Summary
Recent advances in biomedical machine learning demonstrate great potential for data-driven techniques in health care and biomedical research. However, this potential has thus far been hampered by both the scarcity of annotated data in the biomedical domain and the diversity of the domain's subfields. While unsupervised learning is capable of finding unknown patterns in the data by design, supervised learning requires human annotation to achieve the desired performance through training. With the latter performing vastly better than the former, the need for annotated datasets is high, but they are costly and laborious to obtain. This review explores a family of approaches existing between the supervised and the unsupervised problem setting. The goal of these algorithms is to make more efficient use of the available labeled data. The advantages and limitations of each approach are addressed and perspectives are provided.
Keywords: machine learning, data labeling, data value, active learning, self-supervised learning, semi-supervised learning, data annotation, zero-shot learning
The bigger picture
As machine learning models become more complex, requirements for large annotated datasets grow. Annotating data for machine learning applications is especially challenging in the biomedical domain as it requires domain expertise of highly trained specialists to perform the annotations. Several strategies to either increase efficiency of label utilization or improve the annotation process have been proposed by the machine learning community. In this review we explore these strategies, including semi-supervised learning, active learning, data augmentation, transfer learning, self-supervision, weak-supervision, and zero- or few-shot learning. We show successful examples of research that has applied these strategies to multi-modal biomedical data. We conclude that raising awareness of these strategies in the biomedical community may contribute to further adoption of machine learning techniques in this research field.
Introduction
Learning a task is easier when you have examples. In the absence of examples, humans can leverage other strategies to learn new material, such as generalization based on examples from similar tasks or trial and error. Most of these strategies aim to make efficient use of the available examples, if there are any. Such strategies work best when the data (or examples) come from a representative and unbiased sampling of the underlying data landscape.
In life sciences and health care, finding labeled data that sample the whole distribution is a major challenge. For example, in microscopy, data scarcity, cross-equipment compatibility, resolution limitations, and image quality are challenges in building consensus-labeled datasets.1,2 Another example is related to the drug discovery domain; protein-compound interaction datasets are limited to highly studied proteins or compounds leading to an oversampling of privileged protein-compound pairs.3 In health care, access to patient data and finding consensus-labeled electronic health records (EHR) remain a huge challenge.4,5 Furthermore, the population is often not sampled with a fair or representative distribution, leading to bias in the available datasets.6,7 Another area of focus in life sciences and health care is finding information in scientific publications,8 where labeled data are long-tailed due to the flexible queries and differences in research topic popularity. Accelerated by COVID-19 research,9 the field is experiencing a huge increase in the number of publications, making information retrieval a major bottleneck in keeping up with literature.10
The lack of labeled data is a critical challenge that must be overcome to train supervised learning models in the biomedical field. Some solutions to deal with this problem have been reviewed in biomedical imaging11 and clinical text data analysis.12 In this paper, we summarize several machine learning strategies that can be used with no or limited labeled data, with a special focus on life sciences and health-care-related domains. These strategies are blurring the boundary between supervised and unsupervised learning.
Machine learning (ML) approaches are traditionally separated into supervised and unsupervised paradigms.13 Additionally, many researchers single out reinforcement learning as a third paradigm, which is not discussed in this review. In the supervised paradigm, the machine learning algorithm learns how to perform a task from data manually annotated by a person. It is worth mentioning that for the sake of this review we define supervision as manual, although such definition is strict. This is aimed to concretely define assisted approaches to labeling in the respective sections below. In contrast, the unsupervised paradigm aims to identify the patterns within the data algorithmically, i.e., without human help (Figure 1). Supervised learning delivers more desirable results for automation, aiming to mimic human behavior, but it falls short in scalability, due to the laborious and often expensive process of data annotation.14,15 Unsupervised learning can also be used to automate some tasks (e.g., anomaly detection), but it usually performs much more poorly than supervised learning because it is not guided by manually annotated data. Unsupervised learning is therefore more suited to data exploration tasks such as clustering and association mining.
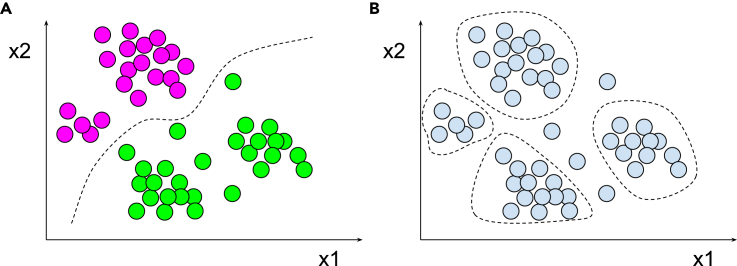
Figure 1.
A schematic depiction of supervised and unsupervised machine learning approaches
(A) An example of a supervised approach. Here, data points (circles) in ×1 and ×2 dimensions are labeled in magenta and green categories, allowing the model (dashed line) to be fitted.
(B) An example of an unsupervised approach. Here an algorithm attempts to detect patterns (clusters; dashed line) in unlabeled data (light blue circles).
For simplicity of graphical illustration, all the data points are depicted in a 2D space of ×1 and ×2. However, similar concepts apply to n-dimensional space. Dashed line represents an abstraction for a model or a decision boundary.
The scarcity of annotated data hinders the use of supervised learning in many practical use cases. This is especially pronounced for the application of higher expressive capacity models used in representation learning and deep learning (DL). Such models are much more data-hungry (often by a factor of a 100) compared with feature-generation-based ML.16, 17, 18 This means that even more annotated examples are required to learn relevant representations from the data than in traditional ML.
In this paper, we review some techniques that have been proposed by the ML community to address the annotated data scarcity problem. First, we discuss the value each data point brings to the resulting model. Next, we discuss Semi-supervised Learning and Active Learning, which fall between the supervised and the unsupervised paradigms by leveraging both labeled and unlabeled data. Data Augmentation and Self-supervised Learning generate reliable annotated data in an automatic fashion that can later be used by classical supervised learning models. Transfer Learning and Zero/One/Few-Shot Learning techniques are able to leverage models pre-trained for a similar context (Table 1). Finally, Weakly Supervised Learning techniques learn predictive models from inexpensive weak labels that may be wrong. Table 2 summarizes the references to biomedical applications for each technique and Table 3 depicts which approaches are relevant according to the amount of labeled and unlabeled data available.
Table 1.
Examples of zero-shot learning by formatting text data to fit models
Table 2.
References to biomedical applications
| Approach | Biomedical applications |
|---|---|
| Supervised | |
| Semi-supervised | |
| Active learning | |
| Data augmentation | |
| Transfer learning | |
| Self-supervised | |
| Few/one/zero-shot and few-shot learning | |
| Weakly supervised |
Table 3.
Relevant approaches according to the amount of labeled and unlabeled data available
| Amount of available data | Some unlabeled data | No unlabeled data |
|---|---|---|
| Enough labeled data to train a supervised model | Supervised learning | Supervised learning |
| Data augmentation | Data augmentation | |
| Semi-supervised learning | ||
| Active learning | ||
| Some labeled data, but not enough to train a supervised model | Data augmentation | Data augmentation |
| Semi-supervised learning | Transfer learninga | |
| Active learning | ||
| Transfer learninga | ||
| Self-supervised learninga | ||
| Few/one/zero-shot learninga | ||
| No labeled data | Active learning Zero-shot learninga |
Zero-shot learninga |
Large labeled datasets are required for pre-training.
Data value
ML model performance often improves as more data are collected.70 Increasing the size of the dataset serves two purposes simultaneously. First, it provides more information about the problem, making the solution likely to be more general. Second, it improves the performance of complex models. Increasing the dataset size often means an increase in the number of unannotated data points.15 However, in a supervised learning setting, simply collecting observations is often insufficient. To be used as the training data for supervised learning, these observations must often be manually annotated. For instance, ImageNet,14 an image dataset of around 1.3 million individual images, required manually annotating each of these images to belong to one or more of 1,000 classes in a routine and laborious process.71
However, not every data point is created equal. Some data points may be more useful to obtain a representative dataset. For example, for classification problems in cases when labeled data is scarce, it is preferable to have data points closer to the decision boundary (Figure 2). Several strategies for data point valuation have thus far been proposed, ranging from linear classification to decision trees and game theory (Shapley values).72 Purposeful data collection may be achieved only when the goal is clearly defined. Cost-effective ways are therefore designed to weight the data points differently during training by the effective number (expected volume) of samples73 or by focusing on hard examples.74
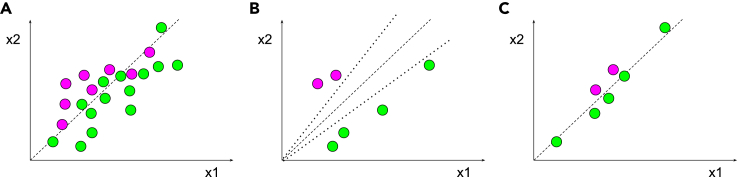
Figure 2.
Example of data point importance for model selection
Magenta and green circles are data points that correspond to respective class labels.
(A) An example of a full dataset with 8 data points belonging to magenta class and 14 belonging to the green class; dashed line represents a selected model.
(B) A subset of six data points selected from the full dataset is less valuable for accurate model definition, as multiple models (e.g., two dotted lines of incorrect models and one dashed line for correct model) can be fitted to this subset, but not the full dataset.
(C) A subset of six data points selected from the full dataset are more valuable for accurate model definition (single dashed line).
For simplicity of graphical illustration, all the data points are depicted in a 2D space of ×1 and ×2. However, similar concepts apply to n-dimensional space. Dashed line represents an abstraction for a model or a decision boundary.
In recent years, a number of approaches to improve efficiency of labeled data utilization have gained traction.17,75,76 Overall, these approaches attempt to improve the efficiency of labeled data utilization by high expressive capacity models. Such models often count millions of trainable parameters and hence require very large labeled datasets to avoid overfitting. Approaches tackling this are therefore indispensable for fields where labeled data is scarce. In this review, the most popular examples are discussed.
Semi-supervised learning
Semi-supervised learning is halfway between supervised and unsupervised learning.77 It trains predictive models from both labeled and unlabeled data (Figure 3A) in order to obtain better models than if they were trained with plain supervised learning on only the labeled data available.
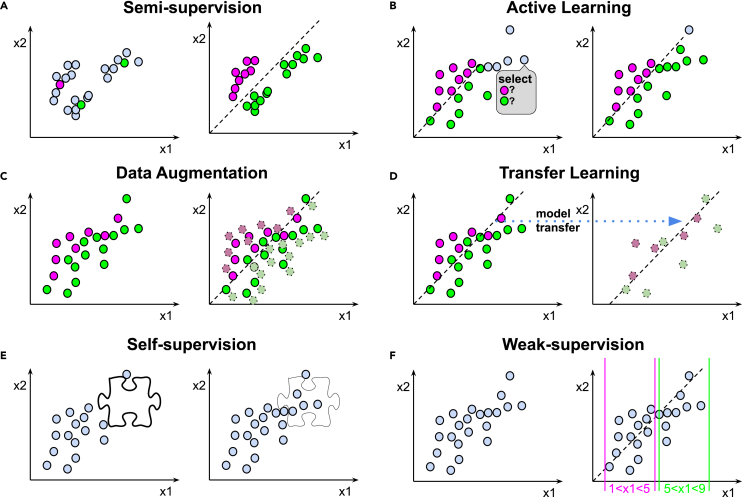
Figure 3.
Strategies between supervised and unsupervised approaches
Magenta and green colors correspond to respective class labels, blue circles represent unlabeled data, dashed lines represent the learned decision boundary.
(A) In a semi-supervised learning approach, clustering and manual annotation of few points is performed.
(B) In an active learning approach, an active request to the user to obtain annotation is performed; here it is depicted in a gray bubble with a “select” call to action.
(C) In the data augmentation approach, light circles with dashed borders represent data points obtained from the original through augmentation (e.g., linear transformation).
(D) A transfer learning approach uses a model pre-trained on one dataset (regular circles) and fine-tuned on another dataset (light circles with dashed borders). Here the trained model parameters transfer is symbolized by the dashed model line “transferred” from left to right as indicated by the blue arrow.
(E) Self-supervised learning approach. Here, a jigsaw in-painting task, which is not related to labels (so-called pretext task) is depicted as an example. The jigsaw pretext task is formulated automatically and allows learning of representations from the data.
(F) Weakly supervised learning approach. Here, magenta and green inequalities represent coarse heuristic rules used for data annotation.
For simplicity of graphical illustration, all the data points are depicted in a 2D space of ×1 and ×2. However, similar concepts apply to n-dimensional space. Dashed line represents an abstraction for a model or a decision boundary.
There is an important prerequisite to obtain better models than supervised learning: the distribution of examples, which unlabeled data will help elucidate, must be relevant for the prediction problem at hand. If this condition is not met, using unlabeled data can degrade the prediction accuracy by misguiding the inference. In practice, semi-supervised learning strategies rely on assumptions about the data.
For instance, the generative semi-supervised method that consists in applying the expectation maximization (EM) algorithm on both labeled and unlabeled data78 relies on the cluster assumption (if points are in the same cluster, they are likely to be of the same class). Another example is transductive support vector machine (SVM),79 which implements the low-density separation assumption (the decision boundary should lie in a low-density region): it maximizes the margin for both labeled and unlabeled data. The reader may refer to Chapelle et al.77,80 for more details about semi-supervised learning assumptions and strategies.
Semi-supervised learning will be most useful whenever there is far more unlabeled data than labeled. This is likely to occur if obtaining data points is cheap, but obtaining the labels costs a lot of time, effort, or money. For instance, semi-supervised learning is particularly suited to the classification of protein sequences.28 Protein sequences are nowadays acquired at industrial speed, but determining the functions of a single protein may require years of scientific work.
Active learning
Active learning81 is an iterative human-in-the-loop process that starts by identifying the unlabeled data points that are considered as the most useful for training a predictive model. At each iteration, unlabeled data points are queried by the active learning strategy and annotated by human experts, and the model is updated with the newly annotated data points (Figure 3B). Similar to semi-supervised learning, active learning aims to efficiently leverage unlabeled data during the learning process for performance promotion, while reducing the human expert workload. Both techniques can be combined to get the best out of the unlabeled data.82
The traditional active learning strategy, uncertainty sampling,83 selects the data points closest to the decision boundary for annotation. The theory is that the model is uncertain about the prediction of these data points, so annotating them will significantly increase the accuracy. Other active learning strategies are more explorative and exploit the structure in data to annotate points from all of the data space.84 Active learning strategies must find the right trade-off between exploitation and exploration, i.e., selecting data points both from sampled and unsampled areas of the data space.
Selecting the right query strategy and model for a given dataset is a real challenge. Indeed, active learning strategies perform differently on different datasets and there is no guarantee that they will outperform random sampling.35,85 Meta-learning is investigated to address this issue:35,86 it leverages reinforcement learning to learn the best active learning strategy.
Active learning is a human-in-the-loop process where the user experience should not be overlooked.87,88 Taking humans into account and ensuring a good user experience is as important as choosing an optimal query strategy to effectively train a performant predictive model with reduced human workload. However, most studies do not evaluate their query strategy in a real-world setting: they leverage oracles that answer the queries automatically from fully annotated datasets and they leave the user experiments for future work. They assess the human workload with the number of manual annotations, but the time spent in the overall annotation process is a much more realistic metric. Indeed, data points do not all have the same cost to be annotated. For instance, the data points queried by uncertainty sampling, close to the decision boundary, are often tricky cases that are more costly to annotate.89 In a real-world setting, even domain experts may not be able to annotate ambiguous data points. Moreover, studies often assume that a single data point is queried and annotated at each active learning iteration, to always select the optimal query. This optimal setting is, however, not workable in real-world annotation systems,90 as the annotators would spend more time waiting for annotation queries (while the predictive model is updated and the next annotation query is computed) than actually annotating data. Finally, most biomedical annotation tasks require specific domain knowledge, and crowdsourcing cannot be leveraged to annotate data at low cost. As annotating data is a cumbersome task, it is critical to show the biomedical experts that their annotations improve the accuracy of the predictive model, so that they continue annotating.18,87 It is, however, not straightforward to provide such feedback:91 most studies leverage a fully annotated validation dataset to assess the performance of the predictive model across iterations, but such a dataset is usually not available when deploying an annotation system.
There is currently little research that focuses on user experience in active learning processes,18,87,92,93 and very few studies assess their method with user experiments.94 Such research would significantly foster the use of this technique in real-world annotation systems.
Active learning strategies are particularly useful to annotate datasets for unbalanced prediction problems where random sampling is not effective. For example, it has been widely applied to automate drug discovery,30,31 where the active learning strategy identifies which experiments to perform next. Active learning is also relevant when crowdsourcing cannot be leveraged because expert knowledge is required to annotate11,35,34 or the data are too sensitive to be shared.
Data augmentation
An early attempt to address the mismatch on availability and requirement of labeled data in DL was data augmentation95 (Figure 3C). Traditional techniques apply label-preserving transformations to the already-annotated data points to increase the amount of training data. For instance, image data can simply be flipped or rotated,17,38 and text data can be slightly modified through deletion of random words and synonym substitution.43,96,97
Generative adversarial networks (GANs)98 can be used to generate a much broader set of augmentations.99 GANs learn the data distribution from some training data and then generate new samples that are as realistic as possible from this distribution. They use two competing neural networks: one that generates new samples from noise and one that discriminates samples as real or synthetic. GANs have been used in medical imaging to generate additional realistic training data.100 For example, Calimeri et al.39 generated magnetic resonance imaging (MRI) slices of the human brain with GANs and human physicians were not able to distinguish the artificially generated examples from the real ones.
Augmentation strategies are often carefully selected and fine-tuned for each individual case, taking domain-specific understanding of medical image data into account to avoid noise and artifacts. For instance, Mok and Chung40 have proposed a new data augmentation technique that generates images with the desired invariance and robustness properties for brain tumor segmentation. They have fine-tuned their GAN to generate images with realistic tumor boundaries. In another study, Gupta and colleagues used image-to-image translation GAN to enrich images with a broader pallet of readouts.101 In contrast to conventional augmentation, however, such an approach did not lead to an increase of individual data points.
In contrast to semi-supervised and active learning techniques, data augmentation strategies are useful even when a large amount of unlabeled data is not available. Data augmentation can be used to address class imbalance by generating more examples of underrepresented classes. For example, Ollagnier and Williams43 have compared several text data augmentation techniques to remedy class imbalance in clinical case classification. In the case of image data, Jin et al.41 have used a GAN to increase the training data of their DL model for pathological lung segmentation of CT scans. They have particularly focused on generating examples where nodules lie on the lung border because these specific cases were previously segmented poorly and underrepresented in the original training dataset.
Transfer learning
Transfer learning is an ML technique where a model that is trained on one task is then repurposed on a second related task102 (Figure 3D). It is not specific to DL, but it has been widely leveraged in this field as the representation layers can be easily shared by different prediction tasks. In practice, the weights of the representation layers are set according to a model trained on a previous task. Then, these weights can be either used for initialization with subsequent update during the training process (fine-tuning) or completely frozen, and only the task-specific layers are trained.
Transfer learning is a relevant solution to deal with the lack of annotated data if you can identify a related task with abundant labeled data or if a pre-trained model is available. It has significantly fostered the use of representation learning in biomedical applications.
Manual feature engineering was often preferred over representation learning in the biomedical field because of the lack of training data. Thanks to transfer learning, generic DL models and generic annotated datasets can be leveraged to train biomedical prediction models with few domain-specific annotated examples. For instance, the natural language model BioBERT103 has been created from the generic model BERT104 through transfer learning to obtain better performance on biomedical text mining tasks. It has been initialized with the weights of BERT (trained on general domain corpora) and trained on biomedical domain corpora. BioBERT has also been further fine-tuned through transfer learning for specific NLP tasks such as biomedical Named Entity Recognition.51 A similar approach was applied to scientific text, clinical notes, and PubMed articles, and yielded SciBERT,52 ClinicalBERT,53 and PubMedBERT.54 Transfer learning is also widely used in computer vision to create biomedical specific models.46 For instance, Cheng et al.47 have leveraged transfer learning from a model pre-trained on ImageNet14 to identify specific patterns from abdominal radiographs with limited training data. Another group48 also used ImageNet pre-trained ResNet-50 for a classification task in micrographs of virus-infected cells. While other examples49 include transfer learning from models trained on, for example, an MNIST dataset,105 overall transfer learning from ImageNet-pre-trained models remains the most-favored approach. Proteomics and genomics benefit from transfer learning as well. For example, Deznabi et al.58 used ProtVec106 to predict kinase-phosphosite association.
Self-supervised learning
Self-supervised learning (Figure 3E) learns relevant representations (or embeddings) without any manual annotation cost.75,107, 108, 109, 110, 111, 112 It relies on a two-step process: (1) a pretext task (or self-supervised task) is used to learn meaningful representations from annotations that are inherent to the data, and (2) the learned representations are leveraged to address the downstream task, the task at hand, with many fewer labeled data.
Pretext tasks themselves do not usually provide any useful application. Their learning objective should be set properly to receive supervision from the data themselves. This way, a large amount of annotated data can be generated automatically to learn high-quality representations. Several pretext tasks have been proposed for images; Doersch et al.113 have proposed a context prediction task where the objective is to predict the relative location of patches extracted from a given image. A similar pretext task consists of solving jigsaw puzzles where the tiles have been generated automatically by splitting images.107,114 Pretext tasks can also rely on generative models. For instance, in-painting115 is a generative pretext task where an arbitrary fraction of an image is removed, and the objective is to reconstruct it from the remaining context. In-painting has recently been demonstrated57 to be capable of predicting unseen fluorescence signals on a single-cell level in confocal microscopy. Colorization116 is another example of a generative pretext task: a gray-scale filter is applied to the input images and the objective is to recover the colors. Pretext tasks for language data are posed more natively, e.g., with next-word prediction117,118 or masked language modeling and next-sentence prediction used by BERT.104
This approach may not immediately make intuitive sense. After all, what practical use would such an ML model present for solving, e.g., a classification problem? However, in the sense of representation learning, performing such a task achieves the main part of the goal: it allows the system to learn meaningful representation from a large dataset. Once done, these representations can be repurposed through transfer learning (weights transfer) and fine-tuning to address an actual task at hand with fewer labeled data. Furthermore, these two steps can be merged into one through shared weights.19,119, 120, 121
Representations learned through self-supervised learning can be leveraged for both supervised and unsupervised downstream tasks. The learned representations can be used directly for unsupervised learning tasks such as clustering122 or similarity computation for search and retrieval tasks.123 The model trained for the pretext task can also be repurposed through transfer learning57 to address supervised tasks with many fewer labeled data. Self-supervised learning bridges the gap between supervised and unsupervised learning by focusing on learning high-quality representations. The pretext task must be chosen carefully to get representations as meaningful as possible for the downstream task. Multi-task learning can also be leveraged to train relevant representations from several pretext tasks.124
Self-supervised learning is proving incredibly powerful across various domains and types of data. This set of techniques refocuses the field on learning high-quality representations bridging domains of generative and discriminative modeling.125
Few/one/zero-shot learning
When there are only a limited number of valuable labeled data points, ML solutions have to adapt and generalize to tackle few-shot or one-shot learning, especially to avoid overfitting.126,127 In some extreme situations, there are no labeled data at all for training (for example, when there are too many labels to annotate), and the model has to deal with unseen labels, leading to a challenge called zero-shot learning.128 The solutions to the few/one/zero-shot learning challenges essentially imitate how humans recognize and learn by analogy when there is limited evidence. These solutions often fall into three strategies: relationship similarity, task reduction, and prior knowledge.
First, in most cases, with proper embedding or vector representation, the few/one/zero-shot learning models can locate the data or labels in the high-dimensional “semantic space”, measure the similarity of their relationship to those in the training set, and provide proper predictions accordingly.129,20 The embedding approach has proved its effectiveness in multiple biomedical domains, such as affinity prediction,58 drug discovery,59 literature indexing,60 image recognition for cancer detection,61,62 and patient clustering.63 To generate suitable embeddings or vector representations in each biomedical domain, self-supervised learning and transfer learning methods can be leveraged.
Second, when there are well-established models in the same domain, it is more straightforward to format data to fit the models than to tune the models to fit the data. For example, with auxiliary sentences, the input and output of a text classification task can be compatible with a question-and-answer model,19 a natural-language-inference model,20 or a cloze model21,22 (Table 1). Last, but not least, prior structured knowledge can provide additional domain knowledge to the models130 to support “reasoning.” For example, the International Classification of Diseases (ICD) hierarchy improves the classification model performance of infrequent labels in electronic medical records (EMR).131
Weakly supervised learning
Many challenging prediction tasks require complex models (i.e., with large numbers of parameters) that need large training datasets. However, it is often difficult to acquire strong supervision such as large fully annotated datasets with perfectly accurate labels due to the high cost of the data-labeling process. In some cases, only weak supervision can be collected, and some training algorithms, called weakly supervised, can handle this imperfect supervision to build predictive models.132 The goal of weak supervision is to train predictive models on large imperfectly labeled training datasets to get better performance than fully supervised models trained on small datasets annotated with perfectly accurate labels.
There are many settings of weak supervision. For instance, distant supervision leverages knowledge bases to derive some weak supervision.133 In other cases, only partial labels, coarse-grained labels, may be available. Only the partial ordering may be provided when learning user preferences over items, or the coarse-grained label “flower" can be provided for a picture of an arum lily instead of spending a consequent amount of time to find the exact taxonomy.134
Multi-instance learning is another setting of weak supervision where the learner receives a set of labeled bags, each containing many instances. In the simple case of multiple-instance binary classification, a bag may be labeled negative if all the instances in it are negative. On the other hand, a bag is labeled positive if there is at least one instance in it which is positive. Multi-instance learning has been applied to drug-activity prediction,64 computer-aided diagnosis,65 and pathology.66
Weak supervision can also rely on labels with a low accuracy coming from rule-based heuristics135,136 (Figure 3F) or crowdsourcing annotations.137 In this setting, each instance can be associated with several labels, and the key technical challenge is how to unify and de-noise them, given that they are each noisy, may disagree with one another, may be correlated, and may have arbitrary (unknown) accuracies that may depend on the subset of the dataset being labeled.136 This type of weak supervision is gaining rapid adoption by the biomedical research community to assist in labeling electronic health records69 or MRI images.67
Conclusions: Biomedical applications
The recent advances in data-intensive computing have been accelerated by improvements in performance due to techniques such as representation learning that address data curation and feature engineering problems. Yet, as we improve the performance, models tend to get larger and the requirement for annotated datasets grows.
The set of techniques described in this review aim to make more efficient use of limited annotated data. These techniques focus on generating synthetic data from existing annotations, automating data annotation processes, or lowering the amount of annotated data needed to train the machine learning systems. Even though they were proposed independently, these methods show increasing overlap. For example, weak supervision and self-supervision in DL ultimately serve the same purpose, exploit similar implicit properties of the learning systems, and often require fine-tuning for the best performance. Weak supervision also has some conceptual overlap with data augmentation: both techniques generate labeled data automatically with a rule-based approach to provide some supervision at low cost. Another example is the similarity between semi-supervision, weak supervision, and active learning: they all leverage unlabeled data to enrich or create an annotated dataset with different levels of involvement from domain experts. While semi-supervised learning pseudo-labels unlabeled data automatically, weakly supervised learning requires domain experts to provide pseudo-labeling rules, and active learning requires even more involvement of domain experts to manually annotate some unlabeled data points. Finally, transfer learning is closely related to self-supervised learning and few/one/zero-shot learning. Self-supervised learning and transfer learning are often applied jointly: self-supervised learning allows the collection of relevant representations at low cost, which are then fine-tuned through transfer learning to answer the problem at hand with fewer annotated data points. As for few/one-shot learning, it often leverages a model previously trained for another task such as transfer learning, while zero-shot learning often leverages a model pre-trained with self-supervised learning without fine-tuning.
Several dawning and established techniques have been omitted from this review due to their broader focus on the way systems learn. They include meta-learning,138 and universal representations.139 The former relates to approaches aimed at optimizing the representation learning algorithms using training meta-data, i.e., learning to learn. Recent work in this field shows further improvement of learning algorithms is still possible.140 The latter seeks to identify the best algorithmic way to obtain reusable (i.e., “universal”) representations valid across multiple domains building upon self-supervised learning approaches. Perhaps it is those, or the methods reviewed in this work, that will one day allow us to stop searching for the labels in a haystack of data.
Acknowledgments
Declaration of interests
The authors declare no competing interests.
Contributor Information
Artur Yakimovich, Email: artur.yakimovich@roche.com.
Anaël Beaugnon, Email: anael.beaugnon@roche.com.
Yi Huang, Email: anael.beaugnon@roche.com.
Elif Ozkirimli, Email: elif.ozkirimli@roche.com.
References
- 1.Sanchez-Garcia R., Segura J., Maluenda D., Carazo J.M., Sorzano C.O.S. Deep Consensus, a deep learning-based approach for particle pruning in cryo-electron microscopy. IUCrJ. 2018;5:854–865. doi: 10.1107/S2052252518014392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wollmann T., Rohr K. Deep Consensus Network: Aggregating predictions to improve object detection in microscopy images. Med. Image Anal. 2021;70:102019. doi: 10.1016/j.media.2021.102019. [DOI] [PubMed] [Google Scholar]
- 3.Xiao X., Min J.-L., Lin W.-Z., Liu Z., Cheng X., Chou K.-C. iDrug-Target: predicting the interactions between drug compounds and target proteins in cellular networking via benchmark dataset optimization approach. J. Biomol. Struct. Dyn. 2015;33:2221–2233. doi: 10.1080/07391102.2014.998710. [DOI] [PubMed] [Google Scholar]
- 4.Ghassemi M., Naumann T., Schulam P., Beam A.L., Chen I.Y., Ranganath R. A review of challenges and opportunities in machine learning for health. AMIA Jt. Summits Transl. Sci. Proc. 2020;2020:191–200. [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith S.D., Tucker M., Bowser B., Calkins G., Chang C.-H.J., Guardino E., Khozin S., Kraut J., You P., Schrag D., Miksad R.A. Generating real-world tumor burden endpoints from electronic health record data: comparison of RECIST, radiology-anchored, and clinician-anchored approaches for abstracting real-world progression in non-small cell lung cancer. Adv. Ther. 2019;36:2122–2136. doi: 10.1007/s12325-019-00970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geneviève L.D., Martani A., Shaw D., Elger B.S., Wangmo T. Structural racism in precision medicine: leaving no one behind. BMC Med. Ethics. 2020;21:17. doi: 10.1186/s12910-020-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obermeyer Z., Powers B., Vogeli C., Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366:447–453. doi: 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 8.Krallinger M., Rabal O., Lourenço A., Oyarzabal J., Valencia A. Information retrieval and text mining technologies for chemistry. Chem. Rev. 2017;117:7673–7761. doi: 10.1021/acs.chemrev.6b00851. [DOI] [PubMed] [Google Scholar]
- 9.Köksal A., Dönmez H., Özçelik R., Ozkirimli E., Özgür A. Vapur: a search engine to find related protein - compound pairs in COVID-19 literature. bioRxiv. 2020 2020.09.05.284224. [Google Scholar]
- 10.Lu Wang L., Lo K., Chandrasekhar Y., Reas R., Yang J., Eide D., Funk K., Kinney R., Liu Z., Merrill W., et al. CORD-19: the covid-19 open research dataset. ArXiv. 2020 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7251955/ [Google Scholar]
- 11.Sahiner B., Pezeshk A., Hadjiiski L.M., Wang X., Drukker K., Cha K.H., Summers R.M., Giger M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019;46:e1–e36. doi: 10.1002/mp.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spasic I., Nenadic G. Clinical text data in machine learning: systematic review. JMIR Med. Inform. 2020 doi: 10.2196/17984. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7157505/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinton G.E., Sejnowski T.J. MIT Press; 1999. Unsupervised Learning: Foundations of Neural Computation; p. 420. [Google Scholar]
- 14.Deng J., Dong W., Socher R., Li L.-J., Li Kai, Fei-Fei Li. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 15.Sorokin A., Forsyth D. 2008 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops. 2008. Utility data annotation with Amazon mechanical Turk; pp. 1–8. [Google Scholar]
- 16.Hinton G.E., Srivastava N., Krizhevsky A., Sutskever I., Salakhutdinov R.R.. Improving neural networks by preventing co-adaptation of feature detectors. ArXiv http://arxiv.org/abs/1207.0580
- 17.Krizhevsky A., Sutskever I., Hinton G.E. In: Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q., editors. Vol. 25. Curran Associates, Inc.; 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105.http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf (Advances in Neural Information Processing Systems). [Google Scholar]
- 18.Sun C., Shrivastava A., Singh S., Gupta A.. Revisiting unreasonable effectiveness of data in deep learning era. ArXiv http://arxiv.org/abs/1707.02968
- 19.Sun Y., Tzeng E., Darrell T., Efros A.A. Unsupervised domain adaptation through self-supervision. ArXiv. 2019 http://arxiv.org/abs/1909.11825 [Google Scholar]
- 20.Yin W., Hay J., Roth D. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) Padó S., Huang R., editors. Association for Computational Linguistics; 2019. Benchmarking zero-shot text classification: datasets, evaluation and entailment approach; pp. 3914–3923. [Google Scholar]
- 21.Schick T., Schütze H. It’s not just size that matters: small language models are also few-shot learners. ArXiv. 2020 http://arxiv.org/abs/2009.07118 [Google Scholar]
- 22.Tam D., Menon R.R., Bansal M., Srivastava S., Raffel C. Improving and simplifying pattern exploiting training. ArXiv. 2021 http://arxiv.org/abs/2103.11955 [Google Scholar]
- 23.Öztürk H., Özgür A., Ozkirimli E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34:i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun S., Dong B., Zou Q. Revisiting genome-wide association studies from statistical modelling to machine learning. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa263. [DOI] [PubMed] [Google Scholar]
- 25.Zrimec J., Börlin C.S., Buric F., Muhammad A.S., Chen R., Siewers V., Verendel V., Nielsen J., Töpel M., Zelezniak A. Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure. Nat. Commun. 2020;11:1–16. doi: 10.1038/s41467-020-19921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisch D., Yakimovich A., Clough B., Wright J., Bunyan M., Howell M., Mercer J., Frickel E. Defining host–pathogen interactions employing an artificial intelligence workflow. eLife. 2019;8:e40560. doi: 10.7554/eLife.40560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas A.M., Ryder P.V., Li B., Cimini B.A., Eliceiri K.W., Carpenter A.E. Open-source deep-learning software for bioimage segmentation. Mol. Biol. Cell. 2021;32:823–829. doi: 10.1091/mbc.E20-10-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weston J., Leslie C., Ie E., Zhou D., Elisseeff A., Noble W.S. Semi-supervised protein classification using cluster kernels. Bioinformatics. 2005;21:3241–3247. doi: 10.1093/bioinformatics/bti497. [DOI] [PubMed] [Google Scholar]
- 29.Krogel M.-A., Scheffer T. Multi-relational learning, text mining, and semi-supervised learning for functional genomics. Mach. Learn. 2004;57:61–81. [Google Scholar]
- 30.Reker D. Practical considerations for active machine learning in drug discovery. Drug Discov. Today Technol. 2019;32–33:73–79. doi: 10.1016/j.ddtec.2020.06.001. (Artificial Intelligence) [DOI] [PubMed] [Google Scholar]
- 31.Schneider G. Automating drug discovery. Nat. Rev. Drug Discov. 2018;17:97–113. doi: 10.1038/nrd.2017.232. [DOI] [PubMed] [Google Scholar]
- 32.Farid D.M., Nowé A., Manderick B. 25th Belgian-Dutch Conference on Machine Learning (Benelearn) 2016. Combining boosting and active learning for mining multi-class genomic data; pp. 1–2. [Google Scholar]
- 33.Liu Y. Active learning with support vector machine applied to gene expression data for cancer classification. J. Chem. Inf. Comput. Sci. 2004;44:1936–1941. doi: 10.1021/ci049810a. [DOI] [PubMed] [Google Scholar]
- 34.Hoi S.C.H., Jin R., Zhu J., Lyu M.R. Proceedings of the 23rd International Conference on Machine Learning. Association for Computing Machinery; 2006. Batch mode active learning and its application to medical image classification; pp. 417–424. (ICML ’06) [DOI] [Google Scholar]
- 35.De Angeli K., Gao S., Alawad M., Yoon H.-J., Schaefferkoetter N., Wu X.-C., Durbin E.B., Doherty J., Stroup A., Coyle L., et al. Deep active learning for classifying cancer pathology reports. BMC Bioinformatics. 2021;22:113. doi: 10.1186/s12859-021-04047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhari P., Agarwal H., Bhateja V. Data augmentation for cancer classification in oncogenomics: an improved KNN based approach. Evol. Intell. 2021;14:489–498. [Google Scholar]
- 37.Chen J., Mowlaei M.E., Shi X.. Population-scale Genomic Data Augmentation Based on Conditional Generative Adversarial Networks. In: Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. 2020. p. 1–6.
- 38.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 39.Calimeri F., Marzullo A., Stamile C., Terracina G. In proceedings of the International Conference on Artificial Neural Networks; 2017. Biomedical data augmentation using generative adversarial neural networks.https://link.springer.com/chapter/10.1007/978-3-319-68612-7_71 [Google Scholar]
- 40.Mok T.C.W., Chung A.C.S. Learning data augmentation for brain tumor segmentation with coarse-to-fine generative adversarial networks. ArXiv. 2019;11383:70–80. [Google Scholar]
- 41.Jin D., Xu Z., Tang Y., Harrison A.P., Mollura D.J. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Frangi A., Schnabel J., Davatzikos C., Alberola-López C., Fichtinger G., editors. Springer; 2018. CT-realistic lung nodule simulation from 3D conditional generative adversarial networks for robust lung segmentation; pp. 732–740. [Google Scholar]
- 42.Horlava N., Mironenko A., Niehaus S., Wagner S., Roeder I., Scherf N. A comparative study of semi- and self-supervised semantic segmentation of biomedical microscopy data. ArXiv. 2020 http://arxiv.org/abs/2011.08076 [Google Scholar]
- 43.Ollagnier A., Williams H. Vol. 9. 2020. Text Augmentation Techniques for Clinical Case Classification. [Google Scholar]
- 44.Schwessinger R., Gosden M., Downes D., Brown R.C., Oudelaar A.M., Telenius J., Teh Y.W., Lunter G., Hughes J.R. DeepC: predicting 3D genome folding using megabase-scale transfer learning. Nat. Methods. 2020;17:1118–1124. doi: 10.1038/s41592-020-0960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taroni J.N., Grayson P.C., Hu Q., Eddy S., Kretzler M., Merkel P.A., Greene C.S. MultiPLIER: a transfer learning framework for transcriptomics reveals systemic features of rare disease. Cell Syst. 2019;8:380–394. doi: 10.1016/j.cels.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raghu M., Zhang C., Kleinberg J., Bengio S. Transfusion: understanding transfer learning for medical imaging. ArXiv. 2019 http://arxiv.org/abs/1902.07208 [Google Scholar]
- 47.Cheng P.M., Tejura T.K., Tran K.N., Whang G. Detection of high-grade small bowel obstruction on conventional radiography with convolutional neural networks. Abdom. Radiol. N. Y. 2018;43:1120–1127. doi: 10.1007/s00261-017-1294-1. [DOI] [PubMed] [Google Scholar]
- 48.Andriasyan V., Yakimovich A., Petkidis A., Georgi F., Witte R., Puntener D., Greber U.F. Microscopy deep learning predicts virus infections and reveals mechanics of lytic-infected cells. Iscience. 2021;24:102543. doi: 10.1016/j.isci.2021.102543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yakimovich A., Huttunen M., Samolej J., Clough B., Yoshida N., Mostowy S., Frickel E.-M., Mercer J. Mimicry embedding facilitates advanced neural network training for image-based pathogen detection. Msphere. 2020;5 doi: 10.1128/mSphere.00836-20. e00836–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., et al. Identifying medical diagnoses and treatable Diseases by image-based deep learning. Cell. 2018;172:1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Symeonidou A., Sazonau V., Groth P. SEMANTICS Posters&Demos. 2019. Transfer learning for biomedical named entity recognition with BioBERT. [Google Scholar]
- 52.Beltagy I., Lo K., Cohan A. Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) Association for Computational Linguistics; 2019. SciBERT: a pretrained language model for scientific text; pp. 3613–3618.https://www.aclweb.org/anthology/D19-1371 [Google Scholar]
- 53.Huang K., Altosaar J., Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. ArXiv. 2020 http://arxiv.org/abs/1904.05342 [Google Scholar]
- 54.Gu Y., Tinn R., Cheng H., Lucas M., Usuyama N., Liu X., Naumann T., Gao J., Poon H. Domain-specific language model pretraining for biomedical natural language processing. ArXiv. 2021 http://arxiv.org/abs/2007.15779 [Google Scholar]
- 55.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kung S.-Y., Luo Y., Mak M.-W. Feature selection for genomic signal processing: unsupervised, supervised, and self-supervised scenarios. J. Signal. Process. Syst. 2010;61:3–20. [Google Scholar]
- 57.Lu A.X., Kraus O.Z., Cooper S., Moses A.M. Learning unsupervised feature representations for single cell microscopy images with paired cell inpainting. PLoS Comput. Biol. 2019;15:e1007348. doi: 10.1371/journal.pcbi.1007348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deznabi I., Arabaci B., Koyutürk M., Tastan O. DeepKinZero: zero-shot learning for predicting kinase–phosphosite associations involving understudied kinases. Bioinformatics. 2020;36:3652–3661. doi: 10.1093/bioinformatics/btaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altae-Tran H., Ramsundar B., Pappu A.S., Pande V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 2017;3:283–293. doi: 10.1021/acscentsci.6b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mylonas N., Karlos S., Tsoumakas G. 11th Hellenic Conference on Artificial Intelligence. ACM; 2020. Zero-shot classification of biomedical articles with emerging MeSH descriptors; pp. 175–184.https://dl.acm.org/doi/10.1145/3411408.3411414 [DOI] [PubMed] [Google Scholar]
- 61.Kim M., Zuallaert J., De Neve W. Proceedings of the 2nd International Workshop on Multimedia for Personal Health and Health Care. Association for Computing Machinery; 2017. Few-shot learning using a small-sized dataset of high-resolution FUNDUS images for glaucoma diagnosis; pp. 89–92. (MMHealth ’17) [DOI] [Google Scholar]
- 62.Medela A., Picon A., Saratxaga C.L., Belar O., Cabezón V., Cicchi R., Bilbao R., Glover B. 2019 IEEE 16th International Symposium on Biomedical Imaging. ISBI 2019; 2019. Few shot learning in histopathological images:reducing the need of labeled data on biological datasets; pp. 1860–1864. [Google Scholar]
- 63.Ma T., Zhang A. Affinity network fusion and semi-supervised learning for cancer patient clustering. Methods. 2018;145:16–24. doi: 10.1016/j.ymeth.2018.05.020. (Data mining methods for analyzing biological data in terms of phenotypes) [DOI] [PubMed] [Google Scholar]
- 64.Dietterich T.G., Lathrop R.H., Lozano-Pérez T. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell. 1997;89:31–71. [Google Scholar]
- 65.Fung G., Dundar M., Krishnapuram B., Rao R.B. Multiple instance learning for computer aided diagnosis. Adv. Neural Inf. Process. Syst. 2007;19:425. [Google Scholar]
- 66.Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Silva V.W.K., Busam K.J., Brogi E., Reuter V.E., Klimstra D.S., Fuchs T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fries J.A., Varma P., Chen V.S., Xiao K., Tejeda H., Saha P., Dunnmon J., Chubb H., Maskatia S., Fiterau M., et al. Weakly supervised classification of aortic valve malformations using unlabeled cardiac MRI sequences. Nat. Commun. 2019;10:3111. doi: 10.1038/s41467-019-11012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doan M., Barnes C., McQuin C., Caicedo J.C., Goodman A., Carpenter A.E., Rees P. Deepometry, a framework for applying supervised and weakly supervised deep learning to imaging cytometry. Nat. Protoc. 2021:1–24. doi: 10.1038/s41596-021-00549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Callahan A., Fries J.A., Ré C., Huddleston J.I., Giori N.J., Delp S., Shah N.H. Medical device surveillance with electronic health records. Npj Digit. Med. 2019;2:1–10. doi: 10.1038/s41746-019-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halevy A., Norvig P., Pereira F. The unreasonable effectiveness of data. IEEE Intell. Syst. 2009;24:8–12. [Google Scholar]
- 71.Sambasivan N., Kapania S., Highfill H., Akrong D., Paritosh P., Aroyo L.M. Proceedings of the 2021 CHI Conference on Human Factors in Computing Systems. Association for Computing Machinery; 2021. “Everyone wants to do the model work, not the data work”: data Cascades in High-Stakes AI; pp. 1–15. (CHI ’21) [DOI] [Google Scholar]
- 72.Tideman L.E.M., Migas L.G., Djambazova K.V., Patterson N.H., Caprioli R.M., Spraggins J.M., Van de Plas R. Automated biomarker candidate discovery in imaging mass spectrometry data through spatially localized shapley additive explanations. Anal. Chim. Acta. 2021:338522. doi: 10.1016/j.aca.2021.338522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cui Y., Jia M., Lin T.-Y., Song Y., Belongie S. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. Class-balanced loss based on effective number of samples; pp. 9268–9277. [Google Scholar]
- 74.Lin H., Gao S., Gotz D., Du F., He J., Cao N. Rclens: interactive rare category exploration and identification. IEEE Trans. Vis. Comput. Graph. 2017;24:2223–2237. doi: 10.1109/TVCG.2017.2711030. [DOI] [PubMed] [Google Scholar]
- 75.Jing L., Tian Y. Self-supervised visual feature learning with deep neural networks: a survey. ArXiv. 2019 doi: 10.1109/TPAMI.2020.2992393. http://arxiv.org/abs/1902.06162 [DOI] [PubMed] [Google Scholar]
- 76.Zhu X.(Jerry) University of Wisconsin-Madison Department of Computer Sciences; 2005. Semi-Supervised Learning Literature Survey.https://minds.wisconsin.edu/handle/1793/60444 [Google Scholar]
- 77.Chapelle O., Scholkopf B., Zien A. Semi-supervised learning (Chapelle, O. et al., Eds.; 2006) [Book Reviews] IEEE Trans. Neural Netw. 2009;20:542. [Google Scholar]
- 78.Titterington D.M., Afm S., Smith A.F., Makov U., others . John Wiley & Sons Incorporated; 1985. Statistical Analysis of Finite Mixture Distributions. [Google Scholar]
- 79.Vapnik V. Vol. 1. Wiley. N. Y.; 1998. p. 2. (Statistical Learning Theory). [Google Scholar]
- 80.van Engelen J.E., Hoos H.H. A survey on semi-supervised learning. Mach. Learn. 2020;109:373–440. [Google Scholar]
- 81.Settles B. Active Learning Literature Survey. CS Technical Reports. 2009;67 https://minds.wisconsin.edu/handle/1793/60660 [Google Scholar]
- 82.Zhu X., Lafferty J., Ghahramani Z. In: Fawcett T., Mishra N., editors. Vol. 3. In proceedings of the Twentieth International Conference on Machine Learning; 2003. Combining active learning and semi-supervised learning using Gaussian fields and harmonic functions; p. 1000. (ICML 2003 Workshop on the Continuum from Labeled to Unlabeled Data in Machine Learning and Data Mining). [Google Scholar]
- 83.Lewis D.D., Gale W.A. In: SIGIR’94. Croft B.W., van Rijsbergen C.J., editors. Springer; 1994. A sequential algorithm for training text classifiers; pp. 3–12. [Google Scholar]
- 84.Dasgupta S., Hsu D. Proceedings of the 25th International Conference on Machine Learning - ICML ’08. ACM Press; 2008. Hierarchical sampling for active learning; pp. 208–215.http://portal.acm.org/citation.cfm?doid=1390156.1390183 [Google Scholar]
- 85.Duros V., Grizou J., Xuan W., Hosni Z., Long D.-L., Miras H.N., Cronin L. Human versus robots in the discovery and crystallization of gigantic polyoxometalates. Angew. Chem. Int. Ed. 2017;56:10815–10820. doi: 10.1002/anie.201705721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bachman P., Sordoni A., Trischler A. In: International Conference on Machine Learning. Precup D., Teh Y.W., editors. PMLR; 2017. Learning algorithms for active learning; pp. 301–310. [Google Scholar]
- 87.Amershi S., Cakmak M., Knox W.B., Kulesza T. Power to the people: the role of humans in interactive machine learning. AI Mag. 2014;35:105–120. [Google Scholar]
- 88.Wagstaff K. In: Proceedings of the 29th International Conference on Machine Learning (ICML-12) Langford J., Pineau J., editors. Omnipress; 2012. Machine learning that matters; pp. 529–536. (ICML ’12) [Google Scholar]
- 89.Settles B., Craven M., Friedland L. In: Krishnapuram B.R., Yu S., Oksana Y., Bharat Rao R., Carin L., editors. Vol. 1. Curran Associates; 2008. Active learning with real annotation costs. (Proceedings of the NIPS Workshop on Cost-Sensitive Learning). [Google Scholar]
- 90.Settles B. Active Learning and Experimental Design Workshop in Conjunction with AISTATS 2010. JMLR Workshop and Conference Proceedings; 2011. From theories to queries: active learning in practice; pp. 1–18.http://proceedings.mlr.press/v16/settles11a.html [Google Scholar]
- 91.Kottke D., Schellinger J., Huseljic D., Sick B. Limitations of assessing active learning performance at runtime. CoRR. 2019:10338. abs/1901. [Google Scholar]
- 92.Choi M., Park C., Yang S., Kim Y., Choo J., Hong S.R. Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems. Association for Computing Machinery; 2019. AILA: Attentive interactive labeling assistant for document classification through attention-based deep neural networks; pp. 1–12. [DOI] [Google Scholar]
- 93.Kulesza T., Amershi S., Caruana R., Fisher D., Charles D. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM; 2014. Structured labeling for facilitating concept evolution in machine learning; pp. 3075–3084.https://dl.acm.org/doi/10.1145/2556288.2557238 [Google Scholar]
- 94.Reker D., Bernardes G., Rodrigues T. Evolving and nano data enabled machine intelligence for chemical reaction optimization. 2018. https://chemrxiv.org/articles/Evolving_and_Nano_Data_Enabled_Machine_Intelligence_for_Chemical_Reaction_Optimization/7291205/1
- 95.Dyk D.A.van, Meng X.-L. The Art of data augmentation. J. Comput. Graph. Stat. 2001;10:1–50. [Google Scholar]
- 96.Giridhara P., Mishra C., Venkataramana R., Bukhari S., Dengel A. Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods. SCITEPRESS - Science and Technology Publications; 2019. A study of various text augmentation techniques for relation classification in free text; pp. 360–367.http://www.scitepress.org/DigitalLibrary/Link.aspx?doi=10.5220/0007311003600367 [Google Scholar]
- 97.Wang W.Y., Yang D. Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; 2015. That’s so Annoying‼!: a lexical and frame-semantic embedding based data augmentation approach to automatic categorization of annoying behaviors using #petpeeve tweets; pp. 2557–2563.https://www.aclweb.org/anthology/D15-1306 [Google Scholar]
- 98.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014;27:2672–2680. [Google Scholar]
- 99.Antoniou A., Storkey A., Edwards H. International Conference on Artificial Neural Networks. Springer; 2018. Augmenting image classifiers using data augmentation generative adversarial networks; pp. 594–603. [Google Scholar]
- 100.Yi X., Walia E., Babyn P. Generative adversarial network in medical imaging: a review. Med. Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 101.Gupta L., Klinkhammer B.M., Boor P., Merhof D., Gadermayr M. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. GAN-based image enrichment in digital pathology boosts segmentation accuracy; pp. 631–639. [Google Scholar]
- 102.Pratt L.Y. In: Advances in Neural Information Processing Systems 5. Hanson S.J., Cowan J.D., Giles C.L., editors. Morgan-Kaufmann; 1993. Discriminability-based transfer between neural networks; pp. 204–211.http://papers.nips.cc/paper/641-discriminability-based-transfer-between-neural-networks.pdf [Google Scholar]
- 103.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., Kang J. In: Bioinformatics. Wren J., editor. 2019. BioBERT: a pre-trained biomedical language representation model for biomedical text mining; p. btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Devlin J., Chang M.-W., Lee K., Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. ArXiv. 2019 http://arxiv.org/abs/1810.04805 [Google Scholar]
- 105.Deng L. The mnist database of handwritten digit images for machine learning research [best of the web] IEEE Signal. Process. Mag. 2012;29:141–142. [Google Scholar]
- 106.Asgari E., Mofrad M.R.K. Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS One. 2015;10:e0141287. doi: 10.1371/journal.pone.0141287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim D., Cho D., Kweon I.S. Self-supervised video representation learning with space-time cubic puzzles. Proc. AAAI Conf. Artif. Intell. 2019;33:8545–8552. [Google Scholar]
- 108.Kolesnikov A., Zhai X., Beyer L. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2019. Revisiting self-supervised visual representation learning; pp. 1920–1929.https://ieeexplore.ieee.org/document/8953672/ [Google Scholar]
- 109.Korbar B., Tran D., Torresani L. In: Bengio S., Wallach H., Larochelle H., Grauman K., Cesa-Bianchi N., Garnett R., editors. Vol. 31. Curran Associates, Inc.; 2018. Cooperative learning of audio and video models from self-supervised synchronization; pp. 7763–7774.http://papers.nips.cc/paper/8002-cooperative-learning-of-audio-and-video-models-from-self-supervised-synchronization.pdf (Advances in Neural Information Processing Systems). [Google Scholar]
- 110.Mahendran A., Thewlis J., Vedaldi A. In: Computer Vision – ACCV 2018. Jawahar C.V., Li H., Mori G., Schindler K., editors. Springer International Publishing; 2019. Cross pixel optical-flow similarity for self-supervised learning; pp. 99–116. (Lecture Notes in Computer Science) [Google Scholar]
- 111.Owens A., Efros A.A. Audio-visual scene analysis with self-supervised multisensory features. 2018. https://openaccess.thecvf.com/content_ECCV_2018/html/Andrew_Owens_Audio-Visual_Scene_Analysis_ECCV_2018_paper.html p. 631–648.
- 112.Sayed N., Brattoli B., Ommer B. In: Pattern Recognition. Brox T., Bruhn A., Fritz M., editors. Springer International Publishing; 2019. Cross and learn: cross-modal self-supervision; pp. 228–243. (Lecture Notes in Computer Science) [Google Scholar]
- 113.Doersch C., Gupta A., Efros A.A. Proceedings of the IEEE International Conference on Computer Vision. 2015. Unsupervised visual representation learning by context prediction; pp. 1422–1430. [Google Scholar]
- 114.Noroozi M., Favaro P. In: Computer Vision – ECCV 2016. Leibe B., Matas J., Sebe N., Welling M., editors. Springer International Publishing; 2016. Unsupervised learning of visual representations by solving jigsaw puzzles; pp. 69–84. (Lecture Notes in Computer Science) [Google Scholar]
- 115.Pathak D., Krahenbuhl P., Donahue J., Darrell T., Efros A.A. Context encoders: feature learning by inpainting. 2016. https://openaccess.thecvf.com/content_cvpr_2016/html/Pathak_Context_Encoders_Feature_CVPR_2016_paper.html p. 2536–2544.
- 116.Zhang R., Isola P., Efros A.A. European Conference on Computer Vision. Springer; 2016. Colorful image colorization; pp. 649–666. [Google Scholar]
- 117.Mikolov T., Sutskever I., Chen K., Corrado G., Dean J. Distributed representations of words and phrases and their compositionality. ArXiv. 2013;1310 arXiv:1310.4546. [Google Scholar]
- 118.Mikolov T., Chen K., Corrado G., Dean J. Efficient estimation of word representations in vector space. 2013. https://arxiv.org/abs/1301.3781v3
- 119.Caron M., Touvron H., Misra I., Jégou H., Mairal J., Bojanowski P., Joulin A.. Emerging properties in self-supervised vision transformers. ArXiv Prepr. ArXiv210414294. 2021.
- 120.Chen L., Zhai Y., He Q., Wang W., Deng M. Integrating deep supervised, self-supervised and unsupervised learning for single-cell RNA-seq clustering and annotation. Genes. 2020;11:792. doi: 10.3390/genes11070792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhai X., Oliver A., Kolesnikov A., Beyer L. S4L: self-supervised semi-supervised learning. ArXiv. 2019 http://arxiv.org/abs/1905.03670 [Google Scholar]
- 122.Zheltonozhskii E., Baskin C., Bronstein A.M., Mendelson A. Self-supervised learning for large-scale unsupervised image clustering. ArXiv. 2020 http://arxiv.org/abs/2008.10312 [Google Scholar]
- 123.Gildenblat J., Klaiman E. Self-supervised similarity learning for digital pathology. ArXiv. 2020 http://arxiv.org/abs/1905.08139 [Google Scholar]
- 124.Doersch C., Zisserman A. Proceedings of the IEEE International Conference on Computer Vision. 2017. Multi-task self-supervised visual learning; pp. 2051–2060. [Google Scholar]
- 125.Oord A.van den, Li Y., Vinyals O. Representation learning with contrastive predictive coding. ArXiv. 2019 http://arxiv.org/abs/1807.03748 [Google Scholar]
- 126.Li F.-F., Fergus R., Perona P. One-shot learning of object categories. IEEE Trans. Pattern Anal. Mach. Intell. 2006;28:594–611. doi: 10.1109/TPAMI.2006.79. [DOI] [PubMed] [Google Scholar]
- 127.Miller E.G., Matsakis N.E., Viola P.A. Vol. 1. CVPR 2000 (Cat. No.PR00662); 2000. Learning from one example through shared densities on transforms; pp. 464–471. (Proceedings IEEE Conference on Computer Vision and Pattern Recognition). [Google Scholar]
- 128.Larochelle H., Erhan D., Bengio Y. Proceedings of the 23rd National Conference on Artificial Intelligence - Volume 2. AAAI Press; 2008. Zero-data learning of new tasks; pp. 646–651. (AAAI’08) [Google Scholar]
- 129.Socher R., Ganjoo M., Manning C.D., Ng A. In: Burges C.J.C., Bottou L., Welling M., editors. Vol. 26. Curran Associates, Inc.; 2013. Zero-shot learning through cross-modal transfer.https://papers.nips.cc/paper/2013/hash/2d6cc4b2d139a53512fb8cbb3086ae2e-Abstract.html (Advances in Neural Information Processing Systems). [Google Scholar]
- 130.Lee C.-W., Fang W., Yeh C.-K., Wang Y.-C.F. Multi-label zero-shot learning with structured knowledge graphs. 2018. https://openaccess.thecvf.com/content_cvpr_2018/html/Lee_Multi-Label_Zero-Shot_Learning_CVPR_2018_paper.html p. 1576–1585.
- 131.Rios A., Kavuluru R. Few-shot and zero-shot multi-label learning for structured label spaces. Proc. Conf. Empir. Methods Nat. Lang. Process. 2018;2018:3132–3142. [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou Z.-H. A brief introduction to weakly supervised learning. Natl. Sci. Rev. 2018;5:44–53. [Google Scholar]
- 133.Mintz M., Bills S., Snow R., Jurafsky D. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP. Association for Computational Linguistics; 2009. Distant supervision for relation extraction without labeled data; pp. 1003–1011.https://www.aclweb.org/anthology/P09-1113 [Google Scholar]
- 134.Cabannes V., Rudi A., Bach F. In: International Conference on Machine Learning. Lawrence N., Reid M., editors. PMLR; 2020. Structured prediction with partial labelling through the infimum loss; pp. 1230–1239. [Google Scholar]
- 135.Mann G.S., McCallum A. Generalized expectation criteria for semi-supervised learning with weakly labeled data. J. Mach. Learn. Res. 2010;11:955–984. [Google Scholar]
- 136.Ratner A.J., De Sa C.M., Wu S., Selsam D., Ré C. In: Lee D.D., Sugiyama M., Luxburg U.V., Guyon I., Garnett R., editors. Vol. 29. Curran Associates, Inc.; 2016. Data programming: creating large training sets, quickly; pp. 3567–3575.http://papers.nips.cc/paper/6523-data-programming-creating-large-training-sets-quickly.pdf (Advances in Neural Information Processing Systems). [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang J., Wu X. Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2018. Multi-label inference for crowdsourcing; pp. 2738–2747. [Google Scholar]
- 138.Finn C., Xu K., Levine S. In: Bengio S., Wallach H., Larochelle H., Grauman K., Cesa-Bianchi N., Garnett R., editors. Vol. 31. Curran Associates, Inc.; 2018. Probabilistic model-agnostic meta-learning; pp. 9516–9527.http://papers.nips.cc/paper/8161-probabilistic-model-agnostic-meta-learning.pdf (Advances in Neural Information Processing Systems). [Google Scholar]
- 139.Dvornik N., Schmid C., Mairal J. Selecting relevant features from a multi-domain representation for few-shot classification. ArXiv. 2020 http://arxiv.org/abs/2003.09338 [Google Scholar]
- 140.Hospedales T., Antoniou A., Micaelli P., Storkey A. Meta-learning in neural networks: a survey. ArXiv. 2020 doi: 10.1109/TPAMI.2021.3079209. [DOI] [PubMed] [Google Scholar]