Abstract
Multi-level proteomics aims to delineate proteins at the peptide (bottom-up proteomics), proteoform (top-down proteomics), and protein complex (native proteomics) levels. Capillary electrophoresis-mass spectrometry (CE-MS) can achieve highly efficient separation and highly sensitive detection of complex mixtures of peptides, proteoforms, and even protein complexes because of its substantial technical progress. CE-MS has become a valuable alternative to the routinely used liquid chromatography (LC)-MS for multi-level proteomics. This review summarizes the most recent (2019–2021) advances of CE-MS for multi-level proteomics regarding technological progress and biological applications. We also provide brief perspectives on CE-MS for multi-level proteomics at the end, highlighting some future directions and potential challenges.
Keywords: capillary zone electrophoresis-mass spectrometry, capillary isoelectric focusing-mass spectrometry, multi-level proteomics, proteoform, protein complex
1. Introduction
Proteomics aims to characterize proteins in cells comprehensively as a function of biological conditions, including but not limited to their expression, post-translational modifications (PTMs), interactions, locations, and turnover. (Zhang et al., 2013; Aebersold & Mann, 2016) Mass spectrometry (MS)-based proteomics has become crucial for pursuing better understandings of molecular mechanisms of fundamental cellular processes and disease development. (Aebersold & Mann, 2016) There are three main strategies: bottom-up proteomics (BUP), top-down proteomics (TDP), and native proteomics.
BUP is the most mature and widely used strategy. It identifies proteins through the MS and tandem MS (MS/MS) measurements of peptides derived from the enzymatic cleavages of the proteins, making BUP a peptide-centric approach. TDP deploys MS and MS/MS to measure intact proteoforms directly without enzymatic cleavages. It is a proteoform-centric approach. Proteoforms represent all forms of protein molecules from the same gene because of the gene-level variations, RNA-level alternative splicing, and protein-level PTMs. (Smith et al., 2013) Native proteomics is a protein complex-centric approach, pursuing global characterization of endogenous protein complexes under physiological conditions with MS and MS/MS. (Skinner et al., 2018; Li et al., 2018; Shen et al., 2018) The BUP, TDP, and native proteomics characterize proteins at three different levels, termed multi-level proteomics. Proteoforms from the same gene could have drastically different functions. (Smith & Kelleher, 2018) The majority of proteins in cells form complexes to function. It is vital to measure proteomes in proteoform- and protein complex-specific manners to better understand protein function.
Figure 1 shows the differences of BUP, TDP, and native proteomics regarding proteoform and protein complex characterizations. BUP can achieve high proteome coverage and the draft map of the human proteome reported in 2014 is a perfect example. (Kim et al., 2014) Tens of thousands of peptides containing various PTMs (e.g., phosphorylation and acetylation) can be routinely characterized with accurate PTM localization using the modern BUP workflows. (Bekker-Jensen et al., 2020; Karayel et al., 2020; Hu et al., 2020) However, BUP usually fails to distinguish proteoforms from the same gene because intact proteoform pictures are lost during the enzymatic digestion step, Figure 1. TDP could give a bird’s eye view of all proteoforms in a sample because it directly measures intact proteoforms. It can tell that there are four different proteoforms in the sample and can identify them through MS/MS. However, it can not provide direct evidence of the protein complex because the complex falls apart under the denaturing conditions used in TDP. Two of the main challenges of TDP are the identification of relatively low-abundance proteoforms and accurate localization of PTMs on each proteoform because proteoforms are substantially larger than peptides, resulting in much lower sensitivity and backbone cleavage coverage for proteoform detection and fragmentation compared to BUP. Native proteomics could provide the most accurate delineation of proteome samples because it characterizes proteomes under close to physiological conditions and directly measures protein complexes. However, there are several technical challenges for native proteomics. First, it can only measure the most abundant protein complexes in a complex sample due to its low sensitivity, and actually, it is routinely used to characterize purified protein complexes (Li et al., 2018; Keener et al., 2019; Wörner et al., 2020). Second, it usually requires sophisticated mass spectrometers to enable the measurement of large protein complexes and the identification of each proteoform involved in a protein complex through MS/MS (MS2) or MS3 analysis (Skinner et al., 2018).
Figure 1.
Schematic about the differences of BUP, TDP, and native proteomics regarding proteoform and protein complex characterizations.
The BUP, TDP, and native proteomics have been integrated for the comprehensive and accurate characterization of proteoforms and protein complexes in cells. The identified proteins and PTM localization information from the BUP can be used to construct a sample-specific proteome database for the TDP to identify and comprehensively characterize proteoforms through a better interpretation of the MS and MS/MS spectra of proteoforms. The identified and fully characterized proteoforms from the TDP of a biological sample provide a list of potential proteoforms in a protein complex and facilitate the delineation of the protein complex. There have been several nice examples of integration of multi-level proteomics. (Schaffer et al., 2020; Dai et al., 2017; van de Waterbeemd et al., 2017; Xu et al., 2020) For instance, Schaffer et al. boosted the proteoform characterization in the human Jurkat T lymphocyte cell line via integrating BUP and TDP datasets, and they observed that the BUP data facilitated the PTM localization on proteoforms identified by the TDP. (Schaffer et al., 2020) In another example, Waterbeemd et al. integrated native proteomics and BUP for confirming the binding of a stationary-phase-induced ribosomal-associated protein (SRA) to E. coli 30S ribosome particles. (van de Waterbeemd et al., 2017)
The multi-level proteomics requires high-capacity separation and highly sensitive detection of peptides, proteoforms, and protein complexes because of the high complexity of proteome samples. Reversed-phase liquid chromatography (RPLC)-MS/MS has been the common choice for proteomics. During the last decade, capillary zone electrophoresis (CZE)-MS/MS and capillary isoelectric focusing (cIEF)-MS/MS have been suggested as alternatives to RPLC-MS/MS for proteomics due to their unique features. First, CZE and cIEF can achieve high-resolution separation of peptides, proteoforms, and even protein complexes according to their electrophoretic mobility (μef) and isoelectric points (pIs). Second, CZE and cIEF can be online coupled with electrospray ionization (ESI)-MS using commercially available CE-MS interfaces. Third, CZE-MS has shown 10–100 times better sensitivity than RPLC-MS for the measurement of peptides and proteoforms. (Han et al., 2014; Wang et al., 2012; Faserl et al., 2011; McCool & Sun, 2019) Lastly, the μef of peptides and proteoforms could be predicted accurately, potentially benefiting the identification of proteoforms with PTMs (e.g., phosphorylation and acetylation). (Chen et al., 2021; Chen et al., 2020; Chen et al., 2019; Krokhin et al., 2017; Kim, Zand & Lubman, 2002)
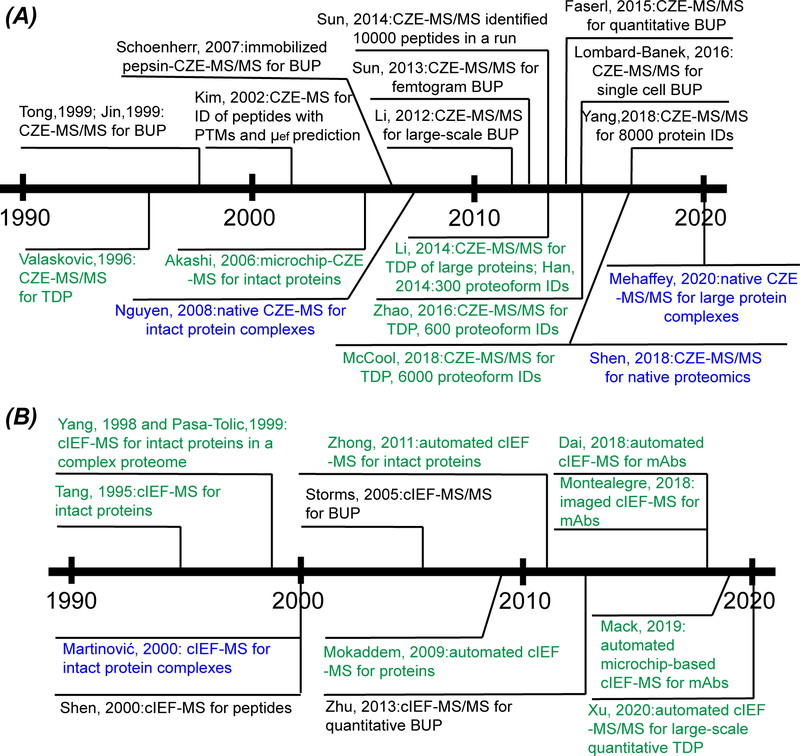
Both CZE-MS and cIEF-MS for proteomics have a long history starting in the 1990s, Figure 2A and 2B. CZE-MS for BUP was pioneered by the Yates and Lubman groups in 1999, providing initial ideas of using CZE-MS for analyzing complex peptide mixtures. (Jin et al., 1999; Tong et al., 1999) From 2000 to 2010, great efforts were made to develop robust and highly sensitive interfaces for coupling CE to MS. (Maxwell & Chen, 2008; Heemskerk, Deelder & Mayboroda, 2014) Many CZE-MS-based BUP studies were also performed during this period. For example, the Lubman group demonstrated the capability of CZE-MS for the characterization of peptides with PTMs and documented great efforts for predicting μef of peptides based on CZE-MS datasets. (Kim, Zand & Lubman, 2002) The Zare group and the Dovichi group coupled online protein digestion using immobilized enzyme microreactors to CZE-MS or MS/MS to boost the throughput of BUP. (Kato et al., 2004; Schoenherr et al., 2007) During the last decade, CZE-MS/MS for BUP has attracted great attentions. (Zhang, Qu & Dovichi, 2018) The Dovichi group made great contributions to the field by boosting the CE-MS interface, sample loading capacity of CZE, and peak capacity for peptide separation to enable large-scale and highly sensitive BUP. (Li et al., 2012; Sun et al., 2013; Sun et al., 2014) The Lindner group demonstrated the first example of large-scale quantitative BUP using CZE-MS/MS. (Faserl et al., 2015) The Nemes group highlighted the value of CZE-MS/MS for mass-limited samples via analyzing single blastomeres isolated from early-stage Xenopus embryos. (Lombard-Banek, Moody & Nemes, 2016) More recently, our research group further improved CZE-MS/MS for BUP regarding overall sensitivity and proteome coverage. (Yang et al., 2018; Chen, Shen & Sun, 2018) CZE-MS/MS for TDP was pioneered by the McLafferty group in 1996, demonstrating the direct coupling of CZE to a FTICR mass spectrometer for analysis of amole amounts of proteins. (Valaskovic, Kelleher & McLafferty, 1996) During 2000–2010, many nice works about CZE-MS for intact protein analysis were done. (Haselberg, de Jong & Somsen, 2010) For example, Akashi et al. documented the coupling of microchip-based CZE to MS for top-down characterization of basic proteins. (Akashi et al., 2006) During the last decade, several research groups have developed CZE-MS/MS methods for large-scale TDP of complex samples due to the improvement of CE-MS interface and mass spectrometer (Shen et al., 2019). Han et al. and Zhao et al. provided effective strategies for boosting the number of proteoform identifications via coupling RPLC fractionation to CZE-MS/MS with the production of 300 and 600 proteoforms from a complex proteome, respectively. (Han et al. 2014; Zhao et al., 2016) Li et al. demonstrated the capability of CZE-MS/MS for the characterization of large proteins. (Li et al., 2014) Our group reported the identification of 600 proteoforms from E. coli in a single CZE-MS/MS run via drastically boosting the sample loading capacity and peak capacity of CZE (Lubeckyj et al., 2017) and documented the identification of nearly 6000 proteoforms from E. coli using a high-capacity size exclusion chromatography (SEC)-RPLC-CZE-MS/MS platform (McCool et al., 2018). Nguyen et al. reported the first study of intact protein complexes under a native condition using CZE-MS in 2008 via establishing an effective sample handling procedure, an efficient CZE separation method, and a reliable coupling of native CZE to native ESI-MS. (Nguyen & Moini, 2008) In 2018, our group documented the application of native SEC-CZE-MS/MS to native proteomics of a complex proteome sample. (Shen et al., 2018) In 2020, Mehaffey et al. demonstrated the potential of native CZE-MS/MS for the delineation of large protein complexes, i.e., E. coli 70S ribosome. (Mehaffey, Xia & Brodbelt, 2020)
Figure 2.
Summary of the history of CZE-MS (A) and cIEF-MS (B) for multi-level proteomics.
cIEF-MS for analysis of intact proteins has been a hot research area for decades, starting in the 1990s. (Hühner, Lämmerhofer & Neusüß, 2015) As shown in Figure 2B, the Lee and Smith groups are the pioneers in this research area and established the basic procedure of coupling cIEF to MS for top-down MS analysis of proteins. (Tang, Harrata & Lee, 1995; Yang et al., 1998; Pasa-Tolic et al., 1999) In 2009 and 2011, automated cIEF-MS systems for intact protein analysis were developed to make the method more straightforward. (Mokaddem, Gareil & Varenne, 2009; Zhong et al., 2011) In 2018 and 2019, several research groups developed automated cIEF-MS or imaged cIEF-MS systems for the characterization of charge variants of monoclonal antibodies (mAbs). (Montealegre & Neusüß, 2018; Dai et al., 2018; Mack et al., 2019). In 2020, our group demonstrated the power of automated cIEF-MS/MS for large-scale qualitative and quantitative TDP of complex proteomes. (Xu et al., 2020) Shen et al. published one of the first studies about high-resolution cIEF separation of complex peptide mixtures for BUP in 2000. (Shen et al., 2000) Storms et al. documented the direct coupling of cIEF to ESI-MS/MS for BUP and demonstrated a carrier ampholyte free cIEF-MS/MS approach for analyzing a complex proteome sample with the identification of over 100 proteins in a single run. (Storms et al., 2005) Zhu et al. employed a mixture of amino acids as carrier ampholyte for cIEF-MS/MS-based quantitative BUP of differentiated PC12 cells and identified over 800 proteins. (Zhu et al., 2013) Interestingly, cIEF-MS has also been evaluated for the characterization of intact protein complexes under a native condition in 2000 and demonstrated the potential of cIEF-MS for native proteomics. (Martinović et al., 2000) We need to highlight that Dr. David M Lubman also made huge contributions in the area of IEF for protein separation and applications in cancer research starting in 2002. (Kachman et al., 2002)
Several comprehensive reviews have been published recently regarding CE-MS for BUP and TDP. (Shen et al., 2019; Zhang, Qu & Dovichi, 2018; Gomes & Yates, 2019; Hühner, Lämmerhofer & Neusüß, 2015) In this review, we focus on the most recent (2019–2021) technical advances and applications of CE-MS for multi-level proteomics.
2. Technological development
The broad recognition and widespread adoption of CE-MS for proteomics are attributed to the continuous improvement in techniques, such as CE-MS interface for better sensitivity and robustness, capillary coating for eliminating analytes’ dead adsorption and electroosmotic flow (EOF), online sample preconcentration methods for better sample loading capacity of CZE, and automation of cIEF-MS for user-friendly operations.
2.1. CE-MS Interface
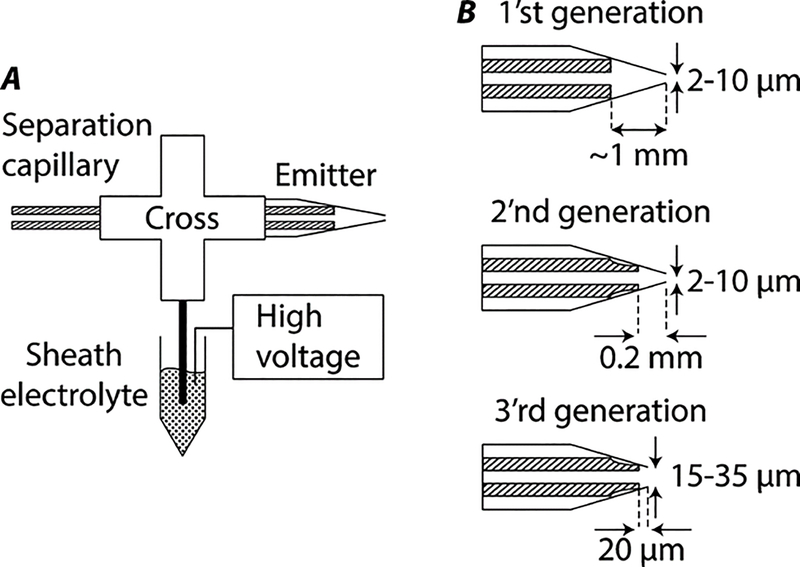
The development of CE-MS interface has been a hot research topic for decades starting from the pioneering work of Smith et al. in 1988. (Smith, Barinaga & Udseth, 1988) The ideal characteristics of a CE-MS interface are highly sensitive, robust, user-friendly, and compatible with different CE-MS conditions. Table 1 summarizes the four commercialized CE-MS interfaces. They are the coaxial sheath flow interface (Agilent) developed by Smith et al. in 1988, (Smith, Barinaga & Udseth, 1988), the sheathless interface using a porous capillary tip as the ESI emitter (CESI, Sciex) developed by Moini in 2007, (Moini, 2007), the glass microfluidic device with integrated CE and CE-MS interface (ZipChip, 908 Devices) developed by the Mellors et al. in 2008, (Mellors et al., 2008), and the electro-kinetically pumped sheath flow interfaces (EMASS-II, CMP Scientific) designed by the Dovichi group in 2010, 2013 and 2015, Figure 3. (Wojcik et al., 2010; Sun et al., 2013; Sun et al., 2015) The commercialized versions of these interfaces are relatively robust, easy to use, and widely adopted. The CESI has no sheath flow, requiring that the separation buffer needs to support ESI. The other three sheath-flow interfaces have better compatibility with various CE-MS conditions due to the sheath buffer. The coaxial sheath flow interface has lower sensitivity than the CESI, ZipChip, and EMASS-II interfaces because it employs a low μL/min flow rate of sheath buffer for ESI, leading to a significant sample dilution. Although the EMASS-II interface also utilizes a sheath buffer, the flow rate of the sheath buffer is very low, ensuring an extremely high sensitivity. For example, Sun et al. reported a low zmole limit of detection (LOD) of peptides using the EMASS-II interface on a Q-Exactive mass spectrometer. (Sun et al., 2013) The sensitivity of CESI and EMASS-II interfaces are most likely comparable. The ZipChip uses a corner of the microfluidic CE chip for ESI, and a high amole LOD of a peptide in a complex background has been estimated recently using the ZipChip system on a Q-Exactive HF mass spectrometer. (Rinas, Jenkins & Orsburn, 2019) Besides the commercialized interfaces, the flow-through microvial interface developed by the Chen group in 2010 is also well recognized and has shown great robustness, sensitivity, and flexibility. (Maxwell et al., 2010) These interfaces have been well reviewed in the literature. (Shen et al., 2019; Ramautar et al., 2012) In 2016, Guo et al. developed a true sheathless CE-MS interface using a metal-coated and tapered capillary as the ESI emitter instead of the porous capillary tip and demonstrated that a hundred CE-MS runs were achieved without significant degradation of interface performance. (Guo et al., 2016) In 2017, Choi et al. presented an improved co-axial sheath flow interface via using a tapered-tip metal emitter and observed stable ESI at the 200–350 nL/min sheath flow, producing a 260-zmol LOD for angiotensin II with an Impact HD quadrupole time-of-flight mass spectrometer in a parallel reaction monitoring mode. (Choi et al., 2017)
Table 1.
Summary of the commercialized CE-MS interfaces.
| CE-MS Interface | sensitivity | robustness | user-friendliness | compatibility with varied CE-MS conditions |
|---|---|---|---|---|
| Coaxial sheath flow interface (Smith, 1988), Commercialized by Agilent | √ | √ | √ | √√ Sheath buffer |
| Sheathless interface using a porous capillary tip as ESI emitter (Moini, 2007), Commercialized by Sciex; CESI | √√√ | √ | √ | √ The separation buffer needs to support ESI. |
| Glass microfluidic device with integrated CE and CE-MS interface (Mellors, 2008), Commercialized by 908 Devices; ZipChip | √√ | √ | √ | √√ Sheath buffer |
| Electro-kinetically pumped sheath flow interfaces (Wojcik, 2010; Sun, 2013 and 2015), Commercialized by CMP Scientific; EMASS-II | √√√ | √ | √ | √√ Sheath buffer |
Figure 3.
Schematic designs of the electrokinetically pumped sheath flow CE-MS interface (A) and its three different generations (B). reproduced from Sun et al. (2015) with permission from American Chemical Society, copyright (2015).
During the last two years, the development of the CE-MS interface has focused on modifications and improvement of the interfaces mentioned above. Sauer et al. developed a new version of the co-axial sheath-flow interface via using a tapered and gold-coated stainless tubing as the ESI emitter. (Sauer, Sydow & Trapp, 2020) The authors showed a slightly improved ESI stability using the gold-coated emitter compared to the stainless emitter and a 2 nM LOD for angiotensin II. Because of the relatively high flow rate of the sheath buffer for ESI (i.e., 3.0 μL/min), the interface’s sensitivity could be limited. Zhang et al. fabricated a new version of the sheathless interface using a gold foil-covered capillary tip as an ESI emitter instead of a porous capillary tip or metal-coated capillary tip. (Zhang et al., 2020) One end of the separation capillary was first etched with a hydrofluoric acid (HF) solution to produce a capillary tip for ESI, and the tip was further covered with a piece of gold foil for electrical contact for ESI. The gold foil was fixed with epoxy resin glue. The sheathless interface produced stable ESI in a wide range of separation buffer flow rates (50–800 nL/min), ensuring a reasonably high sensitivity. The authors documented that the lifetime of the interface was over 180 hours, suggesting good robustness. The fabrication process involving HF etching and fixation of gold foil on the etched capillary tip with glue could limit the user friendliness of the interface. Vermeire et al. created another version of the sheathless interface by making an opening on the separation capillary using CO2 laser for electrical contact. (Vermeire, Van Schepdael & Petersen, 2020) A liquid reservoir was used to cover the opening for two purposes. First, an external voltage can be applied in the reservoir for ESI. Second, a make-up liquid flow at hundreds of nL/min into the separation capillary through the opening from the reservoir can help to maintain stable ESI. However, the complicated fabrication process, nearly porous capillary tip from HF etching for ESI, manual operations of the interface are potential issues regarding robustness and user-friendliness. More recently, Höcker et al. modified the electro-kinetically pumped sheath flow interface by adding another capillary into the pulled glass ESI emitter to further improve the robustness of the system. (Höcker et al., 2021) The purpose of the second capillary is to deliver sheath buffer into the spray emitter to help guide the analytes from the separation capillary for ESI if necessary and for cleaning up the spray emitter to remove sample matrixes that are potentially left in the emitter after sample analysis.
Besides modifications and improvement of previous interfaces, a novel approach for coupling CE to MS was developed by the Holland group in 2020. (Kristoff et al., 2020) The method employed vibrating sharp-edge spray ionization (VSSI) for creating a low flow voltage free CE-MS interface. The interface utilizes a pulled glass probe to transfer vibrations from the piezoelectric source to the liquid pumped out of the separation capillary for ionization. The vibrating probe needs to contact with the liquid from the separation capillary at the capillary orifice, which requires a liquid flow at an nL/min level (i.e., 70–200 nL/min) inside of the separation capillary driven by either an EOF or an external pressure. The system was tested to analyze β-blockers, peptides, and proteins, showing potential for various biological applications.
It is worthy to point out that these new developments of CE-MS interfaces during 2019–2021 discussed above need to be thoroughly evaluated regarding sensitivity, robustness, and compatibility with various CE-MS conditions. Additionally, they can be improved for better user-friendliness.
Several papers about further evaluations of some of the well-known CE-MS interfaces have been published during the last two years. (Wang, Chen & Chen, 2021; Gou et al., 2020; Amenson-Lamar et al., 2019) Wang et al. evaluated the compatibility of the flow-through microvial interface with mass spectrometers from different vendors (Agilent, Sciex, and Thermo Scientific) for CE-MS analysis of amino acids and peptides. (Wang, Chen & Chen, 2021) They demonstrated good compatibility of the interface with various mass spectrometers involved and showed, on average, a 50- and a 10-fold improvement in the signal-to-noise ratio (S/N) of amino acids and peptides compared to the co-axial sheath flow interface. Amenson-Lamar et al. investigated the electrokinetically pumped sheath-flow interface for CZE-MS detection of peptides with ultrahigh sensitivity, documenting the direct detection of 1-zmol angiotensin (about 600 molecules) injected and a 230-ymol LOD for the peptide. (Amenson-Lamar et al., 2019) Gou et al. compared the commercialized electrokinetically pumped sheath-flow interface (EMASS-II) with the coaxial sheath-flow interface for BUP of a complex sample, reporting that the EMASS-II interface based system identified about two times more peptides and proteins. (Gou et al., 2020)
2.2. Coating
For the characterization of large biomolecules (peptides, proteins, and protein complexes) using CZE-MS or cIEF-MS, the inner wall of the separation capillary usually needs to be coated with neutral and hydrophilic polymers, e.g., linear polyacrylamide (LPA). There are at least two purposes. First, the coating can reduce the dead adsorption of the analytes on the inner wall of the capillary, ensuring high separation efficiency and high sensitivity. Second, the neutral polymers immobilized on the inner wall of the capillary eliminate the EOF and slow down the CE separation, providing wide separation windows for mass spectrometers to acquire MS and MS/MS of analytes, which is crucial for large-scale proteomics. For example, hours of separation windows and high separation efficiency have been achieved using CZE-MS or cIEF-MS with LPA-coated separation capillaries for BUP and TDP of complex proteomes, leading to the identification of thousands of protein groups and about 1,000 proteoforms in a single run. (Sun et al., 2014; Busnel et al., 2010; Zhang et al., 2017; Lubeckyj et al., 2017; Lubeckyj et al., 2019; Xu et al., 2020) Many kinds of covalent and dynamic coatings have been developed in the literature, and the coatings developed before 2019 have been comprehensively reviewed. (Gomes & Yates, 2019; Shen et al., 2019; Zhang, Qu & Dovichi, 2018)
During the last two years, several kinds of covalent and neutral coatings have been developed to improve the CE separation of large biomolecules. (Yu et al., 2019; Wang et al., 2019; Meixner, Pattky & Huhn, 2020; Shen et al., 2021) Yu et al. prepared a photosensitive diazotized poly (vinyl alcohol-b-styrene) coating for CZE separation of intact proteins. (Yu et al., 2019) The new coating-based CZE baseline separated a mixture of standard proteins. The EOF mobility had minimal changes after flushing the coated capillary for 10 min with 0.1 M NaOH, 0.1 M HCl, or N-dimethylformamide, indicating good stability of the coating. However, more studies are needed about the stability of the coating for long-time uses and its wide applicability for various complex protein samples. Wang et al. tried to improve the covalent LPA coating by co-polymerizing high-surface-area attapulgite nanoparticles and acrylamide under the assistance of azobisisobutyronitrile initiator and heat. (Wang et al., 2019) The separation capillary with the new coating showed lower EOF mobility than that with the typical LPA coating. The new coating-based CZE-MS demonstrated good stability and reproducibility for the separation of a standard protein mixture. CZE-MS/MS analysis of an E. coli proteome sample using a separation capillary with the new coating achieved a 90-min separation window and identified 300 proteoforms in one run. Meixner et al. created a highly polar, pH-persistent, and covalent poly AAEE (N-acryloylamido ethoxyethanol) coating based on Si-C linkages to AAEE for CZE separations of peptides, proteins, and polyamines in complex samples. (Meixner, Pattky & Huhn, 2020) The EOF mobility of coated capillaries had minor to moderate changes after successively flushing the capillaries with 1M HCl, acetonitrile, and 1M NaOH for 1 hour each, indicating reasonably good stability of the coating. The coated capillary was successfully used for CZE-MS measurements of intact protein samples for 100 hours without special rinsing. More recently, Shen et al. developed a linear carbohydrate polymer (LCP) coating on the inner wall of separation capillaries for boosting the characterization of large proteins and protein complexes using native CZE-MS. (Shen et al., 2021) The LCP coating was prepared by using a newly synthesized sugar monomer (3-O-acryloyl-α/β-D-glucopyranose). Native CZE-MS measurement of monoclonal antibodies (mAbs) using the LCP coating produced a much higher signal intensity of mAbs compared to the typical LPA coating, most likely due to the reduced protein dead adsorption on the capillary inner wall. The data suggest the potential of the LCP-coated capillaries for CZE-MS-based native proteomics.
It has been demonstrated in the literature that polymers with N-H groups could lead to higher protein adsorption compared to that without the N-H groups, most likely because the N-H groups act as hydrogen bonding donors, which causes protein-polymer interaction through hydrogen bonds. (Metzke & Guan, 2008) In general, LPA coating performs well for BUP and TDP of small proteoforms using CE-MS as demonstrated in many publications. (Gomes & Yates, 2019; Shen et al., 2019; Zhang, Qu & Dovichi, 2018) When CE-MS is used for characterizing large proteoforms and protein complexes, its performance could be further boosted substantially by using protein-resistant polymer coatings, as demonstrated by our recent work. (Shen et al., 2021)
2.3. Online sample preconcentration techniques
CZE typically has low sample loading capacity, and about 1% of a total capillary volume can be filled with a sample for analysis to maintain high separation efficiency. For example, a 1-meter-long separation capillary with 50-μm inner diameter (i.d.) has a total volume of 2000 nL, which means only a 20-nL aliquot of a sample is injected for CZE under a typical condition. The low sample loading capacity leads to at least two issues. First, CZE usually has poor concentration LODs due to the limited sample loading volume, which leads to challenges for the analysis of samples with low concentrations. Second, CZE-MS/MS can only be able to identify the most-abundance peptides and proteins in complex proteome samples in proteomics applications because of the limited amounts of peptides and proteins that can be injected into the capillary, limiting the proteome coverage.
When CZE-MS/MS is employed for large-scale proteomics or characterization of low-concentration analytes in samples, usually an online sample preconcentration method is deployed. Many different sample concentration techniques have been developed and evaluated for CZE-MS/MS-based large-scale proteomics, including field-enhanced sample stacking, dynamic pH junction, capillary isotachophoresis (cITP), and solid-phase microextraction (SPME). Different sample preconcentration methods for proteomics have been reviewed in detail recently. (Gomes & Yates, 2019; Shen et al., 2019; Zhang, Qu & Dovichi, 2018)
Several sample-stacking techniques have been developed or further evaluated for CZE-MS analysis of complex samples during the last two years. These techniques can be divided into two categories: electric field-based sample stacking and SPME.
Four electric field-based sample-stacking methods have been investigated. Wells et al. employed electrokinetic supercharging (EKS) for highly efficient sample stacking in CZE-MS analysis of biogenic amines in biological samples. (Wells, Dawod & Kennedy, 2019) The EKS method utilizes a significant electric field difference between the high conductivity leading electrolyte zone and low conductivity water zone in the separation capillary for concentrating analytes at the boundary between the two zones after analytes are electrokinetically injected. The EKS-based method achieved a 5000-fold improvement in sensitivity compared to a typical hydrodynamic injection approach, leading to 10 pM LODs for neurotransmitters. Tang et al. investigated a separation voltage polarity switching transient cITP (PS-tCITP) method for boosting the sample loading capacity of CZE. (Tang et al., 2021; Wu et al., 2019) The basic idea of PS-tCITP is to make analytes move back and forth during the initial ITP in a separation capillary by changing the separation voltage polarity. The goal is to boost the sample loading volume and separation efficiency simultaneously compared to the typical tCITP method. Under an optimized condition, five standard peptides were baseline separated with a sample loading capacity of 100% capillary volume. It is a very promising technique, and more PS-tCITP-MS studies of complex proteome samples will be helpful to demonstrate its capability of advancing CZE-MS-based proteomics.
Dynamic pH junction is another highly efficient sample stacking technique, and it was invented by the Chen group in 2000. (Britz-McKibbin & Chen, 2000) During the dynamic pH junction sample stacking in CZE-MS-based proteomic studies, the analytes (peptides or proteoforms) are concentrated at the moving pH boundary between the basic sample zone and acidic separation buffer. Several papers have shown a microliter scale sample loading capacity with CZE-MS (about 25–75% of the total capillary volume) with the assistance of optimized dynamic pH junction methods for large-scale BUP and TDP. (Zhu et al., 2014; Chen, Shen & Sun, 2017; Lubeckyj et al., 2017; Lubeckyj et al., 2019; Yang et al., 2018) Yan et al. systematically investigated a dynamic pH junction-based CZE-MS method for quantitative analysis of microcystin variants, yielding over two orders of magnitude higher peak heights for four microcystin variants compared to the typical CZE-MS without sample stacking. (Yan et al., 2019) More recently, Wang et al. developed a dynamic pH barrage junction-based CZE-MS and MS/MS method for analysis of amino acids, standard peptides, and mAb digests. (Wang et al., 2020) The dynamic pH barrage junction is a technical variant of the dynamic pH junction, and they have a similar principle. In their design, a separation capillary with a positive charge coating (i.e., polyethyleneimine, PEI) was coupled to an Orbitrap Fusion Lumos mass spectrometer through a flow-through microvial CE-MS interface, Figure 4A. The capillary was first filled with an acidic separation buffer (pH 2.2), followed by sequential injections of a long sample plug (e.g., 10% of the capillary length) in an acidic buffer (pH 2.2) and a basic barrage segment (pH 10.2), before a separation voltage application, Figure 4B. After a high negative potential was applied at the sample injection end, a strong EOF pushed the analytes to flow towards the mass spectrometer, and the positive analyte ions in the acidic sample zone migrated into the basic barrage segment due to the electrostatic propulsion simultaneously. After that, the analytes became negatively charged and moved towards the sample zone under the electric field. The analytes were eventually concentrated at the pH boundary between the sample and basic barrage zones. The optimized CZE-MS/MS system with the dynamic pH barrage junction sample stacking achieved 100% sequence coverage for mAb heavy and light chains via consuming only 9 ng of the protein digest, indicating high sensitivity of the system.
Figure 4.
Schematic designs of the CZE-MS system with the dynamic pH barrage junction stacking and the flow through microvial CE-MS interface (A) and the basic principle of the dynamic pH barrage junction stacking using a PEI-coated separation capillary (B). Reproduced from Wang et al. (2020) with permission from John Wiley and Sons, copyright (2020).
cIEF has also been evaluated as a sample stacking technique recently for native CZE-MS analysis of mAbs. (Shen et al., 2021) Basically, a separation capillary with a neutral coating on its inner wall was first filled with a CZE separation buffer (25 mM ammonium acetate, pH6.8), followed by sequential injections of a short plug of basic buffer (50 mM ammonium acetate, pH 9.5), a mixture of sample and pharmalyte in 10 mM ammonium acetate (pH 6.8), and a short plug of CZE separation buffer. The analytes in the sample zone were first focused according to their pIs in a range of 6.8 and 9.5, followed by typical CZE separation. The native cIEF-assisted CZE-MS resolved three peaks of NIST mAb with a submicroliter sample loading volume. Both the NIST mAb and its homodimer with eight glyco-proteoforms were detected. The data indicated the potential of the method for advancing native proteomics.
Besides the four electric field-based sample-stacking methods, various SPME techniques were developed during the last two years for online concentration of analytes. Pero-Gascon et al. developed a novel SPME-CZE-MS system by coupling a C18-microcartriage for analyte concentration and cleanup to a CZE-MS system via a nanoliter valve (nvSPE-CE-MS). (Pero-Gascon et al., 2020) The new system was used for the analysis of opioid and amyloid beta peptide biomarkers. The nvSPE-CE-MS improved the LODs of the peptide biomarkers by 200 times compared to CE-MS. The nvSPE-CE-MS method allows the independent operations of SPE and CZE-MS and is a useful alternative to regular SPE-CZE-MS for various biological applications. Recently, some new designs of the SPME allow the efficient concentration of analytes from complex samples with high selectivity. Pero-Gascon et al. fabricated an online immunoaffinity SPME-CZE-MS system to analyze serum transthyretin via coupling a microcartridge packed with transthyretin antibody-immobilized magnetic beads to CZE-MS and successfully applied the SPME-CZE-MS method to analysis of transthyretin in human serum. (Pero-Gascon et al., 2019) Pont et al. created a monolithic microcartridge in a capillary and immobilized gold nanoparticles on the monolithic material for selectively concentrating proteins containing thiol, i.e., human transthyretin, because gold nanoparticles have a strong affinity with thiol-containing compounds. (Pont et al., 2020) The microcartridge was integrated into a CZE-MS system for online selective concentration and CZE-MS characterization of human transthyretin. The SPME-CZE-MS method improved the LOD of human transthyretin by 50 times compared to CZE-MS.
2.4. Automated operations of cIEF-MS
The cIEF-MS experiments were usually carried out in a semi-online manner due to the two steps of cIEF (focusing and mobilization). For example, the separation capillary outlet was placed in a catholyte reservoir with a basic buffer for focusing, followed by a manual transfer of the capillary outlet to a CE-MS interface filled with an acidic sheath liquid for mobilization and ESI. The complicated and manual operations became one of the challenges of widespread adoption of cIEF-MS for proteomics. The documentation of “sandwich” injection configuration in 2009 made fully automated cIEF-MS analysis possible. (Mokaddem, Gareil & Varenne, 2009) In 2017, Zhu et al. developed an automated cIEF-MS method based on the “sandwich” injection approach via the electro-kinetically pumped sheath flow interface. (Zhu, Sun & Dovichi, 2017) Dai et al. further systematically investigated the automated cIEF-MS method developed by Zhu et al. and created an improved system for highly efficient separation of mAb charge variants. (Dai et al., 2018) The distal end of separation capillary was placed in the electro-kinetically pumped sheath flow CE-MS interface filled with an acidic sheath buffer (e.g., 20% acetic acid (AA) with 25% acetonitrile). The capillary was first filled with an acidic anolyte (1% formic acid (FA) with 15% glycerol), followed by sequential injections of a short plug of basic catholyte (e.g., 0.2 N NH4OH with 15% glycerol) and a long plug of a mixture of sample and pharmalyte. After moving the injection end of the capillary into a reservoir containing the anolyte, a high electric field was applied across the separation capillary for focusing and separating the analytes according to their pIs. After that, the chemical mobilization occurred automatically when cations from the anolyte (H+) continuously entered the separation capillary and disrupted the pH gradient. More recently, our group developed high-throughput and high-capacity automated cIEF-MS/MS methods according to the “sandwich” injection approach for large-scale TDP using the electro-kinetically pumped sheath flow interface, Figure 5. (Xu et al, 2020) For the high-throughput cIEF-MS/MS method, an 80-cm-long LPA-coated capillary, an acidic sheath buffer containing 0.2% (v/v) FA and 10% (v/v) methanol, an acidic anolyte solution (0.1% (v/v) FA), and a basic catholyte (0.3% (w/w) NH3·H2O, pH 11.8) were used. For the high-capacity method, a 150-cm-long LPA coated capillary and an acidic anolyte solution (5% (v/v) AA) were used and others were the same as the high-throughput method. The high-capacity and automated cIEF-MS/MS enabled the identification of over 700 proteoforms from an E. coli cell lysate in a 2-h run; the high-throughput method identified about 300 proteoforms from the same sample in a 50-min run. The data suggested that the automated cIEF-MS/MS could be a useful analytical tool for large-scale TDP.
Figure 5.
(A) Schematic of the automated cIEF-MS system based on the “sandwich” injection approach. (B) Base peak electropherograms of an E. coli lysate after triplicate analyses by the high-throughput cIEF-MS/MS. (C) Base peak electropherograms of an E. coli lysate after analyses by automated cIEF-MS/MS using an 80-cm LPA-coated capillary and 0.1% FA as the anolyte (high-throughput, red), a 150-cm LPA-coated capillary and 0.1% FA as the anolyte (blue), and a 150-cm LPA-coated capillary and 5% AA as the anolyte (high-capacity, dark cyan). Reproduced from Xu et al. (2020) with permission from American Chemical Society, copyright (2020).
Besides the “sandwich” injection approach, another method for automated cIEF-MS has also been explored by the Chen group in 2018 using the flow-through microvial CE-MS interface. (Wang et al., 2018) Basically, the separation capillary was filled with a mixture of analytes and pharmalyte. The distal end was placed in the CE-MS interface filled with a basic catholyte and the injection end was immersed in an acidic anolyte for focusing. After that, the basic catholyte in the CE-MS interface was replaced automatically by pumping an acidic sheath buffer into the interface for initiating the chemical mobilization step. The automated cIEF-MS method was successfully applied to the analysis of various samples, including peptide pI markers, protein pI markers, and one mAb sample.
3. CE-MS for multi-level proteomics applications
Because of the drastic technical progress, CZE-MS and cIEF-MS have been recognized as useful alternatives to LC-MS for multi-level proteomics. In this part, we summarize their applications in proteomics during the last two years and categorize them into three groups: BUP, TDP, and native proteomics.
3.1. BUP
CZE-MS/MS-based BUP has been widely employed for various biological applications during 2019–2021, including but not limited to highly sensitive characterization of mass-limited biological samples (e.g., single cells), analysis of biopharmaceuticals, measurement of disease-related biomarkers, delineation of protein PTMs, large-scale quantitative proteomics, and peptidomics.
3.1.1. Highly sensitive characterization of mass-limited biological samples
CZE-MS/MS has been well recognized for BUP characterization of mass-limited samples due to its small sample consumption and better sensitivity than RPLC-MS/MS for peptide detection. Lombard-Banek et al. documented the identification and quantification of nearly 800 proteins using CZE-MS/MS from blastomeres isolated from Xenopus laevis early-stage embryos with a consumption of only 5 ng proteome digest. (Lombard-Banek et al., 2019) The CZE-MS/MS was further applied to study the cell lineage of the animal-dorsal (D11) cell in 16-cell embryos to blastomeres in 32-, 64-, and 128-cell embryos with the quantification of hundreds of proteins from blastomeres at every developmental stage studied, demonstrating the high sensitivity of the CZE-MS/MS system for BUP. The data demonstrated significantly higher protein heterogeneity across blastomeres at the 128-cell stage compared to the 16-cell stage, Figure 6A. More recently, Lombard-Banek et al. also presented multi-omics (proteomics and metabolomics) analyses of single blastomeres in early-stage Xenopus embryos with the label-free detection of 150 metabolites and 738 proteins. (Lombard-Banek et al., 2021)
Figure 6.
(A) Hierarchical-cluster-analysis heat map of quantified proteins across blastomeres from 16-cell, 32-cell, 64-cell, and 128-cell Xenopus embryos. Blastomeres from the same embryonic stage are grouped together and blastomeres from the 128-cell stage show significantly higher protein abundance heterogeneity compared to that from the 16-cell stage. Protein examples with quantifiable cell heterogeneity in the 16-cell and 128-cell blastomeres are marked with asterisks (*). Reproduced from Lombard-Banek et al. (2019) with permission from American Chemical Society, copyright (2019). (B) Illustration of nanoRPLC-CZE-MS/MS for orthogonal and high-capacity separations of peptides. Eluates from nanoRPLC are collected every several minutes, followed by dynamic pH junction-based CZE-MS/MS analysis.
To boost the proteome coverage from mass-limited samples using CZE-MS/MS, coupling nanoflow liquid-phase fractionation to CZE-MS/MS is essential. In 2018, our group developed a nanoflow RPLC (nanoRPLC)-CZE-MS/MS system via the electro-kinetically pumped sheath flow interface and the dynamic pH junction sample stacking technique, Figure 6B. (Yang et al., 2018) The dynamic pH junction-based CZE-MS/MS enabled the use of 33% of the available peptide material in each nanoRPLC fraction for measurements. The nanoRPLC and CZE provided orthogonal and high capacity separation of peptides. The nanoRPLC-CZE-MS/MS system identified 7500 proteins and 60000 peptides starting with only 5-μg MCF7 proteome digest using a Q-Exactive HF mass spectrometer. In 2019, we further improved the nanoRPLC-CZE-MS/MS system regarding sensitivity via treating sample vials with bovine serum albumin (BSA) for sample loss reduction, optimizing the nanoRPLC fraction collection, and employing fast CZE separation. (Yang et al., 2019) The new system identified 6500 proteins starting with only 500 ng of MCF7 proteome digest. Coupling an advanced sample preparation method to the improved nanoRPLC-CZE-MS/MS resulted in the identification of nearly 4000 proteins from roughly 1000 HEK293T cells. The data render nanoRPLC-CZE-MS/MS a valuable tool for highly sensitive and large-scale BUP of mass-limited samples.
3.1.2. Analysis of biopharmaceuticals
CZE-MS/MS has been proven as a useful tool for BUP analysis of mAbs. Chen et al. documented a 100% and a 96% sequence coverage for the light and heavy chains of the mAb infliximab using CZE-MS/MS via the flow-through microvial interface and dynamic pH barrage junction sample stacking with the consumption of less than 200 ng protein digest, demonstrating the capability of CZE-MS/MS for sequencing mAbs with high sensitivity. (Cheng et al., 2020) To further boost the separation of mAb digests, Kumar et al. presented offline RPLC fractionation and CZE-MS/MS for nearly complete sequence coverages (99.55% and 98.6%) of heavy and light chains of a mAb, which were significantly better than that from RPLC-MS/MS and CZE-MS/MS alone. (Kumar, Shah & Rathore, 2020) Additionally, the RPLC-CZE-MS/MS approach identified a drastically higher number of peptides containing PTMs, indicating the benefit of better peptide separation for analysis of mAbs.
CZE-MS/MS has also been deployed for BUP analysis of antibody-drug conjugates (ADCs). Fonslow et al. employed the CESI technique for CZE-MS and MS/MS analysis of ADCs, identifying three predominant ligand conjugation sites and determining their rough stoichiometries 73, 14, and 6%. (Fonslow et al., 2020) The data highlighted the value of CZE-MS/MS for analysis of ADCs. Saadé et al. presented a detailed BUP protocol for characterizing the primary structure of ADCs using CZE-MS/MS with the CESI technique. (Saadé et al., 2020) The protocol enabled characterizations of amino acid sequence, glycosylation, and conjugated drug locations on the peptide backbone of ADCs.
Besides mAbs and ADCs, CZE-MS/MS-based BUP has also been employed for the analysis of host cell proteins (HCPs). HCPs are important for the quality of a biotherapeutic product. Kumar et al. employed offline RPLC fractionation and CZE-MS/MS for the identification of HCPs in mAb producing CHO cell line. (Kumar et al., 2021) The offline RPLC-CZE-MS/MS system identified 397 HCPs from the supernatants of CHO cells and outperformed modern RPLC-MS/MS (189 HCPs) and CZE-MS/MS (128 HCPs) regarding the number of identified HCPs.
3.1.3. Analysis of disease-related biomarkers
CZE-MS/MS has been broadly utilized for discovering biomarkers of diseases, including cancer, kidney and cardiovascular disease, heart failure, and IgA nephropathy.
Frantzi et al. utilized CE-MS/MS for quantitatively measuring urinary peptides in hundreds of prostate cancer patient samples and control samples, leading to the identification of 19 peptide biomarkers for distinguishing the cancer patient and control samples. (Frantzi et al., 2019) Identifying single amino acid variants (SAAVs) is vital for understanding tumorigenesis and progression. Tan et al. employed RPLC or nanoRPLC fractionation and dynamic pH junction-based CZE-MS/MS for comprehensive BUP characterization of SAAVs in a PANC-1 cell line, resulting in the detection of 540 SAAVs, which represented the most comprehensive study of SAAVs at that time. (Tan et al., 2020) The two studies highlighted the crucial roles CZE-MS/MS could play in cancer-related research.
CZE-MS/MS was also used for studying other diseases. For example, one recent review paper summarized the recent studies using CZE-MS/MS for quantitatively measuring urinary peptides in human patient samples for discovering biomarkers of kidney and cardiovascular disease. (Latosinska et al., 2020) In one study, Pelander et al. employed CZE-MS and MS/MS for quantitatively comparing the urinary peptide abundance of dogs with and without chronic kidney disease, leading to the detection of 133 differentially excreted peptides. (Pelander et al., 2019) These peptides were used to construct predictive models of chronic kidney disease successfully. Campbell et al. utilized CZE-MS for analyzing 829 human urinary proteome samples, of which 622 samples were from heart failure patients, and 207 samples were from control. (Campbell et al., 2020) A group of detected urinary peptides (HF1) from the analyses were used for diagnosis of heart failure, resulting in comparable diagnostic performance to the B-type natriuretic peptide, which is typically recommended for the diagnosis. Rudnicki et al. analyzed 209 urine proteome samples from patients of immunoglobulin A (IgA) nephropathy (IgAN) with CZE-MS to discover urinary biomarkers for predicting rapid disease progression in IgAN. (Rudnicki et al., 2020) 237 urine peptides were identified as potential biomarkers and were used for predicting IgAN progression, producing an obvious added value for prediction compared to the typical clinical parameters. These studies indicated that CZE-MS could be a valuable tool for discovering biomarkers for disease diagnostics and progression prediction.
3.1.4. Phosphoproteomics
Protein phosphorylation is one key PTMs in cells and involves many cellular processes such as cellular signaling and differentiation. Phosphoproteomics aims to delineate all the protein phosphorylation sites in cells across various biological conditions. Multi-dimensional LC-MS/MS has identified over 50,000 phosphopeptides from a single human cancer cell line (Sharma et al., 2014) and single-shot RPLC-MS/MS has quantified over 20,000 phosphopeptides in 15 minutes (Bekker-Jensen et al., 2020). However, the approached phosphoproteome coverage is still much lower than that predicted in the human proteome (50,000 vs. half a million). Alternative phosphoproteomics approaches to LC-MS/MS will be vital for improving the phosphorylation coverage.
CZE-MS/MS has been evaluated for large-scale phosphoproteomics in the literature and it enabled the identification of over 2000 phosphopeptides from MCF-10A cell line in one run. (Ludwig et al., 2015) CZE-MS/MS outperformed RPLC-MS/MS to analyze mass-limited phosphoproteome samples regarding the number of identified phosphopeptides, and the two methods showed good complementarity for phosphopeptide IDs. More recently, Zhang et al. documented the identification of 4400 phosphopeptides from an enriched phosphopeptide sample of mouse brain using single-shot CZE-MS/MS on an Orbitrap Fusion Lumos mass spectrometer with the consumption of only 220 ng peptides, Figure 7. (Zhang et al., 2019) The data suggested the high sensitivity of CZE-MS/MS for large-scale phosphoproteomics. To boost the phosphoproteome coverage from CZE-MS/MS, our group coupled online strong cation exchange (SCX)-RPLC fractionation to CZE-MS/MS for phosphoproteomics of HCT116 cell line, resulting in the identification of over 11,000 phosphopeptides. (Chen et al., 2019) We observed that phosphopeptides migrated obviously slower than the corresponding unphosphopeptides in our experimental condition due to the charge reduction from phosphorylation. We also showed that μef of singly phosphorylated peptides were predicted with high accuracy (R2 ~0.99), which could be valuable for validating the phosphopeptide IDs. Our large-scale phosphoproteomics study further demonstrated the good complementarity of CZE-MS/MS and LC-MS/MS for phosphoproteomics in terms of phosphopeptide IDs and phosphosite motifs.
Figure 7.
Base peak electropherogram of an enriched phosphopeptide sample from a mouse brain digest analyzed by CZE-MS/MS on an Orbitrap Fusion Lumos Tribrid mass spectrometer. A 1-meter-long LPA-coated capillary (50 μm i.d.) and the electrokinetically pumped sheath flow CE-MS interface were used. About 220 ng of enriched phosphopeptides were loaded for the analysis. Reproduced from Zhang et al. (2019) with permission from American Chemical Society, copyright (2019).
3.1.5. Large-scale quantitative BUP
CZE-MS/MS has been applied to large-scale quantitative BUP using a stable isotopic labeling approach in 2015. (Faserl et al., 2015) More recently, Yan et al. coupled CZE-MS/MS with dimethyl labeling for quantitative BUP of E. coli samples, revealing minimal deuterium isotope effects in protein quantitation. (Yan et al., 2020) Due to the maturity of CZE-MS/MS technique, it has been coupled with stable isotopic labeling or label-free approach for quantitative proteomics applications in single-cell analysis and disease-related biomarker discovery as discussed in parts 3.1.1 and 3.1.3. There are another two papers related to the topic in 2019 and 2020.
Nanomaterial (NM) protein corona is used to describe the proteins adsorbed on the NMs when they contact biological or environmental fluids. The study of protein corona is important for a better understanding of NM uptake as well as impacts on cells and organisms. Faserl et al. successfully used the CESI technique-based CZE-MS/MS for comprehensive and quantitative characterization of protein corona of different nanomaterials. (Faserl et al., 2019) The CESI-MS showed a high degree of reproducibility and had higher throughput than RPLC-MS, indicating the value of CZE-MS/MS for NM protein corona analysis.
Spermatogonial stem cells have a group of undifferentiated spermatogonia during spermatogenesis, and the group of stem cells can be induced for differentiation by retinoic acid (RA). Recently, our group performed large-scale quantitative BUP of undifferentiated spermatogonia and RA-induced differentiating cells through collaborating with Dr. Yuan Wang at Michigan State University to understand the potential molecular mechanisms of the process. (Chen et al., 2020) We employed nanoRPLC-CZE-MS/MS and TMT6plex chemistry in the study. Nearly 5000 proteins were quantified, and about 500 proteins showed a statistically significant difference in abundance between the undifferentiated and differentiated spermatogonia. We observed that some key enzymes in glycolysis (e.g., HK2, ALDOA, PGK1, PKM, and LDHA) were downregulated in differentiating spermatogonia, consistent with the RNA-seq data. The data documented the power of CZE-MS/MS for deep quantitative proteomics.
3.1.6. Peptidomics
CZE-MS/MS has been frequently used for peptidomic studies for disease-related biomarker discovery, as shown in part 3.1.3. Besides that, it has also been broadly used for the characterization of peptides in basic research. (Lombard-Banek et al., 2019; DeLaney & Li, 2019; Delvaux et al., 2020; Liu et al., 2020; Ozawa et al., 2020; Lamp et al., 2020; Delvaux et al., 2021; Piestansky et al., 2020)
Delvaux et al. employed CZE-ESI-MS for the characterization of cysteine connectivity of disulfide bonds on synthetic and biologically relevant peptides via combinations with ion mobility-MS and theoretical calculations. (Delvaux et al., 2020) The data showed that CZE-MS was able to achieve well separations of disulfide isomers of each peptide studied, suggesting CZE-MS as a useful tool for studying cysteine connectivity of disulfide bonds on peptides. Lamp et al. documented CZE-ESI-MS analysis of basic peptides, i.e., Histatin-5, via the electro-kinetically pumped sheath flow CE-MS interface. (Lamp et al., 2020) Histatin-5 (Hst-5) is a human salivary peptide, and it has antibacterial and antifungal activities. It is a very basic peptide, making the analysis of this peptide and its degradation products using typical LC-MS challenging. CZE-ESI-MS achieved reproducible and quantitative measurements of Hst-5 with low carryover. CZE-MS enabled efficient separations of Hst-5 and its degradation products, which allowed a kinetic study of Hst-5 degradation by the Sap9 protease.
CZE has also been coupled to matrix-assisted laser desorption/ionization (MALDI)-MS for peptidomics. DeLaney et al. coupled CZE to MALDI-MS imaging (MSI) for comprehensive characterization of neuropeptides in brains and sinus glands (SG) of C. borealis crabs. (DeLaney & Li, 2019) The analytes in tissue extracts were separated by CZE, followed by continuous deposition onto a MALDI plate for MSI. The CZE-MSI method detected more than 200 neuropeptides in one run from the brain and SG samples and showed high complementarity to the typical LC-ESI-MS method regarding the detected neuropeptides. The data indicated that CZE-MS could be a valuable analytical tool for comprehensive peptidomics.
3.2. TDP
CE-MS has been well recognized as an emerging tool for top-down MS characterization of complex proteomes and biopharmaceuticals. CZE-MS has been mainly used for these applications, and cIEF-MS is gradually attracting attention.
3.2.1. TDP of complex proteomes
The published papers during 2019–2021 on this topic focus on advancing CE-MS-based TDP regarding sensitivity and proteome coverage, predicting μef of proteoforms, or improving proteoform characterization via employing various gas-phase fragmentation methods.
3.2.1.1. Boosting sensitivity and proteome coverage
Our group developed a highly sensitive CZE-MS/MS method for large-scale TDP of mass-limited samples in 2019, which enabled the identification of hundreds to thousands of proteoforms from complex proteome samples via consumption of tens to hundreds of nanograms of proteins. (Lubeckyj et al., 2019) The method employed a 1.5-meter-long LPA coated capillary, dynamic pH junction sample stacking method, the electro-kinetically pumped sheath flow CE-MS interface, and an Q-Exactive HF mass spectrometer. We quantitatively compared relative abundance of thousands of proteoforms between zebrafish brain cerebellum (Cb) and optic tectum (Teo) regions via consuming hundreds of nanograms of proteins per CZE-MS/MS run, revealing drastic differences in proteoform abundance between the two brain regions. The data highlighted the potency of CZE-MS/MS for TDP of mass-limited samples. We also compared the CZE-MS and nanoRPLC-MS regarding sensitivity for proteoform detection. We concluded that CZE-MS produced comparable S/N of proteoforms to nanoRPLC-MS with 10-times lower sample consumption. (McCool & Sun, 2019) More recently, we also showed that CZE-MS/MS produced comparable numbers of proteoform IDs to nanoRPLC-MS/MS for a histone sample with more than 30-fold less sample consumption. (Chen et al., 2021) The studies demonstrate the better sensitivity of CZE-MS/MS for TDP compared to the widely used nanoRPLC-MS/MS, which also agrees with one previous work. (Han et al., 2014) Additionally, coupling optimized membrane ultrafiltration-based sample preparation to dynamic pH junction-based CZE-MS/MS was demonstrated to be a simple and efficient TDP workflow for different complex proteome samples. (Yang et al., 2020)
Besides CZE-MS/MS, automated and online cIEF-MS/MS using the “sandwich” injection approach has also been employed for large-scale TDP of complex proteomes in 2020. (Xu et al., 2020) Coupling SEC to cIEF-MS/MS identified nearly 2000 proteoforms from the E. coli proteome and quantified thousands of proteoforms between zebrafish male and female brains, revealing sex-dependent proteoform profiles in brains. Interestingly, a significantly higher abundance of several proteolytic proteoforms of pro-opiomelanocortin and prodynorphin in male zebrafish brains were observed, and they could be endogenous hormone proteoforms. The work represents the first large-scale TDP study using cIEF-MS/MS and the first example of studying sexual dimorphism of the brain using TDP.
3.2.1.2. Predicting electrophoretic mobility of proteoforms
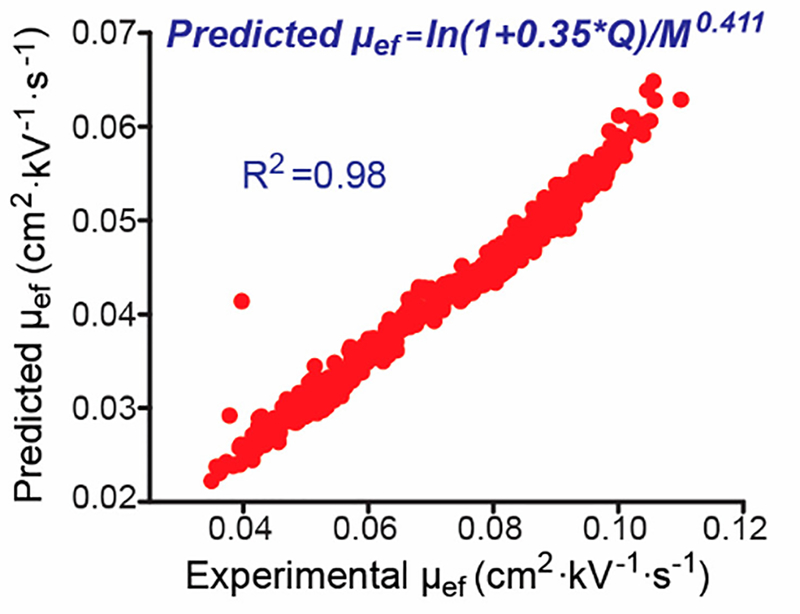
Because CZE separation of proteoforms is simply based on their charge-to-size ratios, it should be relatively easy to predict their μef, which could be useful for further validating the confidence of identified proteoforms from the database search. In 2020, our group showed that the μef of proteoforms without PTMs from an E. coli sample was predicted accurately (R2=0.98) with a simple semiempirical model containing two parameters (Q, charge; M: mass) based on previous peptide works, Figure 8. (Chen et al., 2020) Q was easily obtained by counting the basic amino acid residues (K, R and H) and N-terminus of one proteoform; M was determined by the MS measurement. For proteoforms with single phosphorylation and/or N-terminal acetylation, by simply decreasing the Q by 1 charge unit for one PTM or 2 for both PTMs, the predicted and experimental μef of proteoforms with these PTMs had a good linear correlation (R2=0.92). The μef prediction model with simple modifications also showed good performance for histone proteoforms without PTMs, producing a linear correlation between predicted and experimental μef of those proteoforms (R2=0.98). (Chen et al., 2021) More studies about predicting μef of proteoforms with various PTMs, especially histone proteoforms, will be useful for understanding how PTMs influence μef of proteoforms and employing the predicted μef to evaluate the confidence of proteoform IDs.
Figure 8.
Linear correlation between predicted electrophoretic mobility (μef) and experimental μef of proteoforms without PTMs from E. coli cells identified by CZE-MS/MS. The μef prediction is based on the equation labeled in the figure and the predicted μef relates to the charge (Q) and mass (M) of each proteoform. Reproduced from Chen et al. (2020) with permission from American Chemical Society, copyright (2020).
3.2.1.3. Employing various gas-phase fragmentation methods
Higher-energy collisional dissociation (HCD) is the most widely used approach for fragmentation of proteoforms in TDP. However, it has preferred cleavage sites, leading to challenges for the extensive backbone cleavages of proteoforms using HCD. During the last two years, several other gas-phase fragmentation methods, e.g., electron-capture collision-induced dissociation (ECciD), activated ion-electron-transfer dissociation (AI-ETD), ultraviolet photodissociation (UVPD), and a combination of ETD and HCD (EThcD), were implemented in the CZE-MS/MS-based TDP workflow to achieve more extensive proteoform fragmentation.
In 2019, our group coupled UVPD (213 nm) and AI-ETD to CZE-MS/MS for large-scale TDP of zebrafish brain and E. coli samples, respectively. (McCool et al., 2019; McCool et al., 2019) CZE-UVPD identified 600 proteoforms and 369 proteins from a zebrafish brain sample with the assistance of SEC fractionation and achieved a 75% backbone cleavage coverage for Parvalbumin-7, a 12-kDa protein. (McCool et al., 2019) CZE-AI-ETD produced a higher quality of MS/MS spectra than CZE-HCD and CZE-ETD regarding the number of sequence-informative fragment ions. (McCool et al., 2019) SEC-CZE-AI-ETD yielded over 3000 proteoform IDs from E. coli cells, representing the largest TDP dataset using the AI-ETD method at that time. Very recently, our group developed a novel CZE-MS/MS method via ECciD for intact protein separation and extensive gas-phase fragmentation using an Agilent Q-TOF mass spectrometer. (Shen et al., 2021) The protein positive ions were first cleaved by ECD and further activated/fragmented by CID. The system achieved baseline and reproducible separations of a standard protein mixture and gained about 90% or higher backbone cleavage coverages for lower than 20 kDa proteins (e.g., myoglobin). The results showed the high potential of the CZE-ECciD system for advancing TDP regarding the separation and fragmentation of proteoforms. However, more studies need to be done about improving the CZE-ECciD for large proteoforms (>30 kDa).
In 2020, Gomes et al. applied CZE-MS/MS via the CESI technique with EThcD and UVPD (213 nm) for large-scale TDP of bovine seminal plasma. (Gomes et al., 2020) CZE-MS/MS identified 417 proteoforms from the sample. Combination of CZE-MS/MS and nanoRPLC-MS/MS data from triplicate measurements led to the identification of over 1400 proteoforms (EThcD) and over 2000 proteoforms (213 nm UVPD). The average backbone cleavage coverages of proteoforms from EThcD were higher than that from UVPD (213 nm) (28% vs. 23%).
3.2.2. Analysis of Biopharmaceuticals
CZE-MS and cIEF-MS have been deployed for achieving comprehensive characterization of antibodies and other therapeutic proteins regarding charge variants and PTMs. We highlight some of the most recent advances on this topic here.
3.2.2.1. CZE-MS
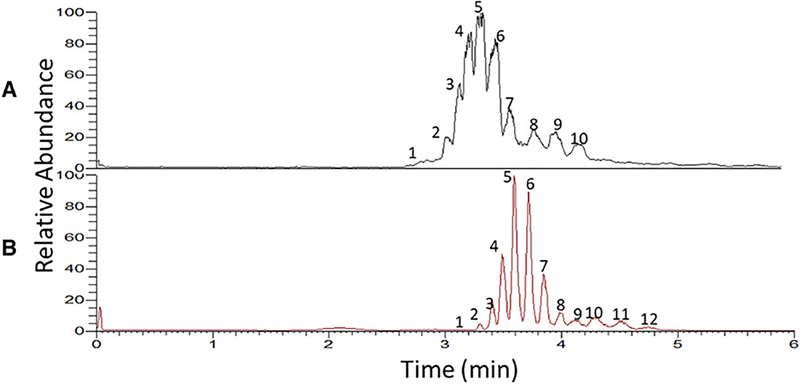
Giorgetti et al. documented the delineation of seven mAbs using CESI technique-based CZE-MS and MS/MS at the intact protein and peptide levels. (Giorgetti et al., 2020) The CZE-MS system allowed the analysis of mAb charge variants and PTMs (i.e., N-glycosylation, K-clip, oxidations or deamidations) within 12 minutes. In another study, Füssl et al. employed the ZipChip-based microfluidic CE-MS system for the analysis of Cetuximab proteoforms. (Füssl et al., 2020) The platform achieved baseline separations of eight different charge variants and detected over 200 Cetuximab proteoforms within 14 minutes. Deyanova et al. also developed a CZE-MS method based on the ZipChip technique for fast and efficient characterization of a highly glycosylated fusion protein, resulting in baseline separations of 12 peaks corresponding to different proteoforms in 6 minutes, Figure 9. (Deyanova et al., 2021) These studies demonstrate the power of CZE-MS for high-throughput and comprehensive characterization of therapeutic proteins.
Figure 9.
CE-MS electropherograms of a fusion protein analyzed by the ZipChip technique with commercially available separation buffer (A), and the optimized separation buffer containing 10% 2-propanol and 0.2% acetic acid (pH 3.2) (B). Reproduced from Deyanova et al. (2021) with permission from John Wiley and Sons, copyright (2021).
Besides the analysis of mAbs, CZE-MS has also been employed for the characterization of bispecific antibodies (BsAbs). Gstöttner et al. showed that CZE-MS using the CESI technique allowed the detailed analysis of free light chains, homo-dimers, heterodimers, and incomplete assemblies of BsAbs. (Gstöttner et al., 2020) With the assistance of hinge region cleavages using enzymes and disulfide-bond reduction, the system separated six different subunits of the BsAbs. More recently, the authors further successfully applied the CZE-MS system to monitor exchange efficiency and stability of in-house produced BsAbs. (Gstöttner et al., 2021)
3.2.2.2. cIEF-MS
cIEF-UV is widely used for monitoring qualities of therapeutic proteins (e.g., mAbs) in industry. Coupling cIEF to MS is a promising approach for informative and comprehensive measurements of therapeutic proteins. Dai et al. developed an automated cIEF-MS method using the “sandwich” injection method based on the electrokinetically pumped sheath flow interface for efficient separations and characterization of charge variants of various mAbs. (Dai et al., 2018) The separation results agreed well with that from cIEF-UV analysis. Dai et al. further applied the automated cIEF-MS method to delineate complex mAb charge variants (i.e., cetuximab) in a middle-up way, resulting in the identification of at least eight different charge variants of cetuximab. (Dai & Zhang, 2018) Wang et al. documented TDP characterization of mAb charge variants using an automated cIEF-MS system based on the flow through microvial interface. (Wang et al., 2018; Wang & Chen, 2019) Four charge variants with 0.05–0.2 pI differences and 13 glycoforms were resolved by consuming only 30 ng of infliximab. Microchip-based cIEF-MS platforms have also been developed for mAb charge variants. Mack et al. developed an automated microchip-based cIEF-MS system for real-time optical monitoring of focusing and mobilization process of cIEF as well as resolving mAb charge variants with high resolution and high throughput (15 minutes each assay). (Mack et al., 2019) Besides a direct coupling of cIEF and MS, cIEF was also coupled to CZE-MS through a nanoliter valve for the characterization of mAb charge variants. (Montealegre & Neusüß, 2018) These studies clearly showed the power of automated cIEF-MS for the delineation of therapeutic mAbs.
3.2.3. Characterization of disease-related protein biomarkers
Detailed and comprehensive analyses of disease-related protein biomarkers are essential for achieving better disease diagnostics and drug development. CE-MS has been proven as a valuable tool for this purpose due to its high resolution and high sensitivity for proteoform separation and detection.
Nyssen et al. developed an efficient and sensitive CZE-MS method based on the CESI technique for the characterization of parathyroid hormone (PTH), which is a common clinical marker. (Nyssen et al., 2019) By using the EKS sample stacking method, the intact and two truncated proteoforms of PTH were resolved and detected at concentrations in a pg/mL range. Pero-Gascon et al. developed an aptamer-based SPME-CZE-MS system via the co-axial sheath flow CE-MS interface for highly selective concentration and highly sensitive characterization of α-Synuclein in blood. (Pero-Gascon et al., 2020) Proteoforms of α-Synuclein have been recognized as potential biomarkers for diagnosis and progression monitoring of Parkinson’s disease (PD). The optimized SPME-CZE-MS system achieved reproducible and quantitative measurements of the α-Synuclein with a LOD of 0.2 μg/mL, which was 100 times lower than the CZE-MS without SPME. The method was also successfully employed for analyses of human samples (healthy controls vs. PD patients). In another study, Stolz et al. developed a powerful CZE-MS and MS/MS method for comprehensive characterization of hemoglobin (Hb) proteoforms due to sequence variants and PTMs extracted from dried blood spot (DBS) samples. (Stolz et al., 2020) Hb proteforoms serve as important biomarkers for diseases, e.g., diabetes and kidney diseases. The CZE-MS method enabled the separations of positional isomers of glycated α- and β-chains of Hb. Very importantly, the quantification data of glycated Hb from CZE-MS agreed well with the data from a clinical routine method. The CZE-MS method was also applied to analyze the dog and cat DBS samples with the discovery of a potentially new sequence variant of the β-chain of Hb in dog (T38 → A). Tie et al. utilized an automated cIEF-MS system for characterizing urinary albumin proteoforms from the membranous nephropathy (MN) patients. (Tie et al., 2020) Distinct patterns of urinary albumin proteoforms from the primary and secondary MN samples were observed, indicating the potential of the cIEF-MS technique for separating different subtypes of MN.
Compared to the traditionally used RPLC-MS, CZE-MS and cIEF-MS have advantages for the top-down MS characterization of protein biomarkers, including better separation resolution (CZE and cIEF) and higher detection sensitivity (CZE) for proteoforms, especially large proteoforms. The accurate prediction of proteoforms’ μef in CZE and the ability of cIEF for accurately determining the pIs of proteoforms are also useful for delineation of protein biomarkers. However, the low sample loading capacity of CZE-MS and the significant signal suppression of proteoforms due to carrier ampholyte in cIEF-MS may limit their performance for the analysis of low-concentration protein biomarker samples and for discovering new and low-abundance protein biomarkers from complex samples.
3.3. Native proteomics
Native proteomics directly characterizes protein complexes and requires high-resolution liquid-phase separation, highly sensitive MS measurement, and extensive gas-phase fragmentation of protein complexes. CZE-MS and MS/MS is a promising tool for native proteomics because CZE can separate protein complexes with high efficiency under native conditions.
The first two reports about CZE-MS/MS for native proteomics were published in 2017 and 2018. (Belov et al., 2017; Shen et al., 2018) Belov et al. documented CZE-MS for highly sensitive characterization of standard protein complexes and a ribosomal isolate from E. coli using the CESI technique. (Belov et al., 2017) Our group developed a native SEC-CZE-MS/MS platform for native proteomics of endogenous protein complexes in E. coli cells via the electrokinetically pumped sheath flow CE-MS interface. (Shen et al., 2018) The platform identified 23 protein complexes in discovery mode.
During the last two years, eight papers on this topic were published and those papers focus on the characterization of mAbs, protein structure and conformation, and large protein complexes.
3.3.1. Analysis of mAbs under native conditions
Le-Minh et al. built a native CZE-MS method via the co-axial sheath flow CE-MS interface for the characterization of conformational heterogeneity and self-association of Infliximab. (Le-Minh et al., 2019) The method detected not only the native and unfolded monomers of Infliximab but also the dimers. Very recently, our group developed a native cIEF-assisted CZE-MS system via a new carbohydrate-based neutral coating, the electrokinetically pumped sheathflow interface, and a quadrupole-time-of-flight (Q-TOF) mass spectrometer. (Shen et al., 2021) The system detected various glyco-proteoforms and homodimer of SigmaMAb. It also resolved eight glyco-proteoforms of NIST mAb at the monomer and dimer levels with a submicroliter sample loading volume. The native cIEF-assisted CZE-MS method showed its capability for high-resolution separation of proteins and protein complexes under native conditions with large sample loading capacity, which is crucial for native proteomics of complex proteomes.
3.3.2. Characterization of protein structure and conformation
Two research groups built novel analytical tools based on native CZE or CZE-MS for studying protein structure and conformation. (Zhang et al., 2019; He et al., 2019; Shen et al., 2019) The Xu group developed a novel mobility capillary electrophoresis (MCE) method for analysis of protein stereo-structures and charges in solution environments through Taylor dispersion and ion mobility analyses. (Zhang et al., 2019) The MCE method was successfully applied to the analysis of charge states and structures of five standard proteins under close to native conditions. They further coupled the MCE method to MS for analyzing structures of proteins and peptides in the liquid phase. (He et al., 2019) The Chen group coupled hydrogen/deuterium exchange (HDX) with native CZE-MS and MS/MS for differential HDX and online separation of different conformers of proteins, followed by MS/MS of each conformer for the identification of protein segments, which underwent conformational changes. (Shen et al., 2019) Using the method for analysis of myoglobin with and without the heme group, the segments of myoglobin undergoing conformational changes in the absence of the heme group were determined.
3.3.3. Analysis of large protein complexes
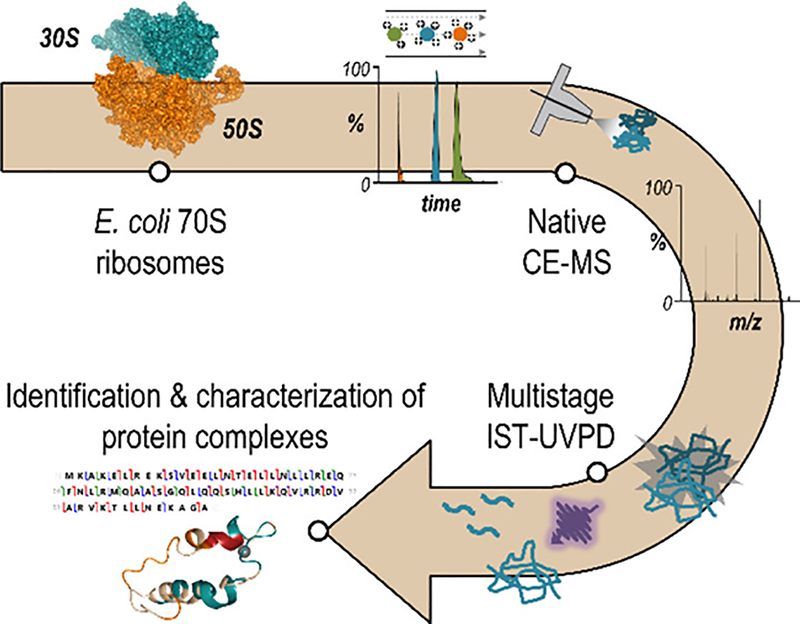
The native CZE-MS/MS studies had focused on relatively small protein complexes. Recently, Mehaffey et al. demonstrated the efficient separation and characterization of E. coli ribosomal protein complexes (30S, 850kDa; 50S: 1450 kDa) using native CZE-MS and UVPD, Figure 10. (Mehaffey, Xia & Brodbelt, 2020) The commercialized electrokinetically pumped sheath flow CE-MS interface (EMASS-II) was used for coupling CE to a Q Exactive UHMR mass spectrometer. By employing front-end collisional activation for disassembling protein complexes into subunits for fragmentation using HCD or UVPD, the system identified 84 proteoforms of 48 known E. coli ribosomal proteins. In another study, Jooß et al. developed a procedure for using native CZE-MS and MS/MS via the CESI technique for the characterization of standard protein complexes with masses up to 800 kDa on three high-end Orbitrap mass spectrometers (Q Exactive-EMR, Q Exactive UHMR, and Orbitrap Eclipse). (Jooß et al., 2021) By adding energies at the front-end of the mass spectrometer for monomer ejection from a large protein complex (i.e., GroEL, ∼800 kDa), five unique proteoforms of the monomer were resolved and four of them were fully characterized by MS/MS. The two studies clearly demonstrate the potency of native CZE-MS and MS/MS for comprehensive characterization of 1 million Da level protein complexes.
Figure 10.
Schematic of the native CZE-MS and MS/MS using UVPD for delineation of E. coli 70S ribosomes. The commercialized electrokinetically pumped sheath flow CE-MS interface (EMASS-II) was used for coupling native CZE to MS. Reproduced from Mehaffey et al. (2020) with permission from American Chemical Society, copyright (2020).
4. Conclusions and Perspectives
CE-MS and MS/MS has shown potency for advancing multi-level proteomics for comprehensive characterization of proteins at the peptide, proteoform, and protein complex levels because of drastic technical progress of CE-MS regarding the CE-MS interface, capillary coating, online sample preconcentration methods, automated operation, and mass spectrometer. CE-MS and MS/MS has been applied in many biological research areas, including but not limited to proteomics of mass-limited samples (i.e., single cells), characterization of biopharmaceuticals (i.e., mAbs), biomarker discovery, and large-scale qualitative and quantitative multi-level proteomics of complex samples.
CE-MS and MS/MS will be extremely useful for at least five research areas. First, CZE-MS/MS will play an essential role in multi-omics (i.e., proteomics and metabolomics) analysis of mass-limited biological samples (i.e., single cells) demonstrated by recent publications. (Lombard-Banek et al., 2021; Huang et al., 2020) However, the widespread adoption of CZE-MS/MS for this research area needs some technical improvements to allow the system to fully use the limited samples for measurements and to further boost the peak capacity for separations. Coupling nanoRPLC fractionation to CZE-MS/MS with highly efficient sample concentration techniques (e.g., dynamic pH junction and SPME) could be a good solution for large-scale multi-omics analyses of mass-limited samples as demonstrated by recent studies. (Yang et al., 2018; Yang et al., 2019) Second, coupling LC to CE-MS/MS will be a useful analytical tool for qualitative and quantitative TDP of complex proteomes with deep proteome coverage for pursuing a better understanding of fundamental biological processes and molecular mechanisms of disease development. Several recent studies have proven this point. (Gomes et al., 2020; Lubeckyj et al., 2019; McCool et al., 2018; Xu et al., 2020) However, the proteome-scale characterization of large proteoforms (>30 kDa) in complex samples is still challenging for TDP. The current MS-based TDP methods need drastic improvement regarding detection sensitivity and backbone cleavage coverage of those large proteoforms. Coupling extensive LC-based intact proteoform fractionation to CZE-MS-based BUP and top-down MS analyses of each LC fractions has been proven as an alternative and effective approach for bettering the delineation of large proteoforms. (Zhu et al., 2003) Third, CE-MS, especially cIEF-MS, for characterization of biopharmaceuticals (i.e., mAbs and ADCs) will still be a hot research area as demonstrated by most recent studies. (Dai et al., 2018; Wang & Chen, 2019; Mack et al., 2019; Füssl et al., 2020) Fourth, CZE-MS and MS/MS for native proteomics will be a new research frontier in proteomics for global and detailed characterization of endogenous protein complexes in cells. However, some technical improvement, e.g., boosting sample loading capacity of native CZE-MS and improving capillary coatings for native CZE separations of large protein complexes, need to be achieved before the routine use of CZE-MS/MS for native proteomics. Lastly, CZE-MS and MS/MS could be a powerful tool for global multi-omics and multi-level proteomics of cells because CZE can achieve highly efficient separations of metabolites, nucleotides, glycans, peptides, proteoforms, and protein complexes. The unique feature of CZE allows CZE-MS/MS for comprehensive analyses of biomolecules in cells at different levels, which is crucial for an accurate understanding of cellular processes.
Acknowledgements
We thank the support from the National Institute of General Medical Sciences (NIGMS) through Grants R01GM125991 and 2R01GM118470. We also thank the support from National Cancer Institute through Grant R01CA247863 and the National Science Foundation through Grant DBI1846913 (CAREER Award).
Abbreviations
- AA
acetic acid
- ADC
antibody-drug conjugate
- AI-ETD
activated ion-electron transfer dissociation
- BSA
bovine serum albumin
- BsAbs
bispecific antibodies
- BUP
bottom-up proteomics
- cIEF
capillary isoelectric focusing
- CZE
capillary zone electrophoresis
- ECciD
electron-capture collision-induced dissociation
- EKS
electrokinetic supercharging
- EOF
electroosmotic flow
- ESI
electrospray ionization
- FA
formic acid
- HCD
higher-energy collisional dissociation
- HCP
host cell protein
- HDX
hydrogen/deuterium exchange
- i.d.
inner diameter
- kDa
kilodalton
- LCP
linear carbohydrate polymer
- LOD
limit of detection
- LPA
linear polyacrylamide
- mAbs
monoclonal antibodies
- MALDI
matrix-assisted laser desorption/ionization
- MCE
mobility capillary electrophoresis
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- MSI
MALDI-MS imaging
- nanoRPLC
nanoflow RPLC
- PS-tCITP
polarity switching transient isotachophoresis
- PTMs
post-translational modifications
- Q-TOF
quadrupole-time-of-flight
- RPLC
reversed-phase liquid chromatography
- SAAV
single amino acid variant
- SEC
size exclusion chromatography
- S/N
signal-to-noise ratio
- SPME
solid-phase microextraction
- TDP
top-down proteomics
- μef
electrophoretic mobility
- UVPD
ultraviolet photodissociation
Biography

Daoyang Chen obtained his Master’s degree in Pharmaceutical Science from the Northeastern University in 2016. Later, he received his Ph.D. degree in Analytical Chemistry from Michigan State University in 2021, advised by Prof. Liangliang Sun. His work focuses on capillary electrophoresis-mass spectrometry method development for bottom-up and top-down proteomics.

Elijah McCool is a PhD candidate in the Chemistry Department, Michigan State University. He received his Bachelor of Science in Chemistry from The University of North Carolina at Chapel Hill. His training is in analytical chemistry and is in his fifth year in Dr. Liangliang Sun’s research group. His current project focuses on coupling capillary zone electrophoresis to mass spectrometry for separation, identification, and characterization of intact proteins in complex proteomes. Direct characterization of intact proteoforms using this platform allows for elucidation of protein-level genetic variation and proteoform specific post-translational modifications that are essential to important biological processes.

Zhichang Yang is PhD student of chemistry at Michigan State University (MSU). He received his B.S. degree in Nankai University and M.S. degree in North Carolina State University. He joined Dr. Liangliang Sun’s research group at MSU in 2017. Since then, he has been focusing on research of developing ultrasensitive platforms for the characterization of proteins in mass-limited samples.

Xiaojing Shen received her Ph.D. degree in Chemistry in 2020 from the Department of Chemistry at Michigan State University under the supervision of Professor Liangliang Sun. Before that, she received her B.S. degree in Chemistry from Nanjing University, China in 2015. She is working for Bio-Techne Corporation as a research scientist currently. Her research project focused on investigating online capillary zone electrophoresis (CZE)-tandem mass spectrometry (MS/MS) for denaturing and native top-down proteomics through improving separation and fragmentation of proteoforms and protein complexes.

Rachele Lubeckyj is a post-doctoral scientist at Zoetis, Inc. Rachele works on multilevel proteomics for characterizing glycohormones and monoclonal antibodies. She received her Ph.D. degree in Analytical Chemistry in 2021 from Michigan State University, advised by Dr. Liangliang Sun. Before joining Zoetis, Rachele’s research primarily focused on using capillary zone electrophoresis-tandem mass spectrometry for highly sensitive top-down proteomics.

Tian Xu is a Ph.D. candidate in the Department of Chemistry at Michigan State University. She obtained her B.S. degree at Minzu University of China and completed M.S. degree at Chinese Academy of Sciences. In 2018, she joined Dr. Liangliang Sun’s lab. Her research work focuses on developing automated capillary isoelectric focusing-mass spectrometry for top-down proteomics.

Qianjie Wang is a Ph.D. student majoring in Chemistry at MSU co-mentored by Dr. Liangliang Sun in Chemistry and Dr. Peter Lundquist in BMB. She graduated from Nanjing University with a Bachelor’s degree in Chemistry working with Dr. Ying Liu. Her current research focused in combining top-down and bottom-up proteomics to study the plastoglobules (lipoprotein particles in chloroplast) proteome and remobilization under different stress.

Liangliang Sun is an Assistant Professor of Chemistry at Michigan State University (MSU). He received his Ph.D. degree in Analytical Chemistry in 2011 from Dalian Institute of Chemical Physics, Chinese Academy of Sciences, advised by Profs. Yukui Zhang and Lihua Zhang. Before joining MSU, he worked with Prof. Norman Dovichi at University of Notre Dame as a postdoctoral fellow and later a Research Assistant Professor (2011–2016). His research lab focuses on developing novel capillary electrophoresis-mass spectrometry methodologies for multi-level proteomics to characterize proteins at the peptide, proteoform, and protein complex levels globally with high throughput, high sensitivity, and single-cell resolution.
Footnotes
Conflict of interest
The authors declared no conflict of interest related to this work.
References
- Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–55. [DOI] [PubMed] [Google Scholar]
- Akashi S, Suzuki K, Arai A, Yamada N, Suzuki E, Hirayama K, Nakamura S, Nishimura Y. Top-down analysis of basic proteins by microchip capillary electrophoresis mass spectrometry. Rapid Commun. Mass Spectrom. 2006;20:1932–8. [DOI] [PubMed] [Google Scholar]
- Amenson-Lamar EA, Sun L, Zhang Z, Bohn PW, Dovichi NJ. Detection of 1 zmol injection of angiotensin using capillary zone electrophoresis coupled to a Q-Exactive HF mass spectrometer with an electrokinetically pumped sheath-flow electrospray interface. Talanta. 2019;204:70–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen DB, Bernhardt OM, Hogrebe A, Martinez-Val A, Verbeke L, Gandhi T, Kelstrup CD, Reiter L, Olsen JV. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 2020;11:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov AM, Viner R, Santos MR, Horn DM, Bern M, Karger BL, Ivanov AR. Analysis of Proteins, Protein Complexes, and Organellar Proteomes Using Sheathless Capillary Zone Electrophoresis - Native Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017;28:2614–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz-McKibbin P, Chen DD. Selective focusing of catecholamines and weakly acidic compounds by capillary electrophoresis using a dynamic pH junction. Anal. Chem. 2000;72:1242–52. [DOI] [PubMed] [Google Scholar]
- Busnel JM, Schoenmaker B, Ramautar R, Carrasco-Pancorbo A, Ratnayake C, Feitelson JS, Chapman JD, Deelder AM, Mayboroda OA. High capacity capillary electrophoresis-electrospray ionization mass spectrometry: coupling a porous sheathless interface with transient-isotachophoresis. Anal. Chem. 2010;82(22):9476–83. [DOI] [PubMed] [Google Scholar]
- Campbell RT, Jasilek A, Mischak H, Nkuipou-Kenfack E, Latosinska A, Welsh PI, Jackson CE, Cannon J, McConnachie A, Delles C, McMurray JJV. The novel urinary proteomic classifier HF1 has similar diagnostic and prognostic utility to BNP in heart failure. ESC Heart Fail. 2020;7:1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Lubeckyj RA, Yang Z, McCool EN, Shen X, Wang Q, Xu T, Sun L. Predicting Electrophoretic Mobility of Proteoforms for Large-Scale Top-Down Proteomics. Anal. Chem. 2020;92:3503–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ludwig KR, Krokhin OV, Spicer V, Yang Z, Shen X, Hummon AB, Sun L. Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Large-Scale Phosphoproteomics with the Production of over 11,000 Phosphopeptides from the Colon Carcinoma HCT116 Cell Line. Anal. Chem. 2019;91:2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Shen X, Sun L. Capillary zone electrophoresis-mass spectrometry with microliter-scale loading capacity, 140 min separation window and high peak capacity for bottom-up proteomics. Analyst. 2017;142(12):2118–2127. [DOI] [PubMed] [Google Scholar]
- Chen D, Shen X, Sun L. Strong cation exchange-reversed phase liquid chromatography-capillary zone electrophoresis-tandem mass spectrometry platform with high peak capacity for deep bottom-up proteomics. Anal. Chim. Acta. 2018;1012:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Yang Z, Shen X, Sun L. Capillary Zone Electrophoresis-Tandem Mass Spectrometry As an Alternative to Liquid Chromatography-Tandem Mass Spectrometry for Top-down Proteomics of Histones. Anal. Chem. 2021;93(10):4417–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang Z, Chang C, Yang Z, Wang P, Fu H, Wei X, Chen E, Tan S, Huang W, Sun L, Ni T, Yang Y, Wang Y. A bioenergetic shift is required for spermatogonial differentiation. Cell Discov. 2020;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Wang L, Rive CM, Holt RA, Morin GB, Chen DDY. Complementary Methods for de Novo Monoclonal Antibody Sequencing to Achieve Complete Sequence Coverage. J. Proteome Res. 2020;19:2700–2707. [DOI] [PubMed] [Google Scholar]
- Choi SB, Zamarbide M, Manzini MC, Nemes P. Tapered-Tip Capillary Electrophoresis Nano-Electrospray Ionization Mass Spectrometry for Ultrasensitive Proteomics: the Mouse Cortex. J. Am. Soc. Mass Spectrom. 2017;28:597–607. [DOI] [PubMed] [Google Scholar]
- Dai J, Lamp J, Xia Q, Zhang Y. Capillary Isoelectric Focusing-Mass Spectrometry Method for the Separation and Online Characterization of Intact Monoclonal Antibody Charge Variants. Anal. Chem. 2018;90:2246–2254. [DOI] [PubMed] [Google Scholar]
- Dai J, Zhang Y. A Middle-Up Approach with Online Capillary Isoelectric Focusing/Mass Spectrometry for In-Depth Characterization of Cetuximab Charge Heterogeneity. Anal. Chem. 2018;90:14527–14534. [DOI] [PubMed] [Google Scholar]
- Dai Y, Shortreed MR, Scalf M, Frey BL, Cesnik AJ, Solntsev S, Schaffer LV, Smith LM. Elucidating Escherichia coli Proteoform Families Using Intact-Mass Proteomics and a Global PTM Discovery Database. J. Proteome Res. 2017;16:4156–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Li L. Capillary electrophoresis coupled to MALDI mass spectrometry imaging with large volume sample stacking injection for improved coverage of C. borealis neuropeptidome. Analyst. 2019;145(1):61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvaux C, Dauvin M, Boulanger M, Quinton L, Mengin-Lecreulx D, Joris B, Pauw E, Far J. Use of Capillary Zone Electrophoresis Coupled to Electrospray Mass Spectrometry for the Detection and Absolute Quantitation of Peptidoglycan-Derived Peptides in Bacterial Cytoplasmic Extracts. Anal. Chem. 2021;93(4):2342–2350. [DOI] [PubMed] [Google Scholar]
- Delvaux C, Massonnet P, Kune C, Haler JRN, Upert G, Mourier G, Gilles N, Quinton L, De Pauw E, Far J. Combination of Capillary Zone Electrophoresis-Mass Spectrometry, Ion Mobility-Mass Spectrometry, and Theoretical Calculations for Cysteine Connectivity Identification in Peptides Bearing Two Intramolecular Disulfide Bonds. Anal. Chem. 2020;92(3):2425–2434. [DOI] [PubMed] [Google Scholar]
- Deyanova EG, Huang RY, Madia PA, Nandi P, Gudmundsson O, Chen G. Rapid fingerprinting of a highly glycosylated fusion protein by microfluidic chip-based capillary electrophoresis-mass spectrometry. Electrophoresis. 2021;42:460–464. [DOI] [PubMed] [Google Scholar]
- Faserl K, Chetwynd AJ, Lynch I, Thorn JA, Lindner HH. Corona Isolation Method Matters: Capillary Electrophoresis Mass Spectrometry Based Comparison of Protein Corona Compositions Following On-Particle versus In-Solution or In-Gel Digestion. Nanomaterials (Basel). 2019;9:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faserl K, Kremser L, Müller M, Teis D, Lindner HH. Quantitative proteomics using ultralow flow capillary electrophoresis-mass spectrometry. Anal. Chem. 2015;87:4633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faserl K, Sarg B, Kremser L, Lindner H. Optimization and evaluation of a sheathless capillary electrophoresis-electrospray ionization mass spectrometry platform for peptide analysis: comparison to liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 2011;83:7297–305. [DOI] [PubMed] [Google Scholar]
- Fonslow B, Jarvas G, Szigeti M, Guttman A. Multilevel Characterization of Antibody-Ligand Conjugates by CESI-MS. Curr. Mol. Med. 2020;20:789–797. [DOI] [PubMed] [Google Scholar]
- Frantzi M, Gomez Gomez E, Blanca Pedregosa A, Valero Rosa J, Latosinska A, Culig Z, Merseburger AS, Luque RM, Requena Tapia MJ, Mischak H, Carrasco Valiente J. CE-MS-based urinary biomarkers to distinguish non-significant from significant prostate cancer. Br. J. Cancer. 2019;120:1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füssl F, Trappe A, Carillo S, Jakes C, Bones J. Comparative Elucidation of Cetuximab Heterogeneity on the Intact Protein Level by Cation Exchange Chromatography and Capillary Electrophoresis Coupled to Mass Spectrometry. Anal. Chem. 2020;92:5431–5438. [DOI] [PubMed] [Google Scholar]
- Giorgetti J, Beck A, Leize-Wagner E, François YN. Combination of intact, middle-up and bottom-up levels to characterize 7 therapeutic monoclonal antibodies by capillary electrophoresis - Mass spectrometry. J. Pharm. Biomed. Anal. 2020;182:113107. [DOI] [PubMed] [Google Scholar]
- Gomes FP, Diedrich JK, Saviola AJ, Memili E, Moura AA, Yates JR 3rd. EThcD and 213 nm UVPD for Top-Down Analysis of Bovine Seminal Plasma Proteoforms on Electrophoretic and Chromatographic Time Frames. Anal. Chem. 2020;92:2979–2987. [DOI] [PubMed] [Google Scholar]
- Gomes FP, Yates JR 3rd. Recent trends of capillary electrophoresis-mass spectrometry in proteomics research. Mass Spectrom. Rev. 2019;38:445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou MJ, Nys G, Cobraiville G, Demelenne A, Servais AC, Fillet M. Hyphenation of capillary zone electrophoresis with mass spectrometry for proteomic analysis: Optimization and comparison of two coupling interfaces. J. Chromatogr. A. 2020;1618:460873. [DOI] [PubMed] [Google Scholar]
- Gstöttner C, Nicolardi S, Haberger M, Reusch D, Wuhrer M, Domínguez-Vega E. Intact and subunit-specific analysis of bispecific antibodies by sheathless CE-MS. Anal. Chim. Acta. 2020;1134:18–27. [DOI] [PubMed] [Google Scholar]
- Gstöttner C, Vergoossen DLE, Wuhrer M, Huijbers MGM, Domínguez-Vega E. Sheathless CE-MS as a tool for monitoring exchange efficiency and stability of bispecific antibodies. Electrophoresis. 2021;42:171–176. [DOI] [PubMed] [Google Scholar]
- Guo X, Fillmore TL, Gao Y, Tang K. Capillary Electrophoresis-Nanoelectrospray Ionization-Selected Reaction Monitoring Mass Spectrometry via a True Sheathless Metal-Coated Emitter Interface for Robust and High-Sensitivity Sample Quantification. Anal. Chem. 2016;88:4418–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang Y, Aslanian A, Bern M, Lavallée-Adam M, Yates JR 3rd. Sheathless capillary electrophoresis-tandem mass spectrometry for top-down characterization of Pyrococcus furiosus proteins on a proteome scale. Anal. Chem. 2014;86:11006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang Y, Aslanian A, Fonslow B, Graczyk B, Davis TN, Yates JR 3rd. In-line separation by capillary electrophoresis prior to analysis by top-down mass spectrometry enables sensitive characterization of protein complexes. J. Proteome Res. 2014;13:6078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselberg R, de Jong GJ, Somsen GW. Capillary electrophoresis-mass spectrometry for the analysis of intact proteins 2007–2010. Electrophoresis. 2011;32:66–82. [DOI] [PubMed] [Google Scholar]
- He M, Luo P, Hong J, Wang X, Wu H, Zhang R, Qu F, Xiang Y, Xu W. Structural Analysis of Biomolecules through a Combination of Mobility Capillary Electrophoresis and Mass Spectrometry. ACS Omega. 2019;4:2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk AA, Deelder AM, Mayboroda OA. CE-ESI-MS for bottom-up proteomics: Advances in separation, interfacing and applications. Mass Spectrom. Rev. 2016;35:259–71. [DOI] [PubMed] [Google Scholar]
- Höcker O, Knierman M, Meixner J, Neusüß C. Two capillary approach for a multifunctional nanoflow sheath liquid interface for capillary electrophoresis-mass spectrometry. Electrophoresis. 2021;42:369–373. [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang B, Weng Y, Sui Z, Zhao B, Chen Y, Liu L, Wu Q, Liang Z, Zhang L, Zhang Y. Bis(zinc(II)-dipicolylamine)-functionalized sub-2 μm core-shell microspheres for the analysis of N-phosphoproteome. Nat. Commun. 2020;11:6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wang Z, Cupp-Sutton KA, Smith K, Wu S. Spray-Capillary: An Electrospray-Assisted Device for Quantitative Ultralow-Volume Sample Handling. Anal. Chem. 2020;92:640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hühner J, Lämmerhofer M, Neusüß C. Capillary isoelectric focusing-mass spectrometry: Coupling strategies and applications. Electrophoresis 2015;36:2670–2686. [DOI] [PubMed] [Google Scholar]
- Jin X, Kim J, Parus S, Lubman DM, Zand R. On-line capillary electrophoresis/microelectrospray ionization-tandem mass spectrometry using an ion trap storage/time-of-flight mass spectrometer with SWIFT technology. Anal. Chem. 1999;71:3591–7. [DOI] [PubMed] [Google Scholar]
- Jooß K, McGee JP, Melani RD, Kelleher NL. Standard procedures for native CZE-MS of proteins and protein complexes up to 800 kDa. Electrophoresis 2021, doi: 10.1002/elps.202000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachman MT, Wang H, Schwartz DR, Cho KR, Lubman DM. A 2-D liquid separations/mass mapping method for interlysate comparison of ovarian cancers. Anal. Chem. 2002;74:1779–91. [DOI] [PubMed] [Google Scholar]
- Karayel Ö, Xu P, Bludau I, Velan Bhoopalan S, Yao Y, Ana Rita FC, Santos A, Schulman BA, Alpi AF, Weiss MJ, Mann M. Integrative proteomics reveals principles of dynamic phosphosignaling networks in human erythropoiesis. Mol. Syst. Biol. 2020;16:e9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sakai-Kato K, Jin H, Kubota K, Miyano H, Toyo’oka T, Dulay MT, Zare RN. Integration of on-line protein digestion, peptide separation, and protein identification using pepsin-coated photopolymerized sol-gel columns and capillary electrophoresis/mass spectrometry. Anal. Chem. 2004;76(7):1896–902. [DOI] [PubMed] [Google Scholar]
- Keener JE, Zambrano DE, Zhang G, Zak CK, Reid DJ, Deodhar BS, Pemberton JE, Prell JS, Marty MT. Chemical Additives Enable Native Mass Spectrometry Measurement of Membrane Protein Oligomeric State within Intact Nanodiscs. J. Am. Chem. Soc. 2019;141:1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zand R, Lubman DM. Electrophoretic mobility for peptides with post-translational modifications in capillary electrophoresis. Electrophoresis 2002;23:782–93. [DOI] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TS, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509:575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoff CJ, Li C, Li P, Holland LA. Low Flow Voltage Free Interface for Capillary Electrophoresis and Mass Spectrometry Driven by Vibrating Sharp-Edge Spray Ionization. Anal. Chem. 2020;92:3006–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin OV, Anderson G, Spicer V, Sun L, Dovichi NJ. Predicting Electrophoretic Mobility of Tryptic Peptides for High-Throughput CZE-MS Analysis. Anal. Chem. 2017;89:2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Shah RL, Ahmad S, Rathore AS. Harnessing the power of electrophoresis and chromatography: Offline coupling of reverse phase liquid chromatography-capillary zone electrophoresis-tandem mass spectrometry for analysis of host cell proteins in monoclonal antibody producing CHO cell line. Electrophoresis. 2021;42:735–741. [DOI] [PubMed] [Google Scholar]
- Kumar R, Shah RL, Rathore AS. Harnessing the power of electrophoresis and chromatography: Offline coupling of reverse phase liquid chromatography-capillary zone electrophoresis-tandem mass spectrometry for peptide mapping for monoclonal antibodies. J. Chromatogr. A. 2020;1620:460954. [DOI] [PubMed] [Google Scholar]
- Lamp J, Ikonomova SP, Karlsson AJ, Xia Q, Wang Y. Online capillary electrophoresis - mass spectrometry analysis of histatin-5 and its degradation products. Analyst. 2020;145:4787–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latosinska A, Siwy J, Faguer S, Beige J, Mischak H, Schanstra JP. Value of Urine Peptides in Assessing Kidney and Cardiovascular Disease. Proteomics Clin. Appl. 2021;15:e2000027. [DOI] [PubMed] [Google Scholar]
- Le-Minh V, Tran NT, Makky A, Rosilio V, Taverna M, Smadja C. Capillary zone electrophoresis-native mass spectrometry for the quality control of intact therapeutic monoclonal antibodies. J. Chromatogr. A. 2019;1601:375–384. [DOI] [PubMed] [Google Scholar]
- Li H, Nguyen HH, Ogorzalek Loo RR, Campuzano IDG, Loo JA. An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nat. Chem. 2018;10(2):139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Champion MM, Sun L, Champion PA, Wojcik R, Dovichi NJ. Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry as an alternative proteomics platform to ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry for samples of intermediate complexity. Anal. Chem. 2012;84(3):1617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Compton PD, Tran JC, Ntai I, Kelleher NL. Optimizing capillary electrophoresis for top-down proteomics of 30–80 kDa proteins. Proteomics. 2014;14:1158–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang XH, Yu Y, Chen HX, Zhou YL, Zhang XX. Snake venom characteristic peptides: novel fingerprints for species identification by sheathless capillary electrophoresis-electrospray ionization-mass spectrometry. Analyst. 2020;145(14):5027–5031. [DOI] [PubMed] [Google Scholar]
- Lombard-Banek C, Li J, Portero EP, Onjiko RM, Singer CD, Plotnick DO, Al Shabeeb RQ, Nemes P. In Vivo Subcellular Mass Spectrometry Enables Proteo-Metabolomic Single-cell Systems Biology in a Chordate Embryo Developing to a Normally Behaving Tadpole (X. laevis). Angew. Chem. Int. Ed. Engl. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard-Banek C, Moody SA, Manzini MC, Nemes P. Microsampling Capillary Electrophoresis Mass Spectrometry Enables Single-Cell Proteomics in Complex Tissues: Developing Cell Clones in Live Xenopus laevis and Zebrafish Embryos. Anal. Chem. 2019;91:4797–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard-Banek C, Moody SA, Nemes P. Single-Cell Mass Spectrometry for Discovery Proteomics: Quantifying Translational Cell Heterogeneity in the 16-Cell Frog (Xenopus) Embryo. Angew. Chem. Int. Ed. Engl. 2016;55:2454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard-Banek C, Yu Z, Swiercz AP, Marvar PJ, Nemes P. A microanalytical capillary electrophoresis mass spectrometry assay for quantifying angiotensin peptides in the brain. Anal. Bioanal. Chem. 2019;411:4661–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeckyj RA, Basharat AR, Shen X, Liu X, Sun L. Large-Scale Qualitative and Quantitative Top-Down Proteomics Using Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry with Nanograms of Proteome Samples. J. Am. Soc. Mass Spectrom. 2019;30:1435–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeckyj RA, McCool EN, Shen X, Kou Q, Liu X, Sun L. Single-Shot Top-Down Proteomics with Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry for Identification of Nearly 600 Escherichia coli Proteoforms. Anal. Chem. 2017;89:12059–12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KR, Sun L, Zhu G, Dovichi NJ, Hummon AB. Over 2300 phosphorylated peptide identifications with single-shot capillary zone electrophoresis-tandem mass spectrometry in a 100 min separation. Anal. Chem. 2015;87:9532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack S, Arnold D, Bogdan G, Bousse L, Danan L, Dolnik V, Ducusin M, Gwerder E, Herring C, Jensen M, Ji J, Lacy S, Richter C, Walton I, Gentalen E. A novel microchip-based imaged CIEF-MS system for comprehensive characterization and identification of biopharmaceutical charge variants. Electrophoresis. 2019;40:3084–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinović S, Berger SJ, Pasa-Tolić L, Smith RD. Separation and detection of intact noncovalent protein complexes from mixtures by on-line capillary isoelectric focusing-mass spectrometry. Anal. Chem. 2000;72:5356–60. [DOI] [PubMed] [Google Scholar]
- Maxwell EJ, Chen DD. Twenty years of interface development for capillary electrophoresis-electrospray ionization-mass spectrometry. Anal. Chim. Acta. 2008;627:25–33. [DOI] [PubMed] [Google Scholar]
- Maxwell EJ, Zhong X, Zhang H, van Zeijl N, Chen DD. Decoupling CE and ESI for a more robust interface with MS. Electrophoresis. 2010;31:1130–7. [DOI] [PubMed] [Google Scholar]
- McCool EN, Chen D, Li W, Liu Y, Sun L . Capillary zone electrophoresis-tandem mass spectrometry using ultraviolet photodissociation (213 nm) for large-scale top-down proteomics. Anal. Methods. 2019;11:2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool EN, Lodge JM, Basharat AR, Liu X, Coon JJ, Sun L. Capillary Zone Electrophoresis-Tandem Mass Spectrometry with Activated Ion Electron Transfer Dissociation for Large-scale Top-down Proteomics. J. Am. Soc. Mass Spectrom. 2019;30:2470–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool EN, Lubeckyj RA, Shen X, Chen D, Kou Q, Liu X, Sun L. Deep Top-Down Proteomics Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry: Identification of 5700 Proteoforms from the Escherichia coli Proteome. Anal. Chem. 2018;90:5529–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool EN, Sun L. [Comparing nanoflow reversed-phase liquid chromatography-tandem mass spectrometry and capillary zone electrophoresis-tandem mass spectrometry for top-down proteomics]. Se Pu. 2019;37:878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehaffey MR, Xia Q, Brodbelt JS. Uniting Native Capillary Electrophoresis and Multistage Ultraviolet Photodissociation Mass Spectrometry for Online Separation and Characterization of Escherichia coli Ribosomal Proteins and Protein Complexes. Anal. Chem. 2020;92:15202–15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixner M, Pattky M, Huhn C. Novel approach for the synthesis of a neutral and covalently bound capillary coating for capillary electrophoresis-mass spectrometry made from highly polar and pH-persistent N-acryloylamido ethoxyethanol. Anal. Bioanal. Chem. 2020;412:561–575. [DOI] [PubMed] [Google Scholar]
- Mellors JS, Gorbounov V, Ramsey RS, Ramsey JM. Fully integrated glass microfluidic device for performing high-efficiency capillary electrophoresis and electrospray ionization mass spectrometry. Anal. Chem. 2008;80:6881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzke M, Guan Z. Structure-property studies on carbohydrate-derived polymers for use as protein-resistant biomaterials. Biomacromolecules. 2008;9:208–15. [DOI] [PubMed] [Google Scholar]
- Moini M Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal. Chem. 2007;79:4241–6. [DOI] [PubMed] [Google Scholar]
- Mokaddem M, Gareil P, Varenne A. Online CIEF-ESI-MS in glycerol-water media with a view to hydrophobic protein applications. Electrophoresis 2009;30:4040–8. [DOI] [PubMed] [Google Scholar]
- Mokaddem M, Gareil P, Varenne A. Online CIEF-ESI-MS in glycerol-water media with a view to hydrophobic protein applications. Electrophoresis 2009;30:4040–8. [DOI] [PubMed] [Google Scholar]
- Montealegre C, Neusüß C. Coupling imaged capillary isoelectric focusing with mass spectrometry using a nanoliter valve. Electrophoresis 2018;39:1151–1154. [DOI] [PubMed] [Google Scholar]
- Nguyen A, Moini M. Analysis of major protein-protein and protein-metal complexes of erythrocytes directly from cell lysate utilizing capillary electrophoresis mass spectrometry. Anal. Chem. 2008;80:7169–73. [DOI] [PubMed] [Google Scholar]
- Nyssen L, Fillet M, Cavalier E, Servais AC. Highly sensitive and selective separation of intact parathyroid hormone and variants by sheathless CE-ESI-MS/MS. Electrophoresis. 2019;40:1550–1557. [DOI] [PubMed] [Google Scholar]
- Ozawa H, Hirayama A, Ishikawa T, Kudo R, Maruyama M, Shoji F, Doke T, Ishimoto T, Maruyama S, Soga T, Tomita M. Comprehensive Dipeptide Profiling and Quantitation by Capillary Electrophoresis and Liquid Chromatography Coupled with Tandem Mass Spectrometry. Anal. Chem. 2020;92:9799–9806. [DOI] [PubMed] [Google Scholar]
- Pasa-Tolic L, Jensen PK, Anderson GA, Lipton MS, Peden KK, Martinovic S, Tolic N, Bruce JE, Smith RD. High throughput proteome-wide precision measurements of protein expression using mass spectrometry. J. Am. Chem. Soc. 1999, 121, 7949–7950 [Google Scholar]
- Pelander L, Brunchault V, Buffin-Meyer B, Klein J, Breuil B, Zürbig P, Magalhães P, Mullen W, Elliott J, Syme H, Schanstra JP, Häggström J, Ljungvall I. Urinary peptidome analyses for the diagnosis of chronic kidney disease in dogs. Vet. J. 2019;249:73–79. [DOI] [PubMed] [Google Scholar]
- Pero-Gascon R, Benavente F, Minic Z, Berezovski MV, Sanz-Nebot V. On-line Aptamer Affinity Solid-Phase Extraction Capillary Electrophoresis-Mass Spectrometry for the Analysis of Blood α-Synuclein. Anal. Chem. 2020;92:1525–1533. [DOI] [PubMed] [Google Scholar]
- Pero-Gascon R, Benavente F, Neusüß C, Sanz-Nebot V. Evaluation of on-line solid-phase extraction capillary electrophoresis-mass spectrometry with a nanoliter valve for the analysis of peptide biomarkers. Anal. Chim. Acta. 2020;1140:1–9. [DOI] [PubMed] [Google Scholar]
- Pero-Gascon R, Pont L, Sanz-Nebot V, Benavente F. On-Line Immunoaffinity Solid-Phase Extraction Capillary Electrophoresis-Mass Spectrometry for the Analysis of Serum Transthyretin. Methods Mol. Biol. 2019;1972:57–76. [DOI] [PubMed] [Google Scholar]
- Piestansky J, Barath P, Majerova P, Galba J, Mikus P, Kovacech B, Kovac A. A simple and rapid LC-MS/MS and CE-MS/MS analytical strategy for the determination of therapeutic peptides in modern immunotherapeutics and biopharmaceutics. J. Pharm. Biomed. Anal. 2020;189:113449. [DOI] [PubMed] [Google Scholar]
- Pont L, Marin G, Vergara-Barberán M, Gagliardi LG, Sanz-Nebot V, Herrero-Martínez JM, Benavente F. Polymeric monolithic microcartridges with gold nanoparticles for the analysis of protein biomarkers by on-line solid-phase extraction capillary electrophoresis-mass spectrometry. J. Chromatogr. A 2020;1622:461097. [DOI] [PubMed] [Google Scholar]
- Ramautar R, Heemskerk AA, Hensbergen PJ, Deelder AM, Busnel JM, Mayboroda OA. CE-MS for proteomics: Advances in interface development and application. J. Proteomics 2012;75:3814–28. [DOI] [PubMed] [Google Scholar]
- Rinas A, Jenkins C, Orsburn B. Assessing a commercial capillary electrophoresis interface (ZipChip) for shotgun proteomic applications. bioRxiv 2019, doi: 10.1101/559591. [DOI] [Google Scholar]
- Rudnicki M, Siwy J, Wendt R, Lipphardt M, Koziolek MJ, Maixnerova D, Peters B, Kerschbaum J, Leierer J, Neprasova M, Banasik M, Sanz AB, Perez-Gomez MV, Ortiz A, Stegmayr B, Tesar V, Mischak H, Beige J, Reich HN; PERSTIGAN working group. Urine proteomics for prediction of disease progression in patients with IgA nephropathy. Nephrol. Dial. Transplant. 2020:gfaa307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadé J, Gahoual R, Beck A, Leize-Wagner E, François YN. Characterization of the Primary Structure of Cysteine-Linked Antibody-Drug Conjugates Using Capillary Electrophoresis with Mass Spectrometry. Methods Mol. Biol. 2020;2078:263–272. [DOI] [PubMed] [Google Scholar]
- Sauer F, Sydow C, Trapp O. A robust sheath-flow CE-MS interface for hyphenation with Orbitrap MS. Electrophoresis. 2020;41:1280–1286. [DOI] [PubMed] [Google Scholar]
- Schaffer LV, Millikin RJ, Shortreed MR, Scalf M, Smith LM. Improving Proteoform Identifications in Complex Systems Through Integration of Bottom-Up and Top-Down Data. J. Proteome Res. 2020;19:3510–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr RM, Ye M, Vannatta M, Dovichi NJ. CE-microreactor-CE-MS/MS for protein analysis. Anal. Chem. 2007;79:2230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, D’Souza RC, Tyanova S, Schaab C, Wiśniewski JR, Cox J, Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell. Rep. 2014;8:1583–94. [DOI] [PubMed] [Google Scholar]
- Shen X, Kou Q, Guo R, Yang Z, Chen D, Liu X, Hong H, Sun L. Native Proteomics in Discovery Mode Using Size-Exclusion Chromatography-Capillary Zone Electrophoresis-Tandem Mass Spectrometry. Anal. Chem. 2018;90:10095–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liang Z, Xu T, Yang Z, Wang Q, Chen D, Pham L, Du W, Sun L. Investigating native capillary zone electrophoresis-mass spectrometry on a high-end quadrupole-time-of-flight mass spectrometer for the characterization of monoclonal antibodies. Int. J. Mass Spectrom. 2021;462:116541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Xu T, Hakkila B, Hare M, Wang Q, Wang Q, Beckman JS, Sun L. Capillary Zone Electrophoresis-Electron-Capture Collision-Induced Dissociation on a Quadrupole Time-of-Flight Mass Spectrometer for Top-Down Characterization of Intact Proteins. J. Am. Soc. Mass Spectrom. 2021, doi: 10.1021/jasms.0c00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Yang Z, McCool EN, Lubeckyj RA, Chen D, Sun L. Capillary zone electrophoresis-mass spectrometry for top-down proteomics. TrAC, Trends Anal. Chem. 2019;120:115644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Berger SJ, Anderson GA, Smith RD. High-efficiency capillary isoelectric focusing of peptides. Anal. Chem. 2000;72:2154–9. [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhao X, Wang G, Chen DDY. Differential Hydrogen/Deuterium Exchange during Proteoform Separation Enables Characterization of Conformational Differences between Coexisting Protein States. Anal. Chem. 2019;91:3805–3809. [DOI] [PubMed] [Google Scholar]
- Skinner OS, Haverland NA, Fornelli L, Melani RD, Do Vale LHF, Seckler HS, Doubleday PF, Schachner LF, Srzentić K, Kelleher NL, Compton PD. Top-down characterization of endogenous protein complexes with native proteomics. Nat. Chem. Biol. 2018;14:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL. Proteoforms as the next proteomics currency. Science. 2018;359:1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL; Consortium for Top Down Proteomics. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Barinaga CJ, Udseth HR. Improved electrospray ionization interface for capillary zone electrophoresis-mass spectrometry. Anal. Chem. 1988; 60: 1948–1952 [Google Scholar]
- Stolz A, Hedeland Y, Salzer L, Römer J, Heiene R, Leclercq L, Cottet H, Bergquist J, Neusüß C. Capillary Zone Electrophoresis-Top-Down Tandem Mass Spectrometry for In-Depth Characterization of Hemoglobin Proteoforms in Clinical and Veterinary Samples. Anal. Chem. 2020;92:10531–10539. [DOI] [PubMed] [Google Scholar]
- Storms HF, van der Heijden R, Tjaden UR, van der Greef J. Optimization of protein identification from digests as analyzed by capillary isoelectric focusing-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;824:189–200. [DOI] [PubMed] [Google Scholar]
- Sun L, Hebert AS, Yan X, Zhao Y, Westphall MS, Rush MJ, Zhu G, Champion MM, Coon JJ, Dovichi NJ. Over 10,000 peptide identifications from the HeLa proteome by using single-shot capillary zone electrophoresis combined with tandem mass spectrometry. Angew. Chem. Int. Ed. Engl. 2014;53:13931–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhu G, Zhang Z, Mou S, Dovichi NJ. Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J. Proteome Res. 2015;14:2312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhu G, Zhao Y, Yan X, Mou S, Dovichi NJ. Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests. Angew. Chem. Int. Ed. 2013;52:13661–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Zhu J, Stemmer PM, Sun L, Yang Z, Schultz K, Gaffrey MJ, Cesnik AJ, Yi X, Hao X, Shortreed MR, Shi T, Lubman DM. Comprehensive Detection of Single Amino Acid Variants and Evaluation of Their Deleterious Potential in a PANC-1 Cell Line. J. Proteome Res. 2020;19:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wu H, Hu JJ, Yu J, Zhang J, Wang C, Yin T, Tang K. On a separation voltage polarity switching transient capillary isotachophoresis method for higher sample loading capacity and better separation performance. Analyst. 2021;146:124–131. [DOI] [PubMed] [Google Scholar]
- Tang Q, Harrata AK, Lee CS. Capillary isoelectric focusing-electrospray mass spectrometry for protein analysis. Anal. Chem. 1995; 67:3515–3519. [Google Scholar]
- Tie C, Liu L, Feng T, Sa R, Xia Q, Liang H, Mao Y. Differential analysis of urinary albumin for membranous nephropathy patients by online capillary isoelectric focusing - Mass spectrometry. J. Proteomics 2020;216:103676. [DOI] [PubMed] [Google Scholar]
- Tong W, Link A, Eng JK, Yates JR 3rd. Identification of proteins in complexes by solid-phase microextraction/multistep elution/capillary electrophoresis/tandem mass spectrometry. Anal. Chem. 1999;71:2270–8. [DOI] [PubMed] [Google Scholar]
- Valaskovic GA, Kelleher NL, McLafferty FW. Attomole protein characterization by capillary electrophoresis-mass spectrometry. Science. 1996;273:1199–202. [DOI] [PubMed] [Google Scholar]
- van de Waterbeemd M, Fort KL, Boll D, Reinhardt-Szyba M, Routh A, Makarov A, Heck AJ. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods. 2017;14:283–286. [DOI] [PubMed] [Google Scholar]
- Vermeire PJ, Van Schepdael A, Petersen NJ. Development of a novel sheathless CE-ESI-MS interface via a CO2 laser ablated opening. Talanta. 2020;214:120853. [DOI] [PubMed] [Google Scholar]
- Wang L, Bo T, Zhang Z, Wang G, Tong W, Da Yong Chen D. High Resolution Capillary Isoelectric Focusing Mass Spectrometry Analysis of Peptides, Proteins, And Monoclonal Antibodies with a Flow-through Microvial Interface. Anal. Chem. 2018;90:9495–9503. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen DDY. Analysis of four therapeutic monoclonal antibodies by online capillary isoelectric focusing directly coupled to quadrupole time-of-flight mass spectrometry. Electrophoresis. 2019;40:2899–2907. [DOI] [PubMed] [Google Scholar]
- Wang L, Cheng J, McNutt JE, Morin GB, Chen DDY. Dynamic pH barrage junction focusing of amino acids, peptides, and digested monoclonal antibodies in capillary electrophoresis-mass spectrometry. Electrophoresis. 2020;41:1832–1842. [DOI] [PubMed] [Google Scholar]
- Wang L, Li Y, Chen D, Chen DDY. Electrospray ionization stability and concentration sensitivity in capillary electrophoresis-mass spectrometry using a flow-through microvial interface. Electrophoresis. 2021;42:360–368. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen D, Lubeckyj RA, Shen X, Yang Z, McCool EN, Qiao X, Sun L. Capillary zone electrophoresis-tandem mass spectrometry for top-down proteomics using attapulgite nanoparticles functionalized separation capillaries. Talanta. 2019;202:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fonslow BR, Wong CC, Nakorchevsky A, Yates JR 3rd. Improving the comprehensiveness and sensitivity of sheathless capillary electrophoresis-tandem mass spectrometry for proteomic analysis. Anal. Chem. 2012;84:8505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SS, Dawod M, Kennedy RT. CE-MS with electrokinetic supercharging and application to determination of neurotransmitters. Electrophoresis. 2019;40:2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis. Rapid Commun. Mass Spectrom. 2010;24:2554–60. [DOI] [PubMed] [Google Scholar]
- Wörner TP, Snijder J, Bennett A, Agbandje-McKenna M, Makarov AA, Heck AJR. Resolving heterogeneous macromolecular assemblies by Orbitrap-based single-particle charge detection mass spectrometry. Nat. Methods. 2020;17:395–398. [DOI] [PubMed] [Google Scholar]
- Wu H, Yi L, Wojcik R, Shi T, Tang K. A separation voltage polarity switching method for higher sample loading capacity and better separation resolution in transient capillary isotachophoresis separation. Analyst. 2019;144:454–462. [DOI] [PubMed] [Google Scholar]
- Xu T, Shen X, Yang Z, Chen D, Lubeckyj RA, McCool EN, Sun L. Automated Capillary Isoelectric Focusing-Tandem Mass Spectrometry for Qualitative and Quantitative Top-Down Proteomics. Anal. Chem. 2020;92:15890–15898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Zhang K, Wang L, Tong W, Chen DDY. Quantitative analysis of microcystin variants by capillary electrophoresis mass spectrometry with dynamic pH barrage junction focusing. Electrophoresis. 2019;40:2285–2293. [DOI] [PubMed] [Google Scholar]
- Yan X, Sun L, Dovichi NJ, Champion MM. Minimal deuterium isotope effects in quantitation of dimethyl-labeled complex proteomes analyzed with capillary zone electrophoresis/mass spectrometry. Electrophoresis. 2020;41:1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lee CS, Hofstadler SA, Pasa-Tolic L, Smith RD. Capillary isoelectric focusing-electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for protein characterization. Anal. Chem. 1998;70:3235–41. [DOI] [PubMed] [Google Scholar]
- Yang Z, Shen X, Chen D, Sun L. Improved Nanoflow RPLC-CZE-MS/MS System with High Peak Capacity and Sensitivity for Nanogram Bottom-up Proteomics. J. Proteome Res. 2019;18:4046–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Shen X, Chen D, Sun L. Microscale Reversed-Phase Liquid Chromatography/Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Deep and Highly Sensitive Bottom-Up Proteomics: Identification of 7500 Proteins with Five Micrograms of an MCF7 Proteome Digest. Anal. Chem. 2018;90:10479–10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Shen X, Chen D, Sun L. Toward a Universal Sample Preparation Method for Denaturing Top-Down Proteomics of Complex Proteomes. J. Proteome Res. 2020;19:3315–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Peng Q, Usman M, Ahmed A, Chen Y, Chen X, Wang Y, Shen Y, Cong H. Preparation of photosensitive diazotized poly (vinyl alcohol-b-styrene) covalent capillary coatings for capillary electrophoresis separation of proteins. J. Chromatogr. A. 2019;1593:174–182. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lou C, Li J, Kang J. A gold foil covered fused silica capillary tip as a sheathless interface for coupling capillary electrophoresis-mass spectrometry. J. Chromatogr. A. 2020;1624:461215. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wu H, Zhang R, Fang X, Xu W. Structure and effective charge characterization of proteins by a mobility capillary electrophoresis based method. Chem. Sci. 2019;10:7779–7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR 3rd. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013;113:2343–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hebert AS, Westphall MS, Coon JJ, Dovichi NJ. Single-Shot Capillary Zone Electrophoresis-Tandem Mass Spectrometry Produces over 4400 Phosphopeptide Identifications from a 220 ng Sample. J. Proteome Res. 2019;18:3166–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Peuchen EH, Dovichi NJ. Surface-Confined Aqueous Reversible Addition-Fragmentation Chain Transfer (SCARAFT) Polymerization Method for Preparation of Coated Capillary Leads to over 10 000 Peptides Identified from 25 ng HeLa Digest by Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry. Anal. Chem. 2017;89:6774–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Qu Y, Dovichi NJ. Capillary zone electrophoresis-mass spectrometry for bottom-up proteomics. TrAC, Trends Anal. Chem. 2018;108:23–37 [Google Scholar]
- Zhao Y, Sun L, Zhu G, Dovichi NJ. Coupling Capillary Zone Electrophoresis to a Q Exactive HF Mass Spectrometer for Top-down Proteomics: 580 Proteoform Identifications from Yeast. J. Proteome Res. 2016;15:3679–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Maxwell EJ, Ratnayake C, Mack S, Chen DD. Flow-through microvial facilitating interface of capillary isoelectric focusing and electrospray ionization mass spectrometry. Anal. Chem. 2011;83:8748–55. [DOI] [PubMed] [Google Scholar]
- Zhu K, Kim J, Yoo C, Miller FR, Lubman DM. High sequence coverage of proteins isolated from liquid separations of breast cancer cells using capillary electrophoresis-time-of-flight MS and MALDI-TOF MS mapping. Anal. Chem. 2003;75:6209–17. [DOI] [PubMed] [Google Scholar]
- Zhu G, Sun L, Dovichi NJ. Simplified capillary isoelectric focusing with chemical mobilization for intact protein analysis. J. Sep. Sci. 2017;40:948–953. [DOI] [PubMed] [Google Scholar]
- Zhu G, Sun L, Keithley RB, Dovichi NJ. Capillary isoelectric focusing-tandem mass spectrometry and reversed-phase liquid chromatography-tandem mass spectrometry for quantitative proteomic analysis of differentiating PC12 cells by eight-plex isobaric tags for relative and absolute quantification. Anal. Chem. 2013;85:7221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Sun L, Yan X, Dovichi NJ. Bottom-up proteomics of Escherichia coli using dynamic pH junction preconcentration and capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry. Anal. Chem. 2014;86:6331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]