Abstract
Despite the fact that eukaryotic cells enlist checkpoints to block cell cycle progression when their DNA is damaged, cells still undergo frequent genetic rearrangements, both spontaneously and in response to genotoxic agents. We and others have previously characterized a phenomenon (adaptation) in which yeast cells that are arrested at a DNA damage checkpoint eventually override this arrest and reenter the cell cycle, despite the fact that they have not repaired the DNA damage that elicited the arrest. Here, we use mutants that are defective in checkpoint adaptation to show that adaptation is important for achieving the highest possible viability after exposure to DNA-damaging agents, but it also acts as an entrée into some forms of genomic instability. Specifically, the spontaneous and X-ray-induced frequencies of chromosome loss, translocations, and a repair process called break-induced replication occur at significantly reduced rates in adaptation-defective mutants. This indicates that these events occur after a cell has first arrested at the checkpoint and then adapted to that arrest. Because malignant progression frequently involves loss of genes that function in DNA repair, adaptation may promote tumorigenesis by allowing genomic instability to occur in the absence of repair.
Cell cycle checkpoints are thought to provide time for DNA repair by delaying cell cycle progress in the face of DNA damage (reviewed in reference 21). Saccharomyces cerevisiae arrests in metaphase for up to 8 h when chromosomes are damaged (e.g., by a double-stranded DNA [dsDNA] break), after which it adapts and continues through the cell cycle (12, 18, 20). Two classes of proteins have been identified that are required for checkpoint adaptation: repair proteins and signaling proteins. Mutations in KU increase the amount of single-stranded DNA that forms at a dsDNA break, thereby increasing the strength of the checkpoint signal and eliminating adaptation (13). Strains in which the casein kinase II specificity subunits (CKB1 or CKB2) are deleted or that contain a special allele of the gene encoding the polo kinase Cdc5p (cdc5-ad) are also unable to adapt to DNA damage arrest (20).
dsDNA breaks can be processed by many mechanisms that result in different outcomes. Archetypal homologous recombination (reattachment of two broken ends using a homologous template) allows error-free repair. However, other, more error-prone outcomes are also seen: (i) nonhomologous end joining results in deletions; (ii) single-strand annealing (SSA) between direct repeats results in deletions; (iii) break-induced replication (BIR) can yield translocations or large gene conversion tracts that cause loss of heterozygosity; (iv) ectopic telomere addition causes terminal truncations; and (v) unrepaired chromosomes may be lost altogether (reviewed in references 4 and 5). Each of these pathways can lead to the loss of genetic information (genomic instability). Which of these scenarios occurs may depend upon where the cell is in the cell cycle when it repairs the damage.
Several variables determine where cells are in the cell cycle when they attempt repair of a dsDNA break: where the cells are in the cycle when they receive the break, the speed with which they repair the break, whether they undergo a checkpoint-mediated arrest in response to the break, and how long they maintain that arrest before adapting. Here, we examine whether some events that lead to genomic instability (e.g., chromosome deletions, translocations, or loss) occur only after cells have first undergone a checkpoint arrest and subsequently have adapted to that arrest. We employed adaptation-defective mutants to show that in the absence of archetypal homologous recombination (which is typically error free), most irradiated cells undergo checkpoint adaptation and only after this adaptation do they undergo BIR, translocations, or chromosome loss. Therefore, checkpoint adaptation serves to increase resistance to DNA damage, but in doing so it allows cells to undergo mutagenic events. Adaptation-defective diploids are particularly sensitive to X rays compared to adaptation-proficient diploids. We suggest that adaptation is more important for achieving maximum radio-resistance in diploids than in haploids, because diploids are able to survive with genetic rearrangements more easily than are haploids. Since adaptation is required to generate many of these rearrangements, adaptation is only seen to increase viability in strains that are able to grow after loss or rearrangement of chromosomes.
MATERIALS AND METHODS
Scoring of X-ray sensitivity, adaptation, chromosome loss, and rearrangements.
All strains were derived from LS20 (additional information is available on request) (18), which has the genotype matΔ cyh2 can1 lys5 ade2 ade3::GalHO trp1 his3 ura3 leu2. Some strains were also leu1::URA3 (see Fig. 3, 4, and 5). The additional chromosome VII was aro2 adh4::HIS3. ckb2, rad51, and rad52 strains were disrupted with LEU2 as indicated. Strains were grown in synthetic media with glucose at 30°C unless noted otherwise. Means and standard deviations were determined for three independent experiments (with the exception of experiments shown below in Fig. 3 [rows 3 and 4] and Table 2 [rows 1 and 2], for which the means and ranges of two experiments are reported). For damage-induced events, frequencies are the number of induced events divided by the number of viable cells. For X-ray sensitivity experiments, cells were sonicated for 7 s on level 2.5 of a Fisher-550 sonicator, plated, and subjected to various doses of X rays. Viability was determined as the number of colonies formed after irradiation, compared to the number of colonies with no irradiation. Haploid MATΔ strains (rad52 cdc5-ad or rad52 CDC5) were transformed with a plasmid encoding the MATα locus and mated to themselves to produce isogenic diploids (20). Diploids were then streaked on 5-fluoroorotic acid (FOA) plates to select against the MATα URA3 plasmid.
FIG. 3.
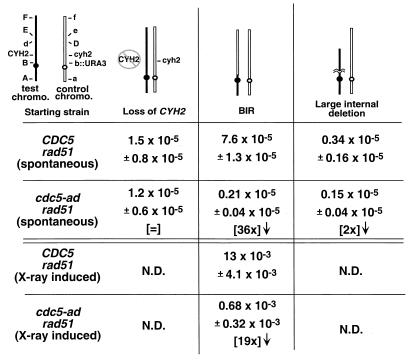
Adaptation is required for spontaneous and damage-induced BIR events. Haploid CDC5 rad51 and cdc5-ad rad51 strains (as in Table 1) were altered to replace the centromere-linked LEU1 gene on the control chromosome with the URA3 gene (shown as B and b::URA3, respectively). Markers A, D, E, and F represent ADE3, ARO2, LYS5, and adh4::HIS3, respectively. Control chromosome DNA is shown in outline. For spontaneous events, strains were plated on medium lacking leucine (selecting for marker B) and containing cycloheximide (selecting against CYH2). The prominent classes of rearrangements are diagrammed. For X-ray induced events, cells were plated on nonselective plates, subjected to 3 kilorads of X rays, allowed to form colonies, and replica plated on cycloheximide plates lacking leucine to screen for BIR events.
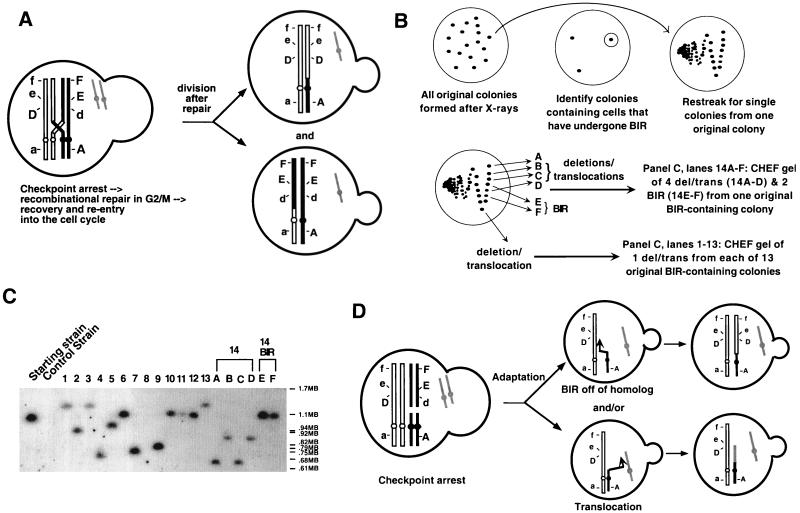
FIG. 4.
Translocations and BIR can both arise from a DNA lesion. (A) This diagram illustrates the expected outcome of a reciprocal recombination event in G2 between sister chromatids of different homologs. Segregation of sisters after mitosis will yield two possible outcomes: (i) both daughters can have markers identical to the starting strain (not shown), or (ii) each daughter can be homozygous for markers distal to the site of recombination (shown). (B) Outline of the experiment, the results of which are shown in panel C. Disomic rad51 CDC5 colonies (from irradiated nonselective plates) were scored for BIR events (as described in Materials and Methods). Colonies found to contain cells that had undergone BIR were restreaked from the original nonselective plate, and the resultant colonies were analyzed genetically for rearrangements of chromosome VII (BIR and/or deletions-translocations, as determined genetically). (C) DNA from the following strains were run on CHEF gels, blotted, and probed for the test chromosome: “starting strain” and “control strain” are marker strains with and without the test chromosome, respectively; 13 translocations-truncations isolated after restreaking 13 independent BIR-containing colonies (one translocation-truncation from each original colony) (lanes 1 to 13); and 4 translocations-truncations (lanes 14A to D) and 2 BIR (lanes 14 BIR E and F) colonies isolated from the same irradiated BIR-containing colony (6 total colonies isolated from one original colony). (D) Model showing three pairs of sister chromatids; the black and outlined sets represent the test and control chromosomes, respectively, and gray represents a different chromosome. Adaptation results in daughters that repair independently.
FIG. 5.
A large region of chromosome VII flanked by two homologous tRNA genes is frequently deleted. (A) Marker strains with (lane 1) or without (lane 2) the control chromosome; 13 strains identified as large internal deletions from rad51 CDC5 (lanes 3 to 15) or rad51 cdc5-ad (lanes 16 to 28) were run on a CHEF gel, blotted, and probed for the test chromosome. (B) DNA from the starting strain or a strain containing a large internal deletion (as in panel A, lane 3) was fluorescently labeled and hybridized to a DNA array as described previously (9). Hybridization ratios are shown for genes on the left arm of chromosome VII. Arrows designate leucine tRNA genes flanking the deletion. (C) Oligonucleotides corresponding to the outside of the deletion were used in a PCR on the strains shown in panel A. M, marker. (D) The putative SSA intermediate between the centromere proximal [TL (CAA) G2] and distal [TL (CAA) G1] tRNA genes flanking the internal deletions mapped in panels A to C. The oligonucleotides used in panel C are indicated as arrows.
TABLE 2.
BIR events in strains with an HO break at the TRP5 locus on chromosome VIIa
| Strain | Viability (%)b | Reciprocal recombination (%)c |
|---|---|---|
| CDC5 | 96 ± 6 | 18 ± 5 |
| cdc5-ad | 80 ± 21 | 15 ± 2 |
| CDC5 rad52 | 89 ± 14 | <0.5 |
| cdc5-ad rad52 | 6.9 ± 1.4 | <0.5 |
| CDC5 rad51 | 52 ± 7 | <0.5 |
| cdc5-ad rad51 | 28 ± 9 | <0.5 |
An HO endonuclease site was integrated into disomic strains at the TRP5 locus (50 kb from the centromere) on the test chromosome VII. RAD51 RAD52, rad51 RAD52, and RAD51 rad52 strains that were either adaptation proficient (CDC5) or deficient (cdc5-ad) were grown overnight in raffinose medium selecting for both copies of chromosome VII. Cells were then spread on plates containing either galactose (which induces the HO endonuclease) or glucose (which does not induce HO).
Viability is given as colonies on galactose/colonies on glucose.
Reciprocal recombination represents the percentage of colonies grown on the galactose plate that were Ade3+ Leu1+ Ura3+ Aro2+ Lys5− His3− CyhR. Since most of these colonies also contained Ade3+ Leu1+ Ura3+ Aro2− Lys5+ His3+ CyhS cells, they are likely reciprocal recombinants.
To examine genomic instability, disomic strains were grown in synthetic media lacking lysine and tyrosine to select for both copies of chromosome VII. Cells were then plated on either complete medium containing cycloheximide, to select for whole chromosome loss, or cycloheximide media lacking leucine, to select for rearrangements (shown below in Fig. 3). Only His3− Lys5− Aro2+ CyhR Ade3− colonies were scored as chromosome loss events. After selection on cycloheximide medium lacking leucine, the selected colonies were analyzed for the ability to grow on media lacking either histidine, tyrosine, lysine, or uracil. The ADE3 locus was analyzed by color. Colonies were also replica plated on FOA plates to select against the URA3 gene product (and thereby the control chromosome). Cells that have copied DNA from the control chromosome to the test chromosome (BIR) should still be able to lose the control chromosome, whereas strains that have lost a portion of the test chromosome (e.g., deletions) should not. FOA-resistant colonies were then checked as described above. (i) BIR events were detected as Ade3+ Leu1+ Ura3+ Aro2+ Lys5− His3− CyhR colonies that papillated on FOA. (ii) Translocation and truncation events (shown below in Fig. 4) had identical markers to BIR events but did not papillate on FOA, indicating that they had lost essential genes on the test chromosome. (iii) Loss-of-CYH2 events were detected as Ade3+ Leu1+ Ura3+ Aro2+ Lys5+ His3+ CyhR colonies. Some colonies papillated on FOA (CYH2 is an essential gene, so null mutations precluded papillation). (iv) Internal deletions were detected as Ade3+ Leu1+ Ura3+ Aro2+ Lys5− His3+ CyhR colonies that did not papillate on FOA. X-ray induction of genomic rearrangements was analyzed by plating cells on complete media, X-irradiating them, allowing colonies to form, and replica plating on cycloheximide plates or on cycloheximide plates lacking leucine. Leu+ CyhR colonies were scored as for spontaneous events.
Zeocin (Invitrogen)- and benomyl-induced chromosome loss was determined by incubating strains (grown to 3 × 106 cells/ml) in 0.5 mg of zeocin/ml or 0.045 mg of benomyl/ml for 14 h at 23°C in synthetic medium lacking lysine and tyrosine. Cells were then plated on complete synthetic plates and scored for chromosome loss by color (scoring ADE3) and loss of LEU1 LYS5 and HIS3. For experiments described in Table 2, cells were grown overnight in synthetic raffinose medium lacking lysine and tyrosine and plated either on glucose plates lacking uracil or on raffinose-galactose plates lacking uracil.
Molecular methods.
Yeast DNA was prepared and contour-clamped homogeneous electric field (CHEF) gels were run as described previously (10). Gels were blotted to nylon filters and probed with a 1,657-bp XhoI/KpnI fragment of ADE3 specific for the test chromosome. DNA was fluorescently labeled for DNA array analysis as described elsewhere (9). DNA arrays (kindly provided by J. DeRisi) were probed as described previously (2). PCR analysis (see Fig. 5C) was performed using the oligonucleotides TGTTCGTAAGCAATAATAAATCAAT and TGTGGTGTATATTGACCCAACGAGT.
To measure the rate of SSA, the HO endonuclease site was integrated between direct repeats of the TRP5 gene, such that TRP5 was disrupted before HO-induced SSA but restored upon repair. This strain was generated by transforming disomic strains with the plasmid pDG1 (described below) linearized with MscI. Strains used to measure HO-induced BIR and reciprocal recombination (see Table 2) were created by introducing the HO endonuclease site at the TRP5 locus (so that no repeats were generated) by cutting the plasmid pDG3 (see below) with BamHI and NsiI and transforming disomic strains with the TRP5-5′/HO/TRP1/TRP5-3′ fragment. Since strains were disomic for chromosome VII, which encodes the TRP5 gene, integrants were analyzed to determine which chromosome VII the construct had targeted. pDG1 was generated by first inserting an ∼1-kb BamHI/XhoI fragment of TRP5 into pRS304. An ∼120-bp fragment containing the HO site (from MATa-stk) was cut from pAR134 with EcoRI (T4 blunted) and PstI. This was inserted into the NsiI/HincII site in the TRP5 gene in pRS304. This generated pDG1. pDG1 was cut with KpnI and SacI to liberate the TRP5/HO fragment, and this fragment was cloned into KpnI/SacI-cut pRS424. The resulting plasmid, pDG2, was cut with NsiI/SnaBI to remove the 2μm sequence. A 400-bp fragment containing the 3′ end of TRP5 was amplified using PCR such that NsiI and SnaBI sites were generated at the fragment ends. This fragment was cloned into the NsiI/SnaBI-cut pDG2 to generate pDG3. All strains used had a galactose-inducible HO endonuclease gene inserted at the ADE3 locus.
RESULTS
Adaptation is important for achieving maximum X-ray resistance in diploids but not haploids.
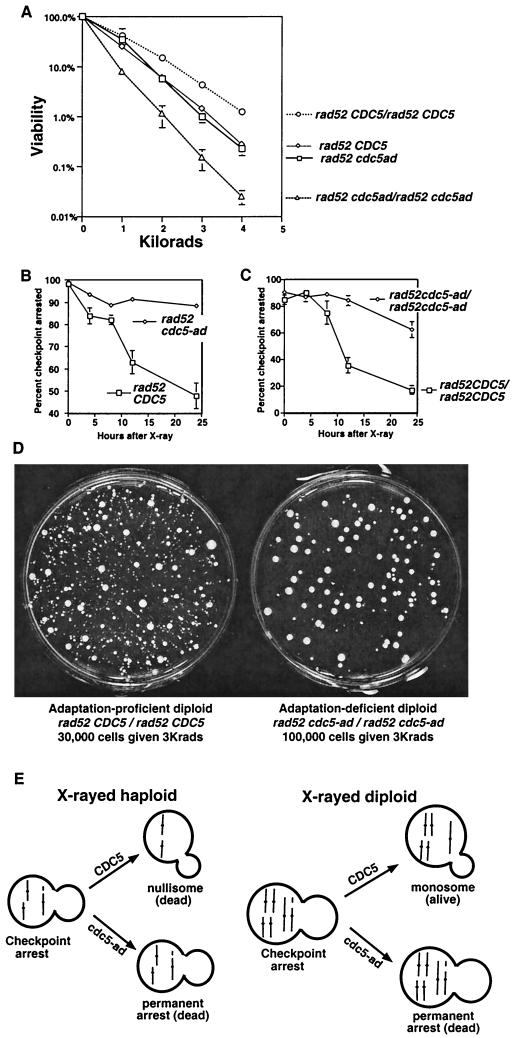
To determine the importance of adaptation in tolerating irreparable DNA damage, we examined the sensitivity of adaptation-proficient (CDC5) and adaptation-deficient (cdc5-ad) cells to X-irradiation in a repair-defective background (rad52) (Fig. 1A). cdc5-ad mutants are unable to adapt to a DNA damage arrest induced by an endonucleolytic break (20) or by X-irradiation (Fig. 1B and C). Despite this, there was no difference in the X-ray sensitivities of rad52 CDC5 and rad52 cdc5-ad haploid strains (Fig. 1A). This was likely because the rad52 CDC5 cells that adapted underwent lethal rearrangements or chromosome loss. We reasoned that this may not be the case in diploids, since they have two copies of each chromosome and are therefore able to tolerate the loss of part or even all of one of their two homologs (Fig. 1E). This is in fact the case; homozygous rad52 CDC5 diploids form many more colonies after X-irradiation than adaptation-defective strains (Fig. 1A). Similar, albeit less dramatic results were seen in RAD52 strains with high X-ray doses; RAD52 cdc5-ad diploids are 3.7 times more sensitive to X rays than RAD52 CDC5 diploids at 45 kilorads. Diploids are also more resistant to DNA damage due to heterozygosity of the MAT genes (8). However, our observations were due to ploidy itself and not due to the genetic difference between haploids and diploids, since the MAT locus was deleted in all strains used in this paper.
FIG. 1.
Adaptation increases X-ray resistance in diploids. (A) Strains were plated and X-irradiated, and all resulting colonies were counted, regardless of their growth rate. Haploid strains (rad52 cdc5-ad or rad52 CDC5) were mated to themselves to produce isogenic diploids (20). (B and C) Cells were irradiated with 3 kilorads, and microcolony assays were performed to determine the percentage of cells that had adapted, as described elsewhere (20). (D) Either 30,000 cells (rad52 CDC5/rad52 CDC5 diploids, left) or 100,000 cells (rad52 cdc5-ad/rad52 cdc5-ad diploids, right) were plated, subjected to 3 kilorads of X-irradiation, and allowed to form colonies. (E) Model showing one broken and two unbroken chromosomes (each line represents two identical sisters) in haploid and diploid strains. Adaptation generates viable but karyotypically altered monosomic diploids.
We suggest that rad52 CDC5 diploids are more resistant to X rays than rad52 cdc5-ad diploids because rad52 CDC5 diploids can form colonies after cells adapt to unrepaired damage. If this is the case, many of the colonies formed after irradiation should be karyotypically altered. Consistent with this, we found that the majority of the colonies that formed on the rad52 CDC5 plates after X-irradiation were sickly (Fig. 1D, left). While many of these colonies were small, almost all were viable; upon restreaking, 20 of 20 of the smallest colonies yielded a very heterogeneous set of colonies, including some that grew with near wild-type growth rates (data not shown). Interestingly, while the adaptation-defective diploid strain formed fewer colonies after irradiation, colonies that did grow appeared much more homogeneous and healthy (Fig. 1D, right). This suggests that cells with chromosome rearrangements that might have gone on to form growth-defective colonies in the wild-type strain instead remained permanently arrested with the damaged chromosome in the adaptation mutant. Colonies formed by the rad52 CDC5 and rad52 cdc5-ad diploids in the absence of irradiation were indistinguishable, as were the irradiated rad52 CDC5 and rad52 cdc5-ad haploids (data not shown).
Checkpoint adaptation precedes chromosome loss.
To examine better whether genomic instability was responsible for the differences seen between the rad52 CDC5 and rad52 cdc5-ad strains shown in Fig. 1, we performed chromosome loss assays on two adaptation-deficient mutants (cdc5-ad and ckb2) (20). These experiments, as well as those shown in all subsequent figures, were performed on disomic strains which harbored a nonessential copy of chromosome VII containing several genetic markers, including ADE3, which is required for the formation of a red pigment, and CYH2, which causes sensitivity to cycloheximide. These experiments allowed us to examine chromosome loss or rearrangements on chromosome VII without loss of essential genes. We found that the frequency with which this chromosome was lost, either spontaneously or after X-irradiation, was strikingly decreased in the adaptation-defective mutants rad52 cdc5-ad and rad52 ckb2 (Table 1). This argues against models in which chromosome loss events are thought to occur either because cells occasionally do not detect the damaged DNA or because the chromosome becomes damaged after the stage of the cell cycle where the checkpoint arrest takes place. Instead, these data suggest that most observed chromosome loss events are preceded by a checkpoint arrest and a subsequent adaptation to that arrest. Similarly, missegregation of minichromosomes correlates with a transient MAD1-dependent checkpoint arrest (23).
TABLE 1.
Loss of chromosome VII in haploid strainsa
| Genotype | Frequency (%) of chromosome loss (mean ± SD)
|
|
|---|---|---|
| Spontaneousb | X-ray inducedc | |
| rad52 CDC5 CKB2d | 1.2 ± 0.21 | 16.3 ± 4.2 |
| rad52 cdc5-ad CKB2d | 0.26 ± 0.07 | 1.7 ± 1.2 |
| rad52 CDC5 ckb2 | 0.29 ± 0.02 | 1.9 ± 1.4 |
| rad51 CDC5 | 0.30 ± 0.071 | 13.8 ± 0.41 |
| rad51 cdc5-ad | 0.073 ± 0.21 | 5.5 ± 1.1 |
Strains used harbored an extra copy of chromosome VII, which contains the CYH2 and ADE3 genes.
Strains were plated onto medium containing cycloheximide, which selects for cells that have lost the CYH2 gene on the extra chromosome (see also Fig. 3).
Cells were plated on nonselective medium and immediately X-irradiated with 2 (rad52 strains) or 3 (rad51 strains) kilorads.
This strain is pictured in Fig. 2.
rad52 mutants have elevated rates of spontaneous chromosome loss (17), probably because these mutants are unable to repair some form of intrinsic DNA damage. To ensure that the effect of adaptation on chromosome loss was not specific to rad52 mutants, we performed similar experiments with strains with deletions for RAD51, a recA homolog required for some forms of recombination, and we found that rad51 cdc5-ad mutants also had a lower frequency of spontaneous and damage-induced chromosome loss than that in rad51 CDC5 strains. rad52 mutants have a higher chromosome loss rate than rad51 mutants, which is probably due to the fact that RAD51 is required for only a subset of RAD52-dependent repair processes. Most chromosome loss events induced by DNA damage in recombination-proficient strains also depended on CDC5. CDC5 and cdc5-ad strains like those in Table 1, except RAD52 RAD51, were grown for 12 h at 23°C in the presence of 500 μg of the DNA-damaging agent zeocin (a bleomycin derivative) per ml and plated nonselectively. In 10 separate experiments, CDC5 strains generated approximately 2.2 times more zeocin-induced chromosome loss events than did cdc5-ad strains (losses per viable cell). This effect is specific to the DNA damage checkpoint, since cdc5-ad strains did not have lower rates of benomyl-induced chromosome loss (not shown). Benomyl induces chromosome loss by disrupting microtubules in the mitotic spindle, evoking a spindle checkpoint-mediated arrest in metaphase. The observation that a smaller percentage of damage-induced chromosome loss events depends upon adaptation in recombination-proficient cells than in recombination-deficient cells (90% in rad52 cells versus 55% in RAD52 cells; Table 1 and described above) suggests that some of the losses seen in RAD52 cells occur after the cell has repaired incorrectly. This RAD52-dependent repair may turn off the damage-signaling pathway, so that adaptation is not required, but yields an unstable (e.g., acentric or dicentric) chromosome. We also monitored spontaneous chromosome loss by colony sectoring (Fig. 2). rad52 CDC5 and rad52 cdc5-ad strains containing an ADE3-marked extra chromosome were plated on nonselective media. Each chromosome loss event was seen as a white sector. Clearly, the rad52 CDC5 strain produced many more sectors than the rad52 cdc5-ad strain, confirming that spontaneous loss events are preceded by checkpoint arrest and adaptation.
FIG. 2.
Adaptation precedes spontaneous chromosome loss. The frequency of spontaneous chromosome loss was determined using haploid strains harboring an extra copy of chromosome VII containing the CYH2 and ADE3 genes. Haploid CDC5 rad52 and cdc5-ad rad52 disomic strains (as in Table 1) were grown on nonselective plates and photographed. Chromosome losses appear as white sectors.
Checkpoint adaptation precedes chromosome translocation and BIR.
While examining chromosome loss rates in rad51 strains, we observed that the spontaneous and X-ray-induced rates of another form of genomic instability, BIR, are also diminished in cdc5-ad. BIR is a recombinational repair event during which only one of the halves of a broken chromosome invades a template such that the entire arm of the broken chromosome is copied from that template (6, 15, 16). This gene conversion event results in loss of heterozygosity over very large sections of the chromosome. Moreover, chromosomes other than homologs can be used for the template, generating nonreciprocal translocations (1) (see also Fig. 4D). This form of recombination is also unusual in that it is independent of the rad51 gene (15), allowing this event to be studied in the absence of competing forms of recombination. To examine adaptation's role in BIR more thoroughly, we generated a disomic strain in which both copies of chromosome VII are marked throughout their length (Fig. 3, “starting strain”). In this strain, the CEN-linked marker LEU1 (marker B) was replaced with the URA3 gene on the control chromosome.
We wished to determine which forms of genomic instability (i) are dependent upon adaptation, (ii) are reduced by adaptation, or (iii) are unaffected by adaptation. To this end, we selected for chromosomal rearrangements that caused cells to lose the CYH2 marker, while retaining the B and b::URA3 markers, and therefore retaining both chromosomes. Several classes of rearrangements were seen. The three most prominent classes were consistent with the rearrangements shown in Fig. 3 as loss of CYH2 (by gene conversion or mutation), BIR, and large internal deletion. In addition to scoring all markers, strains were examined for the loss of essential material on the test chromosome by determining whether the strain was still able to lose the control chromosome. This was measured by the ability to form colonies on FOA, a drug that selects against the URA3 gene located on the control chromosome. BIR events should allow the loss of the control chromosome and its associated markers, since strains that have undergone BIR still contain two copies of all the genetic material on chromosome VII (Fig. 3). In contrast, deletion or truncation events should not allow loss of the control chromosome, since the control chromosome will contain the sole copy of many essential genes in cells that have undergone these events. These analyses showed that spontaneous and X-ray-induced BIR events occurred 36 and 19 times less frequently, respectively, in the rad51 cdc5-ad strain than in rad51 CDC5 cells (Fig. 3).
A reciprocal recombination event could also have generated the arrangement shown as BIR in Fig. 3. This is unlikely, however, since reciprocal recombination events are typically not seen in rad51 mutants (15). A reciprocal recombination would arise from a recombination between homologs in G2 (Fig. 4A, which shows only markers A, D, E, and F), yielding one daughter cell (Fig. 4A, top daughter) with both the original version of the control chromosome (having markers a b::URA3 cyh2 D e f) and the recombined version of the test chromosome (A B cyh2 D e f). This is the same rearrangement seen with BIR. However, with a reciprocal recombination we would expect to find that the colony that grew out of the original irradiated cell would also contain cells that arose from the other daughter cell (Fig. 4A, bottom daughter). This cell would contain the starting version of the test chromosome (A B CYH2 d E F) and a recombined version of the control chromosome (a b::URA3 CYH2 d E F).
We restreaked 26 rad51 CDC5 colonies (from the original irradiated nonselective plate) that contained cells with markers consistent with BIR (Fig. 4B). None of these BIR-containing colonies also had cells with the mirror image event that we would predict from a G2 mitotic recombination. Analysis of the resulting colonies showed that 2 of the 26 BIR-containing colonies yielded, upon restreaking, exclusively colonies that had undergone BIR. Eighteen of 26 colonies yielded a mixture of BIR events and colonies that had lost the same terminal markers (had lost CYH2, E, and F) but were unable to lose the control chromosome. This suggests that in each of these 18 colonies, other events arose from the same X-ray-induced lesions as the BIR events, but they were repaired in such a way that they had lost essential genes on the test chromosome. These could be either terminal truncations at the site of the lesion or nonreciprocal translocations (referred to as translocations-truncations). CHEF gel analysis of 14 translocations-truncations is consistent with their being chromosome rearrangements (Fig. 4C). In lanes 1 to 13 of Fig. 4C, a single translocation-truncation (as determined by genetic analysis) from restreaks of 13 separate original colonies was run on CHEF gels. A terminal truncation eliminating the CYH2 gene would generate a chromosome between 625 and 788 kb. The sizes of the resulting test chromosome for lanes 4, 7, and 14A and C (Fig. 4C) could therefore be terminal truncations or translocations, whereas the larger test chromosomes seen in the remaining lanes probably represent translocations. Most likely, both the BIR event and the translocation-truncation occurred in different daughter cells after adaptation, when the damaged chromosome was brought through subsequent cycles (Fig. 4D). In one case (Fig. 4C, lanes 14A to D and 14 BIR E and F), four colonies with translocations-truncations and two that underwent BIR were isolated from a single original colony grown from an irradiated cell. CHEF gel analysis showed that the four translocation-truncation colonies contained test chromosomes of two discrete sizes and likely represented two events. The finding that two independent translocations-truncations (in addition to one BIR event) had arisen from the same colony suggests that one X-ray-induced lesion was passaged through at least two divisions and processed at least three independent times (two separate truncations-translocations plus at least one BIR). While we were unable to screen for translocations directly, the observation that most BIR-containing rad51 CDC5 colonies also contained translocations allows us to infer that translocations were also reduced in cdc5-ad strains.
Among the 6 remaining colonies of the 26 original colonies examined, 2 showed only truncations-translocations and 2 showed only the starting strain, suggesting that the BIR event initially scored in these 4 colonies represented fewer than about 2% of the cells (about 50 colonies were examined in each restreak). The remaining 2 of the 26 colonies generated a mix of colonies including not only BIR but also marker combinations that are not consistent with any obvious class of rearrangement.
In order to examine directly the DNA break undergoing BIR, we generated a strain containing a single HO endonuclease site at the TRP5 locus of chromosome VII. When an endonucleolytic break was induced at this locus in rad51 CDC5 cells, no BIR was seen. Instead, 52% of the cells formed colonies from which the extra chromosome had been lost (Table 2 and data not shown). When this experiment was performed in RAD51 cells, 15 to 18% of the resulting colonies were homozygous for all markers telomeric to the HO site (Table 2). The majority of these colonies also contained cells with the mirror image recombination product (as in Fig. 4A), suggesting that most of these colonies had undergone a reciprocal recombination. Since we were unable to identify BIR events in rad51 mutants, which would eliminate reciprocal recombination, we were unable to unambiguously score HO-induced BIR events; BIR events in RAD51 strains would be indistinguishable from reciprocal recombinations (which are more common than BIR) in which one daughter cell was inviable. Therefore, we do not yet know whether BIR is also adaptation dependent in RAD51 cells.
A 218-kb region between two leucine tRNA genes is commonly deleted.
To characterize further the internal deletions seen in Fig. 3, 13 isolates from rad51 CDC5 and rad51 cdc5-ad were run on a CHEF gel, blotted, and probed for the test chromosome (Fig. 5A). Interestingly, the majority of the deletion events appear to produce a chromosome of the same size. Most of these events were independent, since the 26 events shown were taken from six independent strains. DNA from one such deletion was compared to DNA from the starting strain by hybridization on a DNA array. A large region of chromosome VII (from YGL041 to YGL158) displayed a 50% reduction in hybridization (Fig. 5B). The junctions of this deletion each contain genes encoding a 99% identical leucine tRNA. To verify that this deletion occurred between these tRNA genes, we amplified the junction by PCR (Fig. 5C) and sequenced the product (results not shown). Given that these events were homology based and independent of RAD51 (11), they probably arose by SSA (Fig. 5D). CDC5 and cdc5-ad had indistinguishable rates of SSA as measured by inducing an HO endonuclease cut between two direct repeats (data not shown; see Materials and Methods).
DISCUSSION
While adaptation to the DNA damage checkpoint has now been clearly documented in response to several forms of damage, including X-irradiation, an endonucleolytic break, and damage generated by the cdc13 mutation (12, 18, 20), the biological consequences of adaptation to this checkpoint had not been previously explored. We found that at least three forms of genomic instability (chromosome loss, translocation, and BIR) occur after cells have arrested at the checkpoint and subsequently adapted. Previous studies have shown that the rate of nonreciprocal translocation and chromosome loss is higher in checkpoint mutants than in wild-type cells (3, 22). Our data address the events which nonetheless appear in wild-type cells and show that these events are preceded by adaptation to a checkpoint arrest.
The X-ray-induced chromosome loss rate we observed is compatible with the hypothesis that chromosome loss is largely responsible for X-ray-induced death in rad52 CDC5 and rad52 cdc5-ad haploids: if a loss rate of 16% seen for chromosome VII (Table 1) is similar for all 16 chromosomes, then approximately 6% (84%16) of cells should not have a loss event. This corresponds to the viability of the haploid at this dose (6% viability at 2 kilorads [Fig. 1A]). In diploids, only 0.4% (84%32) of cells will not have a chromosome loss event. We propose that rad52 CDC5 diploids are viable well above this level (14% at 2 kilorads; Fig. 1A) because they can often form colonies monosomic for the lost chromosome after adapting to the irreparable damage (Fig. 1E). While the initial growth rates of such colonies are quite low, strong selection for subsequent nondisjunctions in the nascent colonies may allow some of these cells to eventually form fast-growing colonies. If rad52 cdc5-ad diploids arrest permanently with a single unrepaired chromosome, their viability at 2 kilorads should be more similar to 0.4%, which it is (1.1% at 2 kilorads; Fig. 1A). These calculations assume that the absolute frequency of events that lead to chromosome loss is equal in CDC5 and cdc5-ad strains.
The effect of adaptation on resistance to DNA damage depends upon whether the damage is repairable and whether the damaged chromosome is essential. When damage is in an essential chromosome and is irreparable (as with the X-irradiated rad52 haploids in Fig. 1), cells die regardless of whether they adapt. When damage is in a nonessential chromosome and is irreparable (as with the irradiated rad52 diploids in Fig. 1), adaptation mutants will have lower viability because they will remain arrested (and therefore eventually die) with damage that is often not inherently lethal. This scenario is seen even more strikingly when an irreparable HO-directed break is induced in a nonessential chromosome in a rad52 strain (as in Table 2 and reference 20). Cells can also be damaged in essential chromosomes in such a way that the damage is fully reversible using the cdc13 mutation. CDC13 encodes a telomere-binding protein that stabilizes telomeric DNA. When temperature-sensitive cdc13 mutants are brought to the nonpermissive temperature, they will suffer damage at all their telomeres. When the strain is brought to the permissive temperature again, it will repair the damaged telomeric DNA. In this case, adaptation mutants will retain their viability better than wild-type cells because they will remain checkpoint arrested until the temperature is lowered and the cells can repair their telomeres (20).
Our finding that adaptation-defective rad51 strains undergo fewer BIR events is consistent with experiments initially characterizing BIR (15). In these experiments, an endonucleolytic break was seen to initiate a BIR event in later cell cycles. Here, we show that adaptation to the DNA damage checkpoint is required for cells to complete BIR. One explanation is that cells must enter S phase in order for the broken chromosome to either invade the donor chromosome or complete the replication of the broken chromosome. The observation that three different rearrangements were seen in a colony that formed from an X-irradiated cell suggests that the break was carried through at least two cell cycles. It is not immediately obvious why cdc5-ad disomes damaged in G1 could not undergo BIR during S phase. It is possible that the majority of double-stranded breaks that initiate BIR are formed when cells pass through S phase with damage, such that cells would need to pass through one S phase just to generate the correct initiating lesion. It had previously been noted that the checkpoint-defective mutant rad9 has a higher rate of nonreciprocal translocations but not reciprocal translocations (3). Given our findings, it is likely that these events represent BIR; if BIR occurs only after cells have continued through the checkpoint, then checkpoint mutants should have a higher BIR frequency.
Induction of an HO break at the TRP5 locus 50 kb from the centromere did not induce rad51-independent BIR in our strain. This is in contrast to published reports examining BIR in response to an HO break at the MAT locus of chromosome III (15). This suggests that the efficiency of BIR may vary considerably depending upon where the initiating lesion is in the genome. As seen previously, induction of an HO break in a nonessential chromosome leads to death in the cdc5-ad rad52 double mutant. The viability seen here was slightly higher than that seen previously (6.9% ± 1.4% versus 1% ± 1%) (Table 2 and reference 20). The number reported here is likely a slight overestimate because in these experiments HO is not induced before plating, so that many cells incur a break after having first divided once or twice on the plate. This will increase the viability two- to fourfold. Interestingly, rad51 cdc5-ad mutants are not as sensitive to break induction as rad52 cdc5-ad mutants (Table 2). It may be that rad51 strains are able to repair this break in such a way that the checkpoint signaling is shut down but the nearby centromere is destroyed (e.g., by SSA).
Our findings that spontaneous DNA damage seen in recombination-deficient mutants (rad51 and rad52) induces a checkpoint response have also been suggested in mammals. Mice with deletions of RAD51 or the RAD51-associated tumor suppressor BRCA1 die early in development; however, this phenotype is partially rescued by the deletion of the mammalian checkpoint gene p53 or (for BRCA1) p21 (7, 14). While BRCA1 is essential during mouse embryogenesis, the BRCA1 gene is lost during tumorigenesis in some familial breast cancers. In yeast, loss of the RAD51 pathway increases the incidence of BIR, possibly by eliminating competing pathways (15). Given that BRCA1 may be involved in the RAD51 repair pathway (19), it is possible that BIR (which leads to loss of heterozygosity across an entire chromosome arm) may be induced in BRCA1− preneoplastic cells, contributing to their tumorigenicity.
ACKNOWLEDGMENTS
We thank J. Bachant, A. Murray, C. Nugent, A. Page, and members of the Toczyski lab for advice and suggestions and J. DeRisi for assistance with the DNA array.
We also acknowledge institutional support from the UCSF Cancer Center and support from NIH grant GM59691-01.
REFERENCES
- 1.Bosco G, Haber J E. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. . (Erratum, 282:1421.) [DOI] [PubMed] [Google Scholar]
- 3.Fasullo M, Bennett T, AhChing P, Koudelik J. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol Cell Biol. 1998;18:1190–1200. doi: 10.1128/mcb.18.3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Featherstone C, Jackson S P. DNA double-strand break repair. Curr Biol. 1999;9:R759–R761. doi: 10.1016/S0960-9822(00)80005-6. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 6.Haber J E. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 7.Hakem R, de la Pompa J L, Elia A, Potter J, Mak T W. Partial rescue of Brcal (5-6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 8.Heude M, Fabre F. a/alpha-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics. 1993;133:489–498. doi: 10.1093/genetics/133.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes T R, Roberts C J, Dai H, Jones A R, Meyer M R, Slade D, Burchard J, Dow S, Ward T R, Kidd M J, Friend S H, Marton M J. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 10.Iadonato S P, Gnirke A. RARE-cleavage analysis of YACs. Methods Mol Biol. 1996;54:75–85. doi: 10.1385/0-89603-313-9:75. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov E L, Sugawara N, Fishman-Lobell J, Haber J E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer K M, Haber J E. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 13.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R D, Haber J E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 14.Lim D S, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrow D M, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortimer R K, Contopoulou R, Schild D. Mitotic chromosome loss in a radiation-sensitive strain of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:5778–5782. doi: 10.1073/pnas.78.9.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 19.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Association of BRCAI with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 20.Toczyski D P, Galgoczy D J, Hartwell L H. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 21.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 22.Weinert T, Hartwell L H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells W A, Murray A W. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]