Graphical abstract
Keywords: Aryl hydrocarbon receptor, PAS proteins, Environmental sensors
Abstract
Proteins containing PER-ARNT-SIM (PAS) domains are commonly associated with environmental adaptation in a variety of organisms. The PAS domain is found in proteins throughout Archaea, Bacteria, and Eukarya and often binds small-molecules, supports protein-protein interactions, and transduces input signals to mediate an adaptive physiological response. Signaling events mediated by PAS sensors can occur through induced phosphorelays or genomic events that are often dependent upon PAS domain interactions. In this perspective, we briefly discuss the diversity of PAS domain containing proteins, with particular emphasis on the prototype member, the aryl hydrocarbon receptor (AHR). This ligand-activated transcription factor acts as a sensor of the chemical environment in humans and many chordates. We conclude with the idea that since mammalian PAS proteins often act through PAS-PAS dimers, undocumented interactions of this type may link biological processes that we currently think of as independent. To support this idea, we present a framework to guide future experiments aimed at fully elucidating the spectrum of PAS-PAS interactions with an eye towards understanding how they might influence environmental sensing in human and wildlife populations.
1. Introduction
The Ah receptor (AHR) is a prototype PER-ARNT-SIM (PAS) domain containing protein that is well known for its role in the adaptive metabolism and physiological consequences of a variety of structurally related trace extended aromatic compounds (TEACOPS) and halogenated-dibenzo-p-dioxins (“dioxins”). In this perspective, we provide an overview of AHR signal transduction, presenting this protein as a prototype small molecule sensor with roles in both environmental adaptation and normal physiology. We begin with a brief discussion of the larger family of PAS domain proteins to emphasize the concept that these proteins display similar functions in a variety of microorganisms, animals, and plants. We then move to a description of AHR signaling as a representation of the idea that PAS domains display multiple characteristics; as a site for homotypic dimerization between two PAS proteins, as a site for heterotypic interactions with distinct output proteins and cellular chaperones, and as a site for small molecule binding. We then provide evidence for the idea that the AHR senses both environmental and endogenous signals and then close this perspective with the possibility that additional PAS dimers and higher order PAS-PAS interactions are still undiscovered and that these interactions may explain the pleiotropy of many environmental sensing pathways.
2. The PAS domain
2.1. PAS diversity
Organisms require mechanisms to adapt to environmental change. The PAS, steroid/nuclear receptor, LacI, Gal, and MarR protein families are examples of ligand-responsive sensors that play essential roles in metabolic adaptation [[1], [2], [3], [4], [5], [6], [7]]. In these systems, sensor proteins often recognize an environmental or metabolic stimulus and initiate signal transduction events to induce physiological change, often through alterations in gene expression. In many organisms, this is occurs through an environmental signal that induces a conformational change and/or post-translational modification of the sensor protein. In turn, this activated sensor influences gene expression through a variety of mechanisms, including increased concentration of the sensor protein at genomic regulatory sites or through phosphorelays that influence the levels of downstream transcription factors at genomic elements. Through such pathways, sensing systems can orchestrate the expression of multiple genomic targets involved in environmental adaptation.
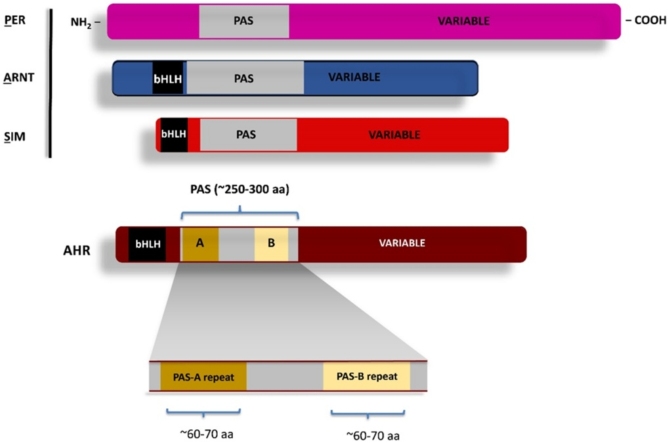
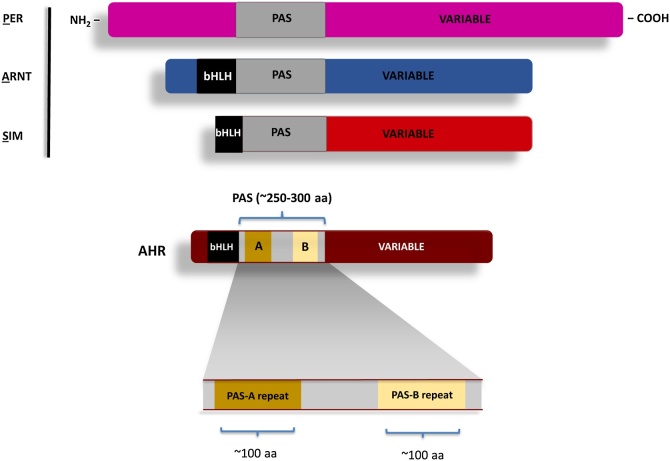
The PAS domain was first defined based upon sequence alignments of the fruit fly Per and Sim, as well as the human ARNT gene products, with its name arising from the acronym of the first letter of each founding member (PER-ARNT-SIM) [[8], [9], [10]]. This initial definition of the PAS domain encompassed approximately 250–300 amino acids and harbored two degenerate internal repeats, known as PAS-A and PAS-B (Fig. 1). In more recent publications, these internal PAS-A, PAS-B repeats of approximately one hundred amino acids in length each are now also commonly referred to as “PAS domains.” By either definition, the PAS domain is found in Archaea, Bacteria and Eukarya, commonly in tandem repeats, and often encoded within a protein that plays a role in the adaptive responses to environmental change [5]. In plants, the PAS domain is often referred to as “Light, Oxygen, Voltage” (LOV) domain. In part, this LOV nomenclature has arisen form the observation that in some plants, the PAS/LOV domain has been shown to bind flavin and act as a photosensor to mediate blue light-induced phototropism [5,11].
Fig. 1.
Two definitions of the PAS Domain: A: Representation of the founding PER-ARNT-SIM proteins and the boundary of the larger ∼250-300 amino acid domain as originally described [64,65,182]. Schematic maps of the PER-ARNT-SIM proteins with the bHLH domains in black. B: Representation of a generic PAS protein with the PAS domain represented as repeats, PAS-A and PAS-B of approximately 100 amino acids each.
2.2. PAS domain functions
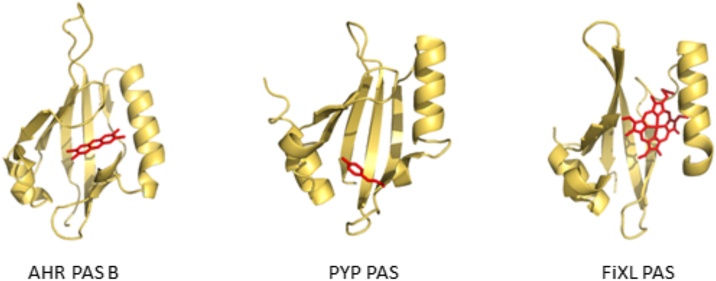
The PAS domain is encoded in sensor proteins found in all kingdoms of life. This domain functions through multiple mechanisms, as a site for small molecule binding and as a site for homotypic (i.e., PAS-PAS) or heterotypic (PAS-other) protein-protein interactions. One highly conserved role of a PAS domain is as a site for small molecule binding and sensory function (Fig. 2) [5,12]. In microbial systems, the PAS domain is found in sensor proteins such as the photoactive yellow protein (PYP) and FixL [[13], [14], [15], [16]]. In the PYP protein, the PAS domain covalently binds 4-hydroxycinnamic acid as a chromophore, while in FixL the PAS domain holds heme as part of an oxygen-binding site. In these two examples, the PAS domain directly functions as a sensor for either light or oxygen to regulate phototropism or nitrogen fixation, respectively. Importantly, modelling of the PAS-B domain of the AHR suggests this domain binds its prototype ligand, 2,3,7,8-tertrachlorodibenzo-p-dioxin within this same PAS repeat fold (Fig. 2) [[17], [18], [19]].
Fig. 2.
Structure of selected PAS repeat domains. Left, predicted structure of the AHR with its ligand 2,3,7,8-tetrachlordibenzo-p-dioxin [17]. Center, photoactive yellow protein (PYP) with its covalently bound chromophore, 4-hydroxycinnamic acid [186]. Right, the FixL with liganded Heme [187]. Models are presented using PyMOL software version 2.3.4 (Schrodinger, Inc, New York).
In addition to binding chromophores, PAS domains can also serve as a protein-protein interaction surface. One example of a homotypic interaction (where two PAS domains interact) is the PAS transcription factor Aureochrome 1a from Phaeodactylum tricornatum (PtAu1a), where the PAS domains of two PtAu1a proteins homodimerize in the presence of light [20]. This dimerization induces a conformational change that enhances affinity of a linked DNA-binding domain for genomic elements regulating targeted gene expression. The result is a light induced transcriptional response system that uses PAS to both bind the chromophore and support dimerization between protein partners.
Another example of interactions of PAS domains from different proteins, comes from the fungal White Collar-1 (WC-1) and White Collar-2 (WC-2) proteins that form the “White Collar Complex” (WCC) [21]. The WC-1 protein binds a flavin within its PAS domain. Absorption of blue light by this chromophore triggers a conformational change in the structure of the WCC and induces novel DNA binding characteristics [21,22]. Another PAS protein, “Vivid” (VVD), also participates in this light sensing/circadian adaptation pathway [23]. In the dark VVD exists as a homodimer with each monomer binding a flavin. When VVD absorbs light, a conformational change exposes the PAS domain of VVD at the same time as the PAS domain of WC-1 is opened up [24]. This transient unpairing of dimers allows a new dimerization in which VVD replaces WC-2 as a partner of WC-1, creating a transcriptionally inert complex [25]. Through this feedback mechanism the amount of active WCC and VVD modulates the light response allowing the organism to respond to incremental levels of light rather than only light versus dark [26].
Interactions of PAS with different domains (i.e., PAS-nonPAS) also serve to influence signal transduction in response to environmental stimuli. Examples of such heterotypic interactions include; 1) The interaction of the PASB domain of the human NCOA1 transcriptional coactivator with the LXXLL motif within the STAT6 transcription factor and 2) The interaction of the PASB domain of the mammalian CLOCK protein with its CRY regulator [27,28]. There are several important examples of heterotypic interactions of PAS domains occur intramolecularly in sensor biology. One example of such an interaction is the PAS domain of the light activated transcription factor El222 and its covalently linked DNA binding motif [29].
3. The ARYL hydrocarbon receptor as a model mammalian PAS sensor
3.1. History
The adaptive metabolism of polycyclic aromatic hydrocarbons (PAHs) has long been an area of scientific investigation due to the carcinogenicity of many congeners and their widespread dispersion in the environment [30]. Sources of PAHs from human activity include combustion of wood and fossil fuels for energy and heat, tobacco smoking, use of coal tar sealants and asphalt, coal liquefying plants, coke and aluminum production, barbecuing, and smoking or charring of food over fire. Natural emissions of PAHs are also significant and include, wildfires, petroleum seepage, coal deposits, and volcanic activities [31,32]. Similarly, structurally related halogenated-dioxins, -dibenzofurans and biphenyls are also environmental contaminants that commonly arise from human activity and sometimes natural sources [33,34]. Thus, organisms have been in contact with environmentally ubiquitous TEACOPS for millions of years, with the use of fire and human industrial activity only serving to increase exposure of certain populations in recent millennia.
Given their widespread occurrence, toxicity and carcinogenicity, PAHs like benzo[a]pyrene (BAP) served as early prototypes in studies of xenobiotic metabolism, bioactivation and detoxification. This research revealed that a collection of cytochromes-P450 dependent monooxygenases (P450 s) named “aryl hydrocarbon hydroxylase” (AHH), had a major influence on the biological half-lives of PAHs in many species [35,36]. Approximately fifty years ago, it was observed that this PAH metabolic system, was inducible by its substrates [37]. Due to parallels with the LacI system, this “appearance” of an AHH enzymatic activity in the presence of its substrate PAHs, was commonly referred to as “induction” [38,39]. Reports of inducibility were followed by the proposition that a “binding species” or a “receptor” for PAHs mediates the upregulation of AHH activity and led to the introduction of the idea that this system may have arisen as a protection against the toxicity of PAHs, as well as structurally related phytoalexins, and TEACOPS in the environment [33,[40], [41], [42], [43]].
The study of a genetic polymorphism in mice that influenced a strain’s inducibility upon response to PAHs led to the identification of the Ah locus and the designation that strains were either “responsive” (prototype strain C57BL/6 or B6) or “nonresponsive” (prototype strain DBA/2 or D2) [44,45]. Genetic studies identified a single autosomal locus as primarily responsible for this differential responsiveness, giving rise to the nomenclature: Ahb (b from B6) to define the responsive allele and Ahd (d from D2) to define the nonresponsive allele. More recent genomic nomenclature alters this locus designation to Ahr instead of Ah and it is this terminology we will use through the remainder of this review (i.e., Ahrb vs Ahb).
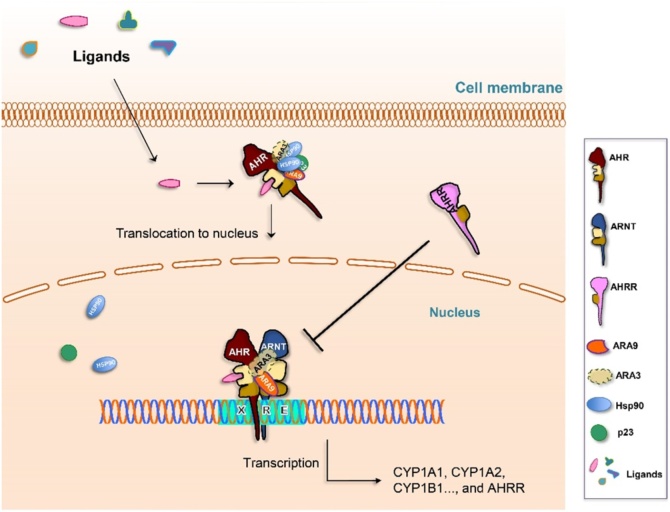
Our understanding of metabolic adaptation to PAHs was aided by the development of radioligands of the AHR [42,46]. These reagents, derived from the radiolabelling of PAHs or the more potent dioxin structure, led to the discovery of a binding protein or “receptor” within target cells and tissues [47]. These studies also revealed that “nonresponsive” Ahrd mice are more appropriately described as “hyporesponsive” as they mount an inductive response, although much less robustly, than the Ahrb strains. Biochemical characterization of this binding species revealed that it was a soluble protein associated with chaperones such as the 90 kDa heat shock protein (HSP90), with later studies adding the smaller co-chaperones such as ARA9 (aka XAP2 or AIP), p23, and possibly ARA3 (aka NS1BP) to this complex (Fig. 3, Fig. 4, see below) [[48], [49], [50], [51], [52], [53], [54], [55]].
Fig. 3.
Signal Transduction by the AHR: See text for details.
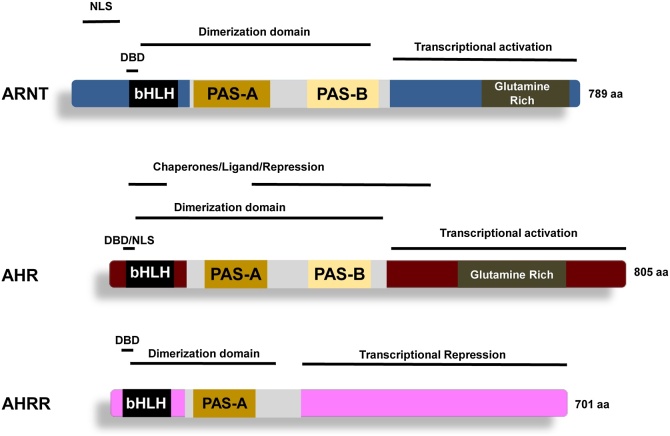
Fig. 4.
Domain map of the AHR and AHRR: Functional domain maps of the ARNT, AHR and AHRR proteins. Not all mapped functional domains are depicted. Domain regions are approximate and are derived from a number of representative biochemical studies. See text for details [74,77,78,80,108,[188], [189], [190]].
Radioligands also led to the development of competitive binding assays that allowed structure-activity studies to explain agonist potency. These studies provided evidence that dioxin ligands that bind with higher affinity to the receptor site are more potent inducers of the target P450 s [56]. The subsequent observation that this binding affinity segregated with the Ahrd and Ahrb polymorphism in mice provided the final formal evidence for the existence of an AHR [57].
The cloning of the P450 encoding genes that comprised AHH activity, i.e., CYP1A1, CYP1A2, and possibly CYP1B1, as well as the identification of the genomic cis-elements that controlled their expression in response to PAH exposure, led to the discovery of the regulatory enhancers linked to the induction phenomenon [36,[58], [59], [60]]. Specifically, early experiments demonstrated that the consensus sequence, GCGTG (sometimes also defined as TNGCGTG), was bound by a heterodimeric complex and controls the upregulation genes like CYP1A1 in response to PAH and dioxin ligands [59,[61], [62], [63]]. This genomic enhancer element goes by many names: xenobiotic response element (XRE, which we will use here), dioxin response element (DRE), and Ah response element (AHRE).
3.2. Discovery of mammalian PAS proteins
A watershed moment in our understanding of adaptive metabolism, and PAS proteins writ large, arose from a somatic-cell genetics approach aimed at identification of gene products essential for the ligand-induced, AHR-dependent, induction of CYP1A1 in cell culture. This work resulted in the molecular cloning of a molecule called ARNT (“aryl hydrocarbon receptor nuclear targeter”) that was essential for the AHR to gain a high affinity for the nuclear compartment and chromatin upon ligand binding [8]. Sequence analysis of ARNT provided two important observations. First, as noted above, the ARNT protein displays homology with the Sim and Per gene products of the fruit fly (D. melanogaster) allowing the initial definition of the PAS homology domains (Fig. 1) [64,65]. Second, homology searches revealed ARNT (as well as SIM, but not PER) harbor a bHLH domain immediately N-terminal to PAS. Such bHLH domains had been previously observed in partners of dimeric transcription factors such as MyoD and Myc [66]. This concept led to the proof that ARNT was a part of a protein complex with that directly bound enhancer elements within the genome [62,67,68].
The use of the photoaffinity ligand, 2-azido-3-iodo-7,8-dibromodibenzo-p-dioxin allowed the purification of the AHR from B6 cytosol, as well as its primary amino acid sequence and antibodies. In turn, this led to the ultimate cloning of the corresponding cDNA and structural gene [46,67,[69], [70], [71], [72]]. Of central importance was that the cDNA encoded the second mammalian member of the PAS family and harbored an adjacent bHLH domain (Fig. 4). Taken in sum, these observations suggested a dimeric partnership between the AHR and ARNT lead to the signal transduction model described in Fig. 3 and discussed more below. If we add the AHR as an additional founding member in chordates, then of the original four PAS proteins described, all harbor two PAS repeats (denoted PAS-A and PAS-B), three out of four (SIM, ARNT and AHR) harbor a bHLH domain immediately N-terminal to the PAS repeats, yet, the C-terminal halves display much lower sequence homology (Fig. 4) [65,73].
4. Molecular biology of the aryl hydrocarbon receptor
4.1. Protein-protein interactions
The observation that two bHLH-PAS proteins, the AHR and ARNT, are essential for CYP1A1 induction led to the demonstration that signaling is dependent upon an AHR-ARNT heterodimer with specificity for the XRE sequence [62,68,74]. With this concept in hand, functional domain maps of the AHR and ARNT were rapidly developed (Fig. 4) [[74], [75], [76], [77], [78], [79], [80]]. Based upon the recognition that bHLH-domains commonly act in dimeric pairs of transcription factors to position basic-alpha helices within the major groove of enhancer DNA, it was quickly demonstrated that the basic region helix found in the bHLH of both the AHR and ARNT would be required for binding to the XRE [67,74,77,[80], [81], [82]] (Fig. 3, Fig. 4).
The PAS-A domain in both the AHR and ARNT, was found to play an essential role in signaling. Molecular and crystallographic studies support the idea that cooperation between the HLH and PAS-A domains is the primary support for dimerization between these two proteins. In turn, these interactions position the basic alpha helix within the major groove of DNA for sequence specific contacts at XREs. Thus, a clear role for this domain is as a PAS-PAS dimerization surface, contributing to AHR-ARNT interaction and formation of a competent transcriptional complex [74,77,[83], [84], [85]].
While some data exists to support the idea that the PAS-B domain is important in AHR-ARNT dimerization, a greater body of evidence documents its role in heterotypic interactions by the AHR. In support of a role for such protein-protein interactions is the early observation that this region of the AHR represses AHR-activity [74,86,87], and the parallel mapping studies demonstrating Hsp90 binds to this same repressing region of PAS-B [77,79,88]. A common interpretation of this data is that a chaperone complex (possibly a dimer of Hsp90, and the cochaperones ARA9, p23 and perhaps ARA3) is holding the PAS-B domain in a conformation that can accept ligand and prevent dimerization with ARNT or inappropriate contacts with other proteins in the cytosol that might lead to its aggregation and inactivation [51,53,[89], [90], [91]]. Once ligand binds, conformational changes are induced which weakens or reorganizes the chaperone complex, simultaneously revealing, nuclear localization and possibly ARNT-dimerization motifs within the AHR.
4.2. Ligand binding
Despite extensive information available regarding the pharmacology of AHR ligands, there remains a gap in our understanding of the structure of the AHR ligand-binding domain (LBD) due to the lack of crystal structure for its PAS-B. The LBD of the AHR was initially mapped by covalently binding an [[125I]-labeled photoaffinity ligand followed by CNBr cleavage and micro-sequencing which revealed the labeled peptide fragment was coincident with PAS-B [67]. This overlap of the LBD with the PAS B domain was supported by a number of subsequent molecular studies [74,77,92]. More recent mutagenesis, and homology model-derived structures, also indicate PAS-B as being important for ligand binding and provide a preliminary view of this PAS-B-ligand-bound structure and provide evidence that a V375A polymorphism within this region explains much of the variability in ligand responsiveness observed across the mouse Ahrb and Ahrd mouse strains (Fig. 2) [[17], [18], [19],[93], [94], [95], [96]].
While the N-terminal halves of the AHR and ARNT appear to play central roles in chaperone interactions, dimerization, DNA binding and ligand induced transformation, the C-terminal halves of these factors appear to harbor regions that influence expression of genomic targets once the complex is bound to chromatin [77,78,86,88,97]. In this regard, multiple subdomains in the C-terminus of the AHR are often reported, with the idea that each confers weak transactivation potency alone, but act synergistically. One possible consequence of this multiplicity is that it may enable the AHR to interact with a variety of transcription factors and activate transcription from a variety of promoters [61,86].
4.3. Variation and polymorphism
Molecular examination of the AHR open reading frame from the human, mouse and rat AHR explains the receptor molecular weight variation observed both within and across species. In large part, this size difference, as documented by western blot analysis and photoaffinity labelling of receptors, is due to altered termination codon usage in the final exon (exon 11) of the structural gene [43,71]. While still a debated concept, the marked difference in molecular weight of the AHR observed across species was initially thought of as evidence that AHR structure may be more evolutionarily responsive to the environmental niche of a given species, as compared to genes encoding receptors for endogenously generated hormones [43,98]. While such a model is difficult to prove, it is an intriguing proposition, with early evolutionary analyses providing some support and arguments against this idea [40,99].
The observation that Ahr polymorphisms in rodent models (e.g., the V375A polymorphism in mice, described above, or the splice junction polymorphism observed in rats, described in detail elsewhere [100]) can lead to alterations in ligand response, spurred considerable interest in whether polymorphisms leading to hyper- or hypomorphic Ahr alleles might be common in human populations [101,102]. While investigations of SNPs (single nucleotide polymorphisms) can harbor significant biases reflecting the geography or population of focus, interrogation of the public database, dbSNP [103], indicates that common SNPs (defined here as greater than 1% in a population) appear in humans at frequencies similar to loci encoding most other nuclear receptors [101].
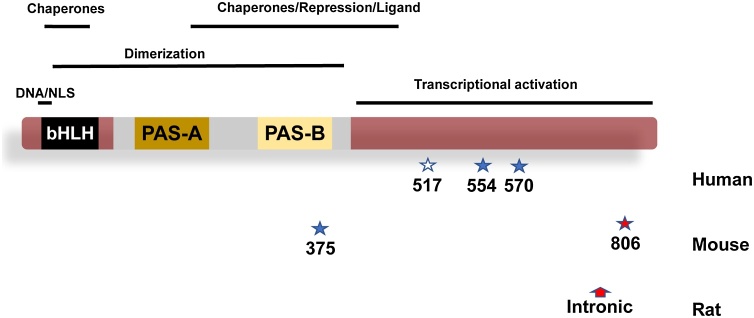
Of the recorded SNPs resulting in nonsynonymous (missense) alterations within the Ahr gene, only a few, e.g., P517S, R554 K and V570I, are commonly reported in various human populations, while nonsynonymous and intronic SNPS are much more common [[104], [105], [106]]. In one study of a Japanese population, a number of novel rare genetic variants were detected (K17 T, K401R, N487D, and I514T) in addition to the more common R554 K allele [107]. Although this study did not include a functional analysis, it provides one example of the diversity of rare, potentially important, missense variants in unique populations. In a second study, haplotypes of the three more common SNPs noted above were examined for their functional impact on AHR signaling in vitro [104]. Interestingly, the initial studies indicated that haplotypes corresponding to I570 and K554 yield a hypomorphic receptor, perhaps due to an attenuated transactivation domain or decreased stability in vivo. In contrast to the missense SNPs, known synonymous SNPs and SNPs within introns and other noncoding regions, number over a hundred in human poulations. While less obvious, nonsynonymous and intronic SNPs have the potential to influence AHR signaling through impacts on splicing, mRNA stability or even codon usage. While the importance of intronic SNPS leading to hypomorphic Ahr alleles in the rat is well documented, a parallel in humans has not yet been reported [100]. A summary of functionally important polymorphisms in the AHR open reading frames across species in presented in Fig. 5.
Fig. 5.
Important Polymorphisms in the AHR open reading frame: Top: Generalized domain identifiers for the AHR as mapped by multiple laboratories (see text). Middle: Structure of a generic AHR open reading frame from human. Bottom: Known results of common nonsynonymous SNPs of potential functional importance that found in human mouse and rat. Dark blue stars denote nonsynonymous SNPS. Open star represents a more rare nonsynonymous SNP that has been studied in vitro. Red star represents a stop codon (nonsense) that results in alterations in receptor molecular weight in commonly used mouse strains. Red arrow represents the position of a splice site variant in the rat intron between exons 10 and 11, resulting in multiple splice variants and three distinct molecular sizes of AHR in rat models.
5. Aryl hydrocarbon receptor pathways
5.1. The AHR adaptive pathway
It is now possible to describe the classical pathway for the adaptive metabolism of PAHs in humans and most mammals (Fig. 3). Upon entry of PAHs into cells, these ligands bind to the AHR in the cytosol inducing a conformational change in the AHR that loosens associations with its chaperone complex and exposes a nuclear localization sequence (NLS). The ligand-activated receptor complex then translocates into the nucleus, where the AHR sheds or rearranges its cellular chaperones and binds to the constitutively nuclear ARNT proteins through PASA, PASB and HLH domains. The dimerization of the AHR-ARNT heterodimer positions the basic alpha helices to recognize the XREs within the major groove of DNA and this is associated with the recruitment of coactivators and chromatin rearrangements, leading to the transcriptional activation of target genes, such as CYP1A1, CYP1A2, and CYP1B1.
The classic adaptive pathway depicted in Fig. 3 is dependent upon a variety of homotypic and heterotypic protein interactions (e.g., PAS-PAS and PAS-chaperone, respectively). Yet, a considerable body of evidence suggests that the role of protein interactions in AHR biology is more complex, and outputs extend beyond genes encoding xenobiotic metabolizing enzymes as the mechanism depicts in Fig. 3 [108]. While we commonly associate the adaptive pathway with the upregulation of target genes through the dimeric AHR-ARNT binding to XREs, a large body of evidence indicates that the AHR is also involved in the up- and down-regulation of a variety of additional genomic targets, most of which are not typically associated with xenobiotic metabolism [[109], [110], [111]]. Importantly, the mechanisms underlying these “nonclassical’ signaling pathways appears to occur by mechanisms that are distinct from that depicted in Fig. 3. While not entirely elucidated, such mechanisms commonly employ heterotypic protein interactions that link the AHR to other cellular signaling molecules, some examples include the estrogen receptor, E2F and RelA [[112], [113], [114], [115]].
The adaptive pathway described above, appears to be suppressed through a number of mechanisms [116,117]. While there are numerous reported mechanisms, the one most relevant to this perspective on PAS proteins is mediated by the “AHR-Repressor” or AHRR [118]. This protein, which has been shown to dimerizes with ARNT, is a close structural homolog of the AHR but is missing a region corresponding to part of its PAS-B repeat (Fig. 5). One idea is that the AHRR competes for dimerization with ARNT and thereby competes with AHR-ARNT for XRE occupancy [118]. Although not completely understood, the AHRR appears to provide feedback inhibition through three potential mechanisms: competition for ARNT, competition for the XRE, and transcriptional repression of target genes upon XRE binding through direct repression on their promoters [[116], [117], [118], [119]].
5.2. The AHR cognate pathway
While the AHR does influence metabolic adaptation in response to environmental chemicals, this may not be the sole evolutionary driver for its conservation in animal species. In support of an additional “cognate pathway,” observations from animal models null for AHR expression describe a variety phenotypes [[120], [121], [122], [123], [124], [125]]. While early reports of AHR-null phenotypes were subtle and often differed across laboratories and species, a limited list includes alterations in peripheral lymphocyte populations, alterations in gastrointestinal immunity, a patent ductus venosus (DV), reduced litter sizes, and age-dependent cardiac hypertrophy [124,[126], [127], [128], [129]]. Given that the immunological aspects of AHR physiology have garnered considerable recent attention, the reader is referred to some excellent reviews for more insight [130,131].
A variety of studies in recombinant mouse models support the idea that the cognate signaling pathways of the AHR have similarities to the adaptive pathway described above. A few experimental observations in support of this idea include the observation that mutant alleles of two proteins important in the adaptive pathway, the ARNT and ARA9, are also required for normal DV closure [132,133]. Moreover, knock-in mutations at the Ahr locus that ablate the XRE binding by the AHR-ARNT dimer disrupt many aspects of the cognate pathway such as DV closure and lymphocyte development [134,135]. Importantly, the target genes of the cognate pathway are still unclear and may or may not be distinct form the adaptive pathways. In this regard, conditional null alleles indicate that AHR signaling to regulate DV closure or barrier immunity are occurring in cellular compartments not traditionally associated with the adaptive response [131,136,137].
6. Ligands
6.1. Endogenous and cognate ligands of the AHR
While initial attention to the AHR arose from an interest in understanding the response to xenobiotics, there has long been a search for endogenous or cognate ligands. Evidence that AHR-null animal models display such a wide variety of phenotypes gives import to such efforts but does not rule out the possibility that the AHR acts in a ligand-independent manner [128]. Along this line of thought, it is possible that the binding of xenobiotic ligands is an independent role for the AHR, selected for independently throughout evolution. For this and future discussion of this topic, we propose two criteria to define a “cognate ligand.” The first is that the ligand can be found in animal tissues naturally and in an evolutionarily consistent manner. The second is that the ligand must be shown to be functionally linked with the receptor for some essential biological process. To date, we have many interesting ligands that appear to meet the first criteria. Where we have less data is in the proof of a link to a biological function. While, to date, we have no physiologically proven cognate ligand, a variety have been reported and summarized in a number of excellent reviews on the topic [127,131,[138], [139], [140], [141]].
6.2. Proligands
In accordance with numerous structure-activity studies performed over the last decades, an early heuristic to define AHR ligands is that they must fit a planar binding pocket with dimensions of a flat hydrophobic rectangle with dimensions of approximately 14 Å × 12 Å × 5 Å [[142], [143], [144], [145], [146], [147]]. Recent structural predictions based on homology models provide additional insights, although to be confident, we must ultimately await structural elucidation of this binding through approaches such as nuclear magnetic resonance or crystallography [17,19,148,149].
The repeated observation that the AHR displays binding affinity for larger planar aromatic structures lead some to speculate that smaller one- or two-ring aromatic compounds are probably not ligands, even though they might lead to receptor activation in vivo [127]. An important early example of this phenomenon was the natural product indole-3-carbinol (I3C), a plant auxin derived from tryptophan in edible Brassica family plants [150]. Early on, this compound was highly touted as a dietary anticarcinogen and determined to be a powerful dietary activator of AHR-regulated xenobiotic metabolizing enzymes such as the CYP1 monooxygenases. Indole-3-carbinol is now emerging as a paradigm of the proligand concept in AHR biology, which posits that many small aromatic compounds are metabolically, chemically, or spontaneously converted to extended aromatic structures by natural processes [127,151,152]. In the case of I3C, the conversion occurs in the low pH environment of the stomach, where the acid-catalyzed condensation of I3C generates the potent AHR agonist, indolo[3,2,b]carbazole (ICZ) 151,153].
Perhaps more important than the identity of I3C as a proligand is the idea that this process may be a common route of ligand production in normal physiology. In this regard, tryptophan (TRP) is metabolized to the alpha-keto acid indole-3-pyruvic acid (I3P) by at least two enzymes (D-amino acid oxidase and aspartate aminotransferase), and I3P spontaneously condenses into TEACOPS with high AHR binding affinity in vivo [[154], [155], [156]]. The aromatic amino acid TRP is a particularly interesting source of endogenous ligands. This substrate can be metabolically and chemically converted into a variety of polyaromatic structures. For example, the TRP condensation product, 6-formylindolo[3,2-b]carbazole (FICZ), is often considered a top candidate as a cognate ligand of the AHR. This compound is generated by UV irradiation of TRP in the skin and displays an AHR binding affinity in the 10−11 molar range (KD) [152,157]. Other TRP metabolites that show potential as physiological endogenous ligands include the TEACOP products produced spontaneously from the TRP metabolite kynurenine (Kyn) [158,159]. These KYN derived TEACOPs have been shown to display potent agonist activity and may be responsible for aspects of reported immunological activity of Kyn on T-cells [160,161]. Interestingly, other TRP metabolites and condensation products are also candidates as ligands; these include simple indoles, the indigoids and cinnabarinic acid [140,162,163].
6.3. Evidence the AHR requires endogenous activation
Multiple experimental lines provide evidence that the cognate pathway requires endogenous ligand activation. First, more than one lab has reported physiological conditions where XRE-driven reporters are activated under conditions consistent with the presence of an endogenous ligand [111,164]. Second, mouse models hypomorphic for AHR expression can be rescued with pre/perinatal exposure to a remarkably potent agonists such as 2,3,7,8-tetrachlordibenzo-p-dioxin or SU5416 (i.e., DV closure) [132,165]. Third, overexpression of the CYP1A1 gene in the mouse leads to phenotypes similar to AHR-null animals. This last observation being consistent with a model where adaptive metabolic clearance of an endogenous ligand is required for normal barrier immunity in the gut [128].
7. The PAS protein family in mammals
Following the cloning and characterization of ARNT and the AHR from mice, rats and humans, a number of additional important PAS proteins were identified in mammals [73,166]. Among these are the HIF-alphas which dimerize with ARNT (also known as HIF-beta) to regulate the hypoxia response and the CLOCK, ARNTL and PER proteins which play central roles in the maintenance of circadian rhythms [73,[167], [168], [169], [170], [171]]. In each of these cases, like the AHR-ARNT, a dimeric pairing of two distinct PAS proteins drives a central transcriptional response through a cognate enhancer element. Also, like the AHR-ARNT system, each of these systems regulates environmental adaptation and each is also important for essential physiological developmental processes. Importantly, the CLOCK and HIF stories are recorded in detail as the result of the recent Nobel Prizes associated with their discovery [170,172].
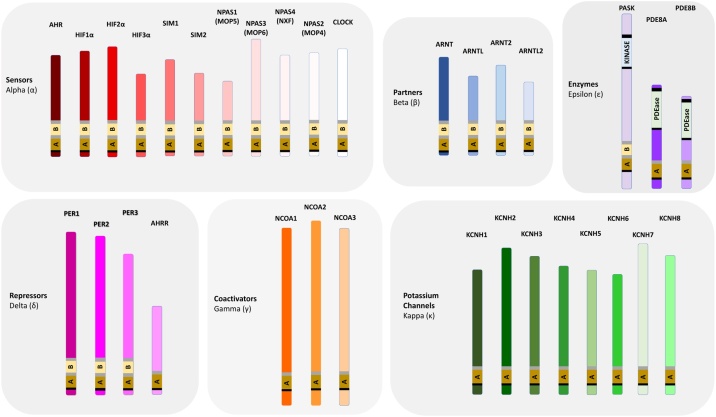
In early attempts at discovery of novel mammalian PAS factors, a variety of nomenclature schemes have been used. For example, in our laboratory, when each newly discovered factor was identified through searches of expressed sequence tags, the unique clones were designated Member of PAS-1, 2, 3…… (MOP1 etc.) [173]. In parallel, the another laboratory employed a similar strategy and their nomenclature denoted the site of expression and number of discovery (e.g., NPAS1, for neuronal PAS 1 etc.) [174]. Additionally, novel bHLH-PAS members have been named based on similarity to existing genes or predicted function [118,[175], [176], [177], [178], [179]]. A recent survey of the human genome reveals this family is quite large, with 33 PAS-domain encoding genes found by homology search or functional cloning (Fig. 6) [73]. Twenty-two of these proteins are documented or presumed to play roles in transcriptional regulation. Nineteen of these twenty-two transcriptional regulators harbor a (bHLH) domain immediately N–terminal to their PAS-A domain. Of the remaining eleven PAS proteins, eight are potassium channels, two are phosphodiesterases, and one is a serine-threonine kinase [73].
Fig. 6.
A proposed functional classification scheme of PAS protein interactions. The alpha class (reds) includes those proteins that often have restricted cell specific expression and bind with beta class proteins, often in response to an environmental stimulus or cellular state. The beta class (blues), are more widely expressed and act as binding partners for proteins from multiple PAS families, especially alpha class. The delta class (pinks) includes proteins that are often considered repressors and harbor deletions of classical domains such as PAS-B or bHLH. The gamma class (orange), include the mammalian coactivators (NCOAs) and are known for their unique role in transcriptional modulation. Finally, we show two PAS protein families that were not discussed in detail in this review; The epsilon class (purple), includes enzymes with PAS domains, while the kappa class (green) includes potassium channels. The bHLH, PAS A and PAS B domains are depicted as A and B respectively in the proteins in which they adhere to motifs as designated by UNIPROT [191].
Based upon our knowledge of AHR-ARNT mediated signal transduction, sequence alignment homology, as well current understanding of the biological function of each PAS family member, our laboratory classifies mammalian PAS members as either alpha-class, beta-class, gamma-class, delta class, or kappa-class (Fig. 6). The alpha-class includes the eleven PAS proteins that commonly display restricted cell type expression patterns, that are most commonly associated with the sensing environmental stimuli or cellular state, and that transduce those signals through an induced pairing with one of four promiscuous beta-class PAS protein partners [168,[180], [181], [182]]. We propose Beta-class to denote PAS proteins that are more widely expressed and often act as essential partners for a broad spectrum of family partners from the alpha-class. In this proposed PAS protein nomenclature, we add the designation of the three known human coactivators (NCOA1, NCOA2 and NCOA3) as Gamma-class because of their unique roles in modulating transcription and because little is currently known about their PAS-PAS interactions. Additionally, we designate as Delta-class, those PAS proteins involved in transcriptional signaling pathways, but that are missing one of the hallmark domains of this family. For example, the AHRR is missing PAS-B and the three PERs are missing a canonical bHLH domain. These structures have led to a general suspicion that Delta-class can be thought of as repressors. Two final classes of mammalian PAS proteins are the Kappa class (for potassium channels) and the Epsilon class (enzymes).
To date, the vast majority of known PAS protein interactions in mammalian systems are homotypic ones, that is, through PAS-PAS domain pairing and HLH domain pairing. This pairing then positions the basic region of the bHLH domain to produce a competent DNA binding structure capable of recognizing cognate enhancers, with each member recognizing one half site of the element (Fig. 3). Of these dimers that directly influence gene expression through cognate enhancer elements, the best understood are alpha-beta class pairings like the AHR-ARNT, HIF1-alpha-ARNT (aka HIF1-beta) or CLOCK-ARNTL (aka bMAL1 or MOP3). While homodimers such as ARNT-ARNT and PER-PER homodimers have been reported, the biological consequences of such interactions through recognition of genomic enhancers is unclear (e.g., ARNT and PER homodimers) [82,183]. Similarly, other non-alpha-beta class interactions have also been reported, but it is currently unclear if such interactions require either partner’s PAS domain for this binding (e.g., NCOA1 AND ARNT) [184].
Given that a complete description of the interactome of all mammalian PAS proteins has not yet been completed, it may still be of values to consider the possibility that more complex networks exist among PAS family members. In this regard, one early idea developed to explain toxicity of highly potent dioxin analogs was that the hyperactivated AHR may induce dimerization with additional bHLH-PAS partners other than ARNT and influence gene expression through unique enhancer elements, or through competition for limiting PAS-partners such as ARNT [72,171,185]. If, as presented above, PAS-PAS interaction surfaces exist in potassium channels, phosphodiesterases, regulators of development, circadian factors, then each of these areas of biology may be influencing each other in unappreciated ways. Put another way, the unknown partnering of PAS domains could represent paths of communication between physiological processes that have net been previously recognized.
8. Summary
The AHR has served as a paradigm for the PAS sensor superfamily and the roles of these proteins in environmental adaptation. When we look back at the research on this ancient domain found in all kingdoms of life, a few common themes emerge. For example, this domain is often associated with environmental sensing mechanisms that may have its origin in the adaptation to “Light, Oxygen and Voltage”, and in many chordates has evolved to mediate adaptive responses to certain xenobiotics, play important roles in barrier immunity, orchestrate midline development, and regulate an organismal response to circadian time. We also see how the PAS domain harbors multiple properties: First as a pocket for the binding of ligands as distinct as heme, 4-hydroxycinnamic acid, and 2,3,7,8-tetrachlorodibenzo-p-dioxin (Fig. 2). Second as a protein-protein interaction surface that dictates both homotypic and heterotypic interactions; With homotypic interactions often generating dimers capable of recognizing target genomic elements, and heterotypic interactions that control levels of signaling and provide a potential link to heterologous output pathways.
Data availability
No data was used for the research described in the article.
Data will be made available on request.
Declaration of Competing Interest
Chris Bradfield reports financial support was provided by National Institutes of Health. Chris Bradfield reports a relationship with National Institutes of Health that includes: funding grants. Chris Bradfield has patent none pending to none.
Acknowledgments
This work was supported by the National Institutes of Health Grants R35ES028377, T32ES007015, T32GM008692, T32CA009135 and P30CA014520. The UW-Madison Science & Medicine Graduate Research Scholars Program, and the National Science Foundation Graduate Research Fellowship under the grant no. DGE-1256259. Thanks to Igor Zhulin, Carrie Partch, Alex Veith, and Robert Bourret for helpful discussions.
Handling Editor: Dr. Aristides M. Tsatsakis
References
- 1.Bondos S.E., Swint-Kruse L., Matthews K.S. Flexibility and disorder in gene regulation: LacI/GalR and hox proteins. J. Biol. Chem. 2015;290(41):24669–24677. doi: 10.1074/jbc.R115.685032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grove A. Regulation of metabolic pathways by MarR family transcription factors. Comput. Struct. Biotechnol. J. 2017;15:366–371. doi: 10.1016/j.csbj.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R.M. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moglich A., Ayers R.A., Moffat K. Structure and signaling mechanism of Per-ARNT-sim domains. Structure. 2009;17(10):1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor B.L., Zhulin I.B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63(2) doi: 10.1128/mmbr.63.2.479-506.1999. 479-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson C.M., Klingeman D.M., Schlachter C., Syed M.H., Wu C., Guss A.M., Brown S.D. LacI transcriptional regulatory networks in Clostridium thermocellum DSM1313. Appl. Environ. Microbiol. 2017;83(5):e02751-16. doi: 10.1128/AEM.02751-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhulin I.B., Taylor B.L. PAS domain S-boxes in archaea, bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 1997;22(9):331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman E.C., Reyes H., Chu F.-F., Sander F., Conley L.H., Brooks B.A., Hankinson O. Cloning of a factor required for activity of the ah (Dioxin) receptor. Science. 1991;252(5008):954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 9.Huang Z.J., Edery I., Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364(6434):259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 10.Nambu J.R., Lewis J.O., Wharton K.A., Jr., Crews S.T. The drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67(6):1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 11.Ogura Y., Tokutomi S., Wada M., Kiyosue T. PAS/LOV proteins: a proposed new class of plant blue light receptor. Plant Signal. Behav. 2008;3(11):966–968. doi: 10.4161/psb.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Repik A., Rebbapragada A., Johnson M.S., Haznedar J.O., Zhulin I.B., Taylor B.L. PAS domain residues involved in signal transduction by the aer redox sensor of Escherichia coli. Mol. Microbiol. 2000;36(4):806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baca M., Borgstahl G.E.O., Boissinot M., Burke P.M., Williams D.R., Slater K.A., Getzoff E.D. Complete chemical-structure of photoactive yellow protein - novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemist. Biochemistry. 1994;33(48):14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 14.Borgstahl G.E.O., Williams D.R., Getzoff E.D. 1.4 Angstrom structure of photoactive yellow protein, a cytosolic photoreceptor - unusual fold, active-site, and chromophore. Biochemistry. 1995;34(19):6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 15.Gong W.M., Hao B., Mansy S.S., Gonzalez G., Gilles-Gonzalez M.A., Chan M.K. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. U. S. A. 1998;95(26):15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monson E.K., Weinstein M., Ditta G.S., Helinski D.R. The FixL protein of rhizobium-meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc. Natl. Acad. Sci. U. S. A. 1992;89(10):4280–4284. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing Y., Nukaya M., Satyshur K.A., Jiang L., Stanevich V., Korkmaz E.N., Burdette L., Kennedy G.D., Cui Q., Bradfield C.A. Identification of the ah-receptor structural determinants for ligand preferences. Toxicol. Sci. 2012;129(1):86–97. doi: 10.1093/toxsci/kfs194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parks A.J., Pollastri M.P., Hahn M.E., Stanford E.A., Novikov O., Franks D.G., Haigh S.E., Narasimhan S., Ashton T.D., Hopper T.G., Kozakov D., Beglov D., Vajda S., Schlezinger J.J., Sherr D.H. In silico identification of an aryl hydrocarbon receptor antagonist with biological activity in vitro and in vivo. Mol. Pharmacol. 2014;86(5):593. doi: 10.1124/mol.114.093369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraccalvieri D., Soshilov A.A., Karchner S.I., Franks D.G., Pandini A., Bonati L., Hahn M.E., Denison M.S. Comparative analysis of homology models of the ah receptor ligand binding domain: verification of structure–function predictions by site-directed mutagenesis of a nonfunctional receptor. Biochemistry. 2013;52(4):714–725. doi: 10.1021/bi301457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heintz U., Schlichting I. Blue light-induced LOV domain dimerization enhances the affinity of aureochrome 1a for its target DNA sequence. eLife. 2021;5 doi: 10.7554/eLife.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froehlich A.C., Liu Y., Loros J.J., Dunlap J.C. White collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297(5582):815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 22.Zoltowski B.D., Schwerdtfeger C., Widom J., Loros J.J., Bilwes A.M., Dunlap J.C., Crane B.R. Conformational switching in the fungal light sensor vivid. Science. 2007;316(5827):1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzen C., Loros J.J., Dunlap J.C. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104(3):453–464. doi: 10.1016/S0092-8674(01)00232-X. [DOI] [PubMed] [Google Scholar]
- 24.Vaidya A.T., Chen C.-H., Dunlap J.C., Loros J.J., Crane B.R. Structure of a light-activated LOV protein dimer that regulates transcription in Neurospora Crassa. Sci. Signal. 2011;4(184):ra50. doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C.-H., DeMay B.S., Gladfelter A.S., Dunlap J.C., Loros J.J. Physical interaction between VIVID and white collar complex regulates photoadaptation in neurospora. Proc. Natl. Acad. Sci. U. S. A. 2010;107(38):16715–16720. doi: 10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dasgupta A., Chen C.-H., Lee C., Gladfelter A.S., Dunlap J.C., Loros J.J. Biological significance of photoreceptor photocycle length: VIVID photocycle governs the dynamic VIVID-white collar complex pool mediating photo-adaptation and response to changes in light intensity. PLoS Genet. 2015;11(5) doi: 10.1371/journal.pgen.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razeto A., Ramakrishnan V., Litterst C.M., Giller K., Griesinger C., Carlomagno T., Lakomek N., Heimburg T., Lodrini M., Pfitzner E., Becker S. Structure of the NCoA-1/SRC-1 PAS-B domain bound to the LXXLL motif of the STAT6 transactivation domain. J. Mol. Biol. 2004;336(2):319–329. doi: 10.1016/j.jmb.2003.12.057. [DOI] [PubMed] [Google Scholar]
- 28.Fribourgh J.L., Srivastava A., Sandate C.R., Michael A.K., Hsu P.L., Rakers C., Nguyen L.T., Torgrimson M.R., Parico G.C.G., Tripathi S., Zheng N., Lander G.C., Hirota T., Tama F., Partch C.L. Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing. eLife. 2020;9:e55275. doi: 10.7554/eLife.55275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nash A.I., McNulty R., Shillito M.E., Swartz T.E., Bogomolni R.A., Luecke H., Gardner K.H. Structural basis of photosensitivity in a bacterial Light-Oxygen-Voltage/Helix-Turn-Helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. 2011;108(23):9449. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alegbeleye O.O., Opeolu B.O., Jackson V.A. Polycyclic aromatic hydrocarbons: a critical review of environmental occurrence and bioremediation. Environ. Manage. 2017;60(4):758–783. doi: 10.1007/s00267-017-0896-2. [DOI] [PubMed] [Google Scholar]
- 31.Lawal A.T. Polycyclic aromatic hydrocarbons. A review. Cogent Environ. Sci. 2017;3(1) [Google Scholar]
- 32.Manzetti S. Polycyclic aromatic hydrocarbons in the environment: environmental fate and transformation. Polycycl. Aromat. 2013;33(4):311–330. doi: 10.1080/10406638.2013.781042. [DOI] [Google Scholar]
- 33.Hahn M.E. The aryl hydrocarbon receptor: a comparative perspective1. This article was invited by guest editors Dr John J. Stegeman and Dr David R. Livingstone to be part of a special issue of CBP on Cytochrome P450 (Comp. Biochem. Physiol. 121 C, Pages 1–412, 1998).1. Comp. Biochem. Physiol. C, Pharmacol. Toxicol. Endocrinol. 1998;121(1):23–53. doi: 10.1016/S0742-8413(98)10028-2. [DOI] [PubMed] [Google Scholar]
- 34.Safe S. Polychlorinated biphenyls (PCBs), Dibenzo-p-Dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit. Rev. Toxicol. 1990;21(1):51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 35.Shimada T., Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 And1B1. Cancer Sci. 2004;95(1):1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nebert D.W., Dalton T.P., Okey A.B., Gonzalez F.J. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279(23):23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 37.Conney A.H. Induction of drug-metabolizing enzymes: a path to the discovery of multiple cytochromes P450. Annu. Rev. Pharmacol. Toxicol. 2003;43(1):1–30. doi: 10.1146/annurev.pharmtox.43.100901.135754. [DOI] [PubMed] [Google Scholar]
- 38.Nebert D.W., Bausserman L.L. Fate of inducer during induction of aryl hydrocarbon hydroxylase activity in mammalian cell culture. II. Levels of intracellular polycyclic hydrocarbon during enzyme induction and decay. Mol. Pharmacol. 1970;6(4):304–314. [PubMed] [Google Scholar]
- 39.Pardee A.B., Prestidge L.S. The initial kinetics of enzyme induction. Biochim. Biophys. Acta. 1961;49(1):77–88. doi: 10.1016/0006-3002(61)90871-X. [DOI] [PubMed] [Google Scholar]
- 40.Aarts J.M.M.J.G., Alink G.M., Scherjon F., MacDonald K., Smith A.C., Nijveen H., Roebroeks W. Fire usage and ancient hominin detoxification genes: protective ancestral variants dominate while additional derived risk variants appear in modern humans. PLoS One. 2016;11(9):e0161102. doi: 10.1371/journal.pone.0161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez F.J., Nebert D.W. Evolution of the P450 gene superfamily:: animal-plant ‘warfare’, molecular drive and human genetic differences in drug oxidation. Trends Genet. 1990;6:182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- 42.Okey A.B. An aryl hydrocarbon receptor odyssey to the shores of toxicology: the Deichmann lecture, international congress of toxicology-XI. Toxicol. Sci. 2007;98(1):5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- 43.Poland A., Glover E. Variation in the molecular mass of the Ah receptor among vertebrate species and strains of rats. Biochem. Biophys. Res. Commun. 1987;146(3):1439–1449. doi: 10.1016/0006-291X(87)90811-4. [DOI] [PubMed] [Google Scholar]
- 44.Nebert D.W., Robinson J.R., Niwa A., Kumari K., Poland A.P. Genetic expression of aryl hydrocarbon hydroxylase activity in the mouse. J. Cell. Physiol. 1975;85(S1):393–414. doi: 10.1002/jcp.1040850407. [DOI] [PubMed] [Google Scholar]
- 45.Poland A.P., Glover E., Robinson J.R., Nebert D.W. Genetic expression of aryl hydrocarbon hydroxylase activity. Induction of monooxygenase activities and cytochrome P1-450 formation by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mice genetically “nonresponsive” to other aromatic hydrocarbons. J. Biol. Chem. 1974;249(17):5599–5606. [PubMed] [Google Scholar]
- 46.Poland A., Glover E., Ebetino F.H., Kende A.S. Photoaffinity-labeling of the Ah receptor. J. Biol. Chem. 1986;261(14):6352–6365. [PubMed] [Google Scholar]
- 47.Poland A., Glover E., Kende A.S. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J. Biol. Chem. 1976;251(16):4936–4946. [PubMed] [Google Scholar]
- 48.Carlson D.B., Perdew G.H. A dynamic role for the Ah receptor in cell signaling? Insights from a diverse group of ah receptor interacting proteins. J. Biochem. Mol. Toxicol. 2002;16(6):317–325. doi: 10.1002/jbt.10051. [DOI] [PubMed] [Google Scholar]
- 49.Carver L.A., Jackiw V., Bradfield C.A. The 90-KDa heat shock protein is essential for Ah receptor signaling in a yeast expression system. J. Biol. Chem. 1994;269(48):30109–30112. [PubMed] [Google Scholar]
- 50.Carver L.A., Bradfield C.A. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 1997;272(17):11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 51.Dunham E.E., Stevens E.A., Glover E., Bradfield C.A. The aryl hydrocarbon receptor signaling pathway is modified through interactions with a kelch protein. Mol. Pharmacol. 2006;70(1):8–15. doi: 10.1124/mol.106.024380. [DOI] [PubMed] [Google Scholar]
- 52.Kazlauskas A., Sundstrom S., Poellinger L., Pongratz I. The Hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol. Cell. Biol. 2001;21(7):2594–2607. doi: 10.1128/mcb.21.7.2594-2607.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kazlauskas A., Poellinger L., Pongratz I. Evidence that the co-chaperone P23 regulates ligand responsiveness of the dioxin (aryl hydrocarbon) receptor. J. Biol. Chem. 1999;274(19):13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 54.Perdew G.H. Association of the Ah receptor with the 90-KDA heat-shock protein. J. Biol. Chem. 1988;263(27):13802–13805. [PubMed] [Google Scholar]
- 55.Pongratz I., Mason G.G.F., Poellinger L. Dual roles of the 90-KDA heat-shock protein HSP90 in modulating functional activities of the dioxin receptor - evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require HSP90 both for ligand-binding activity and repression of intrinsic DNA-binding activity. J. Biol. Chem. 1992;267(19):13728–13734. [PubMed] [Google Scholar]
- 56.Poland A., Knutson J.C. 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22(1):517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 57.Poland A., Glover E. 2,3,7,8-tetrachlorodibenzo-p-dioxin: segregation of toxicity with the Ah locus. Mol. Pharmacol. 1980;17(1):86–94. [PubMed] [Google Scholar]
- 58.Fujii-Kuriyama Y., Mimura J. Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 2005;338(1):311–317. doi: 10.1016/j.bbrc.2005.08.162. [DOI] [PubMed] [Google Scholar]
- 59.Whitlock James P., Jr. Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39(1):103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 60.Nebert D.W., Gonzalez F.J. P450 genes: structure, evolution, and regulation. Annu. Rev. Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- 61.Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35(1)):307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 62.Elferink C.J., Gasiewicz T.A., Whitlock J.P. PROTEIN-DNA interactions at a dioxin-responsive enhancer - evidence that the transformed ah receptor is heteromeric. J. Biol. Chem. 1990;265(33):20708–20712. [PubMed] [Google Scholar]
- 63.Yao E.F., Denison M.S. DNA-sequence determinants for binding o.f. transformed Ah-receptor to a dioxin-responsive enhancer. Biochemistry. 1992;31(21):5060–5067. doi: 10.1021/bi00136a019. [DOI] [PubMed] [Google Scholar]
- 64.Crews S.T. Control of cell lineage-specific development and transcription by BHLH-PAS proteins. Genes Dev. 1998;12(5):607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 65.Crews S.T., Fan C.-M. Remembrance of things PAS: regulation of development by BHLH–PAS proteins. Curr. Opin. Genet. Dev. 1999;9(5):580–587. doi: 10.1016/S0959-437X(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 66.Funk W.D., Ouellette M., Wright W.E. Molecular biology of myogenic regulatory factors. Mol. Biol. Med. 1991;8(2):185–195. [PubMed] [Google Scholar]
- 67.Burbach K.M., Poland A., Bradfield C.A. Cloning of the ah-receptor CDNA reveals a distinctive ligand-activated transcription factor. Proc. Natl. Acad. Sci. 1992;89(17):8185. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reyes H., Reisz-Porszasz S., Hankinson O. Identification of the ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the ah receptor. Science. 1992;256(5060):1193. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 69.Bradfield C.A., Glover E., Poland A. Purification and N-Terminal amino acid sequence of the Ah receptor from the C57BL/6J mouse. Mol. Pharmacol. 1991;39(1):13. [PubMed] [Google Scholar]
- 70.Ema M., Sogawa K., Watanabe N., Chujoh Y., Matsushita N., Gotoh O., Funae Y., Fujii-Kuriyama Y. CDNA cloning and structure of mouse putative Ah receptor. Biochem. Biophys. Res. Commun. 1992;184(1):246–253. doi: 10.1016/0006-291X(92)91185-S. [DOI] [PubMed] [Google Scholar]
- 71.Poland A., Glover E., Bradfield C.A. Characterization of polyclonal antibodies to the Ah receptor prepared by immunization with a synthetic peptide hapten. Mol. Pharmacol. 1991;39(1):20. [PubMed] [Google Scholar]
- 72.Schmidt J.V., Carver L.A., Bradfield C.A. Molecular characterization of the murine Ahr gene - organization, promoter analysis, and chromosomal assignment. J. Biol. Chem. 1993;268(29):22203–22209. [PubMed] [Google Scholar]
- 73.McIntosh B.E., Hogenesch J.B., Bradfield C.A. Mammalian per-arnt-sim proteins in environmental adaptation. Annu. Rev. Physiol. 2010;72(1):625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 74.Dolwick K.M., Swanson H.I., Bradfield C.A. In vitro analysis of ah receptor domains involved in ligand-activated DNA recognition. Proc. Natl. Acad. Sci. 1993;90(18):8566. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antonsson C., Whitelaw M.L., McGuire J., Gustafsson J.A., Poellinger L. Distinct roles of the molecular chaperone HSP90 in modulating dioxin receptor function via the basic helix-loop-helix and pas domains. Mol. Cell. Biol. 1995;15(2):756–765. doi: 10.1128/mcb.15.2.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berg P., Pongratz I. Differential usage of nuclear export sequences regulates intracellular localization of the dioxin (aryl hydrocarbon) receptor. J. Biol. Chem. 2001;276(46):43231–43238. doi: 10.1074/jbc.M105261200. [DOI] [PubMed] [Google Scholar]
- 77.Fukunaga B.N., Probst M.R., Reiszporszasz S., Hankinson O. Identification of functional domains of the aryl-hydrocarbon receptor. J. Biol. Chem. 1995;270(49):29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 78.Jain S., Dolwick K.M., Schmidt J.V., Bradfield C.A. Potent transactivation domains of the Ah receptor and the Ah receptor nuclear translocator map to their carboxyl termini. J. Biol. Chem. 1994;269(50):31518–31524. [PubMed] [Google Scholar]
- 79.Perdew G.H., Bradfield C.A. Mapping the 90 KDa heat shock protein binding region of the Ah receptor. IUBMB Life. 1996;39(3):589–593. doi: 10.1080/15216549600201651. [DOI] [PubMed] [Google Scholar]
- 80.Reisz-Porszasz S., Probst M.R., Fukunaga B.N., Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT) Mol. Cell. Biol. 1994;14(9):6075–6086. doi: 10.1128/mcb.14.9.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murre C., McCaw P.S., Baltimore D.A. New DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56(5):777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- 82.Swanson H.I., Chan W.K., Bradfield C.A. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 1995;270(44):26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 83.Schulte K.W., Green E., Wilz A., Platten M., Daumke O. Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure. 2017;25(7):1025–1033. doi: 10.1016/j.str.2017.05.008. e3. [DOI] [PubMed] [Google Scholar]
- 84.Seok S.H., Lee W., Jiang L., Molugu K., Zheng A.P., Li Y.T., Park S., Bradfield C.A., Xing Y.N. Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Proc. Natl. Acad. Sci. U. S. A. 2017;114(21):5431–5436. doi: 10.1073/pnas.1617035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu D., Potluri N., Kim Y., Rastinejad F. Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol. Cell. Biol. 2013;33(21):4346. doi: 10.1128/mcb.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma Q., Dong L.Q., Whitlock J.P. Transcriptional activation by the mouse Ah receptor - interplay between multiple stimulatory and inhibitory functions. J. Biol. Chem. 1995;270(21):12697–12703. doi: 10.1074/jbc.270.21.12697. [DOI] [PubMed] [Google Scholar]
- 87.Whitelaw M.L., Göttlicher M., Gustafsson J.A., Poellinger L. Definition of a novel ligand binding domain of a nuclear BHLH receptor: co-localization of ligand and Hsp90 binding activities within the regulable inactivation domain of the dioxin receptor. EMBO J. 1993;12(11):4169–4179. doi: 10.1002/j.1460-2075.1993.tb06101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whitelaw M.L., Gustafsson J.A., Poellinger L. Identification of transactivation and repression functions of the dioxin receptor and its basic helix-loop-helix/PAS partner factor Arnt: inducible versus constitutive modes of regulation. Mol. Cell. Biol. 1994;14(12):8343. doi: 10.1128/mcb.14.12.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma Q., Whitlock J.P. A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p -dioxin. J. Biol. Chem. 1997;272(14):8878–8884. doi: 10.1074/jbc.272.14.8878. [DOI] [PubMed] [Google Scholar]
- 90.Petrulis J.R., Perdew G.H. The role of chaperone proteins in the aryl hydrocarbon receptor core complex. Chem. Biol. Interact. 2002;141(1):25–40. doi: 10.1016/S0009-2797(02)00064-9. [DOI] [PubMed] [Google Scholar]
- 91.Wilhelmsson A., Cuthill S., Denis M., Wikström A.C., Gustafsson J.A., Poellinger L. The specific DNA binding activity of the dioxin receptor is modulated by the 90 Kd heat shock protein. EMBO J. 1990;9(1):69–76. doi: 10.1002/j.1460-2075.1990.tb08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ema M., Ohe N., Suzuki M., Mimura J., Sogawa K., Ikawa S., Fujii-Kuriyama Y. Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J. Biol. Chem. 1994;269(44):27337–27343. [PubMed] [Google Scholar]
- 93.Goryo K., Suzuki A., Carpio C.A.D., Siizaki K., Kuriyama E., Mikami Y., Kinoshita K., Yasumoto K., Rannug A., Miyamoto A., Fujii-Kuriyama Y., Sogawa K. Identification of amino acid residues in the Ah receptor involved in ligand binding. Biochem. Biophys. Res. Commun. 2007;354(2):396–402. doi: 10.1016/j.bbrc.2006.12.227. [DOI] [PubMed] [Google Scholar]
- 94.Pandini A., Soshilov A.A., Song Y., Zhao J., Bonati L., Denison M.S. Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry. 2009;48(25):5972–5983. doi: 10.1021/bi900259z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Poland A., Palen D., Glover E. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol. Pharmacol. 1994;46(5):915–921. [PubMed] [Google Scholar]
- 96.Wilson R.H., Carney P.R., Glover E., Parrott J.C., Rojas B.L., Moran S.M., Yee J.S., Nukaya M., Goetz N.A., Rubenstein C.D., Krentz K.J., Xing Y., Bradfield C.A. Generation of an allelic series at the Ahr locus using an edited recombinant approach. Toxicol. Sci. Off. J. Soc. Toxicol. 2021 doi: 10.1093/toxsci/kfab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sogawa K., Iwabuchi K., Abe H., Fujii-Kuriyama Y. Transcriptional activation domains of the Ah receptor and ah receptor nuclear translocator. J. Cancer Res. Clin. Oncol. 1995;121(9):612–620. doi: 10.1007/bf01197779. [DOI] [PubMed] [Google Scholar]
- 98.Hahn M.E. 2002. Aryl Hydrocarbon Receptors: Diversity and Evolution; p. 30. [DOI] [PubMed] [Google Scholar]
- 99.Hubbard T.D., Murray I.A., Bisson W.H., Sullivan A.P., Sebastian A., Perry G.H., Jablonski N.G., Perdew G.H. Divergent Ah receptor ligand selectivity during hominin evolution. Mol. Biol. Evol. 2016;33(10):2648–2658. doi: 10.1093/molbev/msw143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moffat I.D., Roblin S., Harper P.A., Okey A.B., Pohjanvirta R. Aryl hydrocarbon receptor splice variants in the dioxin-resistant rat: tissue expression and transactivational activity. Mol. Pharmacol. 2007;72(4):956. doi: 10.1124/mol.107.037218. [DOI] [PubMed] [Google Scholar]
- 101.Okey A.B., Boutros P.C., Harper P.A. Polymorphisms of human nuclear receptors that control expression of drug-metabolizing enzymes. Pharmacogenet. Genomics. 2005;15(6) doi: 10.1097/01213011-200506000-00001. [DOI] [PubMed] [Google Scholar]
- 102.Harper P., Wong J., Lam M., Okey A. Polymorphisms in the human Ah receptor. Chem. Biol. Interact. 2002;141(1–2):161–187. doi: 10.1016/S0009-2797(02)00071-6. [DOI] [PubMed] [Google Scholar]
- 103.Sherry S., Ward M., Kholodov M., Baker J., Phan L., Smigielski E., Sirotkin K. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong J., Okey A., Harper P. Human aryl hydrocarbon receptor polymorphisms that result in loss of CYP1A1 induction. Biochem. Biophys. Res. Commun. 2001;288(4):990–996. doi: 10.1006/bbrc.2001.5861. [DOI] [PubMed] [Google Scholar]
- 105.KAWAJIRI K., WATANABE J., EGUCHI H., HAYASHI S. Genetic polymorphisms of drug-metabolizing-enzymes and lung-cancer susceptibility. PHARMACOGENETICS. 1995;5:S70–S73. doi: 10.1097/00008571-199512001-00004. [DOI] [PubMed] [Google Scholar]
- 106.Smart J., Daly A. Variation in induced CYP1A1 levels: relationship to CYP1A1, ah receptor and GSTM1 polymorphisms. Pharmacogenetics. 2000;10(1):11–24. doi: 10.1097/00008571-200002000-00003. [DOI] [PubMed] [Google Scholar]
- 107.Fukushima-Uesaka H., Sai K., Maekawa K., Koyano S., Kaniwa N., Ozawa S., Kawamoto M., Kamatani N., Komamura K., Kamakura S., Kitakaze M., Tomoike H., Ueno K., Minami H., Ohtsu A., Shirao K., Yoshida T., Saijo N., Saito Y., Sawada J. Genetic variations of the AHR gene encoding aryl hydrocarbon receptor in a japanese population. Drug Metab. Pharmacokinet. 2004;19(4):320–326. doi: 10.2133/dmpk.19.320. [DOI] [PubMed] [Google Scholar]
- 108.Jackson D.P., Joshi A.D., Elferink C.J. Ah receptor pathway intricacies; signaling through diverse protein partners and DNA-motifs. Toxicol. Res. 2015;4(5):1143–1158. doi: 10.1039/C4TX00236A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nault R., Fader K.A., Harkema J.R., Zacharewski T. Loss of liver-specific and sexually dimorphic gene expression by aryl hydrocarbon receptor activation in C57BL/6 mice. PLoS One. 2017;12(9):e0184842. doi: 10.1371/journal.pone.0184842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boutros P.C., Yan R., Moffat I.D., Pohjanvirta R., Okey A.B. Transcriptomic responses to 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) in liver: comparison of rat and mouse. BMC Genomics. 2008;9:419. doi: 10.1186/1471-2164-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang C.-Y., Puga A. Constitutive activation of the aromatic hydrocarbon receptor. Mol. Cell. Biol. 1998;18(1):525–535. doi: 10.1128/mcb.18.1.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Safe S., Han H., Goldsby J., Mohankumar K., Chapkin R.S. Aryl hydrocarbon receptor (AhR) ligands as selective AhR modulators: genomic studies. Genomic Toxicol. 2018;11–12:10–20. doi: 10.1016/j.cotox.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marlowe J.L., Fan Y., Chang X., Peng L., Knudsen E.S., Xia Y., Puga A. The aryl hydrocarbon receptor binds to E2F1 and inhibits E2F1-induced apoptosis. Mol. Biol. Cell. 2008;19(8):3263–3271. doi: 10.1091/mbc.e08-04-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marlowe J.L., Puga A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell. Biochem. 2005;96(6):1174–1184. doi: 10.1002/jcb.20656. [DOI] [PubMed] [Google Scholar]
- 115.Safe S., Wormke M. Inhibitory aryl hydrocarbon receptor−estrogen receptor α cross-talk and mechanisms of action. Chem. Res. Toxicol. 2003;16(7):807–816. doi: 10.1021/tx034036r. [DOI] [PubMed] [Google Scholar]
- 116.Evans B.R., Karchner S.I., Allan L.L., Pollenz R.S., Tanguay R.L., Jenny M.J., Sherr D.H., Hahn M.E. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 2008;73(2):387–398. doi: 10.1124/mol.107.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.MacPherson L., Ahmed S., Tamblyn L., Krutmann J., Förster I., Weighardt H., Matthews J. Aryl hydrocarbon receptor repressor and TiPARP (ARTD14) use similar, but also distinct mechanisms to repress aryl hydrocarbon receptor signaling. Int. J. Mol. Sci. 2014;15(5):7939–7957. doi: 10.3390/ijms15057939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mimura J., Ema M., Sogawa K., Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of ah (Dioxin) receptor function. Genes Dev. 1999;13(1):20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Karchner S.I., Franks D.G., Powell W.H., Hahn M.E. Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AHR repressor, AHR1, and AHR2. J. Biol. Chem. 2002;277(9):6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- 120.Fernandez-Salguero P., Pineau T., Hilbert D.M., McPhail T., Kimura S., Nebert D.W., Rudikoff S., Ward J.M., Gonzalez F.J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin- binding Ah receptor. Science. 1995;268(5211):722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 121.Harrill J.A., Hukkanen R.R., Lawson M., Martin G., Gilger B., Soldatow V., Lecluyse E.L., Budinsky R.A., Rowlands J.C., Thomas R.S. Knockout of the aryl hydrocarbon receptor results in distinct hepatic and renal phenotypes in rats and mice. Toxicol. Appl. Pharmacol. 2013;272(2):503–518. doi: 10.1016/j.taap.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 122.Li Y., Innocentin S., Withers D.R., Roberts N.A., Gallagher A.R., Grigorieva E.F., Wilhelm C., Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147(3):629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 123.Schmidt J.V., Sutt G., H.-T.; Reddy J.K., SIMONt M.C., Bradfield C.A. Characterization of a murine ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. U. S. A. 1996:6. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thackaberry E.A., Gabaldon D.M., Walker M.K., Smith S.M. Aryl hydrocarbon receptor null mice develop cardiac hypertrophy and increased hypoxia-inducible factor-1alpha in the absence of cardiac hypoxia. Cardiovasc. Toxicol. 2002;2(4):263–274. doi: 10.1385/ct:2:4:263. [DOI] [PubMed] [Google Scholar]
- 125.Veldhoen M., Hirota K., Westendorf A.M., Buer J., Dumoutier L., Renauld J.-C., Stockinger B. The aryl hydrocarbon receptor links t H 17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453(7191):106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 126.Abbott B.D., Schmid J.E., Pitt J.A., Buckalew A.R., Wood C.R., Held G.A., Diliberto J.J. Adverse reproductive outcomes in the transgenic ah receptor-deficient mouse. Toxicol. Appl. Pharmacol. 1999;155(1):62–70. doi: 10.1006/taap.1998.8601. [DOI] [PubMed] [Google Scholar]
- 127.Avilla M.N., Malecki K.M.C., Hahn M.E., Wilson R.H., Bradfield C.A. The Ah receptor: adaptive metabolism, ligand diversity, and the xenokine model. Chem. Res. Toxicol. 2020 doi: 10.1021/acs.chemrestox.9b00476. acs.chemrestox.9b00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schiering C., Wincent E., Metidji A., Iseppon A., Li Y., Potocnik A.J., Omenetti S., Henderson C.J., Wolf C.R., Nebert D.W., Stockinger B. Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542:242. doi: 10.1038/nature21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu Q., Ohsako S., Baba T., Miyamoto K., Tohyama C. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on preimplantation mouse embryos. Toxicology. 2002;174(2):119–129. doi: 10.1016/S0300-483X(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 130.Esser C., Rannug A., Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30(9):447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 131.Esser C., Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol. Rev. 2015;67(2):259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 132.Walisser J.A., Bunger M.K., Glover E., Bradfield C.A. Gestational exposure of Ahr and Arnt hypomorphs to dioxin rescues vascular development. Proc. Natl. Acad. Sci. U. S. A. 2004;101(47):16677–16682. doi: 10.1073/pnas.0404379101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin B.C., Nguyen L.P., Walisser J.A., Bradfield C.A. A hypomorphic allele of aryl hydrocarbon receptor-associated protein-9 produces a phenocopy of the AHR-Null mouse. Mol. Pharmacol. 2008;74(5):1367–1371. doi: 10.1124/mol.108.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bunger M.K., Glover E., Moran S.M., Walisser J.A., Lahvis G.P., Hsu E.L., Bradfield C.A. Abnormal liver development and resistance to 2,3,7,8-Tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-Binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 2008;106(1):83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jin G.-B., Winans B., Martin K.C., Paige Lawrence B. New insights into the role of the aryl hydrocarbon receptor in the function of CD11c+ cells during respiratory viral infection. Eur. J. Immunol. 2014;44(6):1685–1698. doi: 10.1002/eji.201343980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Walisser J.A., Glover E., Pande K., Liss A.L., Bradfield C.A. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc. Natl. Acad. Sci. U. S. A. 2005;102(49):17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stockinger B., Meglio P.D., Gialitakis M., Duarte J.H. The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 2014;32(1):403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 138.Denison M.S., Nagy S.R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43(1):309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 139.Heath-Pagliuso S., Rogers W.J., Tullis K., Seidel S.D., Cenijn P.H., Brouwer A., Denison M.S. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37(33):11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- 140.Hubbard T.D., Murray I.A., Perdew G.H. Indole and tryptophan metabolism: endogenous and dietary routes to ah receptor activation. Drug Metab. Dispos. 2015;43(10):1522–1535. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nguyen L.P., Bradfield C.A. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008;21(1):102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Waller C.L., McKinney J.D. Three-dimensional quantitative structure-activity relationships of dioxins and dioxin-like compounds: model validation and ah receptor characterization. Chem. Res. Toxicol. 1995;8(6):847–858. doi: 10.1021/tx00048a005. [DOI] [PubMed] [Google Scholar]
- 143.Gillner M., Bergman J., Cambillau C., Alexandersson M., Fernstrom B., Gustafsson J.A. Interactions of indolo 3.,2-B carbazoles and related polycyclic aromatic-hydrocarbons with specific binding-sites for 2,3,7,8-tetrachlorodibenzo-p-dioxin in rat-liver. Mol. Pharmacol. 1993;44(2):336–345. [PubMed] [Google Scholar]
- 144.Waller C.L., McKinney J.D. Comparative molecular field analysis of polyhalogenated dibenzo-p-dioxins, dibenzofurans, and biphenyls. J. Med. Chem. 1992;35(20):3660–3666. doi: 10.1021/jm00098a010. [DOI] [PubMed] [Google Scholar]
- 145.Bonati L., Fraschini E., Lasagni M., Modoni E.P., Pitea D. A hypothesis on the mechanism of PCDD biological activity based on molecular electrostatic potential modeling. Part 2. J. Mol. Struct. Theochem. 1995;340(1–3):83–95. doi: 10.1016/0166-1280(95)04171-2. [DOI] [Google Scholar]
- 146.Bonati L., Fraschini E., Lasagni M., Pitea D. Toward a mechanistic understanding of PCDD biological activity based on molecular electrostatic potential modeling. J. Mol. Struct. Theochem. 1994;303(C):43–54. doi: 10.1016/0166-1280(94)80172-x. [DOI] [Google Scholar]
- 147.Fraschini E., Bonati L., Pitea D. Molecular polarizability as a tool for understanding the binding properties of polychlorinated dibenzo-p-dioxins: definition of a reliable computational procedure. J. Phys. Chem. 1996;100(25):10564–10569. doi: 10.1021/jp952888x. [DOI] [Google Scholar]
- 148.Motto I., Bordogna A., Soshilov A.A., Denison M.S., Bonati L. New aryl hydrocarbon receptor homology model targeted to improve docking reliability. J. Chem. Inf. Model. 2011;51(11):2868–2881. doi: 10.1021/ci2001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bisson W.H., Koch D.C., O’Donnell E.F., Khalil S.M., Kerkvliet N.I., Tanguay R.L., Abagyan R., Kolluri S.K. Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 2009;52(18):5635–5641. doi: 10.1021/jm900199u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Loub W.D., Wattenberg L.W., Davis D.W. Aryl hydrocarbon hydroxylase induction in rat tissues by naturally occurring indoles of cruciferous plants. J. Natl. Cancer Inst. 1975;54(4):985–988. [PubMed] [Google Scholar]
- 151.Bradfield C.A., Bjeldanes L.F. Structure—activity relationships of dietary indoles: a proposed mechanism of action as modifiers of xenobiotic metabolism. J. Toxicol. Environ. Health. 1987;21(3):311–323. doi: 10.1080/15287398709531021. [DOI] [PubMed] [Google Scholar]
- 152.Rannug A., Rannug U., Rosenkranz H., Winqvist L., Westerholm R., Agurell E., Grafström A. Certain photooxidized derivatives of tryptophan bind with very high affinity to the ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 1987;262(32):15422–15427. [PubMed] [Google Scholar]
- 153.Bjeldanes L.F., Kim J.Y., Grose K.R., Bartholomew J.C., Bradfield C.A. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc. Natl. Acad. Sci. 1991;88(21):9543. doi: 10.1073/pnas.88.21.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nguyen L.P., Hsu E.L., Chowdhury G., Dostalek M., Guengerich F.P., Bradfield C.A. D-amino acid oxidase generates agonists of the aryl hydrocarbon receptor from d-tryptophan. Chem. Res. Toxicol. 2009;22(12):1897–1904. doi: 10.1021/tx900043s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bittinger M.A., Nguyen L.P., Bradfield C.A. Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol. Pharmacol. 2003;64(3):550. doi: 10.1124/mol.64.3.550. [DOI] [PubMed] [Google Scholar]
- 156.Chowdhury G., Dostalek M., Hsu E.L., Nguyen L.P., Stec D.F., Bradfield C.A., Guengerich F.P. Structural identification of diindole agonists of the aryl hydrocarbon receptor derived from degradation of indole-3-pyruvic acid. Chem. Res. Toxicol. 2009;22(12):1905–1912. doi: 10.1021/tx9000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wincent E., Amini N., Luecke S., Glatt H., Bergman J., Crescenzi C., Rannug A., Rannug U. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-Formylindolo[3,2-b]carbazole is present in humans *. J. Biol. Chem. 2009;284(5):2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- 158.Julliard W., Fechner J.H., Mezrich J.D. The aryl hydrocarbon receptor meets immunology: friend or foe? A little of both. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Seok S.-H., Ma Z.-X., Feltenberger J.B., Chen H., Chen H., Scarlett C., Lin Z., Satyshur K.A., Cortopassi M., Jefcoate C.R., Ge Y., Tang W., Bradfield C.A., Xing Y. Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR) J. Biol. Chem. 2018;293(6):1994–2005. doi: 10.1074/jbc.RA117.000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mezrich J.D., Fechner J.H., Zhang X., Johnson B.P., Burlingham W.J., Bradfield C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010;185(6):3190. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cheong J.E., Sun L. Targeting the IDO1/TDO2–KYN–AhR pathway for cancer immunotherapy – challenges and opportunities. Trends Pharmacol. Sci. 2018;39(3):307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 162.Lowe M.M., Mold J.E., Kanwar B., Huang Y., Louie A., Pollastri M.P., Wang C., Patel G., Franks D.G., Schlezinger J., Sherr D.H., Silverstone A.E., Hahn M.E., McCune J.M. Identification of cinnabarinic acid as a novel endogenous aryl hydrocarbon receptor ligand that drives IL-22 production. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0087877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nishiumi S., Yamamoto N., Kodoi R., Fukuda I., Yoshida K., Ashida H. Antagonistic and agonistic effects of indigoids on the transformation of an aryl hydrocarbon receptor. Arch. Biochem. Biophys. 2008;470(2):187–199. doi: 10.1016/j.abb.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 164.Willey J.J., Stripp B.R., Baggs R.B., Gasiewicz T.A. Aryl hydrocarbon receptor activation in genital tubercle, palate, and other embryonic tissues in 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsivelacZmice. Toxicol. Appl. Pharmacol. 1998;151(1):33–44. doi: 10.1006/taap.1998.8444. [DOI] [PubMed] [Google Scholar]
- 165.Mezrich J.D., Nguyen L.P., Kennedy G., Nukaya M., Fechner J.H., Zhang X., Xing Y., Bradfield C.A. SU5416, a VEGF receptor inhibitor and ligand of the AHR, represents a new alternative for immunomodulation. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0044547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kewley R.J., Whitelaw M.L., Chapman-Smith A. The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 167.Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., Takahashi J.S., Weitz C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 168.Kewley R.J., Whitelaw M.L., Chapman-Smith A. The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 169.Reppert S.M., Weaver D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 170.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Young M.W. Time travels: a 40-year journey from drosophila’s clock mutants to human circadian disorders (nobel lecture) Angew. Chem. Int. Ed. 2018;57(36):11532–11539. doi: 10.1002/anie.201803337. [DOI] [PubMed] [Google Scholar]
- 173.Hogenesch J.B., Chan W.K., Jackiw V.H., Brown R.C., Gu Y.Z., PrayGrant M., Perdew G.H., Bradfield C.A. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 1997;272(13):8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 174.Zhou Y.D., Barnard M., Tian H., Li X., Ring H.Z., Francke U., Shelton J., Richardson J., Russell D.W., McKnight S.L. Molecular characterization of two mammalian BHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. U. S. A. 1997;94(2):713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sun Z.S., Albrecht U., Zhuchenko O., Bailey J., Eichele G., Lee C.C. RIGUI, a putative mammalian ortholog of the drosophila period gene. Cell. 1997;90(6):1003–1011. doi: 10.1016/S0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 176.Shearman L.P., Zylka M.J., Weaver D.R., Kolakowski L.F., Reppert S.M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19(6):1261–1269. doi: 10.1016/S0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 177.Fan C.-M., Kuwana E., Bulfone A., Fletcher C.F., Copeland N.G., Jenkins N.A., Crews S., Martinez S., Puelles L., Rubenstein J.L.R., Tessier-Lavigne M. Expression patterns of two murine homologs of drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of down syndrome. Mol. Cell. Neurosci. 1996;7(1):1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- 178.Ema M., Morita M., Ikawa S., Tanaka M., Matsuda Y., Gotoh O., Saijoh Y., Fujii H., Hamada H., Kikuchi Y., Fujii-Kuriyama Y. Two new members of the murine sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell. Biol. 1996;16(10):5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Probst M.R., Fan C.-M., Tessier-Lavigne M., Hankinson O. Two murine homologs of the drosophila single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein. J. Biol. Chem. 1997;272(7):4451–4457. doi: 10.1074/jbc.272.7.4451. [DOI] [PubMed] [Google Scholar]
- 180.Bersten D.C., Sullivan A.E., Peet D.J., Whitelaw M.L. BHLH–PAS proteins in cancer. Nat. Rev. Cancer. 2013;13(12):827. doi: 10.1038/nrc3621. [DOI] [PubMed] [Google Scholar]
- 181.Gu Y.Z., Hogenesch J.B., Bradfield C.A. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 182.McIntosh B.E., Hogenesch J.B., Bradfield C.A. Mammalian per-arnt-sim proteins in environmental adaptation. Annu. Rev. Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 183.Zylka M.J., Shearman L.P., Levine J.D., Jin X., Weaver D.R., Reppert S.M. Molecular analysis of mammalian timeless. Neuron. 1998;21(5):1115–1122. doi: 10.1016/S0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]
- 184.Beischlag T.V., Wang S., Rose D.W., Torchia J., Reisz-Porszasz S., Muhammad K., Nelson W.E., Probst M.R., Rosenfeld M.G., Hankinson O. Recruitment of the NCoA/SRC-1/P160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 2002;22(12):4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gradin K., McGuire J., Wenger R.H., Kvietikova I., fhitelaw M.L., Toftgård R., Tora L., Gassmann M., Poellinger L. Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol. Cell. Biol. 1996;16(10):5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Anderson S., Crosson S., Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallogr. D Biol. Crystallogr. 2004;60(6):1008–1016. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 187.Miyatake H., Mukai M., Park S.-Y., Adachi S., Tamura K., Nakamura H., Nakamura K., Tsuchiya T., Iizuka T., Shiro Y. Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies 11 edited by K. Nagai. J. Mol. Biol. 2000;301(2):415–431. doi: 10.1006/jmbi.2000.3954. [DOI] [PubMed] [Google Scholar]
- 188.Sakurai S., Shimizu T., Ohto U. The crystal structure of the AhRR–ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription. J. Biol. Chem. 2017;292(43):17609–17616. doi: 10.1074/jbc.M117.812974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ikuta T., Eguchi H., Tachibana T., Yoneda Y., Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J. Biol. Chem. 1998;273(5):2895–2904. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- 190.Murray I.A., Patterson A.D., Perdew G.H. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer. 2014;14(12):801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.UniProt https://www.uniprot.org/ (Accessed 24 October 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.
Data will be made available on request.