Abstract
Background:
The measurement of plasma concentrations of retinol binding protein is a component of nutritional assessment in neonatal intensive care. However, serial testing in newborns is hampered by the limited amount of blood that can be sampled. Limitations are most severe with preterm infants, for whom close monitoring may be most important.
Methods:
We developed an assay to quantify retinol binding protein using trypsin digestion and liquid chromatography-tandem mass spectrometry, which requires a serum or plasma volume of 5 microliters. Additionally, we validated the method according to current recommendations and performed comparison with a standard nephelometry platform in clinical use.
Results:
The assay demonstrated linearity from below 1 mg/dL (0.48 μM) to more than 20 mg/dL (9.7 μM), and an imprecision of 11.8% at 0.43 mg/dL (0.21 μM). The distribution of results observed with the new method was different when compared with nephelometry.
Conclusion:
Liquid chromatography-tandem mass spectrometry facilitated testing a smaller sample volume, thereby increasing the ability to monitor key nutritional markers in premature infants. The differences in results compared with a commercially-available nephelometric assay revealed questionable results for lower concentrations by immunoassay.
Keywords: retinol binding protein, liquid chromatography-tandem mass spectrometry, LC-MS/MS, nutrition, vitamin A
1. INTRODUCTION
Nutritional status is a major determinant of long-term outcomes for neonates under intensive care [1]. Status is evaluated by combining anthropometry (assessments of body weight, length, and other indices) with laboratory testing, the latter for insight into the particular status of electrolytes, proteins, vitamins, and other components. Serial testing, however, is hampered by the volume of blood required for testing [2], particularly in preterm infants for whom close monitoring may be critical. Even assays traditionally considered “sensitive” (e.g., immunological, chromatographic) may require several hundred microliters or more of serum or plasma per analysis. Due to the potential harm of repeated surveillance blood collections, efforts are needed to reduce sample volume requirements.
Fortunately, liquid chromatography-tandem mass spectrometry (LC-MS/MS) is becoming more and more sensitive, which has enabled the interrogation of analytes from small fluid volumes in clinical laboratories [3–4]. Transitioning from conventional approaches may also provide other advantages, such as improved specificity [5] and shorter run times [6]. As a result, the application of LC-MS/MS is now routine for endocrinology, toxicology, therapeutic drug monitoring, among others [7]. Protein quantification by LC-MS/MS, however, is still considered an area for specialized laboratories, perhaps due to perceived challenges in sample preparation and peptide analysis. A more complete repository in the literature of the standard operating procedures that are currently in use may help other institutions take advantage of these technical innovations.
Here we present an LC-MS/MS method for quantitative measurement of plasma retinol binding protein (RBP), a marker of vitamin A sufficiency [8]. RBP is responsible for the transport of vitamin A (primarily as retinol) to tissues [9]. The RBP-vitamin A complex circulates bound to transthyretin (TTR), which prevents its glomerular filtration. In addition to preventing its excretion, vitamin A also stimulates RBP secretion from the liver [10]. Because RBP contains a single vitamin A binding site, under “normal” circumstances circulating RBP and vitamin A ratios approach a 1:1 ratio [11,12]. However, in cases of vitamin A deficiency the molar ratio of vitamin A to its binding protein can decrease, potentially indicating a need for vitamin A supplementation (suggested threshold ≤ 0.6 in the neonatal intensive care unit). Although it is a negative acute phase reactant, RBP is also a marker of hepatic synthetic function with a half-life 12 hours [13]. Conventional assays for RBP, as with those for direct measurement of vitamin A (e.g., HPLC), require several hundred microliters of sample volume per test, posing a challenge for users in neonatal intensive care. Our goal was to develop an assay using a minute sample volume and compare results with an existing nephelometry platform.
2. MATERIALS AND METHODS
2.1. Reagents
A complete list of chemicals and reagents used for sample preparation and LC-MS/MS are provided in the Standard Operating Procedure (see Supplementary Material). Purified RBP was purchased from Bio-Rad Laboratories (Hercules, CA). Concentration was determined by amino acid analysis performed at UC Davis (Molecular Structure Facility). Custom unlabeled and stable isotope-labeled RBP-specific tryptic peptides with sequences FSGTWYAMAK, YWGVASFLQK, and LIVHNGYC[+57]DGR were purchased from EZBiolab (Carmel, IN) and New England Peptide (Gardner, MA).
2.2. Peptide Selection and MS Acquisition Setup
Peptides for the quantification of RBP were chosen based on established principles [14–15]. In addition to factors such as peptide length, non-ideal amino acids, good precision and stability, selection criteria also included sequence alignment (via BLAST search) proteotypic to human but not with proteomes contained in diluent sera (e.g., chicken and/or salmon). The RBP amino acid sequence in FASTA format (UniProtKB database, entry P02753) was analyzed in Skyline-daily (v3.1.1.8980, University of Washington) [16]. Predicted RBP tryptic peptide precursor ion isolation lists and collision energies were generated for a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher) running Xcalibur™ Software. Purified recombinant RBP was digested with trypsin for 1 h (n= 3 replicates) and analyzed using nanoflow LC parallel reaction monitoring mass spectrometry to generate a spectral library in Panorama [17]. This was imported into Skyline-daily for peptide evaluation [14]. Observed peptides in tryptically digested male and female human sera measured in triplicate that met acceptability criteria of <5 ppm mass accuracy and dotp value for ion ratios >0.9 of spectral library peptide fragment ion intensities from purified recombinant RBP were further evaluated for preliminary peptide stability by re-analyzing these samples after 48 hours at 7 °C.
Optimal transitions and voltage settings for selected reaction monitoring (SRM) determined by direct infusion of RBP peptides (FSGTWYAMAK, YWGVASFLQK, and LIVHNGYC[+57]DGR) on a Waters Xevo TQ-S triple quadrupole mass spectrometer running MassLynx automated MS optimization software. The specificity of the selected peptides was confirmed within calibration material based on analysis of a double blank matrix (fish serum, no internal standard), according to CLSI C62-A criteria (i.e., background peaks absent or < 20% of the peak area for the analyte at the lower limit of measuring interval or ≤ 5% of the IS area at the expected retention time) [18].
2.3. Calibration, controls, and internal standards
An initial set of five-point external standard calibrators were generated by spiking AAA-analyzed purified human RBP protein into fish serum at 0.5, 2.3, 5.6, 7, and 10 mg/dL (0.24, 1.1, 2.7, 3.4, and 4.9 μmol/L). Molar concentrations are based on estimated molecular weight of 20618.07 Da (concentration in μmol/L equals concentration in mg/dL divided by 2.061807). Low and high controls consisted of low and high patient serum pools with target values of 1.0 mg/dL (low, 0.49 μmol/L) and 4.0 mg/dL (high, 1.9 μmol/L), also stored as 40–50 μL aliquots in 500 μL lo-bind snap-cap microcentrifuge tubes at − 20°C for up to 2 years. Calibrator and control aliquots were stored as 40–50 μL in 500 μL lo-bind snap-cap microcentrifuge tubes at − 20°C for up to 2 years. Internal standards (FSGTWYAMAK[13C6 15N2] and LIVHNGYC(+57)DGR[13C6 15N4]) were prepared from 2 mg/mL stock solutions of primary internal standards for the 3 peptides. A working internal standard mixture was prepared at 2–3 μM in solvent and additional calibrators were generated in pooled human serum, as described in the Standard Operating Procedure. New calibrators were assayed against current calibrators on at least 3 separate days to establish set points.
2.4. Sample preparation
Sample preparation is described in detail in the Standard Operating Procedure. In brief, samples (5 μL) were added to individual wells of a 96-well Deep Well Microplate and diluted in ammonium bicarbonate containing internal standard and dithiothreitol (DTT). Diluted samples were denatured with trifluoroethanol (TFE) and alkylated using iodoacetamide (IAA), then further diluted and subjected to trypsin digestion. Digestion was quenched using formic acid. A portion of each sample was further diluted into 2% acetonitrile/0.1% formic acid in water in a new microplate and loaded to the auto-sampler for analysis.
2.5. Liquid Chromatography-Mass Spectrometry.
Chromatography separation was performed on an ACQUITY UPLC system and an HSS T3 Column (10000C0035, 1.8 μm, 2.1 mm X 50 mm, equipped with a VanGuard Pre-column: 100Å, 1.8 μm, 2.1 mm X 5 mm) at 50°C. Solvents consisted of 97.9% Optima LC-MS grade water/2% DMSO/0.1% formic acid (Mobile Phase A) and 97.9% Optima LC-MS grade methanol/2%DMSO/0.1% formic acid (Mobile Phase B). Injection volume was 20 μL. Tandem mass spectrometry was performed on a Xevo TQ-S triple quadrupole mass spectrometer. LC-MS/MS setup, including transitions for MRM are described in the Standard Operating Procedure.
2.6. Assay Validation
Assay validation studies are detailed in the Supplementary Material. In brief, the suitability of commercial fish serum as a surrogate matrix for human serum was verified by analyzing human and fish sera before and after addition of equal quantities of purified RBP (3 spike levels). The assay was then characterized by identifying the analytical measurement interval, imprecision, interferences, matrix effects, ion suppression, carryover, sample types, stability (sample and peptide), method comparison (vs. nephelometry, Siemens ProSpec), and reference range. For studies involving residual human specimens, the Human Subjects Division of our institution has determined that the use of leftover, de-identified clinical samples for method development, method validation, and quality improvement is not considered human subjects research.
3. RESULTS
3.1. Assay Development
An LC-MS/MS method was deployed using 5 μL of human serum or plasma (30 μL minimum collection). Transitions for SRM were selected based on in silico tryptic digestion of RBP, identifying 11 peptides for inclusion in the isolation list. We excluded 3 of these 11 peptides based on high-resolution mass spectrometric analysis because the mass accuracy and/or ion ratios did not meet acceptability criteria. From the remaining 8 peptides, 3 were excluded due to digestion variability and stability. Of 5 left, one was excluded based on length (i.e., difficult to synthesize) and a second was excluded after double blank analysis revealed a peptide endogenous to fish serum that interfered with quantification. The 3 remaining peptides – FSGTWYAMAK (FSG), YWGVASFLQK (YWG), and LIVHNGYC[+57]DGR (LIV) -- were evaluated in subsequent experiments. YWG was ultimately excluded based on observed poor recovery in fish serum (data not shown). Absolute quantification was then determined as an average of the concentrations determined from FSG and LIV separately. Representative chromatograms for the FSG and YWG peptides appear in Figure 1.
Figure 1. Representative Chromatograms.
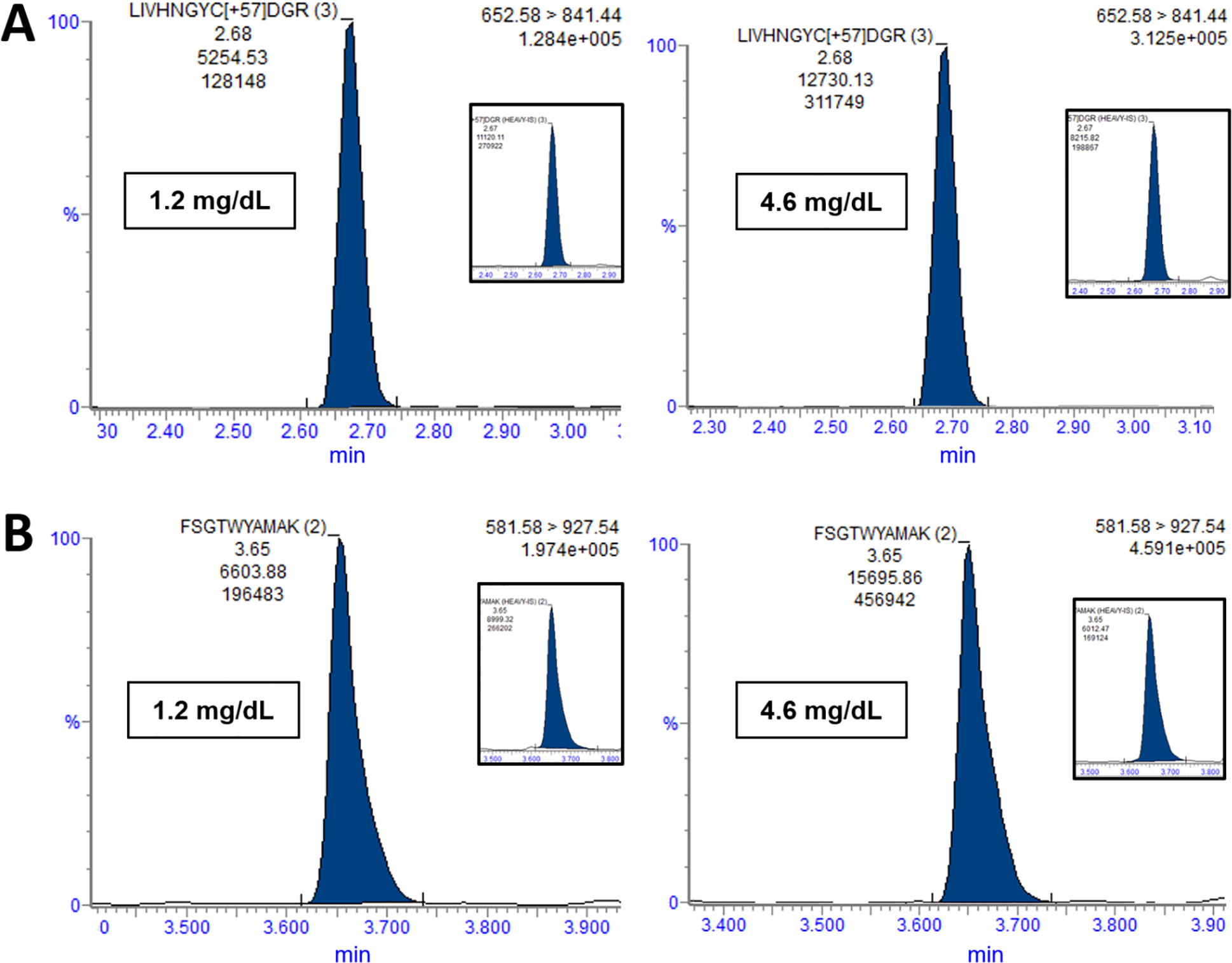
(A) Chromatograms normalized to 100% intensity are shown for the LIV peptide (+3 charge state) for two samples, including a single patient sample (1.2 mg/dL, 0.58 μmol/L), and a population pool (4.6 mg/dL, 2.2 μmol/L) comprising thousands of human sera. (B) Chromatograms are shown for the FSG peptide (+2 charge state). Each inset contains the chromatogram for the corresponding internal standard.
3.2. Suitability of Fish Serum as a Surrogate Calibration Matrix
Interference was observed in a putative negative control (chicken serum) despite no apparent overlap in chicken proteins with selected proteotypic peptides based on BLAST searching. Once further analysis determined that there was no similarity in peptide sequence when compared with fish RBP, we chose fish serum instead as a surrogate matrix for calibrator preparation in subsequent experiments. The average of selected proteotypic peptides, FSGTWYAMAK and LIVHNGYCDGR for RBP, demonstrated measured increments within ±20% between human and fish sera spiked with equal quantities of purified human RBP as shown in Supplemental Table 1 and Supplementary Figure 1.
3.3. Assay Characteristics
The assay was linear over the tested range (0.6 mg/dL to 20.5 mg /dL, or 0.29 μmol/L to 9.9 μmol/L, as shown in Figure 2 and Supplemental Table 2). Imprecision (%CV) was less than 15% for each sample tested at or below 0.5 mg/dL (0.24 μmol/L, Supplemental Table 3). Lower limit of quantitation (or lower limit of the measuring interval, LLMI) was thus set to 0.5 mg/dL. Limit of detection (LOD) was 0.08 mg/dL (0.04 μmol/L, Supplemental Table 4). Imprecision was further investigated in a separate 5×5 repeatability study, consisting of five replicates on each of five days (n = 25, Supplemental Table 5). When calculating the RBP result based on the average of both peptides, %CV was ≤15% at all levels. In interference testing, protein (up to 12 mg/dL), triglycerides (up to 1000 mg/dL), and bilirubin (up to 20 mg/dL) each introduced bias within ±5% (summarized in Supplemental Table 6). However, hemolysis caused a negative, proportional bias, which was more pronounced for the LIV peptide than for FSG (further detailed in Supplemental Tables 7 and 8). Mixing studies demonstrated adequate linearity as shown in Supplemental Table 9 and Supplemental Figure 2. T-infusion showed no appreciable ion suppression (Supplemental Figure 3). Carryover studies indicated that there was <0.4% between-sample contamination (Supplemental Table 10). Of note, at physiologically-relevant concentrations, carryover using this method would be well below the method LLMI. In a tube-type study, sera collected into gold top serum separator tube and plasma collected into Li-heparin-anticoagulated serum separator tubes differed by <10% from sera collected with no gel or preservatives (Supplemental Table 11). However, plasma collected into EDTA-anticoagulated tubes exhibited 14 to 21% under-recovery (Supplemental Table 12). Addition of calcium to the samples appeared to rescue the effect, but was not validated as a modification of the assay. Sample stability studies demonstrated bias within +/−20% for most conditions tested (Supplemental Table 13). Stability testing of peptide digests demonstrated +/−10% bias for most conditions evaluated, suggesting that the digests are stable for reinjection (Supplemental Table 14). For a set of 200 serum samples from an ambulatory pediatric population (ages 12 months to 16 years), the 95th percentile reference interval for the group of was determined to be 1.9–5.3 mg/dL (0.92 to 2.6 μmol/L) using a non-parametric percentile method (Supplemental Table 15 and Supplemental Figure 4).
Figure 2. Linear Range.
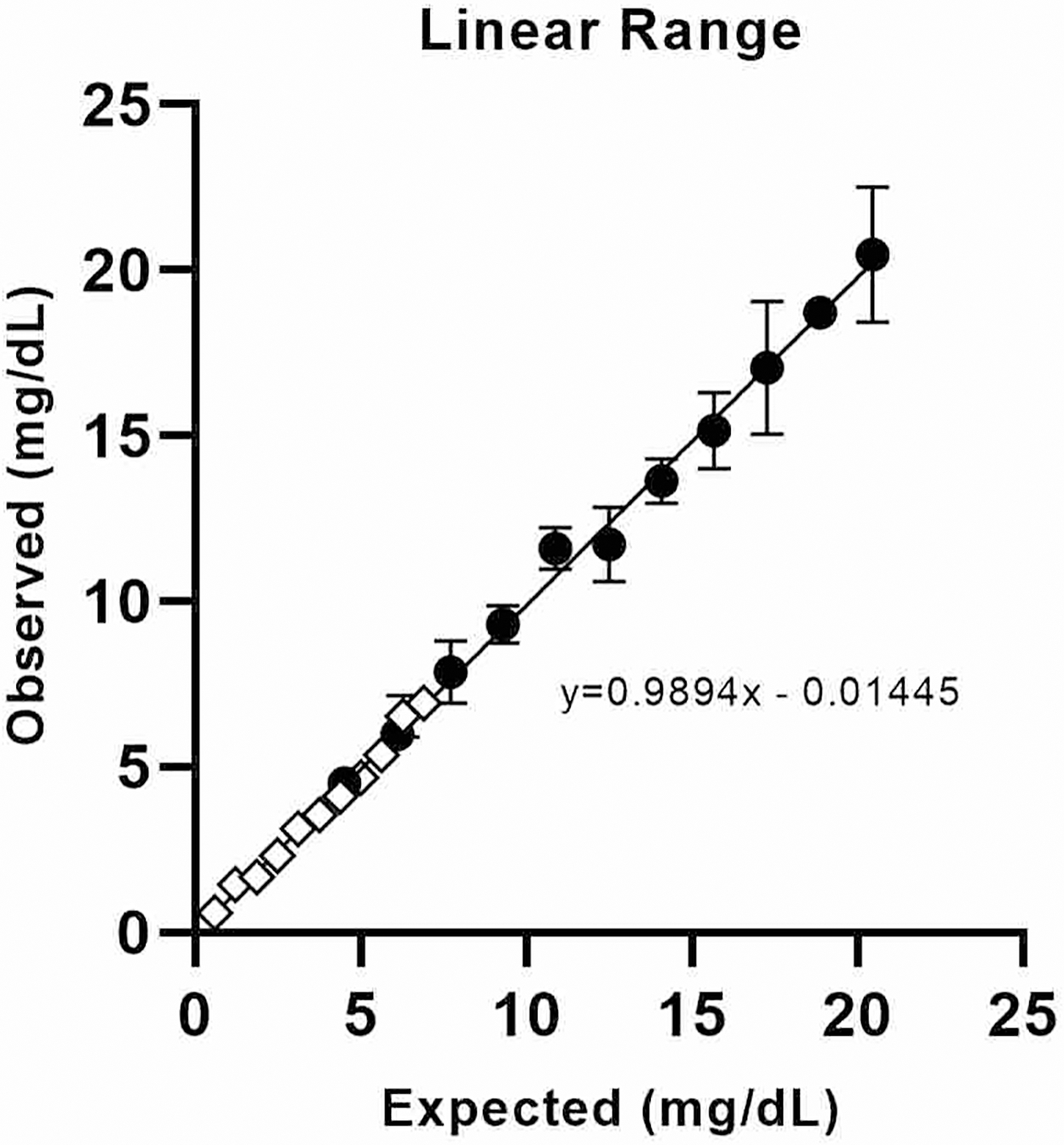
The analytical measurement interval was assessed using two overlapping mixing studies, a high range set (circles) and a low range set (diamonds). Concentrations in mg/dL can be estimated in μmol/L by dividing by 2.061807.
3.4. Comparison with Nephelometry Platform
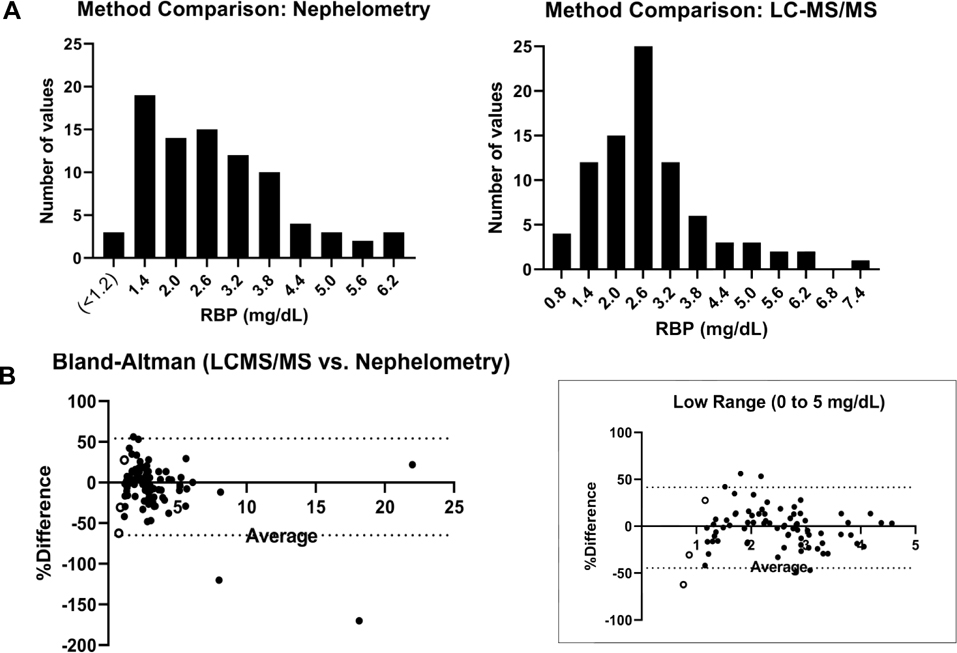
Complete results from the method comparison (n = 89) appear in Supplemental Table 16. Two results were predicted to be falsely elevated by immunoassay (likely secondary to lipemia), which estimated the RBP concentrations of 33.6 and 12.8 mg/dL (16 and 6.2 μmol/L) compared to the LCMS estimations of 2.7 and 3.2 mg/dL (1.3 and 1.6 μmol/L), respectively. The methods correlated with a Pearson R 0.96 when excluding the two extreme outliers and defaulting values less than 1.2 mg/dL (0.58 μmol/L) by nephelometry to 1.0 mg/dL (0.49 μmol/L). However, the distributions of results differed overall (as portrayed in Figure 3A), with the nephelometric assay showing an asymmetric distribution with a sharp drop off in results at the lower tail. The LC-MS/MS assay showed a more symmetric result distribution in analysis of the same samples. A Bland-Altman plot is provided in Figure 3B.
Figure 3. Method Comparison.
(A) Result distributions are shown comparing results less than 8.0 mg/dL (3.9 μmol/L) across the nephelometry and LC-MS/MS platforms (bin width 0.6 mg/dL, or 0.29 μmol/L). (B) Bland-Altman analysis is shown, comparing the LC-MS/MS method relative to nephelometry (calculation: (100*(LC-MS/MS - Nephelometry)/average) vs. average.). The left plot includes the two high outliers on the immunoassay platform and both plots contain the 3 nephelometry results reported as <1.2 mg/dL (< 0.58 μmol/L, open circles), defaulted to 1.0 mg/dL to allow plotting. Concentrations in mg/dL can be estimated in μmol/L by dividing by 2.061807.
DISCUSSION
Despite the growing presence of LC-MS/MS in clinical laboratories, its use for protein quantification remains limited [7]. This is reflected in the most longstanding guidelines (CLSI C62-A) for the design, development, and performance verification of LC-MS/MS-based assays, which are focused primarily on the analysis of small molecules using triple quadrupole instruments [18,20]. Other applications and instrument configurations will require different considerations. More specifically, for the measurement of proteins, the generation and monitoring of peptides introduces additional sources of variability that must be carefully addressed. Fortunately, as new clinical methods are becoming more common in the literature, a broader consensus is developing, which will lead to greater harmonization [21–22]. We have presented a novel LC-MS/MS method for the quantitative measurement of RBP. Sample preparation was straightforward, not requiring immunoaffinity enrichment to achieve adequate sensitivity [23–24]. Based on a sample size of only 5 μL, the method offers more appropriate testing for premature infants and neonates.
Development of this assay demonstrated potential pitfalls that could be encountered when making quantitative measurements using proteotypic peptides. In particular, the selection of peptides when designing transitions cannot be based on amino acid sequence and length alone. Because peptides show unpredictable differences in precision, recovery, and interference, they must be selected empirically. For example, fish serum was selected as a diluent for our calibration matrix, but it demonstrated low level interference with one of candidate proteotypic peptides. Another peptide (LIV) was specifically affected by hemolysis. Adverse findings may not invalidate the use of a particular peptide, but these complications must be documented for troubleshooting in the future (particularly when comparing against other LC-MS/MS-based approaches). Of note, in selecting peptides for SRM, those containing modifications are often avoided due to concerns for variability in reduction and alkylation. Here, a peptide (LIV) was used, which contained an acetylated cysteine and demonstrated good performance during validation. This suggests that acetylation of cysteines can be consistent enough for quantitative analyses and thus, Cys-containing peptides should not necessarily be excluded from selection during SRM method development a priori.
One of the most important findings in our evaluation of the method was the difference in distribution of results when we compared the new method to a nephelometric platform. While the distribution of results by LC-MS/MS was somewhat skewed to the right, the distribution of results from nephelometry appeared to simply drop off near the low end. This cannot be explained by the stated limit of quantitation of the assay (1.2 mg/dL, 0.58 μmol/L), since only 3 results were apparent as less than 1.2 mg/dL by nephelometry. It is unknown what the distribution of RBP results should be for the population tested and we cannot be certain that the measurements by LC-MS/MS are more accurate on the low end based only on the data obtained during our validation; however, a more normal distribution might be expected for a moderately-high abundance vitamin-binding protein [14]. Our findings by nephelometry are consistent with a paper by Kanakoudi et al. [25], who had previously reported almost two decades ago on the concentrations of 10 proteins (including RBP) in term and preterm infants. Based on nephelometry results, these authors noted that most of the proteins studied did not show a Gaussian-like distribution in infants whereas this pattern reversed in adults. Notably, however, the concentrations of the proteins studied increased with age, which may have affected the appearance of the distributions if accuracy and precision of the assay differed at lower and higher concentrations. This may be a subject for future investigation as more users bring protein mass spectrometry online in clinical laboratories.
In addition to the potential errors seen with nephelometry at the low end of the normal range, other issues with immunoassays may contribute to the irreproducibility of the results of clinical research [26–27] and must be considered when assessing whether the methods we employ accurately reflect biology. For example, unanticipated effects of antibody specificity led to a severely flawed explanation for observed differences in plasma concentrations of 25-hydroxyvitamin D between races [28–29]. Mass spectrometry has helped provide clarity in this and other cases [30–31]. This may be due in part to the necessary emphasis on the careful definition of the measurand when deploying LC-MS/MS-based assays. For instance, the application of LC-MS/MS to hemoglobin A1c (HbA1c) measurement ultimately gave rise to accuracy-based HbA1c testing, focusing on a specific, unambiguous definition for the analyte. This was not previously achievable on other analyzers given the underlying heterogeneity contained within the A1c peak observed in ion-exchange chromatography [32].
With respect to irreproducibility of results in clinical studies, RBP is no stranger to controversy that may be method driven [33]. In addition to being a nutritional biomarker, RBP has been proposed as a causative factor in insulin resistance and type 2 diabetes mellitus [34–35]. Specifically, circulating RBP has been proposed to selectively reduce GLUT4 glucose transport in adipose tissue, which appears to underlie peripheral insulin resistance. However, studies have variably supported [36–37] and refuted [38–40] the relationship between RBP and insulin resistance in humans, and overall the precise relationship remains unclear [41]. Importantly, the methods have relied on antibody-based technologies. Whether differences in the applied methods can explain the different findings remains to be seen and the introduction of LC-MS/MS-based approaches may help.
5. CONCLUSION
This new method reduces the sample volume need for laboratory-based nutritional assessment in premature and young infants, which was made possible through the application of LC-MS/MS. This study can be used as a model for clinical laboratories interested in developing protein LC-MS/MS assays and may provide a technique by which the relationship between RBP and insulin resistance could be further explored.
Supplementary Material
6. ACKNOWLEDGEMENTS
This work was partially funded by the UW Nutrition Obesity Research Center P30 DK035816 and UW Diabetes Research Center P30 DK017047. Minh D. Hoang (University of Washington) provided experimental data.
7. DECLARATIONS
Our laboratory has received grant and equipment support from Waters, Inc.
Abbreviations:
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Pereira-da-Silva L, Virella D, Fusch C, Nutritional Assessment in Preterm Infants: A Practical Approach in the NICU., Nutrients 11 (2019). 10.3390/nu11091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Madan A, Kumar R, Adams MM, Benitz WE, Geaghan SM, Widness JA, Reduction in Red Blood Cell Transfusions Using a Bedside Analyzer in Extremely Low Birth Weight Infants, J. Perinatol 25 (2005) 21–25. 10.1038/sj.jp.7211201. [DOI] [PubMed] [Google Scholar]
- [3].Saito J, Tanzawa A, Kojo Y, Maruyama H, Isayama T, Shoji K, Ito Y, Yamatani A, A sensitive method for analyzing fluconazole in extremely small volumes of neonatal serum, J. Pharm. Heal. Care Sci 6 (2020) 14. 10.1186/s40780-020-00170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kjellin LL, Dorsey G, Rosenthal PJ, Aweeka F, Huang L, Determination of the antimalarial drug piperaquine in small volume pediatric plasma samples by LC–MS/MS, Bioanalysis 6 (2014) 3081–3089. 10.4155/bio.14.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Conklin SE, Knezevic CE, Advancements in the gold standard: Measuring steroid sex hormones by mass spectrometry, Clin. Biochem 82 (2020) 21–32. 10.1016/j.clinbiochem.2020.03.008. [DOI] [PubMed] [Google Scholar]
- [6].Phipps WS, Crossley E, Boriack R, Jones PM, Patel K, Quantitative amino acid analysis by liquid chromatography-tandem mass spectrometry using low cost derivatization and an automated liquid handler., JIMD Rep 51 (2020) 62–69. 10.1002/jmd2.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Seger C, Salzmann L, After another decade: LC–MS/MS became routine in clinical diagnostics, Clin. Biochem 82 (2020) 2–11. 10.1016/j.clinbiochem.2020.03.004. [DOI] [PubMed] [Google Scholar]
- [8].Baeten JM, Richardson BA, Bankson DD, Wener MH, Kreiss JK, Lavreys L, Mandaliya K, Bwayo JJ, McClelland RS, Use of serum retinol-binding protein for prediction of vitamin A deficiency: effects of HIV-1 infection, protein malnutrition, and the acute phase response., Am. J. Clin. Nutr 79 (2004) 218–25. 10.1093/ajcn/79.2.218. [DOI] [PubMed] [Google Scholar]
- [9].Li Y, Wongsiriroj N, Blaner WS, The multifaceted nature of retinoid transport and metabolism., Hepatobiliary Surg. Nutr 3 (2014) 126–39. 10.3978/j.issn.2304-3881.2014.05.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Raghu P, Sivakumar B, Interactions amongst plasma retinol-binding protein, transthyretin and their ligands: implications in vitamin A homeostasis and transthyretin amyloidosis., Biochim. Biophys. Acta 1703 (2004) 1–9. 10.1016/j.bbapap.2004.09.023. [DOI] [PubMed] [Google Scholar]
- [11].Smith FR, Goodman DS, Vitamin A Transport in Human Vitamin A Toxicity, N. Engl. J. Med 294 (1976) 805–808. 10.1056/NEJM197604082941503. [DOI] [PubMed] [Google Scholar]
- [12].Lipkin AC, Lenssen P, Hypervitaminosis A in Pediatric Hematopoietic Stem Cell Patients Requiring Renal Replacement Therapy, Nutr. Clin. Pract 23 (2008) 621–629. 10.1177/0884533608327082. [DOI] [PubMed] [Google Scholar]
- [13].Siminkovitch S, Vladimirov B, Prealbumin and Retinol Binding Protein as Screening Tools for Malnutrition, in: Handb. Famine, Starvation, Nutr. Deprivation, Springer International Publishing, Cham, 2017: pp. 1–21. 10.1007/978-3-319-40007-5_54-1. [DOI] [Google Scholar]
- [14].Henderson CM, Lutsey PL, Misialek JR, Laha TJ, Selvin E, Eckfeldt JH, Hoofnagle AN, Measurement by a Novel LC-MS/MS Methodology Reveals Similar Serum Concentrations of Vitamin D-Binding Protein in Blacks and Whites., Clin. Chem 62 (2016) 179–87. 10.1373/clinchem.2015.244541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim H, Sohn A, Yeo I, Yu SJ, Yoon J-H, Kim Y, Clinical Assay for AFP-L3 by Using Multiple Reaction Monitoring–Mass Spectrometry for Diagnosing Hepatocellular Carcinoma, Clin. Chem 64 (2018) 1230–1238. 10.1373/clinchem.2018.289702. [DOI] [PubMed] [Google Scholar]
- [16].MacLean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, Carr SA, MacCoss MJ, Effect of Collision Energy Optimization on the Measurement of Peptides by Selected Reaction Monitoring (SRM) Mass Spectrometry, Anal. Chem. 82 (2010) 10116–10124. 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sharma V, Eckels J, Taylor GK, Shulman NJ, Stergachis AB, Joyner SA, Yan P, Whiteaker JR, Halusa GN, Schilling B, Gibson BW, Colangelo CM, Paulovich AG, Carr SA, Jaffe JD, MacCoss MJ, MacLean B, Panorama: a targeted proteomics knowledge base., J. Proteome Res 13 (2014) 4205–10. 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].CLSI. Liquid Chromatography-Mass Spectrometry Methods. 1st ed. CLSI guideline C62. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- [19].Armbruster DA, Pry T, Limit of blank, limit of detection and limit of quantitation., Clin. Biochem. Rev 29 Suppl 1 (2008) S49–52. [PMC free article] [PubMed] [Google Scholar]
- [20].Lynch KL, CLSI C62-A: A New Standard for Clinical Mass Spectrometry, Clin. Chem 62 (2016) 24–29. 10.1373/clinchem.2015.238626. [DOI] [PubMed] [Google Scholar]
- [21].Carr SA, Abbatiello SE, Ackermann BL, Borchers C, Domon B, Deutsch EW, Grant RP, Hoofnagle AN, Hüttenhain R, Koomen JM, Liebler DC, Liu T, MacLean B, Mani DR, Mansfield E, Neubert H, Paulovich AG, Reiter L, Vitek O, Aebersold R, Anderson L, Bethem R, Blonder J, Boja E, Botelho J, Boyne M, Bradshaw RA, Burlingame AL, Chan D, Keshishian H, Kuhn E, Kinsinger C, Lee JSH, Lee S-W, Moritz R, Oses-Prieto J, Rifai N, Ritchie J, Rodriguez H, Srinivas PR, Townsend RR, Van Eyk J, Whiteley G, Wiita A, Weintraub S, Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach., Mol. Cell. Proteomics 13 (2014) 907–17. 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].CLSI. Quantitative Measurement of Proteins and Peptides by Mass Spectrometry. 1st ed. CLSI guideline C64. Wayne, PA: Clinical and Laboratory Standards Institute; 2021. [Google Scholar]
- [23].Grant RP, Hoofnagle AN, From lost in translation to paradise found: Enabling protein biomarker method transfer by mass spectrometry, Clin. Chem 60 (2014) 941–944. 10.1373/clinchem.2014.224840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Neubert H, Shuford CM, V Olah T, Garofolo F, Schultz GA, Jones BR, Amaravadi L, Laterza OF, Xu K, Ackermann BL, Protein Biomarker Quantification by Immunoaffinity Liquid Chromatography–Tandem Mass Spectrometry: Current State and Future Vision, Clin. Chem 66 (2020) 282–301. 10.1093/clinchem/hvz022. [DOI] [PubMed] [Google Scholar]
- [25].Kanakoudi F, Drossou V, Tzimouli V, Diamanti E, Konstantinidis T, Germenis A, Kremenopoulos G, Serum concentrations of 10 acute-phase proteins in healthy term and preterm infants from birth to age 6 months., Clin. Chem 41 (1995) 605–8. http://www.ncbi.nlm.nih.gov/pubmed/7536645. [PubMed] [Google Scholar]
- [26].Begley CG, Ellis LM, Raise standards for preclinical cancer research, Nature. 483 (2012) 531–533. 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- [27].Picha B, Thompson M, Vondriska TM, Keep “reproducibility” in context, Nature. 485 (2012) 41–41. 10.1038/485041d. [DOI] [PubMed] [Google Scholar]
- [28].Hollis BW, Bikle DD, Vitamin D–Binding Protein and Vitamin D in Blacks and Whites, N. Engl. J. Med 370 (2014) 878–881. 10.1056/NEJMc1315850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hoofnagle AN, Eckfeldt JH, Lutsey PL, Vitamin D-Binding Protein Concentrations Quantified by Mass Spectrometry., N. Engl. J. Med 373 (2015) 1480–2. 10.1056/NEJMc1502602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hoofnagle AN, Wener MH, The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry., J. Immunol. Methods 347 (2009) 3–11. 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu H, Wong L, Yong S, Liu Q, Lee TK, Achieving comparability with IFCC reference method for the measurement of hemoglobin A1c by use of an improved isotope-dilution mass spectrometry method, Anal. Bioanal. Chem 407 (2015) 7579–7587. 10.1007/s00216-015-8961-2. [DOI] [PubMed] [Google Scholar]
- [32].Peterson KP, Pavlovich JG, Goldstein D, Little R, England J, Peterson CM, What is hemoglobin A1c? An analysis of glycated hemoglobins by electrospray ionization mass spectrometry., Clin. Chem. 44 (1998) 1951–1958. [PubMed] [Google Scholar]
- [33].Graham TE, Wason CJ, Blüher M, Kahn BB, Shortcomings in methodology complicate measurements of serum retinol binding protein (RBP4) in insulin-resistant human subjects., Diabetologia 50 (2007) 814–23. 10.1007/s00125-006-0557-0. [DOI] [PubMed] [Google Scholar]
- [34].Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB, Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes., Nature 436 (2005) 356–62. 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- [35].Graham TE, Yang Q, Blüher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson P-A, Smith U, Kahn BB, Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects., N. Engl. J. Med 354 (2006) 2552–63. 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- [36].Cho YM, Youn B-S, Lee H, Lee N, Min S-S, Kwak SH, Lee HK, Park KS, Plasma retinol-binding protein-4 concentrations are elevated in human subjects with impaired glucose tolerance and type 2 diabetes., Diabetes Care 29 (2006) 2457–61. 10.2337/dc06-0360. [DOI] [PubMed] [Google Scholar]
- [37].Kowalska I, Straczkowski M, Adamska A, Nikolajuk A, Karczewska-Kupczewska M, Otziomek E, Górska M, Serum retinol binding protein 4 is related to insulin resistance and nonoxidative glucose metabolism in lean and obese women with normal glucose tolerance., J. Clin. Endocrinol. Metab 93 (2008) 2786–9. 10.1210/jc.2008-0077. [DOI] [PubMed] [Google Scholar]
- [38].Ulgen F, Herder C, Kühn MC, Willenberg HS, Schott M, Scherbaum WA, Schinner S, Association of serum levels of retinol-binding protein 4 with male sex but not with insulin resistance in obese patients., Arch. Physiol. Biochem 116 (2010) 57–62. 10.3109/13813451003631421. [DOI] [PubMed] [Google Scholar]
- [39].Gómez-Ambrosi J, Rodríguez A, Catalán V, Ramírez B, Silva C, Rotellar F, Gil MJ, Salvador J, Frühbeck G, Serum retinol-binding protein 4 is not increased in obesity or obesity-associated type 2 diabetes mellitus, but is reduced after relevant reductions in body fat following gastric bypass., Clin. Endocrinol. (Oxf) 69 (2008) 208–15. 10.1111/j.1365-2265.2007.03156.x. [DOI] [PubMed] [Google Scholar]
- [40].Lewis JG, Shand BI, Frampton CM, Elder PA, Scott RS, Plasma retinol-binding protein is not a marker of insulin resistance in overweight subjects: a three year longitudinal study., Clin. Biochem 41 (2008) 1034–8. 10.1016/j.clinbiochem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- [41].Lee J-W, Im J-A, Park KD, Lee H-R, Shim J-Y, Lee D-C, Retinol binding protein 4 and insulin resistance in apparently healthy elderly subjects., Clin. Chim. Acta 400 (2009) 30–2. 10.1016/j.cca.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.