Abstract
To define the role of Irx4, a member of the Iroquois family of homeobox transcription factors in mammalian heart development and function, we disrupted the murine Irx4 gene. Cardiac morphology in Irx4-deficient mice (designated Irx4Δex2/Δex2) was normal during embryogenesis and in early postnatal life. Adult Irx4Δex2/Δex2 mice developed a cardiomyopathy characterized by cardiac hypertrophy and impaired contractile function. Prior to the development of cardiomyopathy, Irx4Δex2/Δex2 hearts had abnormal ventricular gene expression: Irx4-deficient embryos exhibited reduced ventricular expression of the basic helix-loop-helix transcription factor eHand (Hand1), increased Irx2 expression, and ventricular induction of an atrial chamber-specific transgene. In neonatal hearts, ventricular expression of atrial natriuretic factor and α-skeletal actin was markedly increased. Several weeks subsequent to these changes in embryonic and neonatal gene expression, increased expression of hypertrophic markers BNP and β-myosin heavy chain accompanied adult-onset cardiac hypertrophy. Cardiac expression of Irx1, Irx2, and Irx5 may partially compensate for loss of Irx4 function. We conclude that Irx4 is not sufficient for ventricular chamber formation but is required for the establishment of some components of a ventricle-specific gene expression program. In the absence of genes under the control of Irx4, ventricular function deteriorates and cardiomyopathy ensues.
The atrial and ventricular chambers of the mammalian heart are exquisitely tailored for their precise roles in circulating blood. Unique properties of atrial and ventricular cells, conserved throughout vertebrate evolution, enable the specialized roles that each chamber plays in cardiac function (2, 17, 22, 23, 31, 37, 41). Structurally, atrial myocytes have poorly developed sarcoplasmic reticulums and disorganized sarcomeres compared to ventricular myocytes and contain dense-core secretory granules that are absent in the ventricles. Atrial myocytes display shorter times of contraction and relaxation than their ventricular counterparts, and misexpression of chamber-specific contractile proteins results in abnormal myocardial function (20, 41, 47). Presumably such differences have evolved to accommodate the specific hemodynamic load of each chamber; these differences may also be important for myocardial adaptation to diseases such as hypertension and hypertrophy (12, 13, 17, 28).
The anatomical and functional differences between atrial and ventricular myocardium reflect the expression of specific genes in each chamber. Experiments with chickens suggest that external positional information acts on the precardiac cells in the earliest stages of differentiation, but soon after cardiac differentiation the plasticity of the myocytes is lost, and cardiac cells are irreversibly programmed as atrial or ventricular (40, 55, 56). Subsequently, however, the establishment of chamber-specific gene expression occurs as a gradual and dynamic process throughout embryogenesis. Prior to heart tube formation, expression of ventricle-specific gene myosin light chain 2v (MLC2v) is already regionalized, presumably in the ventricular precursors (34). During heart tube formation and subsequent morphogenetic remodeling to form the mature heart, regionalization of most other transcripts is evident, so that by the time the heart has two atria and ventricles the majority of chamber-specific genes are expressed in their final anatomical compartments (10, 29, 33, 35, 38, 53, 56). Some genes exhibit delayed regionalization; for example, the atrial natriuretic factor (ANF) gene is expressed in both embryonic atria and ventricles but at birth ventricular expression is down-regulated (17, 57). Despite progress in determining the patterns of chamber-specific gene expression during mammalian development, the factors that control the assignment of one gene to its predominant site of expression are not known.
We have recently identified in chickens, mice, and humans a new member of the Iroquois gene family, Irx4, whose cardiac expression is restricted to the ventricles of the developing heart (1, 9). Irx4 is the earliest marker of the ventricular precursors and is expressed in ventricular myocardium during all stages of cardiac development, including during adulthood. Transient misexpression of mouse Irx4 or of a dominant-negative Irx4 molecule in chicken embryos disrupted the chamber-specific expression of cardiac myosin heavy chain genes (1). By virtue of its homology to Iroquois patterning genes and its ventricle-specific expression pattern, Irx4 is a good candidate for a molecule involved in regulating ventricular specification in the developing heart. To fully elucidate its role in heart development and function, we disrupted the murine Irx4 gene. Irx4-deficent mice develop adult-onset cardiac hypertrophy that is preceded by abnormal ventricular gene expression.
MATERIALS AND METHODS
Generation of Irx4-targeted and transgenic mice.
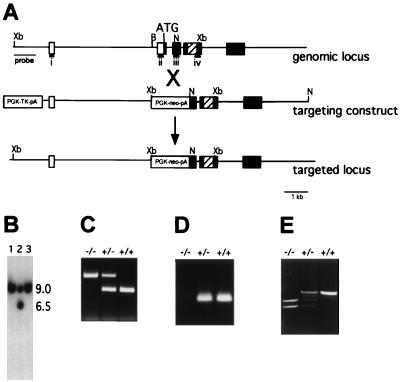
A genomic clone comprising the Irx4 gene was isolated by screening a 129/SvJ mouse genomic phage library with the Irx4 cDNA. A replacement targeting construct was constructed (see Fig. 1); this resulted in the deletion of a 650-bp fragment that includes the 3′ end of exon 2 (including the translation initiation codon), intron 2, and part of exon 3. The thymidine kinase gene driven by the PGK promoter was inserted at the end of the 3′ region of homology. The targeting construct was electroporated into 3 × 107 C1 embryonic stem cells (30). Three separate embryonic stem cell lines were injected into mouse blastocysts; one chimeric mouse transmitted the targeted allele through the germ line. Irx4Δex2/Δex2 and Irx4Δex2/+ mice were maintained on a mixed (SvJ × BlackSwiss) background. Genotyping was performed by PCR using three primers designed to amplify the wild-type and mutant alleles; primer sequences are available upon request. Reverse transcription-PCR (RT-PCR) was performed using primer pairs i (GCGGGCCGGCTCTTTCCTG) and iv (AGTTCTAGCTCCTTGTCGTCTTTG) or ii (CCCGGCATGTCCTACCCGCAGTTT) and iii (GCAGGCCCGGAATCAGCCAGTGTG). SMyHC3-HAP transgenic mice were generated as previously described (54) and were crossed with Irx4Δex2/Δex2 mice.
FIG. 1.
Targeted disruption of Irx4. (A) Diagram of the Irx4 genomic locus, targeting construct, and targeted locus. Open boxes, untranslated sequences; solid boxes, coding sequences; hatched boxes, homeodomain-coding sequences. Only relevant restriction enzyme sites are shown. (B) Southern blot of XbaI-digested embryonic stem cell DNA using a probe external to the targeting construct, showing a targeting event (lane 2) as evidenced by two bands representing the endogenous 9-kb allele and the 6-kb targeted allele. (C) PCR identification of wild-type and targeted alleles in Irx4+/+ (+/+), Irx4Δex2/+ (+/−) and Irx4Δex2/Δex2 (−/−) mice. (D) RT-PCR of mouse heart RNA for all three genotypes using primers internal to the deletion (ii and iii in panel A). (E) RT-PCR of mouse heart RNA for all three genotypes using primers external to the deletion (i and iv in panel A). Xb, XbaI; B, BamHI; N, NotI.
Physiological measurements.
Echocardiography of adult mice was performed as previously described (36) using a Sonos 5500 (Hewlett-Packard) with a 12-MHz transducer. Conscious systolic blood pressure was measured by tail cuff using a Visitech BP2000. Mice were acclimatized to the instrument twice a day for 5 days; sequential measurements were acquired twice a day for 3 days. In vivo left ventricle (LV) physiological measurements and electrophysiological analysis were performed as previously described (4, 24). All physiological analyses were done blinded to the genotypes of the animals.
Analysis of gene expression.
In situ hybridization of whole embryos was performed as previously described (44). In situ hybridization on paraffin sections was done using a modification of the whole-mount protocol. Northern blots were prepared and hybridized according to standard protocols, using cDNA or oligonucleotide probes. Blots were quantitated using a phosphorimager (Molecular Dynamics) and normalized to the signal from a GAPDH probe. Fold increases are reported as means of three to five individual samples and are significantly different from control values at P values of <0.05. Oligonucleotide probes corresponding to α-skeletal actin, α-myosin heavy chain (αMHC), β-myosin heavy chain (βMHC), phospholamban, serca2, MLC1a, MLC1v, MLC2a, and MLC2v were synthesized according to previously published sequences (36). cDNA probes are listed below. Alkaline phosphatase (AP) staining of embryos was done as previously described (54).
cDNA probes.
The cDNA probes used were αMHC (35), βMHC (35), Chisel (R. P. Harvey, unpublished data), COUP-TFII (42), dHand (49), eHand (49), FOG-2 (51), Hermes (25), Irx4 (9), Irx2 (referred to as Irx6 in reference 15) (7), MLC1a (35), MLC1v (35), MLC2a (33), MLC2v (38), MLC3f(32), Msg1 (19), and Tbx5 (10). ANF, BNP, BMP10, and FGF12 cDNAs were cloned by PCR amplification of reverse-transcribed heart RNA using primers based on the published mouse sequences (GenBank accession no. K02781, D16497, AF101033, and AF020738, respectively). Mouse Irx1 and Irx5 cDNAs were obtained from a mouse embryonic heart cDNA library (Stratagene) that was screened with the Irx4 cDNA. Mouse Irx3 cDNAs were identified initially in a library screen; the probe used here was obtained as an expressed sequence tag (GenBank accession no. AI154095).
RESULTS
Mice with a targeted disruption of Irx4 were generated by homologous recombination in embryonic stem cells (Fig. 1). The first coding exon and part of the second coding exon of Irx4 were eliminated, resulting in targeted allele designated Irx4Δex2 (Fig. 1A to D). One-fourth of the offspring of Irx4Δex2/+ animals were Irx4Δex2/Δex2 animals, thereby indicating that Irx4 is not essential for viability. There was no increase in mortality in Irx4Δex2/Δex2 animals compared to that of wild-type mice. Northern blot and RT-PCR analyses revealed that two transcripts are still transcribed at normal levels from the Irx4Δex2 allele. The sequencing of RT-PCR products generated using oligonucleotide primers outside the deleted region (Fig. 1E; see Materials and Methods) defined the structure of the Irx4 transcripts produced from the Irx4Δex2 allele. Two transcripts that contained Irx4 sequences were identified. The 5′ ends of these transcripts contained exon 1 of the Irx4 gene, the 3′ end of the PGKneo gene, and sequences from Irx4 exon 3; all of Irx4 exon 2 and part of exon 3 were deleted. All of these transcripts lacked a ribosome binding site and initiation codon (data not shown). We concluded that the Irx4Δex2 allele does not encode a functional Irx4 protein.
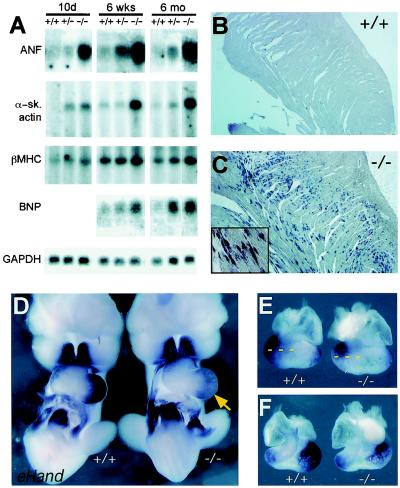
We assessed cardiac gene expression in the hearts of Irx4Δex2/Δex2 embryos and 10-day-, 6-week-, and 24-week-old animals by Northern blot analysis or in situ hybridization. At all time points examined, Irx4Δex2/Δex2 and wild-type LVs contained equal amounts of α-cardiac myosin heavy chain (MHC), phospholamban, serca2, MLC1a, MLC1v, MLC2a, MLC2v, and Tbx5. In contrast, at 10 days postbirth ANF and α-skeletal actin mRNA levels were higher by factors of 5.2 ± 0.6 and 5.8 ± 1.3 in Irx4Δex2/Δex2 hearts than in wild-type hearts, respectively (Fig. 2A). At 6 weeks, βMHC mRNA levels were also increased (by a factor of 2.1 ± 0.2 versus the wild-type level). By 24 weeks, Irx4Δex2/Δex2 hearts contained increased levels of ANF (factor of 6.7 ± 1.3), BNP (factor of 4.3 ± 0.6), α-skeletal actin (factor of 5.4 ± 0.8), and βMHC (factor of 2.7 ± 0.15) mRNAs compared to mRNA levels in wild-type hearts (Fig. 2A). Heterozygous Irx4Δex2/+ animals exhibited intermediate increases of these mRNAs, suggesting an inverse dose relationship between Irx4 levels and RNA expression. In situ hybridization of an ANF-specific probe to sections of adult Irx4Δex2/Δex2 myocardium demonstrated an uneven distribution of ANF, with ANF mRNA localized mainly in the trabecular zone, an area of normal ANF expression in fetal, but not postnatal, life (Fig. 2B and C). No difference in ANF expression between wild-type and Irx4Δex2/Δex2 mice was observed in embryonic day 13.5 (E13.5) fetal hearts (data not shown).
FIG. 2.
Gene expression in wild-type (+/+) and Irx4Δex2/Δex2 (−/−) mice. (A) Northern blot analysis of gene expression of ventricular RNA for wild-type, heterozygous (+/−), and Irx4Δex2/Δex2 mice aged 10 days, 6 weeks, or 6 months. Representative signals for ANF, α-skeletal actin, βMHC, BNP, and GAPDH (as a loading control) are shown. (B and C) Expression of ANF by in situ hybridization on longitudinal sections of wild-type (B) and Irx4Δex2/Δex2 (C) hearts at 6 months of age showing increased ANF transcript levels in Irx4Δex2/Δex2 ventricles. (D to F) eHand expression in E10.5 embryos viewed ventrally (D) or in hearts dissected from E10.5 embryos viewed from the back (E) or the front (F). The arrow indicates lower eHand mRNA levels in Irx4Δex2/Δex2 embryonic hearts. Dashed lines provide a comparison of the domains of eHand expression.
To determine if Irx4 regulates the developmental expression of chamber-specific transcription factors implicated in cardiac development or of other chamber-specific genes, in situ hybridization of whole-mount Irx4Δex2/Δex2 embryos (E10.5 to E11.5) was performed with cDNA probes. The expression patterns of Chisel, COUP-TFII, dHand, FGF12, FOG-2, Hermes, Msg1, BMP10, Tbx5, MLC1a, MLC1v, MLC2a, MLC2v, αMHC, and βMHC in wild-type and Irx4Δex2/Δex2 embryonic hearts were comparable (data not shown). However, expression of the basic helix-loop-helix (bHLH) transcription factor eHand, which at E10.5 to E11.5 predominates in the LV and part of the right ventricle (RV) (5, 49), was altered (Fig. 2D to F). eHand expression in Irx4Δex2/Δex2 embryos was diminished in the anterior and ventral regions of the developing LV (n = 4).
To further explore the role of Irx4 in directing chamber-specific gene expression, we mated Irx4Δex2/+ mice to transgenic mice expressing human AP under the control of the slow myosin heavy chain 3 (SMyHC3) promoter. The SMyHC3 gene is the quail homolog of the chicken atrial myosin heavy chain gene, and the transgenic mice express AP robustly in developing atria but not in ventricles (54). Heterozygous SMyHC3-HAP/Irx4Δex2/+ mice were mated to Irx4Δex2/+ mice to generate SMyHC3-HAP/Irx4Δex2/Δex2 embryos. At E9.5, the atrial chamber-specific transgene was expressed in the presumptive LV as well as the atria of SMyHC3-HAP/Irx4Δex2/Δex2 embryos (Fig. 3), showing derepression of the SMyHC3-HAP transgene in a portion of the ventricles. At E10.5 and E12.5, marked AP staining was detected in both the LV and RV (Fig. 3). However, AP expression was nonuniform and restricted to the LV and RV free walls at E10.5; by E12.5 the entire RV expressed the reporter gene, but transgene expression was excluded from the region to the left of the interventricular septum. These data imply a role for Irx4 and other factors in regulating chamber-specific gene expression in the early embryo; Irx4 can function to repress atrial gene expression within developing ventricular chambers.
FIG. 3.
SMyHC3-HAP transgene expression in wild-type (+/+) and Irx4Δex2/Δex2 (−/−) embryos at E9, E10.5, and E12.5. Hearts were removed from E12.5 embryos for better visualization. a, atrium; v, ventricle; lv, LV; rv, RV.
Members of the Iroquois gene family in Xenopus laevis and Drosophila melanogaster are functionally interchangeable and partially redundant (3, 11, 18, 26, 27). To determine whether other Iroquois family members compensated for the lack of Irx4 in mutant mice, we attempted to identify additional Iroquois genes exhibiting cardiac expression. An E10 embryonic heart cDNA library was screened with the Irx4 cDNA, and three additional Iroquois genes were identified: Irx1, Irx2, and Irx5. Expression of Irx1, -2, -3, and -5 was assessed in wild-type and Irx4Δex2/Δex2 hearts. Only Irx1, Irx2, and Irx5 were expressed in the heart (Fig. 4). Irx1 and Irx2 were detectable in a subset of cells near the interventricular groove (Fig. 4C, D, F, and G). Irx5 was present in both atria and ventricles but was excluded from the atrioventricular junction (Fig. 4A, B, and D). Although the levels of Irx1 and Irx5 in the mutant hearts were not significantly altered (data not shown), increased Irx2 expression was observed in Irx4Δex2/Δex2 hearts (Fig. 4F and G; n = 3).
FIG. 4.
Cardiac expression of Irx family genes. Irx5 is robustly expressed in both atria and ventricles of E9.5 (A) and E10.5 (B and E) embryos. Expression of Irx1 was detected in a subset of ventricular cardiocytes at E10.5 (arrow in panel C, bracket in panel D). Irx2 is also expressed in a pattern that overlaps Irx1 (F and G; red arrows in panel F); Irx2 expression is increased in E10.5 Irx4Δex2/Δex2 embryonic hearts (F and G) a, atrium; v, ventricle; rv, RV; lv, LV.
To determine the consequences of Irx4 deficiency on postnatal cardiac structure and function, histopathologic and hemodynamic studies were performed. The hearts from 10-day-old Irx4Δex2/Δex2 and Irx4Δex2/+ pups were indistinguishable from those of wild-type pups (assessed by morphology, heart weight-to-body weight ratios, and histology; data not shown). Chamber-specific analyses demonstrated right atrium (RA) enlargement in Irx4Δex2/Δex2 mice at 6 weeks of age, with an average increase in RA weight/body weight ratio of 51% compared to that for their wild-type or heterozygous littermates (n = 6; P < 0.02), a finding suggestive of RV dysfunction. Hearts from mature Irx4Δex2/Δex2 mice, age 24 weeks, exhibited significant increases in the ratios of each chamber weight to body weight compared to those from wild-type or heterozygous mice (left atrium, +44%; RA, +57%; LV, +18%; RV, +32%; n = 7; P < 0.05 for each). In vivo assessments confirmed LV hypertrophy (Table 1) in adult mutant mice; wall thickness was greatest in homozygous Irx4Δex2/Δex2 mice, but LV hypertrophy was evident in heterozygous Irx4Δex2/+ mice compared to wild-type mice. Despite increased wall thickness, light microscopy revealed normal myocardial histology without fibrosis in adult heterozygous and homozygous mice. Immunohistochemistry and electron microscopy revealed no pro-ANF secretory granules (data not shown), an ultrastructure unique to atrial cardiocytes (17).
TABLE 1.
Echocardiographic parameters for wild-type, heterozygous Irx4Δex2/+ mice, and homozygous Irx4Δex2/Δex2 micea
| Mouse age (wks) | Genotype (n) | HR (min−1) | LVAW (mm) | LVPW (mm) | LVEDD (mm) | LVESD (mm) | LVFS (%) | VcF (circumferences/s) |
|---|---|---|---|---|---|---|---|---|
| 6 | Wild type (6) | 559 ± 41 | 0.78 ± 0.02 | 0.78 ± 0.02 | 3.20 ± 0.06 | 1.51 ± 0.12 | 53 ± 3 | ND |
| Irx4Δex2/+ (4) | 551 ± 17 | 0.87 ± 0.01 | 0.86 ± 0.01 | 3.00 ± 0.11 | 1.33 ± 0.05 | 55 ± 2 | ND | |
| Irx4Δex2/Δex2 (6) | 566 ± 22 | 0.90 ± 0.06 | 0.90 ± 0.06 | 3.22 ± 0.09 | 1.65 ± 0.15 | 48 ± 3 | ND | |
| 24–30 | Wild type (6) | 547 ± 41 | 0.85 ± 0.04 | 0.85 ± 0.04 | 3.37 ± 0.17 | 1.51 ± 0.20 | 55 ± 5 | 10.7 ± 1.2 |
| Irx4Δex2/+ (5) | 525 ± 27 | 1.00 ± 0.04∗ | 1.00 ± 0.04∗ | 3.20 ± 0.20 | 1.46 ± 0.14 | 55 ± 2 | 9.4 ± 0.4 | |
| Irx4Δex2/Δex2 (7) | 453 ± 24 | 1.06 ± 0.03∗ | 1.06 ± 0.03∗ | 3.65 ± 0.15 | 2.22 ± 0.22∗# | 40 ± 4∗# | 7.1 ± 0.6∗ |
∗, significant difference (P < 0.05) compared with wild-type mice; #, significant difference (P < 0.05) compared with Irx4Δex2/+ mice. HR, heart rate; LVAW, LV anterior wall thickness; LVEDD, LV end-diastolic diameter; LVFS, LV fractional shortening; LVPW, LV posterior wall thickness; LVESD, LV systolic diameter; VcF, velocity of fiber shortening; ND, not determined.
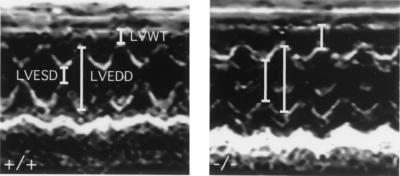
Cardiac echocardiography demonstrated abnormal ventricular function in Irx4Δex2/Δex2 mice including increased end-systolic dimensions, reduced fractional shortening, and diminished velocity of fiber shortening compared to wild-type mice (Table 1 and Fig. 5). In vivo physiological measurements confirmed echocardiographic findings and showed increased end-systolic volumes and decreased ejection fractions (data not shown) in Irx4Δex2/Δex2 mice at 10 and 24 weeks of age. Blood pressure and electrical parameters were normal in Irx4Δex2/Δex2 mice (data not shown). We conclude that Irx4 deficiency adversely effects ventricular function and causes a cardiomyopathy characterized by myocardial hypertrophy, chamber dilation, and systolic dysfunction.
FIG. 5.
Altered ventricular dimensions and function in 6-month-old wild-type and Irx4Δex2/Δex2 mice. M-mode echocardiography shows increased LV wall thickness (LVWT) and LV end-systolic diameter (LVESD) but normal LV end-diastolic diameter (LVEDD) in Irx4Δex2/Δex2 mice (right) compared to those for wild-type animals (left).
DISCUSSION
We have shown that mice with a targeted disruption of the ventricle-specific homeodomain gene Irx4 exhibit aberrant ventricular gene expression and maturity onset cardiomyopathy. Decreased ventricular eHand expression and derepression of an atrial chamber-specific transgene in Irx4-targeted embryos indicate a role for Irx4 in some, but not all, aspects of ventricle-specific gene expression and patterning during heart development. Inappropriate postnatal ventricular expression of ANF, α-skeletal actin, and βMHC in Irx4Δex2/Δex2 mice suggests that Irx4 participates in lifelong maintenance of the ventricular phenotype. While not essential for ventricular chamber formation, Irx4 is required for normal ventricular function.
The response of eHand and of the SMyHC3-HAP transgene in Irx4Δex2/Δex2 embryos indicates that Irx4 controls specific aspects of ventricle-specific gene expression in the developing mouse heart. Aberrant eHand expression in the Irx4Δex2/Δex2 embryo may indicate that Irx4 functions in a manner analogous to Iroquois proteins in Drosophila and Xenopus that establish boundaries of proneural bHLH expression (3, 26, 27) or simply reflects a role for Irx4 in maintaining increased eHand expression levels. The regulation of eHand gene expression in heart development has not been well defined; however it has been shown that eHand expression is decreased in mice lacking Nkx2-5 or FOG-2 (5, 50, 52). Irx4 expression is reduced in mice lacking Nkx2-5 (9) but not in FOG-2-deficient mice (52), suggesting that parallel pathways regulate cardiac eHand expression. This is the case in Drosophila, where parallel pathways involving the Drosophila homologs of Irx4 (Iroquois genes) and FOG-2 (u-shaped) are responsible for the regulation of proneural bHLH genes (16, 26). The role of eHand in cardiomyocytes is unclear and appears not to be related to normal cardiac differentiation, but instead is likely to be related to growth and the looping of the myocardium (21, 45, 46).
Our observation that lack of Irx4 results in the derepression of ANF in the ventricles shortly after birth indicates that Irx4 is a key negative modulator of ANF. Irx4 is expressed in the postnatal ventricular myocardium (9), supporting a role in regulating gene expression after birth as well as in the embryo. The regulation of the ANF gene in cardiac development is complex (17, 48, 57). ANF is initially expressed in the RV precursors, after which its expression appears in the atrial precursors. Subsequent to chamber formation, ANF expression remains strong in the atria and the trabecular region of the ventricles. After birth, ventricular ANF expression decreases to less than 1% of atrial levels. Irx4 is therefore likely to be involved in repressing ANF expression in the ventricular myocardium after birth. We believe that the increased ANF expression in Irx4-deficient mice is independent of the development of cardiomyopathy in these animals, which only becomes physiologically apparent at 6 weeks of age and which is functionally and morphologically measurable at 6 months of age. Despite aberrant ANF expression, we note that ventricular myocytes lacking Irx4 do not express ANF granules as do atrial cells; presumably these ventricular cells lack molecular factors and/or cellular machinery required to produce these secretory granules.
The SMyHC3 gene is the quail ortholog of chicken AMHC1, which previously we have shown to be repressed by Irx4 (1). SMyHC3 elements promote atrial chamber-specific transgenic expression in mice (54); although there is no mammalian ortholog of SMyHC3, the mechanisms for transcriptional regulation of chamber-specific expression appear to have been conserved during myosin gene evolution. It is not known if Irx4 directly binds to SMyHC3 regulatory elements. The DNA-binding site of Irx4 has not been defined; however a bipartite AT-rich binding site in the achaete-scute regulatory sequence has been defined for Drosophila Iroquois protein araucan (26). Since the SMyHC3 promoter region does not contain such a sequence, we suggest either that Irx4 has different DNA-binding specificity than its Drosophila counterparts or that Irx4 acts via protein-protein interactions as do other three-amino-acid length extended class homeodomain proteins (39, 43).
Our data implicate other molecules in specification of the ventricular phenotype. Some are likely to be Iroquois gene family members that incompletely compensate for Irx4 deficiency in Irx4Δex2/Δex2 mice. In Drosophila and Xenopus, Iroquois genes are known to be redundant and functionally interchangeable (3, 11, 18, 26, 27). A deletion of at least two of the Drosophila Iroquois genes araucan, caupolican, and mirror is required to cause a morphological defect, and deletion of all three results in more-profound abnormalities (11, 18, 26). We have shown that besides Irx4 three additional Iroquois genes are expressed in the developing mouse heart. Two of these, Irx1 and Irx2, are expressed in an overlapping pattern in a subset of ventricular cells on the left of the interventricular groove. Additionally, embryonic ventricular Irx2 expression is increased in Irx4Δex2/Δex2 mice. It is intriguing that the sites of expression of Irx1 and Irx2 colocalize with regions where SMyHC3 transgene induction does not occur in Irx4Δex2/Δex2 embryos. In addition we and others (6, 14, 15) have identified a novel Iroquois gene, Irx5. Although widely expressed in both ventricles and atria, Irx5 is excluded from the atrioventricular junction and the outflow tract. Collectively these observations suggest that combinatorial interactions between several Iroquois transcription factors refine the spatial regulation of cardiac gene expression. It is noteworthy in this regard that Irx3 has been shown to play a role in a combinatorial process of neuronal precursor definition in concert with other homeodomain proteins (8).
Our previous studies in which Irx4 function was disrupted with a putative dominant-negative Irx4 molecule in fact indicated a role for Irx4 in ventricle-specific gene expression but not in ventricular morphogenesis (1). While these experiments clearly demonstrated that a dominant-negative molecule could modulate chamber-specific gene expression in a vertebrate heart, they were greatly limited due to multiple technical issues. The chicken cardiac myosin heavy-chain genes are the only chamber-specific genes identified in chicken hearts to date and do not have mammalian orthologs; thus it is difficult to anticipate the response of mammalian chamber-specific genes to similar experimental manipulations. Furthermore, the dominant-negative molecule used in these experiments is predicted to interfere with the actions of multiple Irx family proteins. In addition, the chicken embryos did not survive and are not well suited for physiological measurements of cardiac function; therefore we were not able to address the functional consequences of the manipulation. Also, the timing of viral misexpression is only adequate to disrupt Irx4 function much later than the initiation of Irx4 expression in the developing heart. Thus, using gene targeting, we have been able to address the functional consequences of Irx4 deficiency and now have a useful tool to elucidate the molecular pathways regulated by Irx4.
Previously described etiologies of cardiomyopathy in mice and humans have involved contractile proteins, cytoskeletal proteins, or signaling molecules. The development of cardiomyopathy in Irx4Δex2/Δex2 mice reveals a novel transcriptional pathway in the regulation of ventricular function. We speculate that this cardiomyopathy indicates that Irx4-deficient myocytes lack normal functional properties, thus leading to decompensation when subjected to ventricular load. While it is recognized that cardiac pathologies, in particularly cardiomyopathies, cause ventricular expression of ANF, BNP, βMHC, and α-skeletal actin (12, 13, 17), the expression of ANF and α-skeletal actin in postnatal ventricular Irx4Δex2/Δex2 myocytes is far in advance of cardiac dysfunction, indicating that Irx4-mediated repression of these (and presumably other) genes is important for physiological ventricular function. Identification of other genes regulated by Irx4 and other Iroquois family members should offer further insights into the differences between atrial and ventricular myocytes.
ACKNOWLEDGMENTS
This work was supported by the Howard Hughes Medical Institute (D.F., C.L.C., J.G.S., C.E.S.), the American Heart Association (B.G.B.), the Medical Research Council of Canada/Canadian Institutes of Health Research (B.G.B., M.L.K.B., A.J.B.), the National Institutes of Health (Z.-Z.B., N.R., J.G.S., C.E.S.), the Heart and Stroke Foundation of Ontario (M.L.K.B., A.J.B.).
We thank M. Giewat and J. Vatner for technical assistance, and J. O. Mudd for help with the blood pressure monitoring. We are also grateful to R. Beddington, M. Buckingham, K. Chien, P. Gruss, R. Harvey, C.-C. Hui, S. Izumo, P. Krieg, G. Lyons, S. Orkin, D. Srivastava, S. Tevosian, and S. Tsai for cDNA probes and to C.-C. Hui and R. Harvey for sharing data prior to publication.
REFERENCES
- 1.Bao Z-Z, Bruneau B G, Seidman J G, Seidman C E, Cepko C L. Irx4 regulates chamber-specific gene expression in the developing heart. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- 2.Bass A, Stejskalova M, Ostadal B, Samanek M. Differences between atrial and ventricular energy-supplying enzymes in five mammalian species. Physiol Res. 1993;42:1–6. [PubMed] [Google Scholar]
- 3.Bellefroid E J, Kobbe A, Gruss P, Pieler T, Gurdon J B, Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berul C I, Christe M E, Aronovitz M J, Seidman C E, Seidman J G, Mendelsohn M E. Electrophysiological abnormalities and arrhythmias in alpha MHC mutant familial hypertrophic cardiomyopathy mice. J Clin Investig. 1997;99:570–576. doi: 10.1172/JCI119197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biben C, Harvey R P. Homeodomain factor Nkx2–5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- 6.Bosse A, Stoykova A, Nieselt-Struwe K, Chowdhury K, Copeland N G, Jenkins N A, Gruss P. Identification of a novel mouse Iroquois homeobox gene, Irx5, and chromosomal localization of all members of the mouse Iroquois gene family. Dev Dyn. 2000;218:160–174. doi: 10.1002/(SICI)1097-0177(200005)218:1<160::AID-DVDY14>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Bosse A, Zulch A, Becker M-B, Torres M, Gomez-Skarmeta J-L, Modolell J, Gruss P. Identification of the mammalian Iroquois gene family with overlapping expression during development of the early nervous system. Mech Dev. 1997;69:169–181. doi: 10.1016/s0925-4773(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 8.Briscoe J, Pierani A, Jessell T M, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 9.Bruneau B G, Bao Z Z, Tanaka M, Schott J J, Izumo S, Cepko C L, Seidman J G, Seidman C E. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2–5 and dHand. Dev Biol. 2000;217:266–277. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- 10.Bruneau B G, Logan M, Davis N, Levi T, Tabin C J, Seidman J G, Seidman C E. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 11.Cavodeassi F, Diez Del Corral R, Campuzano S, Dominguez M. Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development. 1999;126:4933–4942. doi: 10.1242/dev.126.22.4933. [DOI] [PubMed] [Google Scholar]
- 12.Chien K R, Knowlton K U, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 13.Chien K R, Zhu H, Knowlton K U, Miller-Hance W, van-Bilsen M, O'Brien T X, Evans S M. Transcriptional regulation during cardiac growth and development. Annu Rev Physiol. 1993;55:77–95. doi: 10.1146/annurev.ph.55.030193.000453. [DOI] [PubMed] [Google Scholar]
- 14.Christoffels V M, Keijser A G, Houweling A C, Clout D E, Moorman A F. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224:263–274. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- 15.Cohen D R, Cheng C W, Cheng S H, Hui C C. Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech Dev. 2000;91:317–321. doi: 10.1016/s0925-4773(99)00263-4. [DOI] [PubMed] [Google Scholar]
- 16.Cubadda Y, Heitzler P, Ray R P, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Bold A J, Bruneau B G. Natriuretic peptides. In: Fray J C S, editor. Handbook of physiology. III. Oxford, United Kingdom: Oxford University Press; 2000. pp. 377–409. [Google Scholar]
- 18.Diez del Corral R, Aroca P, Gomez-Skarmeta J-L, Cavodeassi F, Modolell J. The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes Dev. 1999;13:1754–1761. doi: 10.1101/gad.13.13.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunwoodie S L, Rodriguez T A, Beddington R S P. Msg1 and mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 20.Fewell J G, Hewett T E, Sanbe A, Klevitsky R, Hayes E, Warshaw D, Maughan D, Robbins J. Functional significance of cardiac myosin essential light chain isoform switching in transgenic mice. J Clin Investig. 1998;101:2630–2639. doi: 10.1172/JCI2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firulli A B, McFadden D G, Lin Q, Srivastava D, Olson E N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- 22.Fishman M C, Chien K R. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- 23.Fishman M C, Olson E N. Parsing the heart: genetic modules for organ assembly. Cell. 1997;91:153–156. doi: 10.1016/s0092-8674(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 24.Georgakopoulos D, Christe M E, Giewat M, Seidman C M, Seidman J G, Kass D A. The pathogenesis of familial hypertrophic cardiomyopathy: early and evolving effects from an alpha-cardiac myosin heavy chain missense mutation. Nat Med. 1999;5:327–330. doi: 10.1038/6549. [DOI] [PubMed] [Google Scholar]
- 25.Gerber W V, Yatskievych T A, Antin P B, Correia K M, Conlon R A, Krieg P A. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech Dev. 1999;80:77–86. doi: 10.1016/s0925-4773(98)00195-6. [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Skarmeta J L, del Corral R D, de la Calle-Mustienes E, Ferre-Marco D, Modolell J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- 27.Gomez-Skarmeta J L, Glavic A, de la Calle-Mustienes E, Modolell J, Mayor R. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. EMBO J. 1998;17:181–190. doi: 10.1093/emboj/17.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottshall K R, Hunter J J, Tanaka N, Dalton N, Becker K D, Ross J, Jr, Chien K R. Ras-dependent pathways induce obstructive hypertrophy in echo-selected transgenic mice. Proc Natl Acad Sci USA. 1997;94:4710–4715. doi: 10.1073/pnas.94.9.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber P J, Kubalak S W, Chien K R. Downregulation of atrial markers during cardiac chamber morphogenesis is irreversible in murine embryos. Development. 1998;125:4427–4438. doi: 10.1242/dev.125.22.4427. [DOI] [PubMed] [Google Scholar]
- 30.Hendrickson B A, Conner D A, Ladd D J, Kendall D, Casanova J E, Corthesy B, Max E E, Neutra M R, Seidman C E, Seidman J G. Altered hepatic transport of immunoglobulin A in mice lacking the J chain. J Exp Med. 1995;182:1905–1911. doi: 10.1084/jem.182.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hume J R, Uehara A. Ionic basis of the different action potential configurations of single guinea-pig atrial and ventricular myocytes. J Physiol (London) 1985;368:525–544. doi: 10.1113/jphysiol.1985.sp015874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly R G, Zammit P S, Mouly V, Butler-Browne G, Buckingham M E. Dynamic left/right regionalisation of endogenous myosin light chain 3F transcripts in the developing mouse heart. J Mol Cell Cardiol. 1998;30:1067–1081. doi: 10.1006/jmcc.1998.0705. [DOI] [PubMed] [Google Scholar]
- 33.Kubalak S W, Miller-Hance W C, O'Brien T X, Dyson E, Chien K R. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- 34.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 35.Lyons G E, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McConnell B K, Jones K A, Fatkin D, Arroyo L H, Lee R T, Aristizabal O, Turnbull D H, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen F J, Conner D, Fischman D A, Seidman C E, Seidman J G. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice J. Clin Investig. 1999;104:1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNutt N S, Fawcett D W. The ultrastructure of the cat myocardium. II. Atrial muscle. J Cell Biol. 1969;42:46–66. doi: 10.1083/jcb.42.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Brien T X, Lee K J, Chien K R. Positional specification of ventricular myosin light chain 2 expression in the primitive murine heart tube. Proc Natl Acad Sci USA. 1993;90:5157–5161. doi: 10.1073/pnas.90.11.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passner J M, Ryoo H D, Shen L, Mann R S, Aggarwal A K. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 40.Patwardhan V, Fernandez S, Montgomery M, Litvin J. The rostro-caudal position of cardiac myocytes affect (sic) their fate. Dev Dyn. 2000;218:123–135. doi: 10.1002/(SICI)1097-0177(200005)218:1<123::AID-DVDY11>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Pawloski-Dahm C M, Song G, Kirkpatrick D L, Palermo J, Gulick J, Dorn G W, Robbins J, Walsh R A. Effects of total replacement of atrial myosin light chain-2 with the ventricular isoform in atrial myocytes of transgenic mice. Circulation. 1998;97:1508–1513. doi: 10.1161/01.cir.97.15.1508. [DOI] [PubMed] [Google Scholar]
- 42.Pereira F A, Qiu Y, Zhou G, Tsai M J, Tsai S Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piper D E, Batchelor A H, Chang C P, Cleary M L, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 44.Riddle R D, Johnson R L, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 45.Riley P, Anson-Cartwright L, Cross J C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 46.Riley P R, Gertenstein M, Dawson K, Cross J C. Early exclusion of Hand1-deficient cells from distinct regions of the left ventricular myocardium in chimeric mouse embryos. Dev Biol. 2000;227:156–168. doi: 10.1006/dbio.2000.9864. [DOI] [PubMed] [Google Scholar]
- 47.Sanbe A, Gulick J, Hayes E, Warshaw D, Osinska H, Chan C B, Klevitsky R, Robbins J. Myosin light chain replacement in the heart. Am J Physiol. 2000;279:H1355–H1364. doi: 10.1152/ajpheart.2000.279.3.H1355. [DOI] [PubMed] [Google Scholar]
- 48.Seidman C E, Wong D W, Jarcho J A, Bloch K D, Seidman J G. Cis-acting sequences that modulate atrial natriuretic factor gene expression. Proc Natl Acad Sci USA. 1988;85:4104–4108. doi: 10.1073/pnas.85.11.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srivastava D, Thomas T, Lin Q, Kirby M L, Brown D, Olson E N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka M, Chen Z, Bartunkova M, Yamazaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 51.Tevosian S G, Deconinck A E, Cantor A B, Rieff H I, Fujiwara Y, Corfas G, Orkin S H. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tevosian S G, Deconinck A E, Tanaka M, Schinke M, Litovsky S H, Izumo S, Fujiwara Y, Orkin S H. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 53.Wang G F, Nikovits W, Jr, Schleinitz M, Stockdale F E. A positive GATA element and a negative vitamin D receptor-like element control atrial chamber-specific expression of slow myosin heavy-chain gene during cardiac morphogenesis. Mol Cell Biol. 1998;18:6023–6034. doi: 10.1128/mcb.18.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xavier-Neto J, Neville C M, Shapiro M D, Houghton L, Wang G F, Nikovits W, Stockdale F E, Rosenthal N. A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development. 1999;126:2677–2687. doi: 10.1242/dev.126.12.2677. [DOI] [PubMed] [Google Scholar]
- 55.Yutzey K, Gannon M, Bader D. Diversification of cardiomyogenic cell lineages in vitro. Dev Biol. 1995;170:531–541. doi: 10.1006/dbio.1995.1234. [DOI] [PubMed] [Google Scholar]
- 56.Yutzey K E, Rhee J T, Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development. 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]
- 57.Zeller R, Bloch K D, Williams B S, Arceci R J, Seidman C E. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev. 1987;1:693–698. doi: 10.1101/gad.1.7.693. [DOI] [PubMed] [Google Scholar]