Abstract
Circular RNAs (circRNAs) have been recently recognized as playing a role in the pathogenesis of vascular remodeling–related diseases by modulating the functions of miRNAs. However, the interplay between circRNAs and proteins during vascular remodeling remains poorly understood. Here, we investigated a previously identified circRNA, circEsyt2, whose expression is known to be upregulated during vascular remodeling. Loss- and gain-of‑function mutation analyses in vascular smooth muscle cells (VSMCs) revealed that circEsyt2 enhanced cell proliferation and migration and inhibited apoptosis and differentiation. Furthermore, the silencing of circEsyt2 in vivo reduced neointima formation, while circEsyt2 overexpression enhanced neointimal hyperplasia in the injured carotid artery, confirming its role in vascular remodeling. Using unbiased protein–RNA screening and molecular validation, circEsyt2 was found to directly interact with polyC-binding protein 1 (PCBP1), an RNA splicing factor, and regulate PCBP1 intracellular localization. Additionally, circEsyt2 silencing substantially enhanced p53β splicing via the PCBP1–U2AF65 interaction, leading to the altered expression of p53 target genes (cyclin D1, p21, PUMA, and NOXA) and the decreased proliferation of VSMCs. Thus, we identified a potentially novel circRNA that regulated vascular remodeling, via altered RNA splicing, in atherosclerotic mouse models.
Keywords: Cardiology, Vascular Biology
Keywords: Atherosclerosis, Cardiovascular disease, Epigenetics
Introduction
Vascular remodeling is a common pathological manifestation of certain cardiovascular diseases (CVDs), such as atherosclerosis (AS), hypertension, and restenosis after angioplasty. In response to hazardous stimulus, the vascular smooth muscle cells (VSMCs) undergo alterations during different biological processes, including phenotypic conversion, proliferation, and apoptosis. These alterations often lead to arterial lumen narrowing or structural abnormalities through vascular remodeling, significantly increasing the risk of life-threatening myocardial infarction and stroke. Therefore, elucidating the underlying molecular mechanisms during VSMC remodeling is critical for improving the diagnosis and treatment of CVDs.
Noncoding RNAs (ncRNAs) constitute the majority of the mammalian transcriptome. Although studies on miRNAs and long ncRNAs (lncRNAs) have markedly increased, major challenges remain regarding their clinical applications as therapeutic agents or targets due to their relatively low specificity, low abundance, poor stability, and poor conservation (1, 2). Circular RNAs (circRNAs) have emerged as a group of ncRNAs with newly recognized importance by virtue of technological advancements. In general, circRNAs are generated by the back‑splicing of the 3′ to 5′ exon ends, giving rise to covalently closed molecules that are highly exonuclease-resistant and cell-type specific with a high degree of cross-species conservation (3, 4). Furthermore, an increasing number of studies indicate their functional importance in many biological and pathological processes, specifically in brain development and cancer (5, 6). Several studies have investigated circRNAs in the context of vascular remodeling, but most are limited to in vitro demonstrations rather than clinically relevant disease models (7, 8).
Additionally, the mechanistic understanding of the role of circRNAs in the remodeled vasculature is incomplete as previous studies have mostly focused on their role in gene regulation by virtue of their function as miRNA sponges (9–12). In this study, we studied circEsyt2 to determine its function in vascular remodeling using in vitro and in vivo mice models. Our study may confirm that this circRNA plays an important role during vascular remodeling, thereby providing new insights that may be useful for future research on CVDs.
Results
Identification of circEsyt2 during vascular remodeling in the aortae.
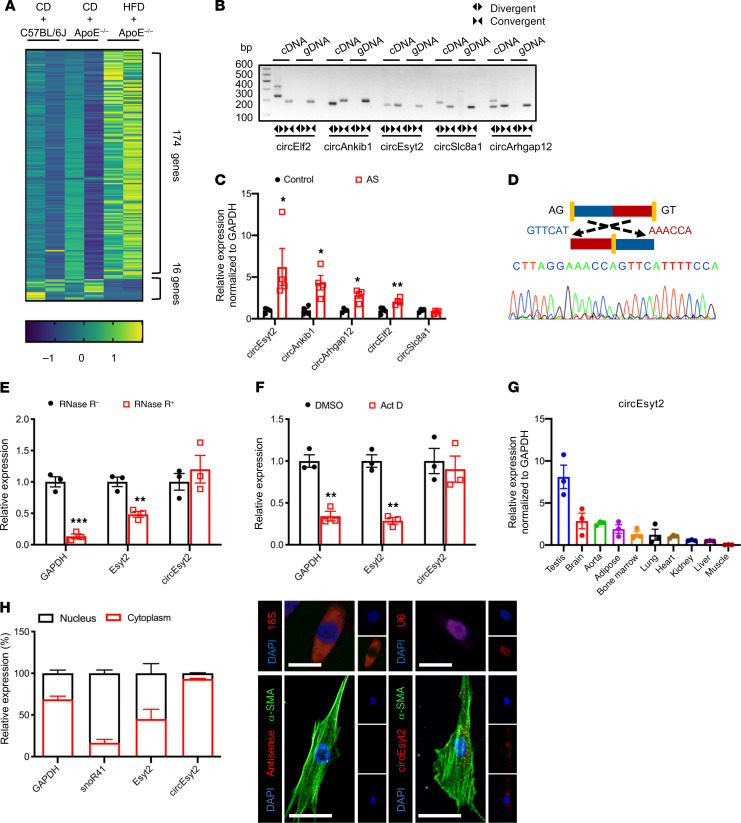
For the identification of vascular remodeling–related circRNAs in atherosclerotic tissues, we used ApoE knockout (ApoE–/–) mice as the model. For the experimental group, we obtained the aortae with plaques from ApoE–/– mice fed a high-fat diet for 3 months. The samples from WT C57BL/6J and ApoE–/– mice fed a regular chow diet were used as the control groups (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI147031DS1). From the RNA-Seq data sets, a total of 20,532 transcripts were initially predicted as circRNAs. Among these, 5149 transcripts contained no less than 2 distinctive back-spliced fragments, which was a prerequisite for their identification as circRNAs (Supplemental Figure 1, B and C). Functional annotation of these transcripts revealed that most of the candidate circRNAs originated from protein‑coding exons, while the rest were derived from introns, 3′-UTRs, 5′-UTRs, and others (Supplemental Figure 1D). The size of the identified circRNAs generally ranged from 100 to 1500 nt (Supplemental Figure 1E). Interestingly, more circRNAs were observed in atherosclerotic tissues than in normal tissues, indicating their potential role in vascular remodeling (Supplemental Figure 1F). Notably, 190 circRNA species were differentially expressed by more than 2-fold between the AS and control samples (P < 0.05, FDR < 0.05), including 174 upregulated and 16 downregulated circRNAs (Figure 1A and Supplemental Figure 1G). For more accurate results, we subjected our raw circRNA‑Seq data to repeated predictions. Using 4 popular algorithms, namely CIRCexplorer, CIRI2, CIRIquant, and Find_circ later (the method we adopted), we obtained a total of 3828 circRNAs, which also included candidate circRNAs previously identified by us (Figure 1D and Supplemental Table 1).
Figure 1. Identification of circEsyt2 based on circRNA profiling of mouse atherosclerotic aortae.
(A) Heat map of differentially expressed circRNAs in atherosclerotic (HFD + ApoE–/–) and control groups (CD + ApoE–/– or CD + C57BL/6J). CD: chow diet. HFD: high fat diet. ApoE–/–: ApoE knockout mice. n = 2. (B) PCR amplification using divergent or convergent primers against complementary DNA (cDNA) or genomic DNA (gDNA) from aortae. (C) qRT-PCR to check for the expression of circRNAs in aortae. *P < 0.05, **P < 0.01 vs. control. n = 4. (D) Confirmation of the backsplicing junction site within the circEsyt2 by Sanger sequencing. (E) mRNA expressions after RNase-R digestion in mouse VSMCs followed by qRT-PCR. **P < 0.01, ***P < 0.001 vs. RNase-R. n = 3. (F) Actinomycin D (Act D) treatment of VSMCs followed by qRT-PCR. Linear mRNA controls: GAPDH and Esyt2. **P < 0.01 vs. DMSO. n = 3. (G) qRT-PCR to check for the expression of circEsyt2 in mouse tissues. n = 3. (H) Subcellular localization of circEsyt2 in VSMCs. Left: qRT-PCR to check for the expression of circEsyt2, Esyt2, linear mRNA in the cytoplasmic and nuclear fractions; GAPDH, cytoplasmic control; snoR41, nuclear control. Right: images of FISH for circEsyt2 (red) and cofluorescence with α-SMA (green) and DAPI (blue). 18S, cytoplasmic control; U6, nuclear control; antisense, negative control. Scale bars: 20 μm. Data are mean ± SEM. Two-sided unpaired t test or Kolmogorov-Smirnov test for C, E, and F.
The top 5 upregulated circRNAs with considerable junction counts among the circRNAs predicted using the 2 strategies are listed in Supplemental Table 2. For validation, we performed real-time reverse transcription–polymerase chain reaction (RT-PCR) using divergent and convergent primers to detect the back-spliced circular (circEsyt2) and canonical linear extended synaptotagmin 2 (Esyt2) transcripts, respectively. As expected, specific amplification products were detected from the cDNA samples, but not from the genomic DNA (gDNA) samples, using the divergent primers (Figure 1B). Further verification based on analyzing the expression of the top 5 upregulated circRNAs in the atherosclerotic and control tissues using quantitative RT-PCR (qRT-PCR) revealed that most circRNAs were significantly upregulated in the atherosclerotic cohort (Figure 1C). The top one circRNA was named as circEsyt2 based on its host gene, Esyt2. Localization of the 5′ to 3′ exon junction within the circEsyt2 was also performed via Sanger sequencing (Figure 1D). In contrast to the controls GAPDH and Esyt2, circEsyt2 was found to be resistant to RNase R digestion (Figure 1E). Furthermore, we discovered that the GAPDH and Esyt2 expression levels were markedly decreased following inhibition of transcription in response to 24-hour treatment with actinomycin D, while the expression of circEsyt2 was unaffected (Figure 1F). These results reveal that circEsyt2 is a genuine circRNA with the expected molecular structure and biochemical features.
Additional studies showed that circEsyt2 was ubiquitously expressed in adult mice, and that the aorta was among the tissues with high levels of circEsyt2 expression (Figure 1G). Furthermore, mouse circEsyt2 exhibited a high degree of sequence conservation, with over 90% homology to its human ortholog (Supplemental Figure 1I). To investigate the expression of circEsyt2 in atherosclerotic vasculature, we employed a combination of FISH and immunofluorescence on the plaque-laden aortic sections from ApoE–/– mice and C57BL/6J control mice. The results showed that circEsyt2 was mainly expressed in the major cell types of the arterial wall, including VSMCs (α-smooth muscle actin, α-SMA+), endothelial cells (CD31+), adventitial fibroblasts (PDGFRα+), and macrophages (CD68+). Notably, circEsyt2 expression was low in C57BL/6J control mice but was significantly and prominently upregulated in the VSMCs of the atherosclerotic plaques (Supplemental Figure 2). This finding was replicated in human coronary arteries, in which the expression of circEsyt2 was much higher in arteries from patients with severe coronary artery disease (CAD) compared with that in arteries from patients with mild CAD, suggesting that circEsyt2 may play a proatherosclerotic role by regulating VSMC function (Supplemental Figure 3). We next investigated the intracellular localization of circEsyt2 in vitro and observed that like most circRNAs, circEsyt2 was widely distributed in the cytoplasm of mouse VSMCs (Figure 1H).
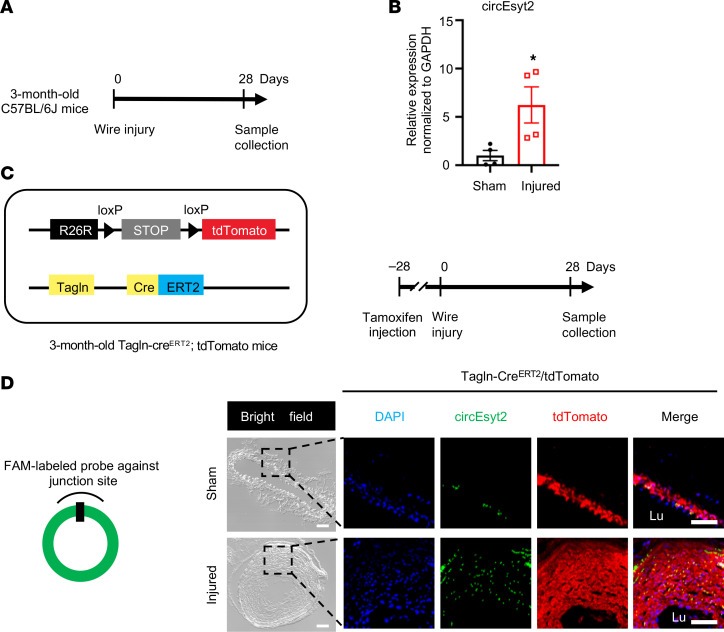
To further determine the relationship between circEsyt2 and the arterial remodeling process, we employed another vascular remodeling model (i.e., carotid artery tissue of the wire injury model), and examined circEsyt2 expression in mouse carotid arteries 28 days following wire injury (Figure 2A). qRT-PCR revealed that the injured arteries had a higher expression of circEsyt2 than the noninjured (sham) arteries (Figure 2B). To illustrate the changes in circEsyst2 expression in the VSMCs of the neointimal hyperplasia animal model, we used a tamoxifen-induced lineage tracing mouse model (Tagln-CreERT2/tdTomato), in which VSMCs (Tagln+) were labeled red using the tdTomato reporter. Compared with the animals in the sham group, animals in the model group exhibited upregulation of circEsyt2 in the VSMCs of injured carotid arteries (Figure 2, C and D).
Figure 2. CircEsyt2 modulates vascular remodeling in vivo.
(A) Schema of the generation of the vascular remodeling model in C57BL/6J mice. Carotid arteries were collected 28 days after wire injury. (B) qRT-PCR to check for the expression of circEsyt2 in carotid arteries. *P < 0.05 vs. sham. n = 4. (C) Schema of the generation of VSMC lineage tracing mice (Tagln-CreERT2/tdTomato) (left) and vascular remodeling model (right). Tamoxifen was injected intraperitoneally 28 days prior to the wire injury of carotid arteries. (D) FISH assay of carotid arteries treated in C. Left: design of FISH probe against circEsyt2. Right: representative fluorescence images of DAPI (blue), circEsyt2 (green) and tdTomato (red). The sham group served as control. Scale bars: 50 μm. Lu, lumen. Data are mean ± SEM. Two-sided unpaired t test for B.
CircEsyt2 controls vascular remodeling by modulating VSMC function.
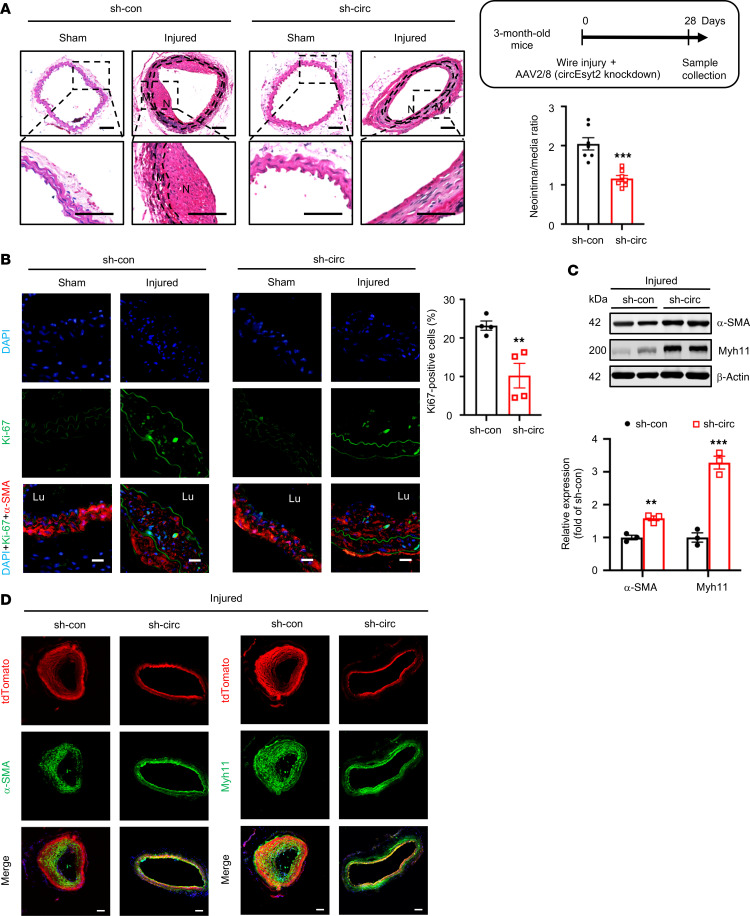
To assess the role of circEsyt2 in vivo, the carotid arteries of C57BL/6J mice were wire-injured and injected with adeno-associated virus 2/8 (AAV 2/8) harboring either control or sh-circEsyt2. The efficacy of the circEsyt2 siRNA‑mediated AAV delivery in vivo was confirmed 28 days after infection using qRT-PCR and FISH combined with immunofluorescence (Supplemental Figure 4, A and B). Histological analysis and quantification of the arterial neointima/media ratio revealed that the inhibition of circEsyt2 expression had no effect on the intima of the uninjured carotid arteries but reduced the neointima formation induced in response to wire injury (Figure 3A). The proliferation of VSMCs (α-SMA+) in the injured group, as detected by Ki67 staining, was inhibited by circEsyt2 knockdown (Figure 3B). In addition, the expression of the phenotype switch markers α-SMA and Myh11 was upregulated in the injured carotid arteries after circEsyt2 knockdown (Figure 3C). Similarly, the expression of the phenotype switch markers was upregulated in the carotid arteries of mice with tamoxifen-induced expression of Tagln-CreERT2/tdTomato after the shRNA‑mediated knockdown of circEsyt2 (Figure 3D). Furthermore, apoptosis was observed to increase in vivo after circEsyt2 knockdown (Supplemental Figure 4C).
Figure 3. Effect of circEsyt2 knockdown on arterial remodeling in vivo.
(A) Wire injury was accompanied by injection of adeno-associated virus 2/8 (AAV2/8) silencing circEyst2 (sh-circ) or control virus (sh-con) in the carotid arteries from C57BL/6J mice. Carotid arteries were collected 28 days after injury. Left: representative H&E staining images. Right: ratio of neointima to media thickness of carotid arteries. ***P < 0.001 vs. sh-con. n = 7. The insert on the right upper side depicts the schema of the experimental design. Note the absence of neointimal in the sham group due to lack of any injurious stimulus. N, neointima. M, media. Scale bars: 50 μm. (B) Immunofluorescence of Ki67 in injured carotid arteries, treated as in A. Left: representative immunofluorescence images. Scale bars:, 20 μm. Right: percentage of Ki67-positive cells in the injured group. **P < 0.01 vs. sh-con. n = 4.(C) Western blotting to check for the expression of α-SMA and Myh11 in injured carotid arteries from C57BL/6J mice, treated as in A. β-actin, protein control. **P < 0.01, ***P < 0.001 vs. sh-con. n = 3. (D) Wire injury was performed in circEsyt2-silenced mice with tamoxifen-induced expression of Tagln-CreERT2/tdTomato. Immunofluorescence staining for α-SMA or Myh11 in carotid arteries 28 days after injury. α-SMA or Myh11 (green), tdTomato (red), and DAPI (blue) were merged. Scale bars: 50 μm. Data are mean ± SEM. Two-sided unpaired t test for A–C.
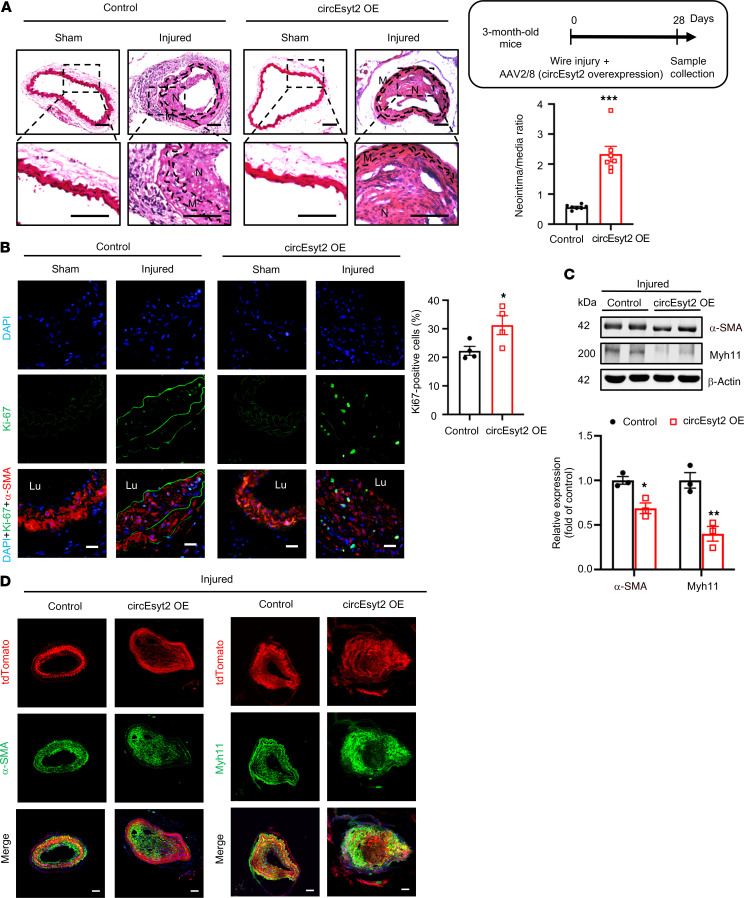
After the knockdown studies, we also performed circEsyt2 overexpression experiments in mice arteries by employing an in vivo AAV delivery approach (Supplemental Figure 4, D and E). Compared with the animals of the control group, increased neointima formation was detected in the wire-injured carotid arteries of the animals in the injured group after circEsyt2 overexpression (Figure 4, A and B). As hypothesized, the decreased expression of the phenotype switch markers α-SMA and Myh11 was evident in animals of the circEsyt2 overexpression group (Figure 4, C and D), while apoptosis was decreased (Supplemental Figure 4F). Overall, these in vivo results confirm the function of circEsyt2 during vascular remodeling in the aortae.
Figure 4. Effect of circEsyt2 overexpression on arterial remodeling in vivo.
(A) Wire injury was accompanied by injection of circEyst2-overexpressing (circEsyt2 OE) or control virus (control) in the carotid arteries from C57BL/6J mice. Carotid arteries were collected 28 days after injury. Left: representative H&E staining images. Scale bars: 50 μm. Right: ratio of neointima to media thickness of carotid arteries. ***P < 0.001 vs. control. n = 7. The insert on the right upper side depicts the schema of the experimental design. (B) Immunofluorescence staining for Ki67 in injured carotid arteries, treated as in A. Left: representative immunofluorescent images. Scale bars: 20 μm. Right: Ki67-positive cells in the injured group. *P < 0.05 vs. control. n = 4. (C) Western blotting to check for the expression of α-SMA and Myh11 in injured carotid arteries from C57BL/6J mice, treated as in A. *P < 0.05, **P < 0.01 vs. control. n = 3. (D) Wire injury was performed in circEsyt2-overexpressed carotid arteries with Tagln-CreERT2/tdTomato under the control of tamoxifen. Immunofluorescence staining for α-SMA or Myh11 in carotid arteries 28 days after injury. α-SMA or Myh11 (green), tdTomato (red), and DAPI (blue) were merged. Scale bars: 50 μm. Data are mean ± SEM. Two-sided unpaired t test for A and C. Kolmogorov-Smirnov test for B.
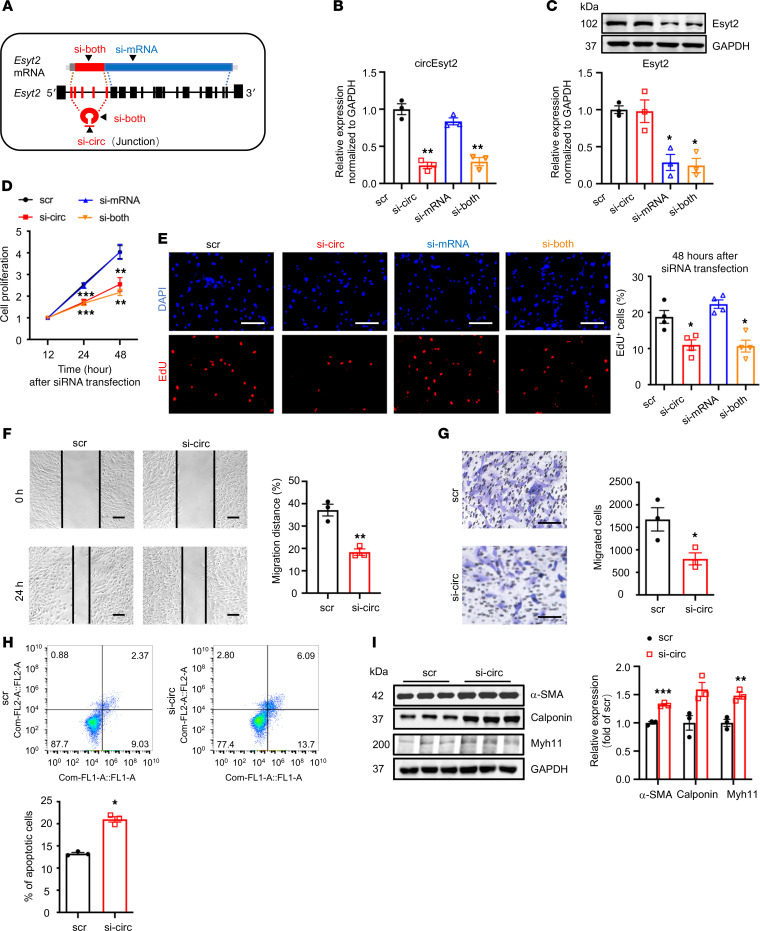
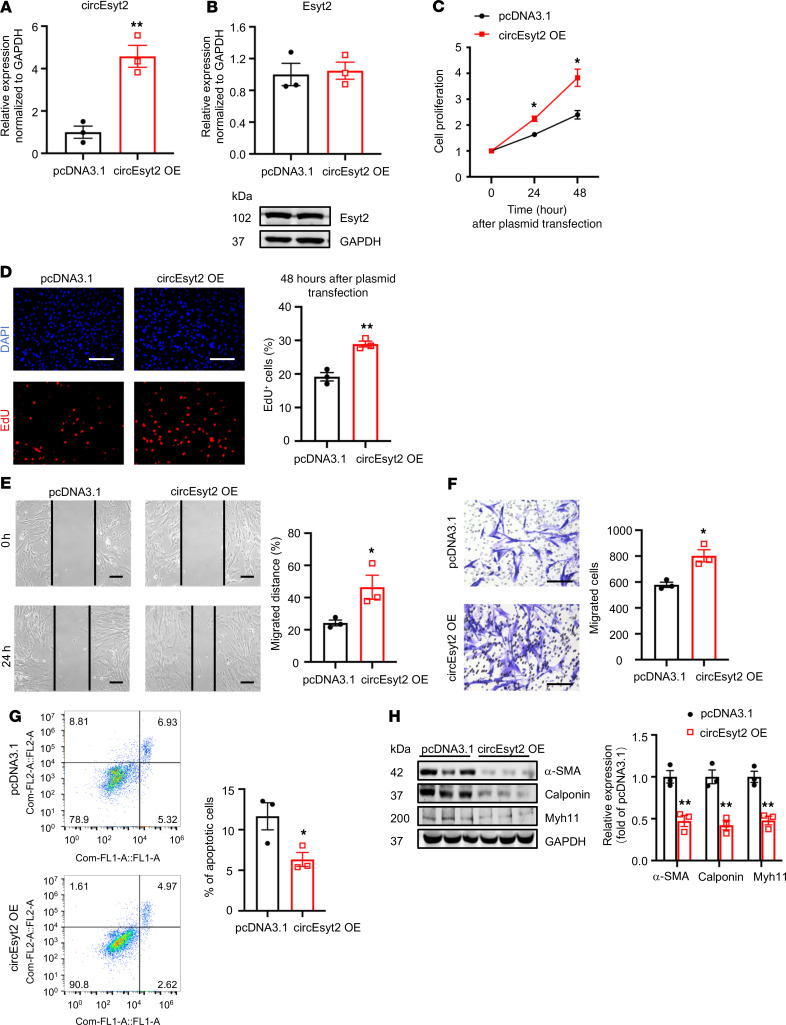
In general, both circular and linear transcripts can be transcribed from the same host gene. To determine their biological roles in vitro, we designed siRNAs that specifically target circEsyt2, the linear Esyt2 transcript, or both (Figure 5A). The specificity and efficacy of the siRNAs were tested in human aortic smooth muscle cells (HASMCs) using qRT-PCR and Western blotting (Figure 5, B and C). Compared with the control, circEsyt2 silencing or cosilencing with Esyt2 significantly reduced the proliferation and migration of HASMCs but enhanced their apoptosis (Figure 5, D–H). In addition, the expression of the differentiation marker genes, namely α-SMA, calponin, and Myh11, was increased after circEsyt2 silencing (Figure 5I). In contrast, Esyt2-specific silencing had no effect on the above-mentioned parameters (Figure 5, D and E). The effects observed in HASMCs after circEsyt2 silencing were completely phenocopied in mouse VSMCs (Supplemental Figure 5), indicating that circEsyt2, independent of Esyt2, can regulate the phenotype switching of VSMCs. Furthermore, compared with the control plasmid–transfected (pcDNA3.1-transfected) group, the gain-of-function experiments performed by transferring the circEsyt2 overexpression plasmid in vitro resulted in the enhanced proliferation and migration but decreased apoptosis and dedifferentiation of HASMCs (Figure 6, A–H).
Figure 5. Effect of circEsyt2 silencing in HASMCs.
(A) Schematic representation of siRNAs designed for targeting circEsyt2 (si-circ), Esyt2 (si-mRNA), or both (si-both). Specifically, si-circ targets the junction formed by back-splicing of exons 6 and 2. Si-mRNA targets exon 7 (human) or exon 17 (mouse). Si-both targets exon 6 (human) or exon 4 (mouse). ex, exon. (B and C) Efficacy of circEsyt2 (si-circ), Esyt2 (si-mRNA) siRNA, or both (si-both) detected by qRT-PCR and Western blotting in HASMCs. Scrambled siRNA (scr) served as control. GAPDH, protein control. *P < 0.05, **P < 0.01 vs. scr. n = 3. (D) Cell counting kit-8 (CCK-8) assay of HASMCs treated with siRNAs, as shown in A for the indicated hours. **P < 0.01, ***P < 0.001 vs. scr. n = 4. (E) EdU incorporation assay of HASMCs, treated as in A for 48 hours. Left: representative immunofluorescence of EdU (red) and DAPI (blue). Scale bars: 100 μm. Right: percentages of EdU-incorporated HASMCs. *P < 0.05 vs. scr. n = 3. (F and G) Migratory ability assessed by wound healing (F) and Transwell assay (G) of circEsyt2-silenced HASMCs. Scale bars: 100 μm. *P < 0.05, **P < 0.01 vs. scr. n = 3. (H) Apoptosis detected by Annexin V–conjugated flow cytometry in circEsyt2-silenced HASMCs. *P < 0.05 vs. scr. n = 3. (I) Western blotting to check for the expression of α-SMA, Calponin, and Myh11 in circEsyt2-silenced HASMCs. *P < 0.05, **P < 0.01, ***P < 0.001 vs. scr. n = 3. Data are mean ± SEM. Two-sided unpaired t test for F–I. Two-way repeated measures ANOVA with LSD’s post hoc test for D. One-way ANOVA test with Dunnett’s T3 post hoc test for B, C, and E.
Figure 6. Effect of circEsyt2 overexpression in HASMCs.
(A and B) Efficacy (A) and specificity (B) of circEsyt2 overexpression plasmid detected by qRT-PCR and Western blotting in HASMCs. **P < 0.01 vs. pcDNA3.1. n = 3. (C) CCK-8 assay of circEsyt2-overexpressed (OE) HASMCs for the indicated hours. *P < 0.05 vs. pcDNA3.1. n = 4. (D) EdU incorporation assay of HASMCs, treated as in C for 48 hours. Left: representative immunofluorescence staining for EdU (red) and DAPI (blue). Scale bars: 100 μm. Right: percentages of EdU incorporated-HASMCs. **P < 0.01 vs. pcDNA3.1. n = 3. (E and F) Migratory ability assessed by wound healing (E) and Transwell assay (F) of circEsyt2-OE HASMCs. Scale bars: 100 μm. *P < 0.05 vs. pcDNA3.1. n = 3. (G) Apoptosis detected by Annexin V–conjugated flow cytometry of circEsyt2-OE HASMCs. *P < 0.05 vs. pcDNA3.1. n = 3. (H) Western blotting to check the expression of α-SMA, Calponin, and Myh11 in circEsyt2-OE HASMCs. **P < 0.01 vs. pcDNA3.1. n = 3. Two-sided unpaired t test for A, B, and D–H. Two-way repeated measures ANOVA with Bonferroni’s post hoc test for C.
CircEsyt2 interacts with the RNA splicing regulator polyC-binding protein 1 (PCBP1) and inhibits its nuclear translocation.
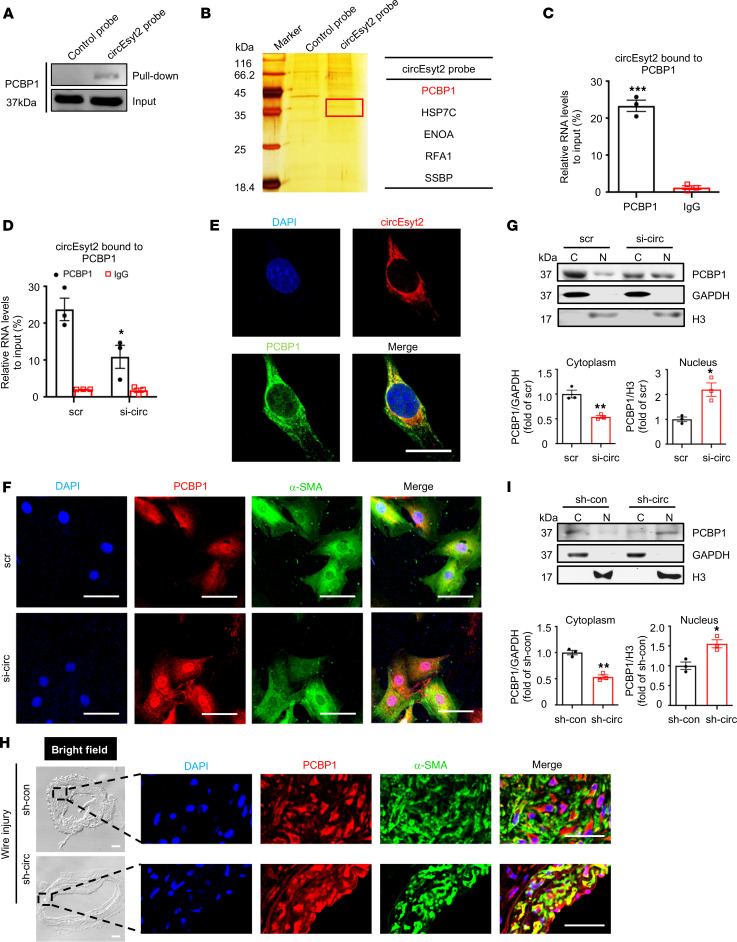
The interplay between circRNAs and RNA‑binding proteins (RBPs) accounts for the key mechanisms underlying their functions. Hence, we used catRAPID to predict the potential binding partners of circEsyt2 (13). Among the predicted circEsyt2-binding proteins, the PCBP family ranked at the top position based on binding probability (Supplemental Table 3). Specifically, a fragment of mouse circEsyt2 with 317–368 nt (or 151–202 nt in the human ortholog) was discovered to interact with a motif of 31–82 aa residues in PCBP1, the main member of the PCBP family (Supplemental Figure 6, A and B). To validate this prediction, we performed RNA pull‑down in circEsyt2-overexpressed HEK293T cells using control and circEsyt2 probes (Figure 7A and Supplemental Figure 6C). The mass spectrometry data revealed that PCBP1 ranked as the top circEsyt2-binding protein with the highest confidence level (Figure 7B and Supplemental Figure 6D). We also performed an RNA immunoprecipitation (RIP) assay and successfully detected circEsyt2 in the PCBP1 immunocomplex of HASMCs (Figure 7C and Supplemental Figure 6E). The specificity of this interaction was supported by the reduced PCBP1 levels in HASMCs transfected with circEsyt2 siRNA (Figure 7D). In addition, immunofluorescence staining for circEsyt2 using the cy3-labeled probe and PCBP1 antibody revealed an overlap between the expression of circEsyt2 and PCBP1 in HASMCs (Figure 7E). Together, these results confirm the physical interaction between circEsyt2 and PCBP1.
Figure 7. CircEsyt2 inhibits the nuclear trafficking of PCBP1 by binding directly to PCBP1.
(A) Western blotting of proteins pulled down by control and circEsyt2 probes in circEsyt2-OE HEK293T cells using the PCBP1 antibody. (B) Identification of circEsyt2-binding proteins. Left: silver staining of pulled-down proteins in circEsyt2-OE HEK293T cells. Right: mass spectrometry showing the main proteins pulled down by the circEsyt2 probe. (C) RIP-qPCR assay confirming the direct binding of PCBP1 to circEsyt2 in HASMCs. ***P < 0.001 vs. IgG. n = 3. (D) RIP-qPCR assay detecting the specific binding of PCBP1 and circEsyt2 in HASMCs by circEsyt2 silencing. Scrambled siRNA (scr) served as control. *P < 0.05 vs. scr. n = 3. (E) FISH of circEsyt2 (red), PCBP1 (green), and DAPI (blue) in HASMCs transfected with circEsyt2 plasmid. Scale bars: 20 μm. (F) Coimmunofluorescence of PCBP1 (red), α-SMA (green), and DAPI (blue) in circEsyt2-silenced HASMCs. Scale bars: 50 μm. (G) Western blotting to check the expression of cytoplasmic (C) and nuclear (N) PCBP1, treated as in F. Cytoplasmic control: GAPDH; nuclear control: histone 3 (H3). *P < 0.05, **P < 0.01 vs. scr. n = 3. (H) Coimmunofluorescence staining for PCBP1 (red), α-SMA (green), and DAPI (blue) in injured carotid arteries of the control (sh-con) and circEsyt2 knockdown (sh-circ) groups. Scale bars: 50 μm (bright field) and 20 μm (immunofluorescence). (I) Western blotting to check the expression of cytoplasmic and nuclear PCBP1 in carotid arteries, treated as in H. *P < 0.05, **P < 0.01 vs. sh-con. n = 3. Data are mean ± SEM. Two-sided unpaired t test for C, D, G, and I.
Furthermore, the regulatory role of circEsyt2 with respect to PCBP1 was observed when the PCBP1 was localized in the nucleus in response to circEsyt2 silencing (Figure 7F). This shift toward nuclear localization of PCBP1 was corroborated by Western blotting of the cytoplasmic and nuclear fractions of HASMCs transfected with circEsyt2 siRNA or scrambled siRNA (Figure 7G). Moreover, coimmunofluorescence staining of PCBP1 and α-SMA in injured mouse carotid arteries revealed the increased accumulation of PCBP1 in the nuclei after circEsyt2 silencing (Figure 7H), which was confirmed by Western blotting of the PCBP1 protein (Figure 7I). These results suggest that the binding of circEsyt2 with PCBP1 promotes its cytoplasmic retention, which may be the underlying molecular mechanism determining its physiological function.
CircEsyt2 facilitates VSMC proliferation by regulating p53β-dependent gene expression.
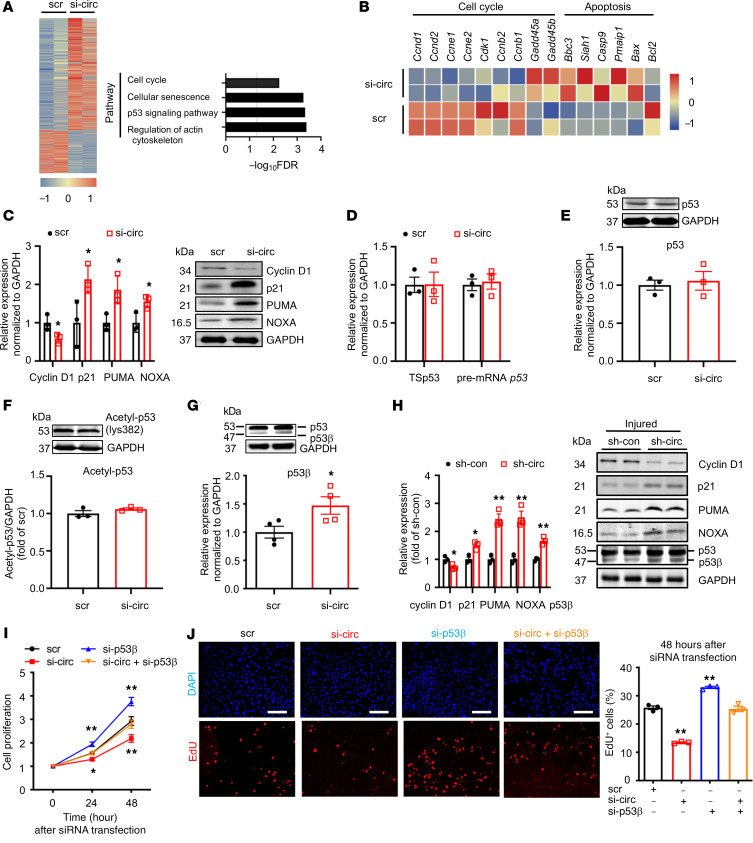
To determine the role of circEsyt2 in gene regulation, we performed RNA-Seq of mouse VSMCs with or without circEsyt2 silencing. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the functional terms of differentially expressed genes were related to the regulation of the actin cytoskeleton, p53 signaling pathway, senescence, and cell cycle (Figure 8A). The analysis of differentially expressed genes strongly correlated to p53 because the expression of p53 target genes (e.g., cyclin D1 [Ccnd1]) decreased, while the expression of p21 (Cdkn1a), PUMA (Bbc3), and NOXA (Pmaip1) increased. Notably, these genes are primarily involved with cell proliferation and apoptosis induction (Figure 8, A–C). However, we did not find any changes in the expression levels of full-length p53 mRNA or protein after circEsyt2 silencing (Figure 8, D and E), nor in the expression of acetylated p53 (an activated form of p53) (Figure 8F). However, we observed that the expression of p53β, one of the p53 splicing variants, was increased both at the mRNA and protein levels after circEsyt2 silencing (Figure 8G), while the expression of other p53 splicing variants, including Δ133p53, Δ40p53, and p53γ, did not change (Supplemental Figure 7, A–C). In contrast, the p53β expression decreased at the mRNA and protein levels after circEsyt2 overexpression (Supplemental Figure 7D). Considering the probability of false priming caused by reverse transcriptase, we further validated our results using strand‑specific qRT-PCR, a significantly improved version with much higher specificity and precision than the standard method (14) (Supplemental Figure 7, E and F). Results showed the increased expression of p53β, p21, PUMA, and NOXA and the decreased expression of cyclin D1 at the mRNA and protein levels in the injured carotid arteries following the in vitro silencing of circEsyt2 (Figure 8H).
Figure 8. CircEsyt2 regulates p53β splicing in HASMCs and mouse VSMCs in vitro.
(A) Heatmap of differentially expressed genes (fold-change > 1.2 and FDR < 0.05) in circEsyt2-silenced mouse VSMCs. KEGG pathway analysis of differentially expressed genes are presented in the right panel. n = 2. (B) Heatmap of p53 target genes. (C) qRT-PCR (left) and Western blotting (right) of Cyclin D1 (Ccnd1), p21(Cdkn1a), PUMA(Bbc3), and NOXA (Pmaip1) expressions in HASMCs after circEsyt2 knockdown (si-circ). *P < 0.05 vs. scr. n = 3. (D) qRT-PCR to check for the expression of total spliced p53 (TSp53) and precursor RNA p53 (pre-mRNA p53) in circEsyt2-silenced HASMCs. n = 3. (E) Western blotting (top) and qRT-PCR (bottom) to check for the expression of p53 in circEsyt2-silenced HASMCs. n = 3. (F) Western blotting to check for the expression of acetylated p53 using an antibody against p53-lys382 in circEsyt2-silenced HASMCs. n = 3. (G) Western blotting to check for the expression of p53 and p53β (top) and qRT-PCR to check for the expression of p53β (bottom) in circEsyt2-silenced HASMCs. *P < 0.05 vs. scr. n = 3. (H) Western blotting to check for the expression of p53β, Cyclin D1, p21, PUMA, and NOXA in circEsyt2-silenced carotid arteries after wire injury. Left, quantification of protein expression (left). Right, representative blot images (right). GAPDH was taken as the control protein. *P < 0.05, **P < 0.01 vs. sh-con. n = 3. (I) CCK-8 assay of HASMCs following silencing circEsyt2 (si-circ) only, p53β (si-p53β) only, or both simultaneously (si-circ+si-p53β). *P < 0.05, **P < 0.01 vs. sh-con. n = 5. (J) EdU incorporation assay of HASMCs, treated as in I for 48 hours. Left: representative immunofluorescence staining of EdU (red) and DAPI (blue). Scale bars: 100 μm. Right: percentage of EdU-incorporated HASMCs. **P < 0.01 vs. scr. n = 3. Data are mean ± SEM. Two-sided unpaired t test for C–H. Two-way repeated measures ANOVA with Least Significant Difference post hoc test for I. One-way ANOVA test with Dunnett’s T3 post hoc test for J.
Functionally, p53β enhances the transcriptional activity of full-length p53 (or p53α) via direct binding, which confers its antiproliferation and proapoptotic effects (15). To study the role of p53β in cell proliferation, we transfected HASMCs with siRNAs targeting circEsyt2, p53β, or both (Supplemental Figure 7, G and H). Consequently, only the p53β silencing resulted in enhanced cell proliferation, while the cosilencing of circEsyt2 and p53β abrogated the proliferation-suppressing effect of circEsyt2 silencing (Figure 8, I and J). Collectively, these data are consistent with the hypothesis that p53β is a downstream target of circEsyt2-mediated RNA splicing and may directly contribute to VSMC proliferation.
CircEsyt2 regulates p53 pre-mRNA splicing via PCBP1.
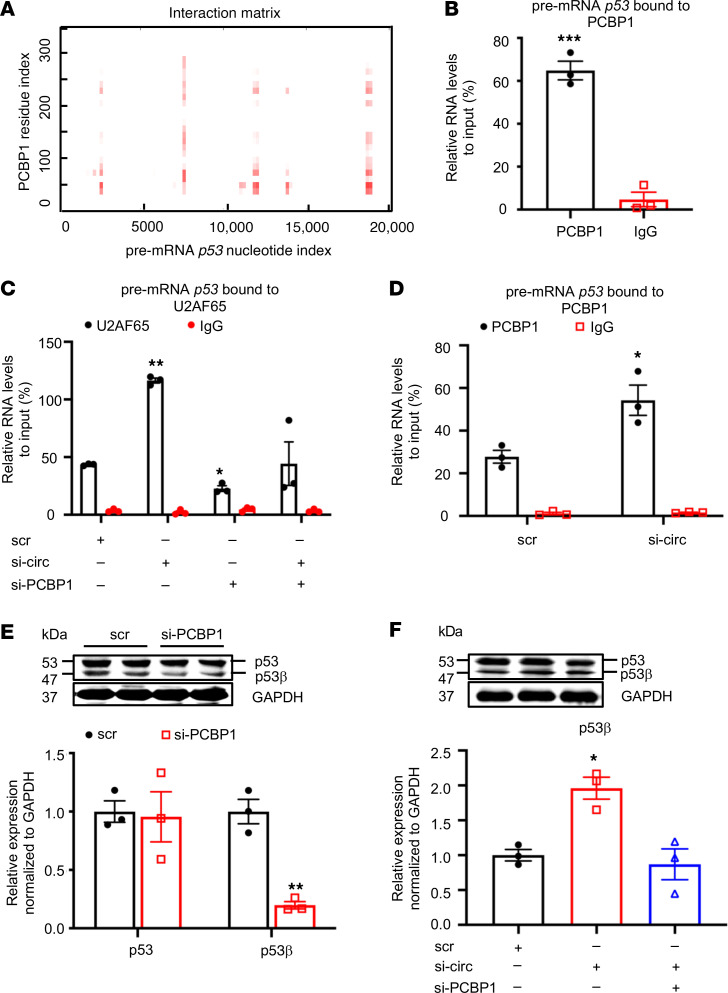
PCBP1 is a member of heterogeneous nuclear ribonucleoprotein (hnRNP) family implicated in mRNA splicing regulation (16, 17). However, its specific splicing target in VSMCs is unknown. Using the same protein–RNA binding prediction program, we discovered multiple regions in the p53 pre-mRNA, a previously unknown splicing substrate for PCBP1, with strong binding probability with PCBP1 (Figure 9A). This interaction was confirmed by the RIP assay using the anti-PCBP1 antibody (Figure 9B and Supplemental Figure 8A). Since spliceosome assembly on pre-mRNA is indispensable for alternative splicing, which is governed by U2 small nuclear RNA auxiliary factor 2 (U2AF65) (17, 18), we first confirmed the colocalization of U2AF65, p53 pre-mRNA, and PCBP1 using FISH and immunofluorescence (Supplemental Figure 8B) and subsequently determine whether circEsyt2 could regulate U2AF65 function through PCBP1 during the splicing process. As expected, the binding between p53 pre-mRNA and U2AF65 was observed, in which the amount of p53 pre-mRNA that was bound to U2AF65 significantly increased after circEsyt2 silencing (Figure 9C). Additionally, PCBP1 may also be involved because the PCBP1 silencing resulted in the reduced amount of p53 pre-mRNA bound to U2AF65 and blunted the increase in p53 pre-mRNA/U2AF65 binding previously induced by circEsyt2 silencing. We further investigated the role of PCBP1 by measuring the interaction between PCBP1 and p53 pre-mRNA and found that circEsyt2 silencing increased the PCBP1/p53 pre-mRNA binding in HASMCs (Figure 9D). These results suggest that circEsyt2 may negatively regulate p53β expression by inhibiting the U2AF65‑mediated PCBP1-dependent splicing. The p53β isoform is a product of alternative splicing generated from the partial retention of intron 9 and consequent premature termination of translation leading to a truncated C-terminus. In PCBP1-silenced HASMCs, the expression of the p53β splicing isoform was significantly decreased (Figure 9E), while the expression levels of full-length p53 mRNA and protein and p53 pre-mRNA were not affected (Supplemental Figure 8, C and D). Moreover, the increased p53β expression in circEsyt2-silenced cells was reversed by the concurrent silencing of PCBP1 (Figure 9F and Supplemental Figure 8, E and F). Taken together, these data verify that circEsyt2 negatively regulates p53β expression, which normally exerts proapoptotic and antiproliferative effects, via the inhibition of PCBP1-dependent splicing in VSMCs.
Figure 9. CircEsyt2 regulates PCBP1-dependent p53β splicing.
(A) Predicted interaction of human PCBP1 and pre-mRNA p53 by catRAPID. The binding sites are depicted in red. (B) RIP-qPCR confirming the direct binding of PCBP1 and pre-mRNA p53 in HASMCs. ***P < 0.001 vs. IgG. n = 3. (C) RIP-qPCR to check for the expression of pre-mRNA p53 pulled down by the U2AF65 antibody in HASMCs with the knockdown of circEsyt2 (si-circ), PCBP1 (si-PCBP1), or both. *P < 0.05, **P < 0.01 vs. scr. n = 3. (D) RIP-qPCR to check for the expression of pre-mRNA p53 pulled-down by PCBP1 antibody in circEsyt2-silenced HASMCs. *P < 0.05 vs. scr. n = 3. (E) Western blotting (top) and qRT-PCR (bottom) to check for the expression of p53 and p53β in PCBP1-silenced HASMCs. **P < 0.01 vs. scr. n = 3. (F) Western blotting (top) and qRT-PCR (bottom) to check for the expression of p53β following the silencing of circEsyt2 (si-circ) only or with PCBP1 (si-circ+si-PCBP1). *P < 0.05 vs. scr. n = 3. Data are mean ± SEM. Two-sided unpaired t test for B, C, and E. One-way ANOVA test with Dunnett’s T3 post hoc test for C and F.
Discussion
Globally, the morbidity and mortality of patients with CVD are increasing every year and have become a major public health problem. The continuous stimulation of risk factors, such as hypertension and hyperlipidemia on target organs (e.g., blood vessels and heart), leads to enhanced cardiovascular modifications, both structural and functional, that are the critical pathophysiological basis for many CVDs, including AS and CAD. The widely used clinical interventions, such as antihypertensive drugs and lipid regulation, have not been able to stop the increasing global incidence of CVDs, and the reasons for this are related to the largely unclear mechanism of vascular remodeling.
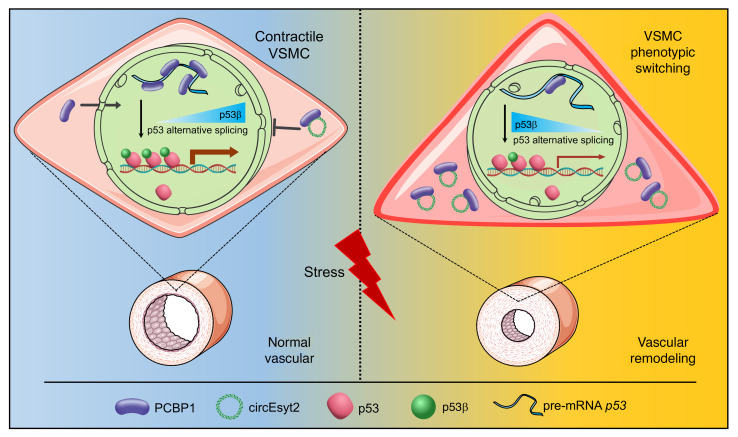
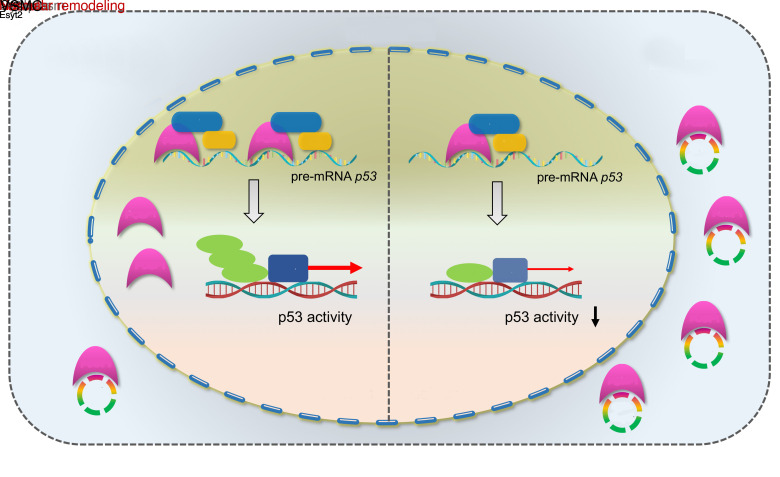
Vascular remodeling, a pathological process governed by genetic and environmental interactions, possess an extremely complex etiology and pathogenesis. However, the in-depth exploration of the roles of ncRNAs, which exhibit multiple mechanisms of gene expression regulation, in vascular remodeling is undoubtedly one of the important breakthroughs in CVD research. In recent years, an increasing number of studies has also revealed the essential regulatory role of circRNAs in the cardiovascular system and resulted in the understanding of heart and related diseases. For instance, profiling of the myocardium has provided a landscape of circRNAs expressed in the various stages of development, species, and conditions of the heart (19, 20). Moreover, multiple circRNAs with important functions have been identified in a wide range of CVDs,like myocardial infarction and heart failure (21–26). Notably, circANRIL was discovered to confer a protective role by inducing apoptosis and inhibiting the proliferation of VSMCs and macrophages via the control of pre-rRNA maturation in AS (27). Other circRNAs, such as circR-284 and circWDR77, were found to regulate cellular phenotypes, such as VSMC proliferation and migration, by acting as miRNA sponges (10, 28). Although circRNAs are being intensively studied, their functions in vivo and molecular mechanisms of action in the pathogenesis of vascular remodeling are still poorly characterized. Gratifying results have been achieved in the course of more in-depth studies regarding the pathogenesis of vascular remodeling, and our discovery of circEsyt2 is a typical example. Using high-throughput screening, our study comprehensively validated the role of circEsyt2 on vascular remodeling at multiple levels—including cells, whole animals, and clinical specimens. Mechanistically, we discovered that the expression of the downstream targets of the p53 pathway were markedly altered after circEsyt2 silencing or overexpression, while circEsyt2 was found to directly bind to and inhibit the nuclear translocation of the splicing regulator PCBP1. Further study revealed that total p53 was not altered, but the expression of its splice isoform P53β was significantly increased. Our findings also demonstrated that circEsyt2 can play a similar role to a “molecular switch” by binding and influencing the nuclear translocation of PCBP1, subsequently regulating the variable splicing of p53 and the production of p53β, and thereby directly influencing the VSMC phenotypic switching and vascular remodeling (Figure 10).
Figure 10. Schematic illustration of the mechanism whereby circEsyt2 regulates p53β splicing via PCBP1.
In general, circEsyt2 binds to PCBP1 in the cytoplasm and sequesters it from nuclear translocation. Reduction of nuclear PCBP1 suppresses U2AF65-dependent p53β splicing, which may alter the expression of p53-regulated proliferation and apoptosis genes. Consequently, VSMCs excessively proliferate, contributing to the development and progression of vascular remodeling.
Besides the function of p53 as a well-known key molecule during cell cycle and apoptosis (29, 30), accumulating evidences reveal that p53 function is not only influenced by its expression and transcriptional activity, but also by its splicing variants (31). For example, alternative promoter usage gives rise to the N-terminal truncated Δ133p53, which can inhibit p53 by suppressing the transactivation of its downstream genes. By contrast, the splicing of intron 9 generates the p53β isoform with a p53-like effect, which is caused by the retention of a part of the intron and a truncated C-terminus due to an early translational termination (15). The roles of the most studied p53 variants, Δ133p53 and p53β, have also been comprehensively revealed in malignancies (32). Moreover, p53β can reportedly increase the expression of p53 targets by facilitating the binding of the p53 protein to the promoter of target genes (33, 34). However, studies regarding the roles of p53 isoforms in CVDs are limited (35). Based on the high‑throughput RNA‑Seq data of HASMCs, we found that circEsyt2 silencing induced a VSMC phenotype (i.e., inhibition of proliferation that is p53β dependent), which subsequently activated the p53 target genes and resulted in replicative inhibition.
Alternative splicing generally requires the assembly of spliceosome on precursor RNA. By binding to the U-rich polypyrimidine tract adjacent to the 5′‑end of the splice acceptor, U2AF65 directly triggers the assembly of U2 spliceosome and stimulates splicing (17). However, splice sites are determined by 2 groups of trans-acting RBPs, namely serine/arginine-rich splicing family proteins and hnRNPs. PCBP1, which belongs to the hnRNPs, enhances splicing activity by recruiting U2AF65 (16). Here, we demonstrated that PCBP1 also positively regulates the alternative splicing of p53 that favors p53β generation via the binding of U2AF65 to the precursor RNA.
In the present study, we identified a circRNA, coined circEsyt2, that was revealed to play an important role in VSMC remodeling through the targeted inhibition of alternative mRNA splicing. Our findings provided new insights that helped expand the current understanding of how circRNAs function in the pathogenesis of CVDs. Driven by potentially novel approaches (e.g., siRNA transfection, antisense oligonucleotide experiments, and CRISPR/cas9 technology), the in vivo manipulation of ncRNAs has been achieved and even produced several prominent clinical breakthroughs (36). Therefore, innovations in epigenetic therapies targeting disease-causing circRNAs are to be expected in the future. Furthermore, mechanistic studies targeting circEsyt2 may provide an excellent strategy to combat vascular remodeling-related diseases.
Methods
Animal models.
C57BL/6J and ApoE–/– mice were used in this study (Vital River). To induce AS, 3-month-old ApoE–/– mice were fed a high-fat diet composed of 16.4% lard oil, 3.6% vegetable oil, and 2% cholesterol for 3 months until there were visible atherosclerotic plaques in the aortae. The C57BL/6J and ApoE–/– mice that were fed with a regular chow diet served as controls. The smooth muscle cell-lineage tracing mice (Tagln-CreERT2/tdTomato) were generated by crossing Tagln-CreERT2 (Cyagen Co.) and tdTomato (Biocytogen Co.) mice and subjecting the 1‑month‑old offspring to intraperitoneal injection of tamoxifen (Sigma, 75 mg/kg) for 5 consecutive days. Wire-induced carotid artery injury was induced in 3-month-old C57BL/6J and tamoxifen-induced Tagln-CreERT2/tdTomato mice (see Carotid artery injury induction and virus injection). The mice were randomly allocated into experimental groups.
Cell culture, RNA interference, transfection, and AAV infection.
The HASMCs and HEK293T cells obtained from the American Type Culture Collection (ATCC) were grown in smooth muscle cell medium (ScienCell) and high‑glucose DMEM containing 10% FBS (Gibco, Thermo Fisher Scientific) and 1% penicillin/streptomycin (Beyotime), respectively. The primary mouse VSMCs extracted from the aorta were purchased from PriCells and cultured in specific smooth muscle cell medium (PriCells). All cells were cultured at 37°C in 5% CO2, and cells between passages 4–10 were used in subsequent experiments.
The siRNAs were synthesized (Ribobio) and transfected at a final concentration of 100 nM into cells diluted in Opti-MEM (Gibco, Thermo Fisher Scientific) using Lipofectamine RNAiMAX (Thermo Fisher Scientific), according to the manufacturer’s protocol.
The circEsyt2 overexpression plasmid was generated using the pcDNA3.1-CMV vector (HANBIO), while the pcDNA3.1 plasmid served as control. A total of 4 μg per well of plasmids were transfected into cells cultured in 6-well plates using Effectene Transfection Reagent (QIAGEN), following the manufacturer’s protocol.
For circEsyt2 silencing in vivo, shRNA specifically targeting circEsyt2 was cloned into the pHBAAV-U6-MCS-CMV-EGFP vector for viral packaging in AAV 2/8 (HANBIO), with the pHBAAV-U6-shControl-CMV-EGFP vector as control. For circEsyt2 overexpression in vivo, the circEsyt2 gene was inserted into the pcDNA3.1-CMV vector, followed by the AAV 2/8 package using the pHBAAV-CMV-circ-EF1-ZsGreen vector (HANBIO). The AAV 2/8 containing only the vector served as control. The oligonucleotide sequences used are listed in Supplemental Table 4.
RNA isolation, treatment, and qRT‑PCR.
Total RNA was extracted from tissues or cells using RNAiso (Takara), according to the manufacturer’s instructions. The RNA concentration was measured using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). For RNase digestion, RNase R (3 U/μg, Epicentre Biotechnologies) was added to a total of 5 μg RNA sample, which was incubated at 37°C for 20 minutes, followed by heat inactivation at 95°C for 3 minutes. Samples without RNase R treatment were considered as control. Then, actinomycin D (Sigma‑Aldrich) was dissolved in DMSO (10 mg/mL, Sigma-Aldrich). To induce transcription blockage, VSMCs cultured in DMEM containing 1% FBS were treated with 2 mg/mL actinomycin D for 24 hours. DMSO‑treated VSMCs were used as control.
For real-time qRT-PCR analysis, cDNA was synthesized from 1–2 μg RNA using PrimeScript RT Master Mix (Takara) by incubating the sample at 37°C for 15 minutes and then at 85°C for 5 seconds. The relative mRNA levels were quantified using SYBR Green Master Mix (Takara) and normalized using GAPDH as internal control. The relative expression levels were calculated using the 2–ΔΔCT method. For small nucleolar RNA R41 (snoR41) quantification, reverse transcription was performed using Bulge-Loop miRNA qRT-PCR Starter Kit (Ribobio), following the manufacturer’s instructions.
For strand-specific qRT‑PCR, 1 μg total RNA was reverse‑transcribed using the Bulge-Loop miRNA qRT-PCR Starter Kit (Ribobio), following the manufacturer’s protocol. However, the RT primers were replaced by those specifically designed against p53, p53β, and GAPDH (Ribobio), followed by the standard qRT‑PCR pipeline mentioned above with minor modifications. The designed GAPDH, p53, and p53β were tested using the specific primers synthesized by Ribobio (Supplemental Table 5).
The gDNA was purified using GenElute Mammalian Genomic DNA Miniprep Kit (Sigma‑Aldrich). For circRNA identification, PCR amplification was carried out using cDNA or gDNA samples, circEsyt2 primers from mouse arterial cDNA, and DreamTaq Green PCR Master Mix (Thermo Fisher Scientific), according to the manufacturer’s instructions. The PCR profile consisted of 35 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute. The PCR products were visualized in 2% agarose gel stained with ethidium bromide. Then, the PCR products were purified and Sanger-sequenced (Sangon) using the forward divergent primer. The primers used are listed in Supplemental Table 5.
Western blotting.
Proteins were lysed in RIPA buffer (Beyotime) containing 1 mM PMSF, incubated on ice for 30 minutes, and centrifuged at 14,000g at 4°C for 30 minutes. The proteins were separated using 10%–15% SDS-PAGE gels and transferred to nitrocellulose membranes (Pall Co.). The membranes were blocked in 5% nonfat dry milk for 1 hour, followed by incubation with primary antibodies at 4°C overnight and then with IRDye 800CW donkey anti-mouse or anti-rabbit antibody (1:10,000, nos. 926-32212 and 926-32213, LI-COR) at room temperature for 1 hour. Protein detection was performed using the Odyssey Infrared Imaging System (LI-COR). The primary antibodies used were as follows: rabbit anti–α-SMA (ACTAT2/SMA, 1:1000, no. 14395-1-AP, Proteintech), rabbit anti-calponin (1:1000, no. 13938-1-AP, Proteintech), mouse anti-myh11 (1:200, no. 6956, Santa Cruz Biotechnology), mouse anti-PCBP1 (HnRNP-E1, 1:500, no. 14523-1-AP, Proteintech), mouse anti-p53 (1:1000, no. 2524, Cell Signaling Technology), mouse anti–acetyl-p53 (lys382) (1:1000, no. 2525, Cell Signaling Technology), mouse anti-p53 (DO-1) (1:200, no. sc-126, Santa Cruz Biotechnology), mouse anti-cyclin D1 (1:1000, no. 60186-1-Ig, Proteintech), rabbit anti-p21 (1:500, no. 27296-1-AP, Proteintech), mouse anti-NOXA (1:500, no. AB5761, Millipore), rabbit anti-PUMA (1:500, no. 55120-1-AP, Proteintech), mouse anti–histone 3 (1:1000, no. ab1791, Abcam), rabbit anti-Esyt2 (1:500, no. bs-11003R, Bioss), rabbit anti–β-actin (1:1000, no. 20536-1-AP, Proteintech), and rabbit anti-GAPDH (1:1000, no. 10494-1-AP, Proteintech).
Cell proliferation assay.
Cells were seeded in 96-well plates (3 × 103 per well) and starved for 2 hours in serum-free DMEM. The HASMCs and VSMCs were transfected with siRNAs or plasmids and subsequently cultured in DMEM containing 1% FBS. Cell growth was evaluated at indicated time points using the cell counting kit-8 (CCK-8) detection assay (10 μL/well, MedChemExpress), following the manufacturer’s instructions. To quantify the number of proliferating cells, EdU incorporation assay was performed 48 hours after siRNA treatment or plasmid transfection using Click-iT EdU Alexa Fluor 594 Imaging Kit (Invitrogen). The cells were incubated with 50 μM EdU for 6 hours before fixation, and images were taken under a fluorescence microscope (Nikon).
Scratch wound and transwell assays.
The HASMCs and VSMCs were initially cultivated in 6-well plates and transfected with siRNAs or plasmids for 24 hours, then cultured in DMEM containing 1% FBS. For the scratch wound healing assay, a sterile pipette tip was used to create a perpendicular line in adherent cells. For the transwell assay, the cells were added to the upper transwell chambers (8 μm pore size, Millipore). After 24 hours, images were taken under a light microscope (Nikon). The cells on the chamber membrane were fixed with 4% paraformaldehyde and stained with crystal violet.
Cell apoptosis assay.
The HASMCs and VSMCs were cultured in 6-well plates and transfected with siRNA or plasmid. After 48 hours, the level of cell apoptosis was visually determined by propidium iodide and Annexin V staining using FITC Annexin V Apoptosis Detection Kit (BD Biosciences). The samples were evaluated using a flow cytometer (BD Biosciences). The data obtained were analyzed using FlowJo software (TreeStar).
Immunofluorescence assay.
Cells grown on EZ slides (Millipore) were washed once with PBS, fixed with 4% paraformaldehyde for 30 minutes, and permeabilized with 0.1% Triton X-100 for 10 minutes. For paraffin-embedded carotid arteries, 4‑μm sections were deparaffinized and subjected to heat-induced antigen recovery using Citrate-EDTA Antigen Retrieval Solution (Beyotime). Freshly collected aortic or carotid arteries were incubated in 4% paraformaldehyde containing 30% sucrose for more than 24 hours at 4°C, and then embedded in an optical cutting temperature compound for cryosection (8 μm). The frozen sections were washed thrice with PBS. For immunostaining, all specimens were blocked with QuickBlock Blocking Buffer (Beyotime) for 30 minutes at 37°C and incubated with primary antibodies at 4°C overnight. The goat anti-mouse Alexa Fluor 488 (1:200, no. A11001, Invitrogen), goat anti-rabbit Alexa Fluor 488 (1:200, A11008, Invitrogen), goat anti-mouse Alexa Fluor 594 (1:200, no. A11005, Invitrogen), goat anti-rabbit Alexa Fluor 594 (1:200, no. A11012, Invitrogen), and goat anti-rabbit IgG (H+L) DyLight 405 (1:100, no. 35551, Thermo Fisher Scientific) secondary antibodies were subsequently added and incubated at 37°C for 1 hour. DAPI (Beyotime) was used to counterstain the nuclei. The primary antibodies included the mouse anti–α-SMA (1:200, no. ab7817, Abcam) or rabbit anti–α-SMA (ACTAT2/SMA, 1:1000, no. 14395-1-AP, Proteintech), mouse anti-GFP (1:100, no. 66002-1-Ig, Proteintech), mouse anti-Myh11 (1:100, no. sc-6956, Santa Cruz), mouse anti-CD31 (1:100, no. BH0190, Bioss), rabbit anti-CD68 (1:100, no. 28058-1-AP, Proteintech; reacted with mouse tissue), mouse anti-CD68 (1:100, no. bs-0649R, Bioss; reacted with human tissue), rabbit anti-PDGFRα (1:100, no. bs-0231R, Bioss), rabbit anti-PCBP1 (1:100, no. 14523-1-AP, Proteintech), and rabbit anti-U2AF65 (1:100, no. 15624-1-AP, Proteintech).
RNA-FISH.
The FISH Kit (RiboBio) was used for the in vitro assay, following the manufacturer’s protocol. After fixation, permeabilization, and blocking, the cells were incubated in prehybridization buffer at 37°C for 30 minutes and then in hybridization buffer with FISH probes at 37°C overnight. The cells were washed thrice with 4× saline sodium citrate (SSC) with 0.1% Tween-20 and then once with 2× SSC and 1× SSC at 42°C for 5 minutes, followed by rinsing with PBS and staining with DAPI (Beyotime). All experimental procedures were performed in the dark. The Cy3-labeled 18S, U6, control, and circEsyt2 probes were designed and synthesized by RiboBio, while the cy3-labeled pre-mRNA p53 probe was obtained from Zoonbio Biotechnology.
In vivo RNA-FISH was performed for the frozen sections of mouse tissues and paraffin-embedded sections of human tissues using FISH Kit (GenePharm), following the manufacturer’s instructions. The biotin-conjugated circEsyt2 probe was obtained from GenePharm. The amplification of the signal was based on the streptavidin-Cy3 or FAM system. Immunofluorescence assay was performed by incubating with the primary antibodies at 37°C for 2 hours and subsequently with the secondary antibodies at 37°C for 1 hour. Fluorescence images were obtained using a laser confocal microscope (Olympus).
RNA pull‑down and mass spectrometry.
RNA pull-down was performed on circEsyt2 overexpression plasmid-transfected HEK293T cells using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. The biotinylated RNA probes for specifically detecting circEsyt2 were designed and synthesized by Ribobio. Two 10-cm dishes with approximately 1 × 107 cells and 50 μL streptavidin beads were used for each sample. For Western blotting and immunoprecipitation (Beyotime), 200 μL cell lysis buffer was added to the cells after washing with cold PBS, supplemented with RNasin Ribonuclease Inhibitor (40 U/μL, Solarbio) and 100× Protease Inhibitor Cocktail Set I (Millipore). The samples were kept on ice for 20 minutes. Then, the lysates were centrifuged at 16,000g for 20 minutes, and the supernatants were collected. The biotinylated RNA probes were incubated with magnetic beads at room temperature for 30 minutes with rotation, followed by the addition of the supernatant and incubation for an additional 1 hour at 4°C with rotation. After washing thrice with the lysis buffer, the RNA-bead complexes were harvested using 50 μL elution buffer and heated in 5× loading buffer at 99°C for 10 minutes.
Before mass spectrometry, silver staining using 20 μL of pulled-down samples separated on 13% SDS-PAGE was performed by GeneCreate. The proteins were alkylated by the addition of 10 mM DL-dithiothreitol, and then digested by incubating with 55 mM iodoacetamide and 1 μg trypsin overnight. The peptides were demineralized and dissolved in 15 μL loading buffer containing 0.1% formic acid and 3% acetonitrile. Liquid chromatography–dual mass spectrometry was performed using the proteomic platform TripleTOF 5600+ (SCIEX). Raw data were analyzed using Protein Pilot Software (SCIEX), with iodoacetamide-alkylation, bias, and background correction as parameters. Following the removal of the common contaminating proteins and their mapping peptides, setting the threshold to confidence greater than 95% and unique peptides greater than 1 revealed a total of 14 and 31 proteins as the control and circEsyt2‑binding proteins, respectively. The identified proteins and peptides were ranked according to the Unused ProtScore, which is considered as the true indicator of protein confidence whose calculation depends on the ranking of the protein in the list of detected proteins. Analysis of the Unused ProtScore was conducted using the Pro Group Algorithm. Mass spectra and data analyses were performed by GeneCreate. The mass spectrometry data were deposited in the iProx database (iProx: IPX0001671000).
CircRNA‑Seq and annotation.
Using TRIzol reagent (Life Technologies), total RNA was isolated from the aortae of 3-month-old C57BL/6J and ApoE–/– mice fed with a regular chow diet and ApoE–/– mice fed with a high‑fat diet for 3 months. The aortae were carefully detached from the aortic arch to the bifurcation of the iliac artery, and its surrounding connective tissues removed. RNA concentration and quality control were determined using Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific). The RNA library was prepared from 2 μg RNA after the removal of rRNA using the Ribo-Zero rRNA Removal Kit (Epicentre Biotechnologies), followed by linear RNA digestion using RNase R (Epicentre Biotechnologies) and fragmentation. The synthesized cDNA was subjected to purification and end-repair, followed by adenylation of the 3′‑end and ligation to adaptors using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). After PCR amplification and purification, the library quality was tested using the TapeStation System (Agilent Technologies). Sequencing was performed on an Illumina HiSeq 3000 platform (Illumina) using the 150‑bp pair-end model. The sequencing data are available in the Gene Expression Omnibus (GEO) of the NCBI database (accession number: GSE133269). The sequencing and data analysis were conducted by Ribobio.
The sequence reads were preprocessed through adaptor trimming and data filtration. The cleaned data were subsequently mapped against the mouse reference genome GRCm38/mm10 using Tophat (37), with the unmapped reads saved for further mapping using Tophat-Fusion (37). The primary results from the alignment were adjusted and filtered based on the “GT/AG” splicing site flanking “back-spliced junction reads,” which denoted circRNA splicing. This method was named as Find_circ later. Expression analysis was conducted by converting the read counts into reads per million (RPM) using HTseq (38). Data showing extreme difference within groups were excluded. Additional algorithms (CIRCexplorer, CIRI2, and CIRIquant) were also adopted to predict circRNAs; data processing was conducted by Ribobio.
The RPM value was calculated as follows: RPM = (number of reads that mapped to circRNAs / total number of reads) × 106. Differentially expressed genes were defined with a threshold of fold change greater than 2, P less than 0.05, and FDR less than 0.05.
RNA-Seq.
The mouse VSMCs transfected with siRNAs for 48 hours were lysed with RNAiso (Takara) and sent to BioWavelet for sequencing. Reverse transcription of RNA samples was conducted at 50°C for 90 minutes and 70°C for 15 minutes in a 20 μL reaction system containing SuperScriptA, SuperScript III reverse transcriptase (Invitrogen), and Superscript III first-strand synthesis buffer (5×) (Invitrogen). The second strand cDNA was synthesized from the products incubated with the reaction buffer and synthesis enzyme mix (Biowavelet) and kept on ice, according to the manufacturer’s guidelines. The products were purified by magnetic separation using Agencourt AMPure XP Beads (1.8×) (Beckman Coulter), and the concentrations were determined using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). The sequencing libraries were prepared and purified using TruePrep DNA Library Prep Kit for Illumina (Vazyme) and PCR with P5/P7 primers (Biowavelet). The reaction system for Tn5 library generation was carried out using 1 ng cDNA incubated at 55°C for 10 minutes. Purified amplicons were obtained using VAHTSTM DNA Clean Beads Kit (Vazyme).
Sequencing was performed using the HiSeq platform (Illumina) with the 150‑bp pair-end model. Reads obtained from RNA‑Seq were filtered and trimmed using Trimmomatic (39), followed by mapping of the cleaned data to the mouse reference genome GRCm38/mm. Gene expression analysis was performed using FeatureCounts (40). Data showing extreme difference within groups were excluded. Fragments per kilobase of exon model per million mapped fragments (FPKM) values were calculated from the read counts for gene expression normalization using the formula: FPKM = fragments that mapped to genes / (mapped reads [millions] × transcript length [kb]). RNA sequencing data have been submitted to the GEO database (accession number: GSE133270). Differentially expressed genes were defined with a threshold of fold change greater than 1.2, P less than 0.05, and FDR less than 0.05. KEGG pathway enrichment analysis was performed for differentially expressed coding genes.
RIP.
The RIP assay was performed using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore), according to the manufacturer’s instructions. The antibody against mouse anti-PCBP1 (no. sc-137249, Santa Cruz Biotechnology) or rabbit anti-U2AF65 (no. 15624-1-AP, Proteintech) was used to immunoprecipitate the target RNAs. In brief, the HASMCs (1–1.5 × 107) were collected with cold PBS and dispersed with RIP lysis buffer containing 200× protease inhibitor cocktail and 400× RNase inhibitors. Then, 50 μL Protein A/G Magnetic Beads were incubated with 5 μg PCBP1 antibody and mouse or rabbit IgG for 30 minutes at room temperature. The lysates were mixed with bead-antibody complexes and incubated with rotation at 4°C overnight. After 6 washings, the proteins were digested by incubating with 10 mg/mL proteinase K at 55°C for 30 minutes, followed by isolation and purification of the coprecipitated and input RNAs using phenol/chloroform/isoamyl extraction and ethanol precipitation. The expression of coprecipitated RNAs was quantified by qRT-PCR. The qRT-PCR products were resolved on a 2% agarose gel stained with ethidium bromide, following the addition of 6× DNA loading buffer (Beyotime).
Cell fractionation.
Cytoplasmic and nuclear proteins were extracted from cells after 24 hours of siRNA transfection in 10‑cm dishes or from carotid arteries (pooling of 8 arteries into one group) using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific), following the manufacturer’s protocol. RNase Inhibitor (40 U/μL, Solarbio) and 100× Protease Inhibitor Cocktail Set I (Millipore) were added to prevent RNA and protein degradation, respectively. The fractions were collected for RNA extraction, followed by qRT-PCR or Western blot analysis.
Carotid artery injury induction and virus injection.
Wire injury of the right common carotid artery was induced in pentobarbital-anesthetized 3-month-old C57BL/6J or Tagln-CreERT2/tdTomato mice, as previously described (41). In brief, the common carotid artery was clearly exposed under a dissecting microscope and ligated with 6-0 silk sutures for the temporary blockage of blood flow. Then, a flexible wire (0.38 mm) was introduced into the lumen and withdrawn 3 times, followed by the intraluminal injection of AAV 2/8 viruses into the common carotid artery at a titer of 5 × 1011 v.g./mL (50 μL per mouse). The cells were allowed to be infected for 30 minutes. The sutures were subsequently released to allow the restoration of blood flow. The uninjured contralateral left carotid artery served as the sham control.
The carotid arteries and aortae were collected after 28 days. Some samples were embedded in wax for histological studies or prepared for frozen cryosection, and the others were used for RNA and protein extraction experiments. Neointimal thickening was visualized by hematoxylin and eosin (H&E) staining and analyzed by determining the ratio of intima to media thickness. Cell proliferation was evaluated by immunofluorescence staining using rabbit anti-Ki67 primary antibody (1:100, no. PA5-19462, Invitrogen) and goat anti-rabbit Alexa Fluor 488 (1:200, no. A11012, Invitrogen) secondary antibody. Cell apoptosis was analyzed by TUNEL assay using the TUNEL Apoptosis Assay Kit (Beyotime). Phenotypic switching was assessed by measuring the expressions of α-SMA or Myh11 and then quantified by Western blotting of injured C57BL/6J carotid arteries or immunostaining of injured Tagln-CreERT2/tdTomato mice.
Oil Red O staining.
The aortae from mice were dissected, incised along their longitudinal axes, and fixed with 4% paraformaldehyde. All samples were washed thrice with PBS, and lipid loading was assessed using the Oil Red O Stain Kit (Solarbio), following the manufacturer’s instructions.
Image analysis.
For the EdU incorporation assay, cells within the whole field were counted using the DAPI-stained nuclei as guides. EdU-positive cells were distinguished by the red fluorescence signal merging with the DAPI signal (blue). The percentage of EdU-positive cells was calculated as the ratio of EdU-positive cells to the total number of cells. Three to five fields of vision were randomly chosen for every group, and cell counting was conducted using a fluorescence microscope at a magnification of 100×. The observer was blinded to the groupings of the specimens.
Statistics.
All data are mean ± SEM. Experiments were repeated at least 3 times. The data were analyzed using GraphPad PRISM software version 8.0 (GraphPad Software, Inc.) and SPSS 23.0 software (SPSS Inc.). First, the Shapiro-Wilk test was used for testing normality. Second, for the comparison of 2 groups, 2-sided unpaired Student’s t test was used for testing normally distributed data when homogeneity of variance was assumed; otherwise, Welch’s t test was used for unequal variances. Third, for the comparison of multiple groups, 1-way or 2-way repeated measures ANOVA was used with LSD, Dunnett’s T3, or Bonferroni’s post hoc test, as indicated in the figure and supplemental figure legends. Finally, Kolmogorov-Smirnov test was used for comparing the data that were not normally distributed. Data with P values less than 0.05 were considered significant.
Study approval.
All experimental procedures involving animals were conducted in accordance with the guidelines for animal care and use of the Third Military Medical University. Human coronary artery sections were provided by the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). Coronary arteries were collected from patients (≥ 65 years old, hypertension‑ and diabetes‑free) receiving heart transplant surgery. According to the outpatient coronary angiographic findings, patients with plaques that narrowed the coronary artery at ≤ 20% were categorized into the mild CAD group, while patients with plaques that narrowed the coronary at ≥ 70% were assigned to the severe CAD group. Detailed information of the coronary artery donors is documented in Supplemental Table 6. The human tissues were harvested following written informed consent from all subjects. All experimental procedures were approved by the Third Military Medical University Ethics Committee and the Research Ethics Committees of the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China).
Author contributions
GW and CYZ conceptualized the study. XG and MT designed the study methodology. XG, MT, NC, PY, ZX, QL, SZ, CZ, and CC conducted the investigations. XG ran the software for the study and wrote the original draft of the manuscript. XG, GW, DZW, YW, PAJ, BZ, and CYZ reviewed and edited the manuscript. CYZ and GW acquired funding for the study. DSJ, XW, ZJ, YX, and CYZ acquired resources for the study. CYZ acted as project administrator. KC, GW, and CYZ supervised the study. The order of co–first authors was determined by the volume of work each contributed to the study.
Supplementary Material
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (82022005, 81970384), the National Key R&D Program of China (2018YFC1312700), Program of Innovative Research Team by National Natural Science Foundation (81721001), Program for Changjiang Scholars and Innovative Research Team in University (IRT1216), Chongqing Talents: Exceptional Young Talents Project (CQYC202005045 cstc2021ycjh-bgzxm0098), and the Technological Innovation Project of Third Military Medical University (2019XQN17).
Version 1. 12/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(24):e147031.https://doi.org/10.1172/JCI147031.
Contributor Information
Xue Gong, Email: 619418088@qq.com.
Miao Tian, Email: 24209841@qq.com.
Nian Cao, Email: lean_chau@163.com.
Peili Yang, Email: yangpl2013@163.com.
Zaicheng Xu, Email: zaichengxu@126.com.
Shuo Zheng, Email: zs197819@163.com.
Qiao Liao, Email: 616407583@qq.com.
Caiyu Chen, Email: ccy19740920@163.com.
Cindy Zeng, Email: xdzeng@ucdavis.edu.
Pedro A. Jose, Email: pjose01@georgetown.edu.
Da-Zhi Wang, Email: dwang@enders.tch.harvard.edu.
Zhao Jian, Email: zhao.j@tmmu.edu.cn.
Yingbin Xiao, Email: xiaoyb@tmmu.edu.cn.
Ding-Sheng Jiang, Email: jds@hust.edu.cn.
Xiang Wei, Email: xiangwei@tjh.tjmu.edu.cn.
Bing Zhang, Email: bingzhang@sjtu.edu.cn.
Yibin Wang, Email: yibinwang@mednet.ucla.edu.
Ken Chen, Email: chenken0205@yahoo.com.
Gengze Wu, Email: wugengze@163.com.
Chunyu Zeng, Email: chunyuzeng01@163.com.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Beermann J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 3.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, et al. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 5.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Zhong Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, et al. circRNA/lncRNA-miRNA-mRNA network in oxidized, low-density, lipoprotein-induced foam cells. DNA Cell Biol. 2019;38(12):1499–1511. doi: 10.1089/dna.2019.4865. [DOI] [PubMed] [Google Scholar]
- 8.Shen L, et al. CircRNA‑0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting miR‑107. Mol Med Rep. 2019;19(5):3923–3932. doi: 10.3892/mmr.2019.10011. [DOI] [PubMed] [Google Scholar]
- 9.Fasolo F, et al. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res. 2019;115(12):1732–1756. doi: 10.1093/cvr/cvz203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, et al. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017;494(1–2):126–132. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 11.Hall IF, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. 2019;124(4):498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, et al. A novel regulatory mechanism of smooth muscle α-Actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res. 2017;121(6):628–635. doi: 10.1161/CIRCRESAHA.117.311441. [DOI] [PubMed] [Google Scholar]
- 13.Agostini F, et al. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29(22):2928–2930. doi: 10.1093/bioinformatics/btt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vashist S, et al. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J Virol Methods. 2012;184(1–2):69–76. doi: 10.1016/j.jviromet.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon H, et al. Modulation of alternative splicing contributes to cancer development: focusing on p53 isoforms, p53β and p53γ. Cell Death Differ. 2014;21(9):1347–1349. doi: 10.1038/cdd.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji X, et al. αCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res. 2016;44(5):2283–2297. doi: 10.1093/nar/gkw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi V, et al. Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-β. Mol Cell. 2016;64(3):549–564. doi: 10.1016/j.molcel.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutandy FXR, et al. In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res. 2018;28(5):699–713. doi: 10.1101/gr.229757.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobi T, et al. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. 2016;14(4):216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werfel S, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Geng HH, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11(3):e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salgado-Somoza A, et al. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24(6):1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7(16):3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du WW, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 27.Holdt LM, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bazan HA, et al. Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circ Cardiovasc Genet. 2017;10(4):e001720. doi: 10.1161/CIRCGENETICS.117.001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guevara NV, et al. The absence of p53 accelerates atherosclerosis by increasing cell proliferation in vivo. Nat Med. 1999;5(3):335–339. doi: 10.1038/6585. [DOI] [PubMed] [Google Scholar]
- 30.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruiswijk F, et al. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 32.Fujita K, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11(9):1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcel V, et al. Modulation of p53β and p53γ expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ. 2014;21(9):1377–1387. doi: 10.1038/cdd.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19(18):2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie N, et al. SRSF1 promotes vascular smooth muscle cell proliferation through a Δ133p53/EGR1/KLF5 pathway. Nat Commun. 2017;8:16016. doi: 10.1038/ncomms16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, et al. Long non-coding RNAs: from disease code to drug role. Acta Pharm Sin B. 2021;11(2):340–354. doi: 10.1016/j.apsb.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, et al. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, et al. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y, et al. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 41.Lindner V, et al. Mouse model of arterial injury. Circ Res. 1993;73(5):792–796. doi: 10.1161/01.RES.73.5.792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.