Abstract
Objective
To evaluate the diagnostic performance of short state homeobox 2 (SHOX2) promoter methylation as biomarker for lung cancer identification through aggregating the open published data.
Methods
We did an electronic search in Pubmed, EMBASE, Ovid, Web of Science, Google Scholar, and the China National Knowledge Infrastructure (CNKI) to identify the publications related to SHOX2 promoter methylation and lung cancer. The diagnostic performance of sensitivity (SEN), specificity (SPE), odds ratio (DOR), and summary receiver operating characteristic (SROC) cure were aggregated by fixed or random effect model. Fagan's nomogram was used to investigate the post‐test diagnostic probability. Deek's funnel plot and line regression test was applied to evaluate the publication bias.
Results
In total, 18 clinical studies about SHOX2 promoter methylation and lung cancer were included in the meta‐analysis. The combined diagnostic SEN, SPE, DOR were 0.63 (95% CI = 0.54–0.70), 0.91 (95% CI = 0.87–0.94), and 16.84 (95% CI = 11.18–25.36) in random effect model respectively. The pooled area under the curve (AUC) of SROC was 0.88 (95% CI = 0.84–0.90). The post‐test probability of lung cancer was 88% and 29% when SHOX2 methylated and unmethylated in humoral components given a pre‐test probability of 50%. Deek's funnel plot and regression test indicated no publication bias (p = 0.62).
Conclusion
SHOX2 promoter methylation in humoral components may be a potential biomarker for lung cancer diagnosis with relative high diagnostic specificity.
Keywords: diagnosis, lung carcinoma, meta‐analysis, methylation
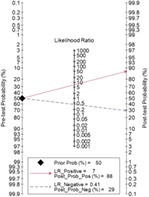
Fagan's nomogram indicated that if SHOX2 methylated in humoral components, the positive post‐test probability of lung cancer was 88%, whereas the negative post‐test probability was 29% when SHOX2 unmethylated in humoral components give a pre‐test probability of 50%.
INTRODUCTION
Genetic abnormality and changes play an important role carcinogens. 1 Epigenetic change is one of the molecular mechanisms that reflects the early stage of tumorigenesis. 2 , 3 As an important way of epigenetics, DNA methylation plays an important role in controlling gene expression, maintaining genomic stability, cell differentiation, and embryonic development. 4 , 5 In addition, DNA methylation is considered as an early event in the process of malignant transformation of cells. 6 , 7 , 8 Therefore, detection of DNA abnormal methylation in tumor cells becomes an important method for early diagnosis of carcinoma.
Lung cancer is the most common carcinoma of human beings and the leading cause of cancer related death globally. 9 The general prognosis of lung cancer is poor because of the lack of effective tools for lung cancer identification in early stages. Recently, studies have shown DNA methylation detection in body fluid such as plasma and bronchoalveolar lavage fluid (BALF) as a promising method for lung cancer early identification. 10 , 11
Short state homeobox 2 (SHOX2), also known as homeobox protein OG12X, is encoded by SHOX2 gene, which encodes 60 amino acid residues representing DNA binding domain. Homeobox proteins are considered as transcription regulators widely expressed in organisms and have complex biological functions. In recent years, it has been reported that the abnormal methylation of SHOX2 gene promoter region was related to the occurrence and development of lung cancer. 12 , 13 The abnormal methylation of this gene may occur in the early stage of lung cancer and is related to the inactivation of this gene after methylation. Studies have also reviewed the occurrence rate of methylation in SHOX2 gene promoter region in lung cancer patients and found that SHOX2 methylation rate was higher in cancer cases than normal controls. The finding indicated that SHOX2 methylation detection may provide promising diagnostic information for lung cancer. 14 , 15
MATERIAL AND METHODS
Electronic publication search
Two authors (X.Z. and X.L.) did an electronic search of Pubmed, EMBASE, Ovid, Web of Science, Google Scholar, and the China National Knowledge Infrastructure (CNKI) to identify the publications related to SHOX2 promoter methylation and lung cancer. The following key words were applied for publication searching: “SHOX2”, “Short stature homeobox 2”, “OG12”, “SHOT”, “OG12X”, “non‐small cell lung cancer”, “lung cancer”, “lung neoplasm”, “carcinoma, non‐small cell lung”, “methylation”, and “hypermethylation”. The references of the included publications were also reviewed to find potential suitable studies.
Study selection criteria
The following study inclusion criteria were applied: (i) the SHOX2 promoter methylation rate was examined in plasma, BALF, or pleural effusion by MSP or other confirmed methods; (ii) studies were published in English or Chinese; (iii) SHOX2 methylation rate in lung cancer cases and controls can be extracted; and (iv) lung cancer diagnosis was confirmed by cytology or pathology.The exclusion criteria included: (i) duplicated publications or data; (ii) SHOX2 methylation was detected in cancer and normal lung tissue; and (iii) autologous controls were applied.
Data extraction
Two reviewers (J.L. and H.H.) made data extraction independently. The extracted data was listed as follows: (i) study ID showed as first author; (ii) time period of the data published; (iii) area the study performed; (iv) methylation rate in lung cancer and controls; (v) tissue type such as plasma, BALF, or pleural effusion; (vi) lung cancer stages of the included cases; and (vii) methylation detection methods.
Statistical methods
All data was managed by STATA12.0 statistical software (Stata Corp LP). The diagnostic sensitivity = true positive/(true positive + false negative), specificity = true negative/(true negative + false positive). The summary receiver operating characteristic (SROC) cure was applied to evaluate the feasibility of SHOX2 promoter methylation as biomarker for lung diagnosis. Fagan's nomogram was used to investigate the post‐test diagnostic probability. Deek's funnel plot and line regression test were used to evaluate the publication bias. Two‐tailed p < 0.05 was considered statistically significant.
RESULTS
Publication searching and main features of the included studies
In total, 766 articles were initially obtained from the relevant electronic databases. Forty‐five duplicate articles or data were removed, and 721 publications were reviewed. After reading the titles and abstracts, 689 studies were eliminated because of obvious unqualification. Finally, 18 clinical studies relevant to SHOX2 promoter methylation as biomarker for lung cancer were included for meta‐analysis (Figure 1). The main features of the 18 studies were demonstrated in Table 1.
FIGURE 1.
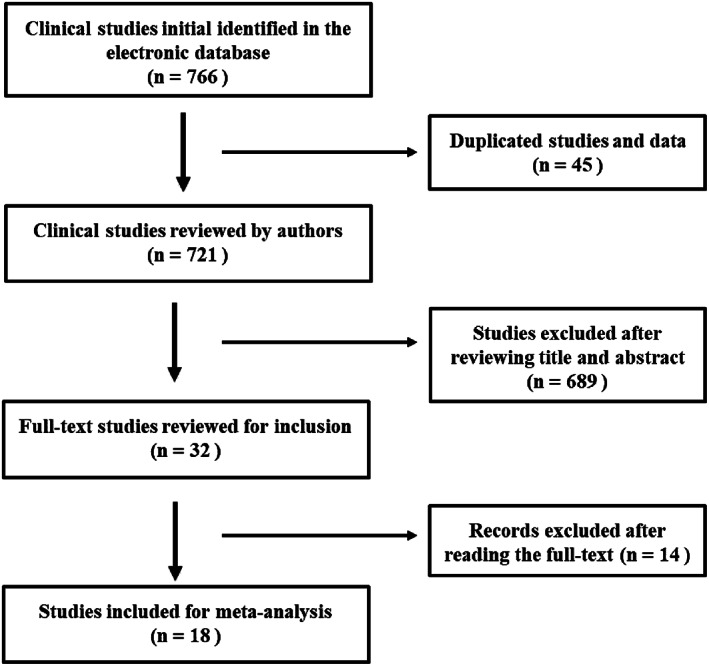
Flow chart of literature retrieval according to inclusion and exclusion criteria
TABLE 1.
General characteristics of the included studies
| Author | Time | Sample size | Cancer | Control | TNM | Tissue | Country | Methods |
|---|---|---|---|---|---|---|---|---|
| (M+/M−) | (M+/M−) | |||||||
| Schmidt 12 | 2010 | 523 | 190/91 | 12/230 | I–IV | BALF | German | MSP |
| Christoph 13 | 2011 | 343 | 112/76 | 16/139 | I–IV | Plasma | German | MSP |
| Konecny 14 | 2016 | 63 | 31/6 | 4/22 | NA | BALF | German | MSP |
| Konecny 14 | 2016 | 59 | 20/11 | 6/22 | NA | Plasma | German | MSP |
| Peter 15 | 2014 | 118 | 48/27 | 1/42 | NA | BALF | German | MSP |
| Dietrich 16 | 2012 | 204 | 78/22 | 4/100 | I–IV | BALF | German | MSP |
| DietrichV 16 | 2012 | 114 | 7/51 | 0/56 | NA | Pleural effusion | German | MSP |
| Peter 17 | 2013 | 719 | 138/138 | 70/373 | NA | Pleural effusion | German | MSP |
| Li 18 | 2014 | 47 | 10/18 | 0/9 | NA | Pleural effusion | China | MSP |
| Zhang 19 | 2016 | 277 | 98/32 | 34/113 | NA | BALF | China | MSP |
| Wang 20 | 2016 | 243 | 79/44 | 8/112 | NA | BALF | China | MSP |
| Ren 21 | 2017 | 253 | 79/44 | 10/120 | I–IV | BALF | China | MSP |
| Rong 22 | 2018 | 48 | 18/20 | 2/8 | III/IV | Plasma | China | MSP |
| Wang 23 | 2018 | 120 | 57/23 | 1/39 | I–IV | Plasma | China | MSP |
| Peng 24 | 2018 | 48 | 18/20 | 2/8 | NA | Plasma | China | MSP‐DHPL |
| Lin 25 | 2019 | 202 | 29/4 | 18/151 | I–IV | BALF | China | MSP |
| Chen 26 | 2019 | 276 | 101/30 | 22/123 | I–IV | BALF | China | MSP |
| Sun 27 | 2020 | 180 | 53/67 | 3/57 | I–IV | Plasma | China | MSP |
| Sun 27 | 2020 | 180 | 74/67 | 4/56 | I–IV | BALF | China | MSP |
| Tian 28 | 2020 | 134 | 54/13 | 12/55 | Early stage | BALF | China | MSP |
| Yang 29 | 2021 | 104 | 32/16 | 14/42 | NA | Plasma | China | MSP |
Abbreviations: BALF, bronchoalveolar lavage fluid; MSP, methylation specific PCR; M+, methylation positive; M−, methylation negative; NA, not available.
Statistical heterogeneity among the included studies
The statistical heterogeneity among studies was evaluated by Q‐test and expressed by I2. There were significant statistical heterogeneity among studies in sensitivity (SEN) (Q = 182.75, I2 = 89.06%, p < 0.01), specificity (SPE) (Q = 89.32, I2 = 77.61%, p < 0.01), and odds ratio (DOR) (I2 = 75.50%, p < 0.01). Therefore, the data of SEN, SPE, and DOR were aggregated in random effect model.
Meta‐analysis
The combined diagnostic SEN, SPE, and DOR were 0.63 (95% CI = 0.54–0.70), 0.91 (95% CI = 0.87–0.94) (Figures 2), and 16.84 (95% CI = 11.18–25.36) (Figure 3) in random effect model. The pooled area under the curve (AUC) of SROC was 0.88 (95% CI = 0.84–0.90) (Figure 4). Fagan's nomogram indicated that if SHOX2 methylated in humoral components, the positive post‐test probability of lung cancer was 88%, whereas the negative post‐test probability was 29% when SHOX2 unmethylated in humoral components given a pre‐test probability of 50% (Figure 5).
FIGURE 2.
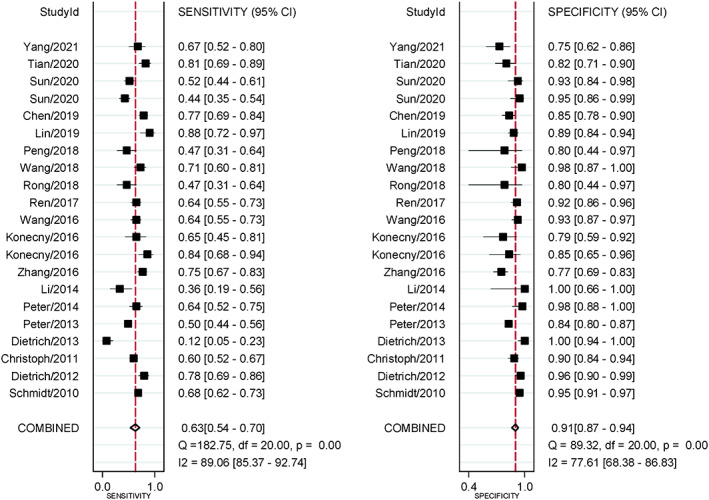
Forest plot for sensitivity and specificity of SHOX2 gene methylation as biomarker for lung cancer diagnosis
FIGURE 3.
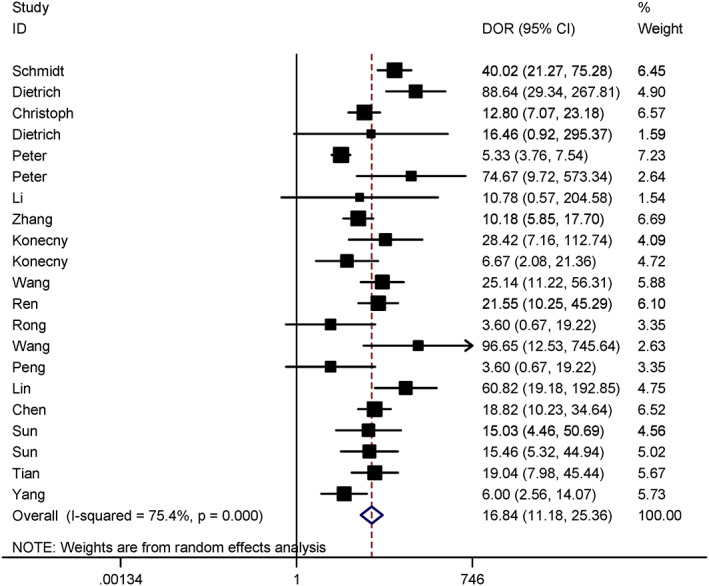
Forest plot for DOR of SHOX2 gene methylation as biomarker for lung cancer diagnosis
FIGURE 4.
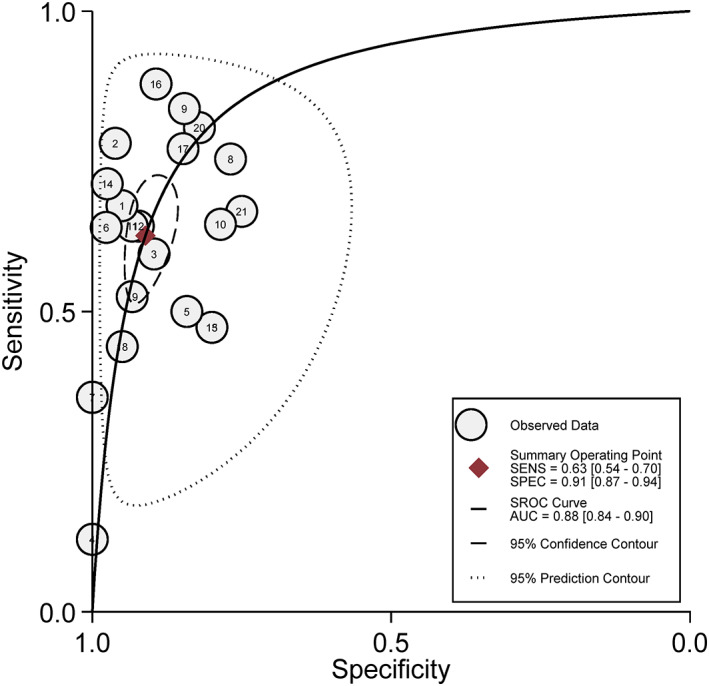
Summary receiver operating characteristic (SROC) cure of SHOX2 gene methylation for lung cancer diagnosis
FIGURE 5.
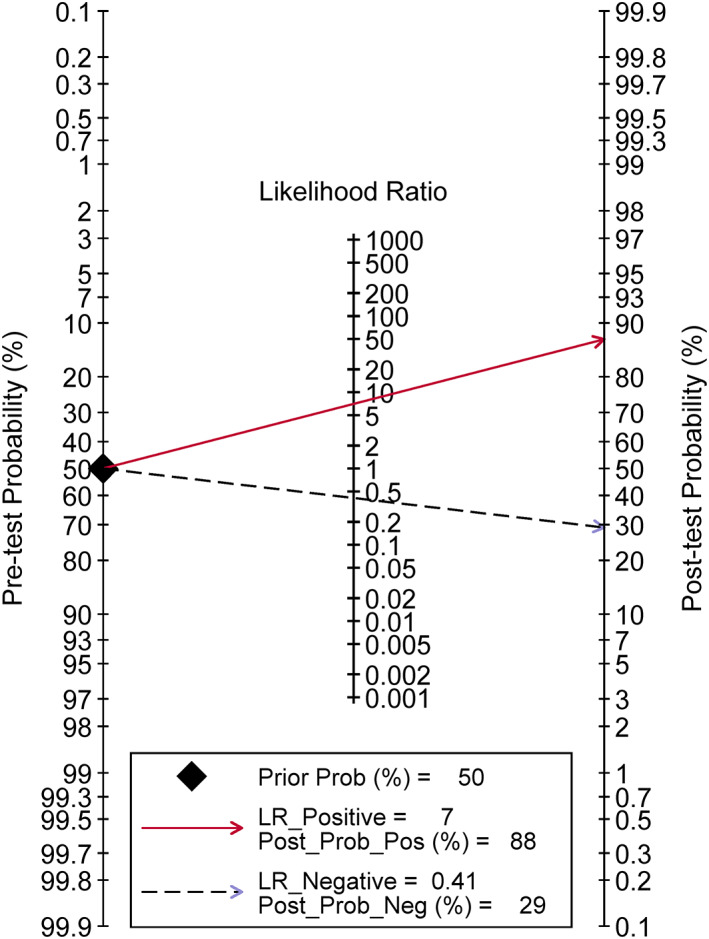
Fagan's nomogram of SHOX2 gene methylation as biomarker for lung cancer diagnosis
Publication bias evaluation
The Deek's funnel plot was general symmetrical and the line regress test did not indicated publication bias (p = 0.62) (Figure 6).
FIGURE 6.
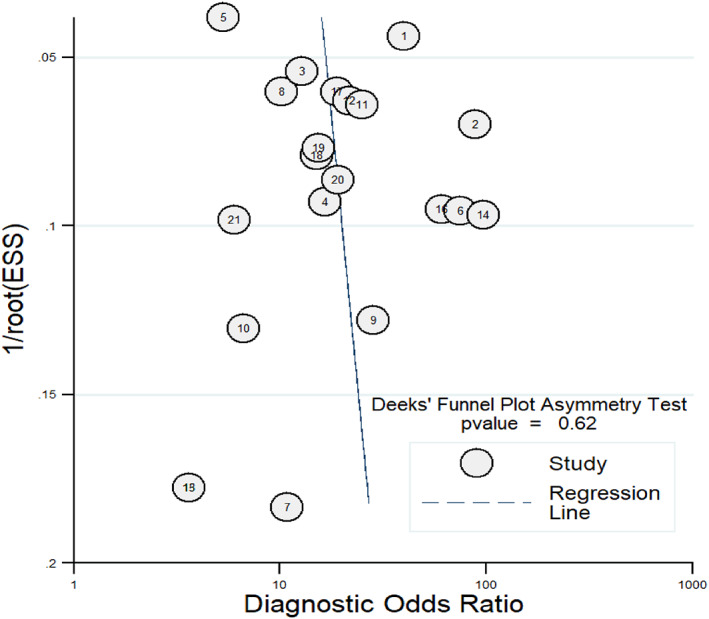
Deek's funnel plot in evaluation the publication bias
DISCUSSION
DNA methylation of gene promoter region, the well‐known mechanism in epigenetic modification, commonly occurs in tumor cells. 30 , 31 DNA methylation of the promoter region is an important way of gene silencing especially for tumor genes. DNA methylation alteration appears to precede apparent malignancy in many cases or at least an early event in the procedure of carcinogenesis. 32 Therefore, methylation can be applied as a promising method for carcinoma diagnosis or recurrence monitoring after effective treatment.
Methylation of SHOX2 in promoter region was widely observed in lung carcinomas. Schmidt et al.12 performed a diagnostic study to investigate whether SHOX2 DNA methylation can be applied as a biomarker for the diagnosis of lung cancer based on bronchial aspirates. The authors found that the lung cancer identification SEN and SPE were 68% (95% CI = 62%–73%) and 95% (95% CI = 91%–97%), respectively. However, Dietrich et al. 16 believed that when using bronchial lavage specimens of SHOX2 methylation as a diagnostic marker, the diagnostic SEN and SPE were 78% and 96%, respectively. Therefore, the diagnostic performance was quite different between publications because of study heterogeneity.
In the present meta‐analysis, we combined 18 studies and pooled the open published data. Combined results showed the diagnostic SEN, SPE, and DOR were 0.63 (95% CI = 0.54–0.70), 0.91 (95% CI = 0.87–0.94), and 16.84 (95% CI = 11.18–25.36), respectively. The lung cancer identification SPE was extremely high, but the SEN was relative low. The low SEN may hinder its clinical application as a lung cancer screening tool. According to Fagan's nomogram, the positive and negative post‐probability of lung cancer was 88% and 29% when SHOX2 methylated and unmethylated in humoral components given a pre‐test probability of 50%. The post‐probability of lung cancer diagnosis was obviously elevated after examination SHOX2 methylation status in body fluid.
Lung cancer is one of the most common malignant tumors in the world. 33 To date, there are few effective and clinically feasible methods for early diagnosis and screening of lung cancer. 34 , 35 , 36 , 37 Although histology and cytology are the gold standard for the diagnosis of lung cancer, patients are often in advanced stages at the time of diagnosis by histology and cytology examination. Therefore, new diagnostic methods with non‐invasive or mini‐invasive with high sensitivity and specificity are urgently needed to improve the diagnosis efficacy and thereafter, decrease the mortality. SHOX2 gene methylation analysis is considered to be a diagnostic method with wide clinical application. SHOX2 gene methylation detection, combined with histology, cytology, and imaging, can improve the identification power of lung cancer and may become an effective tool for lung cancer early diagnosis.
In conclusion, based on the present open published data, SHOX2 promoter methylation in humoral components may be a potential biomarker for lung cancer diagnosis with relative diagnostic specificity. However, because of limitations of statistical heterogeneity and language restriction, high quality diagnostic works are required to further validation our findings. Furthermore, simply relying on detection of SHOX2 gene promoter methylation as the diagnostic reference of lung cancer has limited clinical application value. Therefore, comprehensive judgment should be based on combination of imaging and other diagnostic methods to improve the diagnostic accuracy.
ACKNOWLEDGMENTS
This work was supported by Zhejiang Science and Technology Project of Traditional Chinese Medicine (no. 2021ZB004).
Zhou X, Lu X, Wu H, Liu J, Huang H. Diagnostic performance of SHOX2 promoter methylation as biomarker for lung cancer identification: A meta‐analysis update. Thorac Cancer. 2021;12:3327–3332. 10.1111/1759-7714.14206
Funding information Zhejiang Science and Technology Project of Traditional Chinese Medicine, Grant/Award Number: 2021ZB004
REFERENCES
- 1. Yuan JM, Chan KK, Coetzee GA, Castelao JE, Watson MA, Bell DA, et al. Genetic determinants in the metabolism of bladder carcinogens in relation to risk of bladder cancer. Carcinogenesis. 2008;29:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheng YY, Rath EM, Linton A, Yuen ML, Takahashi K, Lee K. The current understanding of Asbestos‐induced epigenetic changes associated with lung cancer. Lung Cancer (Auckl). 2020;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pruitt K. Molecular and cellular changes during cancer progression resulting from genetic and epigenetic alterations. Prog Mol Biol Transl Sci. 2016;144:3–47. [DOI] [PubMed] [Google Scholar]
- 4. Dhar GA, Saha S, Mitra P, Nag CR. DNA methylation and regulation of gene expression: guardian of our health. Nucleus (Calcutta). 2021;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lianidou ES. Gene expression profiling and DNA methylation analyses of CTCs. Mol Oncol. 2016;10:431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guerrero‐Preston R, Báez A, Blanco A, Berdasco M, Fraga M, Esteller M. Global DNA methylation: a common early event in oral cancer cases with exposure to environmental carcinogens or viral agents. P R Health Sci J. 2009;28:24–9. [PubMed] [Google Scholar]
- 7. Kohonen‐Corish MR, Sigglekow ND, Susanto J, Chapuis PH, Bokey EL, Dent OF, et al. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 2007;26:4435–41. [DOI] [PubMed] [Google Scholar]
- 8. Futscher BW, O'Meara MM, Kim CJ, Rennels MA, Lu D, Gruman LM, et al. Aberrant methylation of the maspin promoter is an early event in human breast cancer. Neoplasia. 2004;6:380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 10. Wang P, Zhao H, Shi R, Liu X, Liu J, Ren F, et al. The role of plasma CDO1 methylation in the early diagnosis of lung cancer. Zhongguo Fei Ai Za Zhi. 2020;23:314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang W, Zhao Y, Huang W, Gao Y, Xu W, Tao J, et al. Non‐invasive diagnosis of early‐stage lung cancer using high‐throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics. 2019;9:2056–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmidt B, Liebenberg V, Dietrich D, Schlegel T, Kneip C, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6:1632–8. [DOI] [PubMed] [Google Scholar]
- 14. Konecny M, Markus J, Waczulikova I, Dolesova L, Kozlova R, Repiska V, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma. 2016;63:246–53. [DOI] [PubMed] [Google Scholar]
- 15. Ilse P, Biesterfeld S, Pomjanski N, Wrobel C, Schramm M. Analysis of SHOX2 methylation as an aid to cytology in lung cancer diagnosis. Cancer Genomics Proteomics. 2014;11:251–8. [PubMed] [Google Scholar]
- 16. Dietrich D, Kneip C, Raji O, Liloglou T, Seegebarth A, Schlegel T, et al. Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int J Oncol. 2012;40:825–32. [DOI] [PubMed] [Google Scholar]
- 17. Ilse P, Biesterfeld S, Pomjanski N, Fink C, Schramm M. SHOX2 DNA methylation is a tumour marker in pleural effusions. Cancer Genomics Proteomics. 2013;10:217–23. [PubMed] [Google Scholar]
- 18. Li SF. Relations pleural effusion SHOX2 gene methylation and lung cancer. Med Health Care. 2014;22(2):3–6. [Google Scholar]
- 19. Zhang Y, Wang ML, Wu J, Shan LH, Zhang JY, Xiong J, et al. Combined detection of SHOX2 and RASSF1A gene methylation in Bronchoalveolar lavage fluid for diagnosis of lung cancer. J Chin Oncol. 2016;22:1032–6. [Google Scholar]
- 20. Wang C. Detection of SHOX2 and RASSF1A Methylation in Bronchoalveolar Lavage Fluid in the Diagnosis of Lung Cancer. Zhejiang, China: Zhejiang Universtity; 2016. [Google Scholar]
- 21. Ren M, Wang C, Sheng D, Shi Y, Jin M, Xu S. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Ann Diagn Pathol. 2017;27:57–61. [DOI] [PubMed] [Google Scholar]
- 22. Rong QP. Diagnostic significance and clinical features of SHOX2 gene methylation in non‐small cell lung cancer. Guangdong Medical College 2016.
- 23. Wang N, Li ZL, Xie X, Shen Y. Analysis of SHOX2 gene methylation by droplet digital PCR in plasma from patients with lung cancer. Chin J Cancer Prev Treat. 2018;25:1241–6. [Google Scholar]
- 24. Peng F, Gao XT, Peng YH, Song ZQ. Clinical significance of Shox2 gene methylation in non‐small cell lung cancer. J Gannan Med Univ. 2018;38:352–7. [Google Scholar]
- 25. Lin LK, Lu CS, Zheng Y, Yang HJ, Zou JB, Wang YN, et al. Application of detection of SHOX2 and RASSF1A gene methylation in bronchoalveolar lavage fluid for the diagnosis of clinical lung cancer. Chin Pract Med. 2019;14:1–3. [Google Scholar]
- 26. Chen RY, Liu Y, Sun T, Liu FH, Ma XH. Value of DNA methylation analysis of SHOX2 and RASSF1A in Bron‐choalveolar lavage fluid in diagnosis of lung cancer. J Zhengzhou Univ (Med Sci). 2019;54:732–6. [Google Scholar]
- 27. Sun YQ, Kong TD, Duan FF, Chen L, Zhou HL, Liu MM, et al. Significance of detection of SHOX2 and RASSF1A gene methylation in peripheral blood and sputum for early diagnosis of lung cancer. Chin J Pract Med. 2020;47(8):9–12. [Google Scholar]
- 28. TIan Q, Zhao J, Liu FF, Luo K, Yin XB, Wang XJ, et al. Application of Shox2 and RASSF1A gene methylation in bronchoalveolar lavage fluid in early clinical diagnosis of lung cancer. Chin J Gerontol. 2020;40:3629–32. [Google Scholar]
- 29. Yang DP, Zhang P, Jin HM, Zhang Y, Zheng JH. Application of combined detection of Shox2, ptger4 gene methylation and SCC in the diagnosis of lung cancer. Lab Med Clin. 2021;18:1101–4. [Google Scholar]
- 30. Wei B, Wu F, Xing W, Sun H, Yan C, Zhao C, et al. A panel of DNA methylation biomarkers for detection and improving diagnostic efficiency of lung cancer. Sci Rep. 2021;11:16782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hong Y, Kim WJ. DNA methylation markers in lung cancer. Curr Genom. 2021;22:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 34. Dong X, Song X, Ding S, Yu M, Shang X, Wang K, et al. Tumor‐educated platelet SNORD55 as a potential biomarker for the early diagnosis of non‐small cell lung cancer. Thorac Cancer. 2021;12:659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu WR, Zhang B, Chen C, Li Y, Ye X, Tang DJ, et al. Detection of circulating genetically abnormal cells in peripheral blood for early diagnosis of non‐small cell lung cancer. Thorac Cancer. 2020;11:3234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kadara H, Tran LM, Liu B, Vachani A, Li S, Sinjab A, et al. Early diagnosis and screening for lung cancer. Cold Spring Harb Perspect Med. 2021;11:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ning J, Ge T, Jiang M, Jia K, Wang L, Li W, et al. Early diagnosis of lung cancer: which is the optimal choice. Aging (Albany NY). 2021;13:6214–27. [DOI] [PMC free article] [PubMed] [Google Scholar]


