Abstract
Identifying the molecular networks that underlie Fibrous Dysplasia (FD) is key to understand the pathogenesis of the disease, to refine current diagnostic approaches and to develop efficacious therapies. In this study, we used the NanoString nCounter Analysis System to investigate the gene signature of a series of nine Formalin Fixed Decalcified and Paraffin-Embedded (FFDPE) bone biopsies from seven FD patients.
We analyzed the expression level of 770 genes. Unsupervised clustering analysis demonstrated partitioning into two clusters with distinct patterns of gene expression. Differentially expressed genes included growth factors, components of the Wnt signaling system, interleukins and some of their cognate receptors, ephrin ligands, matrix metalloproteinases, neurotrophins and genes encoding components of the cAMP-dependent protein kinase. Interestingly, two tissue samples obtained from the same skeletal site of one patient one year apart failed to segregate in the same cluster. Retrospective histological review of the samples revealed different microscopic aspects in the two groups.
The results of our pilot study suggest that the genetic signature of FD is heterogeneous and varies according to the histology and, likely, to the age of the lesion. In addition, they show that the Nanostring technology is a valuable tool for molecular translational studies on archival FFDPE material in FD and other rare bone diseases.
Keywords: Fibrous Dysplasia, Bone disease, Bone biopsy, Nanostring
Highlights
-
•
We used the NanoString technology to analyze Formalin Fixed Decalcified Paraffin Embedded (FFDPE) Fibrous Dysplasia samples.
-
•
We show that Fibrous Dysplasia lesions may have different molecular profiles consistent with its histological heterogeneity.
-
•
NanoString technology is a valuable tool for molecular studies on rare bone diseases by using FFDPE archival material.
1. Introduction
Fibrous Dysplasia of bone (FD) is a genetic skeletal disease that occurs in monostotic or polyostotic forms, the latter being often associated with extra-skeletal disorders in the McCune-Albright Syndrome (Polyostotic FD/MAS; OMIM#174800). FD results from gain-of-function mutations of the GNAS gene (GNAS complex locus; GNAS, OMIM *139320) that arise during embryo development (Riminucci et al., 2007) and usually replace the arginine 201 (R201) in the Gsα transcript with a histidine (R201H) or a cysteine (R201C) (Weinstein et al., 1991). In spite of its early occurrence, the mutation targets the osteoprogenitor cell compartment of the post-natal skeleton (Bianco and Robey, 1999; Riminucci et al., 2007) and severely compromises growing bones causing loss of mechanical strength, deformity and pain (Ippolito et al., 2014; Mancini et al., 2009). The essential pathological features of FD include fibrotic tissue, woven and hypomineralized bone, osteoclasts and blood vessels, which may variably combine to generate site specific histological patterns (Corsi et al., 2017, Corsi et al., 2003; Riminucci et al., 2003, Riminucci et al., 1999). However, it is known that FD lesions may also show additional microscopic aspects (e.g. cartilage, myxoid areas) (Corsi et al., 2006; Tabareau-Delalande et al., 2013) or may develop extensive secondary changes of uncertain significance (e.g. fibroma desmoplastic-like areas), especially in craniofacial bones (Corsi et al., 2020, Corsi et al., 2020). In these cases, substantial deviations from the archetypal FD lesion may occur with consequent diagnostic challenges and prognostic uncertainty. Although the Gsα mutation, which is sufficient to reproduce the core pathology of FD in mice (Saggio et al., 2014), remains the essential genetic marker of the disease, a deeper knowledge of the molecular background is important to improve the specific diagnostic and molecular profiling of FD lesions and histological variants thereof. Previous molecular studies essentially focused on individual genes (Alman et al., 1995; Candeliere et al., 1995; Jia et al., 2011; Kashima et al., 2009; Kumta et al., 2003; Regard et al., 2011; Riminucci et al., 2003; Stanton et al., 1999; Yamamoto et al., 1996), or on restricted panels of selected genes (Kiss et al., 2010) involved in skeletal growth and homeostasis. However, the complex molecular scenario downstream gain-of-function mutations of a pleiotropic gene like Gsα calls for the application of high throughput methods of transcriptomic analysis and, in particular, for methods that allow a tight correlation between molecular profiling and histology. Only two microarray studies on FD are currently available. The first, performed on a pool of fresh pathological samples (Zhou et al., 2014), did not provide any correlation between the molecular data and the actual histological features of the lesions. The second did not use pathological tissue and was conducted on normal osteoprogenitor cells transduced with the Gsα R201C mutation (Raimondo et al., 2020).
The NanoString technology has been proposed as a novel enzyme-free methodology that allows for high throughput analysis of samples with poor extraction quality and low nuclei acid yield (Tsang et al., 2017). For this reason, it is considered as a valid approach to screen potential candidate genes on formalin-fixed paraffin-embedded (FFPE) tissue specimens, including bone(Wang et al., 2019). In this study, the expression level from 770 genes was determined by NanoString technology on a small series of formalin-fixed, decalcified and paraffin-embedded (FFDPE) bone biopsies from seven FD patients.
Although limited, our cohort of samples generated two different FD molecular profiles suggesting that the genetic signature of FD is heterogeneous and varies according to the histology and, likely, to the age of the lesion. More in general, our study demonstrated that the NanoString technology is a valuable tool for molecular translational studies on archival material in FD and other rare bone diseases.
2. Methods
2.1. Patients and tissue samples
Nine archival paraffin blocks collected at the Sapienza University Hospital Policlinico Umberto I of Rome between 2008 and 2017 from seven FD/MAS patients (Pt1-Pt7) were used. The study was approved by the local Ethical Committee (RIF. EC: 5126). Informed consent was obtained from patients or their guardians. Five patients were males and two were females and the age range at tissue harvesting was 6–23 years. The clinical diagnosis was MAS in six patients (four males and two females) and polyostotic FD in one. For two patients two tissue samples were analyzed. The samples were obtained from femur (sample A) and tibia (sample B) for one of them (Pt 1). For the other (Pt 3), both samples (A and B) were obtained from the femur one year apart.
All tissue samples were collected during either diagnostic biopsies or corrective surgeries, immediately fixed in 4% buffered formaldehyde, decalcified with 10% EDTA and routinely processed for paraffin embedding. Mutation analysis was performed in the FD samples from six patients as described previously(Corsi et al., 2006; Riminucci et al., 1999). R201H was detected in three patients (Pt 1, Pt 3 and Pt 7) and R201C in the other three (Pt 2, Pt 4 and Pt 6). A synopsis of the clinical and molecular data of the patients included in the study is reported in Table 1.
Table 1.
Synopsis of the clinical and molecular data of the patients included in the study.
| Id | Gender/agea | Clinical diagnosis | Site of bone biopsy or surgery | Mutation |
|---|---|---|---|---|
| 1 | M/17 and 23 years | MAS | Femur (sample a) and Tibia (sample b) | R201H |
| 2 | M/16 years | MAS | Femur | R201C |
| 3 | M/6 and 7 years | Polyostotic FD | Femur (samples a and b) | R201H |
| 4 | F/16 years | MAS | Femur | R201C |
| 5 | M/8 years | MAS | Femur | NA |
| 6 | F/20 years | MAS | Femur | R201C |
| 7 | M/11 years | MAS | Femur | R201H |
Age at FD tissue harvesting; NA: not available.
2.2. RNA extraction
Total RNA was extracted by RNeasy FFPE kit (Qiagen Germany) according to manufacturer's instructions starting from a minimum of four 20-micron-thick sections from FFDPE samples. RNA quantity and quality were assessed by NanoDrop2000 spectrophotometer (Thermo Fisher Scientific).
2.3. NanoString analysis
The samples that reached the quality standards (A260/A280 higher than 1.7 and A260/A230 higher than 1.8) were analyzed by NanoString using the nCounter PanCancer Profiling Panel (NanoString Technologies). This panel includes 770 genes from 13 canonical pathways including: MAPK, STAT, PI3K, RAS, Cell Cycle, Apoptosis, Hedgehog, Wnt, DNA Damage Control, Transcriptional Regulation, Chromatin Modification, and TGF-β. Since a specific nCounter Panel related to FD is not available to date, we selected this panel as it includes driver genes and gene pathways that, according to our experience, could provide some insights into the molecular features of FD.
Analysis of detected gene counts was performed by employing nSolver Analysis Software 3.0 (NanoString Technologies). First, samples were selected by checking imaging quality controls: percentage of fields of view (FOV) read (higher than 75%), binding density (between 0.05 and 2.25), positive control linearity (higher than 0.95), and positive control limit of detection (higher than 2). For samples that passed imaging quality controls, raw genes counts were normalized on technical controls and housekeeping genes included in the panel. Mean count of negative controls plus two standard deviations (SDs) were subtracted from each gene count to eliminate negative background. Then, normalization on positive controls was conducted by multiplying the count of each gene for a correction factor (CF) calculated, for each sample, as the ratio between the quadratic mean of positive controls counts and the mean of quadratic means in all samples.
To perform CodeSet Content normalization on housekeeping genes, ten reference genes were selected among those available in the panel based on the lowest coefficients of variation (CV = ratio between mean counts and SDs across all samples). Finally, CodeSet Content normalization was performed by multiplying genes counts for a further CF calculated on reference genes as described for positive technical controls. Expression data are available at NCBI GEO: GSE176243.
After the normalization processes, counts were log2 transformed and used for downstream analyses. To perform cluster analysis, we chose the hierarchical agglomerative clustering procedure, which has a bottom-up approach. This procedure was employed using Ward's method and a Euclidean distance metric. Ward's method, essentially, looks at cluster analysis as an analysis of variance problem, starting out with n clusters of size 1 and continuing until all the observations are included into one cluster. Hierarchical clustering of samples and genes was performed with the “heatmap.2” R function by employing “ggplot” and “dendextend” packages. Each cell was colored to reflect quantitatively the relative expression, in NanoString counts, to assist in visualizing expression changes.
2.4. Pathway enrichment and Gene Ontology (GO) analysis
BioPlanet 2019, WikiPathways 2021 and KEGG 20121 enrichment analysis of differential gene expression in addition to Gene Ontology (GO) for biological processes, molecular function and cellular component were performed with the publicly available tool, EnrichR (http://amp.pharm.mssm.edu/Enrichr), that provides access to various gene-set libraries. We considered pathways and Gene Ontology terms as enriched if their p-value was lower than 0.05 and sorted them by combined score ranking. The combined score is a combination of the p-value and z-score calculated by multiplying the two scores as follows: CS = log(p)z where p is the p-value computed using Fisher's exact test, and z is the z-score computed to assess the deviation from the expected rank. The plots show the significance [−log(p-value)] of each gene set from the selected library versus its odds ratio. Each point represents a single geneset. Larger blue points represent significant terms (p-value lower than 0.05) while smaller gray points represent non-significant terms.
The search tool for retrieval of interacting genes (STRING) (https://string-db.org) database, which integrates both known and predicted PPIs, has been applied to predict functional interactions of proteins (Szklarczyk et al., 2019). To search for potential interactions between overexpressed genes, the STRING tool was employed, species limited to “Homo sapiens” and an interaction score > 0.4. Cytoscape software version 3.6.1 was used to visualize the PPI network and to detect the highly connected regions of the network, Markov Clustering (MCL) algorithm was used with default parameters. In networks, nodes correspond to proteins and edges represent interactions.
2.5. Retrospective evaluation of the histological samples
Four-micron-thick FFDPE sections were stained with hematoxylin and eosin (HE) for transmitted light microscopy or with Picrosirius red for analysis with polarized light microscopy. TRAP staining was performed to highlight mono- and multi-nucleated osteoclasts as described previously (Corsi et al., 2020, Corsi et al., 2020). The microscopic review was performed by three pathologists (MDSV, AC, MR) with reference to the FD histological patterns and sub-classification previously reported (Tabareau-Delalande et al., 2013).
Bone volume in each biopsy (Bone Volume/Tissue Volume BV/TV) was quantified on H&E stained sections according to the guidelines of the American Society of Bone and Mineral Research (Dempster et al., 2013). For the comparison of BV/TV between the two groups, the two-sided unpaired Students' t-test was employed. Statistical significance was established at p-value lower than 0.05.
3. Results and discussion
3.1. NanoString analysis revealed different FD molecular clusters
We isolated total RNA from 9 archival FFDPE FD bone lesions [two samples (A and B) from Pt1 and Pt3 and one sample from Pt2, Pt4, Pt5, Pt6 and Pt7]. The RNA yield was adequate in all cases and an RNA input between 150 ng and 250 ng was used for subsequent NanoString analysis. Two samples (Pt1 A and Pt6) were excluded due to degradation of RNA as detected by preliminary analysis of raw data performed by nSolver™ software. In fact, they presented Quality Check flag for “content normalization” indicating an effective reduction in quality (fragmentation) of the input RNA.
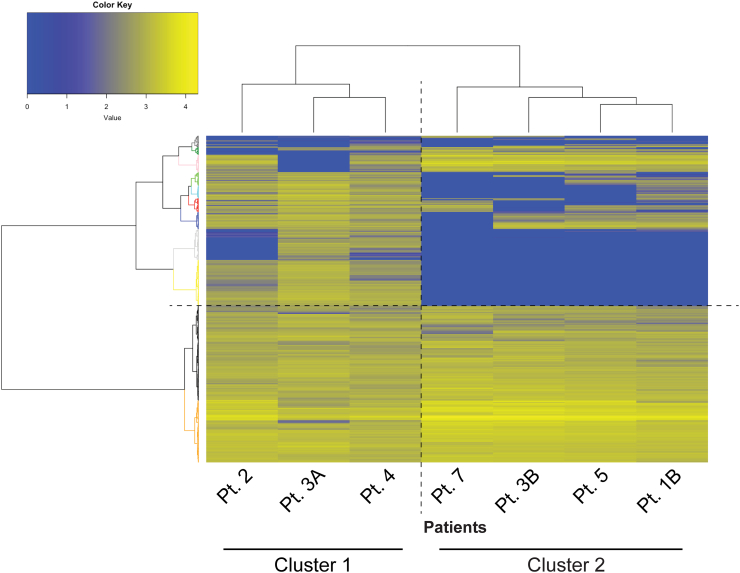
Unsupervised clustering analysis demonstrated that the remaining 7 FD samples could be clearly partitioned into two groups (defined FD cluster 1 and FD cluster 2) with distinct patterns of gene expression (Fig. 1). Interestingly, the two tissue samples obtained from the same skeletal site of Pt3 one year apart during the progression of the disease did not segregate in the same group.
Fig. 1.
Gene profiling of FD patients' biopsies performed with NanoString technology Heat map generated from the analysis of 770 genes in bone biopsies from seven FD patients. Clustering analysis of transcripts in 7 FD samples (in NanoString counts), clearly displayed two distinct groups: FD cluster1 which was separated from the other group defined as FD cluster2. The relative abundance of transcripts is indicated by the color (yellow, high; blue, low).
To confirm the reliability of the Nanostring approach, we randomly selected 5 genes that resulted highly expressed and 5 genes that showed low levels of expression and tested their expression levels by quantitative PCR (qPCR) on two frozen FD bone samples. As reported in Fig. S1, the results of the qPCR analysis were consistent with the data obtained by Nanostring. The sequence of the primers used for qPCR is reported in Table S2.
3.2. Differentially expressed genes and pathway enrichment
FD cluster 1, which included samples from Pt2, Pt3 (sample A) and Pt4, was characterized by the upregulation of 189 genes (Table S1). These included: growth factors; components of the Wnt signaling system; interleukins and some of their cognate receptors; ephrin ligands; matrix metalloproteinases and neurotrophins. Interestingly we also observed upregulation of genes encoding components of the cAMP-dependent protein kinase as the catalytic subunit gamma (PRKACG) and the type I-beta regulatory subunit (PRKAR1B) (Table S1).
Some of these genes may have a pathogenetic role in the disease. For example, WNT3a, which stimulates the proliferation activity and reduces the differentiation and apoptosis of immature osteoprogenitor cells (Boland et al., 2004), could be involved in the expansion of the FD immature osteogenic tissue. FGF8 enhances extracellular matrix degradation, likely one of the mechanisms through which Sharpey's fibers are formed, acting in association with other molecules as MMP3 and IL1 (Uchii et al., 2008) which were also found to be expressed in the same sample cluster. IL1a, IL1b, IL7, IL11and IL23 stimulate osteoclastogenesis (Ponzetti and Rucci, 2019) and could contribute, along with RANKL (de Castro et al., 2019; Palmisano et al., 2019), to the inappropriate osteoclast generation observed in FD. We found of particular interest the brain-derived neurotrophic factor (BDNF) gene. This neurotrophin stimulates RANKL secretion by osteoprogenitor cells in patients with multiple myeloma (Ai et al., 2012; Sun et al., 2012) and is a well-recognized mediator of pain in some bone diseases (Bratus-Neuenschwander et al., 2018; Gowler et al., 2020; Simão et al., 2014). Accordingly, BDNF could be a potential candidate gene that links excess bone resorption and bone pain in FD patients. PPI network for the 189 genes of FD cluster 1 was constructed and used to identify clusters of highly interconnected nodes. In this way we were able to underline pathways dysregulated in FD pathogenesis, confirming what we deduced from single genes overexpression data and comparing our results with two microarray-based analyses that were previously performed on Fibrous Dysplasia using tissue and cell samples (Raimondo et al., 2020; Zhou et al., 2014). Fig. S3 shows the 4 significant clusters that were found in the FD cluster 1 PPI network analysis using Markov Clustering (MCL) approach. In the cluster A.1 (Fig. S3), the most significant biological processes and pathways were associated with immunity and inflammatory responses, including cytokine-mediated signaling pathways, cytokine-cytokine receptor interaction, as well as JAK-STAT signaling pathway. This agrees with what reported in (Raimondo et al., 2020) regarding the upregulation of cytokines and chemokines in FD, suggesting an activation of immune cell trafficking and inflammatory phenomena in FD patients.
Cluster A.2 (Fig. S3) was enriched in signaling pathways regulating pluripotency of stem cells, osteoblast differentiation and related diseases. This enrichment relates several deregulated genes we found in FD cluster 1 and functional categories to FD pathological traits. Cluster A.3 (Fig. S3) was enriched in the cAMP signaling pathway: in fact, in fibrous dysplastic bone the increased expression of cAMP by the mutated lesional cells is known to be associated with abnormal osteoblast differentiation and formation of defective bone. WNT signaling pathways, known to regulate bone mass and development (Liu et al., 2008), are enriched in cluster A.4.
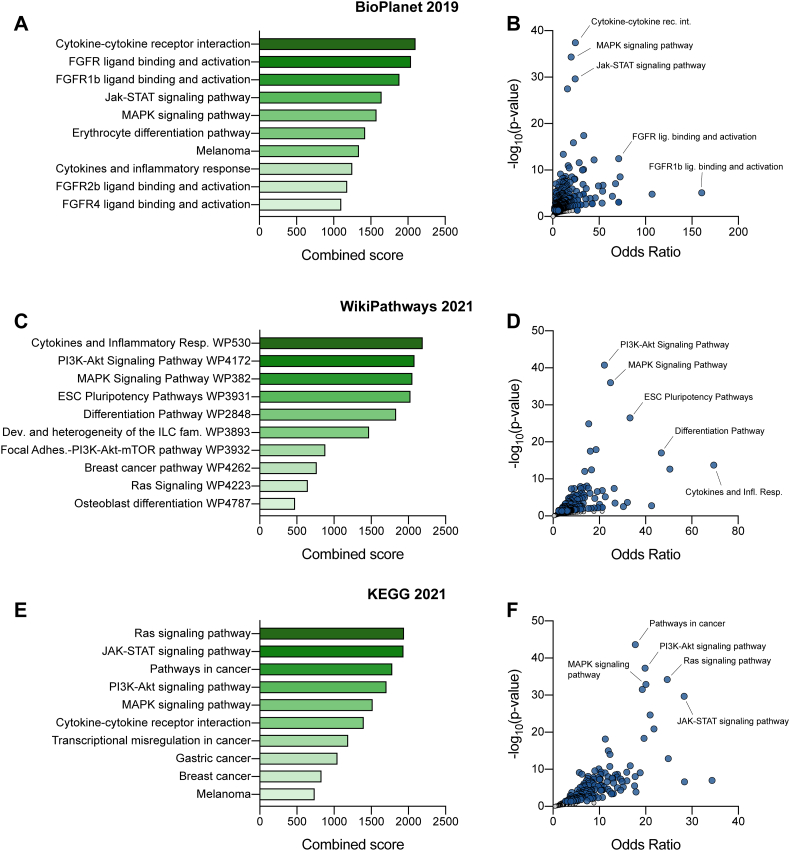
Moreover, functional enrichment analyses, including BioPlanet 2019, WikiPathways 2021 and KEGG 2021 databases at Enrichr (http://amp.pharm.mssm.edu/Enrichr), demonstrated that Cytokines and inflammatory response, PI3K-AKT and MAPK signaling pathways, and FGFR ligand binding and activation were the top enriched pathways (Fig. 2).
Fig. 2.
Pathway enrichment and Gene Ontology (GO) analysis pathway enrichment analysis. Bioplanet 2019 (A, B), WikiPathways 2021 (C, D) and KEGG 2021 (E, F) pathway enrichment analyses of the 188 differentially expressed genes. Pathways in A, C, E are sorted by combined score ranking. Plots in B, D, F show the significance [−log10(p-value)] of each gene set from the selected library versus its odds ratio. Each point represents a single geneset. Blue points represent significant terms (p-value lower than 0.05) while gray points represent non-significant terms.
3.3. Correlation between molecular clusters and FD histology
Retrospective evaluation was performed with reference to the histological subtyping proposed by Flore Tabareau-Delalande et al. who distinguished conventional and unconventional histological patterns of FD (Tabareau-Delalande et al., 2013). All lesions that segregated in the FD cluster 1 [Pt2, Pt3 (sample A) and Pt4] met the criteria of conventional FD. In these samples, histological sections were occupied by a highly vascularized fibrotic tissue and haphazardly distributed pathological bone trabeculae with a woven structure and well evident Sharpey's fibers. In all cases, osteoclasts were promptly recognized in close contact with the FD bone. Increase number and thickness of FD bone trabeculae and heterotopic (i.e., within the fibrotic tissue) osteoclasts were observed in the samples from Pt4 and Pt2, respectively.
In contrast, the samples that segregated in the FD cluster 2 [Pt1 (sample B), Pt 3 (sample B), Pt5 and Pt7] shared an unconventional fibro-involutive pattern. A reduction in the amount of bone, even though not statistically significant was observed compared to FD molecular cluster 1 (BV/TV, mean ± SD, molecular cluster 1 vs molecular cluster 2, 28.87 ± 3.07 vs 23.52 ± 11.83). Focal areas of conventional FD with rare, if any, osteoclasts in contact with FD bone trabeculae were detected in samples from Pt 3 (sample B), Pt5 and Pt7.
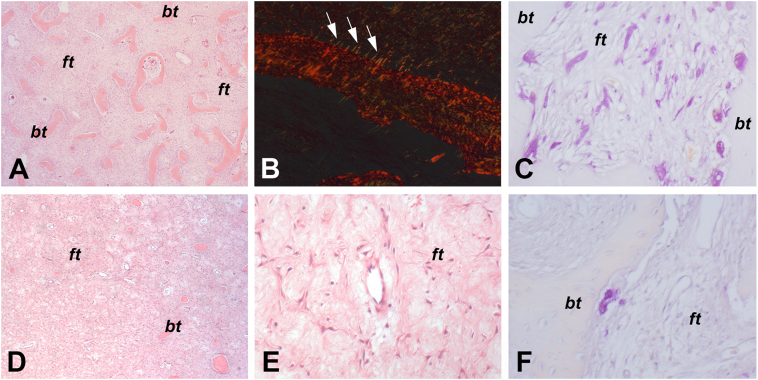
Representative histological images of the FD lesions segregating in the FD molecular cluster 1 [from Pt 3 (sample A)] and in FD molecular cluster 2 [from Pt 1 (sample B)] are illustrated in Fig. 3. Additional histological features of both clusters are reported in Fig. S2.
Fig. 3.
Representative histological images from FD lesions segregating in molecular cluster 1 [A–C, Pt 3 (sample A)] and in molecular cluster 2 [D–F, Pt 1 (sample B)]. The lesions included in molecular cluster 1 consisted of a highly vascularized fibrotic tissue (ft) and haphazardly distributed bone trabeculae (bt) (A) and showed well evident Sharpey's fibers (arrows in B) and a large amount of orthotopic (i.e., along the bone surfaces) and heterotopic (i.e., within the fibrotic tissue) osteoclasts (red staining in C). In contrast, the lesions included in molecular cluster 2 were characterized by a larger amount of fibro-myxomatous tissue with sparse bone trabeculae (D and E) and a very low number of orthotopic osteoclasts. A, D and E: hematoxylin and eosin. B: Picrosirius red. C and F: TRAP. A, C, D, E and F: transmitted light microscopy. B: polarized light microscopy.
Interestingly, the two samples obtained from Pt3 one year apart did not segregate in the same group. The changes in the overall histological and molecular features of these two samples could result, at least in part, from age-dependent changes in the biology of the FD tissue. For example, we have previously shown that a reduction in the number of mutated osteoprogenitor cells occurs within FD lesions over time eventually leading to a quiescent state (Kuznetsov et al., 2008).
4. Conclusions
This is a very preliminary study with multiple limitations, as the reduced number of pathological samples and the use a NanoString nCounter Kit not specific to investigate bone diseases. Nonetheless, our results reveal that the genetic signature of FD, as its histology, is heterogeneous and likely age-dependent and provide some relevant data for future molecular translational research. Furthermore, they indicate that the NanoString technology is a valuable tool for molecular studies on archival histological FFDPE material from rare bone diseases.
The following are the supplementary data related to this article.
Supplementary figures
FD_cluster1 upregulated genes
Sequence of the primers used for qPCR
Funding
Mara Riminucci and Alessandro Corsi received funding from University of Pennsylvania Orphan Disease Center in partnership with the Fibrous Dysplasia Foundation (MDBR-19-110-FD/MAS) and Sapienza University of Rome (RM11916B839074A8, RP11715C7C4DC57A, RM118164289636F0). Domenico Raimondo received funding from Sapienza University of Rome (RP11715C7CC232F4).
CRediT authorship contribution statement
Conceptualization and methodology: A.P., E.M., B.P., A.d.F., E.S., S.D., I.C., M.D.S.V., E.I.; Data analysis and validation: E.M., A.P., B.P., A.d.F., D.R. and M.R.; Original draft preparation: A.P., D.R. and M.R.; Writing-review and editing: M.R., B.P., A.C. and D.R.; Conceptualization: A.P., M.R. and D.R.; Supervision and project administration: M.R., A.C, and D.R.; Funding acquisition: M.R. and D.R.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Mara Riminucci, Email: mara.riminucci@uniroma1.it.
Domenico Raimondo, Email: domenico.raimondo@uniroma1.it.
References
- Ai L.S., Sun C.Y., Zhang L., Zhou S.C., Chu Z.B., Qin Y., Wang Y.D., Zeng W., Yan H., Guo T., Chen L., Yang D., Hu Y. Inhibition of BDNF in multiple myeloma blocks osteoclastogenesis via down-regulated stroma-derived RANKL expression both in vitro and in vivo. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alman B.A., Naber S.P., Terek R.M., Jiranek W.A., Goldberg M.J., Wolfe H.J. Platelet-derived growth factor in fibrous musculoskeletal disorders: a study of pathologic tissue sections and in vitro primary cell cultures. J. Orthop. Res. 1995;13:67–77. doi: 10.1002/jor.1100130111. [DOI] [PubMed] [Google Scholar]
- Bianco P., Robey P.G. Diseases of bone and the stromal cell lineage. J. Bone Miner. Res. 1999 doi: 10.1359/jbmr.1999.14.3.336. [DOI] [PubMed] [Google Scholar]
- Boland G.M., Perkins G., Hall D.J., Tuan R.S. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J. Cell. Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- Bratus-Neuenschwander A., Castro-Giner F., Frank-Bertoncelj M., Aluri S., Fucentese S.F., Schlapbach R., Sprott H. Pain-associated transcriptome changes in synovium of knee osteoarthritis patients. Genes (Basel) 2018;9 doi: 10.3390/genes9070338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeliere G.A., Glorieux F.H., Prud'homme J., St.-Arnaud R. Increased expression of the c-Fos proto-oncogene in bone from patients with fibrous dysplasia. N. Engl. J. Med. 1995;332:1546–1551. doi: 10.1056/nejm199506083322304. [DOI] [PubMed] [Google Scholar]
- Corsi A., Collins M.T., Riminucci M., Howell P.G.T., Boyde A., Gehron Robey P., Bianco P. Osteomalacic and hyperparathyroid changes in fibrous dysplasia of bone: core biopsy studies and clinical correlations. J. Bone Miner. Res. 2003;18:1235–1246. doi: 10.1359/jbmr.2003.18.7.1235. [DOI] [PubMed] [Google Scholar]
- Corsi A., De Maio F., Ippolito E., Cherman N., Robey P.G., Riminucci M., Bianco P. Monostotic fibrous dysplasia of the proximal femur and liposclerosing myxofibrous tumor: which one is which? J. Bone Miner. Res. 2006;21:1955–1958. doi: 10.1359/jbmr.060818. [DOI] [PubMed] [Google Scholar]
- Corsi A., Fadda M.T., Terenzi V., Valentini V., Riminucci M. Secondary desmoplastic fibroma-like tissue changes in mandibular fibrous dysplasia: clinicopathological and molecular study of a case. Br. J. Oral Maxillofac. Surg. 2020;58:96–98. doi: 10.1016/J.BJOMS.2019.09.006. [DOI] [PubMed] [Google Scholar]
- Corsi A., Ippolito E., Robey P.G., Riminucci M., Boyde A. Bisphosphonate-induced zebra lines in fibrous dysplasia of bone: histo-radiographic correlation in a case of McCune–Albright syndrome. Skelet. Radiol. 2017;46:1435–1439. doi: 10.1007/s00256-017-2698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi Alessandro, Palmisano B., Spica E., Di Filippo A., Coletta I., Dello Spedale Venti M., Labella R., Fabretti F., Donsante S., Remoli C., Serafini M., Riminucci M. Zoledronic acid in a mouse model of human fibrous dysplasia: ineffectiveness on tissue pathology, formation of “giant osteoclasts” and pathogenetic implications. Calcif. Tissue Int. 2020;107:603–610. doi: 10.1007/s00223-020-00752-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro L.F., Burke A.B., Wang H.D., Tsai J., Florenzano P., Pan K.S., Bhattacharyya N., Boyce A.M., Gafni R.I., Molinolo A.A., Robey P.G., Collins M.T. Activation of RANK/RANKL/OPG pathway is involved in the pathophysiology of fibrous dysplasia and associated with disease burden. J. Bone Miner. Res. 2019;34:290–294. doi: 10.1002/jbmr.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster D.W., Compston J.E., Drezner M.K., Glorieux F.H., Kanis J.A., Malluche H., Meunier P.J., Ott S.M., Recker R.R., Parfitt A.M. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR histomorphometry nomenclature committee. J. Bone Miner. Res. 2013 doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowler P.R.W., Li L., Woodhams S.G., Bennett A.J., Suzuki R., Walsh D.A., Chapman V. Peripheral brain-derived neurotrophic factor contributes to chronic osteoarthritis joint pain. Pain. 2020;161:61–73. doi: 10.1097/j.pain.0000000000001694. [DOI] [PubMed] [Google Scholar]
- Ippolito E., Farsetti P., Boyce A.M., Corsi A., De Maio F., Collins M.T. Radiographic classification of coronal plane femoral deformities in polyostotic fibrous dysplasia. Clin. Orthop. Relat. Res. 2014;472:1558–1567. doi: 10.1007/s11999-013-3380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S., Yu J., Zhang D., Zheng P., Zhang S., Ma L., Liu G., Li S. Expression and regulation of amphiregulin in Gsα-mutated human bone marrow stromal cells of fibrous dysplasia of mandible. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2011;111:618–626. doi: 10.1016/j.tripleo.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Kashima T.G., Nishiyama T., Shimazu K., Shimazaki M., Kii I., Grigoriadis A.E., Fukayama M., Kudo A. Periostin, a novel marker of intramembranous ossification, is expressed in fibrous dysplasia and in c-fos-overexpressing bone lesions. Hum. Pathol. 2009;40:226–237. doi: 10.1016/j.humpath.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Kiss J., Balla B., Kósa J.P., Borsy A., Podani J., Takács I., Lazáry Á., Nagy Z., Bácsi K., Kis A., Szlávy E., Szendroi M., Speer G., Orosz L., Lakatos P. Gene expression patterns in the bone tissue of women with fibrous dysplasia. Am. J. Med. Genet. A. 2010;152:2211–2220. doi: 10.1002/ajmg.a.33559. [DOI] [PubMed] [Google Scholar]
- Kumta S.M., Huang L., Cheng Y.Y., Chow L.T.C., Lee K.M., Zheng M.H. Expression of VEGF and MMP-9 in giant cell tumor of bone and other osteolytic lesions. Life Sci. 2003;73:1427–1436. doi: 10.1016/S0024-3205(03)00434-X. [DOI] [PubMed] [Google Scholar]
- Kuznetsov S.A., Cherman N., Riminucci M., Collins M.T., Robey P.G., Bianco P. Age-dependent demise of GNAS-mutated skeletal stem cells and “normalization” of fibrous dysplasia of bone. J. Bone Miner. Res. 2008;23:1731–1740. doi: 10.1359/jbmr.080609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Kohlmeier S., Wang C.-Y. Wnt signaling and skeletal development. Cell. Signal. 2008;20:999–1009. doi: 10.1016/j.cellsig.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F., Corsi A., De Maio F., Riminucci M., Ippolito E. Scoliosis and spine involvement in fibrous dysplasia of bone. Eur. Spine J. 2009;18:196–202. doi: 10.1007/s00586-008-0860-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano B., Spica E., Remoli C., Labella R., Di Filippo A., Donsante S., Bini F., Raimondo D., Marinozzi F., Boyde A., Robey P., Corsi A., Riminucci M. RANKL inhibition in fibrous dysplasia of bone: a preclinical study in a mouse model of the human disease. J. Bone Miner. Res. 2019;34:2171–2182. doi: 10.1002/jbmr.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetti M., Rucci N. Updates on osteoimmunology: what's new on the cross-talk between bone and immune system. Front. Endocrinol. (Lausanne) 2019 doi: 10.3389/fendo.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo D., Remoli C., Astrologo L., Burla R., Torre M.La., Vernì F., Tagliafico E., Corsi A., Giudice S.Del, Persichetti A., Giannicola G., Robey P.G., Riminucci M., Saggio I. Changes in gene expression in human skeletal stem cells transduced with constitutively active Gsα correlates with hallmark histopathological changes seen in fibrous dysplastic bone. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard J.B., Cherman N., Palmer D., Kuznetsov S.A., Celi F.S., Guettier J.M., Chen M., Bhattacharyya N., Wess J., Coughlin S.R., Weinstein L.S., Collins M.T., Robey P.G., Yang Y. Wnt/β-catenin signaling is differentially regulated by Gα proteins and contributes to fibrous dysplasia. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20101–20106. doi: 10.1073/pnas.1114656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riminucci M., Collins M.T., Fedarko N.S., Cherman N., Corsi A., White K.E., Waguespack S., Gupta A., Hannon T., Econs M.J., Bianco P., Robey P.G. FGF-23 in fibrous dysplasia of bone and its relationship to renal phosphate wasting. J. Clin. Invest. 2003;112:683–692. doi: 10.1172/JCI18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riminucci M., Liu B., Corsi A., Shenker A., Spiegel A.M., Robey P.G., Bianco P. The histopathology of fibrous dysplasia of bone in patients with activating mutations of the Gs alpha gene: site-specific patterns and recurrent histological hallmarks. J. Pathol. 1999;187:249–258. doi: 10.1002/(SICI)1096-9896(199901)187:2<249::AID-PATH222>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Riminucci M., Saggio I., Robey P.G., Bianco P. Fibrous dysplasia as a stem cell disease. J. Bone Miner. Res. 2007;22:P125–P131. doi: 10.1359/JBMR.06S224. [DOI] [PubMed] [Google Scholar]
- Saggio I., Remoli C., Spica E., Sacchetti B., Robey P.G., Holmbeck K., Cumano A., Boyde A., Bianco P., Riminucci M. Constitutive expression of gsar201c in mice produces a heritable, direct replica of human fibrous dysplasia bone pathology and demonstrates its natural history. J. Bone Miner. Res. 2014;29:2357–2368. doi: 10.1002/jbmr.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão A.P., Mendonça V.A., De Oliveira Almeida T.M., Santos S.A., Gomes W.F., Coimbra C.C., Lacerda A.C.R. Involvement of BDNF in knee osteoarthritis: the relationship with inflammation and clinical parameters. Rheumatol. Int. 2014;34:1153–1157. doi: 10.1007/s00296-013-2943-5. [DOI] [PubMed] [Google Scholar]
- Stanton R.P., Hobson G.M., Montgomery B.E., Moses P.A., Smith-Kirwin S.M., Funanage V.L. Glucocorticoids decrease interleukin-6 levels and induce mineralization of cultured osteogenic cells from children with fibrous dysplasia. J. Bone Miner. Res. 1999;14:1104–1114. doi: 10.1359/jbmr.1999.14.7.1104. [DOI] [PubMed] [Google Scholar]
- Sun C.Y., Chu Z.B., She X.M., Zhang L., Chen L., Ai L.S., Hu Y. Brain-derived neurotrophic factor is a potential osteoclast stimulating factor in multiple myeloma. Int. J. Cancer. 2012;130:827–836. doi: 10.1002/ijc.26059. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C.V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabareau-Delalande F., Collin C., Gomez-Brouchet A., Decouvelaere A.V., Bouvier C., Larousserie F., Marie B., Delfour C., Aubert S., Rosset P., De Muret A., Pagès J.C., De Pinieux G. Diagnostic value of investigating GNAS mutations in fibro-osseous lesions: a retrospective study of 91 cases of fibrous dysplasia and 40 other fibro-osseous lesions. Mod. Pathol. 2013;26:911–921. doi: 10.1038/modpathol.2012.223. [DOI] [PubMed] [Google Scholar]
- Tsang H.F., Xue V.W., Koh S.P., Chiu Y.M., Ng L.P.W., Wong S.C.C. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev. Mol. Diagn. 2017 doi: 10.1080/14737159.2017.1268533. [DOI] [PubMed] [Google Scholar]
- Uchii M., Tamura T., Suda T., Kakuni M., Tanaka A., Miki I. Role of fibroblast growth factor 8 (FGF8) in animal models of osteoarthritis. Arthritis Res. Ther. 2008;10 doi: 10.1186/ar2474. (R90-R90) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang O., Jiang Y., Li M., Xia W., Meng X., Xing X. Efficacy and safety of bisphosphonate therapy in Mccune-albright syndrome–related polyostotic fibrous dysplasia: a single-center experience. Endocr. Pract. 2019;25:23–30. doi: 10.4158/EP-2018-0328. [DOI] [PubMed] [Google Scholar]
- Weinstein L.S., Shenker A., Gejman P.V., Merino M.J., Friedman E., Spiegel A.M. Activating mutations of the stimulatory G protein in the McCune–Albright syndrome. N. Engl. J. Med. 1991;325:1688–1695. doi: 10.1056/nejm199112123252403. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Ozono K., Kasayama S., Yoh K., Hiroshima K., Takagi M., Matsumoto S., Michigami T., Yamaoka K., Kishimoto T., Okada S. Increased IL-6 production by cells isolated from the fibrous bone dysplasia tissues in patients with McCune-albright syndrome. J. Clin. Invest. 1996;98:30–35. doi: 10.1172/JCI118773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S.H., Yang W.J., Liu S.W., Li J., Zhang C.Y., Zhu Y., Zhang C.P. Gene expression profiling of craniofacial fibrous dysplasia reveals ADAMTS2 overexpression as a potential marker. Int. J. Clin. Exp. Pathol. 2014;7:8532–8541. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
FD_cluster1 upregulated genes
Sequence of the primers used for qPCR