Abstract
53BP1 is a human BRCT protein that was originally identified as a p53-interacting protein by the Saccharomyces cerevisiae two-hybrid screen. Although the carboxyl-terminal BRCT domain shows similarity to Crb2, a DNA damage checkpoint protein in fission yeast, there is no evidence so far that implicates 53BP1 in the checkpoint. We have identified a Xenopus homologue of 53BP1 (XL53BP1). XL53BP1 is associated with chromatin and, in some cells, localized to a few large foci under normal conditions. Gamma-ray irradiation induces increased numbers of the nuclear foci in a dose-dependent manner. The damage-induced 53BP1 foci appear rapidly (in 30 min) after irradiation, and de novo protein synthesis is not required for this response. In human cells, 53BP1 foci colocalize with Mrel1 foci at later stages of the postirradiation period. XL53BP1 is hyperphosphorylated after X-ray irradiation, and inhibitors of ATM-related kinases delay the relocalization and reduce the phosphorylation of XL53BP1 in response to X-irradiation. In AT cells, which lack ATM kinase, the irradiation-induced responses of 53BP1 are similarly affected. These results suggest a role for 53BP1 in the DNA damage response and/or checkpoint control which may involve signaling of damage to p53.
Double-stranded DNA (dsDNA) breaks are potentially dangerous to cells since they may lead to chromosome breakage and loss of genetic information. However, transient dsDNA breaks are essential to initiate recombination or to solve topological problems at the end of DNA synthesis (24, 58, 63). dsDNA breaks can also occur if replication forks are stalled (e.g., due to base modifications, single-stranded-DNA gaps, or low deoxynucleoside triphosphate pools), and in Escherichia coli, RuvABC Holliday junction resolvase has been shown to catalyze the breakage (53, 68). In general, cells do not enter S or M phase before the DNA lesions are properly repaired due to the action of the DNA damage checkpoint (19). The sensitivity of cancer cells to DNA-damaging agents is explained by the fact that cancer cells have often lost some aspects of checkpoint function, which has provided them with a higher rate of genomic evolution to acquire a growth advantage (18).
The DNA damage checkpoint is a signal transduction cascade that relays information from DNA lesions to components of the cell cycle (reviewed in references 5, 40, and 41). In response to DNA damage, ATM-related protein kinases (ATM, ATR, and possibly DNA-PK) activate the downstream effector checkpoint kinases Chk1 and Chk2 (28, 35, 62). Then Chk2 (homologues are Rad53 in budding yeast and Cds1 in fission yeast) phosphorylates p53 or Cdc25A, which arrests the cell cycle at the G1/S or G2/M boundary, respectively (6, 7, 8, 20, 54). Compared to these downstream events, the molecular mechanism of how DNA lesions activate the kinase cascade is not well understood. Yeast genetics have identified candidate genes that appear to be involved in these early sensing and processing stages. These genes are classified into three groups: yeast homologues of ATM-related kinases that are structurally similar to phosphatidylinositol 3 kinase (Rad3sp/Mec1sc and Tel1; superscripts are sp for fission yeast and sc for budding yeast genes, respectively), putative sliding clamp and clamp loader complexes structurally related to PCNA and RFC (e.g., Rad1sp, Hus1sp, and Rad17sp), and proteins with BRCT domains (Crb2sp/Rad9sc). In response to DNA damage, ATM-related kinases are activated by unknown mechanisms and the putative sliding clamp components are phosphorylated (13, 27, 42). The sliding clamp and its loader are required for the phosphorylation of BRCT proteins, and BRCT proteins are required for phosphorylation and activation of the checkpoint kinases (48). The carboxyl (C) termini of BRCT proteins contain a tandem repeat of BRCT domains, each consisting of ∼95 amino acid residues, that was first identified in BRCA1 (3). In human and in mouse, BRCA1 is thought to function as a counterpart of the BRCT proteins in yeasts. A number of observations indicate that the BRCT domain promotes protein-protein interactions (11, 34, 37). In contrast to their C termini, the amino (N) termini of BRCT proteins are not homologous to each other and the molecular functions of these large N-terminal domains are totally unknown.
Some proteins involved in repair or checkpoint control are relocalized to nuclear foci after DNA damage. These include BRCA1, Rad51, and the Mre11-Rad50-Nbs1 complex (33, 43, 52, 64). Although the functional significance of these damage-induced foci is unknown, the increased local concentration of the proteins facilitates their functions, such as enzymatic and signal transduction processes.
53BP1 is a human BRCT protein that was originally identified by the yeast two-hybrid screen using p53 as a bait (22). The protein consists of 1,972 amino acid residues and has a tandem repeat of the BRCT domain at its C terminus, which shows similarity to the BRCT domain of Crb2sp (48). Although 53BP1 was shown to stimulate p53-mediated transcription in a transient-transfection assay (23), so far there is no evidence implicating this protein in the DNA damage checkpoint and/or repair pathway. To investigate this possibility, we have cloned a Xenopus homologue of 53BP1 and raised antibodies against the protein. As an initial step towards the understanding of the function of 53BP1, we examined the behavior of XL53BP1 in frog cell lines in response to DNA damage. We found that 53BP1 is relocalized to a number of nuclear foci after DNA damage. dsDNA breaks are most effective in inducing this focal phenotype. The focal redistribution of 53BP1 appears to be related to the extent of its phosphorylation, which is also induced by DNA damage. We will discuss a functional involvement of ATM-related kinases in the focal relocalization of 53BP1.
MATERIALS AND METHODS
Cell culture and drug treatments.
A Xenopus cell line (XTC-2) was grown in a solution containing 60% Leibovitz L-15, 10% fetal calf serum, and 5 to 50 μg of gentamicin per ml at room temperature (55). Human cells (HT1080 and the AT cell line AT1BR/11 obtained from the MRC Cell Mutation Unit, University of Sussex) were grown in Dulbecco's modified Eagle's medium, 10% fetal calf serum, and 5 μg of gentamicin per ml at 37°C with 5% CO2. For treatment with caffeine (0.5 M stock solution in H2O) or with wortmannin (10-mg/ml stock solution in dimethyl sulfoxide [DMSO]; Sigma), XTC-2 cells grown on cover slips were preincubated for 1 h in media containing either 16 mM caffeine or 23 μg of wortmannin per ml before the X-ray irradiation. Cycloheximide (50-mg/ml stock solution in ethanol) and actinomycin D were used as described above at 50 and 25 μg/ml, respectively.
Cloning of XL53BP1 and plasmid construction.
Xenopus cDNA encoding a 53BP1 homologue was cloned by low-stringency hybridization. A Xenopus oocyte cDNA library in lambda gt10 (45) was screened with human cDNA encoding from Ser 1618 to Pro 1963 of human 53BP1 (23). Hybridization was performed at 30°C in a solution of 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 50% formamide, 5× Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS) and 8% dextran sulfate for 16 h. The filters (Hybond-N+; Amersham Pharmacia Biotech) were washed twice at room temperature with 2× SSPE–0.1% SDS and twice at 58°C with 1× SSPE–0.1% SDS. The length of the combined sequences of several overlapping clones was 5,387 bp, and the longest open reading frame (ORF) was 1 to 5199 bp, encoding 1,733 C-terminal amino acid residues.
Antibody preparation.
The 1.5-kb HindIII-EcoRI fragment of XL53BP1 cDNA was cloned into the pRSET plasmid (Invitrogen), and the recombinant protein, tagged with six histidines, was expressed in BL21 (DE3) with pLysS. The protein found in an insoluble fraction was run on an SDS-polyacrylamide gel, eluted by diffusion, and used as an antigen to immunize rabbits (1). Poly(A) · poly(U) was used as an adjuvant (21). The corresponding domain of human 53BP1 was expressed and used for the antigen as described above. Antibodies were affinity purified with an antigen column as follows. The insoluble fraction containing approximately 10 mg of the His-tagged recombinant protein was solubilized with 6 M guanidine-HCl and loaded onto 1 ml of Ni-nitrilotriacetic acid agarose (Qiagen). The protein was renatured on the column by applying a 6 to 1 M urea gradient essentially according to the manufacturer's instructions. The peak fractions were pooled and dialyzed against HNG buffer (0.5 M NaCl, 10% glycerol, 10 mM HEPES-NaOH [pH 7.5]) overnight at 4°C. The dialyzed fractions containing the solubilized recombinant proteins were coupled to Affigel 10 (Bio-Rad) by following the instructions supplied by the manufacturer. Approximately 1 mg of the recombinant protein was coupled to 1 ml of the Affigel resin. Affinity column chromatography was performed essentially according to a standard procedure (17). The antibodies were concentrated to ∼1 ml with Centricon 30 (Amicon), dialyzed against an appropriate buffer, and stored at −80°C in aliquots.
Immunoprecipitation and immunoblotting.
XTC-2 or HT1080 cells were rinsed briefly with Marc's modified ringer's solution (MMR; 100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM HEPES-NaOH [pH 7.4] [55]) or with phosphate-buffered saline (PBS), respectively. The dishes were placed on ice and LA buffer (50 mM HEPES-NaOH [pH 7.5], 0.5 mM EDTA, 80 mM β-glycerophosphate, 1 mM Na-vanadate, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], 1 μM lactacystin, 1% NP-40, 50 mM NaF, 400 mM NaCl, 1× protein inhibitor cocktail) was applied at 6.7 μl/cm2. Protein inhibitor cocktail (1,000×) contained 10 mg each of leupeptin, pepstatin A, chymostatin, and antipain per ml in DMSO. Cells were scraped off with a silicon rubber scraper, and the cell lysates were spun for 10 min in a microcentrifuge at 4°C. The supernatants were stored at −80°C. Protein concentrations were estimated by the Bradford assay (4) using bovine serum albumin (BSA) as a standard. Immunoprecipitation was performed as follows. Four hundred fifty microliters of the cell lysates was diluted with 900 μl of dilution buffer (10 mM HEPES-NaOH [pH 7.5], 0.5 mM EDTA, 10 mM β-glycerophosphate, 1 mM Na-vanadate, 0.1 mM PMSF, 0.5 mM DTT, 0.02% NP-40) and spun for 10 min in a microcentrifuge at 4°C. The supernatants were added with 6 μg of the affinity-purified antibodies and incubated for 1 h at 4°C on a rotating wheel. Twenty microliters of proteins A-agarose beads (Gibco BRL) was added, and incubation was continued for 1 h. The beads were washed five times with washing buffer (10 mM HEPES-NaOH [pH 7.5], 0.5 mM EDTA, 10 mM β-glycerophosphate, 50 mM NaF, 100 mM NaCl, 1 mM Na-vanadate, 0.1 mM PMSF, 0.5 mM DTT, 0.02% NP-40). The proteins were eluted with 20 to 50 μl of denaturation buffers appropriate for the following gel electrophoresis. For phosphatase treatments of immunoprecipitates, the resins were washed three times with Tris-buffered saline, resuspended in 15 μl of phosphatase buffer (50 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 5 mM DTT, 0.01% Brij 35, and 2 mM MnCl2) plus 400 U of lambda protein phosphatase (New England Biolabs), and incubated for 30 min at 30°C. The reaction was stopped by adding EDTA to 5 mM and appropriate sample buffers for gel electrophoresis. Proteins were separated on standard Laemmli gels (5 to 12% gradient [29]) or on NuPAGE Tris-acetate gels (3 to 8% gradient; Novex). For Laemmli gels, the proteins were electrotransferred onto nitrocellulose filters with Tris-glycine buffer containing 10% methanol (60). For NuPAGE gels, we followed the protocol supplied by the manufacturer. The filters were blocked with 5% skimmed milk in Tris-buffered saline and probed with 1 μg of the affinity-purified antibodies per ml. Preimmune sera were diluted to 1:1,000 and used for blotting. The antibodies conjugated with peroxidase were used as secondary antibodies. Enhanced chemiluminescence was used for detection.
Immunofluorescence staining.
Essentially the standard procedure was followed (17). XTC-2 cells grown on coverslips were briefly rinsed with MMR and fixed with freshly prepared 3% paraformaldehyde in MMR for 10 min. The cells were permeabilized in 0.5% Triton X-100 in PBS for 2 min at room temperature. After being washed with PBS, the coverslips were blocked in 3% BSA in PBS for 1 h. HT1080 cells were fixed and permeabilized in 50% methanol–50% acetone at −20°C for 2 min. The first antibodies were affinity-purified antibodies diluted to 10 μg/ml in 3% BSA in PBS. Preimmune serum was used at a 1:100 dilution as a negative control. Antibodies were labeled with fluorescein isothiocyanate (FITC) as described previously (17). For double labeling of HT1080 cells, cells were first probed with anti-Mre11 antibodies (Novus Biologicals) that are detected by the anti-rabbit immunoglobulin G Cy5. Then the samples were fixed with paraformaldehyde, followed by blocking and probing with anti-53BP1 antibody–FITC. The coverslips were mounted in 85% glycerol–2.5% propyl gallate–2.5 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml. Double labeling with anti-53BP1 antibody and bromodeoxyuridine was performed as follows. XTC-2 cells on cover glasses were incubated in the presence of 10 μg of bromodeoxyuridine per ml for 2 h, fixed, and stained with anti-XL53BP1 antibody as described above using secondary antibodies conjugated with rhodamine. After the final wash, the cells were fixed again, rinsed with PBS twice, and treated with 50,000 U of DNase I per ml in a solution containing 150 mM NaCl, 10 mM sodium acetate (pH 5.2), 10 mM MgCl2, and 100 μg of BSA per ml for 30 min at room temperature. The cover glasses were rinsed three times with PBS and probed with monoclonal antibodies against bromodeoxyuridine (Roche Molecular Biology), which was then detected by FITC-labeled secondary antibodies. Images were recorded with either a conventional microscope (Zeiss Axioskop microscope equipped with a VYSIS image recording system) or a confocal microscope (Leica).
Nucelotide sequence accession number.
The sequence data for XL53BP1 are available from the DDBJ, EMBL, and GenBank databases under accession no. AJ298224 and XLA298224.
RESULTS
Isolation of a Xenopus 53BP1 homologue, XL53BP1.
We cloned a cDNA encoding a Xenopus 53BP1 homologue by low-stringency hybridization. The human cDNA sequence encoding the C-terminal 345 amino acid residues was amplified by PCR and used as a probe to screen a Xenopus oocyte cDNA library. The longest clone obtained in the first screening encoded only 60% of the expected full length, and another screening was performed using the 5′ end of the first-round clone as a probe. The combined sequence of the overlapping clones had an open reading frame of 1,737 amino acid residues but still lacked the N terminus (N.B., human 53BP1 is 1,972 amino acids long). The carboxyl-terminal 1,100 amino acid residues of XL53BP1 shares 57% identity and 80% similarity with human 53BP1. The N-terminal half of XL53BP1 shows low similarity to the human protein (data not shown). One common feature of the N-terminal halves of human and Xenopus 53BP1 is the presence of SQ/TQ motifs spread throughout the region (22 motifs for the human and 9 for the Xenopus 53BP1). SQ/TQ motifs are the preferred phosphorylation sites of members of the ATM-related kinase family (25). In addition, both proteins are rich in acidic amino acid residues (with a pI of 4.0 for the human and a pI of 4.3 for the Xenopus 53BP1).
53BP1 exists as an oligomeric protein complex.
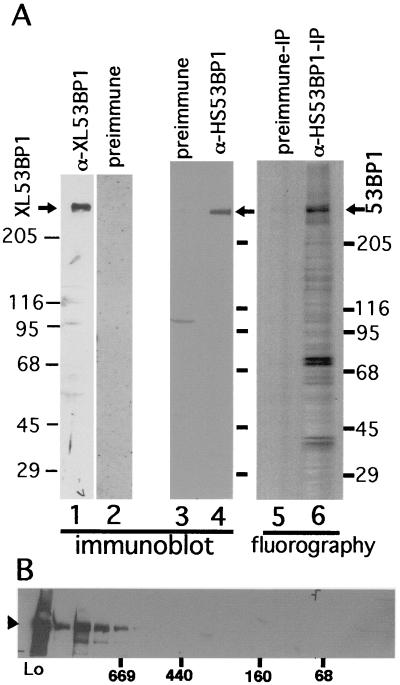
We raised antibodies against the C-terminal 500 amino acid residues of XL53BP1 and those of human 53BP1, both of which were produced in E. coli. Affinity-purified anti-XL53BP1 and anti-HS53BP1 antibodies recognized a single protein of ∼300 kDa by immunoblotting (Fig. 1A, lanes 1 and 4). Preimmune sera showed only very weak reactions with proteins in the extracts (lanes 2 and 3; note that, after the affinity purification, those minor bands disappeared). Thus, in SDS-polyacrylamide gel electrophoresis Xenopus and human 53BP1 appeared to migrate significantly more slowly than expected from the primary sequence. Slower migration of 53BP1 was previously reported for human 53BP1, in which its identity was confirmed by hemagglutinin epitope tagging (23).
FIG. 1.
(A) Verification of antibodies against 53BP1 and immunoprecipitation. Cell lysates were prepared from Xenopus cells (XTC-2), (lanes 1 and 2) or from HeLa cells (lanes 3 and 4), run on an SDS-polyacrylamide gel, and subjected to immunoblotting. Affinity-purified anti-XL53BP1 (Xenopus cells; lane 1) or anti-HS53BP1 (human; lane 4) antibodies react with single proteins in the extracts. Preimmune sera were used as negative controls (lanes 2 and 3). Lane 6, fluorography of immunoprecipitate (IP) with anti-HS53BP1 antibody from 35S-labeled HeLa cell extracts lane 5, immunoprecipitation with preimmune serum control. (B) Gel filtration of HeLa cell extracts. The extract (Lo) was loaded on a Superdex 200 (10/30) column, and the fractions were analyzed by immunoblotting with anti-HS53BP1 antibodies. Positions where the molecular mass markers were eluted are indicated below the gel in kilodaltons.
Anti-HS53BP1 antibodies precipitated the protein with an apparent molecular mass of 300 kDa from HeLa cell extracts labeled with [35S]methionine-cysteine (Fig. 1, lane 6). There also appeared to be several other proteins that coimmunoprecipitated with 53BP1. The preimmune serum control precipitated no detectable proteins (lane 5). Separation of the HeLa cell extracts by a gel filtration column (Superdex 200) showed that 53BP1 was eluted in fractions larger than 669 kDa (approximately 1 MDa) (Fig. 1B). These results suggested that 53BP1 existed as an oligomeric protein complex.
XL53BP1 is associated with nuclear foci that are induced by various DNA-damaging agents.
Some of the proteins involved in the DNA damage repair checkpoint are known to change their localization in response to DNA damage. To determine whether this is the case for 53BP1, we examined the cellular localizations of XL53BP1 under differing conditions. The Xenopus cell line XTC2 was fixed and stained with affinity-purified anti-XL53BP1 antibodies. XL53BP1 showed an exclusively nuclear localization in interphase cells (Fig. 2A). Cells stained with preimmune serum showed little signal under the experimental conditions (Fig. 1B; images were recorded under the same sensitivity setting for panels A to E). In mitosis, XL53BP1 appeared to be dissociated from mitotic chromosomes and was dispersed throughout the cytoplasm (data not shown; see Fig. 5 for human cells). The nuclear XL53BP1 appeared to exist as two populations. One was distributed homogeneously over the chromatin, and the other was associated with a few large foci inside the nucleus. Since the preimmune serum control did not show any chromatin signal, the overall chromatin staining was due to XL53BP1 and not nonspecific background. Often the bright foci were adjacent to nucleoli (Fig. 2A; the nucleolus appears as a dark hole with the XL53BP1 stain). On average, one nucleus had 2.1 ± 3.4 XL53BP1 foci. Approximately half of the nuclei did not have any XL53BP1 foci. Double labeling with anti-XL53BP1 antibody and with bromodeoxyuridine for S-phase nuclei indicated that there was no obvious relationship between the presence of bright XL53BP1 foci and S phase (data not shown).
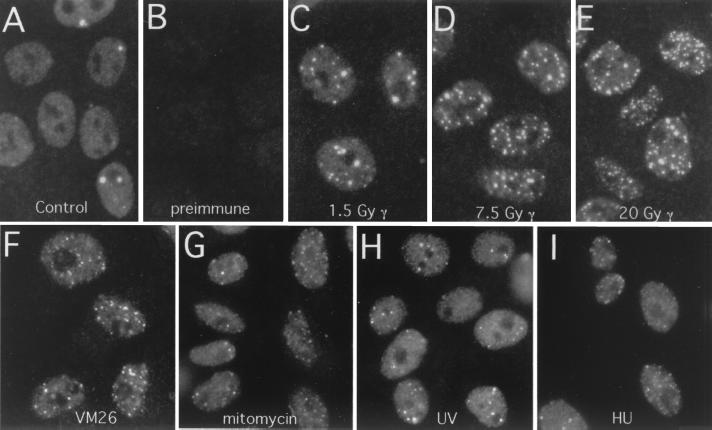
FIG. 2.
XL53BP1 localizes to nuclear foci, which increase in number after DNA damage. Xenopus XTC-2 cells were immunostained with anti-XL53BP1 antibodies. (A) In control cells, there were two types of XL53BP1 populations. One was homogeneously distributed over chromatin, and the other was associated with large foci. (B) Staining with preimmune serum showed very little background. (C to E) After exposure to γ rays, XL53BP1 formed increased numbers of smaller foci dispersed throughout the nucleus. γ-Ray dosages were 1.5 Gy (C), 7.5 Gy (D), and 20 Gy (E). (F to I) XL53BP1 foci were also induced by various DNA-damaging agents. Cells were subjected to 3.3 μg of VM26 per ml (F), 60 μg of mitomycin C per ml (G), 300 J of UV per m2 (H), and 10 mM hydroxyurea (HU) (I). Cells were fixed for staining at 2 h postirradiation or at 16 h after addition of the chemicals.
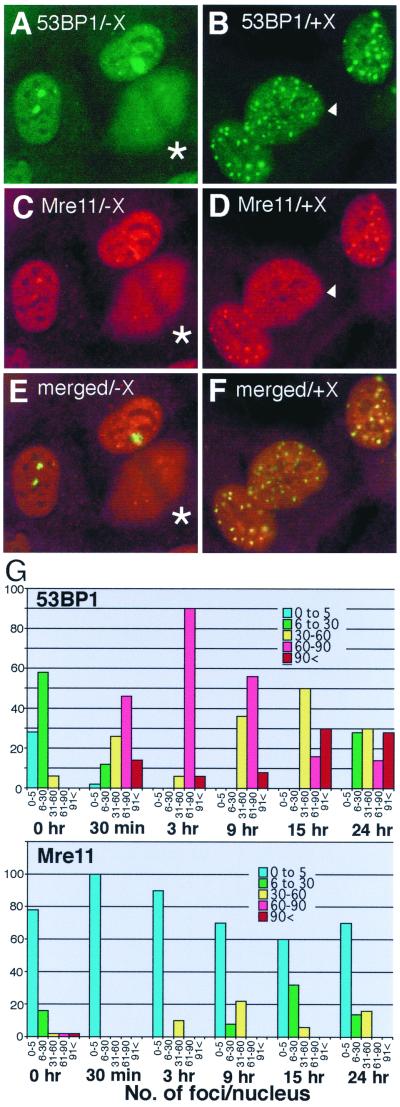
FIG. 5.
Colocalization of 53BP1 foci and Mre11 foci in human HT1080 cells. HT1080 cells untreated (A, C, and E) or exposed to 84 Gy of X rays (X) (B, D, and F) were fixed at 15 h postirradiation and double stained with Cy5 for Mre11 (C and D; pseudo-colored in red) and with FITC for 53BP1 (A and B; green). (E and F) Merged images of panels A and C and of B and D, respectively. Colocalization of green foci and red foci yields yellow signals. The cell marked by an arrowhead in panels B and D had 53BP1 foci but not discrete Mre11 foci. The cells marked by asterisks in panels C and D were in mitosis, and 53BP1 and Mre11 were dissociated from mitotic chromosomes and spread throughout the cytoplasm. (G) Kinetics of 53BP1 and Mre11 focal formation. HT1080 cells were either untreated (0 h) or irradiated with X rays, fixed at the indicated times postirradiation, and stained independently for either 53BP1 or Mre11. Optical sections were recorded by confocal microscope. The number of 53BP1 foci (upper graph) or Mre11 (lower graph) foci per nucleus was counted in 50 cells for each sample. Nuclei were categorized as having 0 to 5 foci (blue bars), 6 to 30 foci (green bars), 31 to 60 foci (yellow bars), 61 to 90 foci (red bars), and >91 foci (brown bars) at each time point. The y axis indicates the percentage of each category at each time point. For Mre11, only large, well-distinguished foci were scored (e.g., the nucleus marked with an arrowhead in panel D was scored as negative for Mre11 foci). Thus, the number of Mre11 foci may have been underestimated. Note that 53BP1 foci form early at 30 min postirradiation, when Mre11 foci were not observed at all.
We next investigated the effects of ionizing irradiation on the localization of XL53BP1. XTC-2 cells were exposed to increasing doses of γ rays, fixed after 2 h of incubation, and stained for XL53BP1 (Fig. 2C to E). Exposure to γ rays caused the appearance of an increased number of smaller XL53BP1 foci inside the nuclei in most of the cells. We performed optical sectioning using a confocal microscope and scored the number of XL53BP1 foci per nucleus for 100 cells. The number of XL53BP1 foci increased in a dose-dependent manner (Fig. 3A).
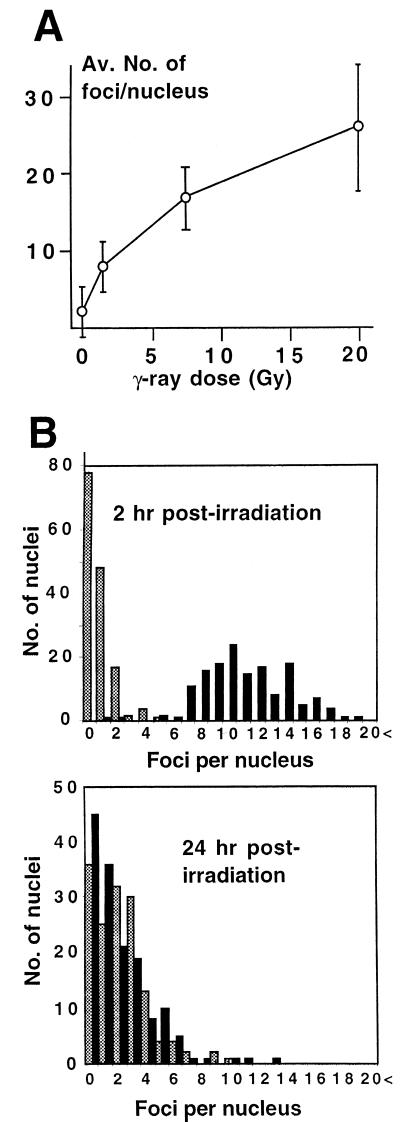
FIG. 3.
(A) γ-Ray-dose-dependent increase of XL53BP1 focal number. The average number of foci per nucleus was plotted versus radiation dose. Error bars indicate standard deviations. (B) Recovery of the XL53BP1 focal phenotype after γ-ray irradiation. XTC-2 cells were either untreated (stippled bars) or irradiated with 5 Gy of γ rays (black bars) and fixed for XL53BP1 staining at 2 h or at 24 h postirradiation. Optical sections were recorded by confocal microscope. The number of XL53BP1 foci per nucleus was counted in 150 cells for each sample. At 2 h postirradiation, the nuclei had 10.9 ± 3.2 XL53BP1 foci (upper graph, black bars). At 24 h postirradiation, the average focus number per nucleus decreased to 1.99 ± 2.15 (lower graph, black bars), which was equivalent to that of the untreated control cells (stippled bars).
We examined the localization of XL53BP1 in cells treated with other DNA-damaging agents (Fig. 2F to I). The results of the treatments examined were as follows: VM26 inhibited topoisomerase II activity by stabilizing the covalent dsDNA break protein intermediate (Fig. 2F) (9), mitomycin C cross-linked interstrand DNA (Fig. 2G), UV induced pyrimidine dimer formation (Fig. 2H) and hydroxyurea reduced the cellular deoxynucleoside triphosphate concentration through inhibiting ribonucleotide reductase (Fig. 2I). It is worth noting that each agent may generate various other lesions as secondary or tertiary effects. We observed that ionizing irradiation (X or γ rays) and VM26 treatments were most effective at inducing the formation of numerous smaller XL53BP1 foci (Fig. 2C to F; see also Fig. 5). The other treatments were less effective and induced smaller and fewer 53BP1 foci (Fig. 2G to I).
The XL53BP1 foci dissociate upon recovery from DNA damage.
In order to determine the fate of the XL53BP1 foci after the lesions were repaired, we examined the XL53BP1 localization 24 h postirradiation (26, 39). XTC-2 cells were exposed to 5 Gy of γ rays. In separate experiments, more than 95% of the cells survived at this dose of irradiation (data not shown). We noted that XTC-2 cells were significantly more resistant to γ-ray irradiation than mouse or human cells for unknown reasons. (Only 20% of mouse cells survived under the same conditions. This might be partly due to the fact that the Xenopus genome has ancient tetraploid features.) The cells were fixed at 2 h or at 24 h postirradiation and stained for XL53BP1. The number of foci per nucleus was counted in 150 cells and scored (Fig. 3B, black bars). Nonirradiated cells served as a control (stippled bars). At 2 h postirradiation, the average number of XL53BP1 foci per nucleus increased to 10.9 ± 3.2 from 0.73 ± 0.98 in the nonirradiated control (Fig. 3B, upper graph). At 24 h postirradiation, the number decreased to 1.99 ± 2.15 foci per nucleus. This number was indistinguishable from the value (2.13 ± 1.95) for the nonirradiated control at the same time point (lower graph). These results indicated that the XL53BP1 foci assembled transiently in response to DNA damage and that they dissociated at later times, presumably after the lesions were repaired.
XL53BP1 is relocalized to the damage-induced foci: de novo synthesis is not required.
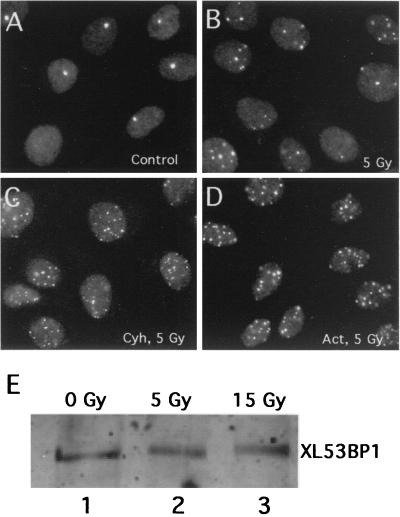
Is the induction of XL53BP1 foci in response to DNA damage due to an increased level of protein expression? To answer this question, we examined whether the inhibition of transcription or translation would block the assembly of XL53BP1 foci. XTC2 cells were exposed to 5 Gy of γ rays in the presence of either cycloheximide or actinomycin D, and after 2 h of incubation the cells were stained for XL53BP1. As shown in Fig. 4, the damage-induced XL53BP1 foci assembly occurred efficiently in the presence of these inhibitors (compare Fig. 4B to D). There were slight increases in the numbers of XL53BP1 foci per nucleus after irradiation in the presence of these inhibitors, although the reason for this was unknown. In a control experiment, there was very little incorporation of35S-labeled amino acids into proteins in the presence of 50 μg of cycloheximide per ml (data not shown). To further confirm these results, we compared the amounts of XL53BP1 in cells exposed or not exposed to γ rays by quantitative immunoblotting (Fig. 4E). We did not observe any change of the XL53BP1 levels before and after exposure to γ rays (compare lanes 1, 2, and 3). These results indicated that the increased number of XL53BP1 foci in cells after ionizing irradiation was due to relocalization of the protein that was already there and that de novo synthesis was not required.
FIG. 4.
Neither inhibitors of transcription nor translation blocks the assembly of XL53BP1. XTC-2 cells were irradiated with 5 Gy of γ rays in the presence of 50 μg of cycloheximide (Cyh) per ml (C) or 25 μg of actinomycin D (Act) per ml (D). (A) Nonirradiated control. (B) Cells irradiated without the inhibitors. (E) the amount of XL53BP1 did not increase after γ-ray irradiation. XTC-2 cells were exposed to 5 or 15 Gy of γ rays, and at 2 h after irradiation, cell extracts were prepared for immunoblotting with anti-XL53BP1 antibody. Each slot was loaded with the same amount of proteins.
Colocalization of 53BP1 foci with Mre11 foci in human cells.
As described above, XL53BP1 relocalized to nuclear foci transiently in response to DNA damage. In the absence of DNA damage, the protein did not appear to change its localization in a cell cycle-dependent manner except in M phase. This behavior of XL53BP1 is quite similar to that of the Rad50-Mre11-Nbs1 complex in human cells (33, 38). To investigate the spatial relationship between 53BP1 and Mre11, we monitored the kinetics of 53BP1 and Mre11 focus assembly in response to DNA damage in the human fibrosarcoma cell line HT1080. HT1080 cells were either untreated or exposed to 84 Gy of X rays, fixed at 15 h postirradiation and doubly stained with anti-53BP1 (detected by FITC) and anti-Mre11 (detected by Cy5) antibodies (Fig. 5A to F). Increased numbers of smaller 53BP1 foci were induced in response to X-ray irradiation (Fig. 5B). At this later time postirradiation, in 40% of cells, Mre11 localized to discrete foci as previously reported (Fig. 5D) (33). Mre11 foci showed almost 100% colocalization with 53BP1 foci in these cells (Fig. 5F, a merged image of panels B and D). In untreated HT1080 cells, there were smaller numbers of 53BP1 foci and the cells had Mre11 foci at the corresponding sites, although the Mre11 signals were not as prominent as 53BP1 signals (Fig. 5A, C, and E). It is worth noting that bleedthrough of the FITC and Cy5 signals to the opposite channel was minimal under the experimental conditions (data not shown).
In Fig. 5G, the number of 53BP1 foci (upper graph) or Mre11 foci (lower graph) per nucleus were counted for 50 nuclei at each time point after X-ray irradiation using a confocal microscope and categorized as indicated, and the percentages of each category were plotted. The average number of 53BP1 foci in untreated HT1080 cells was 12.9 ± 10, which was higher than the number in Xenopus XTC-2 and AT cells for unknown reasons (see Table 1). Some HT1080 cells had very few foci and some had more (5 to 30 foci/nucleus) under normal conditions (see Fig. 8D). At 30 min postirradiation, the number of 53BP1 foci increased to 62 ± 24. In contrast, at this early stage of postirradiation Mre11 did not show discrete focal distribution (Fig. 5G, lower graph) (at 30 min there were no countable foci). This result appeared to be consistent with the previous observation by Nelms et al. (38), who had shown that Mre11 is recruited to chromatin in the nuclear volume, where the DNA lesions and repair synthesis occur as early as 30 min after the irradiation though the protein does not show focal distribution under a standard microscope. As reported, discrete Mre11 foci were observed only at later stages of postirradiation (9, 15, and 24 h) (33). Here we scored only large discrete Mre11 foci. Since Mre11 staining had a higher overall chromatin signal than did 53BP1 staining, it was more difficult to count convincingly the smaller Mre11 foci (e.g., the nucleus marked with an arrowhead in Fig. 5D was scored as negative for Mre11 foci; note that the counting was performed with cells stained singly either with anti-53BP1 or with anti-Mre11 antibody). So in Fig. 5G, the number of Mre11 foci was likely to be underestimated when compared with the number of 53BP1 foci, which always showed more contrast than did the overall chromatin staining. Thus, in human cells, 53BP1 focus assembly was induced at quite an early stage in response to DNA damage.
TABLE 1.
Slight delay in 53BP1 focus formation in AT cells and in XTC-2 cells in the presence of wortmannin after X-ray exposure
| Cell type | Avg no. of 53BP1 foci per nucleus at indicated time after X-ray irradiation:
|
||
|---|---|---|---|
| 0 h | 30 min | 3 h | |
| HT1080 | 12.9 ± 10.3 | 62.1 ± 24.9 | 76.5 ± 10.4 |
| AT | 0.82 ± 1.34 | 23.5 ± 13.2 | 67.7 ± 21.6a |
| XTC-2 | 1.5 ± 1.5 | 54 ± 18 | 73 ± 13 |
| XTC-2 + wortmannin | 1.5 ± 1.5 | 30 ± 16 | 68 ± 14 |
At 6 h.
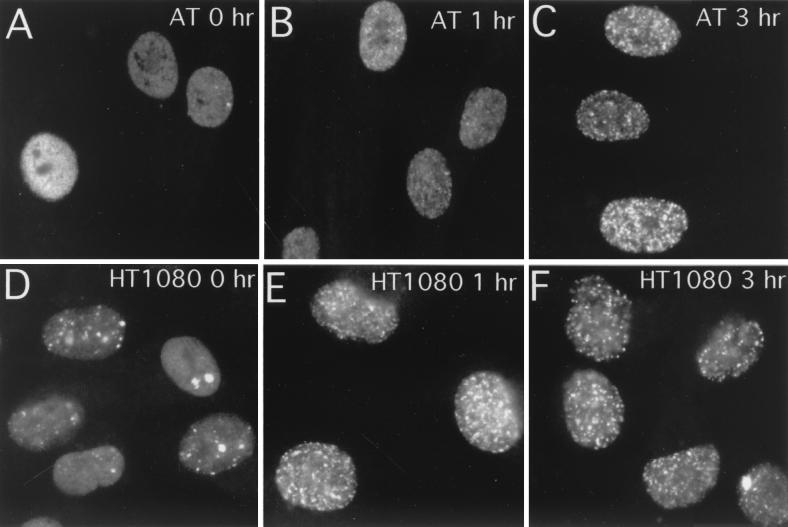
FIG. 8.
AT cells show a slight delay in 53BP1 focus formation in response to X-ray irradiation. AT cells (A to C) or HT1080 cells (D to F) were either untreated (A and D) or exposed to X rays, fixed at 1 h (B and E) or at 3 h (C and F) postirradiation, and stained for 53BP1.
Hyperphosphorylation of XL53BP1 in cells irradiated with X rays.
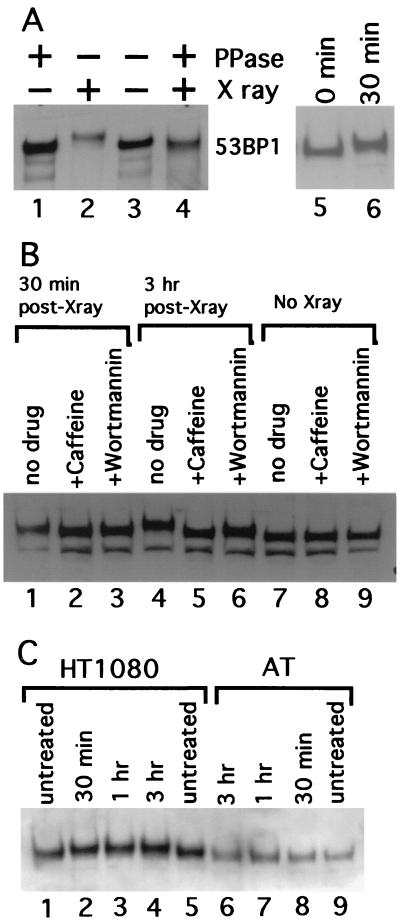
We examined whether XL53BP1 was phosphorylated in response to ionizing irradiation, as this has been reported for other checkpoint proteins like BRCA1, Crb2, Nbs1, and Rad9sp (10, 31, 65, 67). XL53BP1 from cells irradiated with X rays migrated more slowly than the XL53BP1 from untreated cells on SDS-polyacrylamide gel electrophoresis (Fig. 6A, lanes 2 and 3). This retardation was due to phosphorylation, since this slower migration was eliminated by treatment of the immunoprecipitated XL53BP1 with protein phosphatase (Fig. 6A, lane 4). The shift was already visible at 30 min postirradiation (lanes 5 and 6; see also Fig. 6C, lanes 1 and 2, for human cells) and became more pronounced at 3 h (Fig. 6B, lanes 1, 4, and 7). Note that there was no significant increase in the XL53BP1 levels after the X-irradiation.
FIG. 6.
Phosphorylation of XL53BP1 in cells irradiated with X rays. (A) XL53BP1 was immunoprecipitated from XTC-2 cells either untreated (lanes 1 and 3; − X ray) or exposed to X rays (lanes 2 and 4; + X ray) at 2 h postirradiation. Half of the samples were treated with protein phosphatase (lanes 1 and 4; + PPase). The proteins were run on a gel and immunoblotted with anti-XL53BP1 antibodies. XL53BP1 of irradiated cells migrated more slowly than that of nonirradiated cells (lanes 2). This retardation was lost if the sample was phosphatased (lane 4). Lanes 5 and 6 indicate that the band retardation was already seen at 30 min postirradiation. (B) Caffeine blocks the phosphorylation of XL53BP1. XTC-2 cells were either untreated (lanes 7 to and 9) or exposed to X rays (lanes 1 to 6) in the absence of drugs (lanes 1, 4, and 7) or in the presence of 16 mM caffeine (lanes 2, 5, and 8) or 23 μM wortmannin (lanes 3, 6, and 9). Extracts were prepared from irradiated cells at 30 min (lanes 1, 2, and 3) and at 3 h (lanes 4 to 6) post-X-ray exposure. The extent of the band retardation was significantly reduced in the presence of caffeine (e.g., compare lanes 4 and 5). Wortmannin had a similar effect but with lower efficiency (lane 6). The same amount of protein was loaded per slot. The lower bands were degradation products of XL53BP1. (C) AT cells show a reduction in 53BP1 phosphorylation in response to X-ray irradiation. Extracts were prepared from untreated cells (lanes 1, 5, and 9) or from cells irradiated with X rays, at 30 min (lanes 2 and 8), at 1 h (lanes 3 and 7), and at 3 h (lanes 4 and 6) postirradiation. In control ATM+ HT1080 cells (lanes 1 to 5), 53BP1 showed slower migration after X-ray irradiation (lanes 2 to 4). In AT cells that are ATM kinase defective (lanes 6 to 9), the retardation was greatly reduced and the 53BP1 band showed only a slight retardation at 1 and 3 h compared with that of the untreated control (compare lanes 6 and 7 to 9). 53BP1 from the untreated HT1080 cells ran slightly more slowly than that from AT cells for unknown reasons. Samples were loaded in an order to see the shifted and the nonshifted bands next to each other.
Next we investigated the effects of known inhibitors of ATM-related kinases on the hyperphosphorylation of XL53BP1 induced by X-ray irradiation. XTC-2 cells were preincubated in the presence of either 16 mM caffeine or 23 μM wortmannin for 1 h and then irradiated with X rays. At 30 min and at 3 h postirradiation, extracts were prepared and run on a SDS-polyacrylamide gel for immunoblotting (Fig. 6B). Caffeine was shown to be a potent ATM-related kinase inhibitor (2, 16, 49). The 50% inhibitory concentrations of ATM, ATR, and DNA-PK to caffeine were 0.2, 1.1, and 10 mM, respectively (49). In the presence of 16 mM caffeine, most of the activities of ATM-related kinases were expected to be blocked. Wortmannin irreversibly inhibits certain members of the PI3K family, and 23 μM wortmannin blocks the activities of ATM and DNA-PK but not that of ATR in intact cells (50) (in cells, 50% inhibitory concentrations of the kinases were 5.8, 3.6, and 100 μM, respectively). We found that the slower migration of XL53BP1 was almost completely eliminated in the extracts prepared from cells irradiated in the presence of 16 mM caffeine (Fig. 6B, lanes 2, 5, and 8). Wortmannin showed a similar effect but was less effective (Fig. 6B, lanes 3, 6, and 9). In the presence of 23 μM Wortmannin, XL53BP1 migrated slower than that from cells without irradiation or from caffeine-treated cells but faster than that from cells that had not been treated with inhibitors after X-ray irradiation (Fig. 6B, lanes 3, 6, and 9). However, it was possible that the inhibitory effects of these drugs on the phosphoryaltion of XL53BP1 were not necessarily through ATM-related kinases, especially given the relatively high concentration of caffeine used.
To investigate further the possible implication of ATM kinase, we examined the band retardation of 53BP1 before and after the irradiation of AT cells that were deficient for ATM kinase. The extent of the 53BP1 band retardation was significantly reduced, although the band shift was not completely abolished (Fig. 6C, lanes 6 to 9; compare with the results for control HT1080 cells, which are ATM+). Due to technical reasons, the amount of total protein we could load per slot for AT cells was approximately half that of HT1080 cells; therefore, the 53BP1 bands were slightly fainter in AT cell samples. These results suggested that ATM, and possibly its related kinases, were implicated in the hyperphosphorylation of XL53BP1 in response to the DNA damage induced by X-ray irradiation.
Caffeine inhibits the focal redistribution of XL53BP1 in cells irradiated with X rays.
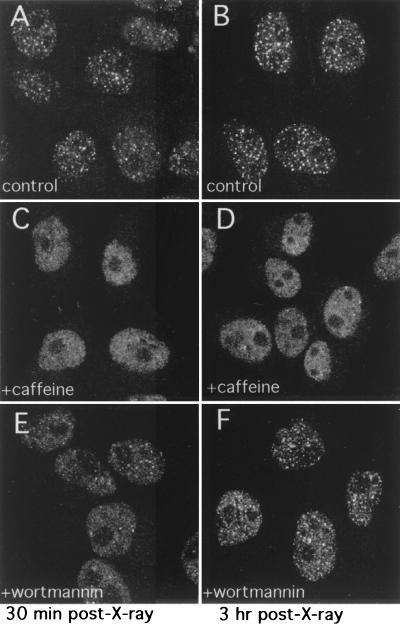
Given that the postirradiation phosphorylation of XL53BP1 was reduced in the presence of either caffeine or wortmannin, we were interested in examining whether the focal redistribution of XL53BP1 would also be affected by the presence of these drugs during damage induction. XTC-2 cells were treated as described above, fixed, and stained for XL53BP1 (Fig. 7). In control cells without inhibitor treatment, the focal redistribution of XL53BP1 was already visible at 30 min postirradiation, and at 3 h, it was prominent (Fig. 7A and B, respectively). However, in the presence of 16 mM caffeine, the protein stayed distributed mostly homogeneously over the chromatin even after 3 h postirradiation and the numbers of 53BP1 foci assembled were significantly lower (Fig. 7C and D). It is worth noting that this concentration of caffeine was four times higher than that usually required to block the DNA damage checkpoint (51). At a lower concentration of caffeine (i.e., 4 mM), we did not observe any delay of 53BP1 focus formation after X-ray irradiation (data not shown). Wortmannin showed a moderate effect, and the focal redistribution of XL53BP1 appeared to be slightly delayed in the early stages (Fig. 7E; see also Table 1) of postirradiation. Later, at 3 h postirradiation, the assembly of XL53BP1 foci was nearly as efficient as that of the no-drug control (F).
FIG. 7.
XL53BP1 focus formation is diminished or delayed in the presence of caffeine or wortmannin, respectively. Cells were treated as described for Fig. 6. XTC-2 cells were exposed to X rays either in the absence of any drugs (A and B) or in the presence of 16 mM caffeine (C and D) or 23 μM wortmannin (E and F) and fixed at 30 min (A, C, and E) or at 3 h (B, D, and F) postirradiation for XL53BP1 stain.
A slight delay of 53BP1 focal redistribution was also observed in AT cells (Fig. 8). In HT1080 (ATM+) cells, 53BP1 focus formation was clearly observed 1 h after X-ray irradiation (Fig. 8E). Whereas, in AT (ATM−) cells at 1 h postirradiation, fewer and less distinct 53BP1 foci had formed (Fig. 8B). By 3 h postirradiation, the AT cells had managed to assemble discrete 53BP1 foci (Fig. 8C). We scored the number of countable 53BP1 foci per nucleus by optical sectioning with a confocal microscope (Table 1). Counting was performed by two independent investigators, and we obtained consistent results. At 30 min postirradiation, more than 60 foci were counted on average in HT1080 nuclei. In contrast, only 23.5 foci were counted per AT cell nucleus on average at 30 min. After 3 h we did not observe a significant difference in numbers of foci between HT1080 and AT cells. A slight delay in XL53BP1 focus formation was also observed in XTC-2 cells in the presence of wortmannin (30 versus 54 foci in the no-drug control after 30 min). These results suggested that ATM kinase might be implicated in the rapid response of 53BP1 focus formation after ionizing irradiation; however, the kinase was not essential for focal assembly.
DISCUSSION
53BP1, a member of the BRCT protein family, is hyperphosphorylated and relocalizes to a number of nuclear foci in response to DNA damage. These foci also contain the Mre11-Rad50-Nbs1 complex. 53BP1, however, concentrates in foci in 30 min, which is significantly earlier than the time any obvious concentration of Mre11 occurs in the postirradiation period (9 to 15 h after irradiation [33]). There were several proteins that coimmunoprecipitated with 53BP1 (Fig. 1A). However, we detected neither p53 nor Mre11 in the 53BP1 immunoprecipitate under the experimental conditions (data not shown).
The assembly of 53BP1 foci is not cell cycle stage dependent. In contrast, Rad51 and BRCA1, which also change their subnuclear localization after DNA damage, show a focal distribution in S phase and disperse homogeneously over the nucleoplasm after genotoxic treatments (52, 64). Ionizing irradiation induces Rad51 and BRCA1 focal assembly in cells presumed to be in either G1 or G2 (15, 59, 64), whereas irradiation-induced 53BP1 foci occur in almost all cells except those in mitosis. Rad51, Mre11, or BRCA1 foci are observed in a subset of cells after irradiation (10 to 40%, depending on the protein, cell type, and cell cycle stage [33, 44, 64]).
The histone H2A variant H2AX, which makes up 20% of the total H2A in human cells, is phosphorylated at Ser 139 in response to dsDNA breaks. H2AX phosphorylation has been demonstrated to occur at the sites of lesions by scissoring laser treatment (46, 47). Recently, a comprehensive study has shown that the different types of damage-induced foci, including BRCA1, Rad51, and the Mre11-Rad50-Nbs1 complex foci, all originate from γ-H2AX foci at certain stages of the postirradiation period (44). The associations appear to follow a sequential order, and some of them might also be cell cycle stage dependent. The focally distributed H2AX phosphorylated at Ser 139 (called γ-H2AX) appears immediately after irradiation, peaks by 10 min, and then gradually decreases and almost disappears after 4.5 h (46, 47). Kinetically, the appearance of 53BP1 foci and 53BP1's hyperphosphorylation occur slightly later than the appearance of foci and H2AX's hyperphosphorylation of H2AX but earlier than with the other types of damage-induced nuclear foci. At 30 min postirradiation the focal distribution and the phosphorylation of XL53BP1 are not at their maxima (Fig. 6 and 7). This result might be reasonable since 53BP1 needs to be recruited to the foci after DNA damage and, in contrast, since H2AX is already there at the dsDNA breaks. More than half of the dsDNA breaks are rejoined in an order of minutes, and the rest persist for several hours (26, 39). 53BP1 foci might be implicated in the repair and/or checkpoint control associated with the latter.
53BP1 is hyperphosphorylated in response to ionizing irradiation, and this phosphorylation is significantly reduced in AT cells which are deficient in ATM kinase (Fig. 6C). The data obtained with the inhibitors of ATM-related kinases are consistent with the AT cell results (Fig. 6B). These results suggest that ATM-related kinases are involved in the phosphorylation of 53BP1 in response to ionizing irradiation. Interestingly, caffeine inhibits 53BP1 phosphorylation more efficiently than wortmannin. This is also true for the suppressive effects of caffeine and wortmannin on the relocalization of 53BP1: caffeine blocks and wortmannin slightly delays 53BP1 focus formation (Fig. 7 and Table 1). We observed a slight delay in 53BP1 focal assembly in AT cells after irradiation (Fig. 8 and Table 1). This suggests that ATM kinase might be involved in the rapid response of 53BP1 relocalization to ionizing irradiation but that it is not essential for formation of the foci. Mre11/Rad50 focus formation is reduced in AT cells but not completely abolished (33). BRCA1 focus formation is not different in wild-type cells and in AT cells (10). ATM, ATR, and DNA-PK might have overlapping roles in phosphorylation and in focus formation. During our paper's revision, Xia et al. published a paper showing that ATM kinase phosphorylates XL53BP1 in vitro, which is consistent with our observation of the inhibitors and AT cells (66).
Our results suggest that the formation of 53BP1 foci is controlled through phosphorylation of the protein. In budding yeast, accumulating evidence implicates phosphorylation-regulated protein-protein interactions in the cellular response to DNA damage. dsDNA breaks induce relocalization of Ku, Rap1, and SIR proteins, which are normally associated with telomeres, to the sites of the lesion, and this process is dependent on MEC1sc, an ATM-related kinase, and Rad9sc, a BRCT protein (32, 36). Rad9SC protein multimerizes through its BRCT domain, and the interaction occurs with higher affinity after DNA damage (56). The FHA domain of Rad53sc physically interacts with phosphorylated forms of Rad9sc (12, 14, 57, 61). Given the fact that there is no evidence for the presence of an active transport system in the nucleus at the moment and that RNA and protein factors move by diffusion through the interchromatin space (see reference 30), it might be reasonable to speculate that the assembly and disassembly of repair and checkpoint protein foci are regulated through changing of the affinities of protein-protein interactions by phosphorylation (see also reference 44).
The results presented here implicate 53BP1 in DNA damage repair or checkpoint control in vertebrate cells; however, direct evidence is still missing. A genetic approach (i.e., disruption of the 53BP1 gene in DT40 or ES cells) will be required to investigate these possibilities directly.
ACKNOWLEDGMENTS
We are grateful to Steve B. Coade for AT cells, Sigenobu Tone for the BrdU labeling protocol, and Mitsuhiro Yanagida for his advice. We thank Kevin Hardwick, Ciaran Morrison, Takashi Toda, and Bill Earnshaw for comments on the manuscript. We also thank colleagues in the ICMB for discussion and support.
The Wellcome Trust supported this work.
REFERENCES
- 1.Adachi Y, Laemmli U K. Identification of nuclear pre-replication centers poised for DNA synthesis in Xenopus egg extracts: immunolocalization study of replication protein A. J Cell Biol. 1992;119:1–15. doi: 10.1083/jcb.119.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasina A, Price B D, Turenne G A, McGowan C H. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 3.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Carr A M. Piecing together the p53 puzzle. Science. 2000;287:1765–1766. doi: 10.1126/science.287.5459.1765. [DOI] [PubMed] [Google Scholar]
- 6.Caspari T. How to activate p53. Curr Biol. 2000;10:R315–R317. doi: 10.1016/s0960-9822(00)00439-5. [DOI] [PubMed] [Google Scholar]
- 7.Chaturvedi P, Eng W K, Zhu Y, Mattern M R, Mishra R, Hurle M R, Zhang X, Annan R S, Lu Q, Faucette L F, Scott G F, Li X, Carr S A, Johnson R K, Winkler J D, Zhou B B. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- 8.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G L, Yang L, Rowe T C, Halligan B D, Tewey K M, Liu L F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984;259:13560–13566. [PubMed] [Google Scholar]
- 10.Cortez D, Wang Y, Quin J, Elledge S J. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 11.Critchlow S E, Bowater R P, Jackson S P. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 12.Durocher D, Henckel J, Fersht A R, Jackson S P. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 13.Edwards R J, Bentley N J, Carr A M. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 14.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 15.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in syanaptonemal complex. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall-Jackson C A, Cross D A, Morrice N, Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 18.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 19.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 20.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 21.Hovanessian A G, Galabru J, Riviere Y, Montagnier L. Efficiency of poly(A) · poly(U) as an adjuvant. Immunol Today. 1988;9:161–162. doi: 10.1016/0167-5699(88)91288-1. [DOI] [PubMed] [Google Scholar]
- 22.Iwabuchi K, Bartel P L, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwabuchi K, Li B, Massa H F, Trask B J, Date T, Fields S. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 24.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 25.Kim S T, Lim D S, Canman C E, Kastan M B. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 26.Kodym R, Hörth E. Determination of radiation-induced DNA strand breaks in individual cells by non-radioactive labelling of 3′ OH ends. Int J Radiat Biol. 1995;68:133–139. doi: 10.1080/09553009514551031. [DOI] [PubMed] [Google Scholar]
- 27.Kostrub C F, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumagai A, Guo Z, Emami K H, Wang S X, Dunphy W G. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lewis J D, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 31.Lim D S, Kim S T, Xu B, Maser R S, Lin J, Petrini J H, Kastan M B. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 32.Martin S G, Laroche T, Suka N, Grunstein M, Gasser S M. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 33.Maser R S, Monsen K J, Nelms B E, Petrini J H. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka S, Huang M, Elledge S J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 36.Mills K D, Sinclair D A, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 37.Nash R A, Caldecott K W, Barnes D E, Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 38.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H J. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 39.Nunez M I, Villalobos M, Olea N, Valenzuela M T, Pedraza V, McMillan T J, Ruiz de Almodovar J M. Radiation-induced DNA double-strand break rejoining in human tumor cells. Br J Cancer. 1995;71:311–316. doi: 10.1038/bjc.1995.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurse P. Checkpoint pathways come of age. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 41.O'Connell M J, Walworth N C, Carr A M. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10:296–302. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- 42.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park M S, Knauf J A, Pendergrass S H, Coulon C H, Strniste G F, Marrone B L, MacInnes M A. Ultraviolet-induced movement of movement of the human DNA repair protein, Xeroderma pigmentosum type G, in the nucleus. Proc Natl Acad Sci USA. 1996;93:8368–8373. doi: 10.1073/pnas.93.16.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paull T T, Rogakou E P, Yamazaki V, Kirchgessner C U, Gellert M, Bonner W M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 45.Rebagliati M R, Weeks D L, Harvey R P, Melton D A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 46.Rogakou E P, Boon C, Redon C, Bonner W M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–915. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogakou E P, Pilch D R, Orr A H, Ivanova V S, Bonner W M. DNA double-strand breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 48.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkaria J N, Busby E C, Tibbetts R S, Roos P, Taya Y, Karnitz L M, Abraham R T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 50.Sarkaria J N, Tibbetts R S, Busby E C, Kennedy A P, Hill D E, Abraham R T. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 51.Schlegel R, Pardee A B. Caffein-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- 52.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 53.Seigneur M, Bidnenko V, Ehrlich D S, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 54.Shieh S Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. . (Erratum, 14:750.) [PMC free article] [PubMed] [Google Scholar]
- 55.Smith J C, Tata J R. Xenopus cell lines. Methods Cell Biol. 1991;36:635–654. [PubMed] [Google Scholar]
- 56.Soulier J, Lowndes N F. The BRCT domain of the S. cerevisiae checkpoint protein Rad9 mediates a Rad9-Rad9 interaction after DNA damage. Curr Biol. 1999;9:551–554. doi: 10.1016/s0960-9822(99)80242-5. [DOI] [PubMed] [Google Scholar]
- 57.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 58.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 59.Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tell-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 63.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X, Ranganathan V, Weisman D S, Heine W F, Ciccone D N, O'Neill T B, Crick K E, Pierce K A, Lane W S, Rathbun G, Livingston D M, Weaver D T. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 66.Xia, Z., J. C. Morales, W. G. Dunphy, and P. B. Carpenter. Negative cell cycle regulation and DNA-damage inducible phosphorylation of the BRCT protein 53BP1. J. Biol. Chem., in press. [DOI] [PubMed]
- 67.Zhao S, Weng Y-C, Yuan S-S F, Lin Y-T, Hsu H-C, Lin S-C, Gerbino E, Song M, Zdzienicka M Z, Gatti R A, Shay J W, Ziv Y, Shiloh Y, Lee E Y-H P. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 68.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a recA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]