Graphical abstract
Keywords: LOV domain, Allostery, AsLOV2, Hydrogen bond network, Machine learning
Abstract
The Light-Oxygen-Voltage 2 (LOV2) domain of Avena Sativa phototropin 1 (AsLOV2) protein is one of the most studied domains in the field of designing photoswitches. This is due to the several unique features in the AsLOV2, such as the monomeric structure of the protein in both light and dark states and the relatively short transition time between the two states. Despite that, not many studies focus on the effect of the secondary structures on the drastic conformational change between the light and dark states. In this study, we focus on the role of A’ helix as a key player in the transition between both states using various computational tools as: 1.5 s molecular dynamics simulations for each configuration, Markov state model, different machine learning techniques, and community analysis. The impact of the A’ helix was studied on the atomistic level by introducing two groups of mutations, helicity enhancing mutations (T406A and T407A) and helicity disrupting mutations (L408D and R410P), as well as on the overall secondary structure by using the community analysis. Maintaining the N-terminal hydrogen bond network was found to be essential for the transition between the two states. Via in-depth hydrogen bonding and contact analysis we were able to identify key residues (Thr407 and Arg410) involved in the functional conformational switch and their impact on the overall protein dynamics. Moreover, the community analysis highlighted the significant role of the sheets in the overall protein allosteric process.
1. Introduction
The Per-Arnt-Sim (PAS) superfamily is widely present in animals, plants, and prokaryotes as the sensors of developmental signals to facilitate the response and the adaptation to different environmental stimulus [1].
The Light-Oxygen-Voltage 2 domain of Avena Sativa phototropin 1 (AsLOV2) exhibits monomeric structure in both light and dark states [2], [3]. This structural simplicity, especially when compared to other oligomeric PAS-containing proteins, makes AsLOV2 the model protein for studying the PAS superfamily [4], [5], [6], [7].
AsLOV2 undergoes an allosteric activation upon exposure to blue light. This activation is initiated by the formation of a covalent bond between the flavin mononucleotide (FMN) ring involving a carbon atom, C(4a), and cysteine C450, resulting in the formation of cysteinyl-flavin adduct [8]. This covalent bond triggers a drastic conformational change in the overall protein structure. On the other hand, the regeneration of the dark state correlates with the breaking of the metastable photo-induced covalent bond, which is strongly influenced by the local environment [9]. The transitions from the dark state to the light state are rather fast with response time around 10 s. The breaking of the photo-induced covalent bond is spontaneous, but the transitions from the light state to the dark state are much slower with half-lives ranging from seconds to minutes [10]. Due to the possibility to reliably induce specific conformational changes in AsLOV2 using light, AsLOV2 is one of the most studied and utilized proteins in the field of designing optogenetic switches [11], [12], [13], [14], [15].
Specifically, past studies have identified the most characteristic conformational changes in AsLOV2 to be localized in the termini. In fact, it has been experimentally and computationally observed that the J helix (located at the C-terminal of the protein) undocks from the LOV core of the protein upon light absorption by the flavin [16], [4], [7], [2]. The light-induced flexibility of the AsLOV2 C-terminal plays a main role in the correlation between light-state and protein functional conformational changes [2].
In addition to the carboxyl terminal, the amino terminal (A’ helix) was found to be another key player for this conformational change. Unfolding of the A’ helix is followed by the undocking and the unfolding of the J helix which is 15 Å apart from the photo-induced covalent bond [16]. A’ helix conformational change is independent in the presence of the J helix, as the removal of the J helix does not affect the response of the A’ helix to light [16], [17]. On the other hand, the J helix did not undergo any conformational change when A’ helix was removed [16], [17]. The signaling propagation from the A’ helix to the J’ helix takes place through the -sheets separating the PAS core from the J helix [18], [19], [20], [21]. Therefore, it is important to study the role of the secondary structures found in the AsLOV2 that might assist the functional conformational changes of J helix.
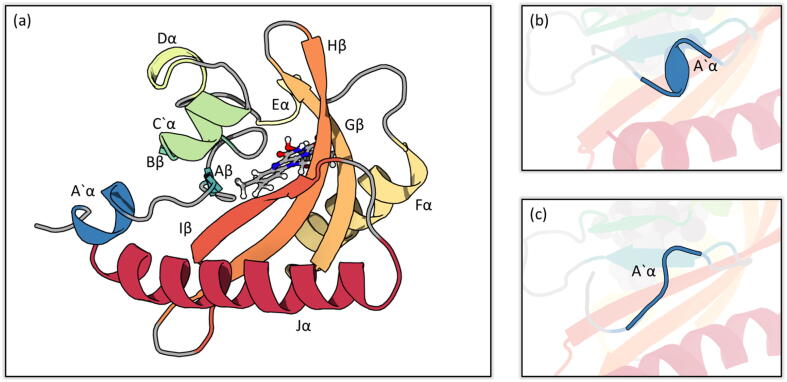
An extensive mutational analysis of AsLOV2 protein, including 100 mutations, was carried out by Zayner et al. to study the relation between the sequence of the protein and its function [4]. The helicity of the A’ helix affected the overall response of the AsLOV2 to light (Fig. 1). Four mutations were found to affect the helicity of the A’ helix as well as its function in different ways. Mutations T406A and T407A stabilized the helical structure of the A’ helix by substituting the two solvent-exposed residues with alanine. R410P destabilized the helical structure by the introduced proline residue. L408D also destabilized the helical structure of A’ helix, resulting in its unfolding, due to the burial of charged acidic residue that substituted the inward-facing core residue [16], [4].
Fig. 1.
Structural representation of AsLOV2. a) Overall AsLOV2 structure. The secondary structures are colored progressively from blue to red starting the N-terminal A’ helix. The chromophore is illustrated in ball and sticks; b) Folded state of A’ helix in the dark state; c) Unfolded state of A’ helix in the light state. Renderings generated using 3D Protein Imaging [47].
In addition to the changes in helicity associated with the mutations, the photocycle time was found to be affected in three of these four mutations. T406A and T407A mutations resulted in faster photo cycle. R410P mutation resulted in slower photo cycle. The changes in the helicity and the photocycle time associated with the mutations were attributed to the changes in hydrogen bonding patterns [4]. However, the mechanistic reason behind this remains unclear [4]. Therefore, these mutants represent valuable systems of investigation to gain insights into the mechanics of the allosteric conformational switch of this protein.
Molecular dynamics (MD) simulations represent the primary tool to investigate the conformational changes and the propagation of the allosteric signal through the allosteric network within the protein. In recent years, various studies in which MD simulations data were analyzed using advanced computational techniques, such as Machine Learning and Markov State Modeling, successfully described allosteric mechanisms of other PAS-containing proteins at atomistic level [22], [23].
Studying the functional role of the A’ helix at the atomistic level is essential to delineate the allosteric mechanism of AsLOV2. In this study, we addressed this problem using an ensemble of computational tools.
MD simulations were employed to investigate the allosteric mechanisms of AsLOV2 native states and mutants. The effect on the overall protein conformation and dynamics was investigated with the focus on the overall helicity of the A’ helix. The native states include the both light and dark native states obtained from the crystal structures. Because the crystal structures are mostly limited to proteins in their equilibrium states, little information could be obtained related to the transitions between dark and light states of AsLOV2. Taking advantages of computational studies, proteins in certain functionally important but unstable states could be simulated to gain insight into transition processes related to protein functions.
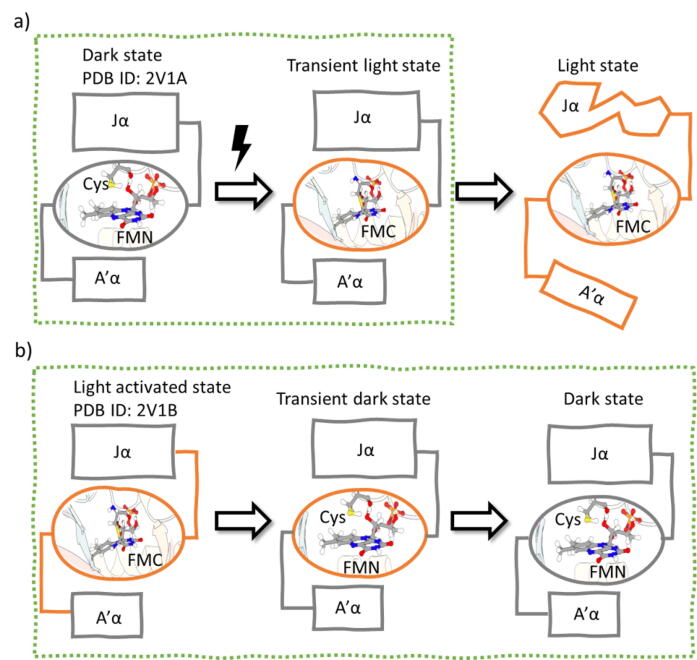
Following previous studies using this strategy [24], [25], two transient states were constructed and subjected to simulations as the following to explore AsLOV2 conformational space and dynamical properties coupled with the transitions between its dark and light states. To mimic AsLOV2 in the moment when its dark state is excited by the blue light with photo-induced covalent bond and before conformational transition to its light state structure, the transient light state could be constructed by forming the photo-induced covalent bond in the AsLOV2 dark state structure (Fig. 2a). Similarly, to mimic AsLOV2 in the moment when its light state loses photo-induced covalent bond and before conformational transition to its dark state structure, the transient dark state could be constructed by breaking the photo-induced covalent bond in the AsLOV2 light state structure (Fig. 2b). In addition to the wild type, transient light and transient dark states were also constructed for four AsLOV2 mutants (T406A, T407A, L408D and R410P) and subjected to simulations in this study.
Fig. 2.
Schematic representation of signal propagation upon light absorption in AsLOV2. a) Formation of transient light state from dark state crystal structure. In the dark state, the A’ helix is docked to the LOV core and the J helix is fully folded. Upon light absorption, FMN forms a covalent bond with the proximal Cys450. At this stage, the folding of the protein remains intact, but local hydrogen bond interactions in the vicinity of FMN (A’ and J helices) change. This is called transient state. With more time, the light perturbation causes the undocking of A’ helix and the unfolding of J helix. The latter is the light state. b) Formation of transient dark state from light activated crystal structure. To build the transient dark state, the covalent bond between the flavin and the proximal cystein is broken. The allosteric responses from protein are highlighted in orange boxes. The states subjected to the investigation in the present study are framed in the green rectangles.
A structural investigation via hydrogen-bond analysis of the A’ helix and contact map between different secondary structures illustrated the critical hydrogen bonds responsible for the structural integrity and the docking of the A’ helix. This information, together with an analysis of the conformational space, protein kinetics, and residues’ importance enables us to comprehensively delineate the role of A’ to the allosteric photo-switch of AsLOV2.
2. Materials and methods
2.1. Molecular dynamics simulations
The crystal structures from Protein Data Bank (PDB) [26] of both the native light state (PDB ID: 2V1B) and the native dark state (PDB ID: 2V1A) of AsLOV2 were obtained. The covalent bond between C(4a) of Flavin Mononucleotide (FMN) ring and cysteine C450 of AsLOV2 is formed in the light state but absent in the dark state.
Because J helix remains folded, the crystal structure (2V1B) of the light state of AsLOV2 lacks the associated conformational changes in J helix upon the formation of the photo-induced covalent bond. As the focus of this study is the mechanism of the signal propagation from the photo-induced covalent bond through A’ helix [16], the observation of fully unfolded J helix in the simulation is not mandated.
In order to explore more conformational space and to investigate the impact on the protein with regard to the covalent bond, two new protein configurations were constructed. The transient light state (ts-light state) was created by forming the photo-activated covalent bond between C(4a) of FMN ring and cysteine C450 of AsLOV2 in the dark state conformation. The transient dark state (ts-dark state) was created by removing this photo-activated covalent bond in the light state conformation.
The force field parameters of the molecular dynamics simulations for the FMN were obtained from a previous work [27]. Four mutations, including T406A, T407A, L408D and R410P (numbering of the residues started by residue 403 not residue 1), were introduced into the transient states [16], [4]. Thus, total of 12 simulation systems were constructed.
The simulation systems were prepared using CHARMM package version 41b1 [28] along with OpenMM to run the molecular dynamics simulation on GPU [29]. The preparation of the system started with adding hydrogen atoms followed by solvating the system using TIP3P water model [30]. Then, the constructed system was neutralizated by adding sodium cations and chloride anions. The subsequent step was the minimization of the overall structure using the steepest-descent method. The entire system was heated to raise the temperature from 0 K to 300 K. Consequently, three independent 10 ns of isothermal-isobaric ensemble (NPT) simulations were applied to the system followed by 0.5 s of canonical ensemble simulation (NVT) for each simulation at 300 K. There are total of 1.5 s simulation for each state and 18 s simulations for all the states. SHAKE constraint for hydrogen covalent bond was employed. Particle mesh Ewald (PME) method was used to account for the electrostatic interactions [31].
MDTraj [32] was used to calculate root-mean-square-deviation (RMSD) and root-mean-square-fluctuations (RMSF). RMSD indicates the conformational change of the protein during the MD simulations with regard to the first frame of each trajectory. RMSF indicates the fluctuations of the atoms during the MD simulations with regard to an averaged structure. RMSD analysis was performed to assess the stability of the simulations.
| (1) |
where N is the number of atoms, is the position of atom is the coordinate of atom i in the reference structure, and U is the best-fit rotational matrix to align a given structure onto the reference structure. The RMSF was calculated as
| (2) |
where T is the number of frames.
2.2. Time-structure independent component analysis (tICA)
tICA method implemented in MSMBuilder [33] was used as the dimensionality reduction method for the generated molecular dynamics trajectories. tICA focuses on finding the slowest-relaxing degree of freedom. The tICA is performed by solving a generalized eigenvalue problem as
| (3) |
C is the covariance matrix,
| (4) |
is the time-lagged covariance matrix with a certain lag time ,
| (5) |
where denote the time average [34], [35]. K and F are eigenvalue and eigenvector matrices, respectively.
Featurization step based on the C distances was employed to transform the Cartesian coordinates into the corresponding biophysical features. This implies that the C distances were used to construct appropriate collective variables to describe the microstates followed by tICA [36].
2.3. Markov state model analysis
Markov state model analysis was carried out using MSMBuilder [33]. Markov state models are a framework to analyze dynamical systems. Its application consists of dividing the molecular conformational space into individual states and building a transition probability matrix for these states. The transition probability matrix is calculated by counting the number of transitions between different pair of states summed over the entirety of the simulations. From the MSM transition probability matrix, Perron-cluster cluster analysis [37] was used to cluster the microstates (groups of conformational states used to build the MSM) into macrostates. This is achieved by taking into consideration that the transitions among microstates belonging to the same macrostate should be faster than the transitions among macrostates [38], [39], [40].
2.4. Machine learning model and community analysis
One-vs-One (OvO) random forest method implemented in scikit-learn python package [41] was employed to build classification models based on macrostates in Markov State Model. Random forests are ensemble learning methods for classification and other tasks that operate by constructing a multitude of decision trees. Random forest methods overcome the over fitting problem often suffered by decision tree models. OvO random forest is based on multi-classification, where different classifiers are used to differentiate different pairs of target data set.
The OvO random forest classification model was subjected to machine learning based community analysis, which was developed to divide residues into different groups by minimizing the total of the feature importance for pair-wised C distances within each group while maximizing the total of the feature importance for pair-wised C distances across different groups [23]. The feature weights calculated using OvO random forest were used to distinguish between the distribution of the different states where lower feature weight indicates lower distinguishability [23]. Kernighan-Lin algorithm [42] was used to search for the local minimum value.
2.5. Distance distribution, hydrogen bonding and definition of the secondary structure of protein analysis
Distance distribution analysis was applied using MDTraj [32] where the distances between the desired pair of atoms are calculated in each frame of the MD simulations. Moreover, MDTraj was used to calculate the hydrogen bonding based on Baker-Hubbard method [43]. Baker-Hubbard methods is used to identify hydrogen bonds based on cutoffs in distance (2.5 Å) and angle (120°) between the hydrogen bond acceptors (N and O) and hydrogen bond donor [43]. In addition, identification of the secondary structure of the protein based on the definition of Define Secondary Structure of Proteins (DSSP) was carried out using MDTraj [32], where a secondary structure is assigned to each residue in each frame of the MD simulations.
3. Results
3.1. Molecular dynamics simulations
As the mutations introduced to the A’ helix showed the same trend with regard to the RMSD values (Figure S1a, S1b), the atomic fluctuations were assessed using RMSF to gain a better insight into the impact of the mutations at the atomistic level. Similar to the RMSD calculations, RMSF was calculated over the time course of 500 ns, where the first frame of the simulation of each configuration was the reference structure.
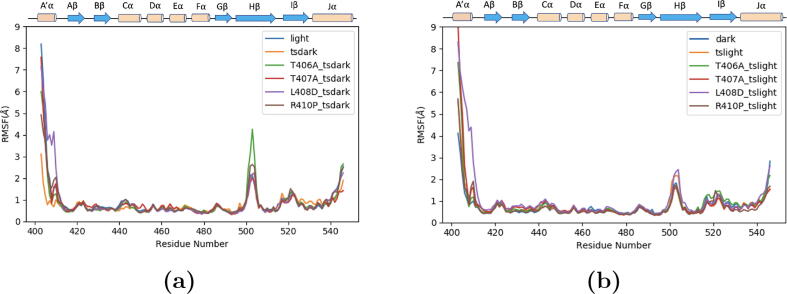
The N-terminal, the C-terminal, the H strand (residues 487 to 507) and the I strand (residues 509 to 517) displayed significant conformational changes as shown by the high fluctuations. The trajectories of the light state configurations (maximum RMSF value of 4 Å) (Fig. 3a) displayed higher atomic mobility in the H strand region than in the dark state configurations (maximum RMSF value of 2 Å) (Fig. 3b). This agrees with the experimental data using temperature-dependent Fourier transform infrared (FTIR) technique [16]. This implies that the H-strand undergoes a more drastic conformational change in the light state than in the dark state. This is due to the essential role of the -sheets in the allosteric mechanism of AsLOV2, where the undocking of the J helix is preceded by a conformational change signal transmitted from the A’ helix via the -sheets [16]. Although only the undocking of the C-terminal J helix was observed in the MD simulations but not its unfolding, this observation indicates that the structural rearrangements of the H are likely required for the conversion of the dark state into the light state.
Fig. 3.
RMSF of the simulation for each configuration. Only one of the three independent simulations for each configuration is shown for clarity. a) RMSF for the wild type light state, transient dark states of both wild type and four mutants; b) RMSF for the wild type dark state, transient light states of both wild type and four mutants.
In addition, the configurations of the helicity-enhancing mutations, especially the mutant T406A in the transient light state configuration, were associated with higher fluctuations (Fig. 3b) than the configurations of the helicity-disrupting mutations. The higher fluctuations in the H strand in the configurations of the helicity-enhancing mutations could be explained with the faster photocycle accompanied with these mutations. The faster photocycle indicates that the allosteric signal could be propagated faster and more effectively through the -sheets indicated by the higher fluctuations [16], [4].
As the allosteric mechanism of AsLOV2 depends on the conformational change of the entire structure; the sum of the RMSF values was calculated for each state. The dark state configuration was found to be the least flexible compared to the other states, while the light state configuration was found to be the most flexible (Figure S2). This agrees with the fact that the allosteric activation of AsLOV2 upon exposure to blue light would ultimately result in undocking and unfolding of both termini. A more flexible light state compared to the dark state suggests that we are capturing the initial stage of the undocking process indicated by the enhanced dynamics of the termini [16], [4], [20]. This is also consistent with the higher RMSD values of the light state configurations compared to that of the dark states (Figure S1a,S1b). The difference of the RMSF of each residue between each structure and the dark state as the least flexible state was calculated (Figure S3). All states show high fluctuations in the A’ helix, except for the tsdark state (Figure S3d). The structures of R410P mutation in both transient states have less fluctuations in A’ helix compared to the other structures. However, the structures of L408D mutation in both transient states showed similar fluctuations to the helicity-enhancing mutations (T406A and T407A) and to the light state. The difference in the fluctuation between the two helicity-disrupting mutations (L408D and R410P) could be one of the reasons behind the difference in the length of the photocycle associated with both mutants [16], [4].
3.2. Structural insights into the role of A’ helix in the AsLOV2 allostery
In this section we investigate the structural changes correlated with different light states and mutations. Because unfolding of A’ helix is a key change of protein structure upon light excitation, we applied definition of secondary structure of protein analysis (DSSP analysis [44]) to determine the secondary structural changes caused by the mutations.
3.2.1. The effect of mutations on the helicity of the A’ helix investigated using Definition of Secondary Structure of Protein (DSSP)
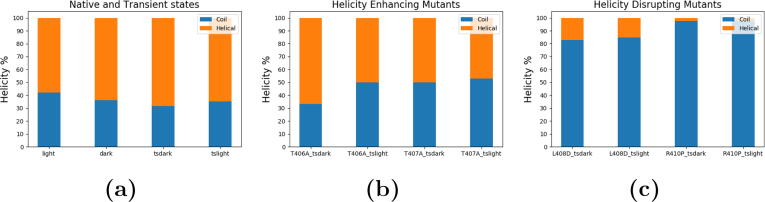
The structure of A’ helix in both the light and transient dark state configurations was found to exhibit less helical structure compared to the dark and transient light state (Fig. 4a). This agrees with the allosteric mechanism suggested by Zayner et al. [16] that the A’ helix undergoes undocking and unfolding in the light state. T406A and T407A mutations maintained the helical structure of the A’ helix in both transient light and transient dark states configurations (Fig. 4b). On the other hand, A’ helix loses its helicity in both transient light and dark state configurations of L408D and R410P mutations (Fig. 4c). This agrees with the far-UV circular dichroism (CD) amplitude change observed for the light state of these mutants [16], [4]: the mutations T406A and T407A enhanced the helicity of the termini; the L408D and R410P mutations resulted in the decrease of the helicity of the termini. [16], [4].
Fig. 4.
Helicity of the A’ helix in AsLOV2 over the course of simulations of all the configurations. The secondary structure assignment is based on Define Secondary Structure of Proteins (DSSP) model [44]. a) Native and transient states; b) Helicity-enhancing mutants; c) Helicity-disrupting mutants.
3.2.2. Changes in the patterns of the hydrogen bonds. The reason behind the changes in the helicity of the A’ helix
As hydrogen bonds are the main intramolecular force to form the helical structure, we focus on the hydrogen bonds of the A’ helix as the next step of our structural investigation. Baker-Hubbard hydrogen bonding analysis [16], [43] was employed to investigate the difference in the hydrogen bonding related to the change of the helicity of the A’ helix. Networks of hydrogen bonds associated with the residues of the A’ helix to maintain the helical structure were identified and listed in Table S2.
The analysis showed that the hydrogen bonding network differs significantly between the native dark and the native light states.
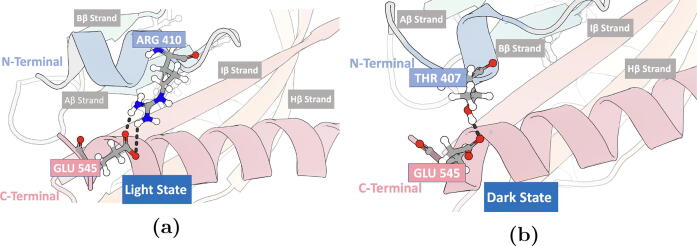
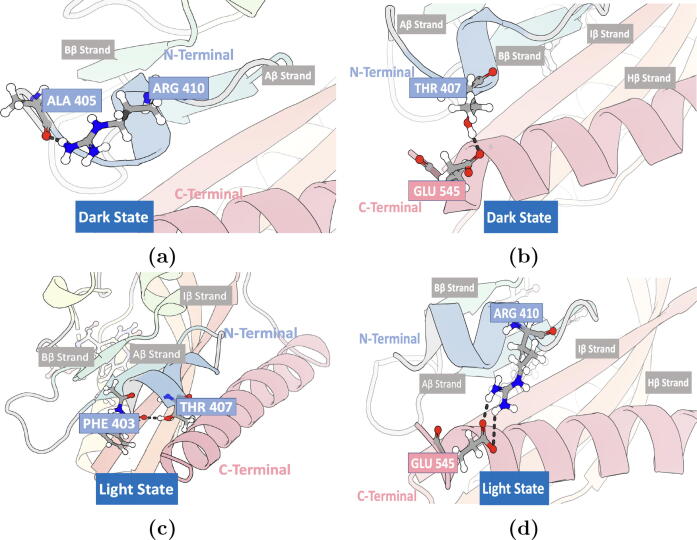
In the native dark state, residue Thr407 in the A’ helix forms hydrogen bonds with residue Glu545 in the J helix (Fig. 5). Residues Ala405 and Arg410 form a local hydrogen bond within the A’ helix (Figure S5a). In the simulation of the native light state, the conformational change leads to the formation of hydrogen bonds between residues Arg410 and Glu545 (Fig. 5) and between residues Thr407 and Ala542 (Figure S5b).
Fig. 5.
N-terminal (A’ helix) and C-terminal (J helix) hydrogen bonds. Both Arg410 (A’ helix) in the light state and Thr407 (A’ helix) in the dark state form a hydrogen bond with Glu545 (residue in the J helix). a) Hydrogen bond between Arg410 and Glu545 in the light state; b) Hydrogen bond between Thr407 and Glu545 in the dark state configuration. Renderings generated using 3D protein imaging [47].
In the simulations, both transient dark (tsdark) and transient light (tslight) states display hydrogen bond networks somewhat different from both the native light and the native dark states (Table S2). The tsdark state has hydrogen bonds between residues Arg410 and Glu545 and between residues Thr407 and Ala542 (Figures S5c), which are both seen in the native light state. In the tslight state (Figure S5d), residue Arg410 forms a hydrogen bond with Glu545, similar to the native light state (Fig. 5). The tslight state has a hydrogen bond between residues Thr407 and Phe403, also seen in the native light state. These comparisons demonstrate that the tsdark and tslight states do provide information about the actual transition processes between native dark and light states.
In the transient dark state of the T406A mutant (T406A tsdark), hydrogen bonds between A’ and J helices are lost. But the local hydrogen bonds within A’ remain (Table S2 and Figure S5e). In the transient light state of the T406A mutant (T406A tslight), a hydrogen bond forms between Thr407 and Glu545 (Figure S5f), which is also seen in the native dark state (Figure S5a). The transient dark and light states of the T407A mutant (T407A tsdark and T407A tslight) display similar hydrogen bond patterns to each other, with formed hydrogen bond between residue Arg410 in A’ helix and Glu545 in J helix (Figure S5g and S5h). These similarity or lack of changes between tsdark and tslight states are in the agreement with the fact that the T406A and T407A are helicity enhancing mutations.
On the contrary, both transient dark and transient light states of L408D mutant (L408D tsdark and L408D tslight) lost significant portion of the hydrogen bond network found in the native structures, leading to the loss of the A’ helicity (Table S2 and Figures S6a and S6b). The mutant R410P display the most significant loss of hydrogen bond network associated with the A’ and J helices and consequently the complete loss of A’ helicity (Table S2 and Figures S6c and S6d).
3.2.3. Cross correlation analysis to study secondary structures
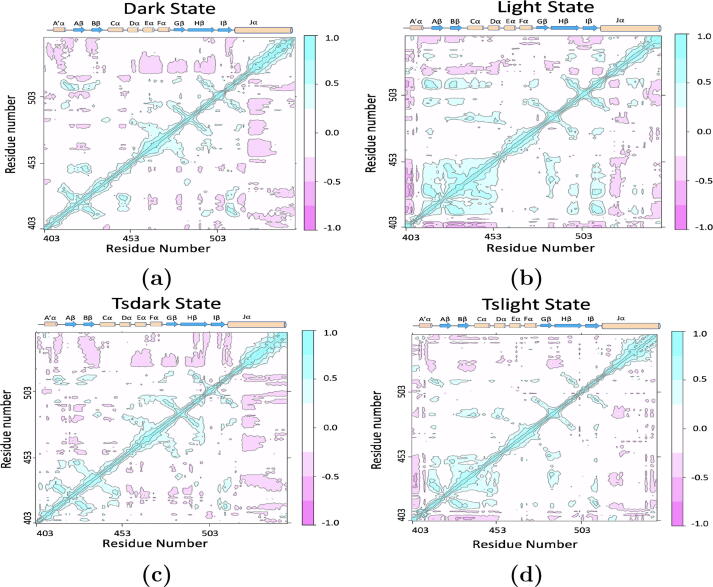
To further investigate the role played by the folded J helix in the light states of AsLOV2, cross-correlation analyses for the residues were carried out based on the simulations in this study (Fig. 6). A’ and J helices show strong negative correlation to each other, but less correlation to other parts of the protein in the dark state (Fig. 6a). On the contrary, A’ and J helices show strong negative correlation to other parts of the protein, including A, B, H strands and C helix, but much less correlation between them in the light state (Fig. 6b).
Fig. 6.
Cross-correlation among the residues of AsLOV2 using Bio3D package [48]. a) Dark state; b) Light state; c) tsdark state; d.) tslight state.
Interestingly, the J helix also displays significant negative correlation with the bulk of the protein but only weak negative correlation with the A’ helix in the transient dark (tsdark) state (Fig. 6c). In the same state, A’ helix does not show correlation with the bulk of the protein. In the transient light (tslight) state, A’ helix displays strong negative correlation with the bulk of the protein, especially A, B strands and C helix (Fig. 6d). On the other hand, the J helix only shows slight negative correlation with the bulk of the protein, including the A’ helix.
For the helicity enhancing mutants T406A and T407A, their tsdark states display much less correlations among secondary structures (Figures S7a and S7c), and their tslight states display similar or even enhanced (in T407A) correlation among secondary structures (Figures S7b and S7d). The helicity disrupting mutant L408D tsdark state does also show negative correlation, although relatively weak, between A’ helix and A, B strands as well as C, J helices (Figures S7e). L408D mutant tslight state shows enhanced correlation among key secondary structures, including A and B strands, and A’, C, and J helices (Figures S7f). Opposite to L408D mutant, the other helicity disrupting mutant R410P displays wider range, although reduced correlations among key secondary structures in its tsdark state (Figures S7g), and much reduced correlations within protein structure in its tslight state (Figures S7h).
3.3. Time-structure independent component analysis and Markov state model analysis
3.3.1. Time-structure independent component analysis (tICA)
tICA for all the trajectories was carried out using the pairwise alpha carbon (C) distances of the backbone to investigate the conformational space explored by the different configurations. tICA was carried out in two different ways to elicit the effect of the helicity on the overall protein allostery. The first tICA model includes the trajectories of only the native states and their transient structures and is referred to as native states model. The second tICA model includes all the trajectories of the native states and their transient states, and the transient states of all mutants, and is referred to as all states model. Because the dark and light states of mutants display similar behavior to the dark and light native states, they are not included in the all state model to simplify the analysis.
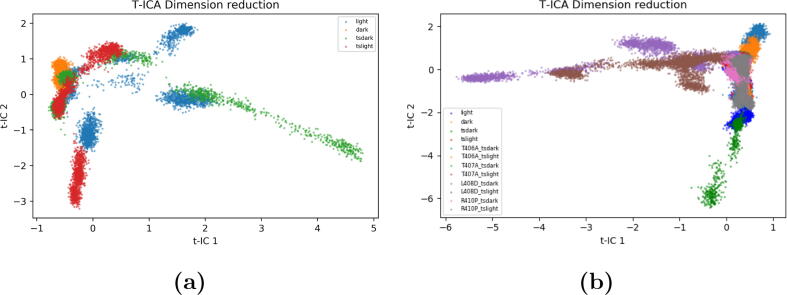
The distribution of the simulation of each state is projected onto a two-dimensional surface defined by the first two components generated from the native state tICA model (Fig. 7a). The native light state explored more conformational space than the native dark state (Fig. 7a). This reflects that the light state configuration is more flexible and likely less stable than the native dark state configuration, which was also supported by the higher RMSD values of the light state. The transient configurations explore more conformational space than the native configurations (Fig. 7a). Because the full interconversion between the dark and light states was not observed in the simulations of either the transient dark state or the transient light state, the simulations of these two transient states do not share similar conformational status as they sampled different conformational space and provide insight into different stages of AsLOV2 allosteric mechanism.
Fig. 7.
tICA analysis based on the C distances (each color represents a total of 1.5 s obtained from the three independent simulations of each configuration). a) tICA analysis of the trajectories of the native light, the native dark, the transient light and the transient dark states; b) tICA analysis of all the trajectories including the native states, transient states, and mutants.
In the all states tICA model, the distributions of both transient dark and transient light states deviate from the distributions of native dark and native light states (Fig. 7b). The distributions of transient dark and transient light states are also significantly different from each other, indicating wide conformational spaces accessible by AsLOV2 during the transition processes between its dark and light states.
3.3.2. Markov state model analysis
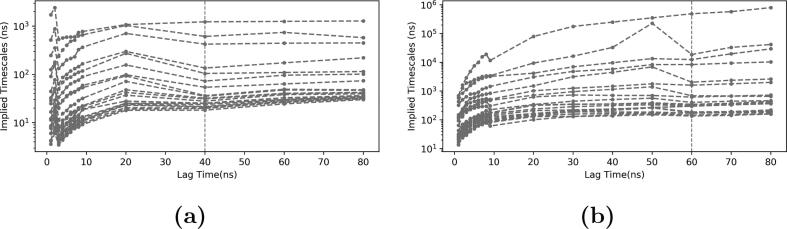
After applying the tICA, 200 microstates were generated using k-means clustering analysis for both tICA models. In each model, the microstates were used to build Markov state model (MSM). The transition probabilities were calculated among the microstates using different lag times. The appropriate lag time was selected based on the convergence of the estimated relaxation timescale. As seen in Fig. 8, the plots are converging with lag time longer than 40 ns, which was chosen to build MSM for the native states model (Fig. 8a). Similarly, a timescale of 60 ns was chosen for the all states model (Fig. 8b).
Fig. 8.
Implied time scale based on the 200 microstates generated using k-means clustering analysis of all the trajectories. The order of lines in each plot from top indicates the number of macrostates in the Markov state model represented by each line. a) The native state model; b) The all state model.
A total of 7 macrostates were identified in the native states model (Fig. 8a) macrostates were identified in the all states model. The helicity-enhancing mutations and helicity-disrupting mutations resulted in additional three macrostates compared to the native states and their transient states (Fig. 8b).
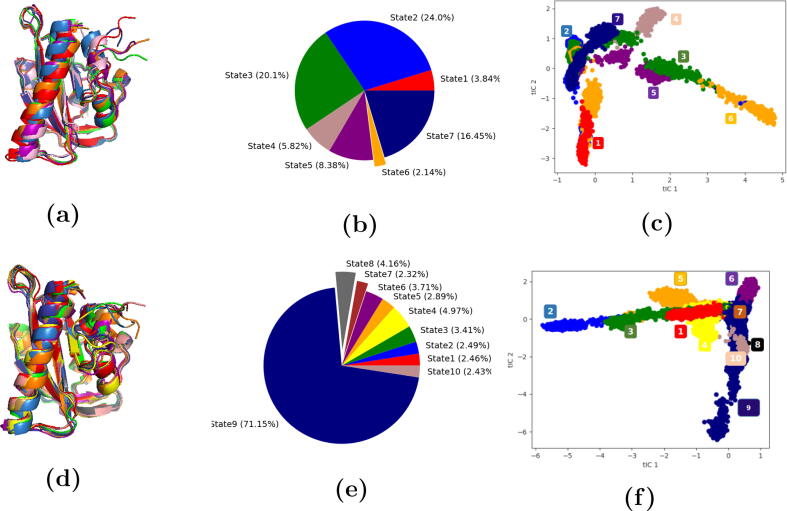
It should be noted that each macrostate does not correspond to specific configurations for the simulation, such as native dark and light states or transient states. The macrostates represent the underlying distribution of the simulations used for the MSM. All the simulations used for the MSM have various contributions to each macrostate as demonstrated in Fig. 10. Macrostates were not chosen from the microstates. To build MSM, microstates, among which the system could transition from one state to another within the chosen lag time, are considered belonging to the same macrostate.
Fig. 10.
Representative structures of the macrostates, Pie chart for the population of each macrostate, distribution of the macrostates. Each macrostate is represented by the same color in all three figures. a) Representative structures for the 7 macrostates from the native states model; b) Pie chart distribution of the 7 macrostates from the native states model; c) Distribution of the 7 macrostates from the native states model; d) Representative structures for the 10 macrostates from the all states model; e) Pie chart representing the 10 macrostates from the all states model; f) Distribution of the 10 macrostates from the all states model.
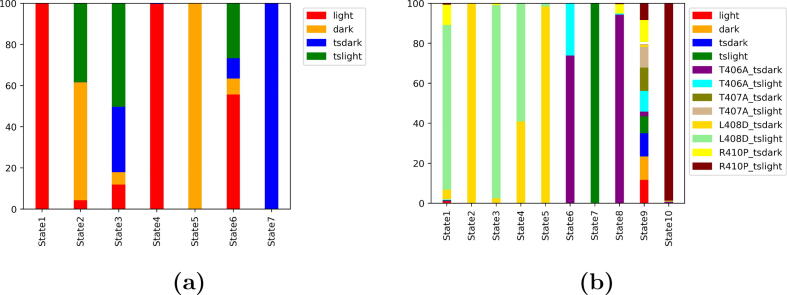
Stacked bar chart distribution, illustrating the configurations composing each macrostate, was plotted to gain better insight into the impact of the mutations. Light state and transient dark state were found to occupy most of the macrostates (State 1, 4 and 7) in the native states model (Fig. 9a). On the other hand, State 5 was composed of the dark state configuration. States 2, 3 and 6 were found to be a combination of the native and transient states configurations.
Fig. 9.
Stacked bar chart distribution of the generated macrostates from MSM analysis. a) The 7 macrostates produced from the native states model; b) The 10 macrostates produced from the all states model.
When the trajectories of all the states were included in the MSM analysis, the configurations of the mutant L408D in both transient states were found to be occupying most of the macrostates (states 1, 2, 3, 4 and 5) (Fig. 9b). This reflects that replacing Leu with Asp resulted in drastic conformational changes in the structure of the AsLOV2. This could be due to the significant change in the N-terminal hydrogen bond network observed in the both states of the L408D mutant (Table S2). This suggests that the mutant adopts a different allosteric route. State 6 and State 8 were mostly occupied by the configurations of T406A mutant in the transient dark state. State 7 was made entirely of the transient light state configuration. State 9 was found to be a combination of the different configurations. State 10 was formed mainly of the mutant R410P in the transient light state (Fig. 9b).
Investigating the states population and their conformational space could provide insights into the free-energy landscapes that determines the allosteric profile of AsLOV2. For the MSM of native state model, states 2, 3 and 6 (combination of the different configurations) were the most abundant states with 57.3% (Fig. 10b). States 1, 4 and 7 (populated by structures containing the photo-excited flavin) were found to be the second most abundant with 32.4%. This implies that the light and transient dark state composing these states were more abundant configurations compared to the dark and transient light state configurations (Fig. 9a). Also, these states occupy most of the conformational space (Fig. 10c). This agrees with the results of the tICA analysis where the light state is more flexible than the dark state and explores more conformational space (Fig. 7a). These results indicate that the dark state is characterized by high free-energy barrier, and is proposed to be linked to the hydrogen bond network of the A’ helix, responsible for keeping the folding of the C-terminal and the anchoring of J helix. The photoactivation of the flavin is thus the trigger necessary for the interconversion from the states 1, 4, and 7 to metastable state 5, accompanied by the enhance in AsLOV2 structural dynamics, and loss of terminal folding.
When all the trajectories were included in the MSM analysis, state 9 composed of all the configurations (Fig. 9b) is the most abundant with 71.2% (Fig. 10e). Furthermore, this state explores most of the conformational space, except for that occupied by states 1, 2, 3, 4 and 5 (Fig. 10f). Based on the states population analysis (Fig. 10), the latter states were exclusively populated by L408D mutant, while state 9 was populated by the remaining structures. This supports the hypothesis that L408D mutant carries an alternative allosteric mechanism, in which losing the helical structure of the A’ helix was likely due to changing the hydrogen bond network in the termini, especially the N-terminal.
States 6 and 8 were composed mainly of the T406A mutant in the transient dark state (Fig. 9b). The conformational space explored by these two states significantly overlaps with state 9, which suggested a similarity in the allosteric mechanism (transition from light to dark states) (Fig. 10f). This implies that enhancing the helicity of the A’ helix associated with this mutation (Fig. 4b) resulted in an alternative N-terminal hydrogen bond network. In addition, hydrogen bonds with Glu545 located in the J helix are maintained as key interactions in the native and transient light states (Table S2). State 10 was mainly composed of the configuration of R410P mutant in the transient light state (Fig. 9b). This mutation disrupts the helicity of the A’ helix (Fig. 4c), results in an alternative N-terminal hydrogen bond network (Table S2), and explores different conformational space (Fig. 10f).
3.4. Machine learning model and community analysis
3.4.1. One-vs-one (OvO) random forest
OvO random forest method was employed to build the classification models against macrostates from MSM. The pairwise distances between the C of amino acids are the features used to build the machine learning models. With 144 residues, there are a total of 10,296 pairwise distances. During the MD simulations, frames were saved every 0.5 ns. 1.5s simulation leads to 3000 frames being saved. Therefore, for the overall 18 s simulations of all sample configurations, 36,000 frames were saved and used to construct the machine learning models. Each frame is labelled by the macrostate it belongs to.
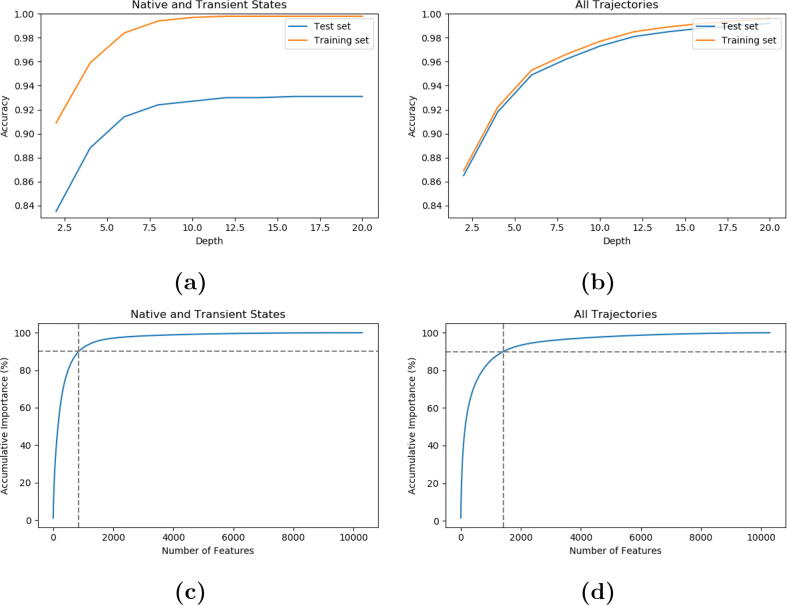
The accuracy of the best OvO random forest model (depth = 12) when the trajectories of the native and transient states were involved in building the model was associated with accuracy of 99.9% in the training set and 93.0% in the test set (Fig. 11a). The accuracy of OvO random forest model when all the trajectories were included (depth = 14) was 98.9% in the training set, and 98.5% in the test set (Fig. 11b).
Fig. 11.
One-vs-One (OvO) Random Forest (RF) models using the 10,296 pairwise C distances. a) Classification accuracy of OvO RF models with different depths for the native states model. OvO RF with depth equals 12 was selected for the natives states model; b) Classification accuracy of OvO RF models using different depths for the all states model. OvO RF with depth 14 was selected for the all states model; c) Cumulative importance for the native states model; d) Cumulative importance for the all states model.
OvO random forest model serves as an effective tool to account for the structural differences. For the native states model, 853 features out of 10,296 pairwise C distances account for the 90% distinguishablity (Fig. 11c). For the all states model, 1,419 features out of 10,296 pairwise C distances account for 90% distinguishablity (represented by the gray horizontal line) (Fig. 11d).
3.4.2. Community analysis investigating the importance of entire secondary structures
Machine learning based community analysis was carried out based on the OvO random forest classification models. In community analysis, the residues are divided into different groups by maximizing the feature importance of the pair-wised C distances across different communities while minimizing the feature importance of the pair-wised C within each group [23].
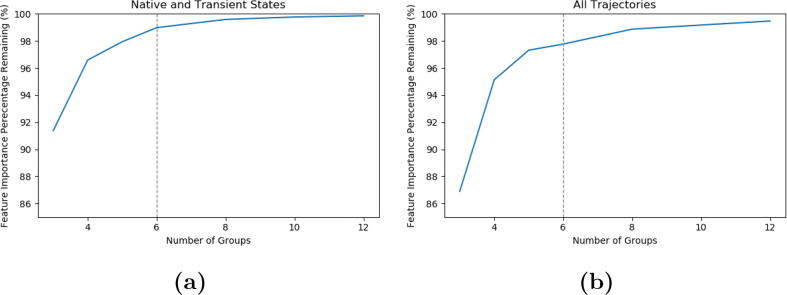
For native state model, six communities were constructed and associated with 98.9% total feature importance among communities (Fig. 12a). Similarly for the all states model, six communities were constructed and associated with 97.8% total feature importance among communities (Fig. 12b).
Fig. 12.
Community analysis of AsLOV2 based on one-vs-one random forest model. a) Community analysis based on the native states model; b) Community analysis based on the all states model.
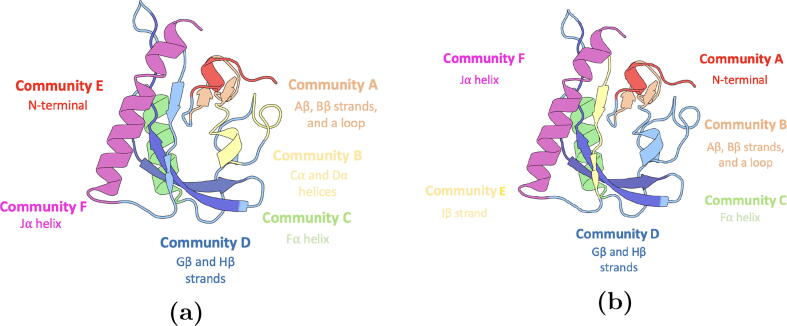
For the native states model (Fig. 13a), Community A consists of A strand, loop between A and B strands, and B strand. Community B consists of C helix and D helix. Community C consists of F helix. Community D includes G and H strands. Community E consists of the N-terminal including A’ helix. Community F consists of J helix.
Fig. 13.
Illustration of communities generated by community analysis of AsLOV2 based on one-vs-one random forest model. The communities consisting the same secondary structures in both cases have the same color for comparison. a) Six communities constructed based on the native states model; b) Six communities constructed based on the all states model.
For the all states model (Fig. 13b), Community A consists of the N-terminal and the A’ helix. Community B consists of A, loop between A and B strands, and B strand. Community C consists of F helix. Community D consists of G and H strands. Community E consists of I strand. Community F consists of the J helix.
When community analysis was applied on both native states model and all states model, A and B strands were among the identified essential secondary structures. This suggests their importance in both models, despite the different configurations involved in the analysis in both cases. This could be attributed to the role of the A and B strands in controlling the movement of the A’ helix [2]. Moreover, the N-terminal, A’ and J helices were among the identified essential communities in both cases. This reflects their importance in the photo-activation of AsLOV2 [16], [4]. F helix and G strand were also identified as important communities in both cases.
The cumulative importance between the different communities were calculated as measurement of the correlation between them. This provides a better insight into the allosteric mechanism and the role of the A’ helix as a secondary structure not as individual residues.
For the native states model, the correlation between Community E (N-terminal A’ helix) (Fig. 13a) and the rest of the protein accounted for 59.39% of the total feature importance. Specifically, this Community E displays the significantly high correlation with Communities A, F, C, and D which are ordered based on their total feature importance with Community E (Table 1).
Table 1.
Cumulative feature importance between each machine learning community pair based on the native states model.
| Features | Commu.A | Commu. B | Commu. C | Commu. D | Commu. E | Commu. F |
|---|---|---|---|---|---|---|
| Commu. A | 0.387% | 4.467% | 3.886% | 2.64% | 17.524% | 2.71% |
| Commu. B | 1.416% | 5.958% | 4.996% | 5.945% | 5.294% | |
| Commu. C | 0.982% | 2.183% | 11.167% | 2.71% | ||
| Commu. D | 1.077% | 10.867% | 2.971 % | |||
| Commu. E | 1.262% | 13.889% | ||||
| Commu. F | 0.46% |
3.4.3. Cumulative importance between the different communities obtained from community analysis
For the all states model, the correlation between the N-terminal A’ helix (Community A) and the rest of the protein accounts for 61.93% of overall feature importance. The highest cumulative feature importance occurs between Communities A and D (A, H, and G strands) as 24% (Table 2). Considering that H in Community D has high RMSF, this clearly reflects the role of the sheets in the protein dynamics and supports the hypothesis that the undocking of the J helix is preceded by a conformational change in the sheets [16].
Table 2.
Cumulative feature importance between each machine learning community pair based on the all states model.
| Features | Commu.A | Commu. B | Commu. C | Commu. D | Commu. E | Commu. F |
|---|---|---|---|---|---|---|
| Commu. A | 1.467% | 12.867% | 9.685% | 24.023% | 5.126% | 10.232% |
| Commu. B | 0.57% | 5.144% | 2.848% | 3.053% | 4.348% | |
| Commu. C | 0.547% | 3.985% | 3.407% | 4.123% | ||
| Commu. D | 0.656% | 3.708% | 2.286% | |||
| Commu. E | 0.665% | 2.934% | ||||
| Commu. F | 0.559% |
Moreover, the significant cumulative feature importance between Communities A and B (13%) indicates that the movement of the A’ helix was not only controlled by A and B strands as indicated by Zayner et al. ([16], [4]) but also by the loops connecting the three secondary structures with each other.
In both models, C-terminal forms Community F and displays high cumulative feature importance with N-terminal (Table 1, Table 2) and implies the role of the both termini as key players in the allosteric mechanism of the AsLOV2 [16], [17], [8].
4. Discussion
A’ helix has an essential role in the allosteric activation of the AsLOV2 upon exposure to blue light. It mediates the propagation of the allosteric signal from the amino terminal to the carboxy terminal (J helix), which becomes undocked and loses its secondary structure [16], [17], [8]. However, the underlying mechanism for the functional role of the A’ helix has not been fully revealed. Thus, we studied the role of A’ helix at the atomistic level by introducing two classes of mutations: helicity-enhancing mutations and helicity-disrupting mutations. In addition, the importance of the overall secondary structure was investigated using community analysis to investigate the importance of the A’ helix as a whole. The different analyses helped to gain insight into the role of the A’ helix in the conformational and dynamical changes associated with the interconversion between the light and the dark states.
Light state and transient dark state were found to be more flexible than the dark state as indicated by their higher RMSF and by occupying more conformational space as seen in the tICA analysis. This is because the light states undergo a conformational change upon photo-activation and involve the unfolding of both termini. Moreover, these two states were found to occupy most of the macrostates (States 1, 2 and 4) when the trajectories of the native and the transient states were involved in the MSM analysis (the natives states model), confirming the flexible nature induced by light absorption. It is worth to note again that the true light state of AsLOV2 does not have well-defined structures of unfold C terminal (as J helix in the dark state) and unfold N terminal (as A’ helix in the dark state). Therefore, the light state of AsLOV2 could not be represented by a single structure. Although the available crystal structure of AsLOV2 (PDB ID: 2V1B) could not be considered as an accurate representation of the true light state of AsLOV2, it is the direct experimental observation of AsLOV2 under constant exposure to the blue light.
It was found that replacing threonine with alanine in T406A and T407A mutants is associated with stabilization the helix (A’ helix). In L408D mutant, on the contrary, the inward facing leucine was replaced with carboxylic acid decreasing the stability of the helix [16] and affecting the overall hydrogen bond network in the A’ helix. This was supported by the data obtained from DSSP analysis and Baker-Hubbard hydrogen bonding analysis (Fig. 4).
The N-terminal hydrogen bond network is different between the light and the dark states, as well as between the helicity-enhancing mutants and helicity-disrupting mutants. This difference highlights the importance of Thr407 and Arg410 in the allosteric mechanism of the AsLOV2. The undocking and the unfolding of the A’ helix was mainly correlated to the changes in the hydrogen bonds formed by these two residues. Based on this, it is proposed that the initial steps in the photo-activation of the AsLOV2 are involved with breaking the hydrogen bonds between Ala405 and Arg410 and between Thr407 and Glu545, followed by formation of a hydrogen bond between Phe403 and Thr407. These changes in the N-terminal hydrogen bond network within the A’ helix are followed by changes in the hydrogen bond between the A’ helix and the J helix, where Arg410 forms a hydrogen bond with Glu545 in the J helix (Fig. 14). These newly formed hydrogen bonds with the J helix could be the reason behind its undocking and unfolding of the latter. This proposed mechanism was supported by the fact that R410P mutant lacks all these hydrogen bonds.
Fig. 14.
Hydrogen bonds critical to the unfolding of the A’ helix in the AsLOV2. a) The hydrogen bond between Ala405 and Arg410 in the dark state; b) The hydrogen bond between Thr407 and Glu545 in the dark state; c) The hydrogen bond between Phe403 and Thr407 in the transient light states; d) The hydrogen bond between Arg410 and Glu545 in the native light, the transient light, and the transient dark states.
The importance of maintaining the hydrogen bond network in the A’ helix can be related to maintaining the helicity of the overall secondary structure. The simulation confirms that helicity enhancing mutations (T406A and T407A) reserve the N-terminal hydrogen bond network of the A’ helix while the helicity disrupting mutations (L408D and R410P) lead to the opposite effect. Interestingly, the role of A’ helix is conserved in other LOV domains such as Vivid, where the N-terminal helix is essential in the conversion of the dark state into the light state [5].
Moreover, it was suggested that the allosteric mechanism of LOV2 domains depends mainly on the changes in the hydrogen bond network [16]. Recent study found the formation of transient bond between N414 and Q513 was essential for the propagation of the signal from the N-terminal to the C-terminal upon photo-activation of AsLOV2 [24]. As comparison, our study suggests that two related hydrogen bonds, between N414 and Q513, and between N414 and D515 may play a role in the signaling process (Figure S8). In the dark state, a hydrogen bond forms between N414 and Q513 (Figure S9a). This hydrogen bond also exists in the light state, but is relatively weaker with higher distribution in longer distance range (Figure S9b). No hydrogen bond forms between residues N414 and D515 in the dark state (Figure S9c). However, such hydrogen bond between N414 and D515 is observed in the light state (Figure S9d). As comparison, distributions of hydrogen bond between N414 and Q513 in the tslight states are different from the dark state and more similar to the light state (Figure S9a). But the hydrogen bond between N414 and Q513 in the tsdark states are similar to the light state (Figure S9b). The similar trend is also observed for the hydrogen bond between N414 and D515. This shows that these two hydrogen bonds are susceptible to the structural perturbations. Therefore, their interplay could play a role in the allosteric signaling of AsLOV2 by bridging the light perturbation from the chromophore core (Q513) to the -sheets proximal to the helical termini (N414 and D515).
The essential role of the N-terminal, A’ helix and J helix in the allosteric mechanism of AsLOV2 was further demonstrated at the level of the secondary structure using community analysis. Through the community analysis, it was shown that N-terminal including the A’ helix has majority of correlation importance percentage (61.93%) with the other communities. This verifies that the importance of the A’ helix lies in its interaction with the other secondary structures or communities. This also supports the role of the A’ helix as mediators for propagating the input signal to signaling outputs [21], [45], [46].
Other key secondary structures including B strand, F helix, and the loop between A and B strands were identified as important secondary structures for allostery in community analysis.
Based on the better understanding of the difference in the hydrogen bonding patterns in A’ helix in the natives, transient, and the mutated states, better optogenetic switches could be developed. For example, the residual activity of AsLOV2 in the dark state was one of the issues with employing the protein in the design of the optogenetic switches. This residual activity was the result of the partial unfolding of the J helix in the dark state. As Zayner et al. found that T406A and T407A mutations led to decreasing the dynamics and enhancing the helicity of the J helix [16], [4], these two mutants with their helicity enhancing effect could be used to decrease the residual effect in the dark state. Based on our study, it is suggested that these two helicity-enhancing mutations strengthen the helical structure of the A’ helix, attenuating the conformational changes in the J helix by the formation of salt bridge between Arg410 and Glu545.
Overall, through a comprehensive computational protocol, we aimed to reveal the crucial role of the A’ helix in the allosteric process of AsLOV2, and provided not only a detailed mechanistic explanation of its biologically relevant interactions but also its effects on the overall protein conformational landscape, as well as its interaction with other secondary structures.
5. Conclusion
In AsLOV2, the A’ helix plays a pivotal role in propagating the allosteric perturbation from the LOV core to the functional J helix, which undergoes an unfolding process. Extensive molecular dynamics simulations were carried out to examine the effect of four mutations (T406A, T407A, L408D, and R410P) introduced into the A’ helix compared to the native light and dark states as well as the transient structures.
The N-terminal hydrogen bonds in the helicity-enhancing mutations (T406A and T407A) are different compared to the helicity-disrupting mutations (L408D and R410P). The change in the hydrogen bonding pattern was correlated with the overall conformational space of protein revealed through the tICA and MSM analyses. It is revealed that Thr407 and Arg410 are key residues for the transitions between the light and dark states due to their hydrogen bonds with Glu545.
The machine learning based community analysis sheds light on the importance of the N-terminal including the A’ helix, J helix, and -sheets and reveals their central role in the allosteric mechanism of AsLOV2. Specifically, the initial mechanism of J helix undocking and unfolding is involved with breaking the hydrogen bond between Ala405 and Arg410 and between Thr407 and Glu545, followed by formation of a hydrogen bond between Phe403 and Thr407.
In conclusion, the change in the N-terminal hydrogen bond network facilitates the unfolding of the A’ helix, which mediates the subsequent conformational changes in the allosteric activation of AsLOV2.
CRediT authorship contribution statement
Mayar Tarek Ibrahim: Investigation, Formal analysis, Visualization, Writing - original draft. Francesco Trozzi: Visualization, Writing - review & editing. Peng Tao: Conceptualization, Supervision, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Research reported in this paper was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R15GM122013. The computational time was provided by the Center for Research Computing at Southern Methodist University
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.csbj.2021.11.038.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gu Yi-Zhong, Hogenesch John B., Bradfield Christopher A. The pas superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40(1):519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Halavaty Andrei S., Moffat Keith. N-and c-terminal flanking regions modulate light-induced signal transduction in the lov2 domain of the blue light sensor phototropin 1 from avena sativa. Biochemistry. 2007;46(49):14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 3.Guntas Gurkan, Hallett Ryan A., Zimmerman Seth P., Williams Tishan, Yumerefendi Hayretin, Bear James E., et al. Engineering an improved light-induced dimer (ilid) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci. 2015;112(1):112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zayner Josiah P., Antoniou Chloe, French Alexander R., Hause Jr Ronald J., Sosnick Tobin R. Investigating models of protein function and allostery with a widespread mutational analysis of a light-activated protein. Biophys J. 2013;105(4):1027–1036. doi: 10.1016/j.bpj.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoltowski Brian D., Schwerdtfeger Carsten, Widom Joanne, Loros Jennifer J., Bilwes Alexandrine M., Dunlap Jay C., Crane Brian R. Conformational switching in the fungal light sensor vivid. Science. 2007;316(5827):1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levskaya Anselm, Weiner Orion D., Lim Wendell A., Voigt Christopher A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461(7266):997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harper Shannon M., Neil Lori C., Gardner Kevin H. Structural basis of a phototropin light switch. Science. 2003;301(5639):1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 8.Crosson Sean, Moffat Keith. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci. 2001;98(6):2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandre Maxime T.A., Arents Jos C., van Grondelle Rienk, Hellingwerf Klaas J., Kennis John T.M. A base-catalyzed mechanism for dark state recovery in the avena sativa phototropin-1 lov2 domain. Biochemistry. 2007;46(11):3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 10.Swartz Trevor E., Corchnoy Stephanie B., Christie John M., Lewis James W., Szundi Istvan, Briggs Winslow R., Bogomolni Roberto A. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biolog Chem. 2001;276(39):36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 11.Mart Robert J., Meah Dilruba, Allemann Rudolf K. Photocontrolled exposure of pro-apoptotic peptide sequences in lov proteins modulates bcl-2 family interactions. ChemBioChem. 2016;17(8):698–701. doi: 10.1002/cbic.201500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niopek Dominik, Benzinger Dirk, Roensch Julia, Draebing Thomas, Wehler Pierre, Eils Roland, Di Ventura Barbara. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun. 2014;5:4404. doi: 10.1038/ncomms5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyman Susan K., Guan Tinglu, Bednenko Janna, Wodrich Harald, Gerace Larry. Influence of cargo size on ran and energy requirements for nuclear protein import. J Cell Biol. 2002;159(1):55–67. doi: 10.1083/jcb.200204163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Yi I., Frey Daniel, Lungu Oana I., Jaehrig Angelika, Schlichting Ilme, Kuhlman Brian, et al. A genetically encoded photoactivatable rac controls the motility of living cells. Nature. 2009;461(7260):104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miralles Francesc, Posern Guido, Zaromytidou Alexia-Ileana, Treisman Richard. Actin dynamics control srf activity by regulation of its coactivator mal. Cell. 2003;113(3):329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 16.Zayner Josiah P., Antoniou Chloe, Sosnick Tobin R. The amino-terminal helix modulates light-activated conformational changes in aslov2. J Mol Biol. 2012;419(1–2):61–74. doi: 10.1016/j.jmb.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zayner Josiah P., Mathes Tilo, Sosnick Tobin R., Kennis John T.M. Helical contributions mediate light-activated conformational change in the lov2 domain of avena sativa phototropin 1. ACS Omega. 2019;4(1):1238–1243. doi: 10.1021/acsomega.8b02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto Atsushi, Iwata Tatsuya, Sato Yoshiaki, Matsuoka Daisuke, Tokutomi Satoru, Kandori Hideki. Light signal transduction pathway from flavin chromophore to the helix of arabidopsis phototropin1. Biophys J. 2009;96(7):2771–2778. doi: 10.1016/j.bpj.2008.12.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennis John T.M., Van Stokkum Ivo H.M., Crosson Sean, Gauden Magdalena, Moffat Keith, van Grondelle Rienk. The lov2 domain of phototropin: a reversible photochromic switch. J Am Chem Soc. 2004;126(14):4512–4513. doi: 10.1021/ja031840r. [DOI] [PubMed] [Google Scholar]
- 20.Nash Abigail I., Ko Wen-Huang, Harper Shannon M., Gardner Kevin H. A conserved glutamine plays a central role in lov domain signal transmission and its duration. Biochemistry. 2008;47(52):13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Möglich Andreas, Ayers Rebecca A., Moffat Keith. Structure and signaling mechanism of per-arnt-sim domains. Structure. 2009;17(10):1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Hao, Trozzi Francesco, Zoltowski Brian D., Tao Peng. Deciphering the allosteric process of the phaeodactylum tricornutum aureochrome 1a lov domain. J Phys Chem B. 2020;124(41):8960–8972. doi: 10.1021/acs.jpcb.0c05842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Hongyu, Dong Zheng, Verkhivker Gennady, Zoltowski Brian D., Tao Peng. Allosteric mechanism of the circadian protein vivid resolved through markov state model and machine learning analysis. PLoS Computat Biol. 2019;15(2):e1006801. doi: 10.1371/journal.pcbi.1006801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iuliano James N., Collado Jinnette Tolentino, Gil Agnieszka A., Ravindran Pavithran T., Lukacs Andras, Shin Seung Youn, Woroniecka Helena A., Adamczyk Katrin, Aramini James M., Edupuganti Uthama R., et al. Unraveling the mechanism of a lov domain optogenetic sensor: A glutamine lever induces unfolding of the j helix. ACS Chem Biol. 2020;15(10):2752–2765. doi: 10.1021/acschembio.0c00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trozzi Francesco, Wang Feng, Verkhivker Gennady, Zoltowski Brian D., Tao Peng. Dimeric allostery mechanism of the plant circadian clock photoreceptor zeitlupe. PLoS Comput Biol. 2021;17(7):e1009168. doi: 10.1371/journal.pcbi.1009168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman Helen M., Bhat Talapady N., Bourne Philip E., Feng Zukang, Gilliland Gary, Weissig Helge, Westbrook John. The protein data bank and the challenge of structural genomics. Nat Struct Biol. 2000;7(11):957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 27.Freddolino Peter L., Gardner Kevin H., Schulten Klaus. Signaling mechanisms of lov domains: new insights from molecular dynamics studies. Photochem Photobiol Sci. 2013;12(7):1158–1170. doi: 10.1039/c3pp25400c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks Bernard R., Brooks III Charles L., Mackerell Jr Alexander D., Nilsson Lennart, Petrella Robert J., Roux Benoıˇt, Won Youngdo, Archontis Georgios, Bartels Christian, Boresch Stefan, et al. Charmm: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eastman Peter, Pande Vijay. Openmm: A hardware-independent framework for molecular simulations. Comput Sci Eng. 2010;12(4):34–39. doi: 10.1109/MCSE.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen William L., Chandrasekhar Jayaraman, Madura Jeffry D., Impey Roger W., Klein Michael L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 31.Essmann Ulrich, Perera Lalith, Berkowitz Max L., Darden Tom, Lee Hsing, Pedersen Lee G. A smooth particle mesh ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 32.McGibbon Robert T., Beauchamp Kyle A., Harrigan Matthew P., Klein Christoph, Swails Jason M., Hernández Carlos X., Schwantes Christian R., Wang Lee-Ping, Lane Thomas J., Pande Vijay S. Mdtraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys J. 2015;109(8):1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrigan Matthew P., Sultan Mohammad M., Hernández Carlos X., Husic Brooke E., Eastman Peter, Schwantes Christian R., Beauchamp Kyle A., McGibbon Robert T., Pande Vijay S. Msmbuilder: statistical models for biomolecular dynamics. Biophys J. 2017;112(1):10–15. doi: 10.1016/j.bpj.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwantes Christian R., Pande Vijay S. Improvements in markov state model construction reveal many non-native interactions in the folding of ntl9. J Chem Theory Comput. 2013;9(4):2000–2009. doi: 10.1021/ct300878a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Hernández Guillermo, Paul Fabian, Giorgino Toni, Fabritiis Gianni De, Noé Frank. Identification of slow molecular order parameters for markov model construction. J Chem Phys. 2013;139(1):07B604_1. doi: 10.1063/1.4811489. [DOI] [PubMed] [Google Scholar]
- 36.Naritomi Yusuke, Fuchigami Sotaro. Slow dynamics in protein fluctuations revealed by time-structure based independent component analysis: The case of domain motions. J Chem Phys. 2011;134(6):02B617. doi: 10.1063/1.3554380. [DOI] [PubMed] [Google Scholar]
- 37.Deuflhard Peter, Weber Marcus. Robust perron cluster analysis in conformation dynamics. Linear Algebra Appl. 2005;398:161–184. [Google Scholar]
- 38.Prinz Jan-Hendrik, Wu Hao, Sarich Marco, Keller Bettina, Senne Martin, Held Martin, Chodera John D., Schütte Christof, Noé Frank, et al. Markov models of molecular kinetics: Generation and validation. J Chem Phys. 2011;134(17):174105. doi: 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 39.Pande Vijay S., Beauchamp Kyle, Bowman Gregory R. Everything you wanted to know about markov state models but were afraid to ask. Methods. 2010;52(1):99–105. doi: 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGibbon Robert T., Schwantes Christian R., Pande Vijay S. Statistical model selection for markov models of biomolecular dynamics. J Phys Chem B. 2014;118(24):6475–6481. doi: 10.1021/jp411822r. [DOI] [PubMed] [Google Scholar]
- 41.Pedregosa Fabian, Varoquaux Gaël, Gramfort Alexandre, Michel Vincent, Thirion Bertrand, Grisel Olivier, Blondel Mathieu, Prettenhofer Peter, Weiss Ron, Dubourg Vincent, et al. Scikit-learn: Machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 42.Lin Shen, Kernighan Brian W. An effective heuristic algorithm for the traveling-salesman problem. Oper Res. 1973;21(2):498–516. [Google Scholar]
- 43.Baker E.N., Hubbard R.E. Hydrogen bonding in globular proteins. Progr Biophys Mol Biol. 1984;44(2):97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W., Sander C. Dssp: definition of secondary structure of proteins given a set of 3d coordinates. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 45.Key Jason, Scheuermann Thomas H., Anderson Peter C., Daggett Valerie, Gardner Kevin H. Principles of ligand binding within a completely buried cavity in hif2αpas-b. J Am Chem Soc. 2009;131(48):17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayers Rebecca A., Moffat Keith. Changes in quaternary structure in the signaling mechanisms of pas domains. Biochemistry. 2008;47(46):12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasello Gianluca, Armenia Ilaria, Molla Gianluca. The protein imager: a full-featured online molecular viewer interface with server-side hq-rendering capabilities. Bioinformatics. 2020;36(9):2909–2911. doi: 10.1093/bioinformatics/btaa009. [DOI] [PubMed] [Google Scholar]
- 48.Grant Barry J., Skjaerven Lars, Yao Xin-Qiu. The bio3d packages for structural bioinformatics. Protein Sci. 2021;30(1):20–30. doi: 10.1002/pro.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.