challenge studies. PPM, TD, and NP wrote the manuscript, with feedback from all authors.
Supplementary Material
Acknowledgments
We thank Thomas Gallagher and Susan Weiss for their comments and suggestions. Our study was made possible by a grant from the National Institute on Drug Abuse (NIDA), NIH (DP2DA051912) a grant from the Emerging and Re-Emerging Pathogens Program (EREPP, to PPM), as well as NIH grant R01AI150672 (to JMR).
Version 1. 10/08/2021
In-Press Preview
Version 2. 12/15/2021
Electronic publication
Funding Statement
Our study was possible with a grant from the National Institute on Drug Abuse (NIDA, DP2DA051912) and a grant from the Emerging and Re-Emerging Pathogens Program (EREPP) to P.P.M.
Footnotes
Conflict of interest: PPM is a task force advisor to the Illinois Department of Public Health (IDPH) on SARS-CoV-2 vaccines in the state of Illinois.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(24):e151969.https://doi.org/10.1172/JCI151969.
See the related Commentary at Family matters for coronavirus disease and vaccines.
Contributor Information
Tanushree Dangi, Email: tanushree.dangi@northwestern.edu.
Nicole Palacio, Email: NicolePalacio2017@u.northwestern.edu.
Sarah Sanchez, Email: SarahSanchez2025@u.northwestern.edu.
Mincheol Park, Email: mincheol.park@northwestern.edu.
Lavanya Visvabharathy, Email: lavanya.visvabharathy@northwestern.edu.
Thomas Ciucci, Email: Thomas_Ciucci@urmc.rochester.edu.
Igor J. Koralnik, Email: igor.koralnik@northwestern.edu.
Justin M. Richner, Email: richner@uic.edu.
Pablo Penaloza-MacMaster, Email: ppm@northwestern.edu.
References
- 1.Krammer F, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;384(14):1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebinger JE, et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat Med. 2021;27(6):981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amanat F, et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. 2021;184(15):3936–3948. doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aydillo T, et al. Immunological imprinting of the antibody response in COVID-19 patients. Nat Commun. 2021;12(1):3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisht H, et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci U S A. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, et al. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J Virol. 2005;79(5):2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassert M, et al. mRNA induced expression of human angiotensin-converting enzyme 2 in mice for the study of the adaptive immune response to severe acute respiratory syndrome coronavirus 2. PLoS Pathog. 2020;16(12):e1009163. doi: 10.1371/journal.ppat.1009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhi Y, et al. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335(1):34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callaway E. Could new COVID variants undermine vaccines? Labs scramble to find out. Nature. 2021;589(7841):177–178. doi: 10.1038/d41586-021-00031-0. [DOI] [PubMed] [Google Scholar]
- 10.Anderson EM, et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184(7):1858–1864. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saunders KO, et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594(7864):553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob-Dolan C, et al. Coronavirus-specific antibody cross reactivity in rhesus macaques following SARS-CoV-2 vaccination and infection. J Virol. 2021;95(11):e00117-21. doi: 10.1128/JVI.00117-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sagar M, et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest. 2021;131(1):143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penaloza-MacMaster P, et al. Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J Virol. 2013;87(3):1373–1384. doi: 10.1128/JVI.02058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dangi T, et al. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep. 2021;36(10):109664. doi: 10.1016/j.celrep.2021.109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palacio N, et al. Early type I IFN blockade improves the efficacy of viral vaccines. J Exp Med. 2020;217(12):e20191220. doi: 10.1084/jem.20191220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat Commun. 2020;11(1):4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver EA. Dose effects of recombinant adenovirus immunization in rodents. Vaccines (Basel) 2019;7(4):E144. doi: 10.3390/vaccines7040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollner CJ, et al. A dengue virus serotype 1 mRNA-LNP vaccine elicits protective immune responses. J Virol. 2021;95(12):e02482-20. doi: 10.1128/JVI.02482-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Q, et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat Commun. 2021;12(1):776. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao YF, et al. Protective efficacy of inactivated vaccine against SARS-CoV-2 infection in mice and non-human primates. Virol Sin. doi: 10.1007/s12250-021-00376-w. [published online April 9, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case JB, et al. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020;28(3):465–474. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matchett WE, et al. Nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J Immunol. 2021;207(2):376–379. doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leist SR, et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183(4):1070–1085. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dangi T, et al. Interrogating adaptive immunity using LCMV. Curr Protoc Immunol. 2020;130(1):e99. doi: 10.1002/cpim.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leibowitz J, et al. Coronaviruses: propagation, quantification, storage, and construction of recombinant mouse hepatitis virus. Curr Protoc Microbiol. 2011;Chapter 15(1):Unit 15E.1. doi: 10.1002/9780471729259.mc15e01s21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynisson B, et al. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(w1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borcherding N, et al. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res. 2020;9:47. doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.