Abstract
At the midblastula transition, the Xenopus laevis embryonic cell cycle is remodeled from rapid alternations between S and M phases to become the complex adult cell cycle. Cell cycle remodeling occurs after zygotic transcription initiates and is accompanied by terminal downregulation of maternal cyclins A1 and B2. We report here that the disappearance of both cyclin A1 and B2 proteins is preceded by the rapid deadenylation of their mRNAs. A specific mechanism triggers this deadenylation. This mechanism depends upon discrete regions of the 3′ untranslated regions and requires zygotic transcription. Together, these results strongly suggest that zygote-dependent deadenylation of cyclin A1 and cyclin B2 mRNAs is responsible for the downregulation of these proteins. These studies also raise the possibility that zygotic control of maternal cyclins plays a role in establishing the adult cell cycle.
In Xenopus laevis, the first 12 cell cycles following fertilization occur in the absence of transcription. Maternal mRNAs and proteins that are synthesized and stored in the growing oocyte control these early embryonic cell divisions. Zygotic transcription begins upon completion of the 12th cell cycle and, in Xenopus, is referred to as the midblastula transition (MBT) (25). Prior to the MBT, gene expression is controlled by posttranscriptional mechanisms including regulation of the adenylation state of maternal mRNAs. Polyadenylated mRNAs are recruited into polysomes and translated, while deadenylated mRNAs are released from polysomes and translationally silenced (27).
In Xenopus, adenylation control regulates both progression through meiosis (oocyte maturation) and the transition from the meiotic to the mitotic cell cycle by regulating levels of cell cycle proteins, including cyclins. The mRNAs encoding cyclins A1, B1, B2, and E1 are synthesized during oogenesis and stored untranslated as poly(A)− mRNAs until oocyte maturation (5, 35). In response to progesterone, cytoplasmic polyadenylation elements (CPEs) in the 3′ untranslated region (UTR) trigger polyadenylation and translational activation of these mRNAs, allowing accumulation of cyclin protein and progression through meiosis. After fertilization, cyclin mRNAs are further adenylated and continuously translated (35). However, periodic degradation of cyclin A1, B1, and B2 proteins during each cell cycle allows progression through the first 12 cell divisions (18).
The 12th cell division is completed approximately 6 h postfertilization (p.f.) and is followed by remodeling of the cell cycle between cell cycles 13 and 15 (24). The cell cycle is remodeled from a rapid alternation between DNA synthesis and mitosis to become a cell cycle containing gap phases and checkpoint controls. The mechanism of cell cycle remodeling is not well understood, but it is accompanied by temporally specific degradation of maternal cyclin A1, B2, and E1 proteins (14, 33). We define this event as the terminal disappearance of the maternal cyclin protein.
Because cyclins A1 and B2 are normally degraded during each cell cycle through the ubiquitin-proteasome degradation pathway (7), we hypothesized that translational silencing of their mRNAs via deadenylation leads to the terminal disappearance of these proteins at the time of cell cycle remodeling. In this report, we show that the disappearance of these cyclins is preceded by deadenylation of their mRNAs. Deadenylation is mediated by sequence elements in the 3′ UTRs of these maternal mRNAs and requires zygotic transcription. Our results show that deadenylation of cyclins correlates with cell cycle remodeling, strongly suggesting that deadenylation regulates the early embryonic cell cycle and initiates cell cycle remodeling. This is also the first report of zygotic control of maternal gene expression in Xenopus.
MATERIALS AND METHODS
Chimeric genes and in vitro transcription.
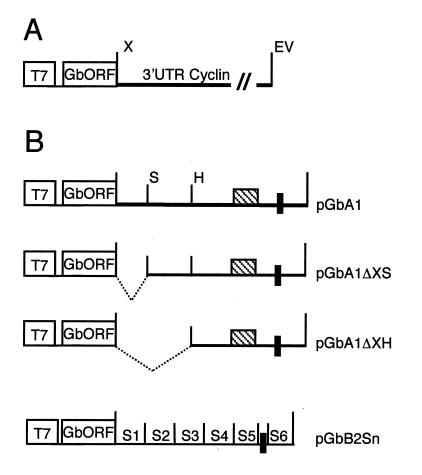
Chimeric genes pGbA1, pGbB1, pGbB2, and pGbE1 were constructed by inserting the 3′ UTR of the indicated cyclin mRNA downstream of the β-globin coding sequence in the plasmid pGbORF/mosEDEN (1). Cyclin 3′ UTRs were generated by PCR, and the XbaI and EcoRV sites were added using the combination of primers and DNA templates shown in Table 1. The vector pGbORF/mosEDEN was digested with XbaI and EcoRV, and a cyclin 3′ UTR was inserted between these sites, replacing mosEDEN (Fig. 1A).
TABLE 1.
Isolation of cyclin 3′ UTRs by PCR
| mRNA | Primer | Accession no. |
|---|---|---|
| Cyclin A1 | 5′ctcatctagaagccttccagagtggacg | X53745a |
| 3′gttcgatatcgagtaaagtcagtttattaaaaacac | ||
| Cyclin B1 | 5′ctcatctagaaggactacgtggcattcc | J03166b |
| 3′gttcgatatccatgttaaaatgagctttattaaaacc | ||
| Cyclin B2 | 5′ctcatctagaaactgttaagtgaccctttcaaagag | J03167b |
| 3′gttcgatatcaaataaaaaatgtgaagttttatttcatactg | ||
| Cyclin E1 | 5′ctcatctagacagtgctttaactctgtgcatcac | Z13966c |
| 3′gttcgatatcaaaaaaaacagctgtcttctaaacagc |
FIG. 1.
Structure of chimeric genes. (A) General structure of the pGb-cyclin 3′-UTR genes. The T7 promoter (T7) and the restriction sites XbaI (X) and EcoRV (EV) are indicated. The β-globin 5′-UTR, open reading frame, and partial globin 3′-UTR sequences are indicated (GbORF). The cyclin 3′ UTRs are complete and contain the hexanucleotide AATAAA. (B) Structure of the mutagenized chimeric genes. The black box represents the nuclear polyadenylation signal, and the striped box corresponds to the CPE. The restriction endonuclease recognition sites used in the deletion analysis of pGbA1 are indicated (S = SpeI; H = HpaI). For pGbB2Sn, mutated sequences S1 to S6 are indicated and have been replaced by the sequence indicated in Materials and Methods.
Chimeric gene pGbA1ΔXS was constructed by deleting the XbaI/SpeI fragment from pGbA1 and recircularizing the vector. Similarly, pGbA1ΔXH was generated by deletion of the XbaI/HpaI fragment from pGbA1. The XbaI and HpaI sites were blunt ended, and the vector was recircularized (Fig. 1B).
Substitution mutants of the GbB2 construct were made using PCR-based linker scanning mutagenesis (3). Six substitution mutants of 30 nucleotides (nt), each covering the complete 180-nt cyclin B2 3′ UTR in the context of pGbB2, were constructed. The deleted sequences were replaced by the sequence GATCTGAGTCTCTAAGCTAGCTAATAC. The nuclear polyadenylation signal or hexanucleotide (AAUAAA) was not mutated, as it is required for cytoplasmic polyadenylation (8). The two CPEs were mutated one at a time. The remaining CPE should still mediate polyadenylation of the injected mRNA (Fig. 1B). The pGbORF/mosEDEN chimeric gene was provided by H. B. Osborne (Universite de Rennes I, Rennes, France) (29).
For in vitro transcription, plasmids were linearized with EcoRV. 32P-labeled, capped mRNAs were synthesized according to the recommendations of the manufacturer (Stratagene).
Embryos and microinjections.
Embryos were obtained according to standard protocols (16) and maintained in 0.1× Marc's modified Ringer's medium at 23°C. For Western blot analysis, five embryos were collected at the times specified, and protein was extracted as described previously (14). For mRNA microinjections, two-cell embryos were injected with 18.4 nl of in vitro-transcribed capped mRNA in water (50,000 cpm/μl; 0.25 to 1 fmol/μl), with the addition of 50 ng of α-amanitin per embryo when indicated. For adenylation, Northern blotting, and poly(A) tail analyses, five embryos were collected at the specified times and processed for RNA extraction as described previously (13) or by following the TRIZOL protocol (Gibco-BRL). The cell cycle stages of the embryos were determined according to the work of Nieuwkoop and Faber (26). In addition, where indicated, the detection of the zygote-specific transcript GS17 (20) was performed.
Immunoanalyses.
One embryo equivalent was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes, which were successively probed with each of the indicated antibodies as described previously (15). Bands were visualized by enhanced chemiluminescence. Polyclonal antibodies to Xenopus cyclins were kindly provided by J. L. Maller (University of Colorado Health Sciences Center, Denver).
Poly(A) tail analysis.
Total RNA was extracted from embryos injected with radiolabeled, capped chimeric mRNAs and resolved by electrophoresis on 4% polyacrylamide-urea gels as described previously (2). Fixed and dried gels were analyzed by autoradiography or phosphorimaging (Molecular Dynamics). The number of replicate experiments performed is indicated in the figure legends. For quantification, poly(A)− mRNA was defined as the radioactive signal present in a window ranging from A0 to A20 as defined by the RNA size markers. The total signal was defined as the signal ranging from A0 to the top of the signal.
The ligation-mediated poly(A) test (PAT) was performed as described previously (34). Reverse transcription was performed using oligo(dT) and an oligo(dT) anchor (5′GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT). PCR was performed on the resulting cDNA with the oligo(dT) anchor and a cyclin 3′-UTR-specific primer: cyclin A1, 5′CTCATCTAGAAGCCTTCCAGAGTGGACG; cyclin B2, 5′CCTGGGGTACCACTCAGCACTATTAC; and cyclin E1, 5′TAAGGTGACAGAGTTACAAGGGTG. PCR conditions for cyclin A1 were one cycle of 94°C for 180 s, 49°C for 60 s, and 72°C for 90 s; 35 cycles of 94°C for 60 s, 49°C for 60 s, and 72°C for 90 s; and 72°C for 7 min. PCR conditions were similar for all other primers except for changing the annealing temperature (cyclin B2, 53°C, and cyclin E1, 47°C).
PCR products were resolved on 2% Nusieve (FMC) or low-melting-point (Gibco-BRL) agarose gels and visualized by ethidium bromide staining. The specificity of PCR amplification was verified by restriction endonuclease digestion using AvaII for the cyclin A1 product, NheI for the cyclin B2 product, and DraIII for the cyclin E1 product.
RESULTS
The disappearance of cyclin A1 and B2 proteins is zygote dependent.
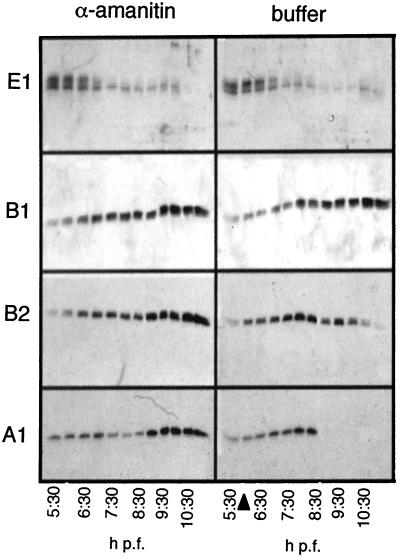
Cyclins A1 and B2 disappear after the initiation of transcription at the MBT (15), suggesting that their disappearance may be mediated by zygotically expressed genes. To determine if zygotic transcription is necessary for cyclin disappearance, we injected one-cell embryos with the transcriptional inhibitor α-amanitin. Injected embryos were sampled every 30 min between 5 and 11 h p.f., and cyclin levels were assessed by Western blot analysis. The same membrane was probed successively with specific antibodies against cyclins A1, B1, B2, and E1. Inhibition of transcription was assayed by Northern blot analysis for the zygote-specific transcript GS17 (20). As shown previously (15), the majority of cyclin E1 disappeared by 7 h p.f. (stage 9), independent of the activation of zygotic transcription (Fig. 2). In contrast (Fig. 2, lower panels), the degradation of cyclins A1 and B2 strictly depended upon zygotic transcription. In control embryos, cyclin A1 was degraded after 8.5 h (stage 9.5/10), whereas in α-amanitin-injected embryos, it accumulated until at least 11 h p.f. Likewise, cyclin B2, which normally disappears approximately 10 h p.f., also accumulated until at least 11 h p.f. in the presence of α-amanitin. Cyclin B1 was detected through 11 h p.f. both in the presence and in the absence of α-amanitin (Fig. 2, second row), indicating that this protein is not zygotically regulated and may not play a role in cell cycle remodeling. These data show that a zygotic product is necessary for the terminal disappearance of maternal cyclin A1 and B2 proteins.
FIG. 2.
Terminal disappearance of cyclin A1 and B2 proteins during cell cycle remodeling is dependent on zygotic transcription. Western blots for the cyclins indicated depict protein levels during embryogenesis. Embryos were injected with 50 ng of α-amanitin to inhibit the onset of zygotic transcription (left panels), harvested every 30 min at the indicated times after fertilization, and processed for protein extraction. One embryo equivalent was loaded per lane. The same blot was stripped and reprobed for each cyclin. Stage 8 embryos are indicated by an arrowhead.
Deadenylation of maternal cyclin A1 and B2 mRNAs precedes disappearance of their encoded proteins.
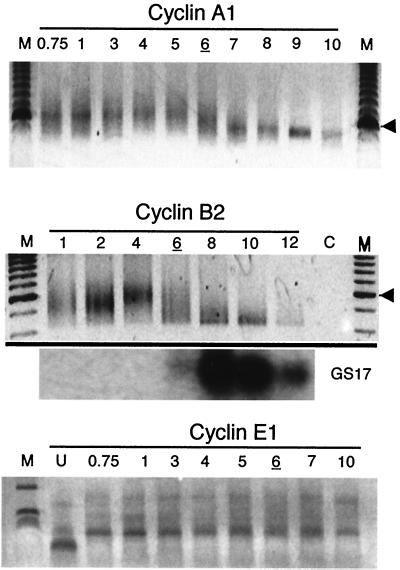
During each of the first 12 cell cycles, A and B cyclins are continuously synthesized and periodically degraded. Therefore, we reasoned that the terminal disappearance of cyclin A1 and B2 proteins may result from a block in the translation of their mRNAs. Deadenylation triggers translational silencing of numerous mRNAs in Xenopus oocytes and embryos (21, 30, 42), and deadenylated mRNAs are unstable in blastula-stage embryos (2, 13). Therefore, we asked if deadenylation of cyclin mRNAs precedes or accompanies disappearance of cyclin A1 and B2 proteins. To determine the adenylation status of cyclin mRNAs, we performed a reverse transcriptase PCR-based PAT (34). PAT for cyclin mRNAs was performed on total RNA isolated from an embryo time course (Fig. 3). A PCR product is present only if the mRNA tested contains a poly(A) tail long enough (12 residues) to serve as a template for hybridization with the oligo(dT) anchor. The expected size of the PCR product for a cyclin A1 mRNA with a minimal poly(A) tail of 12 residues is around 510 nt. The cyclin A1 PCR product migrated as a diffuse band from approximately 500 to 700 nt in 1-h embryos, which corresponds to a poly(A) tail of approximately 200 A's. These results agree with previous measurements of poly(A) tail length for the cyclin A1 mRNA in Xenopus embryos prior to the MBT, as measured by the oligo(dT)-RNase H method (35). The PCR product size remained constant until 6 h p.f., when smaller products appeared, indicating shortening of the poly(A) tail. By 8 h, the product migrated as a discrete band around 500 nt, corresponding to cyclin A1 mRNA with the smallest detectable poly(A) tail. The product size remained constant until 10 h p.f. We and others have shown previously that maternal cyclin A1 is undetectable on Northern blots by 11 h after fertilization (unpublished data) and in stage 10.5 (midgastrula) embryos (17), suggesting that deadenylation leads to destabilization of the mRNA.
FIG. 3.
Cyclin A1 and B2 maternal mRNAs are rapidly deadenylated at the MBT. DNA gel analysis of PAT PCR products is shown for cyclins A1, B2, and E1. Total RNA was extracted from embryos at the indicated hours postfertilization and subjected to the PAT. Lanes M, 100-bp DNA ladder. The arrowheads on the right of the top two panels correspond to the 600-bp marker. Northern blot analysis (GS17) of the cyclin B2 mRNA samples shows the expression of the zygotic transcription-specific mRNA GS17. For cyclin E1 PAT, lane M corresponds to a 1-kb ladder. For each analysis, stage 8 embryos are indicated by the underlined hour postfertilization.
The cyclin B2 PCR product migrated as a diffuse band between 500 and 600 nt at 1 h after fertilization, also indicating a poly(A) tail of about 200 nt, as the minimum size of the predicted PCR product is 415 nt. Similarly to cyclin A1, the cyclin B2 product decreased in size beginning at 6 h p.f. and migrated as a discrete band slightly above 400 nt between 8 and 12 h p.f. For both cyclin A1 and cyclin B2, the decrease in size of the poly(A) tail preceded the decrease in cyclin protein (compare Fig. 2 and 3) and coincided with the onset of zygotic transcription as measured by the appearance of the zygotic mRNA GS17 (Fig. 3, middle panel). In contrast, the PAT of cyclin E1 mRNA showed an increase in polyadenylation during the first hour following fertilization, consistent with accumulation of cyclin E1 protein during this time (14), and a constant poly(A) tail length of about 200 A's thereafter. These results indicate that both cyclin A1 and cyclin B2 mRNAs are rapidly and specifically deadenylated after the onset of zygotic transcription, while cyclin E1 mRNA remains adenylated during the same period. Deadenylation may therefore contribute to downregulation of maternal cyclins A1 and B2 after the MBT but not to the downregulation of cyclin E1.
The 3′ UTRs of cyclin mRNAs specify deadenylation at the MBT.
Sequences controlling cytoplasmic adenylation behavior have primarily been localized to the 3′ UTR of mRNAs. Therefore, we asked if the 3′ UTRs of maternal cyclins could confer specific deadenylation on chimeric mRNAs at the MBT. We took advantage of known CPEs in the cyclin A1, B1, and B2 3′ UTRs (37) and a potential CPE in the cyclin E1 3′ UTR to generate chimeric mRNAs that would be polyadenylated in vivo after their injection into two-cell embryos. Chimeric mRNAs contained the 5′ UTR and the coding sequence of β-globin followed by the complete 3′ UTR of one of the cyclins A1, B1, B2, and E1 (Fig. 1A). Radiolabeled, capped, poly(A)− mRNAs were injected into two-cell embryos. The poly(A) tail size of the injected mRNAs was determined 5 min after injection and every 1 or 2 h thereafter by denaturing gel electrophoresis (Fig. 4).
FIG. 4.
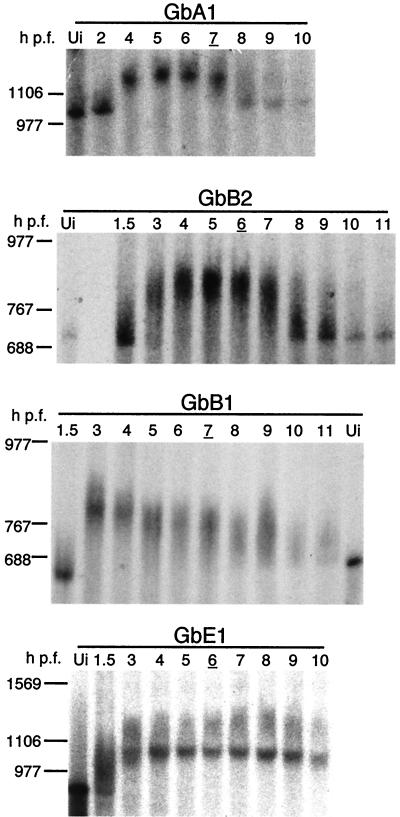
Cyclin A1 and B2 3′ UTRs trigger rapid deadenylation at the MBT. Representative autoradiograms of deadenylation analysis of chimeric cyclin mRNAs are shown. Radiolabeled, poly(A)− GbA1, GbB2, GbB1, or GbE1 mRNAs were injected into two-cell embryos. Total RNA extracted from embryos at the indicated times after fertilization was resolved by denaturing gel electrophoresis. RNA size markers are indicated on the left in bases. The injected mRNA is indicated on the top of each panel. Ui, uninjected poly(A)− mRNA. GbA1, n = 8; GbB2, n = 8; GbB1, n = 4; GbE1, n = 2. Stage 8 embryos are indicated by the underlined times postfertilization.
As expected, because of the presence of the CPEs, about 200 A's were added to all of the injected mRNAs (Fig. 4). This is in accordance with the poly(A) tail length of the maternal cyclin mRNAs as determined by the PAT (Fig. 3). After in vivo polyadenylation, GbA1 and GbB2 mRNAs remained poly(A)+ until 6 h p.f., after which they began to be rapidly deadenylated, accumulating as poly(A)− mRNA by 8 h. At 9 h p.f., the majority of these mRNAs were poly(A)−. In contrast, GbB1 mRNA slowly decreased in size and never accumulated as poly(A)− mRNA. GbE1 mRNA remained polyadenylated until at least 10 h p.f., similar to the adenylation behavior of maternal cyclin E1 mRNA as determined by the PAT (Fig. 3). These results show that the 3′ UTRs of cyclin A1 and B2 mRNAs confer specific and rapid deadenylation on chimeric mRNAs in blastula-stage embryos.
Deadenylation mediated by the cyclin A1 and B2 3′ UTRs is under zygotic control.
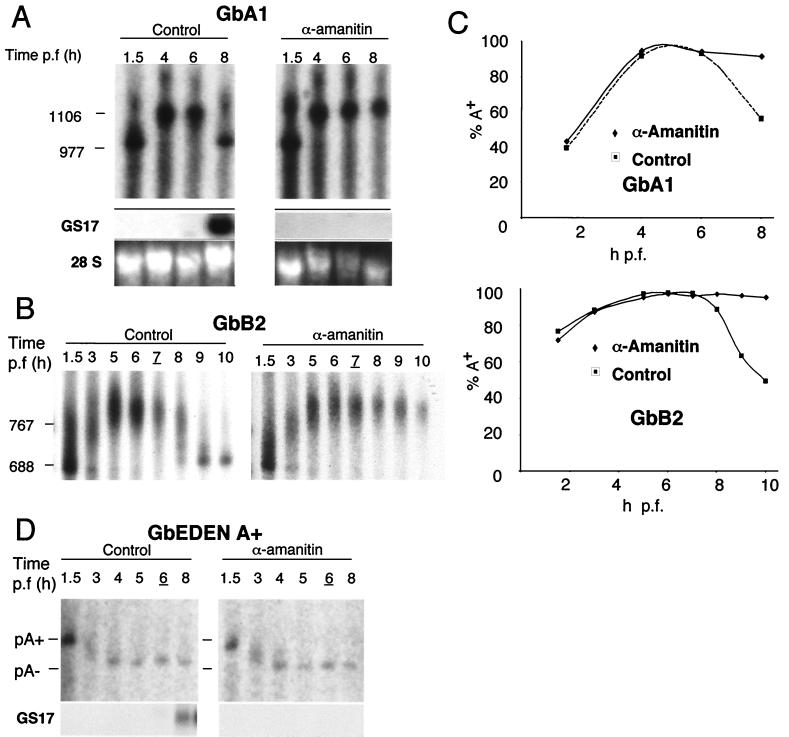
Cyclin A1-B2 mRNA deadenylation occurs after the onset of zygotic transcription (Fig. 3, GS17), and the terminal disappearance of cyclin A1 and B2 proteins requires zygotic transcription (Fig. 2). Therefore, if deadenylation is directly responsible for protein disappearance, inhibition of zygotic transcription should also block deadenylation. To test this prediction, we injected chimeric mRNAs along with the transcriptional inhibitor α-amanitin. In the presence of α-amanitin, the expression of the zygotic transcript GS17 is completely abolished (Fig. 5A), and yet the embryos remain viable for several hours (36). Following injection, the GbA1 and GbB2 mRNAs acquired a poly(A) tail of about 200 A's both in α-amanitin-treated and in control embryos (Fig. 5).This shows that α-amanitin does not affect in vivo polyadenylation. Strikingly, deadenylation of GbA1 and GbB2 mRNAs was completely abolished in α-amanitin-treated embryos (Fig. 5A, compare GbA1 control and α-amanitin-treated embryos at 8 h). As quantitated in Fig. 5C, in the presence of α-amanitin, the percentage of polyadenylated mRNA peaked at approximately 5 h and then remained constant throughout the time course of the experiment for both GbA1 and GbB2. In comparison, in buffer-injected embryos the percentage of polyadenylated mRNA dropped dramatically between 6 and 8 h for GbA1 and between 8 and 10 h for GbB2. To ensure that α-amanitin is not simply stabilizing the poly(A) tail independently of transcription inhibition, we coinjected α-amanitin with the polyadenylated GbORF/mosEDEN mRNA (29). The embryonic deadenylation element (EDEN) contained in this mRNA triggers its rapid deadenylation after injection into fertilized eggs. By 4 h p.f., the mRNA accumulated as poly(A)− RNA either in the absence or in the presence of α-amanitin (Fig. 6). Thus, α-amanitin does not block deadenylation in a manner independent of transcription inhibition. These experiments demonstrate that a zygotic product is required both for protein downregulation of cyclins A1 and B2 (Fig. 2) and for the rapid deadenylation triggered by the 3′ UTRs of their mRNAs (Fig. 5).
FIG. 5.
Deadenylation triggered by the cyclin A1 and B2 3′ UTRs is dependent on zygotic transcription. Shown are autoradiograms of deadenylation analyses of chimeric GbA1, GbB2, and GbEDEN mRNAs in the presence of α-amanitin. (A) Radiolabeled GbA1 mRNA injected in the presence (α-amanitin) or absence (control) of 50 ng of α-amanitin/embryo. RNA was analyzed as indicated in the legend to Fig. 4. Expression of the zygotic gene GS17 and ethidium bromide-stained 28S rRNA is also shown. (B) Radiolabeled GbB2 mRNA injected in the presence (α-amanitin) or absence (control) of α-amanitin. The positions of RNA size markers are indicated to the left of each panel. GbA1, n = 8; GbA1 plus α-amanitin, n = 4; GbB2, n = 8; GbB2 plus α-amanitin, n = 2. (C) Quantification of the percentage of polyadenylated chimeric mRNA at each time point. (D) Radiolabeled GbORF/mosEDEN poly(A)+ mRNA injected in the presence (α-amanitin) or absence (control) of α-amanitin (n = 2). The positions of A65 (pA+) and A0 (pA−) are shown. The expression of the zygotic gene-specific transcript GS17 was monitored by Northern blotting. For each experiment, stage 8 embryos are indicated by underlining of the time postfertilization.
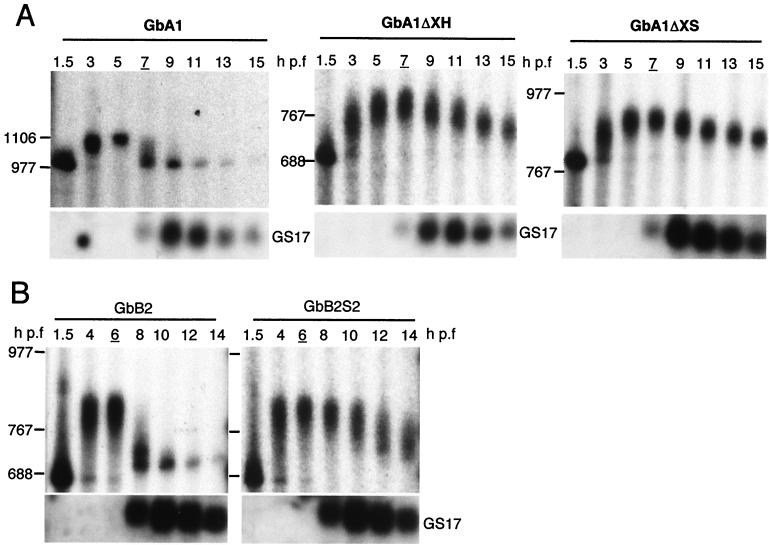
FIG. 6.
Specific regions in the 3′ UTRs of cyclin A1 and B2 mRNAs are required for zygote-dependent deadenylation. (A) Radiolabeled GbA1 mRNA and the deletion mutants GbA1ΔXH and GbA1ΔXS were injected into two-cell embryos. RNAs were analyzed as indicated in the legend to Fig. 4. The expression of the zygotic gene GS17 as determined by Northern blot analysis is shown at the bottom of each panel. (B) Radiolabeled GbB2 mRNA and the substitution mutant GbB2S2 were injected into two-cell embryos and analyzed at the indicated times after fertilization. The positions of RNA size markers are indicated to the left of each panel. GbA1, n = 8; GbA1ΔXH, n = 2; GbA1ΔXS, n = 2; GbB2, n = 8; GbB2S2, n = 2. For each experiment, stage 8 embryos are indicated by the underlined hour postfertilization.
Specific regions in the 3′ UTRs are required for zygote- mediated deadenylation.
Our results show that the 3′ UTRs of cyclins A1 and B2 specify the deadenylation that precedes the disappearance of these cyclin proteins. The ability of the cyclin A1 and B2 3′ UTRs to confer the maternal deadenylation behavior on a chimeric mRNA implies that all of the sequence information required is contained in the 3′ UTR. Several sequence-dependent mechanisms of deadenylation have been described previously for pre-MBT Xenopus embryos (29, 43). To determine if deadenylation at the MBT is due to a known or novel mechanism, we performed substitution and deletion analyses to localize the necessary regions in the 3′ UTRs.
To delimit the necessary region in the 491-bp cyclin A1 3′ UTR, 309 or 200 nt were deleted as depicted in Fig. 1B (pGbA1ΔXH and pGbA1ΔXS). The chimeric mRNAs transcribed from these plasmids were injected into two-cell embryos, and changes in the poly(A) tail length were assayed by denaturing gel electrophoresis as for Fig. 4. The deletions left the two CPEs and the nuclear polyadenylation signal (the hexanucleotide AAUAAA) that are required for polyadenylation (8) intact, and so adenylation of the chimeric transcripts occurred as previously observed (Fig. 6A, 1.5 to 5 h p.f.). Deletion of the first 309 or 200 nt of the 3′ UTR in the context of GbA1 greatly reduced deadenylation (Fig. 6A, GbA1ΔXH and GbA1ΔXS, 7 to 15 h p.f.). Deadenylation of GbA1 was evident by 7 h, while deletion mutants GbA1ΔXH (309 nt) and GbA1ΔXS (200 nt) retained a poly(A) tail until at least 15 h following fertilization. For both of the deletion mutants, a gradual shortening of the poly(A) tail was evident after 7 h, but fully deadenylated transcripts did not accumulate (Fig. 6). The percentage of chimeric mRNAs that are adenylated at each time point is shown in Fig. 7. The maximum percentage of polyadenylated transcripts peaks at 6 h p.f. for full-length GbA1 and both of the deletion mutants. The percentage remains constant for both deleted mRNAs, while the full-length GbA1 is rapidly deadenylated beginning at 7 h. The minimum percentage of adenylated transcripts is seen by 10 h and is comparable to the proportion of polyadenylated mRNA at the first time point. These data show that the sequence information necessary for deadenylation of the cyclin A1 mRNA is contained in the first 200 nt of its 3′ UTR. This 200-nt region contains two AU-rich sequences, UUAUUUAUU and UUAUUUAUAA, that are known to mediate deadenylation in Xenopus embryos before the MBT, i.e., before the initiation of zygotic transcription (43, 46, 47).
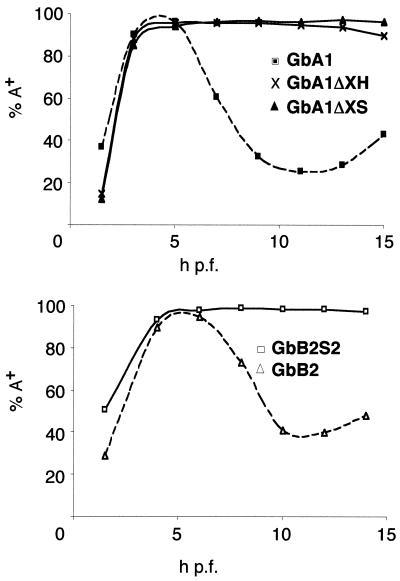
FIG. 7.
Quantification of deadenylation of the GbA1 deletion mutants and the GbB2 substitution mutants. The data collected from the experiments shown in Fig. 6 were quantitated and plotted as the percentage of polyadenylated mRNA for each time point. Upper panel, GbA1 mRNA and the deletion mutants GbA1ΔXH and GbA1ΔXS. Lower panel, GbB2 mRNA and the substitution mutant GbB2S2.
For the cyclin B2 3′ UTR (180 nt), substitution analysis was used to localize the region necessary for deadenylation (Fig. 6B). Substitution mutants of the GbB2 construct were made using linker scanning mutagenesis (3). Six substitution mutants, each with 30-nt substitutions, were constructed to cover the complete 180-nt cyclin B2 3′ UTR in the context of GbB2 (Fig. 1B). As for GbA1 deletion mutants, the nuclear polyadenylation signal was not mutated and the two CPEs were mutated one at a time. Figure 6B shows the results for one mutant (GbB2S2). By 8 h p.f., GbB2 was almost completely deadenylated while GbB2S2 remained polyadenylated. In addition to GbB2S2, another substitution mutant, GbB2S3, also remained polyadenylated after the onset of zygotic transcription. The other substitutions had no significant effect on deadenylation. There are no obvious sequence elements or structures in the 60-nt segment of the cyclin B2 3′ UTR that resemble those previously shown to have a role in deadenylation. The data for GbB2S2 are summarized in the bottom panel of Fig. 7, which shows the constant percentage of polyadenylated GbB2S2 transcripts, compared to the rapid drop in polyadenylated GbB2 transcripts after 6 h p.f. These data show that specific sequence elements are present in the 3′ UTRs of cyclin A1 and B2 maternal mRNAs that are necessary for zygotically mediated deadenylation.
DISCUSSION
In X. laevis, regulation of poly(A) tail length allows for tight translational control during early development from oocyte maturation until the MBT. Our results, along with other recent reports (31, 41), show that control of poly(A) tail length continues after the MBT. We have shown that rapid deadenylation of maternal cyclin A1 and B2 mRNAs coincides with the onset of zygotic transcription and precedes the disappearance of their encoded proteins (Fig. 2 and 3). The 3′ UTR of cyclin A1 or cyclin B2 is sufficient to confer the same deadenylation behavior on a chimeric mRNA, while neither the cyclin B1 nor the cyclin E1 3′ UTR triggers rapid deadenylation (Fig. 4). Further confirming the specificity of deadenylation, we have localized the necessary sequence information to within 60 nt for the cyclin B2 3′ UTR and to within 200 nt for the cyclin A1 3′ UTR (Fig. 5). However, further analyses are needed to characterize the elements sufficient for zygotic deadenylation of these maternal transcripts.
The requirement for zygotic transcription for both disappearance of protein (Fig. 2) and deadenylation of mRNA (Fig. 5) strongly supports a model in which the deadenylation triggered by the cyclin A1 and B2 3′ UTRs leads to translational repression and/or mRNA destabilization, resulting in terminal downregulation of these maternal gene products. There is a difference in timing of the terminal disappearance of cyclin A1 and B2 proteins (9 and 11 h, respectively), but our data show little difference in the deadenylation kinetics of GbB2 and GbA1 mRNAs. This difference may stem from the earlier degradation of cyclin A1 in the embryonic cell cycle. Cyclin B2 is degraded at the end of M phase (38), whereas cyclin A1 is degraded 15 min before (14). After the MBT, the cell cycle becomes asynchronous and lengthens as S phase is extended and gap phases are added. The delay between cyclin A1 and B2 protein degradation may reflect this change. Furthermore, after the MBT, cyclin A1 mRNA is rapidly degraded while cyclin B2 mRNA is more stable (data not shown), which may permit sufficient cyclin B2 protein synthesis to result in the levels of cyclin B2 observed. We do not believe that the inhibition of deadenylation by α-amanitin results from nonspecific stabilization of the poly(A) tail, since deadenylation mediated by the EDEN of c-mos still occurs in these embryos (Fig. 6).
The transcription inhibition in post-MBT cell divisions initially disrupts the cell cycle and ultimately triggers apoptosis in what would be midgastrula embryos (36). First, we observed that, in the presence of α-amanitin, downregulation of cyclin A1 and B2 proteins is prevented. This suggests that the cell cycle is blocked in a phase where cyclin A1 and B2 proteins are stable (before or at the beginning of M phase [14]), and yet the downregulation of cyclin E1 protein still occurs (Fig. 1), showing that not all processes are nonspecifically disrupted at this time. As cyclin protein degradation is a cell cycle stage-specific event, it is not unexpected that such stabilization occurs. If deadenylation is also linked to a specific cell cycle stage, it is possible that blocking cell cycle progression would also block cyclin A1 and B2 deadenylation. In this case, we could say that the block in deadenylation is a consequence of the block in the cell cycle. However, to our knowledge, deadenylation has never been linked directly to a specific cell cycle stage. The other possibilities are that a required component of the cyclin A1-B2 deadenylation machinery is zygotically produced or that this machinery is present in an inactive state before the onset of zygotic transcription and requires an upstream zygotic transcription-dependent event to be activated. Clarification of this issue must await a more complete delineation of the mechanism.
Despite the identical timing and the common requirement for zygotic transcription, it is not clear whether cyclin A1 deadenylation and B2 deadenylation are mediated by the same mechanism. Sequence comparison of the necessary regions of the cyclin A1 and B2 3′ UTRs did not lead to the identification of common elements. We believe that the deadenylation conferred by the cyclin B2 3′ UTR is due to a novel sequence element while that conferred by the cyclin A1 3′ UTR may be mediated by the AU-rich elements (AREs) present in the first 200 nt of its 3′ UTR. In Xenopus, AREs mediate deadenylation just after fertilization and therefore do not require zygotic transcription (43). Thus, if cyclin A1 mRNA deadenylation is ARE mediated, a specific mechanism must prevent rapid deadenylation from fertilization to the MBT. In this case, a zygotic factor would be required to inhibit the protective mechanism after the onset of zygotic transcription (a model is shown in Fig. 8B).
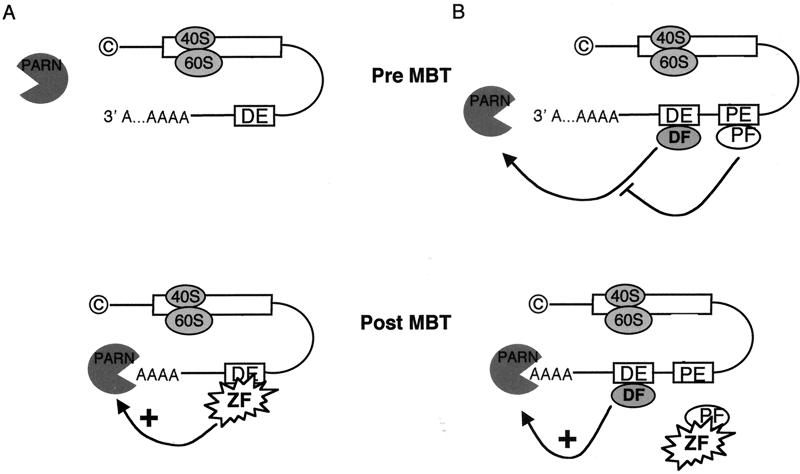
FIG. 8.
Proposed models for zygotically mediated deadenylation of cyclin A1 and B2 mRNAs include a zygotically expressed deadenylation factor (A) or a maternally expressed protective factor (B). © represents the 5′-cap structure. The white box corresponds to the mRNA coding region. DE, deadenylation element; PE, protective element; PF, protective factor; ZF, the unknown zygotic factor that acts either as a deadenylation activator (A) or as an inhibitor of protection mediated by the interaction of the protective factor with the protective element (B).
Figure 8 depicts two models in which a newly synthesized zygotic factor triggers cyclin A1 and B2 deadenylation. These models are based on current knowledge of deadenylation in Xenopus oocytes and/or early embryos and on our results, which are consistent with either model. In the first model, a zygotic factor acts as a positive regulator of deadenylation by targeting the mRNA containing the deadenylation element to the poly(A) exoribonuclease (PARN), which is already present in the embryos (19). Sequence-specific deadenylation is usually mediated by RNA binding proteins. Thus, it is likely that the zygotic factor is an RNA binding protein. Two other alternatives are that the zygotic component is a novel PARN transcribed at the MBT and that the factor is a newly transcribed RNA. This last possibility is exemplified in Caenorhabditis elegans, where the 21 nt let-7 RNA is complementary to 3′-UTR sequences of its target mRNAs and inhibits their expression (32).
In the second model, the deadenylation activity and the specificity factor are present in the pre-MBT embryo but cannot act on the mRNAs because of the presence of a sequence-specific protective factor. In this model, these RNAs should bear a protective element in addition to the deadenylation element. Zygotic transcription would be necessary to inactivate this protective factor. One prediction of this model is that mutagenesis of the protective element alone should lead to the early deadenylation of the chimeric reporter mRNA. In the set of substitutions and deletions that we performed, we never observed such behavior. This may mean that the protective element does not exist or that it was untouched in our set of chimeric mRNAs. Since the CPE and hexanucleotides were always present in our mRNAs to allow in vivo polyadenylation, future work will test if these sequences act as protective elements against zygotic factor-mediated deadenylation. Another possibility is that the deadenylation element and protective element cannot be uncoupled.
Deadenylation has been shown previously to be dependent on and/or facilitated by a 5′ 7-methylguanosine cap and, consistent with this, dependent on active translation of the deadenylated mRNA (6, 11). These requirements also hold true for deadenylation mediated by PARN (6). We do not know if active translation of the chimeric transcripts is necessary for deadenylation, but since these transcripts are capped, contain a coding sequence, and are polyadenylated upon injection into two-cell embryos, it is likely that they are translated. Nevertheless, it must be kept in mind that cap-independent deadenylation does occur (4).
In addition to AREs, other 3′-UTR sequences specifying deadenylation in Xenopus include the EDEN (21, 29), CPE-mediated deadenylation (31), and tra-2 and GLI element (TGE)-mediated deadenylation (40). EDENs are UGUA rich and, along with their binding protein, EDEN-BP, mediate rapid deadenylation of c-mos, Eg-2, and Eg-5 just after fertilization (29). The difference in timing and the absence of UGUA-rich elements in both the cyclin A1 and cyclin B2 3′ UTR suggest that this mechanism is not involved in their deadenylation. UA-rich CPE-mediated deadenylation occurs after the MBT in gastrula-stage embryos and was recently described for maternal lamin B1. This mRNA is adenylated and translationally activated at the MBT and is deadenylated and translationally repressed in gastrula stages (31). In this case, deadenylation does not confer degradation, as the lamin B1 mRNA is stable until late in embryogenesis (stage 22). It is not clear whether CPE binding protein is involved in this process. Our data show that the CPEs are not sufficient for deadenylation of cyclin A1 and B2 mRNA at the MBT, as chimeric mRNAs containing CPEs are not deadenylated (Fig. 6 and 7, GbB2S2, GbA1ΔXH, and GbA1ΔXS). As for the EDEN-mediated deadenylation, the difference in timing and the lack of TGE-like sequences in cyclin A1 or cyclin B2 3′ UTR suggest that TGE-mediated deadenylation is not involved in cyclin A1 and B2 deadenylation.
Another example of posttranscriptional control in post-MBT embryos is that mediated by the 3′ UTR of Xwnt8, a zygotic mRNA. The 3′ UTR of Xwnt8 induces translational repression and degradation of a heterologous transcript at gastrulation, probably by promoting deadenylation (41). The element mediating translational repression has not been identified, but the timing of Xwnt8 downregulation correlates well with that of cyclin B2, raising the possibility that a similar zygotic mechanism is involved. On the other hand, there is no obvious sequence or structural similarity between the Xwnt8 and cyclin B2 3′ UTRs.
Non-sequence-specific or default deadenylation also occurs in Xenopus, during both oocyte maturation and embryogenesis. Default deadenylation acts on mRNAs devoid of CPE (9, 42) and is unable to trigger the complete deadenylation of an injected poly(A)+ mRNA even after 12 h p.f. (2). The slow and incomplete deadenylation of GbB2S2, GbA1ΔXH, and GbA1ΔXS resembles the kinetics of default deadenylation, suggesting that this mechanism may also act on CPE-containing mRNAs when the majority of CPEB protein has been destroyed (12). The default deadenylation described previously for the Xenopus embryo is maternally driven (28), distinguishing it from the zygote-dependent deadenylation described in the current report. This does not rule out the possibility that common components may be involved in the two processes.
The elimination of maternal cyclin A1 and B2 mRNAs and proteins may mediate the transition from the rapid embryonic cell cycle to the highly regulated adult cell cycle. As cyclin A1 disappears, cyclin A2 is upregulated (17). Despite the fact that cyclin A1-Cdk1 and cyclin A2-Cdk1/Cdk2 have roles in both the S and M phases, it is likely that the substrates of Cdk1 and Cdk2 differ (10, 39, 44). The dependence of cyclin A1-B2 deadenylation on zygotic transcription suggests that this mechanism may control the accumulation of cell cycle regulators in somatic cells. This probability is supported by the recent report that, in the adult mammalian cell cycle, cyclins B1 and A2 are controlled at the level of mRNA stability (45). Identification of the sequences and factors involved will allow us to define the role of this novel regulatory mechanism in cell cycle remodeling and in the adult cell cycle. In addition, the zygotic transcription-mediated deadenylation activity that we describe may also silence other maternal mRNAs whose expression is required for early development but detrimental for later stages.
ACKNOWLEDGMENTS
We thank J. Sible, H. B. Osborne, and D. L. Weeks for critical reading of the manuscript and H. B Osborne for pGbORF/mosEDEN and Paul Krieg for pGS17.
This work was supported by a Basil O'Connor award (F 498–758) from the March of Dimes Birth Defects Foundation, a DERC Pilot and Feasibility award (DERC-NIH DK252295), and a Carver Trust Medical Research Initiative grant from the Roy J. Carver Charitable Trust.
REFERENCES
- 1.Audic Y, Omilli F, Osborne H B. Embryo deadenylation element-dependent deadenylation is enhanced by a cis element containing AUU repeats. Mol Cell Biol. 1998;18:6879–6884. doi: 10.1128/mcb.18.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audic Y, Omilli F, Osborne H B. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol Cell Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnhart K M. Simplified PCR-mediated, linker-scanning mutagenesis. BioTechniques. 1999;26:624–626. doi: 10.2144/99264bm06. [DOI] [PubMed] [Google Scholar]
- 4.Brewer G. Characterization of c-myc 3′ to 5′ mRNA decay activities in an in vitro system. J Biol Chem. 1998;273:34770–34774. doi: 10.1074/jbc.273.52.34770. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier S, Couturier A, Chartrain I, Le Guellec R, Beckhelling C, Le Guellec K, Philippe M, Ford C C. Xenopus cyclin E, a nuclear phosphoprotein, accumulates when oocytes gain the ability to initiate DNA replication. J Cell Sci. 1996;109:1173–1184. doi: 10.1242/jcs.109.6.1173. [DOI] [PubMed] [Google Scholar]
- 6.Dehlin E, Wormington M, Korner C G, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshaies R J. The self-destructive personality of a cell cycle in transition. Curr Opin Cell Biol. 1995;7:781–789. doi: 10.1016/0955-0674(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 8.Fox C A, Sheets M D, Wickens M. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- 9.Fox C A, Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- 10.Furuno N, den Elzen N, Pines J. Human cyclin A is required for mitosis until mid prophase. J Cell Biol. 1999;147:295–306. doi: 10.1083/jcb.147.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M, Fritz D T, Ford L P, Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol Cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hake L E, Richter J D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 13.Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102:837–852. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- 14.Hartley R S, Rempel R E, Maller J L. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- 15.Hartley R S, Sible J C, Lewellyn A L, Maller J L. A role for cyclin E/Cdk2 in the timing of the midblastula transition in Xenopus embryos. Dev Biol. 1997;188:312–321. doi: 10.1006/dbio.1997.8647. [DOI] [PubMed] [Google Scholar]
- 16.Heasman J, Holwill S, Wylie C. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 1991;36:213–230. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- 17.Howe J A, Howell M, Hunt T, Newport J W. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- 18.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 19.Korner C G, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieg P A, Melton D A. Developmental regulation of a gastrula-specific gene injected into fertilized Xenopus eggs. EMBO J. 1985;4:3463–3471. doi: 10.1002/j.1460-2075.1985.tb04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legagneux V, Omilli F, Osborne H B. Substrate-specific regulation of RNA deadenylation in Xenopus embryos and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- 22.Minshull J, Blow J J, Hunt T. Translation of cyclin mRNA is necessary for extracts of activated xenopus eggs to enter mitosis. Cell. 1989;56:947–956. doi: 10.1016/0092-8674(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 23.Minshull J, Golsteyn R, Hill C S, Hunt T. The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 1990;9:2865–2875. doi: 10.1002/j.1460-2075.1990.tb07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newport J, Dasso M. On the coupling between DNA replication and mitosis. J Cell Sci Suppl. 1989;12:149–160. doi: 10.1242/jcs.1989.supplement_12.13. [DOI] [PubMed] [Google Scholar]
- 25.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos. I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:673–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 26.Nieuwkoop P D, Faber J. Normal table of Xenopus laevis (Daudin). Amsterdam, The Netherlands: North-Holland Publishing Co.; 1967. [Google Scholar]
- 27.Osborne H B, Richter J D. Translational control by polyadenylation during early development. Prog Mol Subcell Biol. 1997;18:173–198. doi: 10.1007/978-3-642-60471-3_8. [DOI] [PubMed] [Google Scholar]
- 28.Paillard L, Legagneux V, Osborne H B. Poly(A) metabolism in Xenopus laevis embryos: substrate-specific and default poly(A) nuclease activities are mediated by two distinct complexes. Biochimie. 1996;78:399–407. doi: 10.1016/0300-9084(96)84746-8. [DOI] [PubMed] [Google Scholar]
- 29.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne H B. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- 31.Ralle T, Gremmels D, Stick R. Translational control of nuclear lamin B1 mRNA during oogenesis and early development of Xenopus. Mech Dev. 1999;84:89–101. doi: 10.1016/s0925-4773(99)00078-7. [DOI] [PubMed] [Google Scholar]
- 32.Reinhart B J, Slack F J, Basson M, Pasquinelli A E, Bettinger J C, Rougvie A E, Horvitz H R, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 33.Rempel R E, Sleight S B, Maller J L. Maternal Xenopus Cdk2-cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J Biol Chem. 1995;270:6843–6855. doi: 10.1074/jbc.270.12.6843. [DOI] [PubMed] [Google Scholar]
- 34.Salles F J, Richards W G, Strickland S. Assaying the polyadenylation state of mRNAs. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- 35.Sheets D M, Fox C A, Hunt T, Woude G V, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 36.Sible J C, Anderson J A, Lewellyn A L, Maller J L. Zygotic transcription is required to block a maternal program of apoptosis in Xenopus embryos. Dev Biol. 1997;189:335–346. doi: 10.1006/dbio.1997.8683. [DOI] [PubMed] [Google Scholar]
- 37.Stebbins-Boaz B, Hake L E, Richter J D. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart E, Kobayashi H, Harrison D, Hunt T. Destruction of Xenopus cyclins A and B2, but not B1, requires binding to p34cdc2. EMBO J. 1994;13:584–594. doi: 10.1002/j.1460-2075.1994.tb06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strausfeld U P, Howell M, Descombes P, Chevalier S, Rempel R E, Adamczewski J, Maller J L, Hunt T, Blow J J. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 40.Thompson S R, Goodwin E B, Wickens M. Rapid deadenylation and poly(A)-dependent translational repression mediated by the Caenorhabditis elegans tra-2 3′ untranslated region in Xenopus embryos. Mol Cell Biol. 2000;20:2129–2137. doi: 10.1128/mcb.20.6.2129-2137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian Q, Nakayama T, Dixon M P, Christian J L. Post-transcriptional regulation of Xwnt-8 expression is required for normal myogenesis during vertebrate embryonic development. Development. 1999;126:3371–3380. doi: 10.1242/dev.126.15.3371. [DOI] [PubMed] [Google Scholar]
- 42.Varnum S M, Wormington W M. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: a default mechanism for translational control. Genes Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]
- 43.Voeltz G K, Steitz J A. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol. 1998;18:7537–7545. doi: 10.1128/mcb.18.12.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker D H, Maller J L. Role for cyclin A in the dependence of mitosis on completion of DNA replication. Nature. 1991;354:314–317. doi: 10.1038/354314a0. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Caldwell M C, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu N, Chen C Y, Shyu A B. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zubiaga A M, Belasco J G, Greenberg M E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]