Abstract
The Aspergillus nidulans zinc finger transcription factor PacC is activated by proteolytic processing in response to ambient alkaline pH. The pH-regulated step is the transition of full-length PacC from a closed to an open, protease-accessible conformation. Here we show that in the absence of ambient pH signaling, the C-terminal negative-acting domain prevents the nuclear localization of full-length closed PacC. In contrast, the processed PacC form is almost exclusively nuclear at any ambient pH. In the presence of ambient pH signaling, the fraction of PacC that is in the open conformation but has not yet been processed localizes to the nucleus. Therefore, ambient alkaline pH leads to an increase in nuclear PacC by promoting the proteolytic elimination of the negative-acting domain to yield the processed form and by increasing the proportion of full-length protein that is in the open conformation. These findings explain why mutations resulting in commitment of PacC to processing irrespective of ambient pH lead to permanent PacC activation and alkalinity mimicry. A nuclear import signal that targets Escherichia coli β-galactosidase to the nucleus has been located to the PacC zinc finger region. A mutation abolishing DNA binding does not prevent nuclear localization of the processed form, showing that PacC processing does not lead to nuclear localization by passive diffusion of the protein made possible by the reduction in size, followed by retention in the nucleus after DNA binding.
Proteolytic processing activation of transcription factors in response to their cognate environmental signals occurs across distant groups of eukaryotic organisms. pH regulation of gene expression in the mold Aspergillus nidulans is one such example. Here, the key regulatory zinc finger protein PacC activates alkaline genes and represses acidic genes according to the needs imposed by ambient pH, thereby providing the organism with one prerequisite for growing in environments as acidic as pH 2.5 or as alkaline as pH 10.5 (7, 59). Other prototypical members of the group of transcription factors activated by proteolytic processing are the immune and inflammatory response regulator NF-κB (23, 58), the Drosophila melanogaster cubitus interruptus (Ci) zinc finger factor (the transducer of the hedgehog signal) (29, 53), and the sterol regulatory element-binding protein (SREBP), which switches on genes for cholesterol biosynthesis and fat metabolism (5, 6).
The zinc finger transcription factor PacC is synthesized as a 674-residue precursor. At alkaline ambient pH, a signal transmitted to PacC by the orphan pal gene signal transduction pathway (13, 14, 37, 43, 44) results in a conformational change leading to an open conformation in which PacC is accessible to a processing protease (18, 41, 47). This protease removes ∼400 residues from the C terminus, which includes a negative-acting domain. The resulting product (248 to 250 residues) (41) is fully competent in transcriptional regulation through 5′-GCCARG-3′ sites (20) in the promoters of both alkaline (activated by PacC) (17) and acidic (repressed by PacC) (16) genes.
Prototypical NF-κB is a heterodimer of p50 and p65 (RelA) subunits. p50 originates from proteolytic processing of a p105 precursor. As in PacC, a C-terminal moiety in p105 is a cis- and negative-acting domain in the p105/p65 heterodimer. This negative function can be also provided in trans by members of the ΙκB family of inhibitory proteins, which are homologues of the p105 C-terminal moiety and form heterotrimeric complexes with p50 and p65. Both the C-terminal moiety of p105 (4, 25, 51) and the ΙκB proteins (3, 28, 30, 65, 66) preclude the nuclear localization of NF-κB and its binding to DNA. In cubitus interruptus (2) and SREBP (5, 6, 54), the presence of negative-acting domains C terminal to the mature polypeptide also precludes nuclear localization. The fact that in all cases in which the role of the negative-acting domains removable by proteolysis has been investigated such domains preclude nuclear localization underlines the regulatory possibilities offered by the separation into distinct nuclear and cytoplasmic compartments that characterizes eukaryotic cells.
Because translation takes place in the cytoplasm, transcription factors need to be imported into and may also be exported from the nucleus. All passive and active transport into and out of the nucleus occurs through the nuclear pore complexes (NPCs) in the nuclear envelope, which provide a diffusion channel for macromolecules smaller than ∼40 to 60 kDa (45). However, transport of macromolecules through the NPCs is generally energy dependent and requires components of the Ran GTPase system as well as transport (both import and export) receptors and adapter molecules recognizing nuclear localization signals and nuclear export signals in their cargoes (see references 24 and 42 for reviews). The fact that such signals as well as domains involved in their intramolecular masking may be cleaved off after appropriate signaling opens new regulatory possibilities in this category of transcription factors.
We use the genetically amenable system of pH regulation in A. nidulans to understand the mechanisms underlying the regulation of gene expression through transcription factor proteolytic processing. Briefly, two different conformations (“open” and “closed”) of the precursor form and the processed protein participate in pH regulation (18, 41, 47). By preventing the closed-to-open conformation transition (18), mutational inactivation of any of the six palA, -B, -C, -F, -H, and -I genes of the ambient pH signaling pathway prevents proteolytic processing activation (47) and leads to the absence of pacC function (59) and acidity mimicry (1, 7). pacC mutations such as pacC+/-20205, leading to an inability to undergo the conformational change at any ambient pH, result in permanently closed PacC and thereby prevent proteolytic processing and, similarly to pal− mutations, lead to a loss-of-function (pacC+/-) acidity mimicry phenotype (18, 41). Finally, by disrupting interactions between a C-terminal domain of PacC and upstream regions that maintain the closed conformation (18), another class of mutations (pacCc) results in commitment of PacC to the open conformation, consequent processing at any ambient pH (41, 47), and an alkalinity mimicry, gain-of-function phenotype (59). This pacCc class includes missense mutations affecting PacC residues critical for the interactions described above and C-terminal truncation mutations.
Here we address the role of the C-terminal, negative-acting domain of PacC and demonstrate that, by governing the proteolytic removal of this domain, the ambient pH signal regulates the subcellular localization of PacC, which is largely cytoplasmic under acidic conditions. In contrast, an almost exclusive nuclear localization is seen under alkaline conditions, and this correlates with proteolytic processing. A fraction of the full-length form, probably in the open conformation, is able to enter into the nucleus.
MATERIALS AND METHODS
Aspergillus techniques and media.
A. nidulans strains carrying markers in standard use and standard genetic procedures were used (10). Media and phenotype testing for pH regulatory mutations have been described previously (1, 7). Complex PPB broth (41) with 3% (wt/vol) sucrose as a carbon source was used for A. nidulans liquid cultures. This broth was adjusted to an acidic, neutral, or alkaline pH according to reference 47. For strains carrying alcAp-driven transgenes, mycelia were pregrown for 14 to 18 h in acidic PPB medium containing 3% glucose as a carbon source (repressing conditions) and transferred to fresh medium adjusted to different pH values and containing 0.05% glucose and 100 mM threonine (inducing conditions). Mycelia were incubated for a further 8 h before being used for protein extraction, subcellular fractionation, and/or microscopic observation. Alternatively, conidiospores were germinated for 14 h at 37°C in minimal or PPB medium containing 0.05% (wt/vol) glucose and 100 mM threonine, adjusted to different pH values as in reference 47.
Subcellular fractionation procedure and protein extraction.
For subcellular fractionation and protein extraction, we used a modification of a published procedure (63). Washed mycelia were resuspended in buffer LYS, which contained 50 mM Tris-HCl (pH 7.5), 5 mM magnesium acetate, 5 mM EGTA, 3 mM CaCl2, 3 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μM pepstatin, 1 M sorbitol, 7% (wt/vol) Ficoll 400, and 20% glycerol, and lysed with glass beads in a Braun MSK cell disrupter. The lysate was gently mixed in the cold with 2 volumes of buffer CUS (25 mM Tris-HCl [pH 7.5], 5 mM magnesium acetate, 5 mM EGTA, 1 mM DTT, 0.25 mM PMSF, 10% glycerol), laid on top of a 1:1.7 mixture of buffers LYS and CUS, and centrifuged for 7 min at 4,000 rpm in a Sorvall HB4 rotor. The lower phase, containing cell debris, was discarded. The upper phase was loaded on top of a buffer NUC cushion (buffer NUC is 25 mM Tris-HCl [pH 7.5], 1 M sucrose, 5 mM magnesium acetate, 5 mM EGTA, and 10% glycerol) and centrifuged for 15 min at 7,000 rpm in an HB4 rotor. This step resulted in a nuclear pellet with a microsomal fraction sedimenting in the interphase and a cytoplasmic fraction in the upper phase. The nuclear pellet was resuspended in buffer A (25 mM HEPES [pH 7.9], 5 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 1 mM PMSF, 1 μM pepstatin, 0.6 μM leupeptin) with 50 mM KCl, and nuclear protein was extracted with 0.4 M ammonium sulfate. After removal of DNA and debris by centrifugation at 100,000 × g, the cleared nuclear extract was dialyzed against buffer A containing 100 mM KCl. The upper (cytoplasmic) phase was extracted with 0.4 M ammonium sulfate as described above, concentrated by ammonium sulfate precipitation, and desalted through a PD-10 column equilibrated with buffer A containing 100 mM KCl. Mycelial (total) protein extracts were prepared as described previously (47). Protein fractions were stored at −80°C. The presence of nuclei exclusively in the nuclear fraction and their absence from the cytosolic fraction were confirmed by DAPI (4′,6′-diamidino-2-phenylindole) staining.
Plasmids and recombinant strains.
Plasmid pAN 52–1 sGFP (48) (obtained from C. Scazzocchio) contains the coding region of sGFP-TYG (9), previously used to track the subcellular localization of transcription factors in A. nidulans (21, 48). pALC-argB(BglII) was used for the expression of green flourescent protein (GFP) and lacZ fusion proteins under alcAp control. This plasmid (41) contains a frameshift argB− allele that we used to target all of the transforming constructs to the argB2 mutant allele of the recipient strains, which prevented possible position effects on the expression of the transgenes. The recombinant plasmids derived from pALC-argB(BglII) are detailed in Table 1. These were constructed by standard recombinant techniques or by PCR and were transformed (60) into ΔpacC argB2 and ΔpacC palA1 argB2 strains (41), as appropriate. The correct in-frame joining of the fragments and the absence of introduced mutations in PCR-amplified fragments were verified by automatic DNA sequencing. The correct integration of the transforming plasmid in the argB locus was verified by Southern analysis. All recombinant strains carried single-copy integration events, with the exception of p[alcAp::PacC(241–280)::GFP]- and p[alcAp::PacC(5–250)Q155K::GFP]-transformed strains, for which double integration events were the lowest copy number recovered. For experiments involving the latter transgene, a control transformant carrying a double-integration event of the wild-type p[alcAp::PacC(5–250)::GFP] construct was used. The subcellular localization of the encoded fusion protein in this double-copy transformant was indistinguishable from that of its corresponding single-copy transformant. Growth tests under inducing and repressing conditions were used to confirm that the phenotype exclusively resulted from expression of the transgene.
TABLE 1.
Plasmids used in this work and deduced fusion protein products
| Construct | Polypeptide encoded by alcAp-driven transgene |
|---|---|
| p[alcAp::GFP] | sGFPa |
| p[alcAp::GFP::PacC(5–250)] | sGFP-GlySerPro-PacC(5–250)-GlyAsnSerSer |
| p[alcAp::GFP::PacC(5–678)] | sGFP-GlySerPro-PacC(5–678) |
| p[alcAp::PacC(5–678)::GFP] | PacC(5–678)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(5–250)::GFP] | PacC(5–250)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(5–250)Q155K::GFP] | PacC(5–250)Q155K-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(241–280)::GFP] | PacC(241–280)-Gly-sGFP |
| p[alcAp::PacC(5–273)::GFP] | PacC(5–273)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(250–678)::GFP] | PacC(250–678)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(5–173)::GFP] | PacC(5–173)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(66–173)::GFP] | PacC(66–173)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(173–250)::GFP] | PacC(173–250)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(66–250)::GFP] | PacC(66–250)-GlyAsnSerSer-sGFP |
| p[alcAp::PacC(5–234)::lacZ] | PacC(5–234)-SerLeuAlaLeu-LacZ |
| p[alcAp::PacC(169–234)::lacZ] | PacC(169–234)-SerLeuAlaLeu-LacZ |
sGFP is the sGFP-TYG version of the GFP (9).
EMSA and Western blot analysis.
Total, cytoplasmic, and nuclear extracts were analyzed by Western blotting and electrophoretic mobility shift assay (EMSA) as described previously (41). Rat anti-PacC DNA binding domain antiserum (used at 1/4,000) was directed against PacC(5–265) (41). Rabbit anti-PacC C-terminal region antiserum (used at 1/2,000) was directed against residues 529 through 678. Rabbit anti-GFP antiserum (used at 1/3,000) was obtained from Clontech. Rabbit anti-yeast hexokinase antiserum (used at 1/20,000) was obtained from Chemicom and recognizes a protein with the expected electrophoretic mobility of A. nidulans hexokinase. In these cases, peroxidase-coupled goat anti-rabbit (Sigma) antiserum was used as a secondary antibody.
Microscopy.
Mycelial samples taken at appropriate times after induction of transgenes were washed three times in water, and the subcellular distribution of green fluorescence was observed under a Zeiss epifluorescence microscope with 495- and 530-nm excitation and emission filters, respectively. Images were collected with an IPLab Spectrum system and transferred to Adobe Photoshop, version 4.0. Samples stained with DAPI (0.1 μg/ml) were fixed with formaldehyde and washed in phosphate-buffered saline before microscopy.
RESULTS
Interactions between the C-terminal region and upstream regions in the full-length PacC form do not prevent binding to DNA.
In agreement with previous work with A. nidulans (41, 47), extracts of yeast strains expressing either PacC(5–678) or PacC(5–265) proteins (the latter approximating the processed form) contained similar levels of these proteins, as monitored by Western analysis, and similar levels of PacC DNA binding activity, as determined by the amounts of their respective DNA-protein complexes in EMSA (data not shown), which confirms that the negative action of the C-terminal region in the full-length form does not result from prevention of DNA binding. Proteins expressed in yeast were used in this experiment so that the full-length form would not be processed (41).
The processed PacC form is localized in the nucleus.
A plausible explanation for the negative action of the C-terminal region in PacC would be that its presence or absence affects the PacC nucleocytoplasmic distribution. To address this possibility, we used a subcellular fractionation procedure to prepare nuclear and cytoplasmic fractions of A. nidulans in different mutants and in the wild type and examined the distribution of full-length and processed PacC by Western blot analysis (using antiserum against the nearly N-terminal zinc finger region) and by a sensitive EMSA (Fig. 1). Western analysis of the glycolytic, cytosolic enzyme hexokinase was used to confirm that the nuclear fractions were largely free of cytoplasmic contamination (Fig. 1).
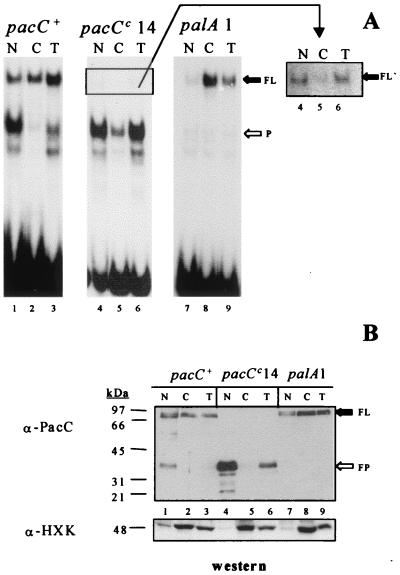
FIG. 1.
Subcellular PacC localization in wild-type and mutant strains in pH regulation. Protein extracts from the nuclear (N) or cytoplasmic (C) fractions or the total cell (T) (see Materials and Methods) of the indicated strains were analyzed by EMSA with a PacC-specific DNA probe (A) or by Western blotting (B) with antisera against the PacC DNA binding domain. Strains were grown on sucrose-MFA for 24 h at 37°C under acidic pH conditions. (A) Five micrograms of each extract was incubated with the DNA probe. FL indicates the protein-DNA complex corresponding to the PacC primary translation product, and P denotes the complex corresponding to the processed PacC form. A longer exposure of the region of the gel including the complex corresponding to the truncated (after residue 492) pacCc14 primary translation product (FL′) is shown on the right. (B) Fifty micrograms of each protein extract was analyzed by Western blotting, both with an anti-PacC (α-PacC) antiserum and with an antihexokinase (α-HXK) antiserum, as indicated.
In a wild-type strain grown under acidic conditions, proteolytic activation of PacC is largely prevented, and the full-length form predominates over the processed form (47) (Fig. 1A and B, lanes 3). This processed form is largely enriched in the nuclear fraction and is virtually undetectable in the cytosol (Fig. 1A and B, lanes 1 and 2). Under acidic conditions, the alkalinity-mimicking pacCc14 mutation results in predominance of the processed PacC form that cofractionates with the nuclear fraction (Fig. 1A and B, lanes 4 to 6). A similar result was obtained for the pacCc50 product (data not shown). The alkalinity-mimicking pacCc50 mutation truncates PacC only ∼13 residues downstream of the deduced processing limit. Taken together, these data strongly suggest that the processed PacC form is predominantly nuclear, which agrees with its function as a transcription factor. Second, they indicate that the pH signal (whose requirement for processing is bypassed by the alkalinity-mimicking pacCc mutation) leads to nuclear PacC localization at least in part by promoting PacC proteolytic processing.
To confirm the nuclear localization of the processed PacC form in vivo, we constructed a transgene encoding a PacC(5–250) protein (approximating the processed form) tagged with GFP in its N terminus (Table 1 and Fig. 2) under the control of the threonine-inducible alcA promoter. This GFP::PacC(5–250) construct and a control construct driving expression of GFP (without PacC sequences) were transformed into a null ΔpacC background in either the presence or the absence (inactivated by the palA1 mutation) of a functional pal gene pathway. To avoid position effects, these and all other further constructs were targeted in single or double copy to the argB locus. The null, strongly acidity-mimicking ΔpacC allele leads to poor conidiation and lack of growth at alkaline pH (59). Expression of GFP::PacC(5–250) restored both conidiation and the ability to grow at alkaline pH in both the ΔpacC and the ΔpacC palA1 backgrounds (data not shown), indicating that the N-terminal GFP tag in this processed PacC does not impair its function. In a control strain expressing GFP without PacC sequences, fluorescence was found throughout the hyphae (Fig. 3A and C). In contrast, in strains expressing the GFP::PacC(5–250) fusion protein cultured at different pH values, the fluorescence localized in the nuclei, regardless of whether the transfer pH was acidic or alkaline or the functional status of the pal pathway (Fig. 3B). Western blotting and EMSA showed that GFP::PacC(5–250) was largely intact under all conditions tested (Fig. 4A and B, right panels). These results agree with those of the subcellular fractionation experiments presented above and demonstrate the preferential and ambient pH signal-independent nuclear localization of the processed PacC form in vivo.
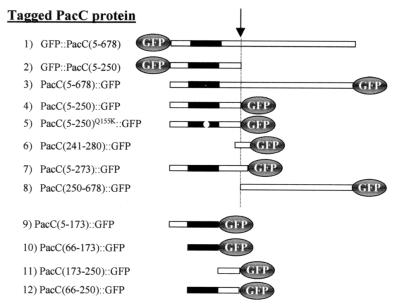
FIG. 2.
Schematic representation of PacC derivatives used in this work as GFP fusion proteins. All fusion proteins were expressed by using the alcohol dehydrogenase I promoter, which is inducible by threonine. The full-length PacC protein (residues 5 to 678; codon 5 is the major translation start codon) is illustrated as an open rectangle, with the zinc finger region shown in black. The approximate position of the processing site in full-length PacC fusion proteins is shown by a dashed line and indicated by an arrow. The fusion proteins are denoted here and throughout the text according to the N-terminal or C-terminal position of GFP and the amino acid coordinates of the corresponding PacC polypeptide.
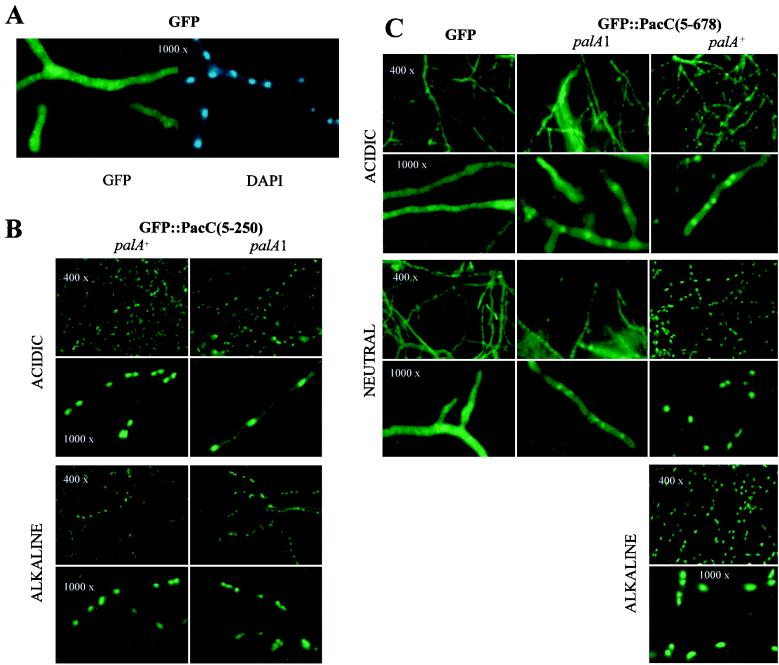
FIG. 3.
Changes in the subcellular localization in vivo of GFP-tagged full-length and processed PacC forms in response to ambient pH changes. Strains were pregrown under acidic, repressing conditions for alcAp and transferred to inducing conditions for 8 h, in media buffered at the indicated pH values. Typical final values were 5.8 (acidic pH), 6.8 (neutral pH), and 7.6 (alkaline pH). (A) Control experiment showing the uniform distribution of GFP across the hyphae (left) compared to the position of nuclei (right, by DAPI staining). Only acidic conditions are shown. This uniform distribution does not change upon transfer to neutral pH (see panel C). Note that this (null pacC) strain does not grow under alkaline conditions. (B) Subcellular localization of GFP::PacC(5–250) in palA+ and palA1 backgrounds. (C) Subcellular localization of GFP::PacC(5–678) under acidic, neutral, and alkaline conditions, in the palA+ and palA1 backgrounds. The palA1 mutation precludes growth of this strain under alkaline conditions. In panels B and C, the original magnifications were ×400 and ×1,000, as indicated.
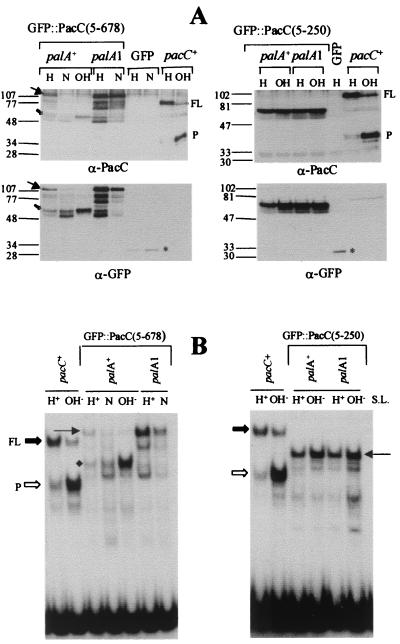
FIG. 4.
Analysis of GFP::PacC(5–250) and GFP::PacC(5–678) fusion proteins in transgenic (ΔpacC) strains. Samples of cultures used in the experiments shown in Fig. 2 were collected and used to prepare whole-cell protein extracts. (A) Western analyses of whole-cell extracts (50 μg of protein in each lane) with antisera against the PacC DNA binding domain (α-PacC) or against GFP (α-GFP). pH growth conditions are indicated on top of each lane, as is the palA+ or palA1 genotype of the corresponding strains. Wild-type control extracts from acidic and alkaline growth conditions are indicated with pacC+. Full-length (FP) and processed (P) PacC are revealed only by the anti-PacC antiserum. The full-length and processed forms of GFP::PacC(5–678), revealed with both the anti-PacC and the anti-GFP antisera, are indicated by solid and open arrows, respectively. An asterisk indicates the position of GFP in the lanes of the corresponding control strain. (B) EMSA analyses of the above extracts with 5 μg of protein and a specific PacC DNA probe. Extracts are as in panel A. Protein-DNA complexes formed by wild-type full-length and processed forms of PacC are indicated, as are the positions of complexes formed by the full-length (thin arrow) and processed (diamond) forms of GFP::PacC(5–678). Note the shift in mobility caused by the GFP tag in these complexes compared to those of untagged PacC.
The full-length form is normally distributed between nuclei and cytosol, but is largely cytosolic in the absence of the pal signal.
In contrast to the preferential nuclear localization of the processed form, the full-length form that predominates in the wild type grown under acidic conditions was found in both the nuclear and the cytosolic fractions (Fig. 1A and B, lanes 1 to 3). The acidity-mimicking palA1 mutation, which interrupted the pH signaling pathway and prevented proteolytic processing, resulted in the almost exclusively cytosolic localization of the full-length form (Fig. 1A and B, lanes 7 to 9). That this effect is not specific to palA− alleles was confirmed by a subcellular fractionation experiment with a palH17 strain, which gave essentially the same results (data not shown). These data showed that in the presence of a functional pal pathway, the full-length PacC form distributes between nuclei and cytosol, but in its absence, this full-length form is predominantly cytosolic. In addition, this indicates that the full-length protein is able to enter the nucleus and that a certain degree of ambient pH signaling (which under the moderately acidic growth conditions used here takes place in the wild type, but not in the palA1 strain) is required for the nuclear localization of a proportion of the full-length form.
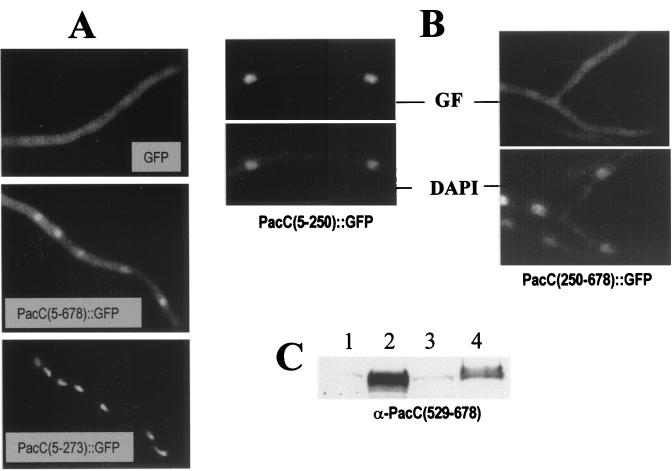
To confirm that a proportion of the full-length form is able to enter the nucleus, we used two fusion proteins in which a GFP tag was attached to the C-terminal residue of PacC. One protein, denoted PacC(5–678)::GFP, corresponds to the full-length PacC protein (Fig. 2 and Table 1). A second, denoted PacC(250–678)::GFP, corresponds to the region of PacC that is removed by proteolytic processing (Fig. 2 and Table 1). PacC(5–678)::GFP can be proteolytically processed (data not shown). Under acidic conditions, the (GFP-tagged) full-length fusion protein predominates, but some processing occurs (data not shown). The resulting processed PacC form is released from the C-terminal moiety of PacC tagged with GFP after residue 678 and therefore is not fluorescent (see the scheme in Fig. 2). Therefore, nuclear green fluorescence would indicate the nuclear localization either of a proportion of the full-length PacC form or of the hypothetical C-terminal polypeptide that would be released after processing. Figure 5A shows that in a strain expressing PacC(5–678)::GFP grown under acidic conditions, fluorescent nuclei were clearly observed over the cytosolic background. This contrasted with strains either expressing GFP alone (Fig. 3A and C and Fig. 5A) or expressing a PacC(250–678)::GFP fusion protein containing the C-terminal moiety (Fig. 5B and C), in which fluorescence was similar throughout the cells. In control strains expressing PacC(5–273)::GFP or PacC(5–250)::GFP (Fig. 2), fluorescence colocalized with nuclei (Fig. 5A and B). Taken together, all of the results presented above strongly indicate that at least part of the full-length form is able to get into the nucleus. No indication of nuclear exclusion of the PacC(250–678)::GFP polypeptide, which might have suggested the presence of a nuclear export signal in PacC residues 250 through 678, was observed.
FIG. 5.
Nuclear fluorescence in a strain expressing PacC(5–678)::GFP. (A) ΔpacC strains expressing GFP, PacC(5–678)::GFP, or PacC(5–273)::GFP proteins from single-copy alcAp-driven transgenes were pregrown for 18 to 20 h under repressing conditions for alcAp and transferred to inducing medium for an additional 7-h incubation at acidic ambient pH, before their green fluorescence was observed under the microscope. (B) Strains expressing PacC(5–250)::GFP (i.e., the GFP-tagged processed PacC form) or PacC(250–678)::GFP (i.e., the GFP-tagged C-terminal moiety removed by processing) were grown at acidic ambient pH under inducing conditions, fixed with formaldehyde, and stained with DAPI. The green fluorescence (GF) and the DAPI channels are shown. (C) The PacC(250–678)::GFP protein is expressed at high levels under the conditions used in panel B. Western analysis was of a protein extract (50 μg) from the following strains (only the relevant portion of the gel is shown). Lanes: 1, PacC(5–250)::GFP strain; 2, PacC(250–678)::GFP strain, 3, GFP strain; 4, a pacC+ strain. The blot was developed with an antiserum raised against the C-terminal region (amino acids 529 through 678) of PacC. The weak cross-reacting band in lanes 1 and 3 is an unspecific band. The band in lane 4 corresponds to full-length PacC. The prominent protein band in lane 2 corresponds to PacC(250–678)::GFP.
In the absence of the pal signal, the region of PacC removed by proteolytic processing prevents the nuclear localization of the full-length form in vivo.
To analyze in vivo the changes in the subcellular localization of PacC under different ambient conditions, we constructed palA+ and palA1 strains showing expression under the control of alcAp a GFP::PacC(5–678) fusion protein (Table 1 and Fig. 2) in a null ΔpacC background. In this fusion protein, GFP is attached to the N terminus of full-length PacC, because this relative arrangement guarantees that the fluorescent tag is not removed by proteolytic processing, which eliminates the PacC C terminus. The phenotype of these strains is indistinguishable from those of the corresponding strains expressing untagged PacC(5–678) with regard to alkaline pH growth and hypostasis of the introduced transgene to the palA1 mutation, indicating that the function of these proteins is under ambient pH control and that the presence of the GFP tag in the N terminus of PacC does not preclude its function.
Under acidic conditions, the subcellular distribution of GFP::PacC(5–678) largely resembled that of GFP (Fig. 3A and C). In contrast to the GFP strain, however, weak fluorescence highlighting the nuclei was reproducibly observed (Fig. 3C). Transfer to either neutral or alkaline conditions dramatically changed this distribution and unequivocally resulted in preferential nuclear localization (Fig. 3C). This dramatic change correlated with proteolytic processing of the GFP::PacC(5–678) primary translation product (Fig. 4A and B, left panels). The palA1 mutation largely prevented this proteolytic processing (Fig. 4A and B, left panels), eliminated the exclusively nuclear fluorescence seen under neutral conditions [Fig. 3C; note that the alcAp-GFP::PacC(5–678) allele is hypostatic to palA1 and the double mutant strain cannot be grown under alkaline conditions], and made the residual nuclear staining seen under acidic conditions less evident (Fig. 3C). These data show that the processing pattern and the subcellular distribution of PacC forms determined by in vitro fractionation studies or by in vivo GFP tagging are coincident. A conclusion of the above experiments is that in the absence of the pal signal, the region of PacC removed by proteolytic processing prevents nuclear localization of full-length PacC and that the processed PacC product [here, GFP::PacC(5–250)] is almost exclusively nuclear.
The processed PacC form contains a nuclear import signal.
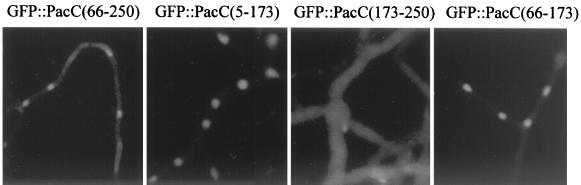
The deduced Mr for GFP::PacC(5–250) is 53.8 kDa. The Mr estimated for this protein by gel filtration chromatography of extracts was 101 kDa (Fig. 6A), which might result from either dimerization or a particularly elongated shape of the fusion protein. In any case, this size is above the limit below which proteins can freely move between cytosolic and nuclear compartments and would suggest that an active transport mechanism is active on the processed PacC form. To address this point, we constructed two alcAp-driven genes encoding fusion proteins between PacC polypeptides and the Escherichia coli lacZ gene product. One encodes a PacC(5–234)::lacZ polypeptide with a predicted Mr of 140 kDa. PacC residues 5 through 234 contain the zinc finger region and lack all major and minor processing sites (E. Díez and M. A. Peñalva, unpublished data). This transgene complements several aspects of the ΔpacC phenotype, strongly indicating that the fusion protein is able to enter the nucleus. Subcellular fractionation experiments confirmed that PacC(5–234)::lacZ is indeed nuclear (Fig. 6B). In contrast, a PacC(169–234)::lacZ fusion protein (deduced Mr of 123 kDa) showed cytosolic localization (Fig. 6B). In vivo localization experiments with equivalent GFP fusion proteins (data not shown) confirmed the subcellular fractionation results. We conclude that PacC residues 5 through 234 contain a nuclear import signal.
FIG. 6.
The processed PacC form contains a nuclear import signal. (A) Protein extracts isolated from a strain expressing GFP::PacC(5–250) were fractionated through a Sephacryl S-200 column, calibrated with protein standards of the indicated Mr. The fusion protein was detected by EMSA, and protein-DNA complexes were quantified by phosphorimaging. wt, wild type; IP, input extract. (B) Subcellular fractionation experiments of mycelia from strains expressing the indicated PacC::lacZ fusion proteins. Nuclear (N), cytosolic (C), and total (T) protein extracts were analyzed by Western blotting with the indicated antibodies. PacC residues 169 through 234 are recognized by the anti-PacC DNA binding domain (DBD) antiserum, which was raised against a polypeptide containing residues 5 through 265.
The zinc finger region of PacC contains a nuclear localization signal whose action is independent of DNA binding.
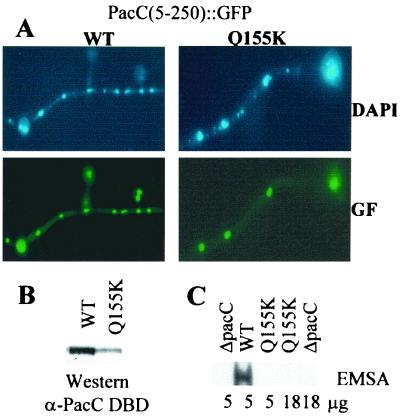
The nuclear import region of PacC(5–250) was delimited by deletion analysis of GFP fusion proteins to residues 66 through 173 (Fig. 7). PacC residues 69 through 168 correspond to the zinc finger region and suffice for high-affinity DNA binding (20). To separate DNA binding from nuclear localization, we used a Gln 155→Lys substitution affecting a critical residue in the recognition α-helix of the third zinc finger that abolishes DNA binding in vitro and leads to a loss-of-function phenotype (20). In common with its parental PacC(5–250)::GFP fusion protein, a mutant PacC(5–250)Q155K::GFP fusion protein localized to the nuclei (Fig. 8A) despite the lack of DNA binding activity shown in vitro by the mutant fusion protein (Fig. 8B and C). This dissociation of DNA binding from nuclear localization based on the phenotype of the Gln 155→Lys substitution agrees with the above conclusion that the processed PacC form (approximately containing residues 5 through 250) contains a nuclear import signal, because it rules out the possibility that its nuclear localization could result from passive diffusion and nuclear retention by DNA binding
FIG. 7.
Delimiting a nuclear localization signal to the zinc finger region. Strains expressing the indicated GFP fusion proteins were grown under inducing, acidic ambient pH conditions and examined by epifluorescence microscopy.
FIG. 8.
A single amino acid substitution abolishing DNA binding does not prevent the nuclear localization of the processed PacC form. Strains expressing the wild type (WT) or a mutant Q155K PacC(5–250)::GFP fusion protein were grown under inducing (also acidic) conditions for 14 h at 37°C and used for epifluorescence microscopy and protein extraction. (A) Samples of the indicated strains were fixed with formaldehyde, stained with DAPI, and analyzed under the microscope in the green fluorescence (GF) and DAPI channels. (B) Protein extracts were analyzed by Western blotting with an antiserum against PacC residues 5 through 265 (α-PacC DBD). (C) Protein extracts (5 or 18 μg, as indicated below the corresponding lanes) were analyzed by EMSA with a PacC-specific probe. The relevant region of the autoradiogram containing the wild-type PacC fusion protein-DNA complex is shown. No binding was detected with the mutant protein carrying the Gln 155→Lys substitution, in agreement with previous work (20).
Nuclear full-length PacC is in the open conformation.
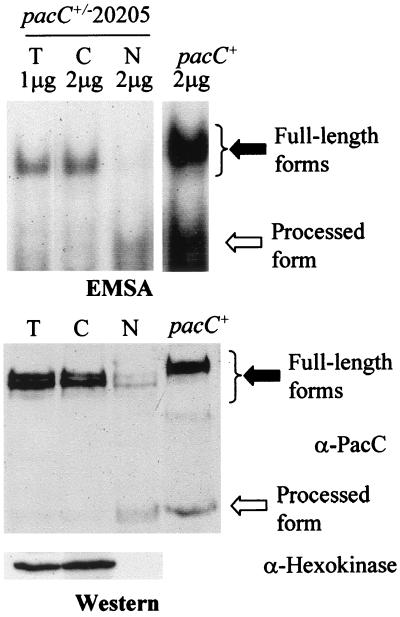
In the presence of the pal signal, a fraction of full-length PacC is localized in the nucleus. This full-length form alternates between two conformations, and the ambient pH signal is required for the closed-to-open conformation transition (18). By removing the interacting C-terminal region of PacC, the truncation mutation in the pacCc14 primary translation product PacC(5–492) results in commitment to the open (protease-accessible) conformation at any ambient pH, leading to proteolytic processing, which results in low levels of the mutant primary translation product (Fig. 1) and bypasses the requirement for the ambient pH signal (18). If the transition to the open conformation were required for the nuclear localization of the full-length form, the pacCc14 primary translation product should be localized in the nucleus. Overexposure of the autorad region corresponding to the pacCc14 full-length product-DNA complex showed that this is indeed the case (Fig. 1A, lanes 4 to 6). In contrast to the pacCc14 product, the pacC+/-20205 product is a prototypic mutant PacC shifted towards the closed conformation. Subcellular fractionation analysis showed that, under neutral growth conditions (in which significant pH signaling occurs) (Fig. 3C), the predominant mutant full-length product is largely excluded from the nuclei (Fig. 9). These data correlate the transition of the PacC primary translation product towards the open conformation with the nuclear localization of a part of the primary translation product.
FIG. 9.
Subcellular localization of PacC forms in a pacC+/-20205 mutant strain. Cytoplasmic (C), nuclear (N), and whole-cell (T) protein fractions were prepared from a pacC+/-20205 strain grown under neutral conditions and analyzed by EMSA or Western blotting, as in Fig. 1. Solid and open arrows indicate the full-length and the processed PacC forms, respectively. The slight increase in mobility of the mutant full-length form complex compared to that of the wild type results from substitution of residues 465 through 540 in PacC by an octapeptide encoded in a different reading frame (41). The pacC+ extract used as a size control corresponds to acidic conditions, to maximize levels of the full-length form.
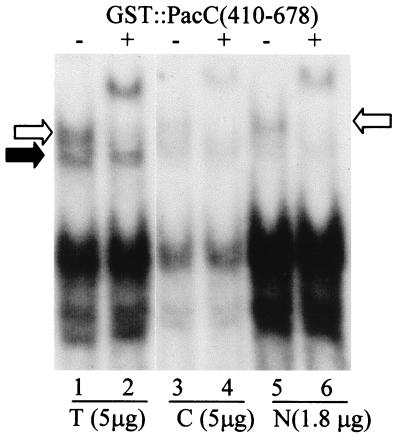
In the open conformation of full-length PacC, two regions between residues 169 and 410 are available for interactions with polypeptides containing the C-terminal residues 529 through 678 (18). This interaction can be monitored by the resulting supershift of the full-length PacC-DNA complex in EMSA (18) (Fig. 10). In the closed conformation, these regions are not available for interaction. This different behavior provides a test for distinguishing between the two conformational forms. We used longer electrophoretic runs to resolve, under alkaline pH conditions, a faster complex corresponding to closed PacC protein unavailable for interactions from a slower complex corresponding to open PacC protein, which is fully supershifted by a purified GST::PacC(410–678) polypeptide (Fig. 10, lanes 1 and 2). This assay unambiguously showed that the full-length protein present in the nuclear fraction is in the open conformation (Fig. 10, lanes 5 and 6).
FIG. 10.
Full-length PacC in the open conformation localizes in the nuclear fraction. Five micrograms of protein from a whole-cell extract (T) and a cytosolic fraction (C) obtained from wild-type cells grown under alkaline conditions and 1.8 μg of the corresponding nuclear extract were analyzed by EMSA. Two micrograms of purified GST::PacC(410–678) was included in the indicated binding mixtures. The open and solid arrows indicate the position of the open and closed full-length PacC-DNA complexes, respectively. The most prominent band corresponds to the protein-DNA complex of the predominant processed form.
DISCUSSION
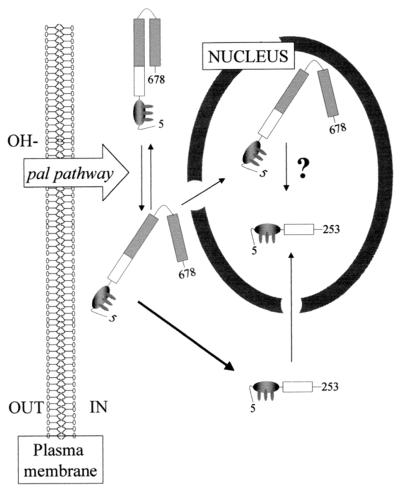
We show here that environmental pH regulates the subcellular localization of the zinc finger transcription factor PacC. In the absence of an ambient alkaline pH signal (for example, in a strain lacking a functional pal pathway), proteolytic processing is prevented and PacC is localized in the cytoplasm. Reception of the ambient pH signal disrupts interactions involving the C-terminal region of PacC and upstream regions and leads to an open (protease accessible) conformation and proteolytic processing (18). The processed PacC form is exclusively found in the nucleus (see the model in Fig. 11).
FIG. 11.
A model for PacC subcellular trafficking in response to ambient pH. Newly synthesized PacC adopts a protease-inaccessible, closed conformation, which is unable to enter into the nucleus. When the growth environment is alkaline, a signal is transmitted by the pal gene pathway. Reception of this signal by PacC switches the protein to an open conformation, in which PacC is accessible to proteolytic processing. We favor a model in which processing takes place exclusively in the cytosolic compartment (see text). The processed protein would be transported into the nucleus, where it activates expression of alkaline genes and represses that of acidic genes. A fraction of full-length PacC, most likely representing protein in the open conformation which escapes processing, is able to enter the nucleus. Because definitive evidence for the cytosolic localization of PacC processing is lacking, the possibility of nuclear processing is indicated by a question mark. However, we note that quick transfer into the nucleus of unprocessed polypeptides in the open conformation would protect them from processing if this were exclusively cytosolic, which would explain why, even with the most extreme C-terminally truncating pacCc alleles under alkaline growth conditions, a fraction of the full-length product appears unprocessed.
In the presence of pH signaling, a proportion of the full-length PacC form is also localized in the nucleus. Our data strongly suggest that the full-length PacC showing nuclear localization corresponds to protein that is the open conformation, but has not yet been processed (Fig. 11). The nuclear localization of a proportion of the full-length form might have implications for our current view of pH regulation because, contrary to our initial molecular model (47), a proportion of the full-length PacC form might be physiologically relevant, since the presence of the C terminus does not preclude DNA binding.
Several possible examples of regulated nuclear transport (26) have been described in transcription factors regulated by proteolytic processing. In all cases, removal of the negative-acting domain in the transcription factor results in nuclear localization of the product. In SREBP, processing releases the transcription factor domain from an endoplasmic reticulum membrane-anchoring domain (5, 6). In cytosolic NF-κB p105/p65 heterodimer (25) or p65/p50/IκB heterotrimer (3, 28, 30, 39, 66), the ankyrin repeat domain masks the nuclear localization signal. However, it has recently been shown that the main function of IκB is that of a nuclear export chaperone (through a nuclear export signal within the N-terminal domain), rather than a cytoplasmic tether, and that the largely cytoplasmic localization of the p65/p50/IκB heterotrimer is due to the predominance of nuclear export over nuclear import (27, 31, 57).
In the Hedgehog signaling pathway, in the absence of the morphogen, the full-length form of Ci is found in a cytoplasmic multiprotein complex (52, 55), from which the processing product is released (2). The processing product, which contains a nuclear localization signal (64), translocates into the nucleus, where it acts as a transcriptional repressor. In addition, processing removes a nuclear export signal (8). Hedgehog signal reception leads to nuclear localization of the full-length transcription factor. A shuttling mechanism, in which the opposite actions of nuclear import and nuclear export mechanisms act through the signals described above, ensures appropriate levels of the nuclear full-length form (8), which has transcriptional activation functions (40, 46). Therefore, three forms of the Ci protein (full-length nuclear, cytoplasmic, and processed nuclear) participate in Hedgehog signaling. We show here that, in marked similarity to Ci, a modified version of the full-length PacC form (corresponding to the open conformation) can get into the nucleus.
The change from the closed to the open conformation of full-length PacC results in commitment for proteolytic processing, which hinders the unambiguous characterization of the subcellular localization of the processing reaction. Two opposite models are a priori possible. In the first, open PacC translocates into the nucleus, where the processing reaction would take place. In the second, processing of open PacC would be cytosolic, and the proportion of full-length PacC translocated into the nucleus would be protected from processing. Although we do not have convincing evidence for either of these models, we favor the cytosolic processing model (Fig. 11) for the following reasons. The pacCc14 allele leads to strong alkalinity mimicry at any ambient pH. It encodes a PacC primary translation product truncated after residue 492 that lacks an interacting region crucial for maintaining the closed conformation. Therefore, even under acidic pH conditions, this protein is in the open conformation, which in turn leads to low levels of primary translation product due to processing. These low levels are hardly detectable by Western blotting, but can be detected by using our sensitive EMSA (Fig. 1A, lanes 4 to 6). This open PacC is mostly present in the nucleus and absent from the cytosol, which might be consistent with protection of this open PacC from cytosolic processing by its compartmentalization into the nucleus.
This work raises interesting questions about the mechanistic bases of the different subcellular localizations of the full-length and processed PacC forms. Our data unambiguously show that the processed PacC form contains a functional nuclear import signal that can target heterologous proteins to the nucleus and strongly indicate that this import signal is localized within the DNA binding domain. While the zinc finger region was sufficient for nuclear targeting of heterologous proteins, the subcellular distribution of all fusion proteins between GFP and PacC polypeptides lacking an intact zinc finger region was indistinguishable from that of GFP. A single-residue substitution abolishing DNA binding does not affect the nuclear localization of a processed form-GFP protein fusion, thus separating nuclear targeting and DNA binding activities. This rules out a model in which the reduction in PacC size resulting from proteolytic processing could lead to preferential nuclear localization by a mechanism involving a combination of passive diffusion and nuclear retention after DNA binding. Using a PacC(250–678)::GFP construct driving high levels of expression of this protein (Fig. 5B), we have been unable to detect the presence of a second nuclear import signal in the region removed by proteolytic processing. In agreement, a bipartite cluster of basic residues between PacC residues 252 and 269 does not act as a nuclear import signal for a PacC(241–280)::GFP fusion protein (data not shown). Therefore, the nuclear import signal mapping to the zinc finger region most likely mediates import of both the processed and the open full-length PacC forms.
While a role of nuclear import in PacC regulation seems clear, we have been unable to obtain evidence for a role of nuclear export. The fact that the subcellular localizations of a GFP::PacC(250–678) fusion protein and of GFP are indistinguishable might suggest that residues removed by processing do not contain a nuclear export signal, which would have led to preferential cytosolic labeling. This conclusion has been verified by confocal fluorescence microscopy (data not shown). A similar GFP-based methodology served to reveal the role of nuclear export in Ci regulation (2, 8). Therefore, the mechanism by which the closed, but not open, full-length PacC is tethered to the cytosol remains elusive. Because the nuclear import signal reported here and the DNA binding domain overlap, the fact that closed PacC is able to bind DNA in vitro suggests that this conformation does not involve masking of the zinc finger region and, by extension, of the nuclear import signal.
Environmental signals (glucose and ambient pH) have been shown to control the nuclear localization of yeast MIG1 (15) and Aspergillus PacC (this work), two key ascomycete wide-domain transcription factors mediating glucose repression of genes for the catabolism of alternative carbon sources and regulating expression of genes for extracellular enzymes, certain permeases, and the biosynthesis of secondary metabolites, respectively. Finally, our work with PacC is relevant for understanding the invasiveness and resistance to alkaline pH in Saccharomyces (22, 34), the adaptation of Candida albicans to changing pH environments (a key factor in its pathogenicity) (11, 12, 49, 50), the synthesis of extracellular metabolites and proteins by fungi (19, 33, 35, 36, 38, 56, 61, 62), and the regulation of aflatoxin biosynthesis (32), all of which are processes regulated by the PacC/RIM101p family of transcription factors.
ACKNOWLEDGMENTS
We thank H. N. Arst for critical reading of the manuscript, C. Scazzocchio for the gift of pAN52–1::sGFP, and E. Reoyo for technical assistance.
We also are grateful for the support of the EU through contracts BIO4-CT96–0535 and QLK3-CT-1999–0729, the CICYT through grant BIO97–348, the Basque Government through a predoctoral fellowship to E.D., and the DGICYT for a postdoctoral contract with E.A.E. and a predoctoral fellowship to J.M.M.
REFERENCES
- 1.Arst H N, Jr, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 2.Aza-Blanc P, Ramirez-Weber F A, Laget M P, Schwartz C, Kornberg T B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 3.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S J. IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 4.Blank V, Kourilsky P, Israel A. Cytoplasmic retention, DNA binding and processing of the NF-κB p50 precursor are controlled by a small region in its C-terminus. EMBO J. 1991;10:4159–4167. doi: 10.1002/j.1460-2075.1991.tb04994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 6.Brown M S, Goldstein J L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by the pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 8.Chen C H, von Kessler D P, Park W, Wang B, Ma Y, Beachy P A. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 9.Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 10.Clutterbuck A J. Aspergillus nidulans. In: O'Brien SJ, editor. Genetic maps. Locus maps of complex genomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 3.71–3.84. [Google Scholar]
- 11.Davis D, Wilson R B, Mitchell A P. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Bernardis F, Mühlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison S H, Negrete-Urtasun S, Mingot J M, Tilburn J, Mayer W A, Goel A, Espeso E A, Peñalva M A, Arst H N., Jr Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol Microbiol. 1998;30:259–264. doi: 10.1046/j.1365-2958.1998.01058.x. [DOI] [PubMed] [Google Scholar]
- 14.Denison S H, Orejas M, Arst H N., Jr Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 15.De Vit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espeso E A, Arst H N., Jr On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol Cell Biol. 2000;20:3355–3363. doi: 10.1128/mcb.20.10.3355-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espeso E A, Peñalva M A. Three binding sites for the Aspergillus nidulans PacC zinc finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J Biol Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 18.Espeso E A, Roncal T, Díez E, Rainbow L, Bignell E, Alvaro J, Suárez T, Denison S H, Tilburn J, Arst H N, Jr, Peñalva M A. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 2000;19:719–728. doi: 10.1093/emboj/19.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espeso E A, Tilburn J, Arst H N, Jr, Peñalva M A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espeso E A, Tilburn J, Sánchez-Pulido L, Brown C V, Valencia A, Arst H N, Jr, Peñalva M A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol. 1997;274:466–480. doi: 10.1006/jmbi.1997.1428. [DOI] [PubMed] [Google Scholar]
- 21.Fernández-Abalos J M, Fox H, Pitt C, Wells B, Doonan J H. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- 22.Futai E, Maeda T, Sorimachi H, Kitamoto K, Ishiura S, Suzuki K. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol Gen Genet. 1999;260:559–568. doi: 10.1007/s004380050929. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh S, May M J, Kopp E B. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 24.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henkel T, Zabel U, van Zee K, Muller J M, Fanning E, Baeuerle P A. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-κB subunit. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 26.Hood J K, Silver P A. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 27.Huang T T, Kudo N, Yoshida M, Miyamoto S. A nuclear export signal in the N-terminal regulatory domain of IκBα controls cytoplasmic localization of inactive NF-κB/IκBα complexes. Proc Natl Acad Sci USA. 2000;97:1014–1019. doi: 10.1073/pnas.97.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huxford T, Huang D B, Malek S, Ghosh G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 29.Ingham P W. Transducing Hedgehog: the story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs M D, Harrison S C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 31.Johnson C, van Antwerp D, Hope T J. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J. 1999;18:6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller N P, Nesbitt C, Sarr B, Phillips T D, Burow G B. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology. 1997;87:643–648. doi: 10.1094/PHYTO.1997.87.6.643. [DOI] [PubMed] [Google Scholar]
- 33.Lambert M, Blanchin-Roland S, Le Louedec F, Lépingle A, Gaillardin C. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol Cell Biol. 1997;17:3966–3976. doi: 10.1128/mcb.17.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W S, Mitchell A P. Proteolytic activation of rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacCabe A P, Orejas M, Pérez-González JA, Ramón D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J Bacteriol. 1998;180:1331–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacCabe A P, van den Hombergh J P T W, Tilburn J, Arst H N, Jr, Visser J. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol Gen Genet. 1996;250:367–374. doi: 10.1007/BF02174395. [DOI] [PubMed] [Google Scholar]
- 37.Maccheroni W, May G S, Martínez-Rossi N M, Rossi A. The sequence of palF, an environmental pH response gene in Aspergillus nidulans. Gene. 1997;194:163–167. doi: 10.1016/s0378-1119(97)00095-4. [DOI] [PubMed] [Google Scholar]
- 38.Madzak C, Blanchin-Roland S, Cordero-Otero R, Gaillardin C. Functional analysis of upstream regulating regions from the Yarrowia lipolytica XPR2 promoter. Microbiology. 1999;145:75–87. doi: 10.1099/13500872-145-1-75. [DOI] [PubMed] [Google Scholar]
- 39.Malek S, Huxford T, Ghosh G. IκBα functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-κB. J Biol Chem. 1998;273:25427–25435. doi: 10.1074/jbc.273.39.25427. [DOI] [PubMed] [Google Scholar]
- 40.Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 41.Mingot J-M, Tilburn J, Díez E, Bignell E, Orejas M, Widdick D A, Sarkar S, Brown C V, Caddick M X, Espeso E A, Arst H N, Jr, Peñalva M A. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol Cell Biol. 1999;19:1390–1400. doi: 10.1128/mcb.19.2.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 43.Negrete-Urtasun S, Denison S N, Arst H N., Jr Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negrete-Urtasun S, Reiter W, Díez E, Denison S H, Tilburn J, Espeso E A, Peñalva M A, Arst H N., Jr Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol Microbiol. 1999;33:994–1003. doi: 10.1046/j.1365-2958.1999.01540.x. [DOI] [PubMed] [Google Scholar]
- 45.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 46.Ohlmeyer J T, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- 47.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 48.Pokorska A, Drevet C, Scazzocchio C. The analysis of the transcriptional activator PrnA reveals a tripartite nuclear localisation sequence. J Mol Biol. 2000;298:585–596. doi: 10.1006/jmbi.2000.3666. [DOI] [PubMed] [Google Scholar]
- 49.Porta A, Ramón A M, Fonzi W A. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol. 1999;181:7516–7523. doi: 10.1128/jb.181.24.7516-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramón A M, Porta A, Fonzi W A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–7530. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice N R, MacKichan M L, Israel A. The precursor of NF-κB p50 has IκB-like functions. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- 52.Robbins D J, Nybakken K E, Kobayashi R, Sisson J C, Bishop J M, Therond P P. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 53.Ruiz i Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 54.Sakai J, Duncan E A, Rawson R B, Hua X X, Brown R S, Goldstein J L. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 55.Sisson J C, Ho K S, Suyama K, Scott M P. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 56.Suárez T, Peñalva M A. Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol Microbiol. 1996;20:529–540. doi: 10.1046/j.1365-2958.1996.5421065.x. [DOI] [PubMed] [Google Scholar]
- 57.Tam W F, Lee L H, Davis L, Sen R. Cytoplasmic sequestration of Rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol. 2000;20:2269–2284. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thanos D, Maniatis T. NF-κB: a lesson of family values. Cell. 1995;24:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 59.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tilburn J, Scazzocchio C, Taylor G G, Zabicky-Zissman J H, Lockington R A, Davies R W. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–211. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- 61.Treton B, Blanchin-Roland S, Lambert M, Lepingle A, Gaillardin C. Ambient pH signalling in ascomycetous yeasts involves homologues of the Aspergillus nidulans genes palF and paIH. Mol Gen Genet. 2000;263:505–513. doi: 10.1007/s004380051195. [DOI] [PubMed] [Google Scholar]
- 62.van den Hombergh J P T W, MacCabe A P, van de Vondervoort P J I, Visser J. Regulation of acid phosphatases in an Aspergillus niger pacC disruption strain. Mol Gen Genet. 1996;251:542–550. doi: 10.1007/BF02173643. [DOI] [PubMed] [Google Scholar]
- 63.van Heeswijck R, Hynes M J. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q T, Holmgren R A. The subcellular localization and activity of Drosophila Cubitus interruptus are regulated at multiple levels. Development. 2000;126:5097–5106. doi: 10.1242/dev.126.22.5097. [DOI] [PubMed] [Google Scholar]
- 65.Zabel U, Baeuerle P A. Purified human IκB can rapidly dissociate the complex of the NF-κB transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- 66.Zabel U, Henkel T, Silva M S, Baeuerle P A. Nuclear uptake control of NF-κB by MAD-3, an IκB protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]