Abstract
The ELL gene encodes an RNA polymerase II transcription factor that frequently undergoes translocation with the MLL gene in acute human myeloid leukemia. Here, we report that ELL can regulate cell proliferation and survival. In order to better understand the physiological role of the ELL protein, we have developed an ELL-inducible cell line. Cells expressing ELL were uniformly inhibited for growth by a loss of the G1 population and an increase in the G2/M population. This decrease in cell growth is followed by the condensation of chromosomal DNA, activation of caspase 3, poly(ADP ribose) polymerase cleavage, and an increase in sub-G1 population, which are all indicators of the process of programmed cell death. In support of the role of ELL in induction of cell death, expression of an ELL antisense RNA or addition of the caspase inhibitor ZVAD-fmk results in a reversal of ELL-mediated death. We have also demonstrated that the C-terminal domain of ELL, which is conserved among the ELL family of proteins that we have cloned (ELL, ELL2, and ELL3), is required for ELL's activity in the regulation of cell growth. These novel results indicate that ELL can regulate cell growth and survival and may explain how ELL translocations result in the development of human malignancies.
Many cellular factors involved in human oncogenesis have been identified as the products of genes at breakpoints of frequently occuring chromosomal translocations. The protein products of some of these genes are transcriptional factors that regulate the general or specific expression of many genes. The ELL gene was initially identified on chromosome 19p 13.1, which undergoes frequent translocation with the trithorax-like MLL (ALL-1, HRX) gene on chromosome 11q23 in acute myeloid leukemia (AML) (47). The 3,968-amino-acid MLL protein contains an N-terminal A-T hook DNA-binding domain, a methyltransferase-like domain near the center of the molecule, and a C-terminal domain with several contiguous zinc fingers (35, 36, 48). Chromosomal translocations involving the MLL gene occur in approximately 80% of infants with AML and acute lymphoblastic leukemia (ALL) and approximately 5% of adult patients with AML as well as up to 10% with ALL (36). These translocations result in fusion of the N-terminal region of the MLL gene product to other cellular gene products to produce chimeric proteins. To date, more than 16 different MLL fusion partner proteins been identified, with the partner proteins of MLL displaying seemingly limited structural similarity (3, 7, 13, 16, 17, 25, 29, 30, 33, 34, 37, 43, 44, 47). The molecular function of MLL is thus far unknown: however, its homology to the Drosophila trithorax protein TRX indicates that MLL serves to regulate and/or maintain homeotic gene expression. Indeed, the production of MLL knockout mice has revealed that MLL is required for maintenance of gene expression during early embryogenesis (52, 53).
The question of whether MLL translocations are oncogenic because of a gain of function of the fusion protein or a loss of function of MLL and/or the partner protein is still unresolved. However, there is evidence to suggest that loss of MLL function alone is not the mechanism. For instance, MLL knockout mice do not develop leukemia, although the development of a number of different tissues, including the hemopoietic compartment, is compromised (15, 52). Additionally, mutations of the MLL gene in AML and ALL have not been found. Transgenic expression of the MLL-AF9 fusion protein in mice (10) and retroviral infection of mouse bone marrow with constructs expressing MLL-ENL and MLL-ELL result in induction of myeloid leukemia (9a, 22). Interestingly, fusion of MLL to the oncogene myc, a chimera not seen in ALL or AML, did not induce this phenotype, indicating that the specific fusion protein produced is important for oncogenesis (8).
While there appears to be little structural similarity between the different MLL fusion proteins, there is now evidence that the proteins may be functionally related. For example, CREB-binding protein (CBP) and p300 are transcriptional coactivators that acetylate histones to mediate gene expression (18), AF4, AF9, and ENL can function as transcription factors (34, 37), and ELL is a transcription factor (38) increasing the catalytic rate of transcription elongation by RNA polymerase II (Pol II). This indicates that altered transcriptional regulation mediated by MLL-containing fusion proteins may be a key molecular event leading to leukemia. Interestingly, it has recently been shown that MLL-ELL, MLL-ENL, and MLL-AF9 but not wild-type MLL can physically interact with GADD34, a growth arrest- and DNA damage-inducible protein, and abrogate GADD34-mediated apoptosis (1). These studies were the first to conclusively demonstrate a gain-of-function phenotype for MLL fused to completely different partner proteins compared to wild-type MLL. While the molecular functions of many of the MLL partner proteins have been identified, their physiological roles are mostly ill defined. As loss of physiological function of the partner proteins may be important for inducing cellular transformation, it is important to determine what cellular function(s) they might regulate in order to understand the events leading to tumorigenesis.
We purified ELL as an 80-kDa Pol II transcription factor that can both increase the catalytic rate of transcription elongation by Pol II and regulate formation of preinitiation complexes by Pol II (38, 39). The Pol II elongation factors fall into at least three functional classes (reviewed in reference 40). The first class prevents pausing or arrest of Pol II and includes P-TEFb, DSIF, and SII. The second class includes the FACT (facilitates chromatin transcription) complex and facilitates transcription elongation through nucleosomes. The third class increases the catalytic rate of transcription elongation and includes TFIIF, elongin, and members of the ELL family. It is presently unclear which members of each class of elongation factors are necessary for correct mRNA synthesis or the stoichiometry of the proposed large complex involving elongation factors and Pol II in vivo. The MLL-ELL fusion proteins contain the first 1,439 amino acids of MLL, containing the A-T hook and methyltransferase-like domains, fused to amino acids 46 to 621 of ELL. ELL mutants lacking the first 50 amino acids can still bind Pol II and are fully active in elongation but do not inhibit promoter-specific initiation (39). In addition to its role in regulating transcription elongation and initiation, ELL may also function as a modulator of gene expression mediated by cellular transcription factors such as p53. ELL has been shown to physically associate with p53 and inhibit both sequence-specific transactivation and sequence-independent transrepression by p53 and to inhibit p53-mediated apoptosis (41). Importantly, the MLL-ELL fusion protein may also bind to p53 and similarly affect the transcription regulation function of p53 (23). Conversely, binding of p53 to ELL results in inhibition of ELL-mediated transcription elongation (41). Although transcriptional activities of the ELL protein have been well characterized in vitro, we know very little about the physiological role of the ELL within mammalian cells.
A limited number of studies have been performed to determine the biological role of ELL. Consistent with its putative oncogenic function, constitutive overexpression of human ELL in RAT1 fibroblast cells resulted in an increase in anchorage-independent cell growth and a decreased dependence on growth factors (20). However, neither morphological change nor focus-forming ability was observed in ELL-expressing cells, indicating that expression of wild-type ELL is not sufficient to induce full neoplastic transformation. In an attempt to determine the effect of expression of human ELL in a human cell line, we expressed ELL in a tetracycline-inducible manner in 293 cells, which do not express endogenous ELL. Human 293 cells are an human embryonic kidney cell line transformed by adenovirus type 5 DNA (11).
In contrast to the previous experiments by Kanda et al. (20), expression of ELL resulted in a decrease in cell growth. However, in support of our observation, it was recently demonstrated that expression of ELL in primary murine myeloid progenitor cells did not result in immortalization of these cells (21a). Analysis of the cell cycle profiles of ELL-expressing cells revealed that there was a dramatic decrease in the G1 population commensurate with a reproducible increase in the percentage of cells in G2/M, followed by a steady increase in the sub-G1population of cells, indicating that these cells were undergoing apoptosis. This was confirmed by assays demonstrating that the ELL-expressing cells underwent morphological changes consistent with programmed cell death. In addition, caspase activation was observed, and inhibition of caspase activity with specific peptide inhibitors or expression of antisense ELL mRNA significantly inhibited ELL-induced cell death. These data show for the first time that expression of wild-type ELL in cells results in the regulation of cell proliferation. It is therefore possible that in addition to the production of MLL-ELL, which might inhibit apoptosis mediated by GADD34, the (11;19)(q23;p13.1) (MLL;ELL) chromosomal translocation may induce tumorigenesis by disrupting in vivo regulatory activities of wild-type ELL.
MATERIALS AND METHODS
Plasmids.
Full-length human ELL was amplified by PCR with ELL-specific primers. The ELL PCR product was inserted into the pTRE expression vector (Clontech, Palo Alto, Calif.) digested with BamHI and EcoRI, and the ELL-pTRE construct was verified by sequence analysis. An antisense ELL expression plasmid was produced by excising the ELL cDNA from ELL-pTRE with BamHI and EcoRI and inserting it into the BamHI and EcoRI cloning sites of pCDNA (Invitrogen, Carlsbad, Calif.).
Transient transfection into 293 Tet Off cells.
293 Tet Off cells (Clontech) were cultured in Eagle's minimal essential medium (EMEM) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 μg of penicillin-streptomycin per ml, and 100 μg of G418 sulfate per ml in the absence of tetracycline. One day prior to transfection, 3 × 105 cells were plated on 100 20-mm dishes. For each plate, 2 μg of ELL/pTRE or pTRE alone was mixed with 12 μl of lipofectamine (Gibco-BRL, Grand Island, N.Y.) in 600 μl of EMEM. This solution was incubated for 45 min, 2.4 ml of EMEM was added, and the cells were then overlaid with this complex solution for 4 h. After 48 h, the cells were harvested.
Stable transfection into 293 Tet Off cells.
293 Tet Off cells were cotransfected with 500 ng of pTK-Hyg vector (Clontech) and ELL/pTRE or pTRE alone as above. After 48 h, selection for stable clones was performed using tetracycline (1 μg/ml) and hygromycin B (100 μg/ml). Positive clones were selected using cloning cylinders and plated into individual flasks. ELL 293 cells are cultured in 90% EMEM with 10% FBS, 2 mM l-glutamine, penicillin-streptomycin (100 μg/ml), G418 sulfate (100 μg/ml) tetracycline (1 μg/ml) and hygromycin B (100 μg/ml). Pooled clones of ELL 293 Tet Off cells were used for functional studies.
Transient transfection of antisense ELL/pCDNA into ELL 293 Tet Off cells.
ELL 293 Tet Off cells were transiently transfected with 2 μg of pCDNA-ELLas or pCDNA alone as above, and cells were grown in the presence or absence of tetracycline for various times before harvesting.
Western blot analysis of expressed ELL.
Transiently transfected 293 Tet Off cells or ELL 293 Tet Off stable cell clones were grown in the presence or absence of tetracycline and lysed in lysis buffer (25 mm HEPES [pH 7.0], 0.25 M NaCl, 2.5 mM EDTA, 0.5 mM dithiothreitol, 10 μg of leupeptin and 1 μg of pepstatin A panel 2 mM phenylmethylsulfonyl fluoride, and 0.1% NP-40) for 30 min on ice. The cell extracts were boiled for 5 min at 95°C, and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% gels and electroblotted onto a nitrocellulose membrane for 1 h at 450 mA. After blocking for 30 min with 5% nonfat dry milk in Tris-buffered saline, blots were incubated with ELL monoclonal antibody or anti poly (ADP ribose) polymerase (PARP, Boehringer Mannheim, Indianapolis, Ind.) monoclonal antibodies overnight at 4°C. Blots were incubated with a 1:10,000 dilution of horseradish peroxidase-coupled goat anti-mouse immunoglobulin antibody (Dako, Carpinteria, Calif.), and immunoreactive proteins were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotechnology, Buckingshire, U.K.).
Cell proliferation and viability assays.
Cells were cultured at 1 × 105 cells/ml in the presence or absence of tetracycline for various times. Trypan blue dye exclusion assays were performed as previously described (19), or cells were analysed by phase contrast microscopy. In all assays, 150 to 300 cells were counted for each data point, and data were calculated as the mean ± standard error of triplicate samples and are representative of at least three separate assays. The number of apoptotic or dead cells (blue cells) was expressed as a percentage of the total cell number. To inhibit the activation of caspases, cells were treated for 30 min with peptidyl fluoromethylketones (zFA-fmk or zVAD-fmk; final concentration, 40 μM) (Enzyme System Products, Dublin, Calif.).
Propidium iodide staining.
Cells (2 × 105) grown in the presence or absence of tetracycline for various lengths of time were washed in phosphate-buffered saline (PBS) and fixed in 50% ice-cold ethanol–PBS (50% vol/vol) for 20 min on ice. Cell were washed with PBS and incubated in propidium iodide solution (69 μM propidium iodide 388 mM sodium citrate, 100 μg of Rnase A per ml) for 15 min at 37°C. Cells were immediately analyzed with a FACscan flow cytometer (BD Pharmingen, San Diego, Calif.).
Immunofluorescence microscopy.
Cells were resuspended in PBS supplemented with 5% bovine serum albumin for slide preparation and developed as described before using monoclonal antibodies generated to either ELL or active caspase 3 (49).
RESULTS
In an early attempt to study the physiological role of ELL, we set out to make a stable cell line constitutively expressing full-length ELL under the control of the Rous sarcoma virus promoter. Although transiently transfected cells expressed ample amounts of ELL, we were unable to select stable cell lines that expressed ELL at high levels (unpublished data). Stable cells selected for constitutive expression of ELL expressed meager amounts of the protein. Furthermore, the selected cells grew very poorly in comparison with mock-transfected cells. We therefore hypothesized that expression of full-length ELL may inhibit growth due to either loss of cell proliferation or induction of apoptosis.
Regulated expression of ELL.
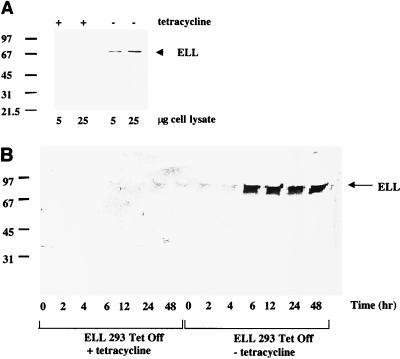
To test the above hypothesis, we constructed the ELL cDNA under the minimal cytomegaloviris (CMV) promoter regulated by the Tet operator and developed ELL 293 Tet Off cell lines expressing full-length ELL under control of the Tet-regulated promoter. Removal of tetracycline from the tissue culture medium for 48 h resulted in the expression of full-length ELL of approximately 80 kDa, as determined by Western blot using an ELL-specific monoclonal antibody (Fig. 1A). It is important to note that ELL in these selected cells is not overexpressed, but rather is produced in amounts that would normally be found in cells expressing ELL. This has been determined by comparing the amount of ELL expressed in 5 × 106 ELL 293 Tet Off cells expressing ELL to the amount expressed by 5 × 106 normal T cells (unpublished data). A time course experiment revealed that expression of ELL in ELL 293 Tet Off cells was detected as little as 6 h following the removal of the tetracycline from the medium (Fig. 1B). Consistent with the nuclear expression of endogenous ELL, Tet-regulated ELL was expressed exclusively in the nucleus of ELL 293 Tet Off cells grown in the absence of tetracycline, as determined by confocal microscopy and Western blot using nuclear lysates (data not shown).
FIG. 1.
ELL expression in the absence of tetracycline. (A) ELL/pTRE was co transfected with the pTK-Hyg selection marker into 293 Tet Off cells to create ELL 293 Tet Off stable cell lines with regulated ELL expression. Cells were incubated in the presence or absence of tetracycline for 48 h. In the presence of tetracycline, the promoter driving ELL expression is inactive. In the absence of tetracycline, the promoter driving ELL expression is activated. Western blot analysis of cell extracts was performed using an anti-ELL monoclonal antibody. The positions of molecular size markers are shown on the left (in kilodaltons), and the presence of a band of approximately 80 kDa in cells grown in the absence of ELL is indicated by an arrow on the right. (B) Time-dependent expression of ELL in ELL 293 Tet Off cells. ELL 293 Tet Off cells were grown in the presence or absence of tetracycline for 0, 2, 4, 6, 12, 24, and 48 h. Cells were harvested, and Western blot analysis was performed using an anti-ELL monoclonal antibody.
Effect of ELL expression on cell proliferation.
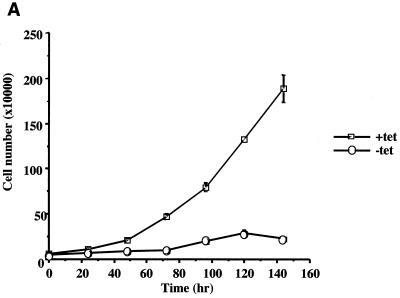
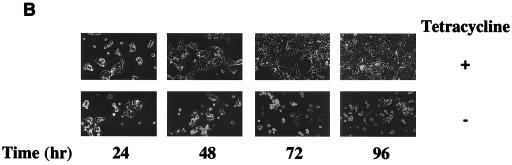
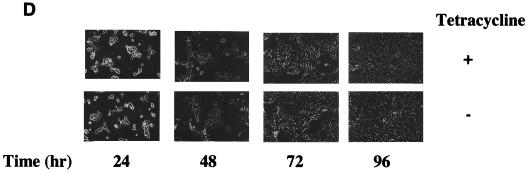
Having produced a cell line capable of expressing ELL in a regulated manner, we next tested the effect of ELL expression on the rate of cell growth. Incubation of ELL 293 Tet Off cells in the absence of tetracycline severely retarded cell proliferation over a 144-h time course compared to cells grown in the presence of tetracycline, as determined by counting cell numbers over time (Fig. 2A). ELL 293 Tet Off cells grown in the absence of tetracycline did not proliferate, and morphological changes were observed, with cells rounding up and detaching from the culture dish, as shown by phase contrast microscopy (Fig. 2B). By contrast, 293 Tet Off cells transfected with empty pTRE vector grew similarly in the presence and absence of tetracycline (Fig. 2C and D).
FIG. 2.
Cell growth in the presence and absence of ELL. ELL 293 Tet Off and 293 Tet Off cells were plated at 10% confluency and incubated in the presence or absence of tetracycline. Each day, cells were photographed and counted. (A) Growth curve and (B) phase contrast microscopy of ELL 293 Tet Off cells grown in the presence and absence of tetracycline. (C) Growth curve and (D) phase contrast microscopy of 293 Tet Off cells grown in the presence and absence of tetracycline.
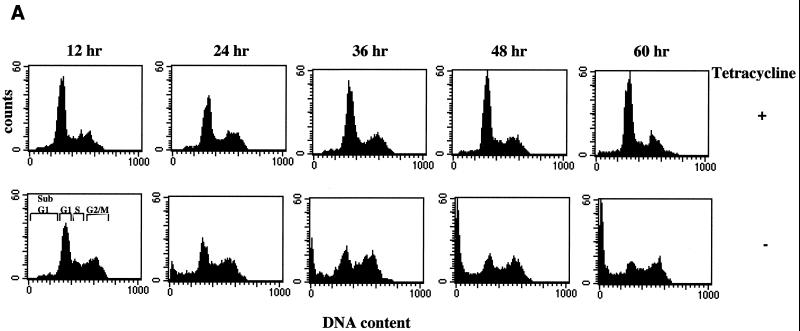
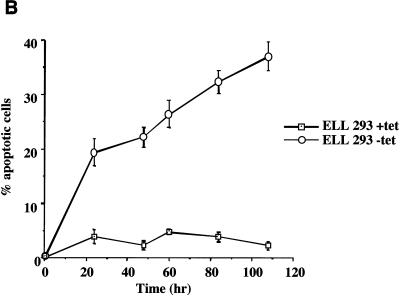
To determine whether ELL expression resulted in altered cell cycle regulation and/or induction of apoptosis, propidium iodide staining and flow cytometry were performed on ELL 293 Tet Off cells grown in the presence or absence of tetracycline (Fig. 3A). Induction of ELL expression resulted in a decrease in the percentage of cells in the G1/G0 phase of the cell cycle and a commensurate increase in the percentage cells in G2/M and sub-G1 (Fig. 3A and Table 1). The presence of sub-G1 cells indicated that these cells had lost or degraded genomic DNA, a phenomenon characteristic of cells undergoing apoptosis. The induction of cell death following removal of tetracycline in ELL 293 Tet Off cells was confirmed by trypan blue dye exclusion assays (Fig. 3B). As can be seen, induction of ELL expression correlated with a time-dependent increase in trypan-positive cells. These data indicate that regulated expression of ELL results in a loss of cell proliferation and induction of apoptosis. It is interesting that there was a consistent increase in the percentage of cells in G2/M following ELL induction, indicating that cells in this phase of the cell cycle were somewhat protected against ELL-mediated apoptosis or that there may be a cell cycle block.
FIG. 3.
Altered cell cycle and induction of cell death as a result of the expression of ELL. (A) Propidium iodide cell cycle analysis was performed on cells cultured at the same initial density for 60 h in the presence (top panels) or absence (bottom panels) of tetracycline. Cell cycle progression of parental 293 Tet Off cells was unaffected by tetracycline withdrawal. Expression of ELL correlated with a loss of the G1/G0 population of cells and an increase in the G2/M and sub-G1/G0 population. (B) Trypan blue dye exclusion assays were performed on ELL 293 Tet Off cells grown in the absence of tetracycline (tet). Induction of ELL resulted in a loss of cell membrane integrity and induction of cell death.
TABLE 1.
Percentage of ELL 293 Tet Off cells in various stages of the cell cycle with and without tetracycline
| Tetracycline | Time (h) | % of cells
|
|||
|---|---|---|---|---|---|
| Sub-G1 | G1 | S | G2/M | ||
| Present | 12 | 4.2 | 55.5 | 14.0 | 26.3 |
| 24 | 4.0 | 55.5 | 10.3 | 30.2 | |
| 36 | 2.9 | 62.6 | 9.4 | 25.1 | |
| 48 | 2.4 | 65.8 | 9.2 | 22.6 | |
| 60 | 3.2 | 63.2 | 9.2 | 24.4 | |
| Absent | 12 | 2.1 | 48.0 | 15.1 | 34.8 |
| 24 | 12.8 | 38.9 | 11.4 | 36.9 | |
| 36 | 17.7 | 31.6 | 12.1 | 38.6 | |
| 48 | 25.0 | 23.9 | 10.7 | 40.4 | |
| 60 | 28.6 | 20.7 | 9.4 | 41.5 | |
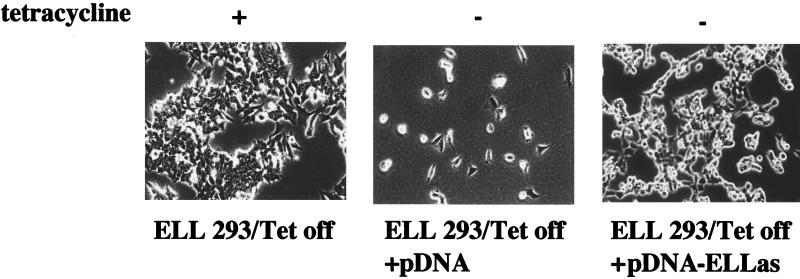
To determine whether the inhibition of cell growth and induction of apoptosis in ELL 293 Tet Off cells grown in the absence of tetracycline was directly related to the expression of the ELL protein, we determined whether the expression of an ELL antisense RNA could inhibit this effect. ELL 293 Tet Off cells were grown in the presence or absence of tetracycline and transfected with either pCDNA-ELLas, containing an ELL antisense cDNA under control of the CMV promoter, or pCDNA alone. Cells were grown for 4 days and then analyzed for the effect of the ELL antisense RNA on the rate of cell growth. As shown in Fig. 4, cells proliferated in the presence of tetracycline (left panel). However, consistent with our earlier results, ELL 293 Tet Off cells transfected with pCDNA and grown in the absence of tetracycline did not proliferate (Fig. 4, middle panel). However, when cells were transfected with the pCDNA-ELLas, containing ELL antisense cDNA under control of the CMV promoter vector, it was reproducibly observed that cell proliferation was restored (Fig. 4, right panel). These data indicate that the decrease in cell growth and alteration of cell viability in the ELL 293 Tet Off cells grown in the absence of tetracycline is a direct effect of expression of ELL in these cells.
FIG. 4.
Expression of ELL antisense mRNA results in reversal of ELL-dependent cell death. ELL 293 Tet Off cells were transiently transfected with pCDNA or pCDNA-ELLas, expressing antisense ELL mRNA, and grown in the absence of tetracycline for 96 h. Transfection of pCDNA-ELLas correlated with an increase in the number of viable cells present following tetracycline withdrawal and a loss of apoptotic morphology. The above picture is a representative colony that survived in the presence of the antisense mRNA.
ELL-induced apoptosis is mediated by caspases.
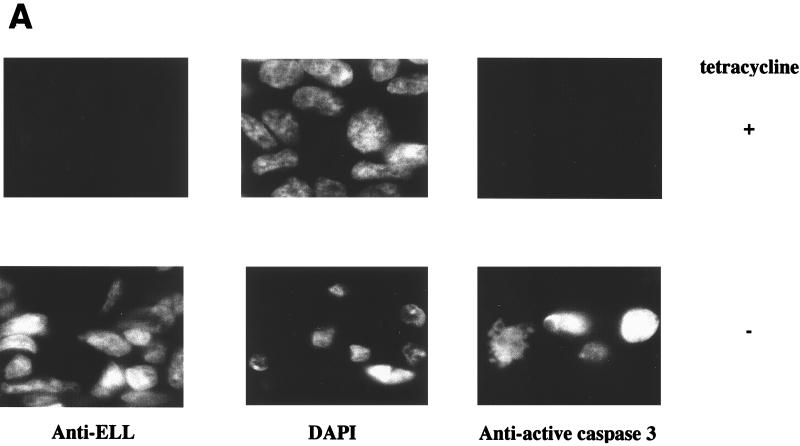
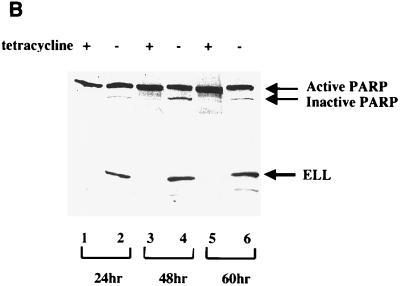
As demonstrated above, induction of ELL expression results in cell death, as determined by the loss of DNA content (Fig. 3A and Table 1) and plasma membrane integrity (Fig. 3B). Apoptosis is mediated by intracellular cysteine proteases (caspases), which are integral components of a highly organized and regulated programmed cell death cascade inducing the morphological changes associated with apoptosis (28). To determine if the expression of ELL results in caspase-dependent apoptotic death, ELL 293 Tet Off cells were grown in the presence or absence of tetracycline for 24 h and analyzed by microscopy for chromatin condensation (a morphological hallmark of apoptosis) and caspase activity. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize DNA and either anti-ELL monoclonal antibody or anti-activated caspase 3 monoclonal antibody. As shown in Fig. 5A, growth of ELL 293 Tet Off cells in the absence of tetracycline results in the expression of ELL in the nucleus coincident with chromatin condensation and activation of caspase 3. In the presence of tetracycline, ELL is not expressed, caspase 3 is not active, and the cell nuclei appear normal. The DNA repair enzyme PARP is a cellular caspase substrate specifically cleaved and inactivated by activated caspase 3 during programmed cell death. We therefore tested the cleavage and inactivation of PARP in response to the regulated expression of ELL over 60 h. Cell lysates were prepared and analyzed by SDS-PAGE and Western blot (Fig. 5B) using antibodies against PARP (top panel) and ELL (bottom panel). Cleavage of PARP from the 118-kDa active form to the 89-kDa inactive form corresponded with induction of ELL expression (Fig. 5B).
FIG. 5.
Expression of ELL induces chromatin condensation and results in the activation of caspase 3. (A) ELL 293 Tet Off cells were grown in the presence or absence of tetracycline for 24 h and analyzed for ELL expression using an anti-ELL monoclonal antibody (left panels), changes to the nucleus via DAPI staining (middle panels), and activation of caspase 3 using a monoclonal antibody specific for the active caspase 3 fragment (right panels). Induction of ELL expression in the nucleus correlated with chromatin condensation and caspase 3 activation. (B) Protein lysates were prepared from ELL 293 Tet Off cells grown in the presence or absence of tetracycline and separated by SDS-PAGE. Western blots were performed using polyclonal anti-PARP (top panel) or monoclonal anti-ELL (bottom panel) antibodies. The positions of ELL and the active 118-kDa and inactive 89-kDa forms of PARP are indicated.
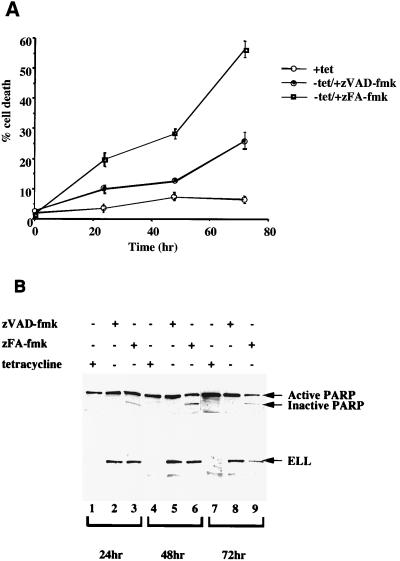
To determine whether activation of caspases in response to expression of ELL is necessary for the induction of apoptosis, we tested the effect of the caspase inhibitor zVAD-fmk on ELL 293 Tet Off cells grown in the absence of tetracycline. As shown in Fig. 6A, removal of tetracycline induced cell death in ELL 293 Tet Off cells, and this death could be significantly inhibited by preincubation of cells with zVAD-fmk but not with the control peptide zFA-fmk. ELL-induced DNA damage was also inhibited by addition of zVAD-fmk, as shown in Table 2, with a significant decrease in the sub-G1 population of cells compared to cells treated with the zFA-fmk control peptide over a 72-h time course. To confirm that zVAD-fmk was indeed inhibiting ELL-induced caspase activation, Western blots to detect PARP cleavage were performed using ELL 293 Tet Off cells grown in the presence or in the absence of tetracycline with zVAD-fmk or zFA-fmk (Fig. 6B). Once again, expression of ELL induced cleavage of PARP, indicating that caspases were activated. Importantly, addition of zVAD-fmk inhibited ELL-induced PARP cleavage, while zFA-fmk had no effect. Taken together, these data demonstrate that expression of ELL induces caspase activation and the activation of these apoptotic molecules is important to mediate apoptosis following ELL's expression in these stable cell lines.
FIG. 6.
Induction of cell death by ELL can be reversed by inhibition of caspase activation. ELL 293 Tet Off cells were treated with the caspase inhibitor zVAD-fmk or the control peptide zFA-fmk and grown in the presence or absence of tetracycline (tet) for 72 h. (A) Induction of cell death was determined by trypan blue dye exclusion assays. Apoptosis was induced in ELL 293 Tet Off cells following tetracycline withdrawal, and this death could be inhibited with zVAD-fmk but not with the control zFA-fmk peptide. (B) Protein lysates were prepared from ELL 293 Tet Off cells treated as above and separated by SDS-PAGE. Western blots were performed using monoclonal anti-ELL (top panel) or polyclonal anti-PARP (bottom panel) antibodies. The positions of ELL and the active (118 kDa) and inactive (89 kDa) forms of PARP are indicated.
TABLE 2.
Inhibition of apoptotic (sub-G1) ELL-expressing 293 Tet Off cells by addition of the zVAD-fmk caspase inhibitor
| Time (h) | Tetracycline | Inhibitor | % of cells
|
|||
|---|---|---|---|---|---|---|
| Sub-G1 | G1 | S | G2/M | |||
| 24 | + | None | 4.9 | 55.4 | 17.3 | 22.4 |
| − | zVAD-fmk | 11.2 | 28.3 | 19.8 | 40.7 | |
| − | zFA-fmk | 21.7 | 22.9 | 19.1 | 36.3 | |
| 48 | + | None | 9.0 | 54.2 | 16.0 | 21.3 |
| − | zVAD-fmk | 17.3 | 34.4 | 18.3 | 30.5 | |
| − | zFA-fmk | 33.1 | 30.5 | 14.5 | 23.7 | |
| 72 | + | None | 4.6 | 54.3 | 18.2 | 22.9 |
| − | zVAD-fmk | 39.9 | 26.5 | 10.7 | 21.9 | |
| − | zFA-fmk | 64.0 | 19.0 | 6.7 | 10.3 | |
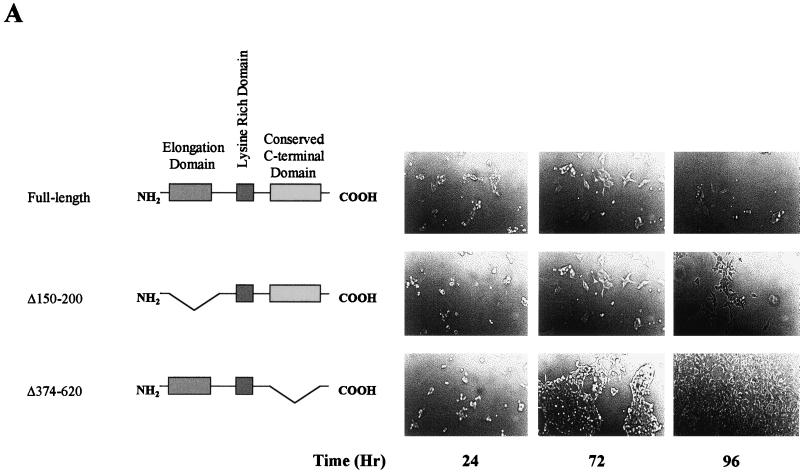
To test the hypothesis that the cellular regulatory activity of ELL is specific and associated with a particular domain of ELL, we generated several mutations within the ELL protein and developed several clonal cell lines with regulated expression of ELL mutant proteins. We reasoned that having such cell lines available, we should be able to define which domain of ELL is required for its cellular growth regulatory activity. Our data presented here (Fig. 7) indicate that the elongation activation domain of ELL is dispensable for the growth-regulatory activity of ELL. However, clonal cell lines expressing ELL missing the conserved C-terminal domain do not demonstrate any slow-growth phenotype (Fig. 7A). We also tested the level of expression of ELL and ELL mutants in these cells (Fig. 7B). This observation indicates that the cellular regulatory activity of ELL in these clonal cell lines is specifically associated with the C-terminal domain of ELL and is not due to a nonspecific ectopic expression of ELL. We have also recently shown that this domain of ELL is highly conserved among the three human ELL proteins that we have cloned (26a). The conservation of the C-terminal domain of ELL is an indication of its physiological importance. We have also demonstrated that this conserved C-terminal domain of ELL is sufficient and essential for the immortalization activity of the MLL-ELL chimera, which is found in patients with leukemia (9a).
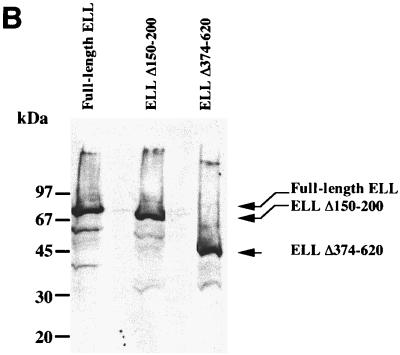
FIG. 7.
Growth comparison among cells expressing full-length ELL and those expressing ELL deletion mutants. (A) Cells expressing full-length ELL, ELL(Δ150–200), or ELL(Δ374–620) were plated at 106 cells per 25-cm2 flask and incubated in the absence of tetracycline. Cells were photographed at 24, 72, and 96 h after plating to record growth. (B) Plating was performed as in panel A, and cells were incubated in the absence of tetracycline and harvested at 72 h after plating. Total cell lysates were prepared and analyzed by SDS-PAGE and Western analysis employing an ELL-specific monoclonal antibody to detect ELL and ELL mutants.
DISCUSSION
Our data demonstrate that expression of ELL results in a loss of the G1 population of cells, induction of caspase activation, and cell death. A role for caspases in mediating ELL-induced cell death was shown by the addition of the broad caspase-inhibitory peptide zVAD-fmk, which significantly inhibited death of cells following ELL expression. One question that now remains and is currently being tested in our laboratories is how the caspases are activated following ELL expression. At present, there are two well-defined pathways leading to caspase-mediated cell death (12). The first involves activation of the so-called death receptor pathway, resulting in caspase activation following binding of ligands such as Fas ligand, tumor necrosis factor, or tumor necrosis factor receptor-like apoptosis-inducing ligand to their cell surface receptors (42). Another arm of the caspase-mediated cell death pathway involves the mitochondria (12). In response to stress stimuli such as serum starvation and addition of chemotherapeutic drugs, the mitochondrial membrane potential is affected, resulting in release of mitochondrial components such as cytochrome c and caspase activation. The death receptor and mitochondrial apoptotic pathways are linked by caspase 8, which not only cleaves and activates caspase 3 but also cleaves and activates Bid, which in turns inserts into the mitochondrial membrane to induce cytochrome c release (9).
The 293 cells contain and express the genes for adenovirus proteins E1A, E1B-19K, and E1B-55K (2, 21). The E1A proteins are transcriptional adaptors that deregulate the cell cycle and, in the absence of the E1B-19K and E1B-55K proteins, induce apoptosis (reviewed in reference 51). The E1B-19K protein is a distant functional homolog of antiapoptotic members of the Bcl-2 family (51). E1B-19K inhibits apoptosis at several levels: it binds proapoptotic members of the Bcl-2 family (6), it binds CED-4 (the homolog of Apaf-1) (14), and it inhibits Fas-mediated apoptosis through Fas-associated death domain sequestration of procaspase 8 (32). The E1B-55K protein inhibits apoptosis by forming a complex with the tumor suppressor p53 and repressing p53-responsive promoters (24, 51). It is not known what effect, if any, these adenovirus proteins have on the apoptosis observed after expression of ELL. ELL-mediated caspase 3 activation and apoptosis could occur via a pathway that is not blocked by E1B-19K and E1B-55K. Alternatively, and perhaps more likely, ELL-induced apoptosis may occur through known pathways, and E1B-19K and E1B-55K are insufficient to block these pathways.
While caspases may be activated in the absence of de novo protein synthesis, there is a link between gene regulation and apoptosis. For example, DNA damage by chemotherapeutic drugs or ionizing radiation results in activation of the p53 tumor suppressor protein, which in turn transcriptionally activates Bax (9). The interferon-responsive transcriptional activator IRF-1 has been shown to induce cell death by transcriptionally upregulating caspases and the Fas receptor (46). Recently, it has been shown that regulation of gene expression on a more general scale by agents such as trichostatin A, sodium butyrate, and suberoylanilide hydroxamic acid, which function as histone deacetylase inhibitors, can induce caspase activation and apoptosis (4, 5, 50). As is the case for ELL, the molecular mechanism(s) underpinning apoptosis induced by the above agents has not been dissected, and the specific genes with altered expression in response to inhibition of histone deacetylases have yet to be identified.
Our preliminary structure-function studies suggest that removal of the elongation activation domain of ELL did not induce cell death but did induce growth suppression (Fig. 7). Furthermore, ELL lacking the C terminus, which is conserved among the ELL family of proteins and plays an essential role in the immortalization of myeloid progenitors, fails to mediate either apoptosis or growth suppression. Comprehensive structure-function analyses are currently being performed to identify the molecular functions of ELL that are necessary and sufficient to induce ELL-mediated growth suppression and/or cell death.
Chromosomal translocations at 11q23 are commonly seen in AML and ALL, resulting in the formation of fusion proteins containing the product of the MLL gene. The production of these fusion proteins is thought to provide a survival or proliferative advantage to myeloid progenitor cells to mediate tumorigenesis. In both ALL and AML, MLL has been shown to be fused to at least 16 different cellular genes, many of which are involved in transcription activation or maintenance of gene expression. Indeed, MLL itself has been reported to be a transcription factor that can regulate the expression of homeobox genes (36). The molecular function of the MLL-ELL fusion protein has not yet been investigated, and while ELL has been demonstrated to play a role in transcription elongation, a physiological function for ELL has not been identified. Our results showing that ELL itself can regulate cell survival indicates that the (t11; 19)(q23; p13.1) (MLL-ELL) translocation may contribute to leukemogenesis in a number ways. First, the loss of function of wild-type MLL could lead to a loss of transcriptional maintenance during morphogenesis, resulting in aberrant cellular development, as demonstrated in MLL knockout mice (15, 52, 53). Second, our studies showing that ELL may regulate apoptosis indicate that a loss of ELL function could result in survival of cells that otherwise may have been destined to die. Additionally, ELL and MLL-ELL can bind to and regulate the transcriptional activities of the p53 tumor suppressor protein (23, 41). Maki and colleagues speculate that altered expression of MLL-ELL or increased in vivo half-life of the fusion protein compared to the wild-type ELL protein may be important for tumor formation, as aberrant expression of MLL-ELL could affect the antitumor activities of p53. Thus, loss of ELL expression and formation of the MLL-ELL fusion protein due to the (11; 19)(q23; p13.1) (MLL; ELL) chromosomal translocation may have at least two consequences: loss of ELL-induced apoptosis, and altered p53 function due to overexpression of MLL-ELL. Indeed it has been demonstrated that p53 mutations occur infrequently in leukemias with 11q23 translocations (26). Therefore, if, as in many other cancers, disruption of p53 function is important for the development of ALL and/or AML, then the overexpression of MLL-ELL may serve this purpose. Finally, the MLL-ELL fusion protein itself is likely to have oncogenic properties, similar to that demonstrated for the MLL-AF9 and MLL-ENL fusion proteins (8, 10, 22). The recent demonstration that MLL fused to ELL, AF9, or ENL can inhibit apoptosis induced by GADD34 while the wild-type MLL protein does not also points to a gain-of-function process as being important for leukemogenesis. Thus, the combination of a loss of MLL and ELL functions and the gain of function resulting from production of the MLL-ELL fusion protein could be sufficient and/or necessary for induction of leukemia following a (t11; 19)(q23; p13.1) (MLL-ELL) translocation.
The molecular mechanisms leading to ELL-induced cell death have yet to be identified and are subject to ongoing experiments in our laboratories. However, our demonstration that the C-terminal domain of ELL, which is conserved among the three ELL proteins that we have cloned (ELL, ELL2, and ELL3), is required for this cellular activity of ELL focuses our studies on the physiological role of this conserved domain of ELL. We are currently testing whether the expression of ELL results in altered expression of genes that are involved in cell death or whether ELL plays a direct role in the regulation of cell growth and survival. The absence of genetically altered mice either devoid of wild-type ELL expression or aberrantly overexpressing ELL with which to study such a physiological function makes the question regarding the role of ELL in regulation of cell growth and survival difficult to answer at present. However, our studies demonstrating that expression of wild-type ELL can induce cell death, coupled with the finding that the ELL locus is frequently disrupted in ALL and AML, raises the possibility that a biological function of ELL may be to regulate cell growth and survival.
ACKNOWLEDGMENTS
We thank Sarah Russell, Mark Smyth, and Trissa Miller for helpful discussions.
This work is supported in part by a grant from the American Cancer Society (RPG-99-218-01-MGO) to A.S. A.S. is an Edward Mallinckrodt, Jr., Young Investigator. R.J. is a Wellcome Trust Senior Research Fellow.
REFERENCES
- 1.Adler H T, Chinery R, Wu D Y, Kussick S J, Payne J M, Fornace A J, Jr, Tkachuk D C. Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 proteins. Mol Cell Biol. 1999;10:7050–7060. doi: 10.1128/mcb.19.10.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiello L, Guilfoyle R, Huebner K, Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293) Virology. 1979;94:460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- 3.Bernard O A, Mauchauffe M, Mecucci C, Van den Berghe H, Berger R. A novel gene, AF-1p, fused to HRX in t(1; 11)(p32; q23), is not related to AF-4, AF-9 nor ENL. Oncogene. 1994;9:1039–1045. [PubMed] [Google Scholar]
- 4.Bernhard D, Ausserlechner M J, Tonko M, Loffler M, Hartmann B L, Csordas A, Kofler R. Apoptosis induced by the histone deacetylase inhibitor sodium butyrate in human leukemic lymphoblasts. FASEB J. 1999;13:1991–2001. doi: 10.1096/fasebj.13.14.1991. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard D, Loffler M, Hartmann B L, Yoshida M, Kofler R, Csordas A. Interaction between dexamethasone and butyrate in apoptosis induction: non-additive in thymocytes and synergistic in a T cell-derived leukemia cell line. Cell Death Differ. 1999;6:609–617. doi: 10.1038/sj.cdd.4400531. [DOI] [PubMed] [Google Scholar]
- 6.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 7.Chaplin T C, Ayton P, Bernard O A, Saha V, Della Valle V, Hillion J, Gregorini A, Lillington D, Berger R, Young B D. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10; 11) translocation in acute leukemia. Blood. 1995;85:1435–1441. [PubMed] [Google Scholar]
- 8.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N, King G, Rabbitts T H. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 9.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou J C. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.DiMartino J F, Miller T, Ayton P M, Landewe T, Hess J L, Cleary M L, Shilatifard A. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood. 2000;96:3887–3893. [PubMed] [Google Scholar]
- 10.Dobson C L, Warren A J, Pannell R, Forster A, Lavenir I, Corral J, Smith A J, Rabbitts T H. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 12.Green D, Kroemer G. The central executioners of apoptosis: caspases or mitochondria? Trends Cell Biol. 1998;8:267–271. doi: 10.1016/s0962-8924(98)01273-2. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Nakamura T, Alder H, Pasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4; 11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Wallen H D, Nunez G, White E. E1B 19,000-molecular-weight protein interacts with and inhibits CED-4-dependent, FLICE-mediated apoptosis. Mol Cell Biol. 1998;18:6052–6062. doi: 10.1128/mcb.18.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess J L, Yu B D, Li B, Hanson R, Korsmeyer S J. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1995;90:1799–1806. [PubMed] [Google Scholar]
- 16.Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard O A. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood. 1997;90:3714–3719. [PubMed] [Google Scholar]
- 17.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 18.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;79:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 19.Johnstone R W, Cretney E, Smyth M J. P-glycoprotein protects leukemia cells against caspase-dependent, but not caspase-independent, cell death. Blood. 1999;93:1075–1085. [PubMed] [Google Scholar]
- 20.Kanda Y, Mitani K, Kurokawa M, Yamagata T, Yazaki Y, Hirai H. Overexpression of the MEN/ELL protein, an RNA polymerase II elongation factor, results in transformation of Rat1 cells with dependence on the lysine-rich region. J Biol Chem. 1999;273:5248–5252. doi: 10.1074/jbc.273.9.5248. [DOI] [PubMed] [Google Scholar]
- 21.Lassam N J, Bayley S T, Graham F L. Tumor antigens of human Ad5 in transformed cells and in cells infected with transformation-defective host-range mutants. Cell. 1978;18:781–91. doi: 10.1016/0092-8674(79)90131-4. [DOI] [PubMed] [Google Scholar]
- 21a.Lavau C, Luo R T, Du C, Thirman M J. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci USA. 2000;97:10984–10989. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavau C, Szilvassy S J, Slany R, Cleary M L. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maki K, Mitani K, Yamagata T, Kurokawa M, Kanda Y, Yazaki Y, Hirai H. Transcriptional inhibition of p53 by the MLL/MEN chimeric protein found in myeloid leukemia. Blood. 1999;93:3216–3224. [PubMed] [Google Scholar]
- 24.Martin M E, Berk A J. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol Cell Biol. 1999;19:3403–3414. doi: 10.1128/mcb.19.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. t(11;22)(q23;q11.2) in acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Megonigal M D, Rappaports E F, Nowell P C, Lange B J, Felix C A. Potential role for wild-type p53 in leukemias with MLL gene translocations. Oncogene. 1998;16:1351–1356. doi: 10.1038/sj.onc.1201637. [DOI] [PubMed] [Google Scholar]
- 26a.Miller T, Williams K, Johnstone R W, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052–32056. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- 27.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 28.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W M, Nowell P C, Croce C M, Canaani E. Genes on chromosomes 4, 9, and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osaka M, Rowley J D, Zeleznik-Le N J. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11; 17)(q23; q25) Proc Natl Acad Sci USA. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parry P, Wie Y, Evans G. Cloning and characterization of the t(X; 11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer. 1994;11:79–84. doi: 10.1002/gcc.2870110203. [DOI] [PubMed] [Google Scholar]
- 32.Perez D, White E. E1B 19K inhibits Fas-mediated apoptosis through FADD-dependent sequestration of FLICE. J Cell Biol. 1998;141:1255–1266. doi: 10.1083/jcb.141.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale R P, Nowell P C, Kuriyama K, Miyazaki Y, Croce C M, Canaani E. Cloning of the ALL-1 fusion partner, the AF-6 gene, involved in acute myeloid leukemias with the t(6;11) chromosome translocation. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- 34.Prasad R, Leshkowitz D, Gu Y, Alder H, Nakamura T, Saito H, Huebner K, Berger R, Croce C M, Canaani E. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc Natl Acad Sci USA. 1994;91:8107–8111. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowley J D. Backtracking leukemia to birth. Nat Med. 1997;4:150–151. doi: 10.1038/nm0298-150. [DOI] [PubMed] [Google Scholar]
- 36.Rowley J D. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 37.Rubnitz J E, Morrissey J, Savage P A, Cleary M L. ENL, the gene fused with HRX in t(11; 19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells. Blood. 1994;84:1747–1752. [PubMed] [Google Scholar]
- 38.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 39.Shilatifard A, Haque D, Conaway R C, Conaway J W. Structure and function of RNA polymerase II elongation factor ELL: identification of two overlapping ELL functional domains that govern its interaction with polymerase and the ternary elongation complex. J Biol Chem. 1997;272:22355–22363. doi: 10.1074/jbc.272.35.22355. [DOI] [PubMed] [Google Scholar]
- 40.Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 41.Shinobu N, Maeda T, Aso T, Ito T, Kondo T, Koike K, Hatakeyama M. Physical interaction and functional antagonism between the RNA polymerase II elongation factor ELL and p53. J Biol Chem. 1999;274:17003–17010. doi: 10.1074/jbc.274.24.17003. [DOI] [PubMed] [Google Scholar]
- 42.Smyth M J, Johnstone R W. The role of TNF in lymphocyte-mediated cytotoxicity. Microsc Res Technol. 2000;50:196–208. doi: 10.1002/1097-0029(20000801)50:3<196::AID-JEMT3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 43.So C W, Caldas C, Liu M M, Chen S J, Huang Q H, Gu L J, Sham M H, Wiedemann L M, Chan L C. EEN encodes a member of a new family of proteins containing an Src homology 3 domain and is the third gene located on chromosome 19p13 that fuses to MLL in human leukemia. Proc Natl Acad Sci USA. 1997;94:2563–2568. doi: 10.1073/pnas.94.6.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J A, Zelezni-Le N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11; 16)(q23; p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taki T, Shibuya N, Taniwaki M, Hanada R, Morishita K, Bessho F, Yanagisawa M, Hayashi Y. ABI-1, a human homolog to mouse Abl-interactor 1, fuses the MLL gene in acute myeloid leukemia with t(10; 11)(p11.2; q23) Blood. 1998;92:1125–1130. [PubMed] [Google Scholar]
- 46.Tamura T, Ishihara M, Lamphier M S, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak T W, Taki S, Taniguchi T. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature. 1995;376:596–599. doi: 10.1038/376596a0. [DOI] [PubMed] [Google Scholar]
- 47.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J. Cloning of ELL, a gene that fuses to MLL in a t(11; 19)(q23; p13.1) in acute myeloid leukemia. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 49.Tollefson A E, Hermiston T W, Lichtenstein D L, Colle C F, Tripp R A, Dimitrov T, Toth K, Wells C E, Doherty P C, Wold W S. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature. 1998;392:726–30. doi: 10.1038/33712. [DOI] [PubMed] [Google Scholar]
- 50.Vrana J A, Decker R H, Johnson C R, Wang Z, Jarvis W D, Richon V M, Ehinger M, Fisher P B, Grant S. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene. 1999;18:7016–7025. doi: 10.1038/sj.onc.1203176. [DOI] [PubMed] [Google Scholar]
- 51.Wold W S M, Chinnadurai G. Adenovirus proteins that regulate apoptosis. In: Cann A J, editor. DNA virus replication. Oxford, U. K: Oxford University Press; 2000. pp. 200–232. [Google Scholar]
- 52.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 53.Yu B D, Hanson R D, Hess J L, Horning S E, Korsmeyer S J. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]