Abstract
Currently, identifying novel biomarkers remains a crucial need for cancer immunotherapy. By leveraging single-cell cytometry data, Greene et al. developed an interpretable machine learning method, FAUST, to discover cell populations associated with clinical outcomes.
Currently, identifying novel biomarkers remains a crucial need for cancer immunotherapy. By leveraging single-cell cytometry data, Greene et al. developed an interpretable machine learning method, FAUST, to discover cell populations associated with clinical outcomes.
Main text
During the development and pathogenesis of biological systems, the cell populations’ composition and physiological state alter continuously across the spatial and temporal dimensions.1 Bulk genomic profiles fail to reveal such heterogeneous and dynamic processes at the molecular and cellular levels. Recently, single-cell techniques for different modalities (e.g., genomic, transcriptomic, and proteomic) have evolved rapidly, providing an unprecedented opportunity to help biologists understand the function and behavior of cells at various conditions. Compared to single-cell sequencing technology processing on average ∼1,000–100,000 cells per sample, single-cell cytometry can reach a higher order up to millions of cells.2 Such high-throughput single-cell resources enable a comprehensive analysis of cell compositions, especially uncovering rare cell populations in complex immune ecosystems. Subsequent investigations combined with treatment and clinical information would significantly promote therapeutic development3 and biomarker discovery.4
A crucial step in cytometry data analysis is to cluster and annotate cell populations in high-dimensional spaces. Previously, biologists identified cell populations manually (termed as “gating”) by using a 2D scatterplot with pairs of predefined markers. However, this strategy suffers from serious reproducibility issues and becomes unpragmatic with the increasing number of markers (up to ∼40–50). Alternatively, automated computational methods have been developed to reduce subjective bias and automate data processing.5 Although computational tools succeed in some cases, several new challenges still arise. For example, the number of clusters (cell populations) is often hard to determine for a large-scale single-cell dataset. Low cluster counts might miss rare cell populations, while high cluster counts might produce false subpopulations. Moreover, since numeric labels are assigned for clusters per independent sample, mapping cluster entities across samples is highly challenging for integrated studies.
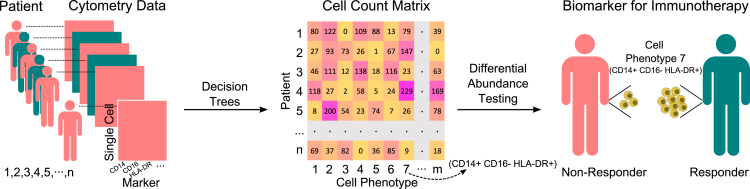
Greene et al. developed an interpretable machine learning method, full annotation using shaped-constrained trees (FAUST), to discover and annotate cell populations from flow and mass cytometry data.6 By deriving the standardized set of thresholds for marker expression, FAUST can construct decision trees with markers as nodes for each sample to determine the phenotype of each cell. FAUST uses input cytometry data matrices with rows and columns corresponding to cells and markers, respectively. The output is a cell count matrix of cell phenotypes across all samples (Figure 1). Each FAUST phenotype is represented as a marker co-expressing pattern, e.g., Marker 1+ Marker 2− Marker 3+.
Figure 1.
Application of FAUST to analyze single-cell cytometry data
FAUST uses decision trees to annotate cell populations. FAUST then carries out statistical testing of differential abundance to identify predictive biomarkers for immunotherapy.
In contrast to other tools, FAUST avoids arbitrarily determining cluster numbers and can integrate multiple samples and studies. Subsequently, FAUST can statistically test the difference of cell abundance between two defined groups, e.g., responders versus non-responders or treatment versus control (Figure 1). The authors showcased that FAUST outperformed other existing methods via simulation and benchmark studies.
Next, Greene et al. demonstrated that FAUST could identify predictive biomarkers for cancer immunotherapy and verify abundance change of cell populations after treatment.6 The authors analyzed a cytometry dataset with 78 longitudinal samples from 27 patients with Merkel cell carcinoma receiving pembrolizumab (anti-PD-1) therapy. First, they tested the abundance difference of each T cell sub-phenotype at the baseline (before treatment) between responders and non-responders and found that four of them were significantly associated with response to treatment. These cell populations together can be considered as effector memory T cells expressing CD28 and PD-1. Consistent with this finding, a study showed that bispecific antibodies targeting CD28 on T cells enhance the clinical outcomes of anti-PD-1 immunotherapy.7 Second, the authors explored the abundance change of CD3+ CD8+ CD4− PD-1-bright phenotype along the course of treatment and observed that its fraction decreases continually after treatment, which may result from blocking PD-1 by pembrolizumab. Similarly, the authors also tested FAUST in several additional cytometry datasets.
Greene et al. also demonstrated that FAUST enables meta-analysis across studies with different marker panels.6 They collected three cytometry datasets with myeloid phenotyping panels, i.e., Merkel cell carcinoma anti-PD-1, FLT3-L+therapeutic Vx, and metastatic melanoma anti-PD-1 trials. Based on the common markers across these datasets, the authors found that CD14+ CD16− HLA-DR+ phenotype has higher abundance for each study in responders than non-responders at baseline.
Currently, most patients still do not respond to cancer immunotherapies.8 Predictive biomarkers can help select patients who might respond before the initiation of therapy to avoid unnecessary treatments. Several studies identified biomarkers from bulk transcriptomic data.9,10 Other studies also exploited single-cell sequencing11 and cytometry4 to identify cell populations associated with therapy response. Given such data with high dimensions and throughputs, user-friendly tools are essential for biologists to draw reliable conclusions, and many tools have been developed recently.5 As cytometry techniques evolve rapidly, we foresee that more software will become available, and a comprehensive benchmark will be necessary to guide scientists to choose the appropriate tool to analyze their data.
Acknowledgments
B.R. and P.J. are supported by the intramural research budget provided by the National Cancer Institute (NCI), part of the National Institutes of Health (NIH).
References
- 1.Haniffa M., Taylor D., Linnarsson S., Aronow B.J., Bader G.D., Barker R.A., Camara P.G., Camp J.G., Chédotal A., Copp A., et al. Human Cell Atlas Developmental Biological Network A roadmap for the Human Developmental Cell Atlas. Nature. 2021;597:196–205. doi: 10.1038/s41586-021-03620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitzer M.H., Nolan G.P. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeves P.M., Sluder A.E., Paul S.R., Scholzen A., Kashiwagi S., Poznansky M.C. Application and utility of mass cytometry in vaccine development. FASEB J. 2018;32:5–15. doi: 10.1096/fj.201700325R. [DOI] [PubMed] [Google Scholar]
- 4.Krieg C., Nowicka M., Guglietta S., Schindler S., Hartmann F.J., Weber L.M., Dummer R., Robinson M.D., Levesque M.P., Becher B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 5.Cheung M., Campbell J.J., Whitby L., Thomas R.J., Braybrook J., Petzing J. Current trends in flow cytometry automated data analysis software. Cytometry A. 2021;99:1007–1021. doi: 10.1002/cyto.a.24320. [DOI] [PubMed] [Google Scholar]
- 6.Greene E., Finak G., D’Amico L.A., Bhardwaj N., Church C.D., Morishima C., Ramchurren N., Taube J.M., Nghiem P.T., Cheever M.A., et al. New interpretable machine learning method for single-cell data reveals correlates of clinical response to cancer immunotherapy. Patterns. 2021;2 doi: 10.1016/j.patter.2021.100372. 100372-1–100372-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waite J.C., Wang B., Haber L., Hermann A., Ullman E., Ye X., Dudgeon D., Slim R., Ajithdoss D.K., Godin S.J., et al. Tumor-targeted CD28 bispecific antibodies enhance the antitumor efficacy of PD-1 immunotherapy. Sci. Transl. Med. 2020;12:eaba2325. doi: 10.1126/scitranslmed.aba2325. [DOI] [PubMed] [Google Scholar]
- 8.Sharma P., Hu-Lieskovan S., Wargo J.A., Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.S., Nair N.U., Dinstag G., Chapman L., Chung Y., Wang K., Sinha S., Cha H., Kim D., Schperberg A.V., et al. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell. 2021;184:2487–2502.e13. doi: 10.1016/j.cell.2021.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., He Y., Wang L., Zhang Q., Kim A., et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]