Abstract
The actinin-associated LIM protein, ALP, is the prototype of a large family of proteins containing an N-terminal PDZ domain and a C-terminal LIM domain. These PDZ-LIM proteins are components of the muscle cytoskeleton and occur along the Z lines owing to interaction of the PDZ domain with the spectrin-like repeats of α-actinin. Because PDZ and LIM domains are typically found in proteins that mediate cellular signaling, PDZ-LIM proteins are suspected to participate in muscle development. Interestingly the ALP gene occurs at 4q35 near the heterochromatic region mutated in facioscapulohumeral muscular dystrophy, indicating a possible role for ALP in this disease. Here, we describe the generation and analysis of mice lacking the ALP gene. Surprisingly, the ALP knockout mice show no gross histological abnormalities and maintain sarcolemmal integrity as determined by serum pyruvate kinase assays. The absence of a dystrophic phenotype in these mice suggests that down-regulation of ALP does not participate in facioscapulohumeral muscular dystrophy. These data suggest that ALP does not participate in muscle development or that an alternative PDZ-LIM protein can compensate for the lack of ALP.
The muscle cytoskeleton comprises a complex protein network that provides cellular structure and permits contraction. Disruptions of the myofiber cytoskeleton underlie several genetic muscular dystrophies, including Duchenne and limb-girdle muscular dystrophies (8). In addition to these dystrophin-related disorders, certain inherited muscular dystrophies are due to mutations in cytoskeletal proteins that do not interact with the dystrophin complex (15, 18). Identification of the responsible genes and clarification of mechanisms that regulate the myofiber cytoskeleton are therefore critical goals.
Recent studies suggest that cytoskeletal elements containing PDZ protein motifs play important roles in skeletal muscle development and disease (9). PDZ domains are ∼80-amino-acid domains that mediate interactions with either C-terminal or internal binding sites on target proteins (10, 12, 17, 27). In skeletal muscle, PDZ domains were first noted in syntrophins and neuronal nitric oxide synthase (5, 6), which are both components of the dystrophin-associated glycoprotein complex. Subsequently, a variety of other PDZ proteins involved in diverse cellular signaling pathways have been identified in skeletal muscle.
One large family of skeletal muscle PDZ proteins comprises a group of gene products that contain an N-terminal PDZ domain and one or more C-terminal LIM domains (21, 23, 32, 33). These PDZ-LIM proteins associate with the actin cytoskeleton via interaction of their PDZ domain with the spectrin-like repeats of α-actinin; this interaction targets these PDZ-LIM proteins to the Z lines (32). The prototypical PDZ-LIM protein in skeletal muscle is the actinin-associated LIM protein (ALP), which is expressed at extremely high levels in skeletal muscle and at much lower levels in cardiac and other tissues (32). Interestingly, the human ALP gene occurs on chromosome 4q35 near the heterochromatic locus that is mutated in facioscapulohumeral muscular dystrophy (FSHD) (1), indicating a possible role for ALP in this disease (32).
To address the consequences of ALP deficiency, we generated ALP knockout mice by homologous recombination in embryonic stem (ES) cells. The ALP mutant mice are viable and fertile and show no apparent muscle abnormalities. Despite the absence of ALP protein, muscle histology appears normal, and muscle sarcolemma is preserved. Furthermore, the actinin-based cytoskeleton is intact in the knockout mice. These data suggest that either ALP is not required for skeletal muscle development and function or an alternative PDZ-LIM protein compensates for the loss of ALP.
MATERIALS AND METHODS
Isolation of ALP genomic DNA and construction of the targeting vector.
PCR primers based on human ALP (32) were used to amplify mouse ALP from first-strand cDNA from skeletal muscle. A bacterial artificial chromosome SV129 mouse genomic library was screened with the ALP cDNA probe, and three bacterial artificial chromosome clones were obtained, each about 100 kb in size. One of these clones was characterized in detail. The genomic region immediately surrounding the first translated exon, which contains 211 bp and encodes amino acids 1 to 30, was sequenced. The targeting construct used the pPNT replacement vector (28) and replaced exon one of ALP with neomycin and was flanked on the two sides by a total of 6.4 kb of genomic DNA (Fig. 1). The 3.5-kb upstream region was PCR amplified, digested with KpnI and BamHI, and subcloned into the KpnI-BamHI sites of the pPNT vector. The 2.9-kb downstream region was PCR amplified, digested with NotI, and inserted into the NotI site.
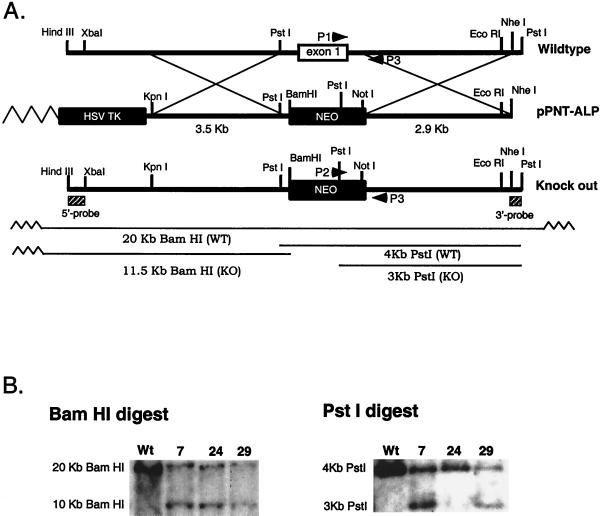
FIG. 1.
Genomic replacement of the mouse ALP exon 1. (A) The restriction maps of the wild-type ALP locus, the pPNT-ALP targeting vector, and the properly targeted locus are shown. Recombination (shown as large Xs) between the targeting vector and the wild-type locus eliminated the HSV TK gene and produced the knockout allele. A BamHI site was engineered to allow the wild-type and knockout genomic loci to be distinguished. (B) Genomic Southern blots of ES cell clones. Use of the 5′ probe for Southern analysis of a BamHI digest yielded a 20-kb band from the wild-type allele and a 11.5-kb band from the mutant allele. Use of the 3′ probe for Southern blots of PstI digests yielded a 4-kb band from the wild-type allele and a 3-kb band from the mutant allele. Clones 7 and 29 show proper targeting in both BamHI (left) and PstI (right) digests.
Generation of ALP-null mice.
The targeting vector was linearized with NheI and ligated to an NheI-compatible cap oligonucleotide as previously described (20) to seal the ends of the linear fragment. This construct was electroporated into JM-1 ES cells (24), which were cultured on neomycin-resistant STO fibroblasts that had been mitotically inactivated by treatment with mitomycin. After electroporation and double-drug selection with G418 and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil, individual cell colonies were picked, expanded, and analyzed by Southern blotting for proper homologous recombination. Two properly targeted ES cell clones (no. 7 and 29) were injected into mouse blastocysts, and the injected embryos were implanted into surrogate mothers. The resulting male chimeric mice were mated to Black Swiss mice (Taconic, Germantown, N.Y.). Germ line transmission was obtained from both injected ES clones.
Genotyping of progeny.
To genotype the mutant mice, tail DNA was purified and analyzed by PCR. About 1 to 2 cm of tail tissue was placed in 0.5 ml of 10 mM Tris-HCl (pH 8.0), 25 mM EDTA, 75 mM NaCl, 1% sodium dodecyl sulfate, and proteinase K (0.5 mg/ml). Digestion was done at 55°C with constant rocking for 16 h. The DNA was extracted with phenol-chloroform, precipitated with ethanol, and redissolved in 100 μl of Tris-EDTA. Each DNA sample was analyzed by two different PCRs: one to detect the targeted exon of ALP and one to detect the neo gene. The ALP PCR used oligonucleotides P1 (5′-TCATCACCAGGGTAGGTGTTTTCC-3′) and P3 (5′-GACCTTTCTGGCTAATGTGGCTGG-3′), whereas the neo PCR used oligonucleotides P2 (5′-GCTAAAGCGCATGCTCCAGACTGC-3′) and P3 (5′-GACCTTTCTGGCTAATGTGGCTGG-3′). The ALP PCR produces a 250-bp fragment, and the neo PCR produces a 290-bp fragment.
Western blotting.
For Western blotting, skeletal muscle was homogenized in 20 volumes (wt/vol) of buffer containing 25 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, and 1 mM phenylmethylsulfonyl fluoride. Protein extracts were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and transferred to polyvinylidine difluoride membranes (Millipore, Bedford, Mass.), which were blocked with 3% bovine serum albumin. Antibodies to ALP or α-actinin (Sigma) were diluted in block solution and were incubated with membranes overnight at 4°C. Western blots were developed using enhanced chemiluminescence as previously described (16). Protein concentration was determined by the Bradford assay (Bio-Rad Laboratories).
Reverse transcription-PCR.
Mice were euthanized by cervical dislocation, and the gastrocnemius and cardiac muscles were isolated. Total RNA was prepared using Trizol reagent (Gibco BRL) and poly(A)+ mRNA was purified from total RNA using Oligotex beads (Qiagen) according to the manufacturer's recommendations. Reverse transcription of mRNA was performed with AMV reverse transcriptase (Roche) and oligo(dT) according to the specifications of the manufacturer. PCR amplification was performed with AmpliTaq Gold (Roche) and the following ALP-specific primers: forward, 5′-ATGATACTGGCTATAGATGGCTTTGGTACG-3′, and reverse, 5′-ATAGTTTAGCGGCCGCTTAAGCTTTGGGGTACAGAGTGAC-3′. These primers lie outside the first coding exon and flank the alternatively spliced region of ALP. After an initial incubation at 94°C for 10 min, 40 cycles of the following heating protocol were carried out: 50°C for 1 min, 72°C for 30 s, and 94°C for 1 min. PCR products were resolved on 0.8% agarose gels containing ethidium bromide, and bands were visualized by UV illumination. The ALP primers produce a 948-bp fragment from skeletal muscle ALP and a 702-bp fragment from cardiac muscle ALP. Control PCRs were carried out with the following tubulin primers: forward, 5′-CACGGGTCTCCAGGGCTTCTT-3′, and reverse, 5′-CATTTCACCATCTGGTTGGCT-3′. PCR conditions were the same as those used for ALP primers except that the extension time was 15 s and 25 cycles were performed.
Immunohistochemistry.
Mice were euthanized with pentobarbital, and gastrocnemius muscle was dissected without tissue perfusion or fixation. Muscle tissue was snap-frozen in a bath of 2-methylbutane cooled with liquid nitrogen. Muscle sections (4- to 10-μm thick) were cut using a cryostat and collected on treated microscope slides (Plus; Fisher). Following hematoxylin and eosin staining, muscle cryosections were evaluated for myofiber size and shape. For immunofluorescent staining, skeletal muscle sections were postfixed in 2% paraformaldehyde in phosphate-buffered saline. Tissues were blocked in phosphate-buffered saline containing 1% normal goat serum. Mouse antibody to α-actinin 2(1 μg/ml) or polyclonal rabbit antibody to ALP (1 μg/ml) was applied to sections overnight at 4°C. For indirect immunofluorescence, secondary goat anti-mouse Cy-3 or donkey anti-rabbit Cy-3-conjugated antibodies were used according to the specifications of the manufacturer (Jackson ImmunoResearch Laboratories).
Serum pyruvate kinase assay.
Blood was obtained from the retroorbital sinus or tail and collected into heparin-treated vials. Pyruvate kinase assays were performed as described (11) by incubating serum with buffer containing 110 mM imidazole-HCl (pH 7.4), 165 mM KCl, 0.19 mM β-NADH, 5.5 mM MgCl2, 5.5 mM ADP, 5.5 mM dithiothreitol, 2.5 U of lactate dehydrogenase (Sigma), and 0.25 mM phosphoenolpyruvate at 30°C and monitoring the loss in absorbance at 340 nm. One unit of pyruvate kinase activity is the amount needed to consume 1 μmol of β-NADH per min. The appropriate institutional review committee approved all experimental protocols.
RESULTS
Generation of ALP-deficient mice.
We chose to delete the first coding exon of ALP, as such a disruption is likely to abolish protein expression. The predicted translation of the deleted exon is identical to residues 1 to 30 of human ALP. Using the PCR, we designed a targeting vector that replaces the targeted first exon with a neomycin cassette. This cassette is flanked on the 5′ side by a 3.5-kb intronic fragment and on the 3′ side by a 2.9-kb intronic fragment. A thymidine kinase cassette was added upstream of the 5′ intronic sequence to permit double-drug selection (Fig. 1A).
The targeting vector was linearized with NheI and was electroporated into mouse ES cells, which were then selected with G418 and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil. Double resistant clones were analyzed by Southern blotting and PCR. For this Southern blotting, genomic DNA from the ES cells was digested with BamHI and probed with a 0.8-kb HindIII/XbaI fragment (Fig. 1B). Among three clones that tested positive for homologous recombination with the 5′ probe, two clones were confirmed as positives by PstI digestion, followed by hybridization using a 3′ genomic probe (a 0.65-kb NheI/PstI fragment; see Fig. 1B). Appropriately targeted clones (2 of 96) were injected into blastocytes, and the resulting chimeric mice were used to breed heterozygous and homozygous mice, which were genotyped by Southern blotting with a 5′ probe and PCR (Fig. 2A). Heterozygous mice were intercrossed, and 138 F2 mice were analyzed, of which 30 (29%) were the wild type, 67 (49%) were heterozygous, and 41 (22%) were homozygous mutants. This indicates that ALP knockouts are born at the predicted Mendelian frequency. Furthermore, both male and female knockout mice are viable, fertile, and indistinguishable in appearance from their littermates.
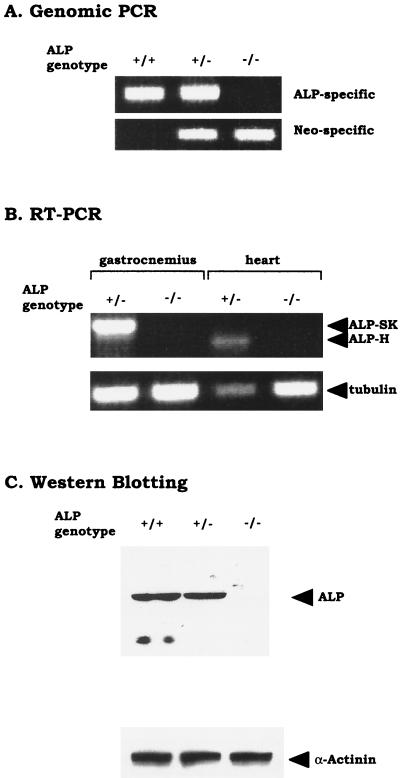
FIG. 2.
Analysis of ALP genotype and gene expression in ALP knockout mice. (A) PCR genotyping of ALP mutant mice. DNA from wild-type (+/+), heterozygous (+/−), or homozygous (−/−) mice was amplified by PCR using an ALP-specific primer (P1), a neo-specific primer (P2), and a common primer (P3). The wild-type ALP allele produced a 250-bp DNA fragment with P1 and P3 (top), and the mutated allele produced a 290-bp DNA fragment with P2 and P3 (bottom). (B) RT-PCR analysis of ALP gene expression. First-strand cDNA from heterozygous (+/−) or homozygous (−/−) mice was amplified by PCR using ALP-specific primers that lie outside the first coding exon and flank the alternatively spliced region. Skeletal muscle ALP (ALP-SK) yielded a 948-bp fragment, and cardiac muscle-ALP (ALP-H) yielded a 702-bp fragment with this primer pair. Primers that amplify a 528-bp DNA fragment of tubulin were used to control for input cDNA. (C) Crude gastrocnemius muscle homogenates from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice were analyzed by Western blotting with antibodies to ALP and α-actinin.
Expression of ALP mRNA was assessed by RT-PCR using primers outside the targeted first exon (Fig. 2B). Amplifications with these primers, which flank the alternatively spliced region of ALP (32), show that ALP mRNA is absent from both skeletal and cardiac muscle of ALP−/− mice. Western blotting was used to assess ALP protein expression in the mutant mice. Homogenates from gastrocnemius muscles of wild-type, ALP+/−, and ALP−/− mice were probed with an antibody raised to a full-length ALP protein. As shown in Fig. 2C, ALP is absent in the knockout mice, although expression of α-actinin protein is preserved.
ALP deficiency does not disrupt skeletal muscle development.
Skeletal muscle from ALP mutant mice appears normal. Because of the proximity of ALP to the chromosomal region mutated in FSHD, we examined muscle histology carefully for signs of dystrophy. In the mdx mouse model of Duchenne muscular dystrophy, histological hallmarks include necrosis, increased fiber degeneration and/or regeneration with central nuclei, and increased variability in muscle bundle size and shape (7). For histological studies, we analyzed hematoxylin and eosin-stained gastrocnemius muscle sections. We found that muscle from ALP mutant mice does not show any of the histopathological changes characteristic of dystrophy, such as variable myofiber sizes and shapes, centralized nuclei, or inflammatory infiltrates (Fig. 3A). As a quantitative measure of myocyte degeneration and/or regeneration, we counted centrally located nuclei. The percentage of central nuclei in ALP mutant mice is <1.0% and not different from wild-type littermates. In mdx mice, >50% of myofibers contain central nuclei.
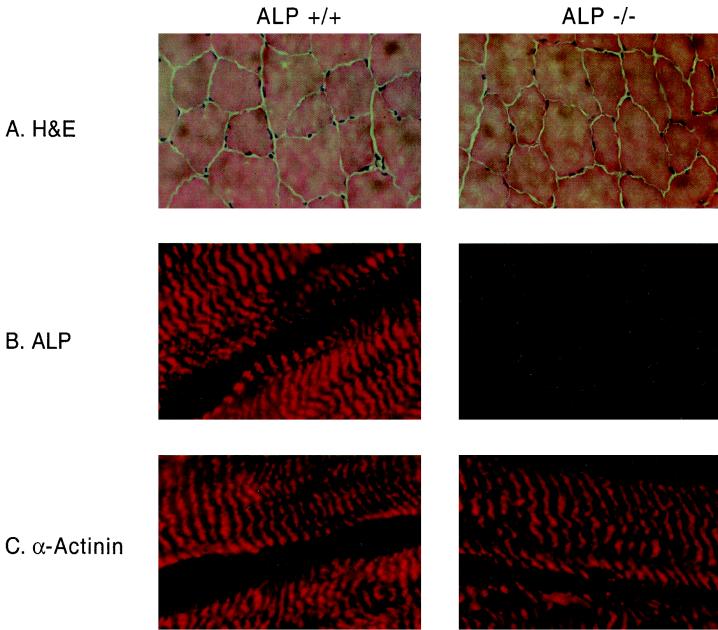
FIG. 3.
Histochemical analysis of ALP knockout mouse muscle. (A) Cryosections of gastrocnemius muscle from ALP+/+ and ALP−/− mice were analyzed with hematoxylin and eosin (H&E) staining. Sections from knockout mouse tissue resembled the wild-types and showed regularly sized and shaped myofibers and predominantly peripherally localized nuclei. (B) Immunofluorescent staining shows that ALP staining at Z lines is abolished in muscle sections from an ALP−/− mouse. (C) The localization of α-actinin at skeletal muscle Z lines remains normal in an ALP−/− mouse.
As ALP associates with α-actinin at the Z lines of skeletal muscle, we stained muscle sections from the knockout mice for α-actinin to evaluate cytoskeletal integrity. In longitudinal sections of wild-type skeletal muscle, ALP closely colocalizes with α-actinin at Z lines. As expected, ALP staining is absent in muscle from ALP−/− mice (Fig. 3B), although α-actinin staining of Z lines appears normal (Fig. 3C).
ALP deficiency does not influence sarcolemmal permeability.
Many muscular dystrophies cause sarcolemmal damage, which results in leakage of muscle enzymes. Serum pyruvate kinase levels are extremely elevated in many muscular dystrophy syndromes and are mildly elevated in FSHD (14). In mdx mice, pyruvate kinase levels rise acutely during the third week of life and remain elevated for at least 6 months (19). To determine whether loss of ALP causes sarcolemmal damage, we assayed serum pyruvate kinase activity of adult wild-type, ALP+/−, and ALP−/− mice. Serum pyruvate kinase levels in the plasma of ALP mutant mice were 0.31 ± 0.07 U/ml, not significantly different from those of the heterozygotes (0.27 ± 0.05 U/ml) or wild types (0.23 ± 0.05 U/ml), suggesting that sarcolemmal integrity is intact in ALP mutant mice. (Results are the means of triplicate assays of plasma samples from 7-month-old mice.)
DISCUSSION
ALP is a newly recognized component of the muscle cytoskeleton and localizes to the Z lines owing to interaction of its PDZ domain with α-actinin. ALP is an extremely abundant protein, as its mRNA represents approximately 0.18% of total mRNA in muscle (22). Expression of ALP is not restricted to skeletal muscle, as it is readily detected in cardiac muscle as well (32). Also, ALP is abundant in certain smooth muscles, as chicken ALP was purified as a major actinin-associated protein from the gizzard (23). Despite the abundance of ALP in muscle, mice lacking ALP show normal muscle development and are without visible muscle abnormalities. Analysis of muscle from these mice demonstrates normal histological parameters. Also, the sarcolemma is not compromised, as serum pyruvate kinase levels are within normal limits. These data suggest that ALP is not required for muscle development.
Recent studies have identified a large family of PDZ-LIM proteins that occur throughout the body. Several PDZ-LIM proteins are expressed in skeletal muscle and occur at the Z lines through a conserved interaction of their PDZ domains with α-actinin (21, 23, 32, 33). Because these other PDZ-LIM proteins are quite similar to ALP, they may compensate for the loss of ALP in the mutant mice.
What is the role for ALP in skeletal muscle? Biochemical and genetic studies have established that PDZ domains typically help assemble signal transduction pathways (10, 12, 17, 27). On the other hand, functions for LIM domains have remained less certain. LIM domains were first characterized in homeodomain transcription factors that determine cellular fate (25). Subsequently, LIM domains have been found in a variety of cytoskeletal proteins that associate with actin (13, 26). The muscle LIM protein (MLP) comprises a pair of LIM domains and, like ALP, is associated with Z lines (2). MLP is a positive regulator of myogenesis, and MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure (3). Whether the LIM domain of ALP has functions similar to the LIM domains of MLP remains uncertain.
Chromosomal mapping studies show that ALP occurs in 4q35, within 7 Mb of the telomeric region that is deleted in FSHD, the most common autosomal dominant muscular dystrophy. Genetic linkage analysis demonstrates that the FSHD locus occurs at 4q35, near the telomere of chromosome 4q (30; M. Upadhyaya, P. W. Lunt, M. Sarfarazi, W. Broadhead, J. Daniels, M. Owen, and P. S. Harper, Letter, Lancet 336:1320–1321, 1990). Both familial and spontaneous FSHD patients often have deletions of an integral number of 3.3-kb tandemly repeated units in this region (29, 31). No transcribed sequences have been found within these repeated units. Taken together, these observations have led to the suggestion that the deletions may mediate position effect variegation and alter the expression of a muscle-specific gene within 4q35 (1).
Studies of ALP expression in FSHD have not demonstrated a reproducible change in diseased muscle (4). However, because FSHD is a dominant disease caused by a heterochromatic mutation, it may be difficult to detect significant changes in steady-state expression levels of the responsible gene. Because of these complex genetics, animal models are needed to ascertain decisively a role for specific gene products in this disease. The normality of ALP knockout mice reported here suggests that down-regulation of ALP expression is unlikely to participate in FSHD. On the other hand, the FSHD mutation may up-regulate nearby transcripts (1), so that analysis of transgenic mice overexpressing ALP or other muscle-specific genes in 4q35 may provide insight.
ACKNOWLEDGMENTS
This work was supported by grants (to D.S.B.) from the National Institutes of Health (R01 NS34822), the Muscular Dystrophy Association, and the Howard Hughes Medical Institute Research Resources Program (76296-549901) and (to R.C.B.) from the American Heart Association Western States Affiliate.
REFERENCES
- 1.Altherr M R, Bengtsson U, Markovich R P, Winokur S T. Efforts toward understanding the molecular basis of facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S32–S38. [PubMed] [Google Scholar]
- 2.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 3.Arber S, Hunter J J, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard J C, Chien K R, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 4.Bouju S, Piétu G, Le Cunff M, Cros N, Malzac P, Pellissier J F, Pons F, Léger J J, Auffray C, Dechesne C A. Exclusion of muscle specific actinin-associated LIM protein (ALP) gene from 4q35 facioscapulohumeral muscular dystrophy (FSHD) candidate genes. Neuromuscul Disord. 1999;9:3–10. doi: 10.1016/s0960-8966(98)00087-x. [DOI] [PubMed] [Google Scholar]
- 5.Brenman J E, Chao D S, Xia H, Aldape K, Bredt D S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 6.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Huang F, Xia H, Peters M F, Froehner S C, Bredt D S. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α-1 syntrophin mediated by PDZ motifs. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 7.Bulfield G, Siller W G, Wight P A, Moore K J. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell K P. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 9.Christopherson K S, Bredt D S. Nitric oxide in excitable tissues: physiological roles and disease. J Clin Investig. 1997;100:2424–2429. doi: 10.1172/JCI119783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven S E, Bredt D S. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 11.Edwards R J, Watts D C. Specific spectrophotometric assay for the M isoenzymes of pyruvate kinase in plasma samples containing mixtures of the muscle (M) and liver (L) isoenzymes. Clin Chem. 1981;27:906–909. [PubMed] [Google Scholar]
- 12.Garner C C, Nash J, Huganir R L. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 13.Gill G N. The enigma of LIM domains. Structure. 1995;3:1285–1289. doi: 10.1016/s0969-2126(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 14.Griggs R C, Mendell J R, Miller R G. Evaluation and treatment of myopathies. F. A. Philadelphia, Pa: Davis Company; 1995. [Google Scholar]
- 15.Hoffman E P, Lehmann-Horn F, Rudel R. Overexcited or inactive: ion channels in muscle disease. Cell. 1995;80:681–686. doi: 10.1016/0092-8674(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 16.Jo K, Derin R, Li M, Bredt D S. Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex. J Neurosci. 1999;19:4189–4199. doi: 10.1523/JNEUROSCI.19-11-04189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornau H-C, Seeburg P H, Kennedy M B. Interaction of ion channels and receptors with PDZ domains. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea J A, Hentati F, Hamida M B, Bohlega S, Culper E J, Amato A A, Bossie K, Oeltjen J, Bejaoui K, McKenna-Yasek D, Hosler B A, Schurr E, Arahata K, de Jong P J, Brown R H., Jr Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 19.McArdle A, Edwards R H, Jackson M J. Time course of changes in plasma membrane permeability in the dystrophin-deficient mdx mouse. Muscle Nerve. 1994;17:1378–1384. doi: 10.1002/mus.880171206. [DOI] [PubMed] [Google Scholar]
- 20.Nehls M, Kyewski B, Messerle M, Waldschütz R, Schüddekopf K, Smith A J, Boehm T. Two genetically separable steps in the differentiation of thymic epithelium. Science. 1996;272:886–889. doi: 10.1126/science.272.5263.886. [DOI] [PubMed] [Google Scholar]
- 21.Passier R, Richardson J A, Olson E N. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev. 2000;92:277–284. doi: 10.1016/s0925-4773(99)00330-5. [DOI] [PubMed] [Google Scholar]
- 22.Piétu G, Alibert O, Guichard V, Lamy B, Bois F, Leroy E, Mariage-Sampson R, Houlgatte R, Soularue P, Auffray C. Novel gene transcripts preferentially expressed in human muscles revealed by quantitative hybridization of a high density cDNA array. Genome Res. 1996;6:492–503. doi: 10.1101/gr.6.6.492. [DOI] [PubMed] [Google Scholar]
- 23.Pomiès P, Macalma T, Beckerle M C. Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J Biol Chem. 1999;274:29242–29250. doi: 10.1074/jbc.274.41.29242. [DOI] [PubMed] [Google Scholar]
- 24.Qiu M, Bulfone A, Ghattas I, Meneses J J, Christensen L, Sharpe P T, Presley R, Pedersen R A, Rubenstein J L. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Garcia I, Rabbitts T H. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 26.Schmeichel K L, Beckerle M C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 27.Sheng M. PDZs and receptor/channel clustering: rounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 28.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 29.van Deutekom J C, Wijmenga C, van Tienhoven E A, Gruter A M, Hewitt J E, Padberg G W, van Ommen G J, Hofker M H, Frants R R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 30.Wijmenga C, Frants R R, Brouwer O F, Moerer P, Weber J L, Padberg G W. Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet. 1990;336:651–653. doi: 10.1016/0140-6736(90)92148-b. [DOI] [PubMed] [Google Scholar]
- 31.Wijmenga C, Hewitt J E, Sandkuijl L A, Clark L N, Wright T J, Dauwerse H G, Gruter A M, Hofker M H, Moerer P, Williamson R, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 32.Xia H, Winokur S T, Kuo W L, Altherr M R, Bredt D S. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Ruiz-Lozano P, Martone M E, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]