Abstract
The term “proteoforms” describes the range of different structures of a protein product of a single gene, including variations in amino acid sequence and post-translational modifications. This diversity in protein structure contributes to the biological complexity observed in living organisms. As the concentration of a particular proteoform may increase or decrease in abnormal physiological states, proteoforms have long been used in medicine as biomarkers of health and disease. Notably, the analytical approaches used to analyze proteoforms have evolved considerably over the years. While ligand binding methods continue to play a large role in proteoform measurement in the clinical laboratory, unanticipated or unknown post-translational modifications and sequence variants can upend even extensively tested and vetted assays that have successfully made it through the medical regulatory process. As an alternate approach, mass spectrometry—with its high molecular selectivity—has become an essential tool in detection, characterization, and quantification of proteoforms in biological fluids and tissues. This review explores the analytical techniques used for proteoform detection and quantification, with an emphasis on mass spectrometry and its various applications in clinical research and patient care including, revealing new biomarker targets, helping improve the design of contemporary ligand binding in vitro diagnostics, and as mass spectrometric laboratory developed tests used in routine patient care.
Keywords: Biomarkers, Protein isoforms, Proteoforms, Mass spectrometry, Post-translational modifications
1. Introduction
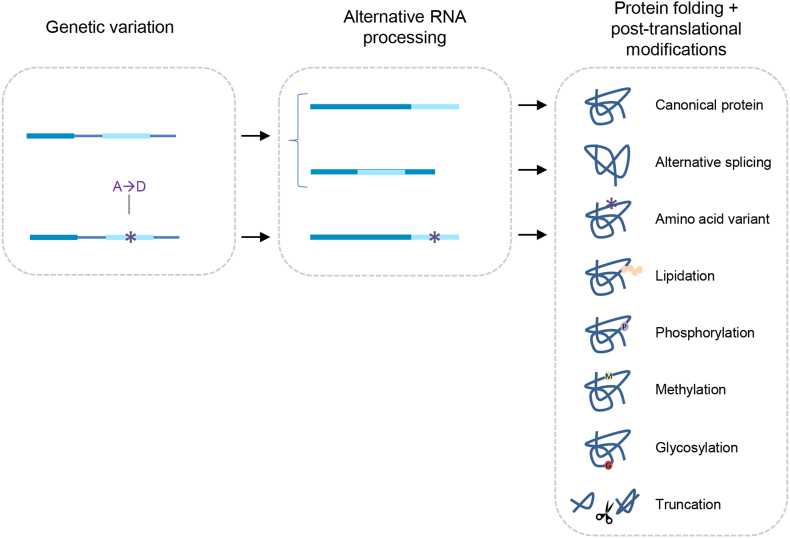
Variation in protein structure based on amino acid sequence and modifications allows for the great biological complexity observed in living organisms; a complexity that far exceeds that of the approximately 20,000 genes in the human genome that encode for proteins [1,2]. The term “proteoforms” is used to encapsulate “all of the different molecular forms in which the protein product of a single gene can be found” [3]. This term, proteoform, is now used to in place of previously used terms like “protein forms”, “protein isoforms”, “protein species” and “protein variants”. The range of proteoforms that can arise from a single protein-coding gene includes genetic variation, alternative splicing of RNA and the addition of post-translational modifications (PTMs) to a canonical protein sequence (Fig. 1) [3]. PTMs add to the diversity of the proteome through the covalent attachment of chemical functional groups, carbohydrates, lipids, peptides, or proteins, and via proteolytic cleavage. PTMs like phosphorylation play an important role in regulating cellular processes like apoptosis and cell growth [4]; ubiquitination tags proteins for enzymatic degradation [5]; glycosylation influences protein folding, conformation and activity [6]; methylation can alter DNA transcription [7]; lipidation affects protein distribution by tethering proteins to cellular membranes [8]; and proteolytic cleavage can activate a protein through processing of a precursor protein or remove a protein through proteolytic degradation [9]. Thus, these modifications can influence both physiological and pathological processes.
Fig. 1.
Proteoforms are generated through three main processes—genetic variation, alternative splicing of RNA, and protein folding and PTMs—to yield a variety of protein structures relative to the canonical protein structure (i.e., the “wild-type” protein).
Although the term proteoform is relatively new, analysis of proteoforms—as biochemical indicators of health or disease—has a longstanding history in laboratory medicine. For example, in 1958, five minor components of hemoglobin A (i.e., hemoglobin A1a-e) were identified via ion-exchange chromatography [10]. Subsequently, in 1968, the structure of hemoglobin A1c was elucidated by glycoprotein synthesis and mass spectrometric analysis, demonstrating hemoglobin A1c to be a glycated proteoform [11]. In 1969, using a combination of agar gel electrophoresis and chromatography, the abnormal band that was found on hemoglobin gels from individuals with diabetes mellitus was identified as hemoglobin A1c [12]. It is now well understood that under conditions of high blood glucose, hemoglobin can undergo non-enzymatic condensation reactions with glucose, including covalent attachment of glucose to the N-terminal valine of the hemoglobin β-chain to form hemoglobin A1c. Identification and characterization of this post-translationally modified proteoform of hemoglobin yielded a new biomarker and analytical approaches, still in use today, for diagnosis and monitoring of diabetes.
Herein, we will review mass spectrometry-related instrumentation, assay design and applications for proteoform analysis in clinical research and patient care including:
-
•
Analytical approaches to proteoform analysis;
-
•
Genotyping and phenotyping;
-
•
Proteolytic processing and degradation;
-
•
Phosphorylation, glycosylation, and glycation; and,
-
•
Endogenous v. exogenous proteoforms.
2. Analytical approaches to proteoform analysis
2.1. Immunoassays
Analytical approaches commonly used in a clinical setting for characterization and quantification of proteoforms have greatly evolved over the years. Historical and contemporary approaches to protein biomarker characterization and quantitation, include two-site immunoassays, nephelometry, immunohistochemistry, chromatography (e.g., gel electrophoresis and liquid chromatography), absorbance detection, and fluorescence in situ hybridization, to name a few. These techniques enabled a specific proteoform or a subset of proteoforms to be identified via strategies such as antibody recognition of the structural change or shift in retention time or electrophoretic migration of the proteoform.
Of the various protein quantitation approaches, the simple two-site sandwich immunoassay is among the most commonly used by clinical laboratories. This approach is generally suitable for use on existing automated analyzers, enables on-demand (v. batch) testing, and can be designed for modest turnaround times. While immunoassays designed to target a specific proteoform can provide robust detection and quantification of biomarkers if designed carefully, the presence of multiple proteoforms can confound immunometric detection as a result of variable cross-reactivity, and positive and negative interference (Fig. 2). For example, immunoassays quantifying thyroid stimulating hormone (TSH), a protein biomarker used to identify and monitor thyroid disorders, have been shown to yield falsely low results in the presence of sequence polymorphisms. After identifying a single case of a clinically euthyroid individual of South Asian descent with a TSH polymorphism causing discordant results on various regulatory-approved TSH immunoassays, a retrospective study examined ∼ 2 million individuals with repeatedly low TSH. From this analysis, twenty individuals of South Asian decent were found with falsely low TSH results due to the same polymorphism as the index case [13]. Samples from these individuals were then tested on 8 different regulatory-approved TSH immunoassays, revealing half of the assays reported (falsely) undetectable TSH. Most of these individuals were originally misdiagnosed with hyperthyroidism. It is suspected that the sequence variant alters the binding ability of either the capture or detection monoclonal antibodies used in the two-site immunoassays, impeding the formation of the ‘sandwich’ and thus inaccurately quantifying the concentration in the sample [13].
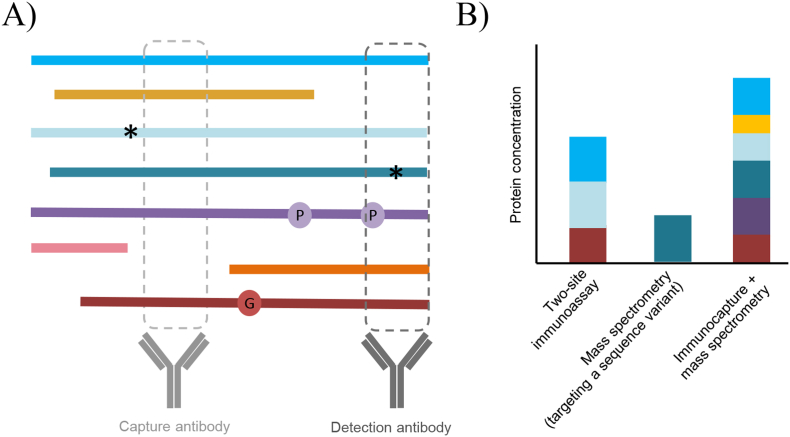
Fig. 2.
Immunometric versus mass spectrometric approaches often yield detection of different proteoforms from the same protein. (A) Two-site (or sandwich) immunoassays use antibodies to detect two short sequences of the target protein; however, proteoforms that share these epitopes cannot be discriminated and those that contain variant sequences like phosphorylation (denoted “P”) or polymorphisms (denoted “*”) may abrogate antibody bindings. Alternatively using mass spectrometry, specific variants can be selected for detection using a combination of chromatographic separation and mass detection, or an immunoprecipitation step can be added during sample preparation to pull down select proteoforms. In this example, the capture antibody is used for immunoprecipitation, and a common proteolytic peptide is targeted by mass spectrometry. (B) The consequence of detection of different epitopes and proteolytic peptides, leads to the detection of different combinations of proteoforms and differences in the total protein concentration measured.
In another example of a proteoform interfering with an immunoassay, a sequence variant in the viral surface antigen protein of the hepatitis B virus (HBV) was found to confound HBV diagnostics [14]. In this case, serum from a dialysis patient was tested via immunoassay for the HBV surface antigen, which yielded a negative result. As a result of the negative HBV test, the usual dialysis center precautions to prevent HBV spread, including dialysis in separate room with separate staff and equipment, were not activated [15]. Six years later the patient was hospitalized for shortness of breath, their HBV status was re-evaluated and yielded a positive result this time. Ultimately the discrepancy was traced back to a sequence variant in the HBV surface antigen protein, whereby the original falsely negative HBV test resulted in the potential exposure to 55 individuals to HBV [14]. A 2012 study investigated the effect of HBV surface antigen sequence variants on 13 clinical immunoassays and found that only two immunoassays could detect all 23 variants screened [16].
In addition to sequence variants, both pathogenic and benign, PTMs have also been demonstrated to interfere with immunometric methods. In 2017, falsely low immunohistochemical staining of programmed death-ligand 1 (PD-L1) – a diagnostic for immune checkpoint therapies in many cancers – was associated with abnormal glycosylation of PD-L1. Examining biopsied tissue from 22 melanoma patients revealed highly glycosylated PD-L1 yielded low estimates of this protein, likely owing to reduced binding of the reagent antibody due to steric interference from the PD-L1 glycans [17]. The downstream impact of this analytical interference on patients is dramatic, as a falsely low concentration of PD-L1 can lead to reduced chances of starting or continuing immunotherapies despite possibly needing the treatment. While sequence variants may directly alter the epitope targeted by a reagent antibody, this case demonstrates that even when the epitope is preserved, neighboring PTMs like glycosylation can confound analytical methods that rely on indirect detection of the proteoform of interest.
On the other hand, when detection of specific proteoforms is not of interest, ligand binding methods can theoretically be designed to detect the “total” concentration of the analyte, i.e., all (or most of) its proteoforms. This is accomplished by targeting an epitope (one-site immunoassay) or epitopes (two-site) immunoassay common to all proteoforms and devoid of theoretical modification sites (Fig. 2). In cases where the assay design did not purposefully consider proteoforms in its development, it may be that whatever the immunoassay is detecting—a specific proteoform, a sub-group of proteoforms, or all proteoforms of a protein—happens to be clinically informative. In such a scenario, knowing the exact composition of the group of proteoforms being detected may not add clinical value. However, as the TSH, HBV, and PD-L1 examples above demonstrate, unanticipated/unknown PTMs and sequence variants can upend even extensively tested and vetted immunoassays that have successfully made it through the regulatory process for use in patient care.
2.2. Mass spectrometry
As the effect of different proteoforms on an immunoassay can be challenging to characterize, utilization of an alternate analytical approach is often desirable. Owing to its excellent molecular selectivity, mass spectrometry has become the preferred analytical approach for proteoforms [18].
Mass spectrometry relies on the detection of molecular ions via their mass-to-charge ratio [[19], [20], [21], [22]]. There are three main components of a mass spectrometer, namely, an ionization source, mass analyzer, and a detector. The ionization source converts the molecules in a sample into charged gaseous ions; common ion sources include matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) [22,23]. The gaseous ions are then transferred to a mass analyzer [22]. These ions are sorted by the mass analyzer, in space or time, based on their mass-to-charge ratios. There are many different types of mass analyzers including quadrupoles (Q), ion trap, time-of-flight (TOF), and Fourier-transform ion-cyclotron resonance (FT-ICR) mass analyzers [23]. Mass spectrometry approaches for proteoform characterization typically utilize one or two phases of mass analysis (although more can be used), with the latter referred to as tandem mass spectrometry. With tandem mass spectrometry, a collision cell is placed between the two mass analyzers to fragment precursor ions, and then the second mass analyzer can serve to separate these fragment ions. The separated ions are then detected by the ion detector in either the time or space domains [22]. The ion detector produces electric signals that are then processed into mass spectra; commonly, the detector used is an electron multiplier [23]. Depending on how the biological sample was prepared and processed, the ions detected may correspond to the target analyte or a portion of the analyte (e.g., proteolytic peptides, released PTMs, fragments from collision induced dissociation or in-source fragmentation) [22]. For a detailed review of the structure of various types of mass spectrometers used for proteomic analysis readers are directed to the following reviews [21,[24], [25], [26], [27]].
Mass spectrometry approaches for proteoform analysis can generally be grouped into two categories, proteolytic digestion (i.e., bottom-up) or intact workflows. Proteolytic digestion workflows perform mass spectrometric analysis on the peptide level and may correlate findings back to the intact protein. To work at the peptide level, proteases–most commonly trypsin–are used to cleave proteins into peptides. For proteases with high cleavage specificity like trypsin, peptides are cut at specific amino acid residues, creating characteristic peptides. This approach relies on detection of a proteotypic peptide (i.e., a peptide unique to that protein in the relevant proteome(s)) of which the mass-to-charge ratio is used as a proxy for the protein [28,29]. On the other hand, intact workflows are devoid of a proteolysis step with the initial mass spectrometric detection targeting the intact protein(s) of interest. This approach allows for full sequence coverage, including detection of protein sequence variations, and detection of PTMs on the whole protein level through the detection of a proteoform profile [30,31].
Mass spectrometry can be used for both characterization (i.e., identifying structural details) and quantification. For the latter, multiple reaction monitoring (MRM, also referred to as selective reaction monitoring; SRM) is a commonly used approach. An MRM can be developed using a tandem mass spectrometer, such as a triple quadrupole mass spectrometer with tandem quadrupole mass filters and a collision cell [20,29,32]. With this instrument, the first quadrupole that filters for a particular ion (precursor), which is then fragmented in the collision cell via collision induced dissociation (CID), generating product ions, which are in turn filtered in the third quadrupole, before reaching the detector. MRM refers to the monitoring of the formation of a set of product ions from the precursor ion of the protein or peptide of interest, which is termed a transition. One or more of these transitions can be selected for quantification purposes, where quantification can be based off peak intensity/area or peak intensity/area relative to an internal standard (e.g., normalized peak area). For quantification, using an internal standard that is a stable isotope labelled version of the target measurand, employing quality control samples, and creating an external calibration curve are all components of best practices for peptide/protein quantification (reviewed in detail in Ref. [33]). Directly detecting the target analyte, and confirming its structure via fragmentation, gives MRM technology a degree of selectivity far exceeding that of immunoassays [29].
Similar to the MRM approach, parallel reaction monitoring (PRM) technologies are also used in hybrid quadrupole mass spectrometers like Q-TOFs and Q-ion traps. In PRM approaches, precursor ions are filtered in the first quadrupole and fragmented in the collision cell, however, unlike in MRM, all product ions are transmitted to the detector [32]. These process leads to a set of transitions (a precursor and one or more product ions) for the target protein [20,32]. This PRM approach is comparable to the MRM approach in terms of selectivity, precision, and linear range, but has a wider dynamic range. Triple quadrupole instruments used for MRM compared to higher resolution instruments used for PRM have average dynamic ranges of 2.2 and 2.9 orders of magnitude, respectively [34]. For more details on PRM approaches readers are directed to the following review [35].
MALDI mass spectrometry approaches rely on a soft ionization technique to ionize molecules via matrix assisted laser desorption. In this workflow the sample of interest is co-crystalized with an excessive amount of an organic solution called the matrix. Then a laser (either ultraviolet or infrared wavelengths) is used to ionize the dried sample, and the ions generated are accelerated and separated based on their mass-to-charge ratio [24]. MALDI is often coupled with TOF or TOF/TOF mass analyzers to allow for single and tandem analyses. These approaches are often used for intact mass analysis, peptide mass fingerprinting, and tissue imaging.
Given the variety of mass spectrometry approaches available, coupled with its high degree of selectivity, one can see why mass spectrometry has become the preferred approach for proteoform characterization and quantification. As the examples described below will reveal, mass spectrometric analysis of proteoforms covers a wide range of clinical applications, from biomarker discovery and characterization, to assisting in biomarker selection and assay design, to direct application of the technology in patient care.
3. Genotyping and phenotyping
Pathogenic mutations, now termed pathogenic variants in medicine, in the human genome that result in alterations to the protein sequence are responsible for many inherited disorders including alpha-1-antitrpysin deficiency (A1AT), cardiovascular disease, Alzheimer's disease (AD), hemoglobinopathies, and hereditary amyloidosis. Typically, identification of these inherited disorders relies on detection of the nucleic acid variant(s) via a variety of molecular techniques such as fluorescence in situ hybridization, PCR, and DNA sequencing. Biochemical phenotyping may be used in conjunction with molecular methods to identify the potential pathology, assess disease severity, or as part of diagnostic algorithms to avoid more costly testing when not warranted [[36], [37], [38]]. For example, in population-based screening for cystic fibrosis, initial measurement of immunoreactive trypsinogen in blood is a test used to determine if sequencing of the CFTR gene should be performed. CFTR encodes for the cystic fibrosis transmembrane conductance regulator (CFTR) protein which normally functions to shuttle chloride in and out of cells. In the case of pathogenic variants, CFTR function is reduced, which can lead to blockages in the pancreatic ducts, resulting in elevated immunoreactive trypsinogen. Such reflexive testing strategies have demonstrated to be cost-effective, while supporting timely diagnosis and intervention [39]. However, if one could combine biochemical phenotyping and molecular genotyping in a cost-effective manner into a single analysis, or further streamline the two, that could yield faster turnaround times, improve laboratory efficiencies, and better serve patients. Fortunately, with the increasing application of mass spectrometry there is the possibility to do just that, and combine phenotyping (e.g., protein quantitation) with genotyping (e.g., detecting variants at the protein sequence level), as described in the following examples.
3.1. Alpha-1-antitrypsin deficiency & alpha-1-antitripsin phenotyping
In alpha-1-antitrypsin (A1AT) deficiency, pathogenic variants in the gene that encodes for A1AT (SERPINA1) result in a decrease, or lack of, functional A1AT protein in circulation. A1AT is a serine protease inhibitor that inactivates neutrophil elastase, protecting the tissues from enzymatic damage during an immune response. In A1AT deficiency, the sub-normal physiological concentration of this protein means failed regulation (inhibition) of endogenous proteases, ultimately resulting in tissue damage, primarily in the lungs and liver [40]. There are numerous SERPINA1 variants associated with A1AT deficiency, with the two most common being the S allele (E288V), which decreases production of A1AT, and the Z allele (E366K), which decreases functional A1AT in circulation via the formation of A1AT aggregates in the liver. There are several analytical methodologies used to aid in the diagnosis of A1AT deficiency, such as quantification of serum A1AT concentration to identify low concentrations of A1AT via immunoassay, isoelectric focusing (IEF) to identify A1AT protein variants, genotyping to identify select pathogenic variants, and DNA sequencing to identify any variants including rare variants [[41], [42], [43]]. Laboratories have developed various test algorithms to diagnose A1AT deficiency given available resources and expertise [41,44]. General approaches include screening using the measurement of A1AT in serum, then for samples with low A1AT concentrations, moving on to more time- and cost-intensive methods such as IEF, genotyping and less commonly, full gene sequencing [41,44,45].
As an example of an alternate approach, a proteolytic digestion mass spectrometry assay for quantification and phenotyping of A1AT in serum has been developed. In this method, serum is digested with trypsin, followed by MRM analysis using liquid chromatography coupled to a triple quadrupole mass spectrometer [46]. In addition to quantification of total A1AT by selecting a single quantotypic tryptic peptide, four additional peptide sequences are monitored to provide greater sequence coverage and enable detection of wild-type versus E288V (S allele) and E366K (Z allele) sequence variants. These variant peptide sequences not only have different masses compared to their wild-type counter-part, but they also elute at a different time from the analytical column, facilitating detection [46]. With the detection of wild-type and variant peptides, the genotypes associated with the most frequent pathologies (i.e., S and Z alleles) are accurately identified. With simultaneous quantification and phenotyping of A1AT, the mass spectrometric assay eliminates the subjective step associated of IEF interpretation.
The financial benefits and operational challenges of incorporating MRM phenotyping versus the traditional IEF method in A1AT deficiency diagnostic algorithm were examined in a retrospective evaluation [47]. A1AT immunonephelometric quantification was combined with phenotyping by either mass spectrometry or the IEF method (genotyping was not performed). MRM phenotyping reduced supply and labor costs for a single high-volume laboratory by 27% and 80%, respectively. However, given that the assay is unable to identify variants beyond S and Z, reflexing to other methodologies, such as IEF, genotyping and sequencing are still required to identify rarer A1AT variants. Of note, both IEF and MRM phenotyping techniques cannot identify null alleles (i.e., absent protein expression). Thus, molecular techniques (e.g., targeted genotyping and/or DNA sequencing) are needed for definitive diagnosis in cases with discordant phenotypic interpretation (e.g., low A1AT concentration and no variant identified). Ultimately, for A1AT deficiency, employing mass spectrometry for phenotyping in populations where S and Z allelic variants are common, is a cost-effective alternative to IEF.
3.2. Cardiovascular risk assessment and apolipoprotein E genotype
The human APOE gene encodes for three common apoE proteoforms due to genetic polymorphisms: E2, E3 and E4. E3 is considered the “wild-type” proteoform, with sequence differences between the common proteoforms occurring at residues 112 and 158, i.e., E2 (Cys112 and Cys158), E3 (Cys112 and Arg158), and E4 (Arg112 and Arg158). Given that humans have two APOE alleles, there are three possible homozygous (E2/E2, E3/E3, and E4/E4) and three possible heterozygous (E2/E3, E2/E4, and E3/E4) phenotypes. While not considered pathogenic variants, apoE2 and apoE4 are associated with increased risk for cardiovascular disease [48]. Similarly, apoE4 is associated with an increased risk of Alzheimer's disease (AD) and other neurodegenerative disorders, although the risk is relatively small as ∼25% of the general population carries an apoE4 allele but not all carriers will develop neurodegenerative disorders [48,49]. In patient care, APOE genotyping is predominantly used in the context of familial dysbetalipoproteinemia, where such individuals are commonly E2/E2 [50,51]. In inherited dysbetalipoproteinemia, the normal function of apoE is impaired leading to decreased clearance of chylomicrons and very-low density lipoproteins, in turn contributing to increased risk of CVD [50,51].
For detection of the E2, E3 and E4 proteoforms, commonly used approaches include phenotyping (via IEF), genotyping (via PCR and melting curve analysis) and full-gene sequencing (via DNA sequencing). Given the cost, complexity and ethical considerations, there have been attempts to use immunoassays for detection of apoE2 [52] and apoE4 proteoforms [53]. However, apoE immunoassays can only detect a single proteoform at a time and thus are unable to discriminate homozygous from heterozygous apoE genotypes. Although identification of apoE2 or apoE4 carriers from non-carriers provides meaningful information for clinicians, this approach is less than ideal. As an alternative to traditional methods, several MRM assays for apoE phenotyping have been described [[54], [55], [56], [57], [58]]. While mass spectrometry assays could theoretically meet these requirements, there are some challenges given the primary structure of apoE.
For identification of sequence variants using a proteolytic digestion mass spectrometry approach, the sequence region(s) of interest must be amenable to proteolytic digestion and yield peptides unique to each proteoform of interest. For example, tryptic digestion of apoE does not yield proteolytic peptides capable of distinguishing apoE3 from apoE2 and apoE4. As a work-around, peptides that contain common sequences between E2 and E3 (“non-apoE4”), and E3 and E4 (“non-apoE2”), along with proteoform-specific peptides (e.g., apoE2 and apoE4) have been used to discriminate apoE phenotypes [58,59]. Using the differential peptide approach, MRM assays are able to detect all apoE proteoforms, thus discriminating homozygous and heterozygous apoE genotypes. For example, one tryptic digestion MRM assay using a liquid chromatography triple quadrupole mass spectrometer demonstrated 100% concordance in comparison to a IEF method, and 98% concordance with genotyping [59]. Furthermore, another MRM method, also using a trypsin digestion workflow, developed simultaneous detection of apoE proteoforms (i.e., E2, E3, E4) and quantification of six apolipoproteins (i.e., apoA-I, apoB, apoC-I, apoC-II, apoC-III, and apoE) using an ultra-high-performance liquid chromatography system coupled to a triple quadrupole mass spectrometer [58]. Thus, for detection of the common proteoforms, mass spectrometry presents an attractive alternative to IEF and genotyping methods.
3.3. Alzheimer's disease and amyloid-β peptide variants
The diagnosis of AD has been shown to be facilitated by the measurement of amyloid β (Aβ) peptides in cerebrospinal fluid (CSF) [60]. Aβ is generated from the endogenous proteolytic cleavage of the transmembrane amyloid precursor protein (APP), resulting in peptides of various lengths, including Aβ peptides spanning residues 1–42 (Aβ42) and 1–40 (Aβ40) (Fig. 3A). The concentration of Aβ42 is highly correlated with AD [61], while the concentration of Aβ40 is not. Aβ40 is however useful in normalizing for potential pre-analytical issues and inter-individual variability in Aβ peptide metabolism [62]. In addition to these two proteolytic forms of Aβ, numerous sequence variants have been identified (Fig. 3A). Many of the variants are pathogenic with high penetrance, resulting in a genetic form of AD, autosomal dominant AD; but there are also benign variants which are not associated with disease. For clinical purposes, a method capable of detecting the key proteolytic proteoforms (Aβ42 and Aβ40) as well as sequence variants is crucial in accurate quantification of total Aβ concentration and, when pathogenic variants are present, identification of autosomal dominant AD.
Fig. 3.
(A) Proteolytic proteoforms of amyloid-β (Aβ)—Aβ40 and Aβ42—are formed by cleavage of the transmembrane amyloid precursor protein (APP) by β- and γ-secretase. In addition to proteolytic proteoforms, nucleic acid variations in the APP gene results in Aβ peptide variants, both pathogenic and benign. Unlike immunoassays, mass spectrometry is able to identify sequence variants as demonstrated in the analysis of CSF from individuals (B) homozygous for wild-type alleles, where only the wild-type peptides (Aβ40 and Aβ42) are observed, and (C) heterozygous for an APP variant, where both wild-type and Aβ variant peptides are observed.
Using two-site immunoassays, individual assays for Aβ42 and Aβ40 have been developed; however, they are unable to identify sequence variants. If the variant occurs outside of the epitope regions of the reagent antibodies the immunoassays are expected to produce a result consistent with the wild-type sequence; however, if the variant occurs within or near the epitopes it may abrogate antibody binding, leading to a falsely low Aβ result. To overcome these analytical shortfalls, an MRM assay was developed for multiplexed quantification of wild-type Aβ peptides (Aβ42 and Aβ40) and identification of Aβ sequence variants [63]. This is an intact method that enriches Aβ peptides via solid phase extraction, without the use of reagent antibodies, followed by Aβ sequence detection and quantification via MRM using a high-performance liquid chromatography triple quadrupole mass spectrometer. Using CSF representative of APP heterozygotes, the method accurately quantified Aβ42 and Aβ40 and identified the presence of sequence variants, thus enabling the diagnosis of autosomal dominant AD (Fig. 3B and C) [63].
3.4. Hemoglobinopathies and hemoglobin variants
Hemoglobinopathies are a group of inherited disorders characterized by abnormalities in the production or structure of hemoglobin (Hb). This includes sickle cell disease caused by amino acid substitutions of the β-globin chain producing defective hemoglobin (e.g., HbS), and the thalassemia syndromes caused by deficiency in the production of α- or β-globin chains. In the context of newborn screening, it is important to identify hemoglobinopathies to enable early intervention and improve outcomes [64]. Several conventional screening methods rely on phenotypic identification of the different hemoglobin proteoforms (e.g., HbF, HbA, HbS, and HbC) by charge separation, including electrophoretic methods and high-performance liquid chromatography [[65], [66], [67]], but they can be cumbersome to implement and yield non-definitive results due to co-migration or co-elution of variants [68,69].
With increased throughput and improved selectivity in mind, several newborn screening labs have turned to mass spectrometry for hemoglobin variant detection [[70], [71], [72]]. These mass spectrometry assays were developed as proteolytic digestion methods to screen for Hb peptides in dried blood spots samples via MRM without prior chromatographic separation. Discrimination between homozygous and heterozygous genotypes was done by calculating the peptide abundance ratio of variants (e.g., HbS) to the wild-type HbA peptide. In comparison with IEF and high-performance liquid chromatography methods, the MRM method demonstrated 100% concordance in identifying HbS, HbC, HbE, and β-globin production defects [71].
Following the implementation of mass spectrometry for newborn screening, a three-year review of testing yielded no discrepancies between the MRM observed genotypes and those determined by genotyping [73]. Although the selectivity of MRM—by detecting only the select variant peptides—avoids misclassification of co-migrating or co-eluting variants in other methodologies, an MRM approach requires prespecifying the mass-to-charge transitions of the target analytes, and thus, any variants not specified in the method (rare, unknown or novel variants) will go undetected. As such, some newborn screening programs have implemented MRM screening for only detection of HbS [72,74]. As most newborn screening programs already use tandem mass spectrometry to screen for other diseases, albeit targeting small molecules, employing the same instrument for sickle cell disease screening provides an attractive alternative to traditional IEF and high-performance liquid chromatography methods.
3.5. Amyloidosis and amyloid typing
Amyloidosis refers to a group of diseases caused by the aggregation of proteins into a characteristic quaternary structure. Numerous proteins have been associated with amyloid formation including immunoglobulin light and heavy chains, transthyretin (TTR), and serum amyloid A [75]. As an example, we will focus on the protein responsible for the most common type of hereditary amyloidosis, transthyretin (TTR). TTR amyloidosis (ATTR) is a systemic disorder caused by aggregation of either the wild-type (wtTTR) or variant (vTTR) form of the TTR protein, which results in cardiomyopathy and/or polyneuropathy [76]. Diagnosis of ATTR is challenging due to symptom heterogeneity, and the desire to avoid heart tissue biopsy to confirm ATTR. Accurate diagnosis of hereditary ATTR is important for prognosis and treatment, as well as for genetic counselling for the patient and family members [77]. Definitive diagnosis of ATTR requires demonstration of amyloid deposits and identification of these deposits of being composed of TTR (i.e., amyloid typing). Traditionally, these two steps have been accomplished via two different experiments: (i) Congo red staining, which identifies the presence of amyloid, and (ii) immunohistochemistry (IHC) which attempts to identify the major proteinaceous component of the aggregate [78]. While Congo red staining is relatively straight forward, IHC has many pitfalls including false positive/false negative staining, subjective interpretation, and is limited by the antibody reagents available [79,80]. Furthermore, IHC methods cannot identify sequence variants that associated with familial forms of amyloidosis.
To overcome these pitfalls, a mass spectrometric approach to amyloid typing has been developed which identifies amyloid-associated peptides (including variant peptides) from Congo red-positive biospecimens for diagnostic evaluation of systemic amyloidosis [[81], [82], [83]]. In this assay, positively stained areas in tissue biopsy specimens and subcutaneous fat aspirates were extracted and digested with trypsin, followed by proteomic analysis via nano-flow liquid chromatography–ion trap mass spectrometry [83]. To identify the predominant protein deposit, amyloid-associated and sequence variant peptides were detected by matching against reference sequences using database search engines (for canonical proteins) and a custom sequence database (for variants) [83]. In biopsy specimens, the diagnostic accuracy improved from 76% to 94% using mass spectrometry to identify the amyloid status of paraffin-embedded tissues as compared to IHC [84]. While in subcutaneous fat aspirates, a slightly modified assay had a sensitivity and specificity of 88% and 96%, respectively [81]. Using this proteomic method, 21 established amyloid types were readily identified in 31 different organs, and amino acid substitutions in cases of hereditary amyloidosis (including vTTR) were also detected with 100% concordance with genetic analysis [83]. Although the majority of common pathogenic variants in amyloidogenic proteins are detected, some are not readily identified including variants that produce isomeric amino acid substitutions or result in an amino acid change not within a detected tryptic peptide sequence. Moreover, given the complexity of this proteomics assay, clinical laboratories would require substantial resources and expertise (e.g., complex instrumentation and bioinformatics pipeline) to implement, and therefore such a method may be best suited for a reference laboratory. With the technical advantages over antibody-based methods, mass spectrometry as described here is now considered to gold standard for amyloid typing with added advantage of identifying cases of hereditary amyloidosis.
3.6. Immunoglobulins and monoclonal gammopathies
Monoclonal gammopathies are detected via identification of an abnormally high concentration of a monoclonal immunoglobulin or a monoclonal light chain (M protein) in plasma or urine. The traditional methods to detect M proteins include gel or capillary electrophoresis, followed by immunofixation (IFE) or immunosubtraction to type the M protein. The presence of an M protein is detected via observation of a ‘spike’ where the monoclonal protein migrates, relative to the much broader distribution noted for normal globulins. As an alternative strategy, mass spectrometric methods have been developed by coupling immunoglobulin enrichment to MALDI-TOF mass spectrometry detection. In this workflow, the M protein is also detected as a visual spike, this time in the MALDI-TOF spectrum, relative the polyclonal background. The MALDI-TOF approach has been demonstrated to have greater analytical sensitivity compared to IFE and combines the previously separate analytical assessments of M protein detection and typing [85]. High resolution mass spectrometry has also been applied in this area, with immunoglobulin enrichment for serum specimens and no enrichment for urine specimens, followed by micro-flow liquid chromatography-ESI-QTOF M protein detection and typing. This high-resolution approach enables patient specific M-protein light and heavy chain molecular mass and typing, and like MALDI-TOF mass spectrometry methods demonstrated greater analytical sensitivity relative to traditional methods [86]. With increased analytical sensitivity, multiplexing detection and typing, ease of interpretation and high-throughput capacity, mass spectrometric methods are anticipated to result in a major shift in the analytical methodologies used by clinical laboratories for monoclonal gammopathies.
4. Proteolytic processing and degradation
Proteins can be proteolytically processed in vivo to alter protein function, induce activation/inactivation, regulate physiological and cellular processes, and as part of the degradation pathway. Given the complex biological consequences of protein cleavage and degradation, in vivo generated proteolytic protein fragments may serve as biomarkers of health and disease; unfortunately, their presence can also confound ligand binding assays targeting related proteoforms.
4.1. Parathyroid hormone
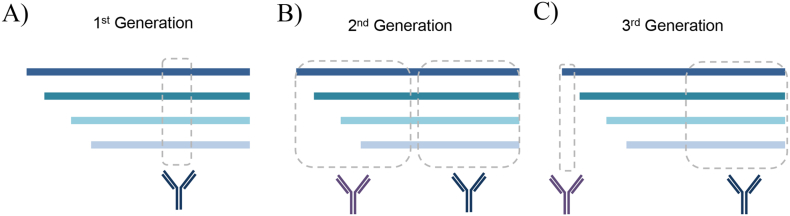
Quantification of parathyroid hormone (PTH) is measured to aid in identifying the cause of hypo- and hypercalcemia, metabolic bone disease, and parathyroid gland tumors. Clinically, PTH is measured via two-site immunoassays; however, in vivo proteolytic cleavage products of PTH can interfere with these assays. Many in vivo fragments of PTH have been identified, including N-terminally truncated proteoforms spanning residues 7–84, 28–84, 34–84, and 38–84, as well as both C and N-terminally truncated proteoforms: 34–77, 38–77, and 45–34 [[87], [88], [89], [90]]. Questions pertaining to such fragments include whether they circulate in high abundance, whether they are active or inactive fragments, and their relation to physiology and clearance (e.g., PTH 1–84 has a half-life of ∼5 min, whereas some fragments have half-lives in the range of 24–36 h) [87,[91], [92], [93]]. To accommodate for the presence of PTH proteoforms, immunoassays for PTH have evolved over the years (Fig. 4); the initial (first generation) radioimmunoassay for PTH utilized an antibody targeting the middle of the PTH sequence, which was subsequently improved through second and third generation immunoassays to target the N-terminus of PTH based on mass spectrometric and biological studies [87,94]. Nonetheless, contemporary PTH immunoassays bind PTH 1–84 and, variably, PTH fragments, which can lead to inconsistencies between immunoassay manufacturers.
Fig. 4.
PTH ligand binding assay evolution. A) The first-generation radioimmunoassays used a single antibody directed against the residues within the sequences 44–68 or 53–84. B) Second-generation immunoassays were two-site sandwich assays with capture antibodies directed against residues 1–34 or 1–38 and detection via residues 39–84 or 53–84. C) The third-generation immunoassays were optimized to detect intact PTH (residues 1–84) using a capture antibody targeting the C-terminus (epitope within residues 39–84) and a detection antibody targeting the N-terminus (residues 1–4).
To aid in selective and accurate quantification of intact PTH, mass spectrometry has been employed to both characterize PTH proteoforms in circulation and quantify select proteoforms. In the seminal application, serum PTH proteoforms were enriched using an antibody targeting the C-terminus; the immunoenriched fraction was then digested with trypsin, separated by nano-flow liquid chromatography and analyzed using MRM on a Q-TOF mass spectrometer [95]. Comparing this MRM method to a clinically available second-generation and third-generation immunoassay, the MRM assay had a negative bias relative to the second generation assay and significant quantification differences compared to the third generation assay [95]. These differences were attributed to antibody cross-reactivity with additional PTH proteoforms or other interferents in both second and third generation immunoassays.
In a recent study examining the primary structure and concentration of PTH fragments, an intact proteomic discovery analysis was used to sequence the PTH fragments circulating in patients with renal failure [90]. This method used immunoenrichment with two anti-PTH antibodies, one against the C-terminus (epitope within residues 44–84) and one against the N-terminus (immunogen/epitope not specified). The enriched samples were separated using liquid chromatography and analyzed using a Q-ion trap hybrid mass spectrometer. This approach revealed abundant PTH fragments including those spanning residues 28–84, 34–77, 34–84, 38–77, 38–84, and 45–34. Most notably the hypothesized PTH 7–84 fragment that was thought to be the primary interferent in the first- and second-generation immunoassays, was not among the abundant proteoforms. Further, when quantifying PTH in individuals with severe renal failure, where increased PTH proteolytic fragmentation is expected, a method comparison between the MRM assay and a third-generation immunoassay demonstrated that the MRM assay had 58% lower PTH 1–84 concentration relative to the immunoassay. While the third-generation immunoassay did not demonstrate significant cross-reactivity with N-terminally cleaved PTH 28–84, 34–84, 37–84, 45–84, or 48–84, it did have nearly 100% cross-reactivity with synthetic PTH 7–84 [90].
Via mass spectrometry, the complexity of PTH proteoforms has been revealed. Moreover, mass spectrometry has helped identified ongoing challenges with immunometric detection of PTH, and their potential impact on clinical management [96].
4.2. Cardiac biomarkers: B-type natriuretic peptides and troponins
Cardiac biomarkers, B-type natriuretic peptide (BNP), N-terminal-pro-BNP (NT-proBNP) and cardiac troponins (cTn) are commonly quantified to assist in the diagnosis, monitoring and risk assessment of heart dysfunction. In the clinical laboratory, these blood biomarkers are measured via two-site immunoassays; however, mass spectrometry is playing a role in identifying proteoform-specific interferences and novel biomarker candidates, as well as assisting with assay harmonization.
In the case of BNP proteoforms, in vivo proteolytic cleavage products of proBNP, BNP and NT-proBNP, are abnormally elevated in heart failure patients as they are released from cardiac myocytes under strain. BNP (residues 1–32) is the bioactive fragment of proBNP (residues 77–108 of proBNP); while NT-proBNP is produced as the inactive form (residues 1–76 of proBNP). Immunoassays for BNP are known to cross-react with BNP breakdown products and proBNP, while NT-proBNP immunoassays are known to cross-react with proBNP [[97], [98], [99]]. To accurately quantify BNP and detect its degradation products, an immunoenrichment intact mass spectrometry method was developed [100]. Using two antibodies targeting the N- and C-termini of BNP (residues 5–13, and 15–25, respectively), plasma BNP proteoforms were extracted for detection and quantification via MALDI-TOF mass spectrometry. Applied to the study of heart failure patients, this mass spectrometry assay demonstrated that intact BNP measured by immunoassays did not correlate with BNP 1–32 but instead correlated best with the BNP fragment spanning residues 5–32, a biologically inactive fragment [101]. These findings were corroborated with the development of a proteolytic digestion workflow, where BNP was digested with trypsin and analyzed by MRM using ultra-high-performance liquid chromatography coupled to a triple quadrupole mass spectrometer [102]. In a comparison using external proficiency samples, the MRM method correlated with but had a negative bias with three different immunoassays [102], which was attributed to differential detection of BNP proteoforms and different calibrants. In general, such mass spectrometric studies have helped elucidate circulating BNP proteoforms and their role in immunoassay interference [103].
In the case of cTn, elevated concentrations of cTnI or cTnT are the current gold standard to diagnose myocardial infarction [104]. cTn's can have extensive PTMs including from phosphorylation and proteolysis, confounding immunoassay quantification [105,106]. The use of different antibodies targeting different cTn sequences by the various in vitro diagnostic manufacturers, results in differences in cTn concentration, owing in large part to differences in cTn proteoforms detected [107]. To illustrate this point, consider the debate on whether cTnT predominantly circulates in its intact form or as proteolytic fragments. To investigate this question, serum from acute myocardial infarction patients was first immunoenriched for cTnT (targeting epitope 146–157) and subjected to western blot analysis, whereby three immunopositive bands were detected at 16, 29, and 37 kDa [106]. The bands were excised subjected to in-gel tryptic digestion and analysis by ultra-high-performance liquid chromatography coupled to a quadrupole-ion trap hybrid mass spectrometer. In the mass spectrometric analysis, detection of fully-tryptic cTnT peptides confirmed the presence of cTnT the immuno-positive bands, and the detection of semi-tryptic cTnT peptides confirm the presence of proteolytic fragments formed in vivo in the 16 and 29 kDa bands. Interestingly, the 29 kDa cTnT proteoforms were found to be the predominant form in serum; however, cTnT immunoassays were designed to detect intact cTnT and thus variably detect this key proteoform [106]. Like the study of BNP proteoforms, characterization of cTn proteoforms is expected to help optimize immunoassay design and contribute to harmonization efforts.
4.3. D-dimer
D-dimer quantification is used in the diagnosis and management of venous thromboembolism, deep vein thrombosis, and disseminated intravascular coagulation [[108], [109], [110], [111]]. D-dimers are formed during fibrinolysis via the breakdown of fibrinogen and fibrin by plasmin. This leads to four fragments, termed X, Y, D, and E, with only fragments originating from fibrin polymers that have undergone factor XIII mediated cross-linking having a covalent bond between two adjacent D domains (i.e., the D-dimer). D-dimers contain two D domains and one E domain, providing a unique target for fibrin-specific degradation product, a structural detail exploited by most immunoassays [112]. There are various ligand binding assay approaches to D-dimer quantification including immunofluorescence, immunoturbidity and chemiluminescence. While D-dimer is routinely quantified via immunoassay in clinical care, this analytical approach has demonstrated cross reactivity with fibrinogen and other fibrinogen breakdown products, leading to falsely elevated estimates of fibrinolysis activity [113,114]. As such, D-dimer is helpful in diagnosing venous thrombosis as it has a good negative predictive value, but cannot be used to diagnose venous thromboembolism due to its poor positive predictive value [115].
To attempt to design an assay specific for D-dimer without the interferences noted for immunoassays, mass spectrometry has been employed [116]. In the mass spectrometry assay workflow, plasma was first depleted of abundant proteins (a step generally to be avoided in quantitative mass spectrometric methods [33]), digested with trypsin, subjected to immunoenrichment targeting the D-dimer peptide spanning residues 305–324, and analyzed by MRM using a high-performance liquid chromatography system coupled to a triple quadrupole mass spectrometer. In preliminary experiments, the mass spectrometric method did not correlate well with a commercial ELISA but correlated better than the ELISA with fibrinolytic activity based on thromboelastography (a measure of blood clotting efficiency). This study highlights the use of mass spectrometry in being able to select for and probe specific proteoforms or groups of proteoforms that may help improve correlation with a physiological parameter, disease or outcome.
4.4. Adrenocorticotropic hormone
Adrenocorticotropic hormone (ACTH, residues 1–39) is a peptide hormone produced by the pituitary gland, which stimulates the adrenals to secrete cortisol. When disorders of the hypothalamic-pituitary-adrenal axis are suspected, ACTH is used as a biomarker to help identify where along the axis the dysfunction is found. Complicating its use as a biomarker, ACTH immunoassays are vulnerable to interference by heterophile antibodies (e.g., endogenous antibodies that bind to reagent antibodies), and ACTH fragments and synthetic analogs [117]. Not only are these assays vulnerable to such interferents but the magnitude and direction of the interference varies based on the targeted epitopes and antibodies used (e.g., monoclonal v. polyclonal), leading to grossly discrepant results between manufacturers based on clinical interpretation of the ACTH concentration [[117], [118], [119]]. To overcome these interferences and resolve discrepant immunoassay results, a mass spectrometry method for quantification of intact ACTH and detection of other ACTH proteoforms was developed [117]. In this intact method, samples were first immunoenriched (targeting residues 9–12), then subjected to high performance liquid chromatography and MRM analysis on a triple quadrupole mass spectrometer. The multiplex method included intact ACTH [[1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39]], cosyntropin (residues 1–24) and corticotropin-like intermediate peptide (CLIP; 18–39). Corroborating assumptions based on clinical phenotypes, a comparison of two regulatory approved clinical immunoassay revealed only one immunoassay correlated with the concentration of biologically active intact ACTH, as revealed by mass spectrometry. Thus, the application of mass spectrometry was able to: 1) selectively detect intact ACTH and various fragments, 2) identify cross-reactivity in an immunoassay, 3) highlight the need for a reference material for ACTH (both mass spectrometric and immunometric assays), and 4) resolve cases where the ACTH immunoassay concentration were discrepant with the clinical picture.
5. Phosphorylation, glycosylation, and glycation
Beyond proteoloytic fragmentation, PTMs such as glycosylation and phosphorylation can also yield challenges for detection and accurate quantification of biomarkers. On the other hand, these PTMs can also be exploited as more sensitive and specific markers of pathological processes and for detection, characterization and quantification of exogenous proteins in circulation.
5.1. Neurodegeneration & tau phosphorylation
Tau is an axonal protein involved in the regulation of microtubule stability. In AD, tau undergoes abnormal hyperphosphorylation and these phosphorylated proteoforms have been shown to be diagnostic biomarkers [120]. CSF and blood tau assays target either total tau (t-tau) or phosphorylated tau (p-tau), with p-tau having greater specificity for AD [[121], [122], [123]]. Challenges for immunometric detection of p-tau proteoforms include cross-reactivity with non-hyperphosphorylated tau [124] and multiplexing limitations restricting the assay to the detection of only one of the many phosphorylation sites.
Based on these limitations, an MRM method using a proteolytic approach was developed for the quantification of tau proteoforms. This was done using immunoenrichment followed by tryptic digestion and micro-flow liquid chromatography-linear ion trap mass spectrometry. Although this method did not quantify p-tau directly, it detected tau proteoforms with and without PTMs, including p-tau-214, p-tau-69, and p-tau-404, demonstrating the capacity for the measurement of multiple proteforms [125]. For p-tau in CSF, several methods have been developed to directly characterize and quantify phospho-proteoforms in biospecimens from individuals with AD. For instance, two methods used enrichment, tryptic digestion and nano-flow liquid chromatography ion trap mass spectrometry. Both methods found greater phosphorylation of tau in AD compared to controls [126,127]. Plasma p-tau proteoforms have also been investigated using a similar proteolytic digestion high resolution mass spectrometry approach. For example, a PRM method found that the plasma tau proteoforms phosphorylated at T217 had the highest diagnostic performance among a group of phophorylation sites tested [128]. These examples for both CSF and blood proteoforms demonstrate the advantages of a mass spectrometric approach in being able to characterize multiple phospho-proteoforms, providing new biomarker candidates with improved diagnostic performance to detect AD pathology which could then be further developed as either ligand binding or quantitative mass spectrometric clinical assays.
5.2. Monoclonal antibody glycoforms
Therapeutic monoclonal antibodies (mAbs), including humanized or chimeric IgG, are now used as therapies in a wide variety of diseases [129]. These mAbs are produced from cell culture, and the glycosylation heterogeneity of different production batches can affect the efficacy, stability, and clearance of the therapeutic product. Traditional methods to characterize PTM patterns for these antibodies include IEF, gel electrophoresis, and western blot [130]. Mass spectrometric methods outperform these traditional methods by providing more structural information, such as glycoform patterns and sites. For example, to examine different approaches for characterization of the glycosylation for therapeutic mAbs, one study compared high-performance liquid chromatography mass spectrometry, MALDI-TOF mass spectrometry, and anion-exchange chromatography. The high-performance liquid chromatography mass spectrometry analysis for the intact protein and MALDI-TOF analysis for the enzymatically-released carbohydrates showed comparable accuracy and reproducibility to anion-exchange chromatography, but mass spectrometric methods provided more structural information compared to traditional methods [131]. In addition to therapeutic antibodies, mass spectrometry has been applied to the study of glycoforms found on human M proteins. For profiling, immunoglobulins were enriched from serum samples using protein A followed by nano-flow liquid chromatography-chip-Q-TOF analysis of the intact M protein. Notably, this method uses a microfluidics chip instead of the traditional liquid chromatography columns. In a proof-of-concept experiment using serum from two multiple myeloma patients pre- and post-treatment, the untargeted high resolution mass spectrometry method detected minimal to moderate changes in M protein glycoform complexity with time [132]. As these examples demonstrate, mass spectrometry is a useful tool for quality control for therapeutic antibodies and may have future applications in characterizing monoclonal gammopathies as well as cryoglobulinemias.
5.3. Glycated hemoglobin
As discussed in the introduction, HbA1c is a glycated form of hemoglobin that serves as a biomarker of glycemic control. Commonly used HbA1c ion-exchange high performance liquid chromatography assays and immunoassays measure both the glycated and non-glycated hemoglobin and calculate the ratio of glycated hemoglobin to total hemoglobin to determine the HbA1c concentration [133]. There are several interferences known with these methods, which can confound the results due to co-elution or cross-reactivity. The interferents, which are method specific, include but are not limited to, unusual concentrations of other hemoglobin fractions, hemoglobin variants, and chemically modified hemoglobin [[134], [135], [136], [137], [138], [139]]. For example, HbF normally accounts for less than 1% of total hemoglobin in adults, but it can rise dramatically in conditions such as sickle cell disease and leukemia, and can be mis-identified as HbA. Chemically modified Hb (e.g., PTMs such as carbamylation and acetylation) are also elevated in conditions such as reduced kidney function, and can be mis-identified as glycated hemoglobin [137].
To overcome issues with both high performance liquid chromatography and interferents associated with hemoglobin proteoforms, a reference method was developed using mass spectrometry. The method utilizes proteolytic digestion with Glu-C followed by high performance liquid chromatography directly coupled to a single stage Q-ESI mass spectrometer [136,140,141]. Glu-C was specifically selected as it releases an N-terminal hexapeptide of the β-chains, containing the key glycation site. This enables direct and selective detection of glycated and non-glycated HbA, without inference from PTMs, sequence variants or other proteoforms, and is the reason why this assay is the gold-standard for HbA1c measurement.
6. Endogenous v. exogenous proteoforms
Peptides and proteins have long been administered as therapeutics in medicine, and taken surreptitiously to boost athletic performance. Take for example the historical use of growth hormone extracted from human pituitary glands compared to present day where synthetic forms are now used. Therapeutic proteins, now most commonly synthesize or expressed to avoid iatrogenic disease transmission, are designed to have similar structures to their endogenous counterpart to maintain functionality and minimize immunogenicity. However, some structural variations may exist whether they be purposeful (e.g., amino acid change to alter pharmacokinetics and/or enable a patent claim) or a function of the production method (e.g., different glycoform patterns based on the expression system). In this section, we will review approaches for characterizing and distinguishing exogenous versus endogenous proteoforms.
6.1. Erythropoietins
Erythropoietin (EPO) is a glycoprotein produced by the kidneys and is involved in the differentiation and development of red blood cells from bone marrow stem cells [142]. Recombinant EPOs (rEPOs) are a group of therapeutic agents used to treat anemia; however, these drugs are also misused by athletes to attempt to improve their performance [143]. As rEPOs are expressed in non-human cell lines (e.g., Chinese hamster ovary cells), the glycoform patterns of rEPOs differ from those generated in humans. These glycoform pattern differences have been exploited by anti-doping labs to detect surreptitious use of rEPO by athletes. The direct doping control test approved by the World Anti-Doping Agency (WADA) uses electrophoresis (e.g., sarkosyl-polyacrylamide gel electrophoresis [SAR-PAGE], IEF) to differentiate EPO glycoform profiles [143]. This method has a lengthy workflow and includes immunopurification, electrophoretic separation by SAR-PAGE, and anti-EPO immunoblotting (Figure 5). In addition, the low mass resolution between the endogenous EPO and some rEPO proteoforms limits the accuracy of the analysis (e.g., epoetin-α and epoetin-β, two different preparations of rEPO, overlap with endogenous EPO) [143,144].
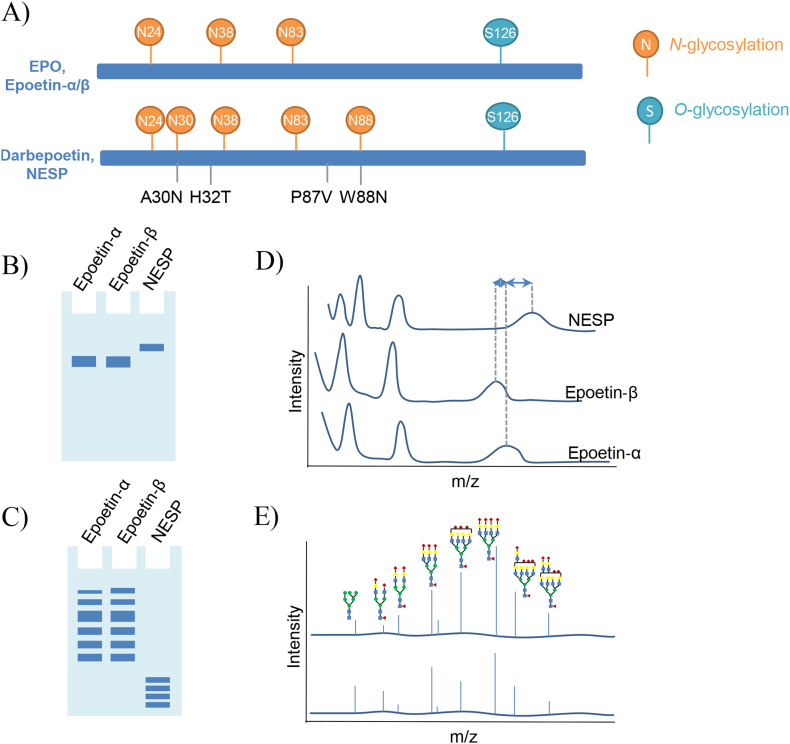
Fig. 5.
Schematic of the detection of the EPO proteoforms using electrophoresis v. mass spectrometry. (A) Structure of EPO and relevant proteoforms and analogues. EPO proteoforms detected using (B) SDS-PAGE or (C) IEF: the position and shape (bandwidth, focused or diffuse) of the band are used for the identification of EPO proteoforms. (D) MALDI-TOF mass spectra of the intact rEPO: EPO proteoforms are identified by the mass-to-charge ratio differences of singly, doubly and triply charged molecular ions. (E) MALDI-TOF mass spectra of EPO N-glycans.
To enable selective detection of rEPO proteoforms (Fig. 5A), namely epoetin‐α, epoetin‐β, and novel erythropoiesis stimulating protein (NESP; an EPO analogue with additional glycosylation sites), a MALDI-TOF mass spectrometry method was developed for intact analysis. Via mass spectrometry, the overlapping epoetin-α and epoetin-β in SAR-PAGE were clearly distinguishable with a 0.4 kDa mass difference at the intact protein level [145]. Looking at the glycoforms specifically, an alternate approach was developed using peptide-N-glycosidase F (PNGaseF) to release the glycopeptides from rEPO products and then analyze the glycans by nano-flow liquid chromatography coupled to a Q-TOF mass spectrometer [146]. Another method applied affinity purification using anti-EPO antibody before glycoforms were released using PNGaseF, followed by MALDI-TOF mass spectrometry glycoform analysis. This method demonstrated improved resolution of glycoform patterns compared to established capillary gel electrophoresis-laser induced fluorescence methods for glycosylation analysis [147]. These patterns identified by mass spectrometry were used to identify proteoforms present, and ultimately EPO versus rEPO. The structural information (i.e., glycoform profiles) collected via mass spectrometry was so detailed, that even differences in product batches could be detected. In addition to glycoform patterns, identification of EPO doping has also relied on differences in protein sequences between endogenous and exogenous EPO. Such methods immunoenrich for EPO proteoforms, then perform tryptic digestion, followed by the identification of relevant proteolytic peptide sequences via nano-flow liquid chromatography-linear ion trap mass spectrometry [148,149]. With better resolution in the detection of EPO proteoforms, mass spectrometric methods have been shown to provide less ambiguous results, as compared to traditional methods, in doping control applications.
6.2. Therapeutic antibodies
Monoclonal antibodies and Fc fusion proteins are on the rise as therapeutic options for a wide range of conditions from arthritis to cancers to viral infections. To guide therapy, that is decreasing or escalating doses of these expensive medicines, therapeutic drug monitoring is employed. Based on the rapid evolution of these drugs and their specialized uses, in vitro diagnostic companies have been slow to keep up with demand for new assays. Moreover, limited assay selection and lack of robust quantitation has left clinical laboratories looking for alternate options.
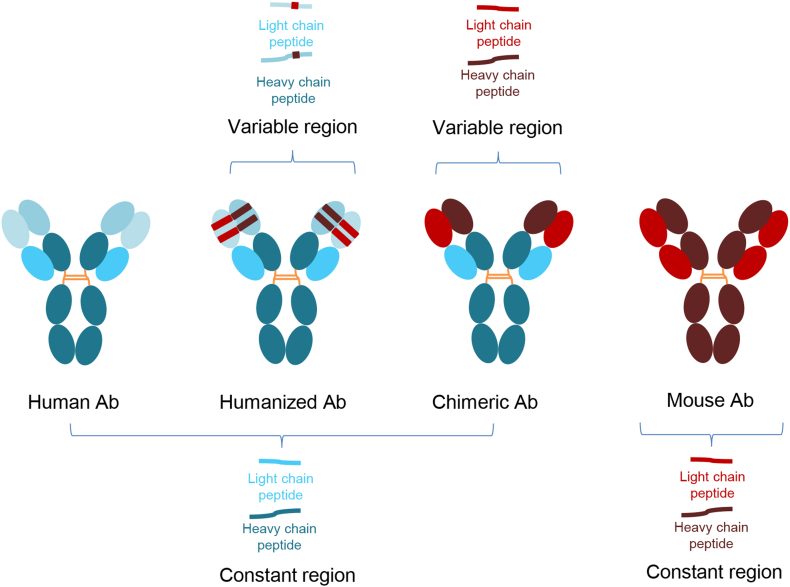
In such a case, a proteolytic digestion method was developed for infliximab [150]. Infliximab is a mouse/human chimeric monoclonal antibody, used in the treatment of chronic inflammatory disease including Crohn's disease, ulcerative colitis, psoriasis and rheumatoid arthritis. Serum was digested with trypsin and then analyzed using high performance liquid chromatography coupled to a triple quadrupole mass spectrometer [150]. Two peptides unique to infliximab, that is, not found in the human proteome were monitored. A streamlined higher-throughput MRM method was then developed for serum and plasma specimens, employing fully automated and simplified sample preparation strategy for routine clinical laboratory testing [151]. In the case of infliximab, a chimeric antibody, a proteolytic digestion approach was used as unique peptides were readily produced and detected from the target measurand. With the newer humanized or fully human biologics, a different strategy is required as there may be no unique proteolytic peptides compared to immunoglobulins in the human proteome (Fig. 6). In these cases, dissociation of light and heavy chains followed by intact mass spectrometric analysis is appropriate (reviewed in detail in Ref. [152]).
Fig. 6.
Variations is therapeutic antibody structure from fully human, humanized, chimeric and mouse. The primary structure dictates the appropriate mass spectrometric approach for therapeutic drug monitoring assays, whether that be an intact or proteolytic digestion workflow.
6.3. Insulins
Exogenous synthetic insulin analogs are used for the treatment of diabetes; however, they can be misused in the setting of accidental or purposeful administration (e.g., Munchausen syndrome, or athletic doping). In routine clinical evaluation, plasma insulin is quantified via immunoassay; however, these methods are known to variably cross-react with endogenous precursor molecules and fragments, as well as, synthetic analogs [153,154]. The interference varies greatly based on the reagent antibodies used and assay design. As such, for forensic investigations and in doping controls, these immunoassays may be of little value.
To selectively identify intact insulin as well as several synthetic analogs a qualitative intact mass spectrometry method was developed [155]. Human plasma was immunoenriched using anti-insulin antibodies, followed by analysis using micro-flow liquid chromatography coupled to a quadrupole ion trap hybrid mass spectrometer. In another method, an MRM approach was used for human insulin, as well as lispro, aspart, and glargine. In a later method, MRM was also used to quantify intact insulins (e.g., Humulin) and four analogs (detemir, aspart, lispro, and glargine), similarly using immunoenrichemnt and analysis by high performance liquid chromatography coupled to a triple quadruple mass spectrometer [156]. Both methods enriched for insulins using antibodies targeting regions common to the proteoforms of interest, followed by liquid chromatography and mass spectrometry to separate, detect and quantify different insulin structures present. Mass spectrometry thus provides unambiguous identification of insulin analogs yielding an approach suitable for forensic and doping control analyses.
7. Limitations of mass spectrometry from a clinical laboratory perspective
Although mass spectrometry has numerous advantages in proteoform detection and quantification, implementation of protein mass spectrometry methods in the clinical laboratory has its challenges. These include familiarization with sample preparation approaches specifically for proteins, the need for highly trained staff to develop methods and operate the equipment, and turnaround time [157]. Also, for many protein measurands the triple quadrupole mass spectrometers commonly available in clinical laboratories (based on historical use for small molecule analysis) can be repurposed for this new application; however, low abundance targets or other complications may necessitate the use of less commonly available equipment, i.e., low flow liquid chromatography systems and high-resolution mass spectrometers.
8. Conclusion
Human proteoforms diversity is essential for normal biological processes; however, this complexity can create challenges for biomarker detection via ligand binding assays. Fortunately, mass spectrometry has developed into a powerful analytical tool for proteoform analysis: (i) in the discovery of novel biomarkers, (ii) in the implementation of qualitative or quantitative assays directly in the clinical laboratory, and (iii) for troubleshooting and refinement of ligand binding methods. Two key strengths of mass spectrometry in the context of proteoforms characterization, detection and quantification include high molecular selectivity and multiplexing capacity. With these advantages, we have seen the translation of several mass spectrometric proteoform assays into the clinical laboratory based on their improved analytical and diagnostic performance over predicate methods. This includes instances where the mass spectrometric method has displaced the predicate method or supplemented the predicate method by become the reference methodology. There is also the ongoing potential for proteoforms research to inform assay refinement or redesign to target a select proteoform or group of proteoforms for improved correlation with pathophysiology or outcome. With a deeper understanding of altered proteoform profiles in disease, coupled with complementary analytical techniques like mass spectrometry, an expanding role for proteoforms in laboratory medicine is anticipated.
Funding
LMF is the recipient of a Frederick Banting and Charles Best Canada Graduate Scholarship from the Canadian Institutes of Health Research. MW is the recipient of a Mitacs Postdoctoral Fellowship co-funded by the Pacific Airway Centre. MLD is the recipient of the Michael Smith Foundation for Health Research (MSFHR) Scholar Award and acknowledges grant funding from Brain Canada through the Canada Brain Research Fund with the financial support of Health Canada, MSFHR, University of British Columbia’s Faculty of Medicine and the Djavad Mowafaghian Centre for Brain Health, Women’s Brain Health Initiative, and the St Paul’s Foundation.
Declaration of competing interest
The authors declare that they have no known financial or personal competing interests that would influence the work outlined in this paper.
References
- 1.International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. Epub 2004/10/22. PubMed PMID: 15496913. [DOI] [PubMed] [Google Scholar]
- 2.Ponomarenko E.A., Poverennaya E.V., Ilgisonis E.V., Pyatnitskiy M.A., Kopylov A.T., Zgoda V.G., et al. The size of the human proteome: the width and depth. Int. J. Analytic. Chem. 2016. 2016 doi: 10.1155/2016/7436849. Epub 2016/05/19. PubMed PMID: 27298622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith L.M., Kelleher N.L. Consortium for top down P. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10(3):186–187. doi: 10.1038/nmeth.2369. PubMed PMID: 23443629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardito F., Giuliani M., Perrone D., Troiano G., Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017;40(2):271–280. doi: 10.3892/ijmm.2017.3036. Epub 2017/06/22. PubMed PMID: 28656226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecker S.H., Goldberg A.L., Mitch W.E. Protein degradation by the ubiquitin–proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006;17(7):1807–1819. doi: 10.1681/asn.2006010083. [DOI] [PubMed] [Google Scholar]
- 6.Goettig P. Effects of glycosylation on the enzymatic activity and mechanisms of proteases. Int. J. Mol. Sci. 2016;17(12):1969. doi: 10.3390/ijms17121969. PubMed PMID: 27898009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore L.D., Le T., Fan G. DNA methylation and it's basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. Epub 2012/07/11. PubMed PMID: 22781841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H., Zhang X., Chen X., Aramsangtienchai P., Tong Z., Lin H. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118(3):919–988. doi: 10.1021/acs.chemrev.6b00750. Epub 2018/01/02. PubMed PMID: 29292991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein T., Eckhard U., Dufour A., Solis N., Overall C.M. Proteolytic cleavage—mechanisms, function, and “omic” approaches for a near-ubiquitous posttranslational modification. Chem. Rev. 2018;118(3):1137–1168. doi: 10.1021/acs.chemrev.7b00120. [DOI] [PubMed] [Google Scholar]
- 10.Allen D.W., Schroeder W.A., Balog J. Observations on the chromatographic heterogeneity of normal adult and fetal human hemoglobin: a study of the effects of crystallization and chromatography on the heterogeneity and isoleucine content. J. Am. Chem. Soc. 1958;80(7):1628–1634. doi: 10.1021/ja01540a030. [DOI] [Google Scholar]
- 11.Bookchin R.M., Gallop P.M. Structure of hemoglobin AIc: nature of the N-terminal β chain blocking group. Biochem. Biophys. Res. Commun. 1968;32(1):86–93. doi: 10.1016/0006-291X(68)90430-0. [DOI] [PubMed] [Google Scholar]
- 12.Rahbar S., Blumenfeld O., Ranney H.M. Studies of an unusual hemoglobin in patients with diabetes mellitus. Biochem. Biophys. Res. Commun. 1969;36(5):838–843. doi: 10.1016/0006-291X(69)90685-8. [DOI] [PubMed] [Google Scholar]
- 13.Drees J.C., Stone J.A., Reamer C.R., Arboleda V.E., Huang K., Hrynkow J., et al. Falsely undetectable TSH in a cohort of South asian euthyroid patients. J. Clin. Endocrinol. Metabol. 2014;99(4):1171–1179. doi: 10.1210/jc.2013-2092. [DOI] [PubMed] [Google Scholar]
- 14.Hendrickson B., Kamili S., Timmons T., Iwen P.C., Pedati C., Safranek T. Notes from the field: false-negative hepatitis B surface antigen test results in a hemodialysis patient. MMWR Morb. Mortal. Wkly. Rep. 2018;67:311–312. doi: 10.15585/mmwr.mm6710a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC Recommendations for preventing transmission of infections among chronic hemodialysis patients. MMWR Recomm. Rep. (Morb. Mortal. Wkly. Rep.) 2001;50(Rr-5):1–43. Epub 2001/05/15. PubMed PMID: 11349873. [PubMed] [Google Scholar]
- 16.Servant-Delmas A., Mercier-Darty M., Ly T.D., Wind F., Alloui C., Sureau C., et al. Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J. Clin. Virol. 2012;53(4):338–345. doi: 10.1016/j.jcv.2012.01.003. Epub 2012/02/03. PubMed PMID: 22296790. [DOI] [PubMed] [Google Scholar]
- 17.Morales-Betanzos C.A., Lee H., Gonzalez Ericsson P.I., Balko J.M., Johnson D.B., Zimmerman L.J., et al. Quantitative mass spectrometry analysis of PD-L1 protein expression, N-glycosylation and expression stoichiometry with PD-1 and PD-L2 in human melanoma. Mol. Cell. Proteomics : MCP. 2017;16(10):1705–1717. doi: 10.1074/mcp.RA117.000037. Epub 2017/05/25. PubMed PMID: 28546465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. Epub 2003/03/14. PubMed PMID: 12634793. [DOI] [PubMed] [Google Scholar]
- 19.Banerjee S., Mazumdar S. Electrospray ionization mass spectrometry: a technique to access the information beyond the molecular weight of the analyte. Int. J. Analytic. Chem. 2012. 2012 doi: 10.1155/2012/282574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freue G.V.C., Borchers C.H. Multiple reaction monitoring (MRM) Circulation: Cardiovascular Genetics. 2012;5(3):378. doi: 10.1161/CIRCGENETICS.111.959528. [DOI] [PubMed] [Google Scholar]
- 21.Kaklamanos G., Aprea E., Theodoridis G. In: Encyclopedia of Food and Health. Caballero B., Finglas P.M., Toldrá F., editors. Academic Press; Oxford: 2016. Mass spectrometry: principles and instrumentation; pp. 661–668. [Google Scholar]
- 22.Urban P.L. Quantitative mass spectrometry: an overview. Philos Trans A Math Phys Eng Sci. 2016;374(2079) doi: 10.1098/rsta.2015.0382. PubMed PMID: 27644965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hossain M. In: Selected Reaction Monitoring Mass Spectrometry (SRM-MS) in Proteomics: A Comprehensive View. Hossain M., editor. Springer International Publishing; Cham: 2020. The mass spectrometer mass spectrometers and its components; pp. 17–52. [Google Scholar]
- 24.Jurinke C., Oeth P., van den Boom D. MALDI-TOF mass spectrometry. Mol. Biotechnol. 2004;26(2):147–163. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 25.Zubarev R.A., Makarov A. Orbitrap mass spectrometry. Anal. Chem. 2013;85(11):5288–5296. doi: 10.1021/ac4001223. [DOI] [PubMed] [Google Scholar]
- 26.Xian F., Hendrickson C.L., Marshall A.G. High resolution mass spectrometry. Anal. Chem. 2012;84(2):708–719. doi: 10.1021/ac203191t. [DOI] [PubMed] [Google Scholar]
- 27.Glish G.L., Burinsky D.J. Hybrid mass spectrometers for tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19(2):161–172. doi: 10.1016/j.jasms.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Fournier M.L., Gilmore J.M., Martin-Brown S.A., Washburn M.P. Multidimensional separations-based shotgun proteomics. Chem. Rev. 2007;107(8):3654–3686. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 29.Wolf-Yadlin A., Hautaniemi S., Lauffenburger D.A., White F.M. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104(14):5860. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catherman A.D., Skinner O.S., Kelleher N.L. Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 2014;445(4):683–693. doi: 10.1016/j.bbrc.2014.02.041. Epub 2014/02/17. PubMed PMID: 24556311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toby T.K., Fornelli L., Kelleher N.L. Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem. 2016;9(1):499–519. doi: 10.1146/annurev-anchem-071015-041550. PubMed PMID: 27306313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidova V., Spacil Z. A review on mass spectrometry-based quantitative proteomics: targeted and data independent acquisition. Anal. Chim. Acta. 2017;964:7–23. doi: 10.1016/j.aca.2017.01.059. [DOI] [PubMed] [Google Scholar]
- 33.CLSI. C64 . 2021. Quantitative Measurement of Proteins and Peptides by Mass Spectrometry. [Google Scholar]
- 34.Peterson A.C., Russell J.D., Bailey D.J., Westphall M.S., Coon J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics : MCP. 2012;11(11):1475–1488. doi: 10.1074/mcp.O112.020131. Epub 2012/08/03. PubMed PMID: 22865924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourmaud A., Gallien S., Domon B. Parallel reaction monitoring using quadrupole-orbitrap mass spectrometer: principle and applications. Proteomics. 2016;16(15–16):2146–2159. doi: 10.1002/pmic.201500543. Epub 2016/05/05. PubMed PMID: 27145088. [DOI] [PubMed] [Google Scholar]
- 36.Park N.J., Morgan C., Sharma R., Li Y., Lobo R.M., Redman J.B., et al. Improving accuracy of tay sachs carrier screening of the non-jewish population: analysis of 34 carriers and six late-onset patients with HEXA enzyme and DNA sequence analysis. Pediatr. Res. 2010;67(2):217–220. doi: 10.1203/PDR.0b013e3181c6e318. PubMed PMID: 19858779. [DOI] [PubMed] [Google Scholar]
- 37.Sontag M.K., Lee R., Wright D., Freedenberg D., Sagel S.D. Improving the sensitivity and positive predictive value in a cystic fibrosis newborn screening program using a repeat immunoreactive trypsinogen and genetic analysis. J. Pediatr. 2016;175:150–158. doi: 10.1016/j.jpeds.2016.03.046. e1. Epub 2016/04/27. PubMed PMID: 27131402. [DOI] [PubMed] [Google Scholar]
- 38.Pasquali M., Yu C., Coffee B. Laboratory diagnosis of galactosemia: a technical standard and guideline of the American college of medical genetics and genomics (ACMG) Genet. Med. 2018;20(1):3–11. doi: 10.1038/gim.2017.172. Epub 2017/10/26. PubMed PMID: 29261178. [DOI] [PubMed] [Google Scholar]
- 39.van den Akker-van Marle M.E., Dankert H.M., Verkerk P.H., Dankert-Roelse J.E. Cost-effectiveness of 4 neonatal screening strategies for cystic fibrosis. Pediatrics. 2006;118(3):896–905. doi: 10.1542/peds.2005-2782. PubMed PMID: 16950979. [DOI] [PubMed] [Google Scholar]
- 40.Kelly E., Greene C.M., Carroll T.P., McElvaney N.G., O'Neill S.J. α-1-antitrypsin deficiency. Respir. Med. 2010;104(6):763–772. doi: 10.1016/j.rmed.2010.01.016. Epub 2010/03/20. PubMed PMID: 20303723. [DOI] [PubMed] [Google Scholar]
- 41.Mattman A., Gilfix B.M., Chen S.X., DeMarco M.L., Kyle B.D., Parker M.L., et al. α-1-antitrypsin molecular testing in Canada: a seven year, multi-centre comparison. Clin. Biochem. 2020;81:27–33. doi: 10.1016/j.clinbiochem.2020.05.001. Epub 2020/05/06. PubMed PMID: 32387440. [DOI] [PubMed] [Google Scholar]
- 42.Abboud R.T., Nelson T.N., Jung B., Mattman A. α-1-antitrypsin deficiency: a clinical-genetic overview. Appl. Clin. Genet. 2011;4:55–65. doi: 10.2147/TACG.S10604. Epub 2011/03/31. PubMed PMID: 23776367; PubMed Central PMCID: PMCPMC3681178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franciosi A.N., Carroll T.P., McElvaney N.G. Pitfalls and caveats in α-1-antitrypsin deficiency testing: a guide for clinicians. Lancet Respir Med. 2019;7(12):1059–1067. doi: 10.1016/S2213-2600(19)30141-9. Epub 2019/07/16. PubMed PMID: 31324540. [DOI] [PubMed] [Google Scholar]
- 44.Snyder M.R., Katzmann J.A., Butz M.L., Wiley C., Yang P., Dawson D.B., et al. Diagnosis of α-1-antitrypsin deficiency: an algorithm of quantification, genotyping, and phenotyping. Clin. Chem. 2006;52(12):2236–2242. doi: 10.1373/clinchem.2006.072991. Epub 2006/10/19. PubMed PMID: 17053153. [DOI] [PubMed] [Google Scholar]
- 45.Miravitlles M., Dirksen A., Ferrarotti I., Koblizek V., Lange P., Mahadeva R., et al. European respiratory society statement: diagnosis and treatment of pulmonary disease. Eur. Respir. J. 2017;50(5) doi: 10.1183/13993003.00610-2017. Epub 2017/11/30. PubMed PMID: 29191952. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y., Snyder M.R., Zhu Y., Tostrud L.J., Benson L.M., Katzmann J.A., et al. Simultaneous phenotyping and quantification of α-1-antitrypsin by liquid chromatography-tandem mass spectrometry. Clin. Chem. 2011;57(8):1161–1168. doi: 10.1373/clinchem.2011.163006. Epub 2011/06/02. PubMed PMID: 21636698; PubMed Central PMCID: PMCPMC3275912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray J.D., Willrich M.A., Krowka M.J., Bobr A., Murray D.L., Halling K.C., et al. Liquid chromatography–tandem mass spectrometry–based α1-antitrypsin (AAT) testing: a retrospective evaluation of a clinical algorithm to detect AAT deficiency. Am. J. Clin. Pathol. 2020;155(4):547–552. doi: 10.1093/ajcp/aqaa149. [DOI] [PubMed] [Google Scholar]
- 48.Mahley R.W. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med (Berl). 2016;94(7):739–746. doi: 10.1007/s00109-016-1427-y. Epub 2016/06/09. PubMed PMID: 27277824; PubMed Central PMCID: PMCPMC4921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verghese P.B., Castellano J.M., Holtzman D.M. Apolipoprotein E in alzheimer's disease and other neurological disorders. Lancet Neurol. 2011;10(3):241–252. doi: 10.1016/S1474-4422(10)70325-2. PubMed PMID: 21349439; PubMed Central PMCID: PMCPMC3132088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Austin M.A., McKnight B., Edwards K.L., Bradley C.M., McNeely M.J., Psaty B.M., et al. Cardiovascular disease mortality in familial forms of hypertriglyceridemia: a 20-year prospective study. Circulation. 2000;101(24):2777–2782. doi: 10.1161/01.cir.101.24.2777. Epub 2000/06/22. PubMed PMID: 10859281. [DOI] [PubMed] [Google Scholar]
- 51.Mahley R.W., Huang Y., Rall S.C., Jr. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J. Lipid Res. 1999;40(11):1933–1949. Epub 1999/12/20. PubMed PMID: 10552997. [PubMed] [Google Scholar]
- 52.Raffaï R.L., McPherson R., Weisgraber K.H., Innerarity T.L., Rassart E., Bersot T.P., et al. Antibody phenotyping test for the human apolipoprotein E2 isoform. Clin. Chem. 2003;49(9):1524–1526. doi: 10.1373/49.9.1524. PubMed PMID: 12928236. [DOI] [PubMed] [Google Scholar]
- 53.Veiga S., Rodríguez-Martín A., Garcia-Ribas G., Arribas I., Menacho-Román M., Calero M. Validation of a novel and accurate ApoE4 assay for automated chemistry analyzers. Sci. Rep. 2020;10(1):2138. doi: 10.1038/s41598-020-58841-7. Epub 2020/02/07. PubMed PMID: 32034174; PubMed Central PMCID: PMCPMC7005722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wildsmith K.R., Han B., Bateman R.J. Method for the simultaneous quantitation of apolipoprotein E isoforms using tandem mass spectrometry. Anal. Biochem. 2009;395(1):116–118. doi: 10.1016/j.ab.2009.07.049. Epub 2009/08/03. PubMed PMID: 19653990; PubMed Central PMCID: PMCPMC2749897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon R., Girod M., Fonbonne C., Salvador A., Clément Y., Lantéri P., et al. Total ApoE and ApoE4 isoform assays in an alzheimer's disease case-control study by targeted mass spectrometry (n=669): a pilot assay for methionine-containing proteotypic peptides. Mol. Cell. Proteomics. 2012;11(11):1389–1403. doi: 10.1074/mcp.M112.018861. Epub 2012/08/23. PubMed PMID: 22918225; PubMed Central PMCID: PMCPMC3494189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M., Chen J., Turko I.V. 15N-Labeled full-length Apolipoprotein E4 as an internal standard for mass spectrometry quantification of apolipoprotein E isoforms. Anal. Chem. 2012;84(19):8340–8344. doi: 10.1021/ac3018873. Epub 2012/09/11. PubMed PMID: 22931427; PubMed Central PMCID: PMCPMC4038910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Morillo E., Nielsen H.M., Batruch I., Drabovich A.P., Begcevic I., Lopez M.F., et al. Assessment of peptide chemical modifications on the development of an accurate and precise multiplex selected reaction monitoring assay for apolipoprotein E isoforms. J. Proteome Res. 2014;13(2):1077–1087. doi: 10.1021/pr401060x. Epub 2014/01/14. PubMed PMID: 24392642. [DOI] [PubMed] [Google Scholar]
- 58.van den Broek I., Romijn F.P., Nouta J., van der Laarse A., Drijfhout J.W., Smit N.P., et al. Automated multiplex LC-MS/MS assay for quantifying serum apolipoproteins A-I, B, C-I, C-II, C-III, and E with qualitative apolipoprotein E phenotyping. Clin. Chem. 2016;62(1):188–197. doi: 10.1373/clinchem.2015.246702. Epub 2015/11/19. PubMed PMID: 26585923. [DOI] [PubMed] [Google Scholar]
- 59.Hirtz C., Vialaret J., Nouadje G., Schraen S., Benlian P., Mary S., et al. Development of new quantitative mass spectrometry and semi-automatic isofocusing methods for the determination of apolipoprotein E typing. Clin. Chim. Acta. 2016;454:33–38. doi: 10.1016/j.cca.2015.12.020. Epub 2015/12/19. PubMed PMID: 26707914. [DOI] [PubMed] [Google Scholar]
- 60.Jack C.R., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., et al. NIA-AA research framework: toward a biological definition of alzheimer's disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. PubMed PMID: 29653606; PubMed Central PMCID: PMCPMC5958625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Formichi P., Battisti C., Radi E., Federico A. Cerebrospinal fluid tau, amyloid-β, and phosphorylated-tau protein for the diagnosis of alzheimer's disease. J. Cell. Physiol. 2006;208(1):39–46. doi: 10.1002/jcp.20602. PubMed PMID: 16447254. [DOI] [PubMed] [Google Scholar]
- 62.Slemmon J.R., Shapiro A., Mercken M., Streffer J., Romano G., Andreasen N., et al. Impact of cerebrospinal fluid matrix on the detection of alzheimer's disease with Aβ42 and influence of disease on the total-aβ42/aβ40 ratio. J. Neurochem. 2015;135(5):1049–1058. doi: 10.1111/jnc.13297. Epub 2015/09/04. PubMed PMID: 26332787. [DOI] [PubMed] [Google Scholar]
- 63.DeMarco M.L., Nguyen Q., Fok A., Hsiung G.R., van der Gugten J.G. An automated clinical mass spectrometric method for identification and quantification of variant and wild-type Amyloid-β 1-40 and 1-42 peptides in CSF. Alzheimers Dement (Amst) 2020;12(1) doi: 10.1002/dad2.12036. Epub 2020/06/30. PubMed PMID: 32617385; PubMed Central PMCID: PMCPMC7327131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frei-Jones M. In: Benign Hematologic Disorders in Children: A Clinical Guide. Kamat D.M., Frei-Jones M., editors. Springer International Publishing; Cham: 2021. Newborn screening for hemoglobinopathies; pp. 313–321. [Google Scholar]
- 65.Eastman J.W., Wong R., Liao C.L., Morales D.R. Automated HPLC screening of newborns for sickle cell anemia and other hemoglobinopathies. Clin. Chem. 1996;42(5):704–710. PubMed PMID: 8653895. [PubMed] [Google Scholar]
- 66.Lorey F., Cunningham G., Shafer F., Lubin B., Vichinsky E. Universal screening for hemoglobinopathies using high-performance liquid chromatography: clinical results of 2.2 million screens. Eur. J. Hum. Genet. 1994;2(4):262–271. doi: 10.1159/000472370. PubMed PMID: 7704556. [DOI] [PubMed] [Google Scholar]
- 67.Campbell M., Henthorn J.S., Davies S.C. Evaluation of cation-exchange HPLC compared with isoelectric focusing for neonatal hemoglobinopathy screening. Clin. Chem. 1999;45(7):969–975. PubMed PMID: 10388471. [PubMed] [Google Scholar]
- 68.Galanello R., Barella S., Gasperini D., Perseu L., Paglietti E., Sollaino C., et al. Evaluation of an automatic HPLC analyser for thalassemia and haemoglobin variants screening. J. Automat. Chem. 1995;17(2):73–76. doi: 10.1155/S1463924695000125. PubMed PMID: 18925016; PubMed Central PMCID: PMCPMC2548064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Higgins T., Mack M., Khajuria A. Comparison of two methods for the quantification and identification of hemoglobin variants. Clin. Biochem. 2009;42(7–8):701–705. doi: 10.1016/j.clinbiochem.2009.01.004. Epub 2009/01/23. PubMed PMID: 19318037. [DOI] [PubMed] [Google Scholar]
- 70.Daniel Y.A., Turner C., Haynes R.M., Hunt B.J., Dalton R.N. Rapid and specific detection of clinically significant haemoglobinopathies using electrospray mass spectrometry-mass spectrometry. Br. J. Haematol. 2005;130(4):635–643. doi: 10.1111/j.1365-2141.2005.05646.x. PubMed PMID: 16098080. [DOI] [PubMed] [Google Scholar]
- 71.Boemer F., Ketelslegers O., Minon J.M., Bours V., Schoos R. Newborn screening for sickle cell disease using tandem mass spectrometry. Clin. Chem. 2008;54(12):2036–2041. doi: 10.1373/clinchem.2008.106369. Epub 2008/10/02. PubMed PMID: 18832473. [DOI] [PubMed] [Google Scholar]
- 72.Moat S.J., Rees D., King L., Ifederu A., Harvey K., Hall K., et al. Newborn blood spot screening for sickle cell disease by using tandem mass spectrometry: implementation of a protocol to identify only the disease states of sickle cell disease. Clin. Chem. 2014;60(2):373–380. doi: 10.1373/clinchem.2013.210948. Epub 2013/10/24. PubMed PMID: 24158758. [DOI] [PubMed] [Google Scholar]
- 73.Boemer F., Cornet Y., Libioulle C., Segers K., Bours V., Schoos R. 3-years experience review of neonatal screening for hemoglobin disorders using tandem mass spectrometry. Clin. Chim. Acta. 2011;412(15–16):1476–1479. doi: 10.1016/j.cca.2011.04.031. Epub 2011/05/04. PubMed PMID: 21569767. [DOI] [PubMed] [Google Scholar]
- 74.Moat S.J., Rees D., George R.S., King L., Dodd A., Ifederu A., et al. Newborn screening for sickle cell disorders using tandem mass spectrometry: three years' experience of using a protocol to detect only the disease states. Ann. Clin. Biochem. 2017;54(5):601–611. doi: 10.1177/0004563217713788. Epub 2017/07/17. PubMed PMID: 28656815. [DOI] [PubMed] [Google Scholar]
- 75.Sipe J.D., Benson M.D., Buxbaum J.N., Ikeda S.I., Merlini G., Saraiva M.J., et al. Amyloid fibril proteins and amyloidosis: chemical identification and clinical classification international society of amyloidosis 2016 nomenclature guidelines. Amyloid. 2016;23(4):209–213. doi: 10.1080/13506129.2016.1257986. Epub 2016/11/24. PubMed PMID: 27884064. [DOI] [PubMed] [Google Scholar]
- 76.Maurer M.S., Bokhari S., Damy T., Dorbala S., Drachman B.M., Fontana M., et al. Expert consensus recommendations for the suspicion and diagnosis of transthyretin cardiac amyloidosis. Circ Heart Fail. 2019;12(9) doi: 10.1161/CIRCHEARTFAILURE.119.006075. Epub 2019/09/04. PubMed PMID: 31480867; PubMed Central PMCID: PMCPMC6736650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gertz M.A. Hereditary ATTR amyloidosis: burden of illness and diagnostic challenges. Am. J. Manag. Care. 2017;23(7 Suppl):S107–S112. PubMed PMID: 28978215. [PubMed] [Google Scholar]
- 78.Schönland S.O., Hegenbart U., Bochtler T., Mangatter A., Hansberg M., Ho A.D., et al. Immunohistochemistry in the classification of systemic forms of amyloidosis: a systematic investigation of 117 patients. Blood. 2012;119(2):488–493. doi: 10.1182/blood-2011-06-358507. Epub 2011/11/21. PubMed PMID: 22106346. [DOI] [PubMed] [Google Scholar]
- 79.Satoskar A.A., Burdge K., Cowden D.J., Nadasdy G.M., Hebert L.A., Nadasdy T. Typing of amyloidosis in renal biopsies: diagnostic pitfalls. Arch. Pathol. Lab Med. 2007;131(6):917–922. doi: 10.1043/1543-2165(2007)131[917:TOAIRB]2.0.CO;2. PubMed PMID: 17550319. [DOI] [PubMed] [Google Scholar]
- 80.Satoskar A.A., Efebera Y., Hasan A., Brodsky S., Nadasdy G., Dogan A., et al. Strong transthyretin immunostaining: potential pitfall in cardiac amyloid typing. Am. J. Surg. Pathol. 2011;35(11):1685–1690. doi: 10.1097/PAS.0b013e3182263d74. PubMed PMID: 21945954; PubMed Central PMCID: PMCPMC4061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vrana J.A., Theis J.D., Dasari S., Mereuta O.M., Dispenzieri A., Zeldenrust S.R., et al. Clinical diagnosis and typing of systemic amyloidosis in subcutaneous fat aspirates by mass spectrometry-based proteomics. Haematologica. 2014;99(7):1239–1247. doi: 10.3324/haematol.2013.102764. Epub 2014/04/18. PubMed PMID: 24747948; PubMed Central PMCID: PMCPMC4077087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vrana J.A., Gamez J.D., Madden B.J., Theis J.D., Bergen H.R., Dogan A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114(24):4957–4959. doi: 10.1182/blood-2009-07-230722. Epub 2009/10/01. PubMed PMID: 19797517. [DOI] [PubMed] [Google Scholar]
- 83.Dasari S., Theis J.D., Vrana J.A., Rech K.L., Dao L.N., Howard M.T., et al. Amyloid typing by mass spectrometry in clinical practice: a comprehensive review of 16,175 samples. Mayo Clin. Proc. 2020;95(9):1852–1864. doi: 10.1016/j.mayocp.2020.06.029. PubMed PMID: 32861330. [DOI] [PubMed] [Google Scholar]
- 84.Gilbertson J.A., Theis J.D., Vrana J.A., Lachmann H., Wechalekar A., Whelan C., et al. A comparison of immunohistochemistry and mass spectrometry for determining the amyloid fibril protein from formalin-fixed biopsy tissue. J. Clin. Pathol. 2015;68(4):314–317. doi: 10.1136/jclinpath-2014-202722. Epub 2015/01/30. PubMed PMID: 25637636. [DOI] [PubMed] [Google Scholar]
- 85.Mills J.R., Kohlhagen M.C., Dasari S., Vanderboom P.M., Kyle R.A., Katzmann J.A., et al. Comprehensive assessment of M-proteins using nanobody enrichment coupled to MALDI-TOF mass spectrometry. Clin. Chem. 2016;62(10):1334–1344. doi: 10.1373/clinchem.2015.253740. Epub 2016/08/18. PubMed PMID: 27540026. [DOI] [PubMed] [Google Scholar]
- 86.Barnidge D.R., Dasari S., Botz C.M., Murray D.H., Snyder M.R., Katzmann J.A., et al. Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. J. Proteome Res. 2014;13(3):1419–1427. doi: 10.1021/pr400985k. Epub 2014/02/11. PubMed PMID: 24467232. [DOI] [PubMed] [Google Scholar]
- 87.Brossard J.H., Cloutier M., Roy L., Lepage R., Gascon-Barré M., D'Amour P. Accumulation of a non-(1-84) molecular form of parathyroid hormone (PTH) detected by intact PTH assay in renal failure: importance in the interpretation of PTH values. J. Clin. Endocrinol. Metab. 1996;81(11):3923–3929. doi: 10.1210/jcem.81.11.8923839. Epub 1996/11/01. PubMed PMID: 8923839. [DOI] [PubMed] [Google Scholar]
- 88.D'Amour P. Circulating parathyroid hormone molecular forms: what we know and what we don't. Kidney Int. Suppl. 2006;(102):S29–S33. doi: 10.1038/sj.ki.5001599. Epub 2006/07/01. PubMed PMID: 16810307. [DOI] [PubMed] [Google Scholar]
- 89.Zhang C.-X., Weber B.V., Thammavong J., Grover T.A., Wells D.S. Identification of carboxyl-terminal peptide fragments of parathyroid hormone in human plasma at low-picomolar levels by mass spectrometry. Anal. Chem. 2006;78(5):1636–1643. doi: 10.1021/ac051711o. [DOI] [PubMed] [Google Scholar]
- 90.Kritmetapak K., Losbanos L.A., Hines J.M., O'Grady K.L., Ulmer C.Z., Vesper H.W., et al. Chemical characterization and quantification of circulating intact PTH and PTH fragments by high-resolution mass spectrometry in chronic renal failure. Clin. Chem. 2021;67(6):843–853. doi: 10.1093/clinchem/hvab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Habener J.F., Segre G.V., Powell D., Murray T.M., Potts J.T. Immunoreactive parathyroid hormone in circulation of man. Nat. New Biol. 1972;238(83):152–154. doi: 10.1038/newbio238152a0. [DOI] [PubMed] [Google Scholar]
- 92.Pines M., Adams A.E., Stueckle S., Bessalle R., Rashti-Behar V., Chorev M., et al. Generation and characterization of human kidney cell lines stably expressing recombinant human PTH/PTHrP receptor: lack of interaction with a C-terminal human PTH peptide. Endocrinology. 1994;135(4):1713–1716. doi: 10.1210/endo.135.4.7925136. Epub 1994/10/01. PubMed PMID: 7925136. [DOI] [PubMed] [Google Scholar]
- 93.Tregear G.W., Van Rietschoten J., Greene E., Keutmann H.T., Niall H.D., Reit B., et al. Bovine parathyroid hormone: minimum chain length of synthetic peptide required for biological activity. Endocrinology. 1973;93(6):1349–1353. doi: 10.1210/endo-93-6-1349. Epub 1973/12/01. PubMed PMID: 4796246. [DOI] [PubMed] [Google Scholar]
- 94.Gao P., Scheibel S., D'Amour P., John M.R., Rao S.D., Schmidt-Gayk H., et al. Development of a novel immunoradiometric assay exclusively for biologically active whole parathyroid hormone 1-84: implications for improvement of accurate assessment of parathyroid function. J. Bone Miner. Res. 2001;16(4):605–614. doi: 10.1359/jbmr.2001.16.4.605. Epub 2001/04/24. PubMed PMID: 11315988. [DOI] [PubMed] [Google Scholar]
- 95.Kumar V., Barnidge D.R., Chen L.S., Twentyman J.M., Cradic K.W., Grebe S.K., et al. Quantification of serum 1-84 parathyroid hormone in patients with hyperparathyroidism by immunocapture in situ digestion liquid chromatography-tandem mass spectrometry. Clin. Chem. 2010;56(2):306–313. doi: 10.1373/clinchem.2009.134643. Epub 2009/12/17. PubMed PMID: 20007860. [DOI] [PubMed] [Google Scholar]
- 96.Smit M.A., van Kinschot C.M.J., van der Linden J., van Noord C., Kos S. Clinical guidelines and PTH measurement: does assay generation matter? Endocr. Rev. 2019;40(6):1468–1480. doi: 10.1210/er.2018-00220. [DOI] [PubMed] [Google Scholar]
- 97.Heublein D.M., Huntley B.K., Boerrigter G., Cataliotti A., Sandberg S.M., Redfield M.M., et al. Immunoreactivity and guanosine 3',5'-cyclic monophosphate activating actions of various molecular forms of human B-type natriuretic peptide. Hypertension. 2007;49(5):1114–1119. doi: 10.1161/hypertensionaha.106.081083. Epub 2007/03/21. PubMed PMID: 17372040. [DOI] [PubMed] [Google Scholar]
- 98.Liang F., O'Rear J., Schellenberger U., Tai L., Lasecki M., Schreiner G.F., et al. Evidence for functional heterogeneity of circulating B-type natriuretic peptide. J. Am. Coll. Cardiol. 2007;49(10):1071–1078. doi: 10.1016/j.jacc.2006.10.063. Epub 2007/03/14. PubMed PMID: 17349887. [DOI] [PubMed] [Google Scholar]
- 99.Luckenbill K.N., Christenson R.H., Jaffe A.S., Mair J., Ordonez-Llanos J., Pagani F., et al. Cross-reactivity of BNP, NT-proBNP, and proBNP in commercial BNP and NT-proBNP assays: preliminary observations from the IFCC committee for standardization of markers of cardiac damage. Clin. Chem. 2008;54(3):619–621. doi: 10.1373/clinchem.2007.097998. Epub 2008/03/04. PubMed PMID: 18310156. [DOI] [PubMed] [Google Scholar]
- 100.Niederkofler E.E., Kiernan U.A., O'Rear J., Menon S., Saghir S., Protter A.A., et al. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Circ Heart Fail. 2008;1(4):258–264. doi: 10.1161/circheartfailure.108.790774. Epub 2009/10/08. PubMed PMID: 19808300. [DOI] [PubMed] [Google Scholar]
- 101.Miller W.L., Phelps M.A., Wood C.M., Schellenberger U., Van Le A., Perichon R., et al. Comparison of mass spectrometry and clinical assay measurements of circulating fragments of B-type natriuretic peptide in patients with chronic heart failure. Circ Heart Fail. 2011;4(3):355–360. doi: 10.1161/circheartfailure.110.960260. Epub 2011/02/05. PubMed PMID: 21292992. [DOI] [PubMed] [Google Scholar]
- 102.Torma A.F., Groves K., Biesenbruch S., Mussell C., Reid A., Ellison S., et al. A candidate liquid chromatography mass spectrometry reference method for the quantification of the cardiac marker 1-32 B-type natriuretic peptide. Clin. Chem. Lab. Med. 2017;55(9):1397–1406. doi: 10.1515/cclm-2016-1054. [DOI] [PubMed] [Google Scholar]
- 103.Xiao P., Li H., Li X., Song D. Analytical barriers in clinical B-type natriuretic peptide measurement and the promising analytical methods based on mass spectrometry technology. Clin. Chem. Lab. Med. 2019;57(7):954–966. doi: 10.1515/cclm-2018-0956. [DOI] [PubMed] [Google Scholar]
- 104.Amsterdam E.A., Wenger N.K., Brindis R.G., Casey D.E., Jr., Ganiats T.G., Holmes D.R., Jr., et al. AHA/ACC guideline for the management of patients with non-ST-elevation acute coronary syndromes: a report of the American college of cardiology/American heart association task force on practice guidelines. Circulation. 2014;130(25):e344–426. doi: 10.1161/cir.0000000000000134. Epub 2014/09/25. PubMed PMID: 25249585. [DOI] [PubMed] [Google Scholar]
- 105.Shi Q., Ling M., Zhang X., Zhang M., Kadijevic L., Liu S., et al. Degradation of cardiac troponin I in serum complicates comparisons of cardiac troponin I assays. Clin. Chem. 1999;45(7):1018–1025. Epub 1999/07/01. PubMed PMID: 10388478. [PubMed] [Google Scholar]
- 106.Streng A.S., de Boer D., van Doorn W.P., Bouwman F.G., Mariman E.C., Bekers O., et al. Identification and characterization of cardiac troponin T fragments in serum of patients suffering from acute myocardial infarction. Clin. Chem. 2017;63(2):563–572. doi: 10.1373/clinchem.2016.261511. Epub 2016/12/13. PubMed PMID: 27940450. [DOI] [PubMed] [Google Scholar]
- 107.Guy M.J., Chen Y.-C., Clinton L., Zhang H., Zhang J., Dong X., et al. The impact of antibody selection on the detection of cardiac troponin I. Clin. Chim. Acta. 2013;420:82–88. doi: 10.1016/j.cca.2012.10.034. Epub 2012/10/26. PubMed PMID: 23107929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cosmi B., Legnani C., Tosetto A., Pengo V., Ghirarduzzi A., Testa S., et al. Sex, age and normal post-anticoagulation D-dimer as risk factors for recurrence after idiopathic venous thromboembolism in the prolong study extension. J. Thromb. Haemostasis. 2010;8(9):1933–1942. doi: 10.1111/j.1538-7836.2010.03955.x. Epub 2010/06/18. PubMed PMID: 20553388. [DOI] [PubMed] [Google Scholar]
- 109.Galle C., Papazyan J.P., Miron M.J., Slosman D., Bounameaux H., Perrier A. Prediction of pulmonary embolism extent by clinical findings, D-dimer level and deep vein thrombosis shown by ultrasound. Thromb. Haemostasis. 2001;86(5):1156–1160. Epub 2002/01/31. PubMed PMID: 11816700. [PubMed] [Google Scholar]
- 110.Tick L.W., Nijkeuter M., Kramer M.H.H., Hovens M.M.C., Büller H.R., Leebeek F.W.G., et al. High D-dimer levels increase the likelihood of pulmonary embolism. J. Intern. Med. 2008;264(2):195–200. doi: 10.1111/j.1365-2796.2008.01972.x. [DOI] [PubMed] [Google Scholar]
- 111.Voves C., Wuillemin W.A., Zeerleder S. International society on thrombosis and haemostasis score for overt disseminated intravascular coagulation predicts organ dysfunction and fatality in sepsis patients. Blood Coagul. Fibrinolysis. 2006;17(6):445–451. doi: 10.1097/01.mbc.0000240916.63521.2e. Epub 2006/08/15. PubMed PMID: 16905947. [DOI] [PubMed] [Google Scholar]
- 112.Riley R.S., Gilbert A.R., Dalton J.B., Pai S., McPherson R.A. Widely used types and clinical applications of D-dimer assay. Lab. Med. 2016;47(2):90–102. doi: 10.1093/labmed/lmw001. Epub 2016/03/27. PubMed PMID: 27016528. [DOI] [PubMed] [Google Scholar]
- 113.Adam S.S., Key N.S., Greenberg C.S. D-dimer antigen: current concepts and future prospects. Blood. 2009;113(13):2878–2887. doi: 10.1182/blood-2008-06-165845. Epub 2008/11/15. PubMed PMID: 19008457. [DOI] [PubMed] [Google Scholar]
- 114.Demple C.E. D-dimer testing and venous thromboembolism: four view points. J. Thromb. Haemostasis. 2005;3(2):377–379. doi: 10.1111/j.1538-7836.2005.01101.x. [DOI] [PubMed] [Google Scholar]
- 115.Lin C., Chen Y., Chen B., Zheng K., Luo X., Lin F. D-dimer combined with fibrinogen predicts the risk of venous thrombosis in fracture patients. Emergency Medicine International. 2020. 2020 doi: 10.1155/2020/1930405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang W., Walker N.D., Zhu L.J., Wu W., Ge L., Gutstein D.E., et al. Quantification of circulating D-dimer by peptide immunoaffinity enrichment and tandem mass spectrometry. Anal. Chem. 2012;84(15):6891–6898. doi: 10.1021/ac301494d. Epub 2012/07/14. PubMed PMID: 22788854. [DOI] [PubMed] [Google Scholar]
- 117.Shi J., Dhaliwal P., Zi Zheng Y., Wong T., Straseski J.A., Cervinski M.A., et al. An intact ACTH LC-MS/MS assay as an arbiter of clinically discordant immunoassay results. Clin. Chem. 2019;65(11):1397–1404. doi: 10.1373/clinchem.2019.306365. [DOI] [PubMed] [Google Scholar]
- 118.Yener S., Demir L., Demirpence M., Mahmut Baris M., Simsir I.Y., Ozisik S., et al. Interference in ACTH immunoassay negatively impacts the management of subclinical hypercortisolism. Endocrine. 2017;56(2):308–316. doi: 10.1007/s12020-017-1268-7. Epub 2017/03/02. PubMed PMID: 28247312. [DOI] [PubMed] [Google Scholar]
- 119.Gleeson H.K., Walker B.R., Seckl J.R., Padfield P.L. Ten years on: safety of short synacthen tests in assessing adrenocorticotropin deficiency in clinical practice. J. Clin. Endocrinol. Metab. 2003;88(5):2106–2111. doi: 10.1210/jc.2002-020969. Epub 2003/05/03. PubMed PMID: 12727962. [DOI] [PubMed] [Google Scholar]
- 120.Funnell C., Feldman H.H., Mackenzie I.R.A., DeMarco M.L. Applying the alzheimer disease ATN diagnostic framework in atypical dementia. Alzheimer Dis. Assoc. Disord. 2020;34(4):357–359. doi: 10.1097/WAD.0000000000000372. PubMed PMID: 33215886. [DOI] [PubMed] [Google Scholar]
- 121.Mattsson N., Zetterberg H., Janelidze S., Insel P.S., Andreasson U., Stomrud E., et al. Plasma tau in alzheimer's disease. Neurology. 2016;87(17):1827–1835. doi: 10.1212/WNL.0000000000003246. Epub 2016/09/30. PubMed PMID: 27694257; PubMed Central PMCID: PMCPMC5089525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hanes J., Kovac A., Kvartsberg H., Kontsekova E., Fialova L., Katina S., et al. Evaluation of a novel immunoassay to detect phosphorylated-tau Thr217 in the CSF to distinguish alzheimer's disease from other dementias. Neurology. 2020;95(22):e3026–e3035. doi: 10.1212/WNL.0000000000010814. Epub 2020/09/24. PubMed PMID: 32973122; PubMed Central PMCID: PMCPMC7734919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barthélemy N.R., Bateman R.J., Hirtz C., Marin P., Becher F., Sato C., et al. Cerebrospinal fluid phosphorylated-tau T217 outperforms T181 as a biomarker for the differential diagnosis of Alzheimer's disease and PET amyloid-positive patient identification. Alzheimer's Res. Ther. 2020;12(1):26. doi: 10.1186/s13195-020-00596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ercan E., Eid S., Weber C., Kowalski A., Bichmann M., Behrendt A., et al. A validated antibody panel for the characterization of tau post-translational modifications. Mol. Neurodegener. 2017;12(1):87. doi: 10.1186/s13024-017-0229-1. Epub 2017/11/21. PubMed PMID: 29157277; PubMed Central PMCID: PMCPMC5697095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McAvoy T., Lassman M.E., Spellman D.S., Ke Z., Howell B.J., Wong O., et al. Quantification of tau in cerebrospinal fluid by immunoaffinity enrichment and tandem mass spectrometry. Clin. Chem. 2014;60(4):683–689. doi: 10.1373/clinchem.2013.216515. Epub 2014/02/24. PubMed PMID: 24566260. [DOI] [PubMed] [Google Scholar]
- 126.Russell C.L., Mitra V., Hansson K., Blennow K., Gobom J., Zetterberg H., et al. Comprehensive quantitative profiling of tau and phosphorylated tau peptides in cerebrospinal fluid by mass spectrometry provides new biomarker candidates. J Alzheimers Dis. 2017;55(1):303–313. doi: 10.3233/JAD-160633. PubMed PMID: 27636850. [DOI] [PubMed] [Google Scholar]
- 127.Barthélemy N.R., Mallipeddi N., Moiseyev P., Sato C., Bateman R.J. Tau phosphorylation rates measured by mass spectrometry differ in the intracellular brain vs. Extracellular cerebrospinal fluid compartments and are differentially affected by alzheimer's disease. Front. Aging Neurosci. 2019;11:121. doi: 10.3389/fnagi.2019.00121. Epub 2019/05/21. PubMed PMID: 31178717; PubMed Central PMCID: PMCPMC6537657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barthélemy N.R., Horie K., Sato C., Bateman R.J. Blood plasma phosphorylated-tau isoforms track central nervous system change in Alzheimer's disease. J. Exp. Med. 2020;217(11) doi: 10.1084/jem.20200861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu R.M., Hwang Y.C., Liu I.J., Lee C.C., Tsai H.Z., Li H.J., et al. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020;27(1):1. doi: 10.1186/s12929-019-0592-z. Epub 2020/01/02. PubMed PMID: 31894001; PubMed Central PMCID: PMCPMC6939334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Administration USAFaD. Q6B specifications: test procedures and acceptance criteria for biotechnological/biological products 1999 [cited 2021 25 may] https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q6b-specifications-test-procedures-and-acceptance-criteria-biotechnologicalbiological-products Available from:
- 131.Siemiatkoski J., Lyubarskaya Y., Houde D., Tep S., Mhatre R. A comparison of three techniques for quantitative carbohydrate analysis used in characterization of therapeutic antibodies. Carbohydr. Res. 2006;341(3):410–419. doi: 10.1016/j.carres.2005.11.024. Epub 2005/12/27. PubMed PMID: 16378604. [DOI] [PubMed] [Google Scholar]
- 132.Jacobs J.F., Wevers R.A., Lefeber D.J., van Scherpenzeel M. Fast, robust and high-resolution glycosylation profiling of intact monoclonal IgG antibodies using nanoLC-chip-QTOF. Clin. Chim. Acta. 2016;461:90–97. doi: 10.1016/j.cca.2016.07.015. Epub 2016/07/22. PubMed PMID: 27458127. [DOI] [PubMed] [Google Scholar]
- 133.Gillery P. A history of HbA1c through clinical chemistry and laboratory medicine. Clin. Chem. Lab. Med. 2013;51(1):65–74. doi: 10.1515/cclm-2012-0548. PubMed PMID: 22992284. [DOI] [PubMed] [Google Scholar]
- 134.Can M., Güven B., Demir Akca A.S. Effect of hemoglobin F and A2 on hemoglobin A1c determined by cation exchange HPLC. J. Lab. Med. 2019;43(5):265–268. doi: 10.1515/labmed-2019-0040. [DOI] [Google Scholar]
- 135.Sterling B., Griffith M.L., Shajani-Yi Z. A patient with discordant hemoglobin A1c results. Clin. Chem. 2020;66(7):990–991. doi: 10.1093/clinchem/hvaa060. PubMed PMID: 32628762. [DOI] [PubMed] [Google Scholar]
- 136.Rohlfing C.L., Connolly S.M., England J.D., Hanson S.E., Moellering C.M., Bachelder J.R., et al. The effect of elevated fetal hemoglobin on hemoglobin A1c results: five common hemoglobin A1c methods compared with the IFCC reference method. Am. J. Clin. Pathol. 2008;129(5):811–814. doi: 10.1309/YFVTUD0GHJF7D16H. PubMed PMID: 18426743. [DOI] [PubMed] [Google Scholar]
- 137.Bry L., Chen P.C., Sacks D.B. Effects of hemoglobin variants and chemically modified derivatives on assays for glycohemoglobin. Clin. Chem. 2001;47(2):153–163. PubMed PMID: 11159762. [PubMed] [Google Scholar]
- 138.Schechter A.N. Hemoglobin research and the origins of molecular medicine. Blood. 2008;112(10):3927–3938. doi: 10.1182/blood-2008-04-078188. PubMed PMID: 18988877; PubMed Central PMCID: PMCPMC2581994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carver M.F., Huisman T.H. International hemoglobin information center variant list. Hemoglobin. 1996;20(3):213. doi: 10.3109/03630269609027930. PubMed PMID: 8854131. [DOI] [PubMed] [Google Scholar]
- 140.Kobold U., Jeppsson J.O., Dülffer T., Finke A., Hoelzel W., Miedema K. Candidate reference methods for hemoglobin A1c based on peptide mapping. Clin. Chem. 1997;43(10):1944–1951. PubMed PMID: 9342017. [PubMed] [Google Scholar]
- 141.Jeppsson J.O., Kobold U., Barr J., Finke A., Hoelzel W., Hoshino T., et al. Approved IFCC reference method for the measurement of HbA1c in human blood. Clin. Chem. Lab. Med. 2002;40(1):78–89. doi: 10.1515/CCLM.2002.016. PubMed PMID: 11916276. [DOI] [PubMed] [Google Scholar]
- 142.Krantz S.B. Erythropoietin. Blood. 1991;77(3):419–434. PubMed PMID: 1991159. [PubMed] [Google Scholar]
- 143.WADA Science WADA technical document – TD2021EPO 2021 [cited 2021 25 may] https://www.wada-ama.org/en/resources/science-medicine/td2021epo Available from:
- 144.Reichel C., Kulovics R., Jordan V., Watzinger M., Geisendorfer T. SDS-PAGE of recombinant and endogenous erythropoietins: benefits and limitations of the method for application in doping control. Drug Test. Anal. 2009;1(1):43–50. doi: 10.1002/dta.10. PubMed PMID: 20355158. [DOI] [PubMed] [Google Scholar]
- 145.Stübiger G., Marchetti M., Nagano M., Reichel C., Gmeiner G., Allmaier G. Characterisation of intact recombinant human erythropoietins applied in doping by means of planar gel electrophoretic techniques and matrix-assisted laser desorption/ionisation linear time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom.: RCM (Rapid Commun. Mass Spectrom.) 2005;19(5):728–742. doi: 10.1002/rcm.1830. PubMed PMID: 15700237. [DOI] [PubMed] [Google Scholar]
- 146.Harazono A., Hashii N., Kuribayashi R., Nakazawa S., Kawasaki N. Mass spectrometric glycoform profiling of the innovator and biosimilar erythropoietin and darbepoetin by LC/ESI-MS. J. Pharmaceut. Biomed. Anal. 2013;83:65–74. doi: 10.1016/j.jpba.2013.04.031. Epub 2013/04/29. PubMed PMID: 23708432. [DOI] [PubMed] [Google Scholar]
- 147.Falck D., Haberger M., Plomp R., Hook M., Bulau P., Wuhrer M., et al. Affinity purification of erythropoietin from cell culture supernatant combined with MALDI-TOF-MS analysis of erythropoietin N-glycosylation. Sci. Rep. 2017;7(1):5324. doi: 10.1038/s41598-017-05641-1. Epub 2017/07/13. PubMed PMID: 28706253; PubMed Central PMCID: PMCPMC5509673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Guan F., Uboh C.E., Soma L.R., Birksz E., Chen J. Identification of darbepoetin alfa in human plasma by liquid chromatography coupled to mass spectrometry for doping control. Int. J. Sports Med. 2009;30(2):80–86. doi: 10.1055/s-0028-1103283. Epub 2009/01/28. PubMed PMID: 19177313. [DOI] [PubMed] [Google Scholar]
- 149.Vogel M., Blobel M., Thomas A., Walpurgis K., Schänzer W., Reichel C., et al. Isolation, enrichment, and analysis of erythropoietins in anti-doping analysis by receptor-coated magnetic beads and liquid chromatography-mass spectrometry. Anal. Chem. 2014;86(24):12014–12021. doi: 10.1021/ac5024765. Epub 2014/12/05. PubMed PMID: 25423444. [DOI] [PubMed] [Google Scholar]
- 150.Willrich M.A., Murray D.L., Barnidge D.R., Ladwig P.M., Snyder M.R. Quantitation of infliximab using clonotypic peptides and selective reaction monitoring by LC-MS/MS. Int. Immunopharm. 2015;28(1):513–520. doi: 10.1016/j.intimp.2015.07.007. Epub 2015/07/25. PubMed PMID: 26210595. [DOI] [PubMed] [Google Scholar]
- 151.van der Gugten J.G., Bressler B., DeMarco M.L. An automated mass spectrometric blood test for therapeutic drug monitoring of infliximab. Clinic. Mass Spectrometry. 2019;12:16–22. doi: 10.1016/j.clinms.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ladwig P.M., Barnidge D.R., Willrich M.A.V. Mass spectrometry approaches for identification and quantitation of therapeutic monoclonal antibodies in the clinical laboratory. Clin. Vaccine Immunol. 2017;24(5) doi: 10.1128/CVI.00545-16. e00545-16. PubMed PMID: 28274937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Owen W.E., Roberts W.L. Cross-reactivity of three recombinant insulin analogs with five commercial insulin immunoassays. Clin. Chem. 2004;50(1):257–259. doi: 10.1373/clinchem.2003.026625. PubMed PMID: 14709671. [DOI] [PubMed] [Google Scholar]
- 154.Heurtault B., Reix N., Meyer N., Gasser F., Wendling M.J., Ratomponirina C., et al. Extensive study of human insulin immunoassays: promises and pitfalls for insulin analogue detection and quantification. Clin. Chem. Lab. Med. 2014;52(3):355–362. doi: 10.1515/cclm-2013-0427. PubMed PMID: 24150210. [DOI] [PubMed] [Google Scholar]
- 155.Thevis M., Thomas A., Delahaut P., Bosseloir A., Schänzer W. Qualitative determination of synthetic analogues of insulin in human plasma by immunoaffinity purification and liquid chromatography-tandem mass spectrometry for doping control purposes. Anal. Chem. 2005;77(11):3579–3585. doi: 10.1021/ac050066i. PubMed PMID: 15924392. [DOI] [PubMed] [Google Scholar]
- 156.Van Der Gugten J.G., Wong S., Holmes D.T. Quantitation of insulin analogues in serum using immunoaffinity extraction, liquid chromatography, and tandem mass spectrometry. Methods Mol. Biol. 2016;1378:119–130. doi: 10.1007/978-1-4939-3182-8_14. PubMed PMID: 26602124. [DOI] [PubMed] [Google Scholar]
- 157.Holmes D.T., Romney M.G., Angel P., DeMarco M.L. Proteomic applications in pathology and laboratory medicine: present state and future prospects. Clin. Biochem. 2020;82:12–20. doi: 10.1016/j.clinbiochem.2020.05.007. [DOI] [PubMed] [Google Scholar]