Summary
Toxoplasma gondii surface antigen 1 (TgSAG1) is a surface protein of tachyzoites, which plays a crucial role in toxoplasma gondii infection and host cell immune regulation. However, how TgSAG1 regulates these processes remains elucidated. We utilized the biotin ligase -TurboID fusion with TgSAG1 to identify the host proteins interacting with TgSAG1, and identified that S100A6 was co-localized with TgSAG1 when T. gondii attached to the host cell. S100A6, either knocking down or blocking its functional epitopes resulted in inhibited parasites invasion. Meanwhile, S100A6 overexpression in host cells promoted T. gondii infection. We further verified that TgSAG1 could inhibit the interaction of host cell vimentin with S100A6 for cytoskeleton organization during T. gondii invasion. As an immunogen, TgSAG1 could promote the secretion of tumor necrosis factor alpha (TNF-α) through S100A6-Vimentin/PKCθ-NF-κB signaling pathway. In summary, our findings revealed a mechanism for how TgSAG1 functioned in parasitic invasion and host immune regulation.
Subject areas: Molecular biology, Immune response, Parasitology
Graphical abstract
Highlights
-
•
TgSAG1 interacts with host protein S100A6 then regulates T. gondii infection
-
•
TgSAG1 could regulate binding vimentin with S100A6 during T. gondii infection
-
•
TgSAG1 regulate TNFα secretion through S100A6-vimentin/PKCθ-NF-κB signaling pathway
Molecular biology; Immune response; Parasitology
Introduction
Toxoplasma gondii (T. gondii) is a zoonotic parasite distributed worldwide that poses a threat to public health (Montoya and Liesenfeld, 2004). As an obligate intracellular protozoan, T. gondii has been found to infect nearly all warm-blooded animals including humans (Dubey and Frenkel, 1972). In immunocompetent hosts, T. gondii infections are subclinical. However, in immunosuppressive patients such as acquired immunodeficiency syndrome (AIDS) patients, tumor patients with chemotherapy, or the patients undergoing organ transplantation, T. gondii infection will result in toxoplasmic encephalitis, retinochoroiditis, and even death (Luft and Remington, 2002). T. gondii has a complex life cycle, felids are the only definitive hosts, while in the intermediate hosts, the parasite undergoes tachyzoite and bradyzoite asexual multiplication, tachyzoites rapidly proliferate and quickly disseminate within the hosts (Montoya and Liesenfeld, 2004). Tachyzoite is the key disease-causing form and and activates a potent host immune response during the acute infection. The surface of the tachyzoite is the main target for host immune responses (Lekutis et al., 2001). T. gondii tachyzoites surface is covered with glycosylphosphatidylinositol (GPI)-anchored antigens, most of which are members of the surface antigen 1 (SAG1) or SAG2 families (Boothroyd et al., 1998; Manger et al., 1998; Tomavo et al., 1989).
The SAG1 family maintains the 12 conserved cysteine residues which have an overall identity of 30% approximately (Lekutis et al., 2001). T. gondii surface antigen 1 (TgSAG1), also known as P30, belongs to the SAG1 family and plays a crucial role in T. gondii for host recognition, attachment, invasion, and immune regulation. Treatment of extracellular parasites with the poly-clonal anti-SAG1 serum inhibited T. gondii's attaching to fibroblasts monolayer by 71%. In the bovine kidney cells pretreated with a mono-clonal antibody 6A8 and C1E3 against TgSAG1, T. gondii attachment was significantly inhibited by 65% (Mineo JR, 1994; Mineo et al., 1993); and another TG05.54 anti-SAG1 monoclonal antibody pretreatment inhibited T. gondii invasion to adult bovine kidney cells was in a dose-dependent manner (Grimwood and Smith, 1996). After BALB/c or C57BL/6 mice were intravenously inoculated with a DNA vaccine derived from TgSAG1 gene, significantly higher levels of total IgG, as well as cytokines including interferon γ (IFN-γ), interleukin-2 (IL-12), interleukin-10 (IL-10), and tumor necrosis factor alpha (TNF-α) were detected in the vaccination group, compared with the control group (Pagheh et al., 2020). However, as a surface antigen, how TgSAG1 functions in recognizing, attaching and invading host cells, and modulating host immunity remains unclear. Till now, only one research reveals that TgSAG1 strongly binds with H. sapiens lysine coil-coiled rich protein (HLY) during T. gondii invasion (Lai and Lau, 2018). To address this issue, we applied the proximity dependent biotin identification (BioID) technology to label host cell proteins that may interact with TgSAG1. BioID is an effective method to screen for physiologically relevant protein interactions, it utilizes a promiscuous biotin ligase being fused to an interesting protein named bait protein, and the bait protein can traffic to an organellar sub-compartment and biotinylate multiple proximal proteins. Then, the biotinylated proteins are purified using streptavidin affinity chromatography and identified by mass spectrometry. This method can identify weak and transient interactions and is amenable to temporal regulation (Roux et al., 2013).
In this study, we adopted the BioID system with TgSAG1 as the bait protein to identify its host interactome. The host S100 Calcium Binding Protein (S100A6) was further identified to interact and co-localize with TgSAG1, besides, as a surface antigen of tachyzoites, TgSAG1 could regulate host cell immunity through S100A6 signaling pathway.
Results
TgSAG1-TurboID-HA fusion was successfully constructed on the T. gondii tachyzoites
We adapted the BioID system to identify putative substrates and interacting proteins of TgSAG1, using TgSAG1 as a bait protein. A recombinant RH strain overexpressing TgSAG1 fused with biotin ligase TurboID followed by C-terminal hemagglutinin (HA) epitope tag was constructed, which was named RH-SAG1-turboID-HA strain (Figure S1A). The recombinant strain was verified by Western blot (WB) using the anti-HA antibody, the result showed that TgSAG1-turboID-HA fusion protein was correctly expressed at the expected size (Figure S1B). The immunofluorescence assay (IFA) result showed the co-localization of the fusion protein and the native SAG1 protein (Figure S1C).
Identification of the host TgSAG1 interactome
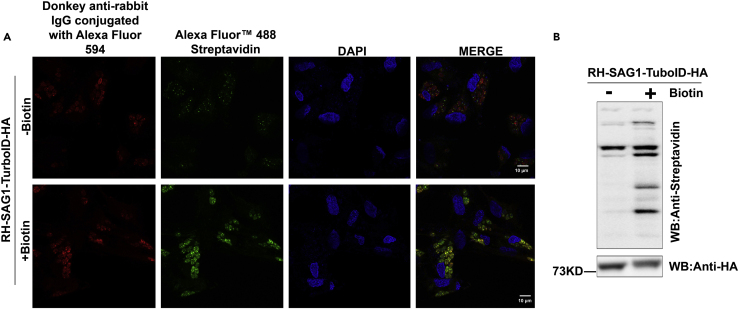
HFFs were infected with the recombinant RH-SAG1-TurboID-HA tachyzoites, and grown in the DMEM with or without D-Biotin addition. The biotinylated proteins were identified with IFA using the probe streptavidin conjugated with Alexa Fluor 488. We found that the recombinant parasites grown in the cells supplemented with exogenous D-Biotin displayed stronger biotinylation activity compared with the parasites in the cells without D-Biotin supplementation (Figure 1A). After that, the cells were harvested and lysed with RIPA buffer containing proteinase inhibitor cocktails, and subjected to affinity purification of biotinylated proteins with streptavidin conjugated magnetic beads. The elutes were analyzed by WB, and the results showed us that, in addition to the proteins common in both groups (with or without D-Biotin addition), some proteins were only present in the recombinant parasites infected cells cultured with D-Biotin supplementation (Figure 1B). Having confirmed the presence of various specific biotinylated proteins in the recombinant parasites infected cells with D-Biotin addition, we scaled up the affinity purification of the biotinylated proteins grown with D-Biotin supplementation and the purified proteins were sent to Beijing Genomics Institute (BGI) for mass spectrometry analysis.
Figure 1.
Detection of the biotinylate proteins in RH-SAG1-TurboID-HA infected HFF cells
(A) The biotinylated proteins were detected with immunofluorescent assay (IFA) in the RH-SAG1-TurboID-HA parasites infected HFF cells with or without addition of D-Biotin. Streptavidin Alexa Fluor 488 (green fluorescence) was used to detect the biotinylated proteins, and SAG1-TurboID-HA was detected with an anti-HA monoclonal antibody and visualized with the anti-rabbit IgG conjugated with Alexa Fluor 594 (red fluorescence) (scale bars, 10 μm).
(B) Western blot (WB) detection of the biotinylated proteins in the RH-SAG1-TurboID-HA infected cells with or without addition of D-Biotin, Streptavidin-HRP was used to detect the biotinylated proteins and anti-HA antibody was used to detect the SAG1-TurboID-HA recombinant protein.
See also Figure S1.
Gene Ontology (GO) terms enrichment analysis of TgSAG1 interacting proteins
The proteomic data generated from the RH-SAG1-TurboID-HA recombinant strain infected cells supplemented with D-Biotin or not were compared. The recombinant parasites infected cells with D-Biotin supplementation yielded 119 specific TgSAG1 interactive proteins (Table S2).
Next, these 119 specific TgSAG1 interactive proteins were analyzed with GO term enrichment, and the result showed us that these proteins were categorized into three groups: biological processes, molecular functions, and cellular components. As shown in Figure S2, in cellular components category, 76 proteins were enriched in membrane proteins and subjected to analysis with string 11.0 database (https://string-db.org/), which builds protein-protein interactions based on experimental evidences and predictions based on genomic context (Von Mering et al., 2005). Some membrane proteins were revealed, including S100 calcium binding protein A6 (S100A6) involved in hepatitis C virus (HCV) infection (Tani et al., 2013), lymphocyte cell surface protein CD26 (DPP4) responsible for Middle East respiratory syndrome coronavirus (MERS-CoV) invasion (Raj et al., 2013), annexin A6 (ANXA6) impairing influenza A virus budding and releasing (Ma et al., 2012), myosin heavy chain 9 (MYH9) functioning as a host cell receptor for porcine reproductive and respiratory syndrome virus (PRRSV) internalization in porcine alveolar macrophages (Hou et al., 2019), cytoskeleton associated protein 4 (CKAP4) involved in SARS-CoV-2 infection and COVID-19 disease severity (Patel et al., 2021), and integrin subunit beta 1 (ITGB1) involved in Rabies virus (RABV) cellular entry (Shuai et al., 2020).
All the proteins mentioned above were the top-ranking membrane proteins, regulating the invasion or infection of intracellular pathogens. The interaction of these six membrane proteins with TgSAG1 was further identified.
Identification of S100A6-TgSAG1 interaction
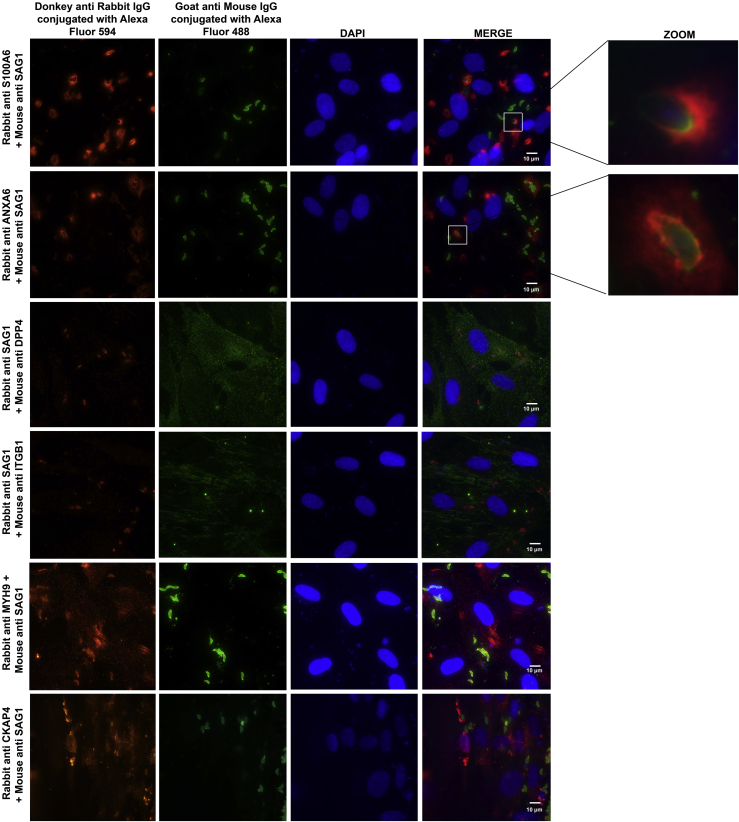
HFF cells were infected with the recombinant RH-SAG1-TurboID-HA tachyzoites and then subjected to IFA. From the result, we observed the co-localization of TgSAG1 with the host protein S100A6 and ANXA6 when T. gondii tachyzoites attached to HFF cells (Figure 2). However, the co-localization of the host proteins DPP4, ITGB1, MYH9, and CKAP4 with TgSAG1 was not shown in T. gondii infected HFF cells.
Figure 2.
Identification of host cell S100A6 and ANXA6's co-localization with TgSAG1 when T. gondii is attaching to host cells
Co-localization of TgSAG1 with its interactive proteins in RH-WT infected HFF cells were visualized by IFA. The primary antibodies, rabbit S100A6, ANXA6, MYH9, CKAP4, and HA-tag antibody, followed by the second antibody Donkey anti-rabbit IgG conjugated with Alexa Fluor 594 were used to detect S100A6, ANXA6, MYH9, and CKAP4, whereas mouse anti DPP4, ITGB1, SAG1 antibody followed by Goat anti-mouse IgG conjugated with Alexa Fluor 488 were used to detect DPP1, ITGB1, and SAG1(scale bars, 10 μm).
See also Figure S2.
ANXA6 and S100A6 belong to Annexins and S100 protein families, respectively. Annexins are made up of a highly alpha-helical core domain that binds calcium ions, allowing them to interact with phospholipid membranes. Several annexins have been shown to bind proteins that belong to the S100 calcium-binding protein family, such as S100A6 and ANXA6 (Cmoch et al., 2011; Rintala-Dempsey et al., 2008). In our protein-protein interaction analysis with string 11.0, S100A6, instead of ANXA6, formed a central hub in the network (Figure S2B). S100A6 also connected with the glycoprotein NMB (GPNMB), AHNAK Nucleoprotein (AHNAK), S100A8, and ADP Ribosylation Factor 4 (ARF4), which are known to play important roles during the infection of the viruses such as hepatitis C virus, hepatitis B virus, and Porcine circovirus (Guo et al., 2019; Sun et al., 2018; Zhang et al., 2017). S100A6 has been shown to play important roles in virus infection, but has not previously been implicated in T. gondii infection; therefore, we took this protein as research target.
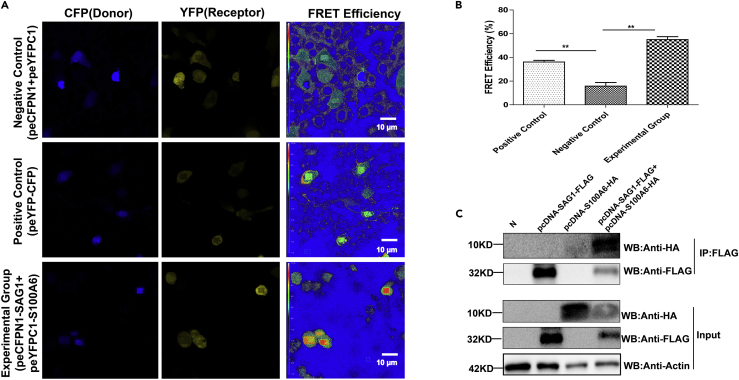
The interaction of the host protein S100A6 and TgSAG1 was identified with FRET. As shown in Figure 3A, the different color bands represent different ranges of FRET efficiencies: the dark blue represents the FRET efficiency of <25%, the green for the FRET efficiency of 25%–50%, and the red for the FRET efficiency range of >50%. The result showed positive FRET signals in the positive controls group (cells transfected with peYFP-CFP) and the experimental group (cells co-transfected with peYFPC1-S100A6 and peCFPN1-TgSAG1), both yielding significantly higher FRET efficiency than the negative control group (cells co-transfected with peYFP and peCFP) (Figure 3B). This finding demonstrated the stable interaction of S100A6 with TgSAG1. This result was further confirmed with the co-immunoprecipitation (co-IP) assay, after we co-transfected pcDNA3.1(+)-TgSAG1-3×FLAG and pcDNA3.1(+)-S100A6-HA plasmids into COS7 cells. The WB result showed that in the dually transfected cells, S100A6 could be readily detected in the immunoprecipitants using anti-Flag antibody as the bait to capture the proteins interacting with TgSAG1 (Figure 3C). Therefore, these results indicated the interaction of TgSAG1 and host S100A6.
Figure 3.
Identification of the interaction of host S100A6 with TgSAG1
(A) Co-localization and Fluorescence resonance energy transfer (FRET) detection of TgSAG1 and S100A6. Localization and co-localization of TgSAG1 and S100A6 are shown in the donor channel (column 1) and the receptor channel (column 2), respectively. The FRET efficiency is shown in column 3, in which a thermal pseudo color-matched FRET signal intensity scale is indicated in each image (scale bars, 10 μm).
(B) Quantitative analysis of FRET efficiency between TgSAG1 and S100A6. Error bars represent the means ± SD of the triplicates. Student's t-tests results were used to detect the difference between the two experimental groups and Negative Control (∗∗p < 0.05).
(C) Lysates of COS7 cells overexpressing TgSAG1 and S100A6 were immunoprecipitated with the anti-Flag antibody. The immunoprecipitants were detected by SDS-PAGE and WB using the antibodies as indicated.
S100A6 expression is important for parasite infection
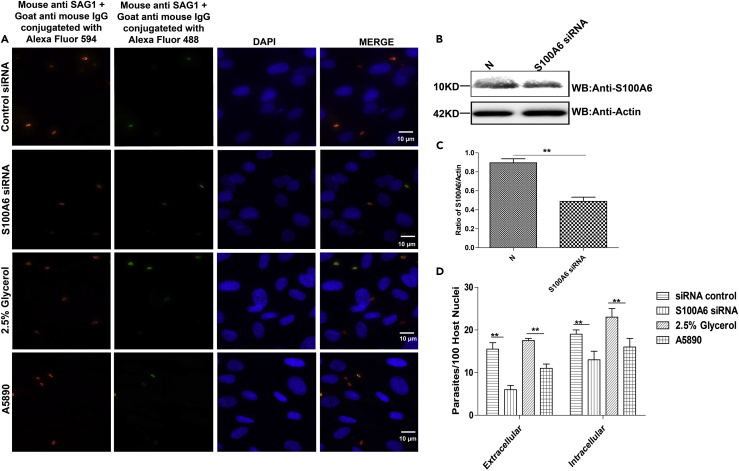
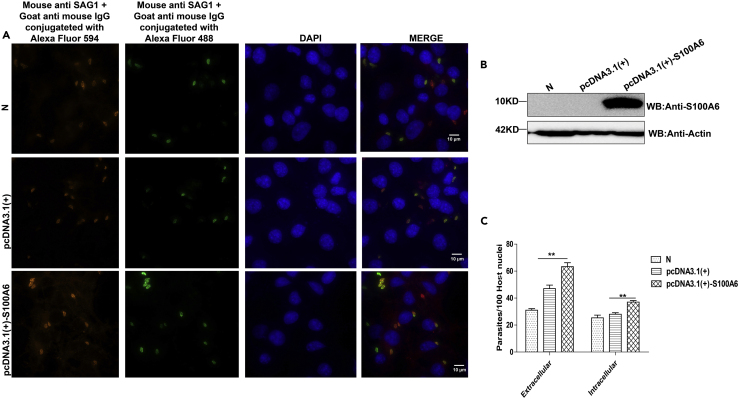
It has been previously reported that disruption of S100A6 expression results in deficient host cell invasion of hepatitis B virus (Tong et al., 2009). Next, to determine the importance of S100A6 during T. gondii invasion, we knocked down S100A6 expression with small interference RNA (siRNA) or blocked its epitopes by using specific antibody A5890 (Figures 4B and 4C), and then the attachment and invasion of the parasite was evaluated by IFA. We found that when S100A6 was knocked down or the immune epitopes were blocked, T. gondii infection efficiency was significantly inhibited compared with that of siRNA control group or 2.5% glycerol treatment group (Figures 4A and 4D). To further determine whether S100A6 could promote the invasion of the parasites or not, we overexpressed S100A6 in COS7 cells. The WB analysis confirmed the overexpression of S100A6 (Figure 5B). The empty vector pcDNA3.1(+) transfected cells exhibited a comparable invasion efficiency with the N group, whereas the S100A6 overexpression cells showed a higher attachment and invasion rate than the N group and pcDNA3.1(+) transfection group (Figures 5A and 5C). These data indicated that S100A6 ccould promote the parasite's infection.
Figure 4.
The S100A6 expression level was highly co-related with T. gondii invasion efficiency
(A) Representative wide-field epifluorescence images (10 × 100) for RH-WT invasion in cells with S100A6 knockdown or epitope blocking with anti-S100A6 antibody A5890, and the other treatments as indicated. The intracellular parasites were labeled red, and the extracellular parasites were labeled both green and red (to appear as yellow) (scale bars, 10 μm).
(B) WB verification of S100A6 knockdown in HFF cells.
(C) Densitometric analysis of the western blotting results with the ImageJ software. The ratios of S100A6/actin in N and S100A6 siRNA group were calculated independently.
(D) Quantitative analysis of T. gondii attachment and invasion efficiency in different treatment groups (presented with parasites/100 host cells). Each experiment was performed in triplicate. Quantification of the mean ± SD and analyzed by t test (∗∗p < 0.05).
Figure 5.
Host cell protein S100A6 promoted T. gondii invasion
(A) Representative wide-field epifluorescence images (10 × 100) for the COS7 cells transfected with the indicated plasmid and infected by RH-WT. The intracellular parasites are labeled red, and the extracellular parasites are labeled both green and red to appear as yellow (scale bars, 10μm
B) Verification of S100A6 overexpression in COS7 cells by WB.
(C) Quantitative analysis of T. gondii attachment and invasion efficiency in different treatment groups (presented with parasites/100 host cells). Each experiment was performed in triplicate. Quantification of the mean ± SD and analyzed by t test (∗∗p < 0.05).
TgSAG1 participated in host cytoskeleton organization during T. gondii invasion
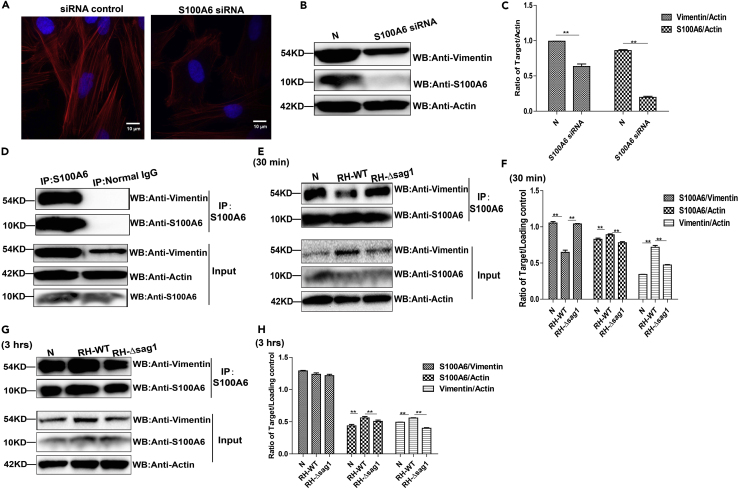
Next, we intended to understand how TgSAG1 regulated the parasitic infection through S100A6 signaling. Here we found that S100A6 knockdown in HFF cells decreased the formation of microtubules (Figure 6A), as well as the intermediate filaments vimentin (Figures 6B and 6C). As a binding partner of numbers of proteins engaged in cytoskeletal organization, S100A6 was observed to interact with vimentin (Figure 6D). T. gondii infection causes the reorganization of host intermediate filaments and microtubules around the parasitophorous vacuole (PV) (Li et al., 2008). Whether TgSAG1 participates in and regulates this organization through S100A6 signaling pathway was examined by co-IP. The results showed that at 30 min post infection, the host cell S100A6-vimentin interaction was significantly prohibited in RH-WT infected cells, compared to RH-Δsag1 infected cells and normal control cells (Figures 6E and 6F). However, this phenomenon didn't persist throughout the infection. At 3 h post infection, the similar binding of vimentin with S100A6 was observed in RH-WT infected and RH-Δsag1 infected cells (Figures 6G and 6H). Data presented here showed that TgSAG1 could regulate the binding between S100A6 and vimentin for cytoskeleton organization during T. gondii invasion to host cells.
Figure 6.
TgSAG1 was involved in host cell cytoskeleton organization during T. gondii invasion
(A) Representative wide-field epifluorescence images of the actin cytoskeleton arrangement (10 × 100) in the S100A6 knockdown HFF cells. Cytoskeleton was labeled with Phalloidin eFluor 570, the nucleus was labeled with DAPI (scale bars, 10 μm).
(B) Detection of vimentin expression in the S100A6 knockdown cells with WB.
(C) Densitometric analysis of the WB result, data was shown with Vimentin/Actin in Normal and S100A6 siRNA groups.
(D) Lysates of HFF cells were immunoprecipitated with the anti-S100A6 antibody. The immunoprecipitants were detected by WB using the indicated antibody.
(E and G) Lysates of HFF cells infected with RH-WT or RH-Δsag1 for 30 min or 3 h were immunoprecipitated with the anti-S100A6 antibody. The immunoprecipitants were detected by WB using the antibody indicated.
(F and H) Densitometric analysis of the WB result, data was shown with S100A6/Vimentin, S100A6/Actin, Vimentin/Actin in the indicated treatment groups. Each experiment was performed in triplicate, data was presented as mean ± SD and statistical analysis was done by t test (∗∗p < 0.05).
See also Figure S3.
TgSAG1 promoted TNF-α expression in vitro
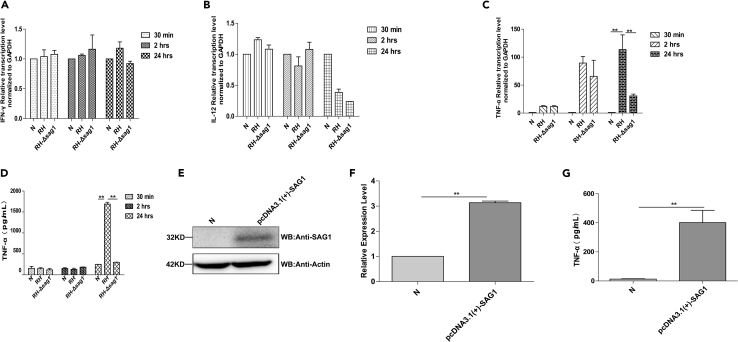
As TgSAG1 is a strong immunogen, we next examined whether TgSAG1 manipulated cytokine secretion in vitro. We generated RH-Δsag1 as Figure S3 described, and then RH-WT or RH-Δsag1 strain was used to infect THP-1 cells. The results showed that no significant difference was observed for the transcription of IL-12p70 and IFN-γ between RH-WT and RH-Δsag1 infected cells (Figures 7A and 7B). However, THP-1 cells infected by RH-WT promoted TNF-α transcription and secretion more than those infected by RH-Δsag1 strain (Figures 7C and 7D). Furthermore, to determine whether TgSAG1 could promote TNF-α expression, we overexpressed of TgSAG1 in RAW264.7 cells (Figure 7E), the results showed us that cells overexpressed TgSAG1 strongly promoted TNF-α production (Figures 7F and 7G). Thus, we concluded that the TgSAG1 could promote TNF-α secretion.
Figure 7.
TgSAG1 promoted TNF-α secretion in THP-1 cells
(A–C) Transcriptional levels of IL-12p70, IFN-γ, TNF-α in RH-WT or RH-Δsag1 infected THP-1 cells were detected by qRT-PCR.
(D) The secretion of TNF-α in RH-WT or RH-Δsag1 infected THP-1 cells were detected by ELISA. Data were presented as pg/mL.
(E) Verification of TgSAG1 overexpression in RAW264.7 cells by WB.
(F and G) the transcription and secretion of TNF-α in TgSAG1 transfected RAW264.7 cells were detected by qRT-PCR and ELISA, respectively. Data were presented as means ± SD combined from three independent experiments and statistical significance were analyzed by t test (∗∗p < 0.05).
TgSAG1 promoted TNF-α secretion through S100A6-Vimentin/PKCθ-NF-κB signaling pathway
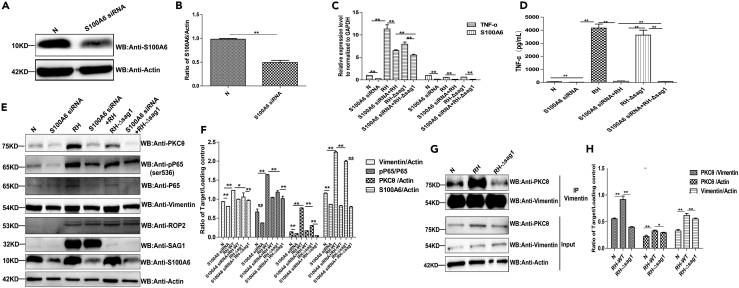
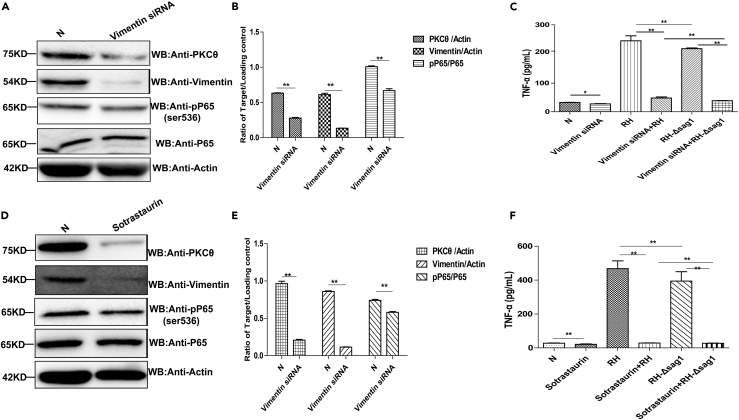
To investigate the mechanism of TgSAG1 on TNF-α secretion, THP-1 cells were transiently transfected with S100A6 siRNA followed by RH-WT or RH-Δsag1 infection. The results showed that in the cells with S100A6 knockdown and infected with RH-WT, TNF-α secretion was significantly inhibited compared with the RH-WT infection group. In addition, the THP-1 cells transfected with S100A6 siRNA followed by RH-Δsag1 infection showed dramatic inhibition of TNF-α secretion, compared with the cells transfected with S100A6 siRNA and infected with RH-WT (Figures 7A–7D). Taken together, these results implied that TgSAG1 promoted TNF-α secretion through S100A6 signaling pathway. Previous findings indicate that vimentin mediated initial activation of NF-κB followed by TNF-α secretion (Cui et al., 2019); however, the underlying signaling pathway from vimentin to NF-κB activation remains unclear. Therefore, we hypothesized that TgSAG1 mediated parasite infection and promoted the TNF-α secretion both through S100A6 and vimentin signaling pathway. WB and densitometric analysis showed that S100A6 knockdown significantly decreased the expression of vimentin and PKCθ. Additionally, higher levels of phosphorylated P65 at ser536, and PKCθ were observed in the THP-1 cells infected with RH-WT than that in the cells infected with RH-Δsag1 strain. It could be concluded that T. gondii increased the expression of phosphorylated P65 at ser536 and PKCθ through TgSAG1 signaling. Furthermore, in S100A6 knockdown cells, the PKCθ expression was largely suppressed by RH-Δsag1 infection compared to RH-WT infection (Figures 8E and 8F). These findings implied that TgSAG1 could increase the expression of PKCθ through S100A6 signaling pathway. Our IP results also showed that RH-WT infection strongly promoted the interaction of vimentin and PKCθ compared with RH-Δsag1 infected cells and non-infected group (Figures 8G and 8H), which indicated that TgSAG1 could enhance the binding of vimentin and PKCθ, and then promoted NF-κB activation and TNF-α secretion.
Figure 8.
TgSAG1 promoted the secretion of TNF-α through S100A6 signaling pathway
(A and B) Verification of S100A6 knockdown in THP-1 cells by WB and densitometric analysis for the WB result.
(C and D) The transcription and secretion of TNF-α in the S100A6 knockdown THP-1 cells with RH-WT or RH-Δsag1 infection were detected by qRT-PCR and ELISA, respectively.
(E) The expression of S100A6, vimentin, PKCθ, P65, and the pP65-ser536 in the S100A6 knockdown cells with RH-WT or RH-Δsag1 infection were detected by WB.
(F) Densitometric analysis of the WB result, the ratio of Vimentin/Actin, pP65/P65, PKCθ/Actin and S100A6/Actin were calculated independently.
(G) Lysates of THP-1 cells infected with RH-WT or RH-Δsag1 for 24 h were immunoprecipitated with the anti-vimentin antibody. The immunoprecipitants were detected by WB using the antibodies as indicated.
(H) Densitometric analysis of PKCθ/Vimentin, PKCθ/Actin, Vimentin/Actin. Statistical data were presented as means ± SD combined from three independent experiments and statistical significance were analyzed by t test (∗∗p < 0.05, ∗p < 0.1).
Additional experiments treating cells with vimentin siRNA or PKCθ inhibitor sotrastaurin (Selleck, USA) followed by RH-WT or RH-Δsag1 infection, consistently showed that the inhibited vimentin and PKCθ expression could restrain the phosphorylation of P65 at ser536 followed by TNF-α secretion (Figures 9A–9F). Taken together, the results suggested that TgSAG1 could enhance the interaction between PKCθ and vimentin through S100A6 signaling pathway, and then activate the NF-κB finally for the TNF-α secretion.
Figure 9.
TgSAG1 promoted the secretion of TNF-α through vimentin/PKCθ-NF-κB pathway
(A) Detection of P65 phosphorylation at ser536 and PKCθ in vimentin knockdown THP-1 cells.
(B and E) The ratios of Vimentin/Actin, pP65/P65 and PKCθ/Actin were calculated by densitometric analysis.
(C) The secretion of TNF-α in the vimentin knockdown cells with RH-WT or RH-Δsag1 infection were detected by ELISA.
(D) Detection of P65 phosphorylation at ser536 and vimentin in PKCθ inhibitor treated THP-1 cells.
(F) The secretion of TNF-α in PKCθ inhibitor treatment cells with RH-WT or RH-Δsag1 infection were detected by ELISA. Data were presented as means ± SD combined from three independent experiments and analyzed by t test (∗∗p < 0.05).
Discussion
In this study, we screened TgSAG1 interactome in physiological condition through SAG1-TurboID expression and streptavidin affinity chromatography and mass spectrometry. Three enzymes for proximity labeling (PL) have been reported: BioID/BioID2, APEX/APEX2 and TurboID/miniTurbo. As for BioID, it's attractive for the simplicity of its labeling protocol, non-toxic labeling conditions, and only biotin needs to be added to initiate tagging. However, the major disadvantage for this enzyme is its slow kinetics, which necessitates 18-24 h or much longer for biotin labeling (Uezu et al., 2016). APEX/APEX2, derived from dimeric soybeans, can be expressed in the reducing cytosolic environment without loss of activity owning to lacking disulfides and calcium binding sites (Rhee et al., 2013). APEX/APEX2 can be used for intracellular specific protein imaging with electron microscope (EM) (Martell et al., 2012). The main advantage of APEX/APEX2 is its speed: proximal proteins can be tagged in 1 min or less, enabling dynamic analysis of protein interaction networks. However, APEX labeling requires H2O2, which is toxic to cells and difficult to be delivered to live organisms without causing severe tissue damage, and limited sensitivity of APEX, precludes its applications (Lam et al., 2015). As for TurboID/miniTurbo, both have much higher activity than BioID/BioID2, enabling proteomic labeling in just 10 min. Furthermore, TurboID can produce more biotinylated products than BioID within the same labeling time (Branon et al., 2018).
The internalization of T. gondii in host cells starts from an initial contact of the cell surface, and then a signal is transduced from the parasite surface to the apex and is immediately followed by progressive internalization (Dubremetz et al., 1998). In this study, by using TurboID infusion with surface antigen SAG1 of T. gondii, the host cell plasma membrane protein S100A6 was found to bind with TgSAG1 (Figures 2 and 3).
S100A6 belongs to the S100 protein family of Ca2+-binding proteins. As a cytoplasm and cell membrane protein, S100A6 has been implicated in the regulation of several cellular functions, such as cell proliferation and cancer development, endothelial cell cycle and senescence, mesenchymal stem cells adhesion, breast cancer cells and neuro cells apoptosis, and so on (Donato et al., 2017). Extracellular role of S100A6 might be exerted through its binding to the transmembrane receptor for advanced glycation end products (RAGE) for signal transduction (Leclerc et al., 2007). In porcine alveolar macrophages infected with highly pathogenic porcine reproductive and respiratory syndrome virus or Haemophilus parasuis, the expression of S100A6 was significantly upregulated (Wang et al., 2012; Zhou et al., 2015). Similarly, we found the protein level of S100A6 was highly co-related to T. gondii infection efficiency that a higher level of S100A6 protein resulted in a higher infection rate, and a lower level of S100A6 resulted in a lower infection rate (Figures 4 and 5).
Because of the limited proteins coded by the small genome of the T. gondii, the host factors will be exploited for its successful parasitism, these factors are defined as host dependency factors (HDF) (Wu et al., 2020). By using large-scale human small interfering RNA screening to identify the host genes hexokinase 2 (HK2) gene is indispensable for T. gondii growth, T. gondii infection increases host HK2 transcription and the protein level, and the parasite growth is restored in cells transfected with HK2 expression plasmids at 3% oxygen condition (Menendez et al., 2015). Our previous research also found that guanosine triphosphatase (GTPase) family protein RhoA and Rac1 could regulate the host cell cytoskeleton reorganization to facilitate T. gondii invasion, cells with RhoA and Rac1 knockdown showed significantly decreased T. gondii infection rates (Na et al., 2013). We have previously reported a screening of host genes essential for T. gondii parasitism by using a genome-wide CRISPR-Cas9 technology, and S100A6 was screened as an HDF (Wu et al., 2020), we proved that S100A6 functioned as a HDF through binding with TgSAG1 to regulate the parasitic infection. However, how this HDF binds with TgSAG1, and then regulates the parasite infection is still vague.
S100A6 knockdown in NIH-3T3 fibroblastic cells causes a reorganization of the actin cytoskeleton with an extensive cortical network of actin filaments and tropomyosin structures, and an increase in the number of focal adhesions at the cell periphery (Breen and Tang, 2003). Being S100A6 binding protein, calcyclin binding protein and Siah-1 interacting protein (CacyBP/SIP) are identified to interact with tubulin, which plays an important role in microtubules rearrangement (Filipek et al., 2008). Vimentin, an interactor of S100A6 identified in our research, belongs to the most widely expressed and highly conserved type III Intermediate filament protein family. This multifunctional protein interacts with large a number of protein and makes it a potential regulator of several different physiological functions such as cell adhesion and migration (Nieminen et al., 2006). In addition, rearrangement of cytosolic vimentin was observed in Cercopithecus aethiops kidney epithelial cells (VERO) during infection of African swine fever virus (Stefanovic et al., 2005). A direct interaction between vimentin and severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein during viral entry was observed in VERO cells (Yu et al., 2016). Significant rearrangement of host cell vimentin was observed in T. gondii infected COS7 cells at 1 h post infection (He et al., 2017). TgSAG1 regulated the association between vimentin and S100A6 during T. gondii infection; therefore, we implied that T. gondii's regulation on the cytoskeleton reorganization partly attributed to TgSAG1 through S100A6 signaling pathway (Figure 6). Acute lymphoblastic leukemias (ALL) have a poor prognosis. This poor prognosis is mainly because of their resistance to TNF-α or upregulated expression of the immune escape factor S100A6 (Tamai et al., 2014). It has been found that TNF-α induces mixed-lineage leukemia (MLL)/AF4-positive acute lymphoblastic leukemia (ALL) cells apoptosis through inhibition of S100A6's function (Tamai et al., 2012). In addition, S100A6 can regulate the expression of TNF-α in THP-1 cells (Figure 8). Vimentin is a key metabolic and functional controller of Regulatory T cells (Tregs) activity. By phosphoproteomic screen, vimentin was considered as a PKCθ phosphorylation target, vimentin disruption augmented PKCθ′s suppressor function (McDonald-Hyman et al., 2018). Vimentin, as an NF-κB regulator, plays a detrimental role in the host defense against Meningitic Escherichia coli K1-induced pathogen invasion, polymorphonuclear neutrophil (PMN) recruitment, blood-brain barrier (BBB) permeability, and neuronal inflammation through modulating the NF-κB signaling pathway (Huang et al., 2016). We showed in our study, TgSAG1 could regulate the binding between vimentin and PKCθ, then regulate the phosphorylation of P65 at ser536, finally promote the expression of TNFα through S100A6 signaling (Figure 9).
Conclusions
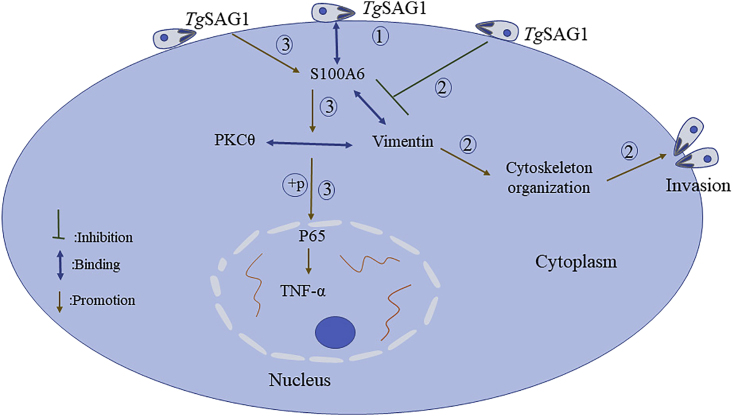
Our successful construction of the recombinant T. gondii RH strain (RH-SAG1-TurboID-HA) enabled us to screen for TgSAG1 interactome under physiological condition, and 119 biotinylated host proteins were identified. Host cell protein S100A6 co-localized with TgSAG1 during T. gondii attached to HFF cells. S100A6 was considered as a host dependent factor, which could promote T. gondii's attachment and invasion to host cells. TgSAG1 could regulate binding with vimentin and S100A6 then for cytoskeleton organization during T. gondii infection. Also, as an immunomodulatory factor, TgSAG1 could regulate the expression of TNFα through S100A6-vimentin/PKCθ-NF-κB signaling pathway (Figure 10).
Figure 10.
The mechanism for TgSAG1 regulating T. gondii invasion and host immunity
As a surface antigen, TgSAG1 could bind host protein S100A6 for attaching to and invading host cells. During T. gondii invasion, TgSAG1 could regulate the interaction between S100A6 and vimentin, then for cytoskeleton organization. In addition, as an immunomodulatory factor, TgSAG1 could regulate the expression of TNFα through S100A6-vimentin/PKCθ-NF-κB signaling pathway.
Limitations of the study
This study showed TgSAG1 could bind host cell protein S100A6 to regulate T. gondii invasion and host immunity. One limitation to this study is not investigating how TgSAG1 binds with S100A6 to regulate T. gondii infection. We have demonstrated that host cell protein ANXA6 co-localized with TgSAG1 during T. gondii attached to host cells, because ANXA6 is also binding partner of S100A6; therefore, further studies are needed to better explore the relationship between ANXA6 and TgSAG1 during T. gondii invasion.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Toxoplasma gondii antibody | Abcam | Cat#ab8313 |
| RRID: AB_306466 | ||
| Anti-DPP4 | Abcam | Cat#ab114033 |
| RRID: AB_10901695 | ||
| Anti-ITGβ1 | Abcam | Cat#ab30394 |
| RRID: AB_775726 | ||
| Anti-MYH9 | Abcam | Cat#ab24762 |
| RRID: AB_2282292 | ||
| Anti-CKAP4 | Thermo Fisher Scientific | Cat#PA5-100059 |
| RRID: AB_2815589 | ||
| Anti-ANXA6 | Thermo Fisher Scientific | Cat#720160 |
| RRID: AB_2609027 | ||
| Anti-vVimentin | Cell Signaling Technology | Cat#D21H3 |
| RRID: AB_10695459 | ||
| Anti-HA tag | Cell Signaling Technology | Cat#3724S |
| RRID: AB_1549585 | ||
| Anti-DDDK | Sigma Aldrich | Cat#F1804 |
| RRID: AB_262044 | ||
| Anti-Actin | Abcam | Cat#ab179467 |
| RRID: AB_2737344 | ||
| Anti-S100A6 | Thermo Fisher Scientific | Cat#PA5-97153 |
| RRID: AB_2808955 | ||
| Anti-NF-κB | Cell Signaling Technology | Cat#8242 |
| RRID: AB_10859369 | ||
| Anti-p NF-κB | Cell Signaling Technology | Cat#3033S |
| RRID: AB_331284 | ||
| Anti-PKCθ | Abcam | Cat#ab76197 |
| RRID: AB_1310589 | ||
| Goat anti-mouse IgG (H+L) secondary antibody, HRP | Abclonal | Cat#AS003 |
| RRID: AB_2769851 | ||
| Goat anti-rabbit IgG (H+L) secondary antibody, HRP | Abclonal | Cat#AS014 |
| RRID: AB_2769854 | ||
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A11001 |
| RRID: AB_2534069 | ||
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A11032 |
| RRID: AB_2534091 | ||
| Donkey anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A32731 |
| RRID: AB_2633280 | ||
| Donkey anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 |
Thermo Fisher Scientific |
Cat#A32740 |
| RRID: AB_2762824 | ||
| Chemicals, peptides, and recombinant proteins | ||
| Sotrastaurin | Selleck | S2791 |
| Lipofectamine™ 3000 Transfection Reagent | Thermo Fisher Scientific | L3000 |
| D-Biotin | Thermo Fisher Scientific | B1595 |
| eBioscience™ Phalloidin eFluor™ 570 | Thermo Fisher Scientific | 41-6559-05 |
| Critical commercial assays | ||
| Human TNF-α kit | MULTI SCIENCES | EK182 |
| Mouse TNF-α kit | MULTI SCIENCES | EK282/3 |
| Experimental models: Cell lines | ||
| THP-1 cells | ATCC | TIB-202 |
| COS-7 cells | ATCC | CRL-1651 |
| HFF cells | ATCC | SCRC-1041 |
| Experimental models: Organisms/strains | ||
| RH-SAG1-TurboID-HA | This paper | N/A |
| RH-Δsag1 | This paper | N/A |
| Oligonucleotides | ||
| Primer for qPCR was shown in Table S1 | This paper | N/A |
| Software and algorithms | ||
| Prism | Graph Pad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | Java | https://imagej.en.softonic.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hong-Juan Peng (floriapeng@hotmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
The human foreskin fibroblast cell line (HFF), mouse monocyte/macrophage cell line RAW264.7 and the Cercopithecus aethiops kidney fibroblast cell line COS7 were obtained from American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagles Medium (DMEM) (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, USA). The human monocyte/macrophage cell line THP-1 (ATCC) were propagated in Roswell Park Memorial Institute 1640 medium (RPMI 1640) (Thermo Fisher Scientific, USA) supplemented with 10% (v/v) FBS, 1% Penicillin-Streptomycin (Thermo Fisher Scientific, USA) and 10 mg/ml gentamicin (Thermo Fisher Scientific, USA) at 37°C and 5% CO2 (Park et al., 2007). All the recombinant T. gondii strains used in this research were derived from the type I RH wild type (RH-WT) strain. Transgenic parasites were obtained by electroporation of recombinant plasmids into RH-WT tachyzoites and selection with 3 μM pyrimethamine (Sigma-Aldrich, USA). All T. gondii strains were maintained by growth in HFF cells. Freshly egressed parasites purified with 3 μm nucleopore filters were used in all experiments.
Method details
Construction of the recombinant T. gondii RH-SAG1-TurboID-HA strain
The recombinant RH strain expressing SAG1-TurboID-HA was constructed with random insertion technique following the processes reported (Striepen et al., 1998). Genomic DNA of RH-WT was extracted using DNeasy Blood & Tissue Kit (Qiagen, Germany). The coding sequence of the SAG1 was amplified with the T. gondii genomic DNA as the template (as there is no insert in sag1 gene), and the TurboID-HA coding sequence was inserted before the SAG1 stop codon. The pyrimethamine resistant gene, dihydrofolate reductase (DHFR) reading frame was inserted downstream of the coding sequence of SAG1-TurboID-HA. The construction of the recombinant plasmid containing the SAG1-TurboID-HA and DHFR reading frames was shown in Figure S1. The recombinant plasmid was electroporated into RH-WT tachyzoites, and the recombinant parasites were screened by culturing in HFFs with the DMEM medium containing 3 μM pyrimethamine (Sigma, Germany). The cloning stable lines of the recombinant parasites were obtained through limited dilution and culturing, and then confirmed by WB for the expression of SAG1-TurboID-HA.
Immunofluorescence assay (IFA)
HFF cells were grown on coverslips in 12-well plates (Corning, China) to 100% confluence and infected with T. gondii for 30 min or 24 hrs, followed by washing with phosphate-buffered saline (PBS) for 3 times to get rid of the unrecruited tachyzoites. The parasite-infected monolayers were fixed with 4% formaldehyde for 15 min at room temperature (RT), and then permeabilized with 100 μM Digitonin (Abcam, England) for 20 min at RT (for invasion efficiency assessment, permeabilization was unnecessary), and blocked in 10% Bovine Serum Albumin (BSA, Thermo Fisher Scientific, USA) for 1 hr at RT. The coverslips were incubated with the primary antibody diluted in 10% BSA overnight at 4°C with gentle shaking, then the coverslips were washed for 3 times with PBS at RT followed by incubation with the secondary antibodies diluted in 10% BSA at RT for 1 hr. After that, coverslips were taken out and rinsed with ddH2O, air-dried, and then mounted with DAPI Fluoromount® (Southern Biotech, USA). The coverslips were visualized and images were captured using an Olympus FLUOVIEW FV1000 confocal laser scanning microscope or with a fluorescence microscope (ECLIPSE Ni, NA = 1.4, Nikon, Japan) at 10×100 magnification.
Affinity purification of the biotinylated proteins
HFF monolayers infected with RH-SAG1-TurboID-HA strain were cultured in DMEM containing 150 μM D-Biotin (Sigma-Aldrich, USA) for 24 hrs was considered as experimental group. While the infected HFF group with no D-Biotin supplementation was regarded as the control group. Then, the infected cells were washed with PBS, and lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 0.2% SDS, 0.5% sodium deoxycholate (DOC), 1% NP-40, 1% Triton X-100) supplemented with complete protease inhibitor cocktail (Roche, Switzerland). The cell lysates were sonicated with a microtip in Sonic Dismembrator JY92-Ⅱ (Ningbo Scieniz Biotechnology Co. Ltd) using a setting at 5 sec pulse, 7 sec interval time, and 2 min total pulse time. The cell lysates were sat on ice for 10 min, then transferred to 2 mL Eppendorf tubes, and centrifuge at 20,000 × g, 4°C for 20 min. The supernatants were transferred to fresh tubes. Next, 50 μL Pierce™ Streptavidin Magnetic Beads (Thermo Fisher Scientific, USA) were added to each supernatant samples, and incubated overnight in a 4°C shaker. Beads were collected and washed twice in RIPA buffer, followed by washing once in buffer 1 (20 mM Tris [pH 7.5], 2% SDS), twice in buffer 2 (0.1% DOC, 1% Triton X-100, 500 mM NaCl [pH 7.5]), once in buffer 3 (0.5% NP-40, 0.5% DOC, 1 mM EDTA, 10 mM Tris [pH 8.1]), twice in buffer 4 (50 mM Tris [pH 7.4], 50 mM NaCl), and twice in PBS. Finally, the beads were resuspended in PBS, and 10% of each suspension sample was transferred to mix with 6×SDS- polyacrylamide gel (SDS-PAGE) loading buffer and subjected to WB with the streptavidin-HRP (Abcam, England) probe. The remaining suspension samples were used for protein analysis with mass spectrometry (Long et al., 2018).
Mass spectrometry and bioinformatics analysis for the biotinylated proteins
The purified proteins bound to streptavidin beads were reduced, alkylated, and digested by sequential addition of trypsin protease. The dried peptide samples were reconstituted with mobile phase A (2% ACN, 0.1% FA), centrifuged at 20,000 × g for 10 min, and the supernatant was collected for following analysis. Separation was performed by Thermo UltiMate 3000 U HPLC. The peptides separated by liquid phase chromatography were ionized by a nanoESI source and then passed to a tandem mass spectrometer Q-Exactive HF (Thermo Fisher Scientific, USA) for data-dependent acquisition (DDA) mode detection. The experimental MS/MS data were aligned with the theoretical MS/MS data from database for protein identification. The databases used in our research included UniProtKB/Swiss-Prot and reference sequence (RefSeq) of NCBI. The whole process starts from converting raw MS data into a peak list and then searching matches in the database. The search results were subjected to strict filtering with quality control. Finally, for the proteins with high expectation values, functional annotation analysis such as Gene Ontology (GO) enrichment (http://www.geneontology.org/), and protein-protein interaction analysis (STRING 11.0, https://www.string-db.org) was performed.
Fluorescence resonance energy transfer (FRET) assay
COS7 cells were cultured on coverslips in a 12-well plate to 70% confluence. The cells were co-transfected with peCFPN1-SAG1 and peYFPC1-S100A6 for the experimental group, peCFP-N1 and peYFP-C1 for the negative control group, and transfected with peYFP-CFP for the positive control. Cells were fixed in 4% paraformaldehyde at 37°C for 30 min at 48 hrs post-transfection, and then washed twice with PBS. The FRET efficiency of the different transfection groups was measured under a confocal laser scanning microscope (FLUOVIEW FV1000; Olympus, Tokyo, Japan) (Elangovan et al., 2003). Ten fields were evaluated and three repetitive experiments were performed independently for statistical analysis.
Co-immunoprecipitation (Co-IP)
COS7 cells were grown in T25 flasks to 70% confluence, and then transfected with pcDNA3.1(+)-S100A6-HA individually or together with pcDNA3.1(+)-SAG1-3×Flag for 48 hrs. The cells were washed for 3 times with PBS and lysed with cell lysis buffer (Beyotime, China) containing 1 mM phenylmethanesulfonyl fluoride (Dingguo Changsheng Biotechnology, China). Cell lysates were centrifuged at 14,000×g, 4°C, for 10 min. The supernatants were collected and incubated with Anti-FLAG® M2 antibody (Sigma Aldrich, USA) for 2 hrs at 4°C with gentle rotation. Then, 20 μL protein A-agarose (Santa Cruz Biotechnology, USA) was added to the mix and incubated overnight at 4°C with gentle rotation. The beads were collected by centrifugation at 500×g for 5 min at 4°C and then resuspended in 6×SDS-PAGE sample buffer. The samples were boiled, loaded onto the gels for SDS PAGE and then analyzed by WB.
Double-antibody sandwich enzyme-linked immunosorbent assay (ELISA)
The cytokine TNF-α secreted by THP-1 or RAW264.7 cells were measured with Double-antibody sandwich ELISA kits (Multisciences, Lianke, Biotech,Co., Ltd, China) following the manufactures’ instruction. Briefly, the supernatant of the cell culture was collected and centrifuged briefly at 10,000×g, 5 min to remove the cell debris. 50 μL of the supernatant was added to the well precoated with TNF-α antibody, followed by addition of 100 μL of the detection antibody to the well. The well was sealed and incubated for 2 hrs with 300 rpm rotation at RT. After that, the well was washed with PBS for 3 times, and incubated with 100 μL Streptavidin-HRP for 45 min at RT. Finally, the optical density (OD) of the well was detected by microplate reader (Tiangen Biotech, Beijing, Co.Ltd, China) at 450 nm absorption wavelength. The results were presented as pg/mL.
Reverse transcription quantitative PCR (RT-qPCR)
The transcription levels of TNF-α, IL-12, Interleukin-10 (IL-10), and IFN-γ, and the knockdown efficiency of S100A6 and vimentin in THP-1 cells were validated by RT-qPCR. The total RNAs was extracted with TRIzol™ Reagent (Thermo Fisher Scientific, USA) following its protocol. Then, the total RNAs was reverse-transcribed to single stranded cDNAs using the TransScript One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen, China). SYBR Green Real-Time PCR Master Mixes (Thermo Fisher Scientific, USA) was used to perform RT-qPCR on a Real-Time PCR cycler (Thermo Fisher Scientific QuantStudio™, USA). Forward (F) and reverse (R) primers used to amplify the selected genes were listed in Table S1. The amplification reactions were performed using the following conditions: 95°C for 2 min followed by 40 cycles of 95°C for 10 s, 56°C for 30 s and 72°C for 30 s. Melting curve analysis was performed using the following conditions: 95°C for 15 s, 60°C for 1 min, 95°C for 15 s. The cycle number of threshold (CT) value was normalized to the internal control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or Actin value. The fold change in the relative gene transcription was calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001), and the calibrator sample was the uninfected or non-transfected N group.
Western blotting
Protein samples were boiled in 6× SDS PAGE loading buffer for 10 min. The samples were loaded to 15% SDS PAGE gels for protein separation, and the protein were transferred to polyvinylidene fluoride (PVDF) membrane. The membrane was blocked in TBST (20 mM Tris-HCl buffer, 5% bovine serum albumin (BSA), 0.1% Tween-20, pH 7.4) for 2 hrs at 37°C with gentle shaking, and then probed with the primary antibody at 4°C overnight. The membrane was washed with TBST for 3 times, followed by incubation with the HRP conjugated secondary antibody for 2 hrs at 37°C with gentle shaking. Finally, the PVDF membranes were visualized by adding the enhanced chemiluminescence (ECL) reaction buffer (Bio-Rad, USA), and imaged with a ChemDocTM Touch Imaging System (Bio-Rad, USA) as recommended by the manufacturer.
T. gondii invasion efficiency assay
HFF and COS7 cells were grown on coverslip in a 12-well plate to about 70% confluence. The HFF cells were then transfected with S100A6 siRNA as shown in Table S1, and COS7 cells were transfected with pcDNA3.1(+)-S100A6 plasmid for 24 hrs. The cells were then infected with T. gondii RH-WT tachyzoites at a multiplicity of infection (MOI) of 10 for 30 min at 37°C, 5% CO2. The cells were then washed with PBS for three times to get rid of the unrecruited tachyzoites, then fixed with 4% paraformaldehyde. The extracellular parasites were detected by staining with mouse anti-SAG1 antibody followed by goat anti-mouse IgG (H+L) conjugated with Alexa Fluor 488. The intracellular and extracellular parasites were detected after cell permeabilization and staining with mouse anti-SAG1 antibody followed by goat anti-mouse IgG (H+L) conjugated with Alexa Fluor 594. Both intracellular (red) and extracellular (yellow) parasites were observed and imaged under a fluorescence microscope (Eclipse Ni, NA = 1.4, Nikon, Japan) at 10 × 100 magnification. The parasites with different fluorescence color in five views from the up, down, left, right and center of the coverslips were counted. Values were represented as “means ± SD” of the number of parasite/100 host cell nuclei from three independent experiments.
Quantification and statistical analysis
Statistical analysis
All experiments were performed in triplicate. Data were represented as means ± SD, unless otherwise indicated. SPSS 20.0 software was used for statistical analyses. Differences between groups were analyzed by using t-test. Values of p < 0.05 were considered statistically significant.
Acknowledgments
The authors are grateful to the participants in this study and the anonymous reviewers and editors for their comments and valuable inputs. This research was supported by National Natural Science Foundation of China (81971954), Science and Technology Planning Project of Guangdong Province (2018A050506038), Key project of Guangzhou science research (201904020011), and the Basic Research Project of Key Laboratory of Guangzhou (202102100001) to HJP.
Author contributions
L.J. Z. performed the experiments, analyzed the data, and wrote the manuscript. J.P generated RH-SAG1-TurboID-HA, RH-Δsag1 T. gondii. M.C., W.H.Z and L.J.Y performed FRET. C.Y.H. suggested study and manuscript revision. H.J.P. performed study conception and designing, supervision of the research group, funding support, and drafting the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103514.
Supplemental information
Data and code availability
The paper does not report original code
Data and any additional information reported in this paper will be shared by the lead contact upon request.
References
- Boothroyd J.C., Hehl A., Knoll L.J., Manger I.D. The surface of Toxoplasma: more and less. Int. J. Parasitol. 1998;28:3–9. doi: 10.1016/s0020-7519(97)00182-3. [DOI] [PubMed] [Google Scholar]
- Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen E.C., Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J. Cell. Biochem. 2003;88:848–854. doi: 10.1002/jcb.10398. [DOI] [PubMed] [Google Scholar]
- Cmoch A., Strzelecka-Kiliszek A., Palczewska M., Groves P., Pikula S. Matrix vesicles isolated from mineralization-competent Saos-2 cells are selectively enriched with annexins and S100 proteins. Biochem. Biophys. Res. Commun. 2011;412:683–687. doi: 10.1016/j.bbrc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Cui Y., Jiang L., Yu R., Shao Y., Mei L., Tao Y. β-carboline alkaloids attenuate bleomycin induced pulmonary fibrosis in mice through inhibiting NF-kb/p65 phosphorylation and epithelial-mesenchymal transition. J. Ethnopharmacol. 2019;243:112096. doi: 10.1016/j.jep.2019.112096. [DOI] [PubMed] [Google Scholar]
- Donato R., Sorci G., Giambanco I. S100A6 protein: functional roles. Cell. Mol. Life Sci. 2017;74:2749–2760. doi: 10.1007/s00018-017-2526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey J.P., Frenkel J.K. Feline toxoplasmosis from acutely infected mice and the development of toxoplasma cysts. J. Protozool. 1972;23:537–546. doi: 10.1111/j.1550-7408.1976.tb03836.x. [DOI] [PubMed] [Google Scholar]
- Dubremetz J.F., Garcia-Réguet N., Conseil V., Fourmaux M.N. Apical organelles and host-cell invasion by Apicomplexa. Int. J. Parasitol. 1998;28:1007–1013. doi: 10.1016/s0020-7519(98)00076-9. [DOI] [PubMed] [Google Scholar]
- Elangovan M., Wallrabe H., Chen Y., Day R.N., Barroso M. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29:58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- Filipek A., Schneider G., Mietelska A., Figiel I., Niewiadomska G. Age-dependent changes in neuronal distribution of CacyBP/SIP: comparison to tubulin and the tau protein. J. Neural. Transm. 2008;115:1257–1264. doi: 10.1007/s00702-008-0062-3. [DOI] [PubMed] [Google Scholar]
- Grimwood J., Smith J.E. Toxoplasma gondii: the role of parasite surface and secreted proteins in host cell invasion. Int. J. Parasitol. 1996;26:169–173. doi: 10.1016/0020-7519(95)00103-4. [DOI] [PubMed] [Google Scholar]
- Guo K., Xu L., Wu M., Hou Y., Jiang Y., Lv J., Xu P., Fan Z., Zhang R., Xing F., et al. A host factor GPNMB restricts porcine circovirus type 2 (PCV2) replication and interacts with PCV2 ORF5 protein. Front. Microbiol. 2019;9:3295. doi: 10.3389/fmicb.2018.03295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Kong L., Zhou L., Xia J., Wei H., Liu M., Peng H. Host cell vimentin restrains Toxoplasma gondii invasion and phosphorylation of vimentin is partially regulated by interaction with TgROP18. Int. J. Biol. Sci. 2017;13:1126–1137. doi: 10.7150/ijbs.21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou G., Xue B., Li L., Nan Y., Zhang L., Li K., Zhao Q., Hiscox J.A., Stewart J.P., Wu C., et al. Direct interaction between CD163 N-terminal domain and MYH9 C-terminal domain contributes to porcine reproductive and respiratory syndrome virus internalization by permissive cells. Front. Microbiol. 2019;10:1815. doi: 10.3389/fmicb.2019.01815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chi F., Peng L., Bo T., Zhang B., Liu L., Wu X., Mor-Vaknin N., Markovitz D.M., Zhou Y.-H. Vimentin, a Novel NF-κB regulator, is required for meningitic Escherichia coli K1-induced pathogen invasion and PMN transmigration across the blood-brain barrier. PLoS One. 2016;11:e162641. doi: 10.1371/journal.pone.0162641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M., Lau Y. Measurement of binding strength between prey proteins interacting with Toxoplasma gondii SAG1 and SAG2 using isothermal titration calorimetry (ITC) Acta Parasitol. 2018;63:106–113. doi: 10.1515/ap-2018-0012. [DOI] [PubMed] [Google Scholar]
- Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., Ting A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc E., Fritz G., Weibel M., Heizmann C.W., Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J. Biol. Chem. 2007;282:31317–31331. doi: 10.1074/jbc.M703951200. [DOI] [PubMed] [Google Scholar]
- Lekutis C., Ferguson D.J.P., Grigg M.E., Camps M., Boothroyd J.C. Surface antigens of Toxoplasma gondii: variations on a theme. Int. J. Parasitol. 2001;31:1285–1292. doi: 10.1016/s0020-7519(01)00261-2. [DOI] [PubMed] [Google Scholar]
- Li L., Li X., Yan J. Alterations of concentrations of calcium and arachidonic acid and agglutinations of microfilaments in host cells during Toxoplasma gondii invasion. Vet. Parasitol. 2008;157:21–33. doi: 10.1016/j.vetpar.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long S., Brown K., Sibley L. CRISPR-mediated tagging with BirA allows proximity labeling in Toxoplasma gondii. Bio Protoc. 2018;8:e2768. doi: 10.21769/BioProtoc.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft B.J., Remington J.S. Toxoplasmic encephalitis in AIDS. Clin. Infect. Dis. 2002;15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- Ma H., Kien F., Maniere M., Zhang Y., Lagarde N., Tse K.S., Poon L.L.M., Nal B. Human annexin A6 interacts with influenza A virus protein M2 and negatively modulates infection. J. Virol. 2012;86:1789–1801. doi: 10.1128/JVI.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manger I.D., Hehl A.B., Boothroyd J.C. The surface of toxoplasma tachyzoites is dominated by a family of glycosylphosphatidylinositol-anchored antigens related to SAG1. Infect. Immun. 1998;66:2237–2244. doi: 10.1128/IAI.66.5.2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., Ting A.Y. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-Hyman C., Muller J.T., Loschi M., Thangavelu G., Saha A., Kumari S., Reichenbach D.K., Smith M.J., Zhang G., Koehn B.H. The vimentin intermediate filament network restrains regulatory T cell suppression of graft-versus-host disease. J. Clin. Invest. 2018;128:4604–4621. doi: 10.1172/JCI95713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez M.T., Teygong C., Wade K., Florimond C., Blader I.J. siRNA screening identifies the host hexokinase 2 (HK2) gene as an important hypoxia-inducible transcription factor 1 (HIF-1) target gene in Toxoplasma gondii-infected cells. mBio. 2015;6:e462. doi: 10.1128/mBio.00462-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo K.L., JR. Attachment of Toxoplasma gondii to host cells involves major surface protein, SAG-1 (P30) Exp. Parasitol. 1994;79:11–20. doi: 10.1006/expr.1994.1054. [DOI] [PubMed] [Google Scholar]
- Mineo J.R., McLeod R., Mack D., Smith J., Khan I.A., Ely K.H., Kasper L.H. Antibodies to Toxoplasma gondii major surface protein (SAG-1, P30) inhibit infection of host cells and are produced in murine intestine after peroral infection. J. Immunol. 1993;150:3951–3964. [PubMed] [Google Scholar]
- Montoya J.G., Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- Na R., Zhu G., Luo J., Meng X., Cui L., Peng H., Chen X., Gomez-Cambronero J. Enzymatically active Rho and Rac small-GTPases are involved in the establishment of the vacuolar membrane after Toxoplasma gondii invasion of host cells. BMC. Microbiol. 2013;13:125. doi: 10.1186/1471-2180-13-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen M., Henttinen T., Merinen M., Marttila-Ichihara F., Eriksson J.E., Jalkanen S. Vimentin function in lymphocyte adhesion and transcellular migration. Nat. Cell. Biol. 2006;8:156–162. doi: 10.1038/ncb1355. [DOI] [PubMed] [Google Scholar]
- Pagheh A.S., Sarvi S., Sharif M., Rezaei F., Ahmadpour E., Dodangeh S., Omidian Z., Hassannia H., Mehrzadi S., Daryani A. Toxoplasma gondii surface antigen 1 (SAG1) as a potential candidate to develop vaccine against toxoplasmosis: a systematic review. Comp. Immunol. Microbiol. Infect. Dis. 2020;69:101414. doi: 10.1016/j.cimid.2020.101414. [DOI] [PubMed] [Google Scholar]
- Park E.K., Jung H.S., Yang H.I., Yoo M.C., Kim C., Kim K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 2007;56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- Patel H., Ashton N.J., Dobson R.J.B., Andersson L., Yilmaz A., Blennow K., Gisslen M., Zetterberg H. Proteomic blood profiling in mild, severe and critical COVID-19 patients. Sci. Rep. 2021;11:6357. doi: 10.1038/s41598-021-85877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially- restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala-Dempsey A.C., Rezvanpour A., Shaw G.S. S100-annexin complexes--structural insights. FEBS. J. 2008;275:4956–4966. doi: 10.1111/j.1742-4658.2008.06654.x. [DOI] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Burke B., May D.G. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 2013;74 doi: 10.1002/0471140864.ps1923s74. [DOI] [PubMed] [Google Scholar]
- Shuai L., Wang J., Zhao D., Wen Z., Ge J., He X., Wang X., Bu Z. Integrin β1 promotes peripheral entry by Rabies virus. J. Virol. 2020;94:e1819. doi: 10.1128/JVI.01819-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Windsor M., Nagata K., Inagaki M., Wileman T. Vimentin rearrangement during african swine fever virus infection involves retrograde transport along microtubules and phosphorylation of vimentin by calcium calmodulin kinase II. J. Virol. 2005;79:11766–11775. doi: 10.1128/JVI.79.18.11766-11775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B., Yingxin He.C., Matrajt M., Soldati D., Roos D.S. Expression, selection, and organellar targeting of the green fluorescent protein in Toxoplasma gondii. Mol. Biochem. Parasitol. 1998;92:325–338. doi: 10.1016/s0166-6851(98)00011-5. [DOI] [PubMed] [Google Scholar]
- Sun L., Li K., Liu G., Xu Y., Zhang A., Lin D., Zhang H., Zhao X., Jin B., Li N., Zhang Y. Distinctive pattern of AHNAK methylation level in peripheral blood mononuclear cells and the association with HBV-related liver diseases. Cancer Med. 2018;7:5178–5186. doi: 10.1002/cam4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai H., Miyake K., Yamaguchi H., Takatori M., Dan K., Inokuchi K., Shimada T. AAV8 vector expressing IL24 efficiently suppresses tumor growth mediated by specific mechanisms in MLL/AF4-positive ALL model mice. Blood. 2012;119:64–71. doi: 10.1182/blood-2011-05-354050. [DOI] [PubMed] [Google Scholar]
- Tamai H., Miyake K., Yamaguchi H., Shimada T., Dan K., Inokuchi K., Expand A. Inhibition of S100A6 induces GVL effects in MLL/AF4-positive ALL in human PBMC-SCID mice. Bone. Marrow Transplant. 2014;49:699–703. doi: 10.1038/bmt.2014.18. [DOI] [PubMed] [Google Scholar]
- Tani J., Shimamoto S., Mori K., Kato N., Moriishi K., Matsuura Y., Tokumitsu H., Tsuchiya M., Fujimoto T., Kato K., et al. Ca(2+)/S100 proteins regulate HCV virus NS5A-FKBP8/FKBP38 interaction and HCV virus RNA replication. Liver Int. 2013;33:1008–1018. doi: 10.1111/liv.12151. [DOI] [PubMed] [Google Scholar]
- Tomavo S., Schwarz R.T., Dubremetz J.F. Evidence for glycosyl-phosphatidylinositol anchoring of Toxoplasma gondii major surface antigens. Mol. Cell. Biol. 1989;9:4576–4580. doi: 10.1128/mcb.9.10.4576-4580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A., Gou L., Lau Q.C., Chen B., Zhao X., Li J., Tang H., Chen L., Tang M., Huang C., et al. Proteomic profiling identifies aberrant epigenetic modifications induced by hepatitis B virus X protein. J. Proteome. Res. 2009;8:1037–1046. doi: 10.1021/pr8008622. [DOI] [PubMed] [Google Scholar]
- Uezu A., Kanak D.J., Bradshaw T.W.A., Soderblom E.J., Catavero C.M., Burette A.C., Weinberg R.J., Soderling S.H. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353:1123–1129. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu C., Fang Y., Liu X., Li W., Liu S., Liu Y., Liu Y., Charreyre C., Audonnet J., et al. Transcription analysis on response of porcine alveolar macrophages to Haemophilus parasuis. BMC Genomics. 2012;13:68. doi: 10.1186/1471-2164-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wei H., Jiang D., Li S., Zou W., Peng H. Genome-wide CRISPR screen identifies host factors required by Toxoplasma gondii infection. Front. Cell. Infect. Microbiol. 2020;9:460. doi: 10.3389/fcimb.2019.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.T., Chien S., Chen I., Lai C., Tsay Y., Chang S.C., Chang M. Surface vimentin is critical for the cell entry of SARS-CoV. J. Biomed. Sci. 2016;23:14. doi: 10.1186/s12929-016-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Ke Y., Zhang L. Interplay between hepatitis C virus and ARF4. Virol. Sin. 2017;32:533–536. doi: 10.1007/s12250-017-4000-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang P., Michal J.J., Wang Y., Zhao J., Jiang Z., Liu B. Molecular characterization of the porcine S100A6 gene and analysis of its expression in pigs infected with highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) J. Appl. Genet. 2015;56:355–363. doi: 10.1007/s13353-014-0260-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The paper does not report original code
Data and any additional information reported in this paper will be shared by the lead contact upon request.