Summary
Seasonal influenza infects between 10 and 50 million people in the United States every year. Accurate forecasts of influenza and influenza-like illness (ILI) have been named by the CDC as an important tool to fight the damaging effects of these epidemics. Multi-model ensembles make accurate forecasts of seasonal influenza, but current operational ensemble forecasts are static: they require an abundance of past ILI data and assign fixed weights to component models at the beginning of a season, but do not update weights as new data on component model performance is collected. We propose an adaptive ensemble that (i) does not initially need data to combine forecasts and (ii) finds optimal weights which are updated week-by-week throughout the influenza season. We take a regularized likelihood approach and investigate this regularizer’s ability to impact adaptive ensemble performance. After finding an optimal regularization value, we compare our adaptive ensemble to an equal-weighted and static ensemble. Applied to forecasts of short-term ILI incidence at the regional and national level, our adaptive model outperforms an equal-weighted ensemble and has similar performance to the static ensemble using only a fraction of the data available to the static ensemble. Needing no data at the beginning of an epidemic, an adaptive ensemble can quickly train and forecast an outbreak, providing a practical tool to public health officials looking for a forecast to conform to unique features of a specific season.
Keywords: Combination forecasting, Forecast aggregation, Influenza, Statistics, Public health
1 |. INTRODUCTION
Every year seasonal influenza costs hospitals time and resources and causes significant loss of life, especially among patients with cardiac disease, previous respiratory illness or allergies, children and the elderly1,2,3,4. During the season’s peak incidence hospitals admit patients beyond capacity, and in severe cases, for greater lengths of stay that postpone lower priority but still important surgeries5,6,7. The total economic burden of influenza outbreaks (including medical costs, lost earnings, and loss of life) in the US alone are estimated to exceed $85 billion annually7.
Accurate forecasts of influenza can help with public health response and mitigate the impact of an outbreak. Public health officials at the national, regional, and state level benefit from the advance warning forecasts can provide by preparing hospitals for increased disease burden8, shifting vaccines to places in need9, and alerting the public10. Forecasts of the intensity, timing, and overall impact of influenza are currently used to help guide targeted prevention and treatment messages to the public, and used to inform public health officials11,12. There are many examples, not just for influenza, where forecasting efforts have improved situational awarness for public health officials13,14,15. Recognizing the value of accurate forecasting, the US Centers for Disease Control and Prevention (CDC) have organized forecasting challenges around seasonal influenza outbreaks11,16,17. These challenges focus on generating weekly forecasts of influenza-like illness (ILI) rates at the national and regional level in the US. ILI is a syndromic classification, defined as a fever plus an additional symptom such as a cough or sore throat. Public health officials regard ILI surveillance data as an important indicator of the trends and severity of a given influenza season18.
The CDC influenza forecasting challenges, called FluSight, have spurred methodological development in the area of influenza forecasting and infectious disease forecasting more broadly. This effort has led multiple research groups to develop models trained on historical ILI data to forecast future incidence across the US. These models include statistical and mechanistic models, among other approaches 19,20,21,22.
Decision makers often request a single unified forecast and several modeling groups have developed multi-model ensemble forecasts of influenza. These models synthesize predictions from a set of individual component models and have shown better predictive ability compared to any single model23,24,25.
Multi-model ensembles aggregate diverse component model predictions into a final ‘meta-prediction’ that is often, but not always, more robust than any single component model26,27. This robustness comes from the ensemble’s mixture of component models. By combining different types of models, the ensemble is freed from having to make a single set of assumptions about the data26,27. In contrast to ensemble modeling approaches like bagging28,29 and boosting30,29 which create a library of similar component models, multi-model ensembles combine distinct component models together, possibly models designed and implemented in completely different modeling paradigms. Applications include weather 31, economics32, energy production33, education34, and infectious disease25.
The multi-model ensembles addressed in this work are related to Bayesian Model Averaging (BMA)35,36,37,38. The fundamental difference between multi-model ensembles and BMA is the assumed data-generating process. BMA assumes a single model from an ensemble generated the entire data stream, and weights are assigned to models proportional to the probability they generated the entire observed data stream35. Multi-model ensembles assume each observation was generated by a convex combination of predictive distributions from the ensemble39,40,41. No one model generated all the data.
This work builds on an existing ensemble implementation25 by developing a new method for combining component models that relies on recently observed in-season data to adaptively estimate a convex combination of models, sometimes referred to as a “linear pool”. Though past work has studied methods for training ensembles from a fixed data set42,43,44 and adapting ensemble weights to streaming data45,46,47, a unique and new challenge in our setting is to assess component model performance over the course of a season from factors such as variability in component model performance over the course of a season or the reliance on noisy, revision-prone observations like weekly ILI surveillance data. Component model performance has been shown to change based on the target that is predicted, when predictions are made over the course of the influenza season, and across different seasons48. Recent ILI observations, because they come from a real-time public health surveillance system, are subject to revisions, which are occasionally substantial and can negatively impact forecast accuracy49,48. Like past work in ensemble forecasting, this work combines a fixed set of probabilistic forecasting models provided by different forecasting teams48 and does not presume to have the capability to refit component model parameters.
To protect the adaptive multi-model ensemble framework from relying too heavily on recent, revision-prone ILI data and on past component model performance that is not necessarily correlated with future model performance, we developed a Bayesian model combination algorithm that uses a prior to regularize ensemble weights. Previous results in this field show equally-weighted ensembles often perform well17,25, and to align with these results, we chose a uniform Dirichlet prior that shrinks model weights towards equal. Our prior is also time dependent. Unlike a typical model where the prior becomes less influential with increasing data, our prior exerts a constant influence on the final model weights. Our model never allows model weights to depend only on recent model performance and revision-prone data. We show our method can be implemented using a variational inference algorithm (devi-MM), and demonstrate that an existing expectation-maximization approach to ensemble weight estimation25 is a special case.
We compared ensembles that assign equal weights to models (EW) and those with possibly unequal but static and unchanging weights throughout a season (static), against our adaptive ensemble (adaptive) which updated component model weights every week. Static ensembles were trained on all cumulative forecasting data before the beginning of each season and were unable to modify their model weights in-season. In contrast, adaptive ensembles were only trained on within-season data—starting the season with no training data—but could modify their weights over time as each additional week of ILI data provided information about component model performance. Comparing static and equal to adaptive ensembles, we highlight: (i) the adaptive model’s ability to buffer against sparse and revision-prone data by using a prior over ensemble weights and (ii) similar, and sometimes improved, performance when comparing adaptive to static models, where adaptive models require substantially less data to train.
The manuscript is structured as follows: Section 2 describes the CDC influenza training data and component models included in all ensembles, defines performance metrics, and develops the methodology for our adaptive ensemble. Section 3 presents results, investigating the prior’s impact on adaptive ensemble performance and compares the adaptive ensemble to static and equally weighted ensembles. Section 4 relates this work to linear pooling algorithms, and discusses our approach in the broader context of data-driven, evidence-based public health decision making.
2 |. METHODS
2.1 |. Data
2.1.1 |. Influenza data
Every week throughout the season, for 10 different Health and Human Services reporting regions (HHS1, HHS2, …, HHS10) and a national average, the CDC publishes surveillance data on the number of patients who visited a sentinel site and the number of those patients that were diagnosed with influenza-like illness (ILI)(See Fig. 1 for ILI over time for all regions and seasons.). Percent ILI is defined as the number of patients presenting with a fever (greater than 100F) plus cough or sore throat divided by the number of all patient visits times one hundred. More than 3,500 outpatient clinics report ILI data to the CDC as part of the ILINet surveillance system. Every week, new ILI data and updates to past ILI data are published.
FIGURE 1.
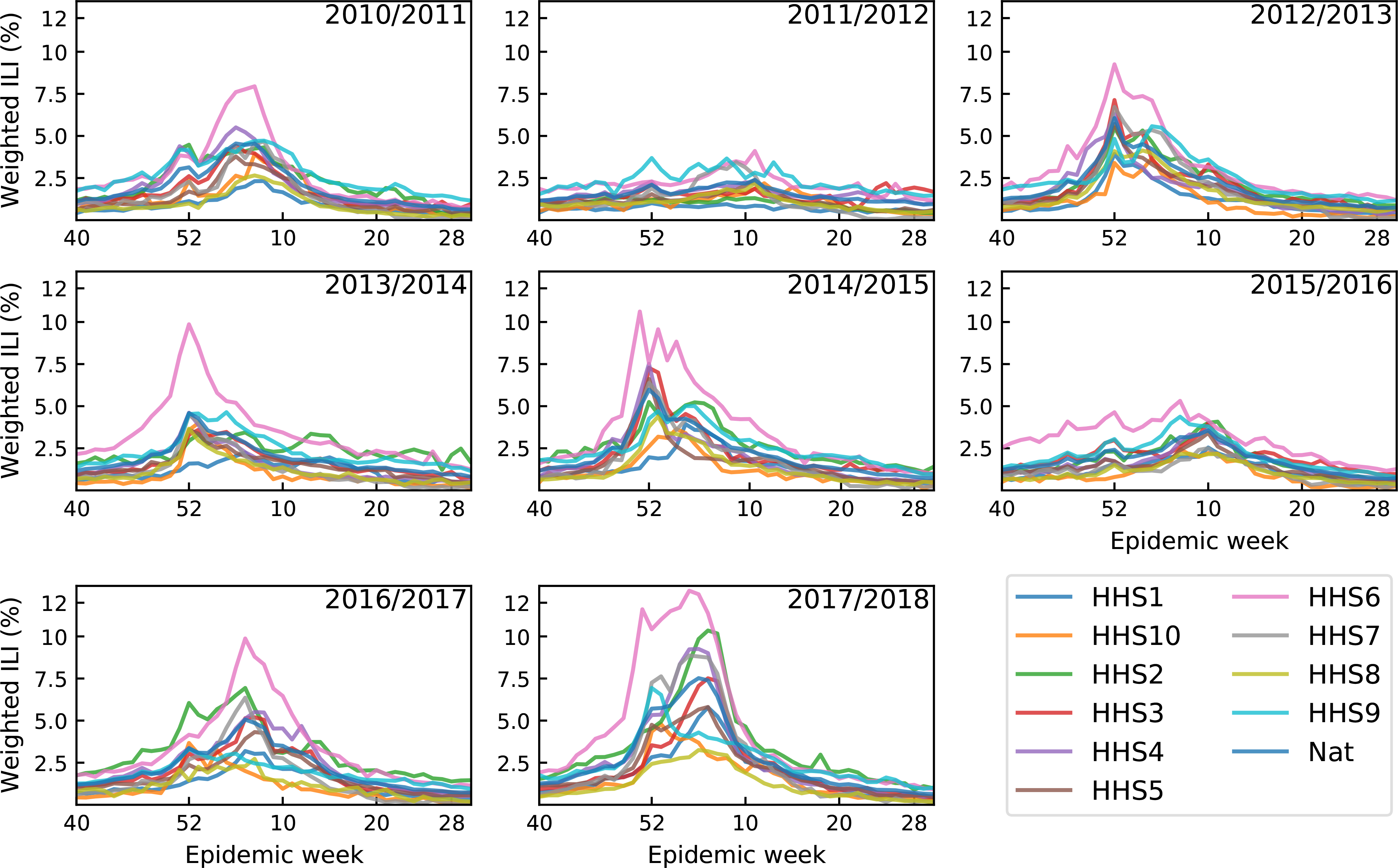
Weighted influenza-like illness by epidemic week for all 10 HHS regions and at the national level, for seasons 2010/2011 up to season 2017/2018, and an example component model probabilistic forecast. Most often, influenza-like illness percents are small at the beginning of the season (epidemic week 40), increase to a single peak, and then decrease. On occasion more than one influenza-like illness peak will form as was the case for the national ILI for season 2017/2018. Probabilistic forecasts attempt to quantify the uncertainity in future ILI
Because of reporting delays, ILI percentages undergo revisions every week after they’re first reported, finalized several weeks after the season ends (See Suppl. G4 for an example of revisions to ILI in the 2017/2018 season). Component model forecasts are updated week-to-week because (i) new ILI data is reported and (ii) all previous ILI values are revised. Revisions are one factor that make predicting future ILI difficult, and when forecast models adjust for this revision process they typically outperform models that do not account for data revisions50. Revisions to past ILI data is one factor that will cause component model performance from past weeks to change that in turn modifies optimal ensemble weight assignments.
2.1.2 |. Forecasting data
The FluSight Network (FSN) is a collaborative group of influenza forecasters that use historical ILI data to build retrospective component models and ensemble forecasts. Forecasts are probabilistic densities over future influenza-like illness percentages and, most often, public health officials are provided with probabilisitic densities, the median, and 50% and 90% prediction intervals12. Teams train their models and generate forecasts as if their model had been applied in real-time across all seasons since 2010∕2011. The resulting collection of forecasts serves as a historical record of how each model would have performed in real-time48.
Probabilistic forecasts for 21 ‘component’ models were submitted to the FSN for the 2018∕2019 season (See http://flusightnetwork.io/ for an interactive visulization of probabilistic forecasts of ILI, citation48, specifically Table 1, for details about each component model, and citation12 for more details on applications of influenza forecasting.). The majority of forecasts were submitted retrospectively (models from teams KoT and CU submitted in real-time), as if they were forecasting in real-time and contending with ILI data revisions. A retrospective forecast that is created during week ω of the influenza season must produce forecasts for week 1 of the season assuming no seasonal data, for week 2 of the season using data avilable during week 2 of the season (noisy week 1 data), and so on. This process prevents a retrospective model from forecasting on revision free influenza data.
TABLE 1.
Description of the 5 ensemble models analyzed.
| Model | Description |
|---|---|
| Equally-weighted (EW) | Assign a weight of 1/21 to every component model |
| Static with no regularization (Static) | Weights are trained on all previous seasons and fixed throughout the target season |
| Adaptive with no regularization (Adaptivenon) | This ensemble begins with no training data and a 0% prior regularization. Weights are trained each week within the target season. |
| Adaptive with optimal regularization (Adaptiveopt) | This ensemble begins with no training data and an optimal prior calculated from the 2010/2011 season. Weights are trained each week within season. |
| Adaptive with over regularization (Adaptiveover) | This ensemble begins with no training data and a prior greater than the optimal. Weights are trained each week within season. |
For the CDC FluSight challenges, component forecast models and ensembles built from them, forecast 7 key targets (Figure 2). These targets are 1, 2, 3, and 4 week-ahead ILI percentages, onset week (defined as the first of three consecutive weeks where the ILI% is above CDC-defined baseline ILI%), the season peak week (the epidemic week with highest ILI percentage), and the season peak percentage (the highest ILI percentage throughout the season). The CDC cannot determine the start of the influenza season and when the peak occurs until mid-summer due to continued revisions to data. This means final performance data on forecasts—the data used for building ensembles—for 3 seasonal targets (baseline onset, season peak week and percentage) can not be compiled until after the season closes. Because our adaptive ensemble is trained on within-season data, only component model forecasts of the 4 week-ahead targets from the FluSight Network will be used for training.
FIGURE 2.
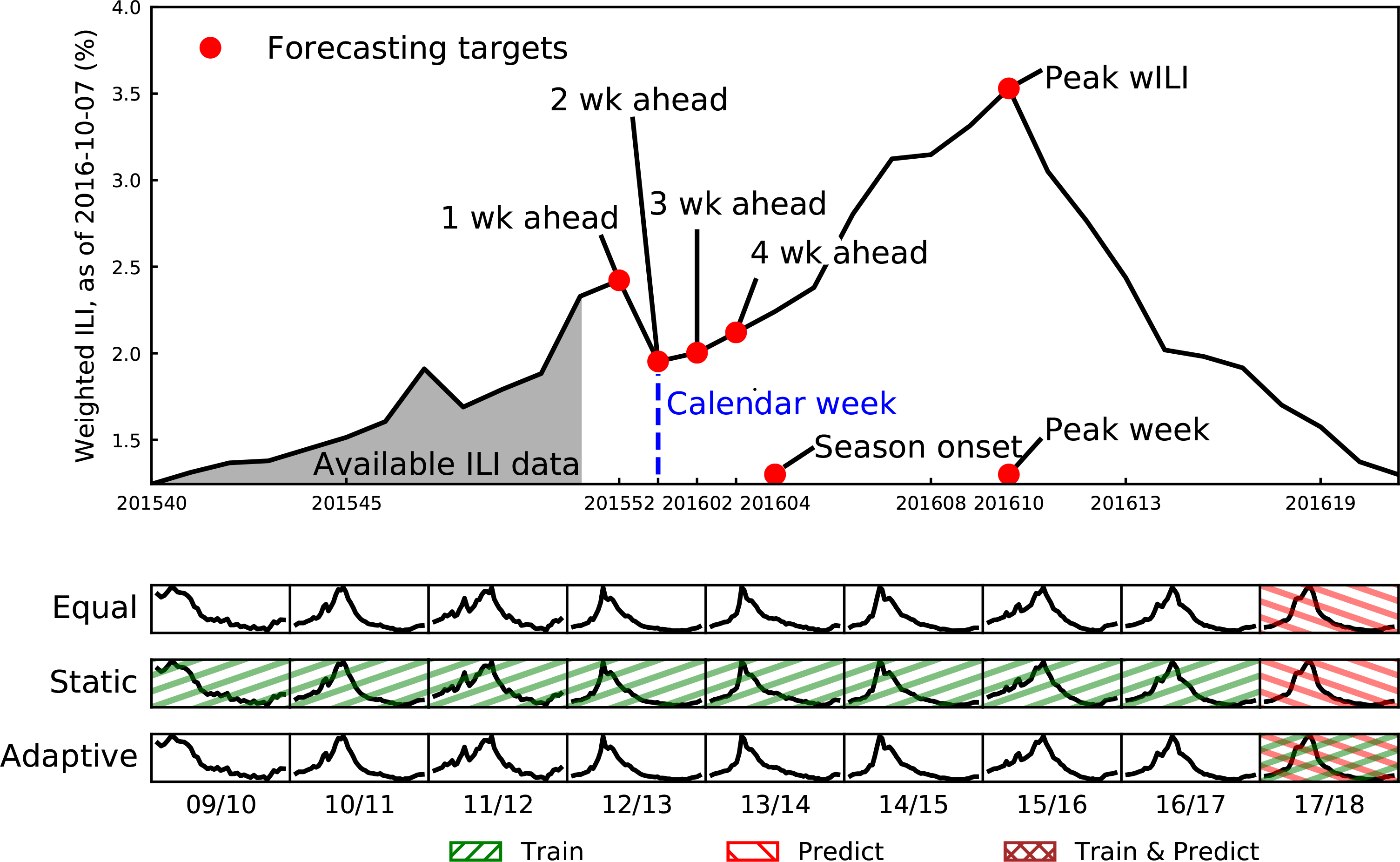
Overview of forecasting targets and data used in model estimation. Top Panel: Zooming in on one region-season illustrates the 7 forecasting targets, divided into week ahead (1, 2, 3, and 4), and seasonal (onset, peak week, peak percentage) targets. Due to delays in reporting at time t, forecasting models only have data available up to t − 2 weeks, making the current calendar week the 2 wk ahead forecast (sometimes titled the nowcast). Bottom Panels: Three ensemble algorithms: equally-weighted, static, and adaptive, take different approaches to training and prediction, illustrated by how they would operate in the 2017/2018 season. Equally-weighted ensembles ignore past ILI data, weighting all component models the same and forecast the next season. Static ensembles train on all previous seasons (green, up-right slanting lines), find optimal weights for all component models, and keeping these weights fixed, forecast the next season (red, down-right slanting lines). Adaptive ensembles train and forecast all in the current season (red and green cross-hatch), ignoring any past data. For adaptive ensembles, every week component model weights are updated on all current within-season data and then used to forecast the next week.
One difficulty with estimating ensemble weights comes from ILI data revisions. An adaptive ensemble must compute weights based on component model performance—the probability component model forecasts place on true ILI values. But past ILI data is revised weekly and changes component model performance in past epidemic weeks. Unlike an ensemble that trains on component models scored on finalized ILI data, an adaptive ensemble must account for ILI revisions because they change past component model performance week to week.
Retrospective component model forecasts and ILI data from 2011∕2012 up to the 2017∕2018 season will be used to compare equal, static, and adaptive ensembles. The 2010∕2011 season will be used as hold-out data to build a prior for the adaptive ensemble. The structure of our training data is shown in Figure 3.
FIGURE 3.
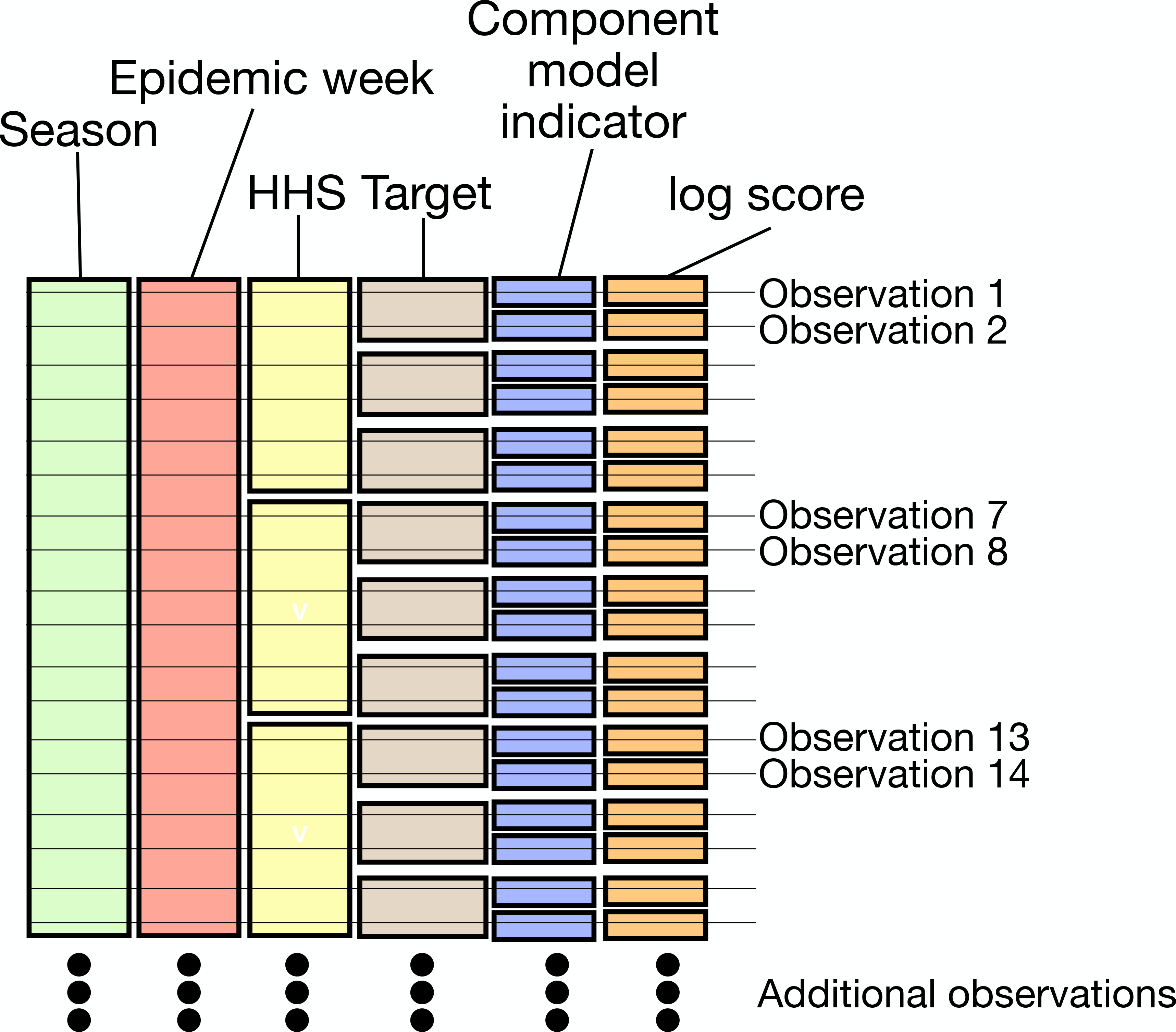
The structure of a single season of training data used by an ensemble model. Within every season and for each epidemic week, component models make forecasts of ILI for 10 HHS regions and at the national level, and for 7 different targets. Component model forecasts are then scored using a metric called the log score. Every epidemic week generates 77 scoreable forecasts per component model.
2.1.3 |. Performance metrics
A useful predictive model must be well-calibrated and accurate, consistently placing high probability on finalized ILI percentages. A proper scoring rule51 widely accepted in the forecasting community is the log score51,48, and defined as the predicted probability placed on a future true (or finally revised) ILI value. Proper scoring rules measure a forecast’s calibration and sharpness52,53 by penalizing a predictive distribution F that does not match the distribution of true values G52. By assigning higher scores for a forecast that matches the true data generating process, a proper score encourages both calibration and sharpness 52,53,54,55.
The CDC adopted a modified log score as their standard scoring metric. Given a model with probability density function f(z) and true ILI value i ∗, both the log score and modified log score can be written
| (1) |
where ω is a small positive number. The traditional log score sets ω = 0% and this is the metric we use to compare ensembles.
In practice, we bin predictive probabilities of ILI percentage from 0 to 13% by 0.1% plus a single bin from 13% to 100%. The proper log score reduces to computing the log of the probability given to the single true ILI bin Models that place high probabilities on the bins containing the observed final ILIs are scored closer to 0 (perfect score), while models that place low probabilities on observed ILI bins are scored closer to −∞ (the worst score). Log scores are truncated to −10 to follow convention from CDC FluSight scoring17.
2.2 |. Ensemble Framework
2.2.1 |. Ensemble model specification
Static and adaptive ensemble algorithms use a mixture model to combine (or average) component models forecasts of influenza-like illness. The probabilistic approach to training these ensembles is similar, but the adaptive ensemble has two principal advantages over the static ensemble. By specifying prior probability that at first places equal weight on component models, the adaptive ensemble can combine component models with no training data. The choice of prior for our adaptive ensemble is meant to discourage erratic shifts when assigning weights to component models as we move week-by-week through an influenza season. The adaptive ensemble will use a likelihood function to learn which component models are performing better than others and update prior weight assignments each week. This is in contrast to the static model. The static model too will use the same likelihood function to find optimal weights for component models, but no prior is included and weights are assigned at the beginning of the season and are fixed. Because weights are fixed the static ensemble requires multiple seasons of training data to find weights the model assumes are optimal over an entire season.
We take two approaches to finding optimal ensemble weights: a maximum likelihood method (for the static ensemble) and a maximum aposteriori Bayesian approach (for the adaptive ensemble). Below we choose to find optimal weights using the EM algorithm and Variational Inference56,57,58 which find stable optima at the price of slow convergence. Alternative methods like a quasi-newton59, or the broader class of sequential quadratic programming algorithms60 could be used to find optimal weights with superior convergence properties but may be less stable than the EM and VI approaches61.
Our general ensemble approach, for both static and adaptive ensembles, assumes a generative model for our data—ILI percentage at time t (yt)—by first sampling a component model m with probability πm, and second, sampling the ILI percentage from the component model m with probability mass function fm(i). (In practice, the submitted probabilistic forecasts fm(i) are probability mass functions defined over discrete bins of possible ILI incidence. These are often discretized versions of probability densities.)
| (2) |
| (3) |
where z(t) is a vector of length M and M is the number of component models included in the ensemble. Each entry in z(t) corresponds to one of M possible component models and assigns the value 1 to the model that generated the observed ILI percentage yt and 0 otherwise. The probability of z = (1,0,…,0) = π1 and the probability of z = (0,1,…,0) = π2 and so on.
We denote probability density selected by z(t) as fz(t). The weights π1,π2, πM are the only parameters we can change. We have no access to how the component models were trained or to parameters of the models, since we only receive a discretized probability mass function for each component model f in our applied setting.
This framework conveniently expresses the probability distribution over possible ILI values as a convex combination of component models by averaging over the random variable z. We define the probability of observing an ILI percentage at time t,
| (4) |
where p [yt|z(t)] is the component model f selected by z(t). The πs are required to be non-negative and sum to one.
This mixture model framework for probabilistic model averaging makes the reasonable assumption that no single model is responsibile for generating ILI values over time. Instead, individual models propose different mechanisms and incorporate different types of data to model how ILI evolves over time. A probabilistic average aims to emphasize models that lead to accurate forecasts (by assigning these models larger weights) and minimize those that lead to poor forecasts. By assigning weights to many different models a mixture approach to ensemble forecasting may reduce noise compared to an individual forecast50,27,29,26,28. Previous work forecasting infleunza-like illness, ebola, dengue, zika, and other infectious diseases have used a probabilistic averaging approach 25,24,12,17,18,19,23,62,63.
This probabilistic averaging approach has become a standard for the field because of the above reasons.
2.2.2 |. Expectation-Maximization method for weight estimation
Though (4) is a convenient way to specify the model, directly optimizing the loglikelihood in this form is difficult. The loglikelihood over all T time points equals
| (5) |
and is a summation over log-sums, where is the observed T ILI percentages. An alternative method to optimize the loglikelihood40,64 considers the loglikelihood over both ILI percentages and the set of hidden indicator variables z(t)—one for each week of data—that decide which component model (from M possible models) was selected to generate data point yt. The hidden variables Z simplify the likelihood to
| (6) |
where z(m,t) equals 1 when the mth model generated the tth ILI observation and 0 otherwise (ie. z(t) = [z(1,t), z(2,t), z(M,t)]), and Z is a M × T matrix of all hidden variables. The Expectation-Maximization (EM) algorithm56 iteratively optimizes a lowerbound on the logliklihood, the expectation over with respect to or to find the component model weights that maximize the loglikelihood (deEM-MM Fig. 2 Alg. 1). The prefix de (degeneracy) refers to this algorithm’s inability to change parameters inside component models and the suffix MM (mixture model) references the assumed data generating process.
2.2.3 |. Variational Inference for weight estimation
While the deEM-MM algorithm find an optimal set of weights, it considers π a fixed vector of parameters. If instead we assume π is a random variable, we can restructure our problem as Bayesian and infer a posterior probability over weights. Our Bayesian framework extends (6) by modeling the posterior probability of π
| (7) |
While many different choices are available for a prior over ensemble weights, p(π), we chose a Dirichlet distribution because it is conjugate to the likelihood. We further decided on a Dirichlet prior that has a single parameter (α) shared across all M models. The scale of the shared parameter (α) governs the influence of the prior distribution on the posterior. A larger value of α shrinks ensemble weights closer towards an equally weighted ensemble. Equal weights are a reasonable first choice for many ensemble settings and especially when performance data has not yet been collected for component models. Smaller values of α allow the model to rely more heavily on the observed data.
Typically the prior is fixed during training. But because in our setting the amount of ILI data is growing, past ILI percentages are revised as we move through a season, and component model performance may change over time, we allow the prior to change over time. A time-dependent prior can act to continually weaken the influence of the (likely to be revised) data on the ensemble weights and consistently pull component model weights towards equal over the course of the season, guarding against several factors such as data revisions that change past component model performance.
We specify the prior as
where α(t) is the parameter (the same across all M component models) that defines the Dirichlet distribution at time t. We chose (t) to be a constant fraction (ρ) of the number of observations on component model performance at time t divided equally amongst all M component models
| (8) |
where N(t) is the number of training data points, M is the number of component models, and ρ is a value between 0 and 1. Throughout the season the prior parameter will grow linearly as the ensemble trains on an increasing number of observations N(t) and regularize component model weights at a constant rate.
We can plug our specific prior in for time t
| (9) |
and find an optimal set of mixture weights by optimizing the following function of π and Z
| (10) |
We note that the first term on the right-hand side of equation (10) is the same as taking the log of the frequentist expression for the likelihood shown in equation (6). Intuitively, we expect a stronger (increasing ρ) even prior to encourage weights across components to move towards 1∕M, uniformly distributing probability mass to every component model.
To estimate the posterior probability over ensemble weights (10), we will use a variational inference algorithm (deVI-MM). Variational inference is similar to the EM algorithm and both can be motivated by introducing a distribution over hidden variables and decomposing the loglikehood into two parts: a lowerbound on the log likelihood and an error term that is the difference between the lowerbound and loglikehood .
Given weights π, hidden variables Z, and fixed data , we first rewrite the posterior over Z and π
Reordering terms and taking the log,
We can express the marginal loglikelihood in terms of the complete loglikelihood (numerator) and log of the posterior distribution over π and Z (denominator). The last step introduces a distribution q over hidden variables Z and π
and integrates over q(Z)
| (11) |
where KL is the Kullback-Leibler divergence65, a non-negative function of q. Since KL is a non-negative function, the first term on the right hand side is a lower bound to the marginal loglikeihood and the second term, the KL term, is the difference between the marginal loglikelihood and the lower bound. Iteratively optimizing this lower bound can be shown to monotonically improve the loglikelihood and converge on the best possible representation (q) of the true posterior distribution over Z, and more importantly, π 56,40.
The EM and VI algorithms diverge on how to choose q: the EM algorithm considers π a fixed set of parameters and chooses q = , zeroing out the above Kullback-Leibler divergence but also assuming can be computed (see40 for theoretical development and48 for an application to infectious disease forecasting). The VI algorithm allows any choice for q. The mean-field approximation, the most common choice for q, factors q into discrete pieces 57,58 and is the approach we take. We chose to factor our distribution over hidden variables into a distribution over Z and distribution over Z, (q,Z) = q(Z) q(π) to separate indicator variables from mixture weights.
This choice yields (see details in Suppl. 1) a q(π) that follows a Dirichlet distribution and indicator variable z(m,t), a single element of z(t), that follows a Bernoulli distribution:
| (12) |
| (13) |
where the responsibility r(m, t), or the probability that model m generated datapoint yt, equals
| (14) |
and the responsibilities summed over component models (m) equals one. The distribution q [z(m, t)] computes the probability ILI value yt was generated by model m. Since the vector z(t) has entries z(1,t), z(2,t), z(M, t) which are each Bernoulli distributed variables required to sum to one, the vector z(t) is Dirichlet distributed.
The Variational approach here can also be interpreted as a generalization of the EM solution. If we fix π to be a non-random parameter, the responsibilities reduce to
| (15) |
the same responsibility function for the EM algorithm48. Both VI and EM algorithms will find a unique global maximum of the log-likelihood (5) due to the convexity of the log-likelihood function (see analysis of convexity in Suppl. 3).
2.2.4 |. How prior impacts ensemble weights
The prior can be recognized as a regularizer. The maximum aposteriori estimate (MAP) for the mth component model can be computed by dividing the weight given to model m by the sum over all weights (sum over m)
| (16) |
where is the total number of ILI values used for training at time t. Though useful for computation, an alternative characterization expresses the MAP as a convex combination of the prior and the percent responsability assigned to component model m. Define the prior percent weight assigned to component model m as . and the percent responsibility assigned to component model m due to the data as . The MAP of π can be reexpressed as
| (17) |
a convex combination of the prior plus responsibility learned from the data. Strengthening the prior, in our case increasing ρ, shifts the MAP estimate away from the data-estimated responsibility and towards the prior. The constant shift, despite increased training data, is a result of our time-dependent prior. Note that this model is likely not a consistent estimator for ρ as we are more interested in forecast accuracy than on inference for ρ. By setting our prior percentage to equal across all models in the ensemble (1∕M), the prior weight ρ interpolates between an equally-weighted ensemble (ignoring past model performance) and a completely data-driven weighting.
2.3 |. The ensemble framework as a doubly mixed model of pseudocounts
The choice of a prior may appear arbitrary, but this prior can be motivated using two techniques: (i) using pseudocounts to reinterpret the Bayesian framework of the adaptive ensemble. and (ii) supposing ILI data is generated by a mixture of an equally weighted ensemble and an ensemble trained on the data.
From our Bayesian framework, we found weights were distributed, from (12), Dirichlet
| (18) |
This distribution over weights can be reinterpreted as psuedocounts. Instead of assuming a single observation is a mixture of M component models, we can assume at time t each component model generated a subset of observations from the total number of ILI observations as follows
| (19) |
| (20) |
where θ is the expected number of ILI values generated by component model m (θ = N(t)πm) and πm is the fraction of ILI values generated by component model m, α parameterizes a prior distribution over pseduocounts (over the θs), N(t) is the number of ILI values at time t and.
If we define N(t) × M variables ILIm,t as the value 1 when component model m generated ILI value t and 0 otherwise, then
The posterior distribution (see40) over θ is
| (21) |
We can draw a parallel between the above model of pseudocounts and the posterior we found in (12). The sum of responsabilities over all ILI values at time t is similar to the number of data points generated by model m, or . Likewise, the parameter that controlled the prior over pseduocounts, α, resembles our prior α(t) from (12).
The Dirichlet distribution in (21) can be parameterized as
| (22) |
where
| (23) |
and we can again draw a parallel between this fration of ILI values generated by component model m and the MAP estimate from our Bayesian framework (16). Our goal now is to find a suitable prior α. We can motivate the choice of a prior by assuming our ensemble of component models will be combined with an equally weighted ensemble.
If we assume the data generation process for ILI values is a mixture of an equally weighted ensemble (fequally) and an ensemble that is a mixture of component models fm as in (2) and (3) (ftrained) then we can write the mixture model with constant weights ρ and (1 − ρ) as
If we assume that the weights ρ will be small relative to 1 then
and so our weights for this doubly mixed model are proportional to
| (24) |
We can guarantee our doubly mixed weights sum to one by dividing each weight by the sum of all weights plus the additional constant
| (25) |
where we used the fact that .
Assume each component model generated N(t)πdoubly mixed of the ILI values on average.
| (26) |
A potential prior that will act like a mixture of an equally weighted ensemble and an ensemble fit via maximum likelihood is then
2.3.1 |. deEM-MM and deVI-MM Algorithms
A comparison of the EM and VI algorithms is shown below in Algorithms 1 and 2. The EM and VI algorithms are similar to one another: both rely on adding hidden variables to the loglikelihood and iteratively maximizing a lower bound. When computing Z, both algorithms need the probability that component model m generated data point i for models 1 to M and data points 1 to T. A key difference is how the EM and VI algorithms approximate the distribution over hidden variables Z. The EM algorithm requires the previous point estimate of ensemble weights. The VI algorithm requires the ensemble weight’s expectation (see Suppl. 2 for details on calculating ). These differences are present in step 7, updating Z, and step 10 updating π. Another difference is in evaluating model fit. Because the EM algorithm chooses as q the exact conditional probability over Z, we can monitor convergence by computing the loglikelihood, log of (6). The choice of q for the VI algorithm allows us to compute a related quantity called the Evidence Lower Bound (ELBO). The ELBO is defined as ELBO = loglikelihood−{log[q(π)] + log[q(Z)]}, or the difference between the loglikelihood and our approximation over hidden variables. When updating the ensemble weights, both algorithms sum over the probability yt belongs to model m, but the VI algorithm adds an additional term to the ensemble weight, the prior.
2.4 |. Experimental design
Five ensemble models will be analyzed in detail (Table. 1). The equally-weighted (EW) ensemble assigns the same weight (1∕M) to all component models. The Static ensemble trains on all past component model forecasts of ILI and assigns weights to component models before the start of the next season. Weights are kept fixed throughout the next season. The adaptive model with three types of regularization, that correspond to three values of ρ, will be studied: 0% regularization (Adaptivenon), ‘optimal’ regularization (Adaptiveopt), and ‘over’ regularization (Adaptiveover). All adaptive ensembles will begin each season with no training data and learn optimal weights over the season. We will consider our adaptive model ‘optimally’ regularized by computing log score statistics for the held-out 2010∕2011 season and choosing the prior that has the highest average log score. The equally weighted, static, and adaptive ensemble generate forecasts of 1, 2, 3, and 4 week ahead influenza-like illness at the national level.
2.4.1 |. Training and scoring component models for ensembles
We created a record of component model scores on revisable ILI data, as if they had been scored in real time, every week throughout each season. Starting each season with no data, adaptive models were trained on component model log scores throughout a season, and our static ensemble was trained on finalized component model log scores from past seasons, not the current season. Using within season component model performance data impacted how the adaptive model was trained in two ways: (i) every epidemic week new ILI data was observed and generated new component model log scores, and (ii) past ILI data was revised, which in turn changes past component model log scores. Ensemble log scores were calculated on ILI percentages reported on EW28, the “final” week used by the CDC FluSight challenges. ILI values at or after EW28 for each season are considered definitive. Based on previous work48, and due to the relatively smaller amount of data available to the adaptive ensemble, we chose to fit a ‘constant weight’ ensemble model—the constant referring to the same weight assigned to a component model for differing targets and regions—for both the static and adaptive approaches.
2.4.2 |. Fitted ensemble models
We computed adaptive ensembles for prior values from close to 0% (10−5 which we will refer to as 0%) to 100% by 1% increments for the 2010∕2011 season. To determine a prespecified ‘optimal prior’, we chose the prior corresponding to the highest average log score. The 2010∕2011 season was removed from all formal comparisons between ensembles. The adaptive ensemble corresponding to the optimal prior was used for formal comparisons.
For each season in 2011∕2012 through 2017/2018, we computed adaptivenon, adaptiveopt, adaptiveover, ensembles using the VI algorithm developed. Static ensemble and equally-weighted ensembles were also fit at the beginning of each season, using only data for prior seasons.
2.4.3 |. Formal Comparisons
To compare ensemble models, we computed the difference between the log score assigned to one ensemble versus another (see17 for a similar regression approach to comparisons). A random intercept model described differences between log scores, averaged over epidemic weeks and paired by season, region, and target.
| (27) |
where D is the difference between log scores assigned to two ensembles: for epidemic week ω, in season s, region r, and for target t. The fixed intercept (average difference in log score between ensemble models) is denoted β0, and season (as), region (br), and target (ct) effects are Normally distributed with corresponding variances.
We fit two random effects models: one comparing the adaptive vs. equally-weighted ensemble, and the second comparing the adaptive vs. static ensemble. The conditional mean, 95% confidence interval, and corresponding p-value are reported. An additional bootstrapped p-value is also reported. Random samples (with replacement) are selected from the set of all season-region-target-epidemic week tuples. For every random sample, a random effects models is fit and conditional means collected for: seasons, regions, and targets. The set of random samples is centered to create a null distribution and compared to our original dataset’s conditional mean. We report the empirical probability a random sample from the centered null distribution exceeds our original dataset’s conditional mean.
3 |. RESULTS
3.1 |. Choosing prior to maximize performance
We chose an optimal prior for the adaptive model by fitting our adaptive model to the 2010∕2011 season for priors from 0% to 100% by 1% and selecting the prior with the highest average log score (Suppl. 7 includes adaptive fits and average log scores for all seasons from 2010∕2011 to 2017∕2018). The average log score for the 2010∕2011 season peaks at a prior of 8% (Fig. 4), and we will consider an adaptive model with 8% prior (ρ= 0.08) our adaptiveopt ensemble. After a prior of 8%, the log score sharply decreases. This decrease in performance suggest ensemble weights are over regularized, and we chose a 20% prior as our adaptiveover ensemble. Finally, a 0% prior was chosen as our adaptivenon ensemble.
FIGURE 4.
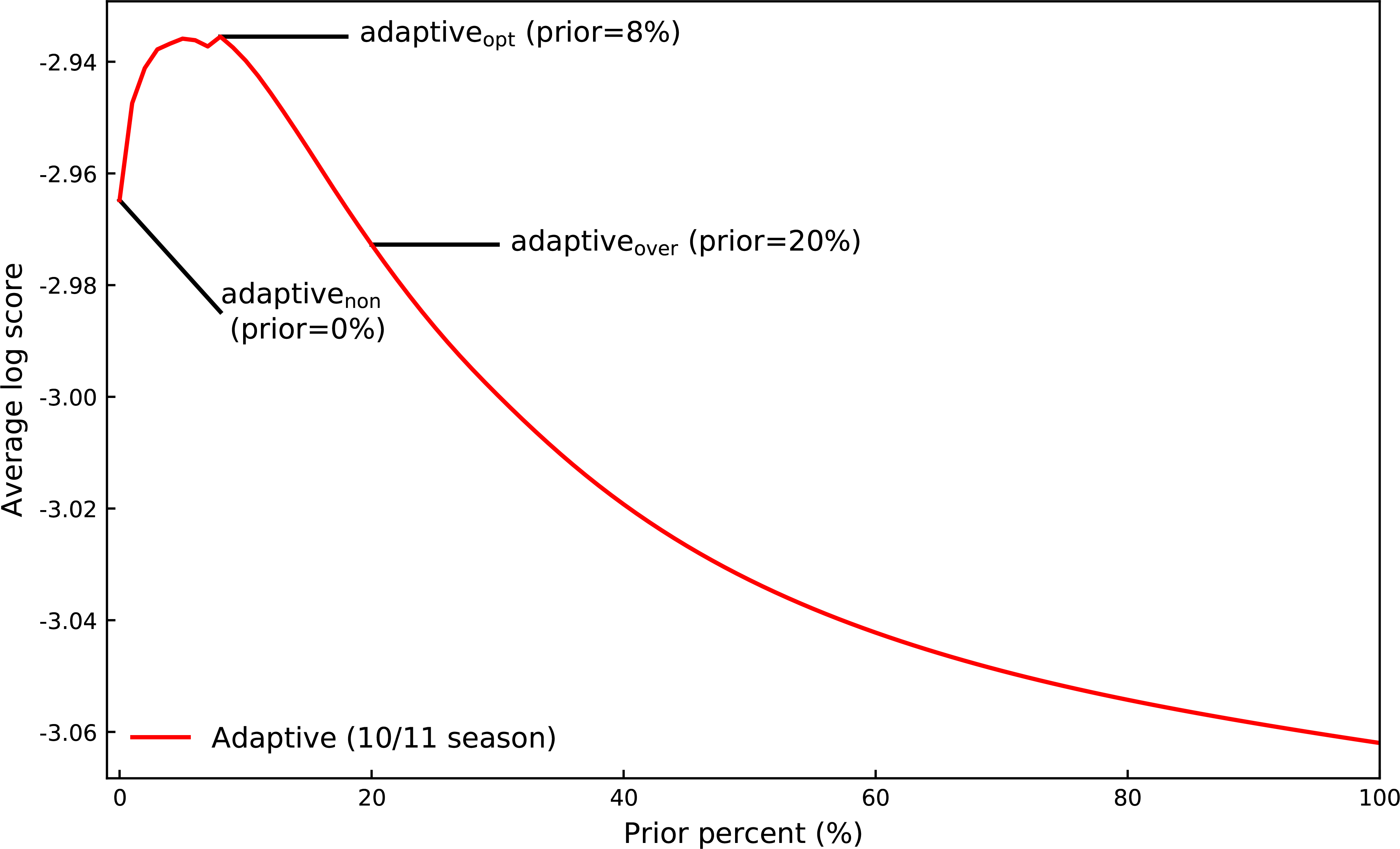
The log score for the 2010/2011 season, averaged over region, target, and epidemic week for priors (ρ) from 0% to 100% by 1% increments. The prior corresponding to the maximum log score (prior (ρ)=8%) was chosen as our adaptiveopt ensemble for formal comparisons to static and equally weighted ensembles.
3.2 |. Prior successfully regularizes ensemble weights
We compared the adaptivenon, adaptiveopt, and adaptiveover ensembles (ρ = 0.00, 0.08, and 0.20 respectively) to investigate how the ensemble weights change with the prior. Adding a prior regularizes component model weights (Fig. 5). Smaller priors yield higher variability in ensemble weights throughout any given season. For example, in 2017/2018 (Fig. 5), this is especially evident for the adaptivenon ensemble, when ensemble weights vacillate between assigning almost all weight to one component model or another. The adaptiveopt and adaptiveover weights, in comparison, do not show as high variability over time. Component model weights for all three ensembles do track one another, with specific model weights moving up and down in unison, albeit in a more muted fashion for the stronger 8% prior (adaptiveopt) and 20% prior (adaptiveover). The patterns shown in Fig. 5 persist in other seasons as well (see Suppl. 4).
FIGURE 5.
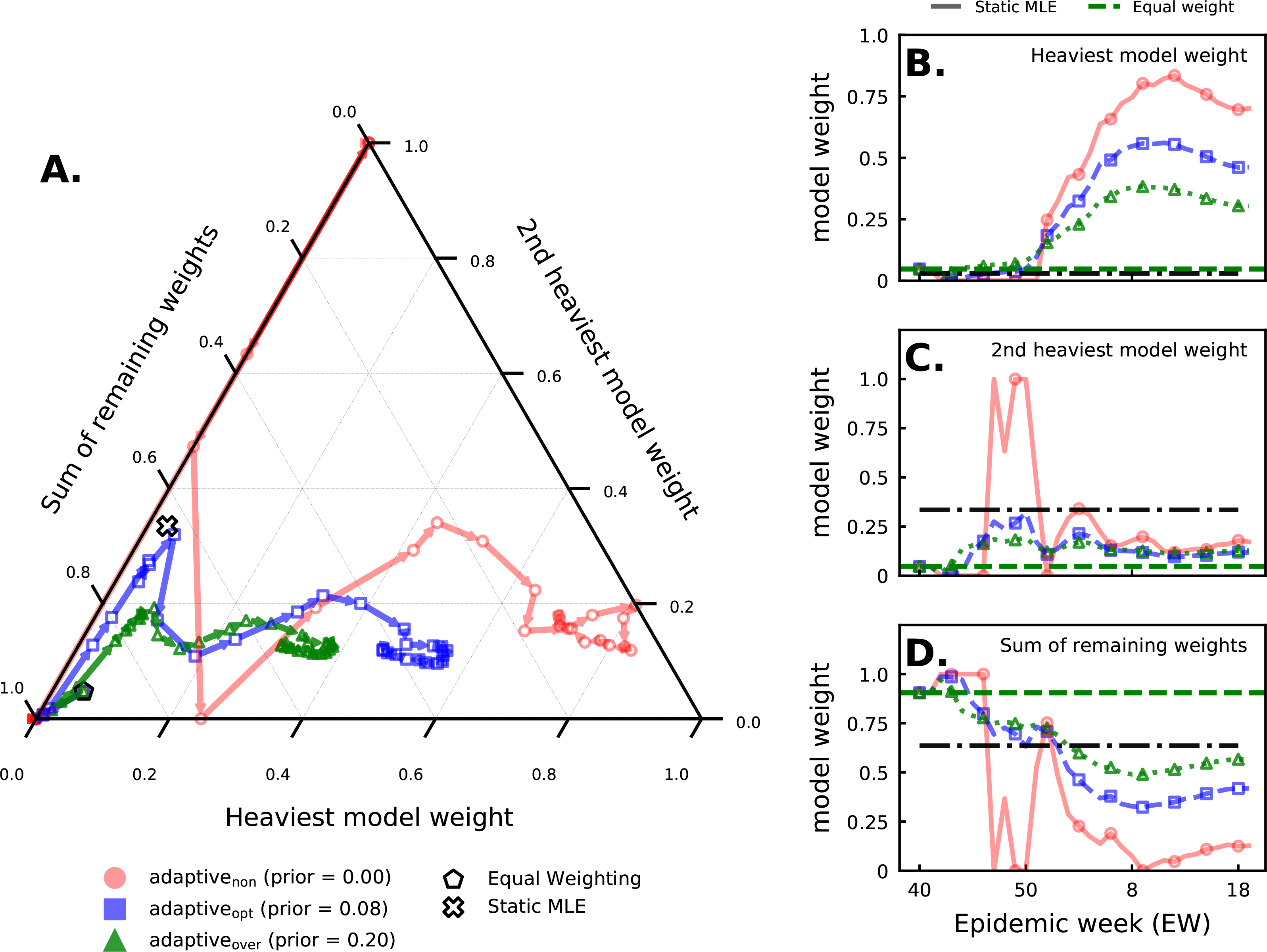
Component model weights plotted over epidemic weeks for all ensembles during the 2017/2018 season. We highlight the weights for the two component models the adaptivenon ensemble assigned the most weight to at the end of the season and compare this to the sum of all other model weights. The component models that were assigned the heaviest and second heaviest model weight are fixed over all epidemic weeks. (A) A simplex plot shows how, for each ensemble, the weights assigned to the top two models and the rest of the models move together across the season. The lines can be seen as a trajectory, beginning at the black pentagon (the equal-weight ensemble) and, following the arrows, migrate through the simplex with each point representing the triple (, , ) of weights estimated in week t. The estimated weights from the static ensemble using all data prior to 2017/2018 (but, unlike the adaptive ensemble, using no data from this season) are represented by the ‘x’. Plots of (panel B) and (panel C) across all weeks t in the 2017/2018 season. The estimates of this model’s weight from the static ensemble and the equal weight (1/21) are shown in horizontal dashed lines. (D) A plot of across weeks t. The estimates of the sum of these models’ weights from the static ensemble and the equal weight (19/21) are shown in horizontal dashed lines.
3.3 |. Comparing adaptive vs. equally-weighted and static ensembles
Adaptive ensembles—starting each season with no training data—consistently outperform equally-weighted ensembles, and show comparable performance to static ensembles (Fig. 6). The adaptiveopt model has higher log scores than either adaptivenon and adaptiveover models, and the EW model. The adaptiveover, unlike the adaptivenon model, always outperforms the EW model, indicating it is better to over- than under-regularize, at least to some degree. The static model—trained on multiple seasons of data—performs the best. Adaptivity improves over assigning equal weights to component models and performs similar to the data-rich static model.
FIGURE 6.
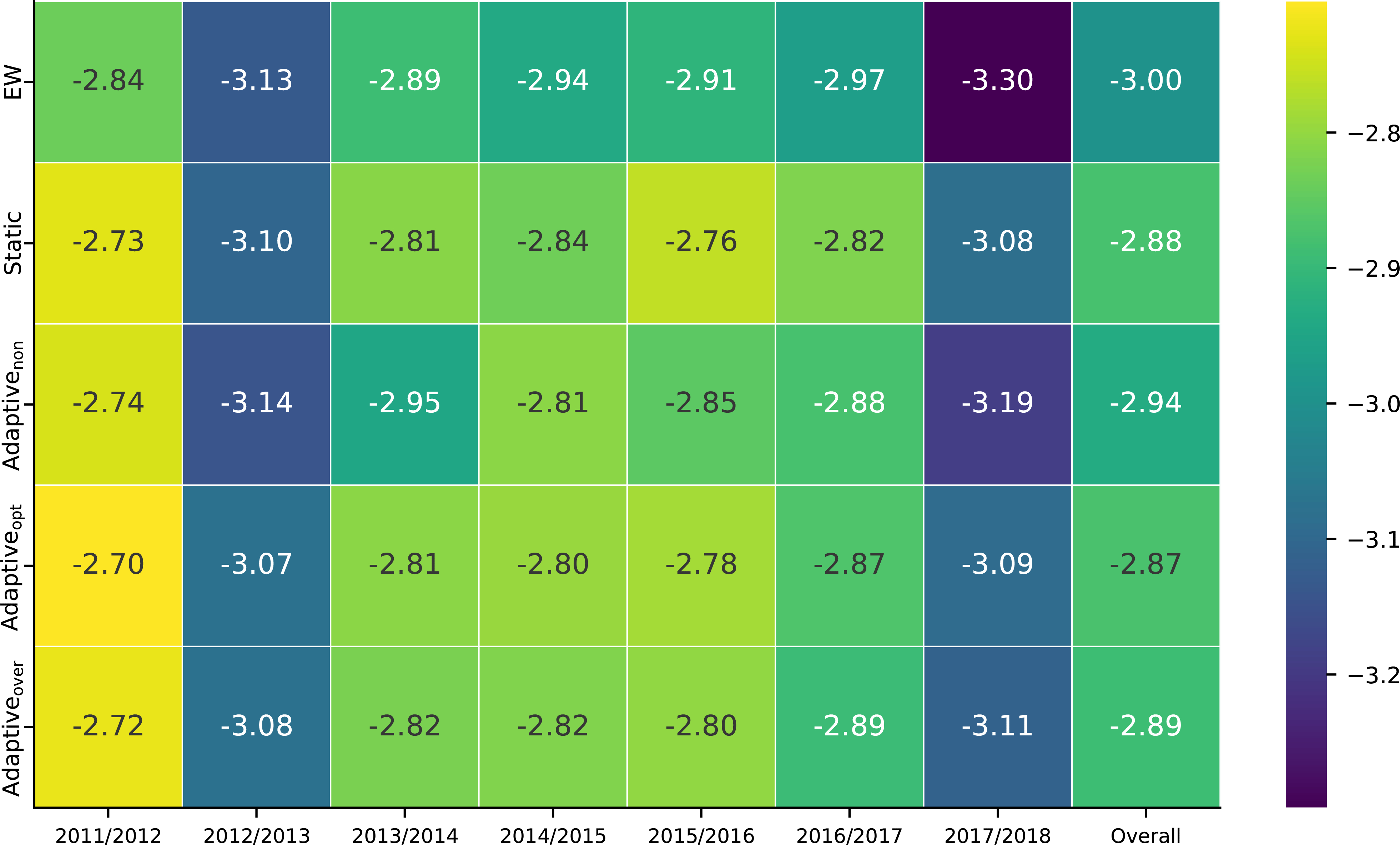
Log score averaged over region and target for ensemble algorithms stratified by season. The 8% prior was prespecified as the optimal prior based on analysis of the excluded 2010/2011 season. All ensembles show variability in performance across seasons. Adaptive and static ensembles outperform the equal weighted ensemble, the adaptive and static ensemble showing similar performance. Adaptive ensembles perform similar to static ensembles despite having less training data in later seasons.
Formal comparisons (see Fig. 7 and regression table 1) demonstrate the adaptiveopt ensemble has statistically higher log scores compared to the EW ensembles, and shows similar performance to static ensembles. The adaptiveopt ensemble has lower log score point estimates against the static models for any particular choice of season, region, and target, but the performance between static and adaptiveopt is not statistically different. EW and adaptiveopt ensembles perform similar for the 2012/2013 season. The 2015/2016, 2016/2017, and 2017/2018 season does show better (but not significant) performance for the static ensemble. This suggests that the static model’s performance comes from training on multiple seasons of data. Differences in performance are less variable when stratified by region (see Fig. 7B), and the difference in log scores between all ensembles decrease as forecasting horizons increase. This reflects the difficulty in forecasting further into the future rather than ensemble performance.
FIGURE 7.
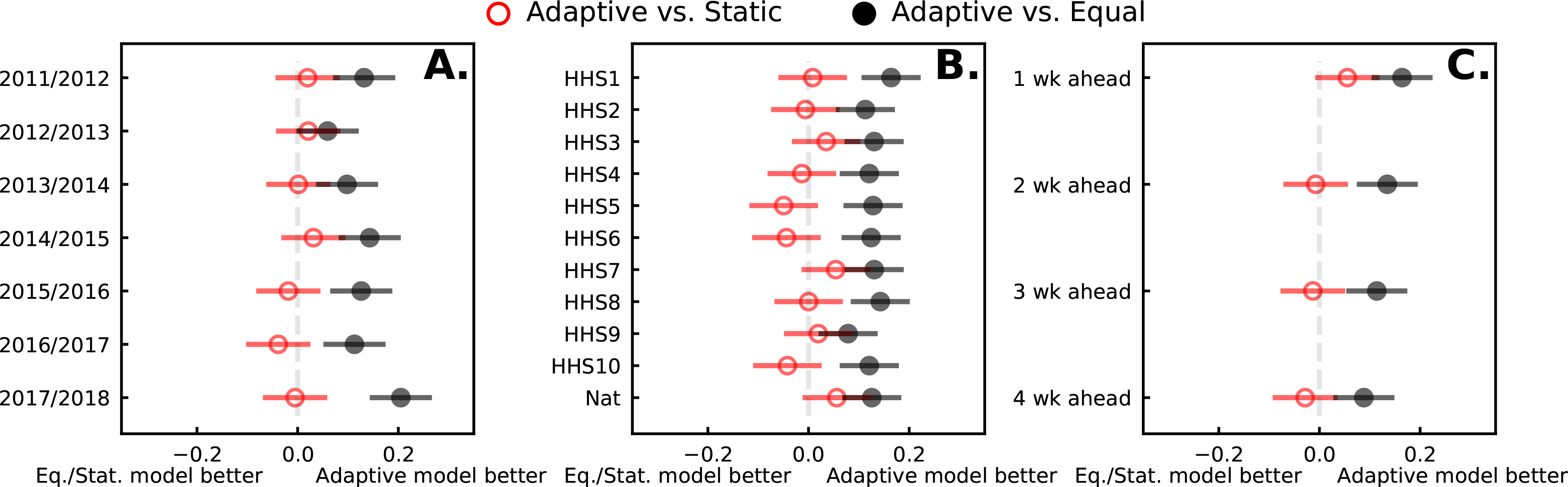
Log scores from equally-weighted, static, and adaptive ensembles within season, region, target, and epidemic week. Estimates and 95% confidence intervals are plotted from two random intercepts models that estimate (i) the difference in log score between adaptive and equally-weighted ensembles and (ii) the difference between adaptive and static ensembles. Positive differences suggest the adaptive ensemble performs better than the reference ensemble. Adaptive ensembles outperform the equally-weighted ensembles, though the qqually-weighted and adaptive ensembles perform similarly in the 2012/2013 season. Adaptive ensembles perform similar to static ensembles with differences in log scores favoring static or adaptive ensembles cannot obtain statistical significance for both parametric and bootstrapped p-values. Adaptive ensembles consistently outperform equally-weighted ensembles, and show similar or slightly improved (but not significant) performance against static ensembles.
In select strata the adaptive ensemble shows a statistically significant higher log score compared to the EW and close to signficiant difference compared to the static ensemble. This significance is not enough to conclude the adaptive ensemble should outperform the static ensemble in most cases, however the data suggest, unsurprisingly, that the static ensemble performs relatively better the more data is has to train on.
3.4 |. It may be better to over regularize than to under regularize
Formal comparisons (see regression tables 2 and 3) show that both the adaptiveover and adaptivenon ensembles outperform the equally weighted ensemble and have similar performance when compared to the static ensemble. Compared to the adaptivenon ensemble, the adaptiveover ensemble shows improved performance (i.e. higher log scores) against the equally weighted and static ensemble.
The adaptiveover has an average increase in log score compared to the equally weighted ensemble between 0.06 to 0.21. The adaptivenon has an average increase between 0.03 to 0.11. The average increases in log score for comparions between the adaptiveover and equally weighted ensemble are all significant. The largest pvalue is 0.03, when comparing the adaptiveover to the equally weighted ensemble in the 2012/2013 season, and all other pvalues are smaller than 0.01. A small fraction of increases in log score for comparisons between the adaptivenon and equally weighted ensemble are significant. The adaptivenon ensemble shows no improvement in HHS6 with an average increase in log score of 0.00 (95CI = [−0.09, 0.09]; pvalue = 0.99). However, the adaptiveover shows a signficant increase in log score in HHS6 (average increase0.07; 95CI = [0.03,0.12]; pvalue < 0.01). Results between the adaptiveover and adaptivenon versus the static ensemble are similar—both ensembles show similar performance when compared to the static ensemble, but compared to the adaptivenon ensemble, differences between the adaptiveover and static ensemble are more positive and have smaller pvalues.
3.5 |. The adaptive ensemble’s ability to optimize weights within season
The adaptive model has the opportunity to outperform the static model by tailoring component model weights within season. Training weights based on component model performance data throughout the season is useful, as is shown by the adaptiveopt model outperforming the EW model. But the adaptive ensemble must accrue several weeks of training data before it can perform similar, and in some cases better, than the static ensemble (Fig. 8).
FIGURE 8.
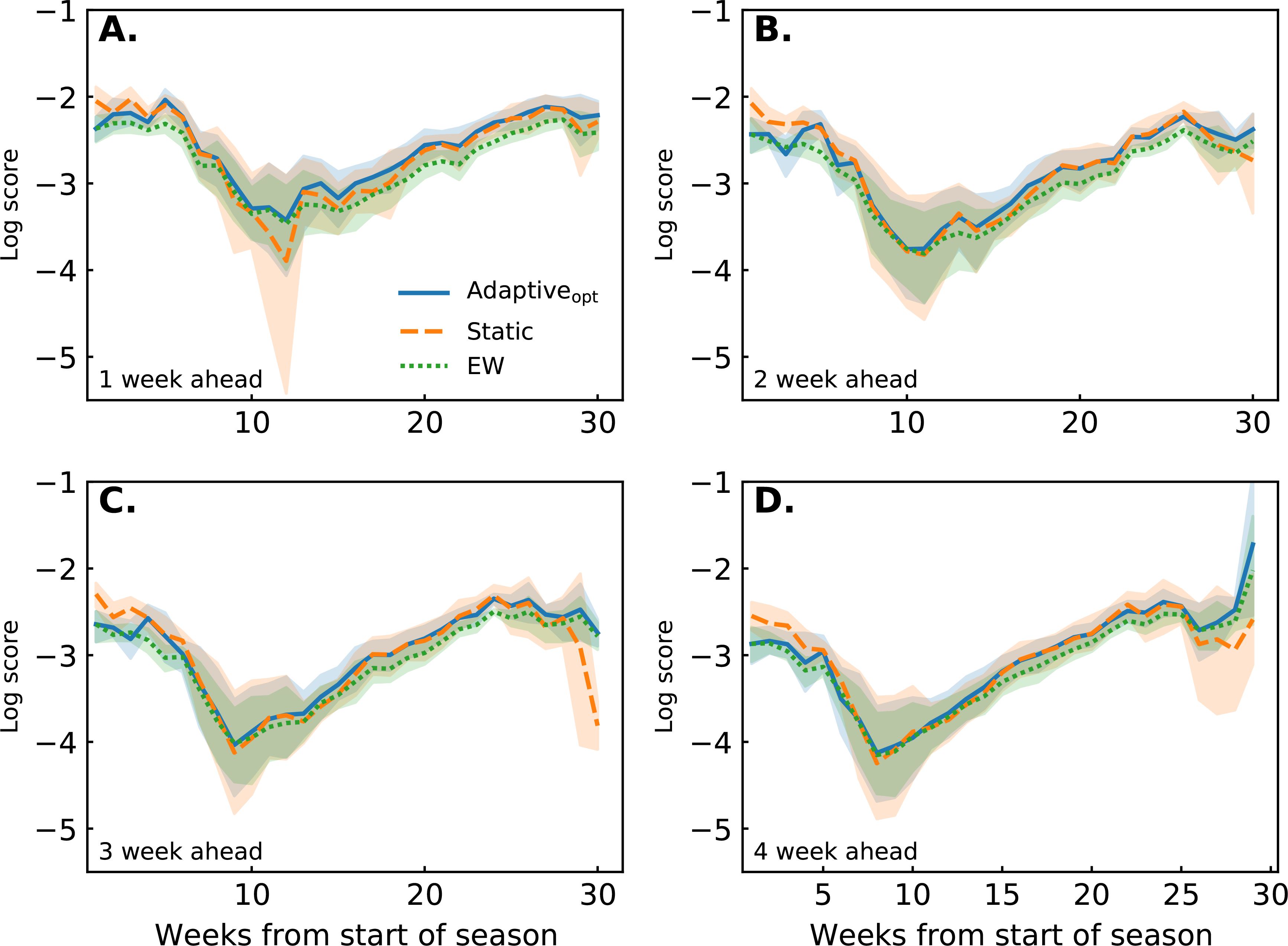
Average log score over epidemic weeks stratified by target for adaptiveopt (blue solid line), static (orange dashed line), and EW (green dotted line) ensembles. Across targets, all models perform better at the beginning and end of the season. Performance decreases at approximately 10 weeks into the season, corresponding with peak ILI values. Comparing ensembles, the adaptiveopt model performs better than the EW model at all time points, the difference between performance increasing throughout the season. At the beginning of the season the adaptiveopt model performs worse than the static model, performing similar or better within 10 weeks. The 1 week ahead target shows the adaptiveopt model outperforming the static model by week 10. The remaining targets show the adaptiveopt model performing similar to the static.
However, the adaptive models also are able to, without any prior data, learn how to create an optimal weighting within a season. While the adaptive models may be penalized early on in a season by not possessing the historical knowledge of which models have performed well, they adjust for this by learning which models are performing well in a particular season. This is illustrated clearly in Fig. 5 where the heaviest model at the end of the season (with 46.2% of the weight for the adaptiveopt ensemble) was assigned only 4.82% of weight at the beginning of the season. This ability to adapt to season-specific model performance appears to provide the adaptiveopt model with a slight advantage over the static model in the middle and end of the season.
4 |. DISCUSSION
We developed a novel algorithm for assigning weights to component models that starts with no training data at the beginning of each season and shows comparable performance to a static ensemble trained on multiple seasons of data. This novel ensemble can combine component models immediately because it requires no training data, and when component model performance data is used, it is specific to the current season’s forecasts. Our model extends a previous ensemble algorithm25 by assuming component model weights are random variables, rather than fixed quantities. This model reassigns component model weights every week, but because component model performance varies within a season due to factors like revisions to past influenza data, we introduced a time-dependent uniform Dirichlet distribution that regularizes ensemble weights. This adaptive ensemble outperforms an equally-weighted ensemble, and performs similarly to a static ensemble that requires multiple years of data to perform well.
It is expected that learning from training data, and the amount available, would contribute to better predictive performance. The equally-weighted model ignores any training data, and similar to an adaptive ensemble with no training data, assigns equal weights. The equally-weighted ensemble performs worst. The next best model, the adaptive ensemble, trains on component model performance within the same season. Even though the static model has multiple seasons of training data available, and does have the best performance, the adaptive ensemble is not far behind. It is difficult to tell whether the similar performance is a shortcoming of the static or adaptive ensemble. Similar performance between ensembles could be because component model performance from past seasons (available to the static ensemble) does not generalize to future seasons. Alternatively, the adaptive ensemble may not be using the within-season training data efficiently and could be leaving behind hidden patterns in the component model forecasting data. But we did find static weights outperforming the adaptive ensemble early on in the season, and if enough seasonal data was available it is possible that the static ensemble could statistically outperform the adaptive ensemble. These results suggest a model could perhaps use the static ensemble weights, when available, as a prior for the adaptive ensemble.
The advantage of the adaptive ensemble over the static ensemble is the ability to combine models without any training data. If an ensemble is needed for a new infectious agent or for a new region where component models have just begun to make forecasts, a static ensemble cannot be trained. An equally-weighted ensemble could be used. But we found an adaptive ensemble like the one we propose can assign equal weights to component models at first and, by learning optimal weighting over time, outperform an equally-weighted ensemble.
Though there may be an optimal mix between an equally-weighted and training data only approach to ensemble models, regularizing ensemble weights increased adaptive ensemble performance. Not only can regularization improve adaptive model performance, but could also improve the performance of the static model (Suppl. 6). Improving performance by regularizing ensemble weights with a time-dependent uniform Dirichlet prior may be applicable to any ensemble weighting scheme.
Our empirical work suggests an optimal prior near 8%, but we expect the optimal prior to vary by season, region, and target. Different priors will likely be optimal for different biological phenomena too. A more flexible prior could improve adaptive ensemble performance: changing the prior percent throughout the season, allowing the prior to depend on performance data from past seasons, or modeling the prior on regional and target information. Instead of limiting the prior as a regularizer, we can include outside information from past seasons, or patterns in influenza data the ensemble cannot learn from training data. If a pattern in data revisions or how component models are assessed is present then novel ensemble models could be built that better model past, present, and future noise when assessing component model performance. Future research will explore different methods of modeling prior ensemble weights.
Our work here connects to Gneiting and other’s work on linear pooling66,67,68,41. Linear pools assume that the data is generated by a combination of component models. Similar to Gneiting’s work, our weights are optimized with respect to the loglikelihood of a generative model, but the loglikelihood is really a different representation of a log score—the log of the probability corresponding to weights π. Our model could be fit by minimizing a generic loss function based on log score, but we found the EM (and VI) algorithms fit our model fast and consistently found a global optimum. Unlike Gneiting, we did not recalibrate the combined predictive distributions, and methods like the beta transformed linear pool66 or shifted linear pool69 could improve predictive performance even further.
This paper has many limitations future work can address: (i) better choice of prior, (ii) accounting for correlated component models, (iii) post-processing or ‘recalibrating’ our combined forecast; (iv) inclusion of seasonal targets; and, (v) handling missing forecasts. Our adaptive ensemble examined how a prior can impact ensemble performance, but we only explored a uniform prior. Future work could study informative priors and how to manipulate priors during the course of a season to improve ensemble performance. Our model also assumed component models made independent forecasts. A more advanced ensemble would examine the correlation structure of the data (region, target), and the component model forecasts. In addition, our ensemble model focused on an optimal set of weights for forecasting, and made no efforts to recalibrate the combined forecast67. It is important to note that this adaptive ensemble can only train on week ahead targets, excluding potentially useful information about a component model’s ability to forecast seasonal targets such as the seasonal peak, peak intensity, and when the influenza season begins. Other ensemble methods may not be limited to training on only week ahead data. Finally, our adaptive ensemble does not yet address how to weight component models that have submitted forecasts for past epidemic weeks, missed forecasts in the middle of the season, and then started submitting forecasts at a later time point.
Recent forecasting of COVID-19 transmission uses an alternative score called the interval score70, highlighting the adaptive ensemble’s dependence on the log score. The deEM-MM and deVI-MM algorithms depend explicitly on the log score as a measure of the probability that each component model generated a given ILI value per epidemic week. For an alternative scoring system to work with the adaptive ensemble, component model scores would first need to be converted to probabilities that each component model generated a given ILI value. We plan to pursue different proposals for how to convert forecast scores to probabilities which would generalize the adaptive ensemble to any scoring rule.
Several factors may impact component model performance and a detailed analysis in the future of potential factors should be performed to address (i) the most influential factors that contribute to component model performance and (ii) estimates of variability in component model performance within a season and across seasons. The association between component model performance and targets, and from season to season should also be explored.
From a public health perspective, officials have been moving away from stand-alone subjective decision making in favor of incorporating data-driven analyses into decisions. This trend has become particularly apparent in infectious disease outbreak management71. Adaptive ensembles, providing real-time forecasts of infectious disease, supports this trend towards evidence based decision making for public health. Combining their expertise with statistical models like our adaptive ensemble, public health officials can work towards impeding outbreaks and changing public health policy. Public health officials often find themselves making decisions in the middle of crises, times when fast effective forecasting is necessary. The adaptive ensemble’s flexibility, reliance only on near-term data, and capacity to track unusual disease trends can support accelerated public health decision making.
Adaptive ensembles—reweighting component models, week by week, throughout a season—can handle sparse data scenarios, admit new component models season by season, and shows similar prescience compared to the more data-heavy static ensemble. This added flexibility makes them an important tool for real-time, accurate forecasts of an outbreak.
5 |. REPOSITORY
The data and code used to train equal, static, and adaptive ensembles can be found at https://github.com/tomcm39/adaptively_stacking_ensembles_for_infleunza_forecasting_with_incomplete_data. Influenza-like illness, component model forecasts, and ensemble performance metrics are published on Harvard’s Dataverse. Links to the data are provided on the above GitHub page.
Algorithm 1.
deEM-MM Algorithm: An Expectation maximization algorithm estimating ensemble weights for a Frequentist adaptive algorithm.
| 1: | input: y1×T, π0, τ |
| 2: | output: π |
| 3: | |
| 4: | ℓℓ ← [] |
| 5: | πM×1 = π0 |
| 6: | for j=1:maxIters do |
| 7: | ZM×T ← π × f(y) |
| 8: | Z ← Z∕colSum(Z) |
| 9: | π ← rowSum(Z) |
| 10: | π ← π∕ sum(π) |
| 11: | ℓℓ[j] ← computeLL(y,π) |
| 12: | if LL[j] - LL[j-1] < τ then |
| 13: | break |
| 14: | end if |
| 15: | return π |
| 16: | end for |
Algorithm 2.
deVI-MM Algorithm: A Variational Inference algorithm estimating ensemble weights for a Bayesian adaptive algorithm. Although the deEM-MM (above) and deVI-MM algorithms have different underlying probability models, they follow similar steps. The major differences between EM and VI algorithms are: how Z is computed (step 7), the prior over π (step 10), and how weights are evaluated (steps 11 and 12).
| 1: | input: y1×T, π0,αM×1, τ |
| 2: | output: π |
| 3: | |
| 4: | ELBO ← [] |
| 5: | ZM×1 = π0 |
| 6: | for j=1:maxIters do |
| 7: | |
| 8: | Z ← Z∕colSum(Z) |
| 9: | π ← rowSum(Z) |
| 10: | π ← π∕ sum(π) + α |
| 11: | ELBO[j] ← computeELBO(y,π) |
| 12: | if ELBO[j] - ELBO[j-1] < τ then |
| 13: | break |
| 14: | end if |
| 15: | return π |
| 16: | end for |
Supplementary Material
6 |. ACKNOWLEDGEMENTS
This work has been supported by the National Institutes of General Medical Sciences (R35GM119582) and the Defense Advanced Research Projects Agency. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS, the National Institutes of Health, or the Defense Advanced Research Projects Agency. We also thank the reviewers whose recommendations and insights led to a more impactful work.
References
- 1.Centers for Disease Control and Prevention and others. Prevention and control of seasonal influenza with vaccines. Recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Early release 2009; 58(Early release): 1–54. [PubMed] [Google Scholar]
- 2.Russell CA, Jones TC, Barr IG, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science 2008; 320(5874): 340–346. [DOI] [PubMed] [Google Scholar]
- 3.Harper SA, Bradley JS, Englund JA, et al. Seasonal influenza in adults and children—diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clinical infectious diseases 2009: 1003–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garten R, Blanton L, Elal AIA, et al. Update: Influenza Activity in the United States During the 2017–18 Season and Composition of the 2018–19 Influenza Vaccine. Morbidity and Mortality Weekly Report 2018; 67(22): 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krumkamp R, Kretzschmar M, Rudge J, et al. Health service resource needs for pandemic influenza in developing countries: a linked transmission dynamics, interventions and resource demand model. Epidemiology & Infection 2011; 139(1): 59–67. [DOI] [PubMed] [Google Scholar]
- 6.Schanzer DL, Schwartz B. Impact of seasonal and pandemic influenza on emergency department visits, 2003–2010, Ontario, Canada. Academic Emergency Medicine 2013; 20(4): 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molinari NAM, Ortega-Sanchez IR, Messonnier ML, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 2007; 25(27): 5086–5096. [DOI] [PubMed] [Google Scholar]
- 8.Reed D, Kemmerly SA. Infection control and prevention: a review of hospital-acquired infections and the economic implications. The Ochsner Journal 2009; 9(1): 27–31. [PMC free article] [PubMed] [Google Scholar]
- 9.Germann TC, Kadau K, Longini IM, Macken CA. Mitigation strategies for pandemic influenza in the United States. Proceedings of the National Academy of Sciences 2006; 103(15): 5935–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughan E, Tinker T. Effective health risk communication about pandemic influenza for vulnerable populations. American Journal of Public Health 2009; 99(S2): S324–S332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biggerstaff M, Alper D, Dredze M, et al. Results from the centers for disease control and prevention’s predict the 2013–2014 Influenza Season Challenge. BMC infectious diseases 2016; 16(1): 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutz CS, Huynh MP, Schroeder M, et al. Applying infectious disease forecasting to public health: a path forward using influenza forecasting examples. BMC Public Health 2019; 19(1): 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipsitch M, Finelli L, Heffernan RT, Leung GM, 2009 H1N1 Surveillance Group R. f. tSC. Improving the evidence base for decision making during a pandemic: the example of 2009 influenza A/H1N1. Biosecurity and bioterrorism: biodefense strategy, practice, and science 2011; 9(2): 89–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chretien JP, Riley S, George DB. Mathematical modeling of the West Africa Ebola epidemic. Elife 2015; 4: e09186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins TA, Siraj AS, Ruktanonchai CW, Kraemer MU, Tatem AJ. Model-based projections of Zika virus infections in childbearing women in the Americas. Nature microbiology 2016; 1(9): 1–7. [DOI] [PubMed] [Google Scholar]
- 16.Biggerstaff M, Johansson M, Alper D, et al. Results from the second year of a collaborative effort to forecast influenza seasons in the United States. Epidemics 2018; 24: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGowan CJ, Biggerstaff M, Johansson M, et al. Collaborative efforts to forecast seasonal influenza in the United States, 2015–2016. Scientific reports 2019; 9(1): 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggerstaff M, Kniss K, Jernigan DB, et al. Systematic assessment of multiple routine and near real-time indicators to classify the severity of influenza seasons and pandemics in the United States, 2003–2004 through 2015–2016. American journal of epidemiology 2017; 187(5): 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaman J, Karspeck A. Forecasting seasonal outbreaks of influenza. Proceedings of the National Academy of Sciences 2012; 109(50): 20425–20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks LC, Farrow DC, Hyun S, Tibshirani RJ, Rosenfeld R. Flexible modeling of epidemics with an empirical Bayes framework. PLoS computational biology 2015; 11(8): e1004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray EL, Sakrejda K, Lauer SA, Johansson MA, Reich NG. Infectious disease prediction with kernel conditional density estimation. Statistics in medicine 2017; 36(30): 4908–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osthus D, Gattiker J, Priedhorsky R, Del Valle SY, others. Dynamic Bayesian Influenza Forecasting in the United States with Hierarchical Discrepancy. Bayesian Analysis 2018. [Google Scholar]
- 23.Yamana TK, Kandula S, Shaman J. Individual versus superensemble forecasts of seasonal influenza outbreaks in the United States. PLoS computational biology 2017; 13(11): e1005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray EL, Reich NG. Prediction of infectious disease epidemics via weighted density ensembles. PLoS computational biology 2018; 14(2): e1005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reich NG, McGowan CJ, Yamana TK, et al. A Collaborative Multi-Model Ensemble for Real-Time Influenza Season Forecasting in the US. bioRxiv 2019: 566604. [Google Scholar]
- 26.Zhou ZH. Ensemble methods: foundations and algorithms. Chapman and Hall/CRC. 2012. [Google Scholar]
- 27.Sewell M Ensemble learning. RN 2008; 11(02). [Google Scholar]
- 28.Breiman L Bagging predictors. Machine learning 1996; 24(2): 123–140. [Google Scholar]
- 29.Dietterich TG. Ensemble methods in machine learning. In: Springer. 2000: 1–15. [Google Scholar]
- 30.Schapire RE. The boosting approach to machine learning: An overview. In: Springer. 2003. (pp. 149–171). [Google Scholar]
- 31.Krishnamurti T, Kishtawal C, LaRow TE, et al. Improved weather and seasonal climate forecasts from multimodel superensemble. Science 1999; 285(5433): 1548–1550. [DOI] [PubMed] [Google Scholar]
- 32.Garratt A, Mitchell J, Vahey SP, Wakerly EC. Real-time inflation forecast densities from ensemble Phillips curves. The North American Journal of Economics and Finance 2011; 22(1): 77–87. [Google Scholar]
- 33.Pierro M, Bucci F, De Felice M, et al. Multi-Model Ensemble for day ahead prediction of photovoltaic power generation. Solar energy 2016; 134: 132–146. [Google Scholar]
- 34.Adejo OW, Connolly T. Predicting student academic performance using multi-model heterogeneous ensemble approach. Journal of Applied Research in Higher Education 2018; 10(1): 61–75. [Google Scholar]
- 35.Raftery AE, Gneiting T, Balabdaoui F, Polakowski M. Using Bayesian model averaging to calibrate forecast ensembles. Monthly weather review 2005; 133(5): 1155–1174. [Google Scholar]
- 36.Steel MF. Bayesian model averaging and forecasting. Bulletin of EU and US Inflation and Macroeconomic Analysis 2011; 200: 30–41. [Google Scholar]
- 37.Madigan D, Raftery AE, Volinsky C, Hoeting J. Bayesian model averaging. In: NA. 1996: 77–83. [Google Scholar]
- 38.Montgomery JM, Nyhan B. Bayesian model averaging: Theoretical developments and practical applications. Political Analysis 2010; 18(2): 245–270. [Google Scholar]
- 39.Minka TP. Bayesian model averaging is not model combination. Available electronically at http://www.stat.cmu.edu/minka/papers/bma.Html 2000: 1–2.
- 40.Bishop CM. Pattern recognition and machine learning. springer. 2006. [Google Scholar]
- 41.Wallis KF. Combining forecasts–forty years later. Applied Financial Economics 2011; 21(1–2): 33–41. [Google Scholar]
- 42.Gneiting T, Raftery AE. Weather forecasting with ensemble methods. Science 2005; 310(5746): 248–249. [DOI] [PubMed] [Google Scholar]
- 43.Foley AM, Leahy PG, Marvuglia A, McKeogh EJ. Current methods and advances in forecasting of wind power generation. Renewable Energy 2012; 37(1): 1–8. [Google Scholar]
- 44.Laan V. dMJ, Polley EC, Hubbard AE. Super learner. Statistical applications in genetics and molecular biology 2007; 6(1). [DOI] [PubMed] [Google Scholar]
- 45.Pari R, Sandhya M, Sankar S. A Multi-Tier Stacked Ensemble Algorithm to Reduce the Regret of Incremental Learning for Streaming Data. IEEE Access 2018; 6: 48726–48739. [Google Scholar]
- 46.Fern A, Givan R. Online ensemble learning: An empirical study. Machine Learning 2003; 53(1–2): 71–109. [Google Scholar]
- 47.Benkeser D, Ju C, Lendle S, Laan v. dM. Online cross-validation-based ensemble learning. Statistics in medicine 2018; 37(2): 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reich NG, Brooks LC, Fox SJ, et al. A collaborative multiyear, multimodel assessment of seasonal influenza forecasting in the United States. Proceedings of the National Academy of Sciences 2019; 116(8): 3146–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osthus D, Daughton AR, Priedhorsky R. Even a good influenza forecasting model can benefit from internet-based nowcasts, but those benefits are limited. PLoS computational biology 2019; 15(2): e1006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks LC, Farrow DC, Hyun S, Tibshirani RJ, Rosenfeld R. Nonmechanistic forecasts of seasonal influenza with iterative one-week-ahead distributions. PLoS computational biology 2018; 14(6): e1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawid AP, Musio M. Theory and applications of proper scoring rules. Metron 2014; 72(2): 169–183. [Google Scholar]
- 52.Gneiting T, Balabdaoui F, Raftery AE. Probabilistic forecasts, calibration and sharpness. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2007; 69(2): 243–268. [Google Scholar]
- 53.Winkler RL. Rewarding expertise in probability assessment. In: Springer. 1977. (pp. 127–140). [Google Scholar]
- 54.Gneiting T, Raftery AE. Strictly proper scoring rules, prediction, and estimation. Journal of the American Statistical Association 2007; 102(477): 359–378. [Google Scholar]
- 55.Garthwaite PH, Kadane JB, O’Hagan A. Statistical methods for eliciting probability distributions. Journal of the American Statistical Association 2005; 100(470): 680–701. [Google Scholar]
- 56.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the royal statistical society. Series B (methodological) 1977: 1–38. [Google Scholar]
- 57.Blei DM, Kucukelbir A, McAuliffe JD. Variational inference: A review for statisticians. Journal of the American Statistical Association 2017; 112(518): 859–877. [Google Scholar]
- 58.Rustagi JS. Variational methods in statistics. In: Academic Press. 1976. [Google Scholar]
- 59.Bertsekas DP. Constrained optimization and Lagrange multiplier methods. Academic press. 2014. [Google Scholar]
- 60.Nocedal J, Wright SJ. Sequential quadratic programming. Numerical optimization 2006: 529–562. [Google Scholar]
- 61.Kim Y, Carbonetto P, Stephens M, Anitescu M. A Fast Algorithm for Maximum Likelihood Estimation of Mixture Proportions Using Sequential Quadratic Programming. Journal of Computational and Graphical Statistics 2020; 29(2): 261–273. doi: 10.1080/10618600.2019.1689985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viboud C, Sun K, Gaffey R, et al. The RAPIDD ebola forecasting challenge: Synthesis and lessons learnt. Epidemics 2018; 22: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson MA, Apfeldorf KM, Dobson S, et al. An open challenge to advance probabilistic forecasting for dengue epidemics. Proceedings of the National Academy of Sciences 2019; 116(48): 24268–24274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy K Machine learning: a probabilistic approach. Massachusetts Institute of Technology 2012: 1–21. [Google Scholar]
- 65.Cover TM, Thomas JA. Elements of information theory. John Wiley & Sons. 2012. [Google Scholar]
- 66.Ranjan R, Gneiting T. Combining probability forecasts. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2010; 72(1): 71–91. [Google Scholar]
- 67.Gneiting T, Ranjan R, others. Combining predictive distributions. Electronic Journal of Statistics 2013; 7: 1747–1782. [Google Scholar]
- 68.Geweke J, Amisano G. Optimal prediction pools. Journal of Econometrics 2011; 164(1): 130–141. [Google Scholar]
- 69.Kleiber W, Raftery AE, Baars J, Gneiting T, Mass CF, Grimit E. Locally calibrated probabilistic temperature forecasting using geostatistical model averaging and local Bayesian model averaging. Monthly Weather Review 2011; 139(8): 2630–2649. [Google Scholar]
- 70.Bracher J, Ray EL, Gneiting T, Reich NG. Evaluating epidemic forecasts in an interval format. PLoS computational biology 2021; 17(2): e1008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rivers C, Chretien JP, Riley S, et al. Using “outbreak science” to strengthen the use of models during epidemics. Nature Communications 2019; 10(1): 3102. doi: 10.1038/s41467-019-11067-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


