Abstract
In neuro-oncology, magnetic resonance imaging (MRI) is a critically important, non-invasive radiologic assessment technique for brain tumor diagnosis, especially glioma. Deep learning improves MRI image characterization and interpretation through the utilization of raw imaging data and provides unprecedented enhancement of images and representation for detection and classification through deep neural networks. This systematic review and quality appraisal method aim to summarize deep learning approaches used in neuro-oncology imaging to aid healthcare professionals. Following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines, a total of 20 low-risk studies on the established use of deep learning models to identify glioma genetic mutations and grading were selected, based on a Quality Assessment of Diagnostic Accuracy Studies 2 score of ≥9. The included studies provided the deep learning models used alongside their outcome measures, the number of patients, and the molecular markers for brain glioma classification. In 19 studies, the researchers determined that the deep learning model improved the clinical outcome and treatment protocol in patients with a brain tumor. In five studies, the authors determined the sensitivity of the deep learning model used, and in four studies, the authors determined the specificity of the models. Convolutional neural network models were used in 16 studies. In eight studies, the researchers examined glioma grading by using different deep learning models compared with other models. In this review, we found that deep learning models significantly improve the diagnostic and classification accuracy of brain tumors, particularly gliomas without the need for invasive methods. Most studies have presented validated results and can be used in clinical practice to improve patient care and prognosis.
Keywords: neuro imagining, glioma classification, accuracy, neuro-oncology, artificial intelligence, deep learning
Introduction and background
Gliomas arise from precursor or glial cells and account for 27% of all tumors and 80% of major brain malignant tumors. They include glioblastoma, astrocytoma, oligodendroglioma, ependymoma, mixed glioma, malignant glioma, and not otherwise specified (NOS) and other rare histology [1]. Cellular invasion, heterogeneous angiogenesis, apoptosis, and cellular proliferation of glioma biology make its quantitative assessment complicated and significantly increase morbidity and mortality [2]. Histopathologic grading of glioma is important to plan the treatment approach, assess the response to treatment, and provide the overall prognosis. Stereotactic brain biopsy allows accurate and definitive diagnosis but is considered an invasive procedure [3].
Magnetic resonance imaging (MRI) serves as the primary contributor to brain tumor diagnosis, staging, treatment, and follow-up. The National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology for Central Nervous System (CNS) Cancers recommends MRI for the evaluation of patients with a primary brain tumor and in the determination of the response to therapy [4]. Preoperative brain MRI is a useful, non-invasive imaging technique for the assessment of the histopathological grade of gliomas. Both dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI) have been used prior to surgery to differentiate the grades of gliomas by using different quantitative parameters; relative cerebral blood volume (rCBV) is the most sensitive parameter [5]. In addition, computer-aided diagnosis (CAD) using intensity-invariant MRI features has been proposed to grade gliomas by using quantitative image features such as histogram moment and texture analyses, which are practical to use in the clinical setting [6]. Moreover, several approaches have been proposed for subjective visual interpretation of malignant glioma. Gutman et al. [7] developed a comprehensive subjective MRI feature called Visually AcceSAble Rembrandt Images (VASARI) to predict overall survival and correlate it with different genomic biomarkers.
Artificial intelligence (AI) is expanding rapidly and evolving in different fields including diagnostic radiology and medical imaging [8]. Machine learning (ML) is a subset of AI that allows systems to automatically learn and gain experience from existing training data and to make predictions about new data by using different algorithms and without explicit programming. Deep learning (DL) is a subfield of ML, and it uses neural networks (NN) that contain many layers to analyze different factors. In radiology and medical imaging, most ML applications rely on supervised forms comprising algorithms trained on “ground truth” labels [9]. These labels may contain different classes of diagnoses, prognoses, or classes existing in one set of images [10]. Both ML and DL methods are being used increasingly in neuro-oncological imaging. DL provides an astonishing improvement in image analysis by using raw data obtained from MRI images to automatically detect, grade, or classify gliomas. DL has become the most widely used approach within the field of ML because it can achieve outstanding results in several complex tasks, similar to and sometimes exceeding those provided by humans [11]. Multiple DL models are currently in use. These include convolutional neural network (CNN), deep Boltzmann machine (DBM), deep neural network (DNN), recurrent neural network (RNN), deep autoencoder (DA), and deep belief network (DBN) [12].
DL has the potential to detect image patterns that usually require the eyes of an experienced neuroradiologist. With the use of magnetic resonance (MR) images, DL is a noninvasive method that can rapidly identify the genetic features of glioma and make predictions regarding the treatment response and future outcome [13]. This systematic review aims to summarize ML and DL approaches used in neuro-oncological imaging to aid healthcare professionals, improve treatment outcomes, and add value to patient care.
Review
Methods
This systematic review was registered with PROSPERO (International Prospective Register of Systematic Reviews) and was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [14]. The literature search was done with the following databases: PubMed, Medline, Cumulative Index to Nursing and Allied Health Literature (CINAHL), Web of Science, and Google Scholar. The search was done for articles in English published between 2005 and 2020, and for articles addressing the clinical application of DL in patients with glioma. The search terms included “deep learning AND glioma” OR “deep learning AND glioblastoma” OR “artificial intelligence AND glioma” OR “artificial intelligence AND glioblastoma” AND “glioma classification” AND “deep learning approaches” OR “artificial intelligence” OR “brain metastasis”.
Eligibility Criteria
All prospective and retrospective studies that examined neuro-oncology patients with glioma, glioma tumor grading, and mutations using MRI and AI, ML, or DL models as a major diagnostic tool were eligible for inclusion. The target population included patients with an established diagnosis of glioma. No restriction was applied to the patient population or age. Articles that examined radiomics and histopathological data without the use of an imaging modality were excluded. Only articles written in English were considered. All study forms were included except letters to the editor and review articles.
Data Extraction
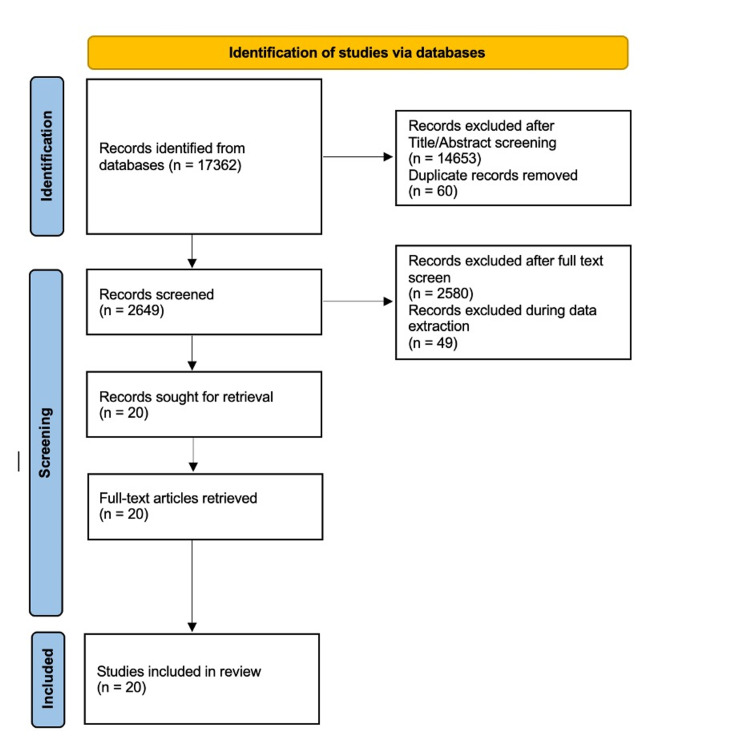
Two reviewers performed the eligibility assessment of the search results by screening titles and abstracts. The review placed a limitation on the presence of a glioma tumor, the intended context for using the model, and the disease outcome of interest. Data were extracted independently by two reviewers using a predefined data extraction sheet. Furthermore, both reviewers cross-checked the extracted data and resolved any disagreements by discussion. The information for study characteristics included author(s), the purpose of the study, the number of patients or exams, the diagnostic model used, and the outcome measures including accuracy, sensitivity, specificity, the dice index, the positive predictive value (PPV), and the negative predictive value (NPV). The PRISMA flowchart showing studies retrieved at each stage of the systematic review is shown in Figure 1.
Figure 1. PRISMA flowchart showing the number of studies retrieved at each stage of the systematic review.
PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
Quality Assessment of the Studies
The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool was used to evaluate the risk of bias, the applicability, and the quality of the studies. All studies with scores greater than 9 during the quality assessment were classified as low risk and included in this review
Data Synthesis
A single contingency table was developed to report the accuracy of each DL model used. Binary diagnostic accuracy data were extracted preferably. Contingency tables of true negatives, false negatives, true positives, and false positives were used to report sensitivity and specificity.
Results
The initial literature review produced 17,362 articles. After title and abstract screening and removal of duplicates, a total of 2,649 articles were retrieved. After the review by two independent reviewers and application of the inclusion and exclusion criteria, 2,580 articles were excluded, and an additional 49 articles were excluded during data extraction. Finally, a total of 20 articles were included in this review.
Characteristics of the Included Studies
Table 1 shows the DL models used in various studies along with their outcome measures, the number of patients or MRI scans, and the molecular markers for brain glioma classification [15-36]. In 19 studies, the researchers determined the DL model accuracy. In six studies [15-20], the authors reported the sensitivity of the DL model used, and in five studies [15,16,18-20], the authors reported the specificity of the DL models. In eight studies, the researchers examined glioma grading by using different DL models compared with other models [19,21-27].
Table 1. Description of deep learning models used and comparison of their outcome measures in neuro-oncology patients.
2D: two-dimensional; 2D-DOST: two-dimensional discrete orthonormal Stockwell transform; 3D: three-dimensional; ANN: artificial neural networks; AUC: area under the curve; CNN: convolutional neural network; DL: deep learning; GB: gradient boosting; HGGs: high-grade gliomas; IDH: isocitrate dehydrogenase; KNN: k-nearest neighbor; LGGs: low-grade gliomas; MGMT: O6-methylguanine-DNA methyltransferase; MLP: multilayer perceptron; NPV: negative predictive value; PPV: positive predictive value; RF: random forest; SVM: support vector machine; T2w: T2-weighted; TS: true segmentation; VGG: visual geometry group; Wndchrm: weighted neighbor distance using a compound hierarchy of algorithms representing morphology.
| No. | Author | Year | Study markers | Best performing DL model used | Sample size | Other DL models used | Outcome measures | Values (DL model) |
| 1 | Akkus et al. [15] | 2015 | Preoperative patients with LGGs segmentation | 2D and 3D segmentation | 30 | STAPLE TS | 3D segmentation | |
| Dice index | 0.89 | |||||||
| Sensitivity | 0.91 | |||||||
| Specificity | 0.99 | |||||||
| 2D segmentation | ||||||||
| Dice index | 0.9 | |||||||
| Sensitivity | 0.92 | |||||||
| Specificity | 0.99 | |||||||
| 2 | Bangalore et al. [16] | 2020 | Predict IDH mutation status | T2-Net | 214 patients (94 IDH mutated and 120 IDH wild-type) | T2W image only network (T2-Net) | T2-net | |
| Accuracy | 97.14% | |||||||
| Sensitivity | 0.97 | |||||||
| Specificity | 0.98 | |||||||
| PPV | 0.98 | |||||||
| NPV | 0.97 | |||||||
| AUC | 0.98 | |||||||
| Multi-contrast network (TS-Net) | TS-net | |||||||
| Accuracy | 97.12% | |||||||
| Sensitivity | 0.98 | |||||||
| Specificity | 0.97 | |||||||
| PPV | 0.97 | |||||||
| NPV | 0.97 | |||||||
| AUC | 0.99 | |||||||
| 3 | Díaz-Pernas et al. [17] | 2021 | Segmentation and classification of brain tumors (meningioma, glioma, and pituitary tumors) | Deep CNN with a multiscale approach | 233 | Classic ML and DL methods | Accuracy | 97.30% |
| 4 | Naser et al. [18] | 2020 | Grade LGGs (grade II vs. III) and tumor detection by segmentation | DL model | 110 | Detection model | Accuracy | 0.92 |
| Sensitivity | 0.92 | |||||||
| Specificity | 0.92 | |||||||
| Grading model | Accuracy | 0.89 | ||||||
| Sensitivity | 0.87 | |||||||
| Specificity | 0.92 | |||||||
| 5 | Zhuge et al. [19] | 2020 | Glioma classification | 3DConvNet | 285 | 2D Mask R-CNN | Accuracy | 96.30% |
| Sensitivity | 93.50% | |||||||
| Specificity | 97.20% | |||||||
| 3DConvNet | Accuracy | 97.10% | ||||||
| Sensitivity | 94.70% | |||||||
| Specificity | 96.80% | |||||||
| 6 | Nalawade et al. [20] | 2019 | Predict IDH mutation status | DenseNet-161 | 260 patients (120 HGGs and 140 LGGs) | Inception-v4 | Inception-v4 slice-wise | |
| Accuracy | 76.10% | |||||||
| Precision | 59.40% | |||||||
| Sensitivity | 59.20% | |||||||
| Specificity | 84.50% | |||||||
| F1 score | 58.20% | |||||||
| Inception-v4 subject-wise | ||||||||
| Accuracy | 64.20% | |||||||
| Precision | 65.80% | |||||||
| Sensitivity | 65.10% | |||||||
| Specificity | 65.10% | |||||||
| F1 score | 64.00% | |||||||
| ResNet-50 | ResNet-50 slice-wise | |||||||
| Accuracy | 89.70% | |||||||
| Precision | 79.30% | |||||||
| Sensitivity | 81.70% | |||||||
| Specificity | 94.10% | |||||||
| F1 score | 81.30% | |||||||
| ResNet-50 subject-wise | ||||||||
| Accuracy | 81.40% | |||||||
| Precision | 81.50% | |||||||
| Sensitivity | 81.50% | |||||||
| Specificity | 81.50% | |||||||
| F1 score | 81.40% | |||||||
| DenseNet-161 | DenseNet-161 slice-wise | |||||||
| Accuracy | 90.50% | |||||||
| Precision | 79.90% | |||||||
| Sensitivity | 83.10% | |||||||
| Specificity | 94.80% | |||||||
| F1 score | 81.30% | |||||||
| DenseNet-161 subject-wise | ||||||||
| Accuracy | 83.80% | |||||||
| Precision | 84.10% | |||||||
| Sensitivity | 83.50% | |||||||
| Specificity | 83.50% | |||||||
| F1 score | 83.50% | |||||||
| 7 | Gutta et al. [21] | 2021 | Predict glioma grade | CNN | 237 | CNN trained with radiomic features alone | Accuracy CNN | 87% |
| GB | Accuracy GB | 64% | ||||||
| RF | Accuracy RF | 58% | ||||||
| SVM | Accuracy SVM | 56% | ||||||
| 8 | Latif et al. [22] | 2021 | Glioma tumor detection | CNN | 65 patients (26 LGGs and 39 HGGs) | MLP classifier | Accuracy | 98.50% |
| KNN classifier | Accuracy | 97.96% | ||||||
| SVM classifier | Accuracy | 90.04% | ||||||
| 9 | Lu et al. [23] | 2020 | Glioma classification | Modified ResNet50 | 193 cases | Modified ResNet | Accuracy | 80.11% |
| CNN | Accuracy | 75.43% | ||||||
| DenseNet | Accuracy | 67.55% | ||||||
| MLP | Accuracy | 63.58% | ||||||
| ResNet50 | Accuracy | 78.59% | ||||||
| 10 | Ahammed Muneer et al. [24] | 2019 | Glioma grade identification (LGG, oligodendroglioma, anaplastic glioma, and glioblastoma multiforme) | VGG-19 CNN | Wndchrm classifier | Accuracy | 92.86% | |
| VGG-19 | Accuracy | 98.25% | ||||||
| 11 | Mzoughi et al. [25] | 2020 | Glioma classification into LGG and HGG | 3D-CNN | 284 | 2D-CNN | Accuracy | 96.49% |
| 12 | Yang et al. [26] | 2018 | Glioma classification | CNN (GoogLeNet) | 113 patients (53 LGGs and 61 HGGs) | AlexNet | Validation accuracy | 0.866 |
| Test accuracy | 0.855 | |||||||
| Test AUC | 0.894 | |||||||
| GoogLeNet | Validation accuracy | 0.867 | ||||||
| Test accuracy | 0.909 | |||||||
| Test AUC | 0.939 | |||||||
| 13 | Khawaldeh et al. [27] | 2017 | Glioma classification | Modified AlexNet | 109 | ConvNet-3 | Accuracy | 85.71% |
| Modified AlexNet | Accuracy | 91.16% | ||||||
| 14 | Chang et al. [29] | 2018 | Predict IDH mutation status | Residual CNN (ResNet) with age incorporation | 496 | Residual CNN (ResNet) with age incorporation | Training accuracy | 87.30% |
| Validation accuracy | 87.60% | |||||||
| Testing accuracy | 89.10% | |||||||
| Residual CNN (ResNet) without age incorporation | Training accuracy | 82.80% | ||||||
| Validation accuracy | 83.00% | |||||||
| Testing accuracy | 85.70% | |||||||
| 15 | Chang et al. [30] | 2018 | Predict MGMT promoter methylation status, IDH1 mutation status, and 1p/19q codeletion status | CNN | 259 patients | Predict MGMT promoter methylation status | Accuracy | 83.00% |
| AUC | 0.81 | |||||||
| Predict IDH1 mutation status | Accuracy | 94.00% | ||||||
| AUC | 0.91 | |||||||
| 1p/19q codeletion status | Accuracy | 92.00% | ||||||
| AUC | 0.88 | |||||||
| 16 | Levner et al. [32] | 2009 | Predict MGMT promoter methylation status | 59 | 2D-DOST + ANN | Accuracy | 87.70% | |
| 17 | Korfiatis et al. [33] | 2017 | Predict MGMT promoter methylation status (no tumor, methylated MGMT, or non-methylated MGMT) | ResNet50 | 155 patients (66 methylated and 89 unmethylated tumors) | ResNet50 | Test accuracy | 94.40% |
| ResNet34 | Test accuracy | 80.72% | ||||||
| ResNet18 | Test accuracy | 76.75% | ||||||
| 18 | Ge et al. [34] | 2018 | Glioma classification (LGGs vs. HGGs) and 1p19q codeletion | Deep CNN | 285 | Deep CNN | Training accuracy | 91.93% |
| Validation accuracy | 93.25% | |||||||
| Test accuracy | 90.87% | |||||||
| Deep CNN | 159 | Deep CNN 1p19q codeletion | Training Accuracy | 97.11% | ||||
| Validation accuracy | 90.91% | |||||||
| Test accuracy | 89.39% | |||||||
| 19 | Rehman et al. [35] | 2020 | Classification of brain tumors (meningioma, glioma, and pituitary tumors) | CNN (fine-tuned VGG-16) | 233 | Fine-tuned GoogLeNet | Accuracy | 98.69% |
| Fine-tuned AlexNet | Accuracy | 97.39% | ||||||
| Fine-tuned VGG-16 | Accuracy | 98.04% | ||||||
| 20 | Matsui et al. [36] | 2020 | Prediction of LGG molecular subtype | DL model | 217 patients with LGG | Prediction of LGG molecular subtype | Accuracy training | 96.60% |
| Accuracy test | 68.70% | |||||||
DL Models Used to Predict the Isocitrate Dehydrogenase Mutation Status
The isocitrate dehydrogenase (IDH) mutation status is an important marker in glioma diagnosis, prognosis, and treatment. Generally, an invasive neurosurgical procedure is required to determine the IDH mutation status. Glioblastomas with an IDH mutation have a significantly improved survival compared with IDH wild-type gliomas [28]. Out of the 20 included studies, the authors of four [16,20,29,30] used different DL models in combination with MR images to predict the IDH mutation status. Nalawade et al. [20] used T2-weighted (T2w) MRI of 120 patients with high-grade gliomas (HGGs) and 140 patients with low-grade gliomas (LGGs) and compared them between three CNN models (Inception-v4, ResNet-50, and DenseNet-161). DenseNet-161 with five-fold cross-validation was found to be the best performing model with few preprocessing steps. It attained a mean slice-wise accuracy, sensitivity, and specificity of 90.5%, 83.1%, and 94.8%, respectively, and a subject-wise accuracy, sensitivity, and specificity of 83.8%, 83.5%, and 83.5%, respectively [20]. Bangalore et al. [16] used multiparametric brain MRI of 214 patients (94 IDH mutated and 120 IDH wild-type) in combination with voxelwise DL that uses either T2w image-only network (T2-Net) or multi-contrast (T2w, fluid-attenuated inversion recovery [FLAIR], and T1 postcontrast) network (TS-net). With minimal data preprocessing, T2-Net and TS-net achieved a mean cross-validation accuracy of 97.14% and 97.12%, respectively. Chang et al. [29] used residual CNN (ResNet) to predict non-invasively the IDH status of glioma from MR images of 496 patients. Incorporation of the age at diagnosis into the model increased the accuracy from 82.8% to 87.3% for the training set, from 83.0% to 87.6% for the validation set, and from 85.7% to 89.1% for the test set. Chang et al. [30] used a CNN model in combination with MRI in 259 patients with glioma to predict the IDH1 mutation status; the model achieved an accuracy of 94.0%.
DL Models Used to Predict the MGMT Promoter Methylation Status
The O6-methylguanine-DNA methyltransferase (MGMT) gene is associated with improved prognosis and a good response to treatment with temozolomide [31]. In three studies [30,32,33], the authors evaluated different DL models to predict the MGMT gene status using MRI. Levner et al. [32] performed a texture analysis of T2, FLAIR, and T1 postcontrast MR images based on two-dimensional discrete orthonormal Stockwell transform (2D-DOST) in combination with artificial neural networks (ANN) to predict the MGMT promoter methylation status in 59 newly diagnosed patients with glioblastoma. The 2D-DOST in conjunction with ANN achieved an accuracy of 87.7%, which is comparable to that of the invasive biopsy technique (approximately 90%). Korfiatis et al. [33] compared three deep CNN architectures (ResNet50, ResNet34, and ResNet18) to evaluate their ability to predict MGMT promoter methylation. ResNet50 performed significantly better than ResNet34 and ResNet18. The test accuracy for ResNet50, ResNet34, and ResNet18 was 94.40%, 80.72%, and 76.75%, respectively. Another study by Chang et al. [30] used a CNN model and MRI of 259 patients with glioma to predict the MGMT promoter methylation status. The CNN accuracy reached 83.0%.
DL Models Used to Predict Glioma Grade or Classification
In eight studies [19,21-27], the researchers reported accurate determination of glioma grade and classification using CNN compared with other techniques. Gutta et al. [21] proposed a deep CNN model in 237 patients to predict glioma grade and compared that to ML models trained by using standard radiomic features alone. The proposed deep CNN model demonstrated an accuracy of 87.0%, outperforming the ML models using radiomic features alone. The top-performing ML model trained with radiomic features alone was gradient boosting (GB), with an average accuracy of 64.0%. Similarly, Latif et al. [22] proposed a four-step CNN technique to classify brain MR images into tumorous and non-tumorous and tested them on 65 cases by using multilayer perceptron (MLP) and achieved an average accuracy of 98.77%. Lu et al. [23] used the CNN ResNet model based on the pyramid dilated convolution to classify gliomas using MRI. The classification accuracy for the modified ResNet was 80.11% compared with 63.58%, 75.43%, 67.55%, and 78.59% for MLP, CNN, DenseNet, and traditional ResNet, respectively. Ahammed Muneer et al. [24] performed automatic glioma grade identification with the weighted neighbor distance using the compound hierarchy of algorithms representing morphology (Wndchrm) classifier and compared that to a 19-layer visual geometry group (VGG-19) deep CNN. The Wndchrm classifier showed a maximum accuracy of 92.86% compared with 98.25% for the VGG-19.
Mzoughi et al. [25] proposed a fully automatic three-dimensional (3D) CNN architecture and volumetric MRI T1 postcontrast sequence to classify brain tumors into LGGs and HGGs. The 3D-CNN model achieved an accuracy of 96.49%. Yang et al. [26] compared the performance of two trained and fine-tuned CNN models (AlexNet and GoogLeNet) in 113 patients with glioma. GoogLeNet showed better performance than AlexNet whether it was trained from scratch or pre-trained models. The validation accuracy, test accuracy, and the test area under the curve (AUC) were 86.7%, 90.9%, and 93.9%, respectively, for GoogleNet compared with 86.6%, 85.5%, and 89.4%, respectively, for AlexNet. Zhuge et al. [19] made another comparison between two CNN models (3DConvNet and two-dimensional [2D] Mask R-CNN) to grade gliomas using conventional MRI. The results showed better performance for the 3DConvNet, with a test accuracy of 0.971 compared with the 2D Mask R-CNN accuracy of 0.963. Khawaldeh et al. [27] proposed a 12-layer CNN model in combination with axial FLAIR MR images. The modified AlexNet model demonstrated a greater accuracy of 91.16% compared with 85.71% for ConvNet. Ge et al. [34] used 2D CNN to assess glioma grading by using two datasets: the first for glioma classification into LGGs and HGGs and the second to identify gliomas with/without 1p19q codeletion. The proposed model showed a high test accuracy of 90.87% for glioma classification and 89.39% for 1p19q codeletion.
DL Models Used to Detect or Classify Gliomas, Meningiomas, and Pituitary Tumors
In two studies [17,35], the researchers evaluated the detection and classification of three types of brain tumors, namely, gliomas, meningiomas, and pituitary tumors, by using deep CNN. Diaz-Pernas et al. [17] presented a fully automatic segmentation and classification model using deep CNN with a multiscale approach and achieved a classification accuracy of 0.973. Rehman et al. [35] evaluated three CNN architectures with data augmentation techniques (AlexNet, GoogLeNet, and VGG-16). The VGG-16 fine-tuned architecture achieved the highest classification and detection accuracy of 98.69%.
DL Models Used for Segmentation of LGGs
MRI segmentation of LGGs is challenging because they rarely enhance after administration of gadolinium. In three studies [15,18,36], the authors investigated the segmentation of LGGs by using deep learning models. Akkus et al. [15] proposed a semi-automated segmentation process using only T2w and optionally postcontrast T1-weighted images and compared that to manual segmentation by three experts. Matsui et al. [36] developed a DL model that was able to predict the molecular subtypes of LGGs by using three different imaging modalities: positron emission tomography (PET), MRI, and computed tomography (CT). The performance of the model combining the three modalities had an accuracy of 96.6% for the training set and 68.7% for the test set. Naser and Deen [18] used T1-precontrast, FLAIR, and T1-postcontrast MR images to grade and segment LGGs. The tumor detection model achieved an accuracy of 0.92 while the grading model achieved an accuracy of 0.89.
Discussion
This systematic review summarizes the DL models used to classify and grade gliomas as well as the status of different molecular biomarkers. In three studies [16,20,29], the authors discussed the role of DNN in IDH1 mutation detection. In two studies [32,33], the authors discussed the MGMT promoter methylation mutation status with different DNN. Ge et al. [34] discussed 1p19q codeletion mutation along with glioma grading and Chang et al. [30] classified gliomas based on their genetic category (MGMT promoter methylation status, IDH1 mutation status, and 1p19q codeletion) using CNN architecture. Considering glioma classification, in several studies [19,21-27], the researchers discussed grading gliomas into low and high grades to adapt the treatment approach appropriately. The authors of two studies [17,35] evaluated the diagnostic accuracy of DNN through cross-validation. In three studies [15,18,36], the researchers specifically focused on LGG classification through CNN models to identify the most accurate and sensitive model for preoperative diagnosis.
In this review, the authors of six studies [16,19,26,27,33,35] compared different DNN to establish an accurate and effective model. Levner et al. [32] used L1-regularized NN and quantitative assessment of tumor texture to predict the MGMT promoter methylation status in 59 newly diagnosed patients with glioblastoma multiforme (GBM). Korfiatis et al. [33] tested three different residual CNN models. ResNet50 (accuracy 94.90%) outperformed ResNet34 (80.72%) and ResNet18 (76.75%). Deepak and Sarath [37] evaluated a DNN model (ResNet) in brain tumor classification using MRI and attained excellent processing with an accuracy of 98.3%. When Liu et al. [38] used the G-ResNet model (global average pooling residual network) to classify brain tumors using ResNet34, they attained 95% accuracy, which is significantly better than the previously used DNN models. Ghosal et al. [39] used the SE-ResNet-101 model to classify three brain tumors (glioma, meningioma, and pituitary tumors) without data augmentation and the proposed CNN attained an accuracy of 89.93%. In two studies [17,35], the authors examined multiple brain tumor classification by using deep CNN models. This approach produced a tumor classification accuracy of 97.3%, higher than the classic ML models [17]. Comparison between DNN (GoogLeNet, AlexNet, and VGG16) was performed and the fine-tuned VGG-16 demonstrated the highest accuracy of 98.69%. Ghosh et al. [40] used improved U-Net with VGG16 architecture cross-validated in patients from The Cancer Genome Atlas Low Grade Glioma (TCGA-LGG) dataset for tumor segmentation. The accuracy was 99.75% for the improved U-Net model, outperforming the basic U-Net model, which had an accuracy of 99.4%.
On the other hand, NN, as identified in other studies, utilizes energy to activate neurons. Only a small number of neurons are active throughout the thought process with the human brain, whereas the neurons that will be used in the future are temporarily unregulated until they are required. Single-task allocation for subsequent neurons reduces communication costs. It is anticipated that ANN will be developed in the future to help complete more multifaceted tasks.
DL approaches have a wider application in the clinical field. In this regard, use-cases of DL networks are employed for conducting medical diagnoses. As discussed previously, this process encompasses prediction, segmentation, classification, and detection. The findings of the reviewed studies show that DL methods can be dominant with respect to other high-performing algorithms. Thereby, it is safe to assume that DL will endure and continue to expand its offerings. The future progression of DL shows more potential in different fields of medicine, specifically in the realm of medical diagnosis. On the other hand, it is currently not clear whether DL can replace the role of clinicians or doctors in medical diagnosis. In this regard, DL can offer better support for professionals in the clinical field. All predictors show a broader aspect of AI and DL in different fields. Conventional approaches to different similarity measures are ineffectual compared with DL. Based on such outcomes, it has been recommended that DNN and DL will succeed, and they will be explored for a myriad of other uses in the near future.
AI could revolutionize all stages of the pathway by which patients with gliomas are managed: the postoperative acute phase; outpatient and oncological care preoperative screening, treatment planning, and diagnosis; and intraoperative tissue analysis and intraoperative workflow analysis. In addition, AI could change how national guidelines and policies are formed and help research into brain tumors as well as therapeutics. In this regard, AI could enhance clinical findings for patients in the future. Several obstacles exist for the development of AI in the field of brain tumors. The collaboration will be fundamental to developing clinically applicable AI as the field quickly diversifies. Such collaboration must emphasize the progression of databases and sources that might be utilized to train additional AI models.
This systematic review represents a simple, precise, and objective article that should contribute to the existing body of literature concerning the use of DL in neuro-oncology. The research outcomes of the included studies offer adequate information and insight into the applications of DL and AI to detect, classify, segment, and diagnose different impairments and diseases in certain anatomical realms of interest. The most important issue regarding ML and clinical medicine that should be taken into consideration is that most of the papers did not perform validation. They either developed models or performed cross-validation. According to the guidelines for developing and reporting ML predictive models in biomedical research, validation is necessary [41]. The application of AI and DL will continue to develop beyond the significant findings that have been shown in imaging gliomas. This may elevate the quality and efficiency of health care in the long term and, therefore, reduce the risk of late diagnosis of extreme diseases. On the other hand, there is still a long road before objective NNs are used widely in medical diagnosis. Finally, it is anticipated that AI will increase the combination of complex reasoning and representation learning in neuroradiological and neurosurgical practice [42,43].
Conclusions
ML and DL models incorporating MRI have been evaluated extensively. They have a significant value in improving the diagnostic and classification accuracy of brain tumors, especially gliomas, without the need for invasive methods. Most studies have presented validated results and can be used in clinical practice to improve patient care and prognosis. Open access to such algorithms is essential to support broader technological progression because ML algorithms have become more advanced. Clinical trials must follow reporting guidelines to ensure robust evidence is collected and to reduce biases as AI platforms associated with brain tumor surgery develop. There remain valid issues about the further implementation of machines in modern neurosurgery when AI promises to enhance patient management. Enhancements in patient findings might be challenged by job replacement, unique neglect, and physician deskilling. Clinician acceptability and stringent patient approval must be considered in the future to ensure that the potential of AI, ML, and DL does not lead to unidentified adverse outcomes.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
Footnotes
The authors have declared that no competing interests exist.
References
- 1.CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Ostrom QT, Gittleman H, Fulop J, et al. Neuro Oncol. 2015;17:0–62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genetics of glioblastoma: a window into its imaging and histopathologic variability. Belden CJ, Valdes PA, Ran C, et al. Radiographics. 2011;31:1717–1740. doi: 10.1148/rg.316115512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.High-grade and low-grade gliomas: differentiation by using perfusion MR imaging. Hakyemez B, Erdogan C, Ercan I, Ergin N, Uysal S, Atahan S. Clin Radiol. 2005;60:493–502. doi: 10.1016/j.crad.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Central Nervous System Cancers, version 3.2020, NCCN Clinical Practice Guidelines in Oncology. Nabors LB, Portnow J, Ahluwalia M, et al. J Natl Compr Canc Netw. 2020;18:1537–1570. doi: 10.6004/jnccn.2020.0052. [DOI] [PubMed] [Google Scholar]
- 5.Diagnostic values of DCE-MRI and DSC-MRI for differentiation between high-grade and low-grade gliomas: a comprehensive meta-analysis. Liang J, Liu D, Gao P, Zhang D, Chen H, Shi C, Luo L. Acad Radiol. 2018;25:338–348. doi: 10.1016/j.acra.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Li-Chun Hsieh K, Chen CY, Lo CM. Comput Biol Med. 2017;83:102–108. doi: 10.1016/j.compbiomed.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 7.MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Gutman DA, Cooper LA, Hwang SN, et al. Radiology. 2013;267:560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deep learning in medical imaging and radiation therapy. Sahiner B, Pezeshk A, Hadjiiski LM, et al. Med Phys. 2019;46:0–36. doi: 10.1002/mp.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Convolutional neural networks: an overview and application in radiology. Yamashita R, Nishio M, Do RK, Togashi K. Insights Imaging. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerging applications of artificial intelligence in neuro-oncology. Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S. Radiology. 2019;290:607–618. doi: 10.1148/radiol.2018181928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. Alzubaidi L, Zhang J, Humaidi AJ, et al. J Big Data. 2021;8:53. doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Applications of deep learning and reinforcement learning to biological data. Mahmud M, Kaiser MS, Hussain A, Vassanelli S. IEEE Trans Neural Netw Learn Syst. 2018;29:2063–2079. doi: 10.1109/TNNLS.2018.2790388. [DOI] [PubMed] [Google Scholar]
- 13.Updates on deep learning and glioma: use of convolutional neural networks to image glioma heterogeneity. Chow DS, Khatri D, Chang PD, Zlochower A, Boockvar JA, Filippi CG. Neuroimaging Clin N Am. 2020;30:493–503. doi: 10.1016/j.nic.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Moher D, Liberati A, Tetzlaff J, Altman DG. Ann Intern Med. 2009;151:264–269. [PMC free article] [PubMed] [Google Scholar]
- 15.Semi-automated segmentation of pre-operative low grade gliomas in magnetic resonance imaging. Akkus Z, Sedlar J, Coufalova L, et al. Cancer Imaging. 2015;15:12. doi: 10.1186/s40644-015-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas. Bangalore Yogananda CG, Shah BR, Vejdani-Jahromi M, et al. Neuro Oncol. 2020;22:402–411. doi: 10.1093/neuonc/noz199. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Díaz-Pernas FJ, Martínez-Zarzuela M, Antón-Rodríguez M, González-Ortega D. Healthcare (Basel) 2021;9:153. doi: 10.3390/healthcare9020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Naser MA, Deen MJ. Comput Biol Med. 2020;121:103758. doi: 10.1016/j.compbiomed.2020.103758. [DOI] [PubMed] [Google Scholar]
- 19.Automated glioma grading on conventional MRI images using deep convolutional neural networks. Zhuge Y, Ning H, Mathen P, Cheng JY, Krauze AV, Camphausen K, Miller RW. Med Phys. 2020;47:3044–3053. doi: 10.1002/mp.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Classification of brain tumor isocitrate dehydrogenase status using MRI and deep learning. Nalawade S, Murugesan GK, Vejdani-Jahromi M, et al. J Med Imaging (Bellingham) 2019;6:46003. doi: 10.1117/1.JMI.6.4.046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Improved glioma grading using deep convolutional neural networks. Gutta S, Acharya J, Shiroishi MS, Hwang D, Nayak KS. AJNR Am J Neuroradiol. 2021;42:233–239. doi: 10.3174/ajnr.A6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brain MR image classification for glioma tumor detection using deep convolutional neural network features. Latif G, Iskandar DN, Alghazo J, Butt MM. Curr Med Imaging. 2021;17:56–63. doi: 10.2174/1573405616666200311122429. [DOI] [PubMed] [Google Scholar]
- 23.The classification of gliomas based on a pyramid dilated convolution resnet model. Lu Z, Bai Y, Chen Y, et al. https://doi.org/10.1016/j.patrec.2020.03.007 Pattern Recognit Lett. 2020;133:173–179. [Google Scholar]
- 24.Glioma tumor grade identification using artificial intelligent techniques. Ahammed Muneer KV, Rajendran VR, K PJ. J Med Syst. 2019;43:113. doi: 10.1007/s10916-019-1228-2. [DOI] [PubMed] [Google Scholar]
- 25.Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. Mzoughi H, Njeh I, Wali A, Slima MB, BenHamida A, Mhiri C, Mahfoudhe KB. J Digit Imaging. 2020;33:903–915. doi: 10.1007/s10278-020-00347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glioma grading on conventional MR images: a deep learning study with transfer learning. Yang Y, Yan LF, Zhang X, et al. Front Neurosci. 2018;12:804. doi: 10.3389/fnins.2018.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noninvasive grading of glioma tumor using magnetic resonance imaging with convolutional neural networks. Khawaldeh S, Pervaiz U, Rafiq A, Alkhawaldeh RS. Appl Sci. 2018;8:27. [Google Scholar]
- 28.IDH1 and IDH2 mutations in gliomas. Yan H, Parsons DW, Jin G, et al. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Chang K, Bai HX, Zhou H, et al. Clin Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Chang P, Grinband J, Weinberg BD, et al. AJNR Am J Neuroradiol. 2018;39:1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Rivera AL, Pelloski CE, Gilbert MR, et al. Neuro Oncol. 2010;12:116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Predicting MGMT methylation status of glioblastomas from MRI texture. Levner I, Drabycz S, Roldan G, De Robles P, Cairncross JG, Mitchell R. Med Image Comput Comput Assist Interv. 2009;12:522–530. doi: 10.1007/978-3-642-04271-3_64. [DOI] [PubMed] [Google Scholar]
- 33.Residual deep convolutional neural network predicts MGMT methylation status. Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. J Digit Imaging. 2017;30:622–628. doi: 10.1007/s10278-017-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks. Ge C, Gu IY, Jakola AS, Yang J. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:5894–5897. doi: 10.1109/EMBC.2018.8513556. [DOI] [PubMed] [Google Scholar]
- 35.A deep learning-based framework for automatic brain tumors classification using transfer learning. Rehman A, Naz S, Razzak MI, Akram F, Imran M. Circuits Syst Signal Process. 2020;39:757–775. [Google Scholar]
- 36.Prediction of lower-grade glioma molecular subtypes using deep learning. Matsui Y, Maruyama T, Nitta M, et al. J Neurooncol. 2020;146:321–327. doi: 10.1007/s11060-019-03376-9. [DOI] [PubMed] [Google Scholar]
- 37.Multi-class brain cancer classification using deep learning convolutional neural network. Deepak VK, Sarath R. https://archives.palarch.nl/index.php/jae/article/view/2697 PalArch J Archaeol Egypt/Egyptol. 2020;17:5341–5360. [Google Scholar]
- 38.Liu D, Liu Y, Dong L. Neural Information Processing. ICONIP 2019. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2019. G-ResNet: improved ResNet for brain tumor classification; pp. 535–545. [Google Scholar]
- 39.Ghosal P, Nandanwar L, Kanchan S, Bhadra A, Chakraborty J, Nandi D. Second International Conference on Advanced Computational and Communication Paradigms (ICACCP) Piscataway, New Jersey: IEEE; 2019. Brain tumor classification using ResNet-101 based squeeze and excitation deep neural network; pp. 1–6. [Google Scholar]
- 40.Improved U-Net architecture with VGG-16 for brain tumor segmentation. Ghosh S, Chaki A, Santosh KC. Phys Eng Sci Med. 2021;44:703–712. doi: 10.1007/s13246-021-01019-w. [DOI] [PubMed] [Google Scholar]
- 41.Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. Luo W, Phung D, Tran T, et al. J Med Internet Res. 2016;18:0. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postsurgical functional outcome prediction model using deep learning framework (Prediction One, Sony Network Communications Inc.) for hypertensive intracerebral hemorrhage. Katsuki M, Kakizawa Y, Nishikawa A, Yamamoto Y, Uchiyama T. Surg Neurol Int. 2021;12:203. doi: 10.25259/SNI_222_2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preliminary development of a prediction model for daily stroke occurrences based on meteorological and calendar information using deep learning framework (Prediction One; Sony Network Communications Inc., Japan) Katsuki M, Narita N, Ishida N, et al. Surg Neurol Int. 2021;12:31. doi: 10.25259/SNI_774_2020. [DOI] [PMC free article] [PubMed] [Google Scholar]