Summary
This protocol illustrates the use of an in vitro assay to study the cleavage of the IMPA1 3′UTR by the endonuclease Ago2 in sympathetic neurons. The procedure includes the preparation of cytoplasmic protein extracts and also describes the synthesis and labeling of the RNA probe. The protocol can be applied to other cell systems, RNA transcripts, and endonucleases to confirm the role of known cleavage site(s) and cleavage proteins, or to investigate new ones.
For complete details on the use and execution of this protocol, please refer to Andreassi et al. (2021).
Subject areas: Molecular Biology, Neuroscience, Chemistry
Graphical abstract

Highlights
-
•
Describes cleavage assay of the IMPA1 3′UTR by the endonuclease Ago2
-
•
Use of radionuclides enables high sensitivity
-
•
Adaptable to other RNA sequences and endonucleases
This protocol illustrates the use of an in vitro assay to study the cleavage of the IMPA1 3′UTR by the endonuclease Ago2 in sympathetic neurons. The procedure includes the preparation of cytoplasmic protein extracts and also describes the synthesis and labeling of the RNA probe. The protocol can be applied to other cell systems, RNA transcripts, and endonucleases to confirm the role of known cleavage site(s) and cleavage proteins, or to investigate new ones.
Before you begin
Post-transcriptional endonucleolytic cleavage is a widespread processing mechanism that controls cytoplasmic mRNA metabolism in a variety of conditions, including miRNA-mediated silencing (Jin et al., 2021), mRNA decay (Harigaya and Parker, 2010), mRNA targeting by viral endonucleases (Rodriguez et al., 2021) and generation of non-coding RNA from mRNA (Andreassi et al., 2018). We have modified and integrated previously published protocols (Miyoshi et al., 2008; Jenal et al., 2012) to develop an in vitro assay to study IMPA1 3′UTR cleavage by the endonuclease Ago2, in the presence of cytoplasmic extracts from sympathetic neurons. The protocol includes preliminary steps to prepare cytoplasmic extracts, and to synthesize and label the RNA probe.
Cytoplasmic extracts from sympathetic neurons
Timing: 5 days
This step describes the preparation of cytoplasmic protein extracts that will be added to the cleavage reaction mix.
Note: Mechanical force and hypotonic cell lysis can be used to separate cytosolic and nuclear proteins fractions (DeCaprio and Kohl, 2019). Please, note that optimization of lysis conditions may be necessary depending on the cell type. We routinely use the NE-PER kit for Nuclear and cytoplasmic extractions as it ensures reproducible results with both cell lines (N2A cells, HEK293 cells, PC12 cells) and primary neurons (cortical neurons, sympathetic neurons).
CRITICAL: From the cell harvest step onwards, use RNAse-free reagents and techniques (Troubleshooting Problem1).
-
1.All protocols for animal studies were approved by the Institutional Animal Care and Use Committees at University College London. Culture 2,500,000–3,000,000 sympathetic neurons dissected from P0/P1 Wistar rats on one collagen-coated 35 mm dish for 5 days (Andreassi et al., 2010). Rinse cells with RNAse-free PBS and scrape them in 1 mL of RNAse-free PBS. Centrifuge the cells at 500×g for 5 min at 22°C–25°C and remove the PBS leaving the rinsed cells as dry as possible.
-
a.Add 100 μL of CERI solution+1:100 Protease inhibitor cocktail added fresh.
-
b.Vortex the tube vigorously on the highest setting for 15 s to fully suspend the cell pellet and incubate on ice for 10 min.
-
c.Add 5.5 μL of CERII solution and vortex for 5 s on the highest setting. Incubate on ice for 1 min.
-
d.Vortex for 5 s on the highest setting and centrifuge for 5 min at maximum speed (∼16,000 × g) in a microcentrifuge at 4°C.
-
e.Immediately transfer the supernatant (cytoplasmic extract) to a clean pre-chilled tube. Add 1.25 μL of RNasin Plus Ribonuclease Inhibitor (40 U/μL stock). To avoid repeated freezing and thawing, store in 35 μL aliquots (sufficient for 3 experiments) at −80°C. Proceed with nuclear protein extraction if necessary as a control for step 2, otherwise discard the nuclear fraction.
-
a.
-
2.
Run an aliquot (generally 5%–10% of the total volume) of the cytosolic (and nuclear, if available) fraction samples on a denaturing SDS-PAGE and perform western blotting for known nuclear and cytoplasmic markers (for example HDAC2 and Impa1) to verify the purity of the fractions (Figure 1).
Figure 1.
Western blotting to check purity of nuclear and cytosolic fractions
Aliquots of proteins purified from nuclear or cytosolic fractions of sympathetic neurons cultured in vitro were separated by PAGE, blotted and probed with antibodies for nuclear or cytosolic protein markers (HDAC2 and Impa1, respectively).
In vitro transcription, de-phosphorylation and 5′end-labeling of the RNA marker and of the probe, and full-length probe purification
Timing: 1–2 days
This section provides a detailed description of 4 preliminary steps (in vitro transcription, de-phosphorylation, labeling and isolation of full length probe by band isolation) necessary to generate the probe that will be used in the cleavage reaction mix, and to label the RNA marker that will serve as size reference when separating the products of the cleavage reaction.
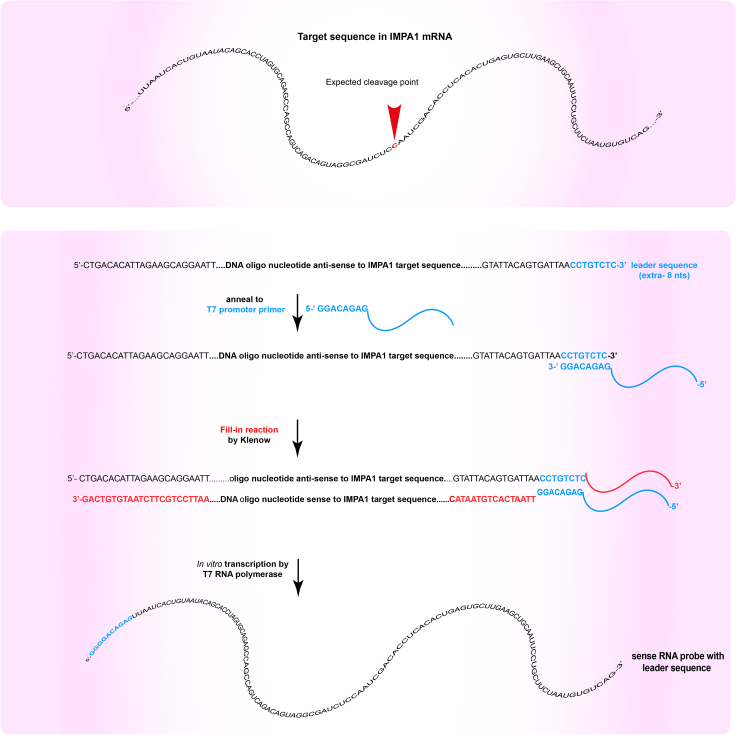
Note: Although any suitable protocol for in vitro transcription will work to generate the RNA probe, we routinely use the mirVana miRNA Probe Construction kit, as we found it to be flexible and very reliable in terms of yield. By using a primer to incorporate the promoter for the T7 RNA polymerase (see below), time-consuming cloning steps are avoided. Below we describe the protocol in its essential steps, and we refer to the manufacturer’s user manual (for more details see the http://tools.thermofisher.com/content/sfs/manuals/fm_1550.pdf). For the RNA transcription, a DNA oligonucleotide about 120 nt long whose sequence encompasses the expected cleavage site should be used. The position of the expected cleavage site within the oligonucleotide sequence should be chosen considering the size of the expected fragments and potential additional restrictions due to the surrounding sequences. If the probe will be uniformly labeled by incorporation of a labeled nucleotide during the in vitro transcription, it is advisable to design the oligonucleotide such that the cleavage site is not centered in the oligo sequence. This will allow both fragments generated after cleavage to be easily resolved by electrophoresis. The sequence of the oligonucleotide should be antisense to the RNA strand to be synthesized and should include the leader sequence CCTGTCTC at its 3′end (Figure 2).
Figure 2.
Oligonucleotide design and steps of in vitro transcription reaction
Schematics of the IMPA1 3′UTR sequence targeted by Ago2 (upper panel), and of the oligonucleotide template, intermediate products and RNA from the in vitro transcription reaction for probe synthesis (lower panel).
-
3.To synthesize the RNA probe, first resuspend the purchased oligonucleotide to 100 μM concentration in H2O using RNAse-free conditions and use it to prepare the dsDNA template for the in vitro transcription. The leftovers of resuspended oligo can be stored at −80°C.
-
a.Thaw, mix by vortexing and keep at 22°C–25°C the following reagents from the mirVana kit: T7 promoter primer, DNA hybridization buffer, 10× Klenow buffer, 10× dNTP mix, nuclease-free water. Keep the tube of Exo-Klenow Enzyme mix at −20°C and do not vortex it. Add the reagents to a PCR tube in the following order:
T7 Promoter Primer 2 μL DNA Hybridization Buffer 6 μL 100 μM oligonucleotide template 2 μL Incubate at 70°C for 5 min and then at 22°C–25°C for 5 min, before adding in this order:10× Klenow Reaction Buffer 2 μL 10× dNTP Mix 2 μL Nuclease-free water 4 μL Exo–Klenow enzyme 2 μL Mix by pipetting, quick spin and incubate at 37°C for 30 min, then proceed with the in vitro transcription, or store at −20°C for several months. -
b.To set up the in vitro transcription reaction, thaw and mix by vortexing 10× transcription buffer, nucleotide solutions (NTPs), and nuclease-free water from the mirVana kit. Keep NTPs always on ice and the 10× transcription buffer at 22°C–25°C. Keep the tube of T7 Enzyme at −20°C and do not vortex it. Prepare the reaction mix at 22°C–25°C by mixing in a PCR tube in the following order:
Nuclease-free water 11 μL dsDNA template (step 3a.) 1 μL 10× transcription buffer 2 μL 10 mM ATP 1 μL 10 mM CTP 1 μL 10 mM GTP 1 μL 10 mM UTP 1 μL T7 RNA polymerase 2 μL Gently pipette up and down several times to mix, then centrifuge briefly to collect the reaction mixture at the bottom of the tube, and incubate the reaction mixture at 37°C for at least 60 min. CRITICAL: Make sure that the transcription buffer is completely thawed and at 22°C–25°C before adding it to the reaction tube that should also be kept at 22°C–25°C. The step is critical to avoid precipitation of the oligonucleotide by the spermidine contained in the buffer.
CRITICAL: Make sure that the transcription buffer is completely thawed and at 22°C–25°C before adding it to the reaction tube that should also be kept at 22°C–25°C. The step is critical to avoid precipitation of the oligonucleotide by the spermidine contained in the buffer. -
c.Digest the dsDNA template by adding:
RNase-free DNase I 1 μL Mix well by pipetting and incubate at 37°C for 10 min. -
d.Add nuclease-free water to 50 μL total volume and purify the probes using G50 columns as per manufacturer’s instructions https://www.cytivalifesciences.com/en/us/shop/molecular-biology/purification/gel-filtration-columns/illustra-probequant-g-50-micro-columns-p-00055#tech-spec-table to eliminate unincorporated nucleotides.
-
a.
-
4.De-phosphorylate the in vitro transcribed RNA probe using Calf Intestinal Phosphatase (CIP) and purify it using a phenol/chloroform extraction followed by ethanol precipitation.
-
a.Dilute CIP to 2.5 U/μL using 1× CutSmart buffer (dispose of unused diluted enzyme) and proceed as follows by mixing:
Purified IVT rxn (step 3.d) 50 μL 10× CutSmart buffer 5.5 μL Diluted CIP 1.1 μL Incubate at 37°C for 30 min. -
b.Remove the CIP by phenol/chloroform extraction by adding to each reaction:
RNAse-free water 75 μL 2.0 M sodium acetate pH 4.3 15 μL Phenol/chloroform 25:24:1 150 μL Mix well by vigorous shaking of the tube by hand, then spin at 17000×g at 22°C–25°C for 5 min in a microcentrifuge. Transfer the aqueous (top) phase to a clean 1.5 mL tube. -
c.Precipitate the RNA by adding
-
a.
| Linear acrylamide | 1 μL |
| 100% ethanol | 750 μL |
Vortex and precipitate at −20°C >30 min STOP point. Spin at the maximum speed at 4°C for 15 min in a microcentrifuge. Carefully remove the supernatant and discard. Wash the pellet once with 1.5 mL of ice cold 70% ethanol. Spin at the max speed at 4°C for 5 min in a microcentrifuge. Carefully withdraw nearly all supernatants, spin for 1 min at max speed to collect the residual ethanol on the wall of the tube and discard the liquid using an extended pipette tip. Resuspend the pellet in 15 μL of nuclease-free water and proceed with labeling STOP point.
-
5.To achieve high sensitivity, the probe is labeled with a radionuclide.Note: The probe can be either uniformly labeled, by including a labeled nucleotide in the in vitro transcription reaction (step 3.b), or labeled at its 5′end. We prefer the latter as the results are generally cleaner and easier to interpret (Figure 3). The amount of radioactive ATP to add to the labeling reaction depends on the reference activity date of the batch used. 1 μL of a fresh batch is usually sufficient. Increase the volume up to 5 μL if the batch is 2 weeks old, adjusting the reaction volume to 25 μL total. For best results, do not use a radioactive batch that is more than 3 weeks old.
 CRITICAL: The protocol involves the use of radiolabeled nucleotides. Strictly follow institutional guidelines for working with radionuclides.
CRITICAL: The protocol involves the use of radiolabeled nucleotides. Strictly follow institutional guidelines for working with radionuclides.-
a.Perform 5′-end labeling using radioactive ATP ([γ-32P ATP] and T4 Polynucleotide Kinase (PNK). Mix by pipetting:
CIP-treated IVT RNA (step 4.c) 15 μL 10× T4 PNK Buffer A 2 μL [γ-32P] ATP ∼25 pmol x μL T4 Polynucleotide Kinase 1 μL Incubate at 37°C for more than 1 h and less than 3 h, then add 1 μL of 0.5 mM EDTA, incubate 2 min at 22°C–25°C and heat-inactivate at 95°C for 2 min. -
b.Eliminate unincorporated nucleotides using G-50 columns as in step 3.d.
-
c.Purify the labeled RNA using 1 mL of TRIzol according to the manufacturer’s protocol https://tools.thermofisher.com/content/sfs/manuals/trizol_reagent.pdf and by adding 1 μL of Linear acrylamide as co-precipitant. Resuspend the pellet in 10 μL of RNAse-free water, add 10 uL of 2× Gel Loading Buffer from mirVana kit (or NEB supplemented with 18 mM EDTA) and either freeze STOP point or proceed with gel purification (step 7).
-
a.
-
6.The RNA marker that will be used as a size reference for the gel electrophoresis needs to be labeled as well.Note: Any RNA marker of suitable size can be used, but it will need to be dephosphorylated (see point 4). For this reason, we prefer to use the Decade Marker system that is ready to be labeled following manufacturer’s instructions https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2F4386532D.pdf=UEkgU2hlZXQ6IERlY2FkZSBNYXJrZXJzIFN5c3RlbQ==. Upon receiving the markers, they should be dissolved by adding 60 μL of nuclease-free water, aliquoted in 1.5 mL tubes (7 μL each) and stored at −80°C. See preparation point 5 for information about the amount of radioactive label that will be used in the labeling reaction.
-
a.To label the RNA marker, use the reagents provided in the Decade Marker systems kit to set up the reaction as follows:
Decade™ Marker RNA (100 ng) +H2O 7 μL 10× T4 PNK Buffer A 1.5 μL Nuclease-free H2O up to 4.5 μL [γ-32P] ATP x μL T4 Polynucleotide Kinase 1 μL Incubate at 37°C for 1 h. -
b.Add to the reaction tube:
Nuclease-free Water 3 μL 10×Cleavage Reagent 2 μL Incubate at 22°C–25°C for 5 min, then add 20 μL of 2× Gel Loading Buffer from mirVANA kit (or NEB supplemented with 18 mM EDTA) and store at −20°C or use.
-
a.
-
7.Separate the probe by denaturing polyacrylamide gel electrophoresis (PAGE) and purify the probe by band isolation.Note:In vitro transcription reactions generally produce minor products of intermediate length that may confound the results of the cleavage reaction. Thus, it is necessary to purify the full-length probe by band isolation using a denaturing polyacrylamide gel. Precast polyacrylamide gels can be used, but the size of the gel is usually not adequate to obtain an optimal band separation. Thus, we recommend pouring home-cast gels.
-
a.Cast a denaturing polyacrylamide gel as follows. Volumes in this protocol are set for the Biometra Maxi-gel system.
-
i.Clean plates and combs with soapy water, dry and rinse with 100% Ethanol. Treat all parts with RNAseZap and rinse with RNAse-free water. Dry and set up the plates for casting the gels according to the manufacturer’s instructions https://manualzz.com/doc/6464039/minigel-twin-user-manual.
-
ii.Prepare an 8% acrylamide-7M urea gel according to the recipe table below. In a 50 mL tube, weigh the urea, add 5× TBE buffer and water to 25.36 mL final volume, vortex and place in a water bath at 42°C vortexing at regular intervals until all crystals are dissolved (about 30 min). Syringe filter with a 0.45 μm filter. Add freshly made APS and TEMED, swirl to mix and pour the gel.Note: To avoid introducing air bubbles, do not completely empty the pipette and make sure to have excess solution. If air bubbles are present, hold the plates at an angle of about 45° and use a pair of scissors, or similar, to gently bang the plates and dislodge the bubbles. Insert the comb slowly, at an angle, to avoid trapping air bubbles in the wells. After polymerizing, the gel can be stored at 4°C for several days, as long as it is kept moist by wrapping it in water-soaked tissue and sealed with cling film.
-
iii.Before loading the samples, it is advisable to pre-run the gel to warm it and to keep the samples denatured. Remove the comb, rinse the wells with water or a running buffer and set up the gel in the running apparatus. Fill the reservoirs with 1× TBE running buffer and pre-run the gel (empty) at 20–25 mA for about 45 min.
-
i.
-
b.Prepare the samples for the gel loading and run electrophoresis.
-
i.While the gel is pre-running, complete the probe purification (step 4.c) or thaw on ice the purified probe stored at −20°C. Aliquot the needed amount of labeled marker. The amount of marker depends on the activity of the batch of the radioactive labeling used. Use 10 μL or more to get an intense labeling upon autoradiography.
-
ii.Heat markers and samples for 3 min at 100°C, incubate on ice for 3 min and quick spin in a cold centrifuge.
-
iii.While heating the samples, flush the gel wells with a syringe and running buffer.
-
iv.After spinning, load both samples and marker on the pre-warmed denaturing gel using long gel loading tips. Leave empty lanes between marker and samples to prevent carry over and over-shadowing of the bands.
-
v.Run at 20–25 mA for 1.5 h. Use the migration pattern of the loading dyes according to the table below, to decide when to interrupt the electrophoresis.
% Acrylamide (denaturing conditions) Blue bromophenol (nt) Xylene cyanol (nt) 8 19 75 12 10 70 15 8 28
-
i.
-
c.Identify the full-length probe by autoradiography.
-
i.Remove the glass plates from the running apparatus.
-
ii.Cool down only one glass side by placing ice on it. This will prevent the gel from detaching from both glass plates when separating them.
-
iii.Open the glass plates, by using a spatula as a lever in between the glass plates, and place a transparency film on the gel. Secure with tape the position of the transparency film to the gel and use a fluorescent sticker for orientation (Figure 4).
-
iv.In a dark room, expose the gel to an X-ray film.Note: The probe is very radioactive, thus an exposure of 30 s-few minutes at 22°C–25°C (or at −80°C if the X-ray cassette has intensifying screens) is usually sufficient to identify the full-length probe that should be the most intense band at the highest size (Figure 5). Do not overexpose as a faint gray band is ideal for cutting and will allow to minimize the amount of acrylamide.
-
v.To cut out the band of the full length probe, superimpose the film and the gel using the fluorescent stickers for reference, label the band on the glass plate, remove the film and transparency and cut with a clean disposable scalpel tightly around the band (Figure 4).
-
vi.Roughly chop the band and transfer it to a 500 μL tube, pierced on the bottom with a 25G needle and placed on top of a 1.5 mL tube. Spin for 1 min at max speed to crush the gel band.
-
vii.Elute the RNA from the gel by incubation with 400 μL of elution buffer from the mirVana Kit for 14–16 h at 22°C–25°C on a rotor wheel. Expose the gel again to make sure that the band isolation was effective (Figure 5). Troubleshooting Problems 2, 3, and 5:Note: Throughout the next step, the use of a Geiger counter will help to make sure that radioactivity signal (i.e. sample) is not lost during the extraction.
-
viii.To precipitate the eluted probe, centrifuge for 1 min at max speed to pellet the gel fragments and collect the supernatant. Add about 3× volumes of 100% ET-OH and incubate for at least 30 min at −20°C.
-
ix.Centrifuge at max speed for 30 min at 4°C, aspirate the supernatant and wash the pellet with ice-cold 75% EtOH. Centrifuge at max speed for 5 min at 4°C and remove the supernatant until the pellet is dry. Dissolve the pellet in 20 μL (or other suitable volume) of nuclease-free water for 5 min at 22°C–25°C. The purified probe can be stored at −20°C for a few days STOP point but it is advisable to use it fresh to minimize radiolysis and decay of the radioactive signal.
-
i.
-
d.Using a liquid scintillation β-counter, count 1 μL of the purified probe and 1 μL of the isotope stock opportunely diluted (i.e., 1:100). Calculate probe concentration as follows:
P = Total activity of the probe = measured cpm/μL probe × dilution factor × total volume of probe S = Total amount of radioactivity used in the labeling reaction = measured cpm/μL ATP stock × dilution factor × volume of ATP stock used in the reaction. If necessary, correct for decay of the signal using the activity reference date of the ATP stock F = Fraction of incorporated radioactivity = P/S A = Amount (pmol) of probe used in the reaction. From the concentration of the stock calculate the amount of ATP in pmol used for the labeling reaction (i.e., if the ATP stock is 3000Ci/mmol, 10mCi/mL, 1 μL ATP stock used in the reaction corresponds to 3.3pmol) Y = Probe yield (pmol) = A × F Concentration of the probe (pM) = (Y/volume of the probe in mL) × 1000
-
a.
Figure 3.
Comparison between total labeling and 5′end-labeling of the probe
(A and B) The probes can be labeled during the in vitro transcription reaction (total labeling, A) or by treatment with T4 PNK (5′end-labeling, B). The latter gives better results as there is a substantial decrease in background signal (asterisk in A). Non-contiguous lanes from the same experiment and autoradiography blot are shown side by side. Gray lines show where irrelevant lanes have been removed.
Figure 4.
Band-isolation of full-length probe
Schematics of the set-up to identify the area of the gel to cut in order of purifying the full length probe.
Figure 5.
Band isolation of full-length probe
(A and B) The band corresponding to the full length probe is identified by its size (120 nt in this example) on the auto-radiography (A) and, after being superimposed to the gel, it is used to mark the area of the gel to cut. After cutting, the gel is exposed to film again (B). Absence of the marked band (dotted oval) suggests successful isolation of the correct band. Non-contiguous lanes from the same experiment and autoradiography blot are shown side by side. Gray lines show where irrelevant lanes have been removed.
Probe concentration varies between purifications, but it is usually in between 0.5 nM and 10 nM.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-IMPA1 (use 1:10000 for western blot) | Abcam | ab184165 |
| Mouse anti-HDAC2 (use 1:1000 for western blot) | Abcam | ab12169 |
| Chemicals,peptides, andrecombinantproteins | ||
| Recombinant Ago2 | Active Motif | 31486 |
| Rat tail collagen | Homemade | n/a |
| PBS 0.0095M(PO4) w/o Ca and Mg | Lonza | BE17-516F |
| Protease Inhibitor Cocktail | Merck | P8340 |
| RNasin Plus Ribonuclease Inhibitor | Promega | N2611 |
| RNAseZap | Merck | R2020 |
| Calf Intestinal Phosphatase (CIP) | New England Biolabs | M0290S |
| Sodium Acetate buffer solution | Merck | S7899 |
| Linear acrylamide | Thermo Fisher Scientific | AM9520 |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) | Fisher Scientific | 15875408 |
| Ethanol | Merck | 32221 |
| γ-32P ATP | PerkinElmer | BLU002A100UC |
| T4 Polynucleotide Kinase | Thermo Fisher Scientific | EK0031 |
| 0.5M EDTA pH 8 | Merck | E7889 |
| TRIzol Reagent | Thermo Fisher Scientific | 15596018 |
| RNA Loading Dye, (2×) | New England Biolabs | B0363S |
| ProtoGel (30% Solution at 37.5:1 Ratio) | GENEFLOW | EC-890 |
| Ammonium persulfate | Merck | A3678 |
| TEMED | VWR | 443083g |
| Trizma base | Merck | T1503 |
| Boric acid | Meck | B6768 |
| Urea | Merck | U5578 |
| HEPES Buffer, 1M Solution | Fisher Scientific | 83264 |
| Potassium hydroxide | Merck | 60377 |
| Potassium acetate | Merck | 60035 |
| Magnesium Acetate | Merck | M5661 |
| DL-Dithiothreitol | Merck | D9779 |
| Yeast tRNA (10 mg/mL) | Thermo Fisher Scientific | AM7119 |
| Experimentalmodels: Celllines | ||
| rat sympathetic neuron extracts | This paper | N/A |
| Experimentalmodels: Organisms/strains | ||
| Wild-type P0/P1 Wistar rats, either sex | N/A | N/A |
| Oligonucleotides | ||
| CTGACACATTAGAAGCAGGAATTGCAGCTTCA AGCACTCAGTGTGAGGTGTCGATTGGAGATCG CCTACTGTCTGACTGGCTCTGCACTAGGTGCT GTATTACAGTGATTAACCTGTCTC |
Sigma-Genosys | N/A |
| Other | ||
| NE-PER™ Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher Scientific | 78833 |
| Decade Markers System | Thermo Fisher Scientific | AM7778 |
| mirVana™ miRNA Probe Construction Kit | Thermo Fisher Scientific | AM1550 |
Materials and equipment
Related to preparation steps 3.d and 5.d
ProbeQuant G-50 Micro Columns Cytiva 28903408
Related to preparation step 7.a and main protocol step 3
Maxi gel system Biometra 010-400
Related to main protocol step 3.f
Grade 3MM Chr Cellulose Chromatography Papers Cytiva 3030-917
Related to preparation step 7.c and main protocol step 3.g
Hyperfilm™ MP Merck GE28-9068-44
Hypercassette Merck GERPN11649
5× TBE buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Trizma base | 89 mM | 54 g |
| Boric acid | 89 mM | 27.5 g |
| 0.5M EDTA pH 8 | 2 mM | 20 mL |
| Total | n/a | 1 L |
Store at RT. After prolonged storage, EDTA may precipitate out of solution.
8% PAA gel
| Reagent | Final concentration | Amount |
|---|---|---|
| 30% solution Acrylamide to Bisacrylamide | 8% | 10.4 mL |
| urea | 7M | 16.8 g |
| 5× TBE | 1× | 8 mL |
| 100mg/mL Ammonium persulfate | 0.5 mg/mL | 0.2 mL |
| TEMED | 0.1% | 0.04 mL |
| Total | n/a | 40 mL |
Before adding ammonium persulfate and TEMED, the solution may be stored for some days in the dark, but we advise to use it immediately to pour the gel.
5× cleavage buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 500 mM Hepes-KOH pH 7.5 | 125 mM | 0.25 mL |
| 1M KOAc | 250 mM | 0.25 mL |
| 250m Mg(OAc)2 | 25 mM | 0.1 mL |
| 1M DTT | 25 mM | 0.025 mL |
| ddH2O | n/a | 0.375 mL |
| Total | n/a | 1 mL |
Stock solutions can be stored at RT for several months. Make the working solution fresh.
CRITICAL: The following reagents are toxic and need to be handled with precautions like stated below:
TRIzol/Phenol Chloroform: Toxic by inhalation and contact. Use PPE and handle in the fume hood.
Ethanol: Flammable.
Acrylamide-bis acrylamide solution (liquid): Toxic and carcinogenic. Use PPE.
TEMED: Toxic by inhalation and contact. Use PPE and handle in the fume hood.
Tris base: Toxic and irritant. Use PPE.
Boric acid: Irritant and carcinogenic. Use PPE.
EDTA: Irritant. Use PPE.
Urea: Use dust-mask.
DTT: Corrosive. Use PPE.
Alternatives: All reagents and resources listed could be sourced by other suppliers or replaced by equivalent items; however, the impact of these substitutions on the protocol performance has not been tested.
Step-by-step method details
Cleavage reaction
Timing: 3–5 h
The in vitro cleavage reaction will result in digestion of the labeled probe by the endonuclease(s).
Note: We have successfully used both commercial and home-made preparations of recombinant Ago2 in the cleavage reaction. To set the reaction conditions, it is advisable to perform preliminary experiments with variable ratios of the amount of probe and enzyme. A 10- to 100 fold-molar excess of the enzyme over the probe and 5 nM maximum probe concentration is normally used (see table below).
| Test 1 | Test 2 | Test 3 | Test 4 | |
|---|---|---|---|---|
| Probe | 0.5 nM | 1 nM | 2.5 nM | 5 nM |
| Recombinant protein | 50 nM | 50 nM | 50 nM | 50 nM |
If using a different amount of recombinant protein, change the amount of probe accordingly.
-
1.In our system, the cleavage reaction required pre-incubation of the probe with cytosolic protein extracts (preparation step 1. e). This step might be omitted if it is not necessary for the cleavage reaction under study. Various control reactions for the specificity of the cleavage site and of the cleavage enzyme can be performed, as suggested in the expected outcomes section.
-
a.To pre-incubate the probe with the cytosolic extracts, dilute the purified probe to the desired concentration and thaw on ice one 10 μL aliquot of cytoplasmic extracts/reaction. To each reaction tube add (Figure 6):
Cytoplasmic extract 10 μL RNAse inhibitor 1 μL Purified probe According to need Incubate at 26°C for 90 min, then move the tubes to ice. -
b.Just before starting the cleavage reaction, thaw the recombinant Ago2 enzyme on ice and dilute it to the desired concentration using a 1× cleavage buffer. Discard diluted unused enzyme. Add to each tube from step a (except the probe only tube) the following and incubate at 26°C for 90 min.
5× cleavage buffer 6 μL RNAse inhibitor 1 μL 0.5 μg/μL yeast RNA 1 μL Diluted recombinant Ago2 1 μL -
c.Stop the reaction by adding 0.5 mL/tube of TRIzol and purify samples.
-
i.Samples can be purified according to TRIzol manufacturer’s protocol https://tools.thermofisher.com/content/sfs/manuals/trizol_reagent.pdf and adding 1 μL of Linear acrylamide as co-precipitant, or stored at −20°C STOP point. Avoid prolonged storage to prevent radiolysis of the samples.
-
ii.After purification, each pellet is resuspended in 20 μL of 2× Gel Loading Buffer from mirVana kit (or NEB supplemented with 18 mM EDTA) and stored at −20°C STOP point or used immediately for PAGE. In case of storage, the samples should be resolved by PAGE in the next few days to prevent radiolysis.
-
i.
-
a.
Figure 6.
Diagram of a typical cleavage reaction.
Denaturing PAGE and auto-radiography
Timing: 4 h
The products of the cleavage reaction will be separated by denaturing PAGE and visualized by auto-radiography.
-
2.Separate the purified products of the cleavage reaction by PAGE.
-
a.Prepare and pre-run a 8% denaturing polyacrylamide gel (see preparation step 7.a). If needed, change acrylamide concentration according to the size of the reaction products to separate.
-
b.Thaw 1 μL of labeled marker (see preparation point 5.a) and add 19 μL of 2× Gel Loading Buffer from mirVana kit (or NEB supplemented with 18 mM EDTA). 1 μL of marker will suffice, as the exposure will be long enough to get a strong signal.
-
c.Denature marker and purified samples from step 2.b for 3 min at 100°C and incubate on ice for 3 min.
-
d.Load marker and samples on the pre-warmed 8% denaturing polyacrylamide gel (see preparation point 7.a).
-
e.Run at 20–25 mA for about 2.5 h. If possible, leave an empty lane in between the marker and samples to prevent carry over and over-shadowing of the bands. Use the migration pattern of the loading dyes (see table in preparation point 7.b) to decide when to interrupt the electrophoresis.
-
f.Remove the glass plates from the running apparatus and cool one of the glass plates by placing ice on it. This will prevent the detachment of the gel from both glass plates when separating them. Open the glass plates by using a spatula as a lever in between and detach the gel from the sides of the glass plates using a scalpel. Wet the gel dropwise with 1–2 mL of RNAse-free water and place on it a piece of 3MM filter paper, roughly cut to size, to lift the gel off the glass plate. Place the gel in between the sheets of a clear polypropylene plastic wallet and use a fluorescent sticker for orientation, if needed.
-
g.Expose to an X-ray film in an X-ray cassette or to a PhosphorImager screen. If using films, it is better to expose at −80°C using intensifier screens. It is also useful to place two films, one on top and one underneath the gel, to get a long and short exposure. Using tape, secure the film underneath the gel to both the cassette and the gel, so that it will not move when opening the cassette to develop the top film. Troubleshooting Problems 3, 4, and 5:
-
h.Develop the top film after 18–24 h exposure. Depending on the intensity of the bands, expose the film underneath the gel for a longer time or place a new film on top of the gel for a shorter exposure. Troubleshooting Problem 6.
-
a.
Expected outcomes
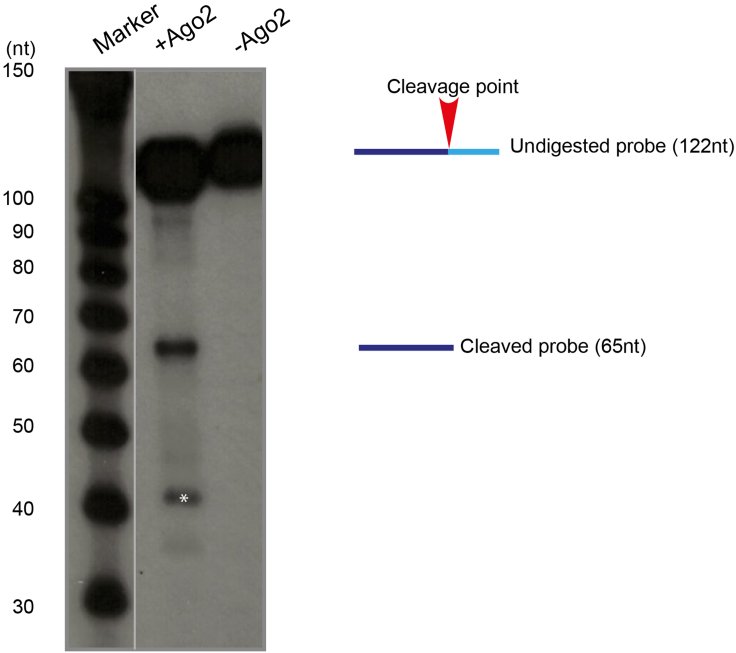
A successful cleavage reaction should show bands at the expected size with little or no background signal (Figure 7). If the enzyme has additional enzymatic activities (such as RNA “trimming” for Ago2), further bands can be observed. The specificity of the cleavage site can be assessed using probes bearing mutations of the cleavage sequence. When testing a known endonuclease for potential cleavage of a new target, a control reaction including a known target for the desired enzyme should be included. Recombinant protein preparations (commercially or home-made generated) should be tested for enzymatic activity on known substrates before performing an experiment on the target of interest. The use of enzymes with defective catalytic activity can further confirm the specific involvement of the endonuclease under consideration in the cleavage reaction and the absence of RNases contaminating the recombinant protein preparations. Finally, the use of probes carrying mutations of the sequence targeted by the endonuclease under study can help to narrow down the exact position of the cleavage site, if not known. When reaction conditions (such as temperature for the cleavage reaction, concentration of enzyme and probe, gel running conditions) are established for the desired enzyme and probe, the protocol can be reproduced very reliably.
Figure 7.
Cleavage of the Impa1 probe gives rise to a fragment of expected size
A 120 nt probe encompassing the cleavage site for Impa1 3′UTR is digested by Ago2 to produce a fragment of 67 nt. Fragments of smaller size (labeled by an asterisk) are due to a further trimming reaction performed by Ago2 after the cleavage reaction.
Limitations
Previous knowledge of the cleavage sequence targeted by the endonuclease is necessary to design the appropriate oligo template for the probe synthesis. If the sequence is not known, other techniques like RML-RT-PCR followed by cloning of the cleaved fragment can help to identify the target sequence (Andreassi et al., 2021).
The protocol relies on the use of nucleotides labeled with 32P. This allows very high sensitivity, however their use is limited by the relative fast decay of this radionucleotide and by the potential hazard for the operator. Training by experienced users and facilities dedicated to the use of radionuclides should be provided before attempting the protocol.
Troubleshooting
Problem 1
Smears in all lanes or no signal. This could be caused by RNAse contamination of solutions or equipment.
Potential solution
The protocol assesses cleavage of RNA substrates. As such, strict RNAse-free conditions should be followed in all steps. Working material (gloves, benches, pipettes…) should be cleaned with RNase inactivating solutions and DNase and RNase free tubes and tips should be used.
Problem 2
No bands in marker and/or sample lanes. This could be caused by failure in the labeling reaction or degradation (see in vitro transcription, de-phosphorylation and 5′end-labeling of the RNA marker and of the probe, and full-length probe purification, see preparation steps 3 and 4)
Potential solution
Use fresh reagents. Prior to labeling, run an aliquot of the marker and/or the in vitro transcribed RNA on a Bioanalyzer to verify integrity.
Problem 3
Double bands in any lane (Figure 8). This could be caused by incomplete denaturation of the samples (see in vitro transcription, de-phosphorylation and 5′end-labeling of the RNA marker and of the probe, and full-length probe purification, preparation step 7.b and denaturing PAGE and auto-radiography, step 3.c).
Figure 8.

Incomplete denaturation of the samples
(A and B) The altered migration pattern caused by incomplete denaturation results in the detection of unexpected bands and the migration of the marker bands at the wrong size (compare A to B).
Potential solution
Before loading on the gel, boil samples longer and make sure to cool them quickly on ice. Pre-warm the gel before loading. Check urea concentration in the gel mix.
Problem 4
Size of the marker bands are as expected but sample bands are at the wrong size, smeared or there are no bands. This could be caused by RNAse contamination of the samples or failure in the probe synthesis (see in vitro transcription, de-phosphorylation and 5′end-labeling of the RNA marker and of the probe and full-length probe purification, preparation steps 3 and 4).
Potential solution
Use fresh reagents. Prior to labeling, run an aliquot of the in vitro transcribed RNA on a Bioanalyzer to verify integrity.
Problem 5
Wobbly bands. This could be caused by poor gel polymerization or the presence of urea in the gel wells (see in vitro transcription, de-phosphorylation and 5′end-labeling of the RNA marker and of the probe, and full-length probe purification, preparation steps 7.a and denaturing PAGE and auto-radiography, 3.a).
Potential solution
Filter the gel solutions before pouring the gel to remove particulate contaminants. Carefully flush the gel wells with the running buffer before loading the samples.
Problem 6
Bands seem to move after longer exposure (Figure 9). This could be due to radioactivity non-specifically bound by the loading buffer dye that keeps diffusing in the gel when it is left too moist before exposure (See denaturing PAGE and auto-radiography, step 3.f).
Figure 9.
Smeared bands due to diffusion of the dye
Appearance of new bands that move during prolonged exposure (labeled by the white asterisks) is due to radioactivity bound nonspecifically by the loading buffer dye that keeps diffusing in the gel when it is left too moist before exposure to X-ray.
Potential solution
Gently blot excess buffer from the gel before exposure or dry the gel before exposing it.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Antonella Riccio (a.riccio@ucl.ac.uk).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We are grateful to Fabricio Loayza-Puch and Reuven Agami for advice and sharing their cleavage protocol. We thank Adolfo Saiardi for helping with Ago2 purification and all members of the Riccio lab for helpful discussions. This work was supported by Wellcome Trust Investigator Awards 103717/Z/14/Z and 217213_Z_19_Z to A.R. and a Wellcome Trust Institutional Strategic Support Fund 2014 to C.A.
Author contributions
C.A. established this protocol and co-wrote the manuscript. A.R. co-wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Catia Andreassi, Email: c.andreassi@ucl.ac.uk.
Antonella Riccio, Email: a.riccio@ucl.ac.uk.
Data and code availability
This study did not generate or analyze datasets/code.
References
- Andreassi C., Zimmermann C., Mitter R., Fusco S., De Vita S., Saiardi A., Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Andreassi C., Crerar H., Riccio A. Post-transcriptional processing of mRNA in neurons: the vestiges of the RNA world drive transcriptome diversity. Front. Mol. Neurosci. 2018;11:304. doi: 10.3389/fnmol.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C., Luisier R., Crerar H., Darsinou M., Blokzijl-Franke S., Lenn T., Luscombe N.M., Cuda G., Gaspari M., Saiardi A., Riccio A. Cytoplasmic cleavage of IMPA1 3′UTR is necessary for maintaining axon integrity. Cell Rep. 2021;34:108778. doi: 10.1016/j.celrep.2021.108778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J., Kohl T.O. Using dounce homogenization to lyse cells for immunoprecipitation. Cold Spring Harb. Protoc. 2019 doi: 10.1101/pdb.prot098574. [DOI] [PubMed] [Google Scholar]
- Harigaya Y., Parker R. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA. 2010;1:132–141. doi: 10.1002/wrna.17. [DOI] [PubMed] [Google Scholar]
- Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kühn U., Menzies F.M., Oude Vrielink J.A.F., Bos A.J., Drost J., Rooijers K., et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Jin S., Zhan J., Zhou Y. Argonaute proteins: structures and their endonuclease activity. Mol. Biol. Rep. 2021;48:4837–4849. doi: 10.1007/s11033-021-06476-w. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Uejima H., Nagami-Okada T., Siomi H., Siomi M.C. In vitro RNA cleavage assay for Argonaute-family proteins. Methods Mol. Biol. 2008;442:29–43. doi: 10.1007/978-1-59745-191-8_3. [DOI] [PubMed] [Google Scholar]
- Rodriguez W., Macveigh-Fierro D., Miles J., Muller M. Fated for decay: RNA elements targeted by viral endonucleases. Semin. Cell Dev. Biol. 2021;111:119–125. doi: 10.1016/j.semcdb.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze datasets/code.