Abstract
TFIIA and TATA-binding protein (TBP) associate directly at the TATA element of genes transcribed by RNA polymerase II. In vivo, TBP is complexed with approximately 14 TBP-associated factors (TAFs) to form the general transcription factor TFIID. How TFIIA and TFIID communicate is not well understood. We show that in addition to making direct contacts with TBP, yeast TAF40 interacts directly and specifically with TFIIA. Mutational analyses of the Toa2 subunit of TFIIA indicate that loss of functional interaction between TFIIA and TAF40 results in conditional growth phenotypes and defects in transcription. These results demonstrate that the TFIIA-TAF40 interaction is important in vivo and indicate a functional role for TAF40 as a bridging factor between TFIIA and TFIID.
Transcription by eukaryotic RNA polymerase II (Pol II) involves the assembly of a preinitiation complex consisting of Pol II and the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (for review, see reference 66). An important step of transcription initiation is the binding of TFIID to the core promoter. TFIID, a multisubunit protein complex that is highly conserved among eukaryotes, is composed of the TATA-binding protein (TBP) and over a dozen TBP-associated factors (TAFs) (reviewed in references 26 and 27). TBP mediates promoter recognition through the sequence-specific binding of the TATA element found at many promoters. The importance of the TBP-TATA interaction is illustrated by many studies which demonstrate that recruitment of TBP is a rate-limiting step at a majority of promoters (12, 17, 37, 39, 46, 50).
In yeast, 13 TAFs are required for viability, indicating essential roles for individual TAFs (75, 77; reviewed in reference 26). However, the precise functional requirements for the TAFs remain unresolved. In vitro biochemical experiments suggest that TAFs function in higher eukaryotic systems as obligatory coactivators essential for activator response (reviewed in references 9 and 84). In contrast, functional inactivation and depletion studies with certain TAFs in yeast cells demonstrate that the expression of many genes are unaffected by TAF loss, although TAF inactivation results in distinct cell cycle phenotypes (2, 55, 59, 60, 62, 86). In addition, disruption of the TFIID complex with a temperature-sensitive mutation in TBP results in gene-specific transcriptional defects (71), and promoter occupancy studies indicate that TAFs are not present on certain transcriptionally active promoters in vivo (45, 49). Promoter-specific requirements for particular TAFs are further illustrated by whole-genome transcriptional profiles. For example, inactivation of TAF145/130 has no effect on the expression of a majority of genes, while transcription of a subset of genes is affected (31). This TAF dependence was mapped to the core promoter (78), indicating important TAF functions in promoter activity in vivo. In contrast to these gene-specific effects, inactivation of several other TAFs, namely TAF17 (2, 59, 61), TAF40 (44), TAF60 and TAF61/68 (59, 62), and TAF23/25 (76), results in dramatic effects on a large fraction of genes transcribed by Pol II. The requirement for these particular TAFs is not yet understood, but it is clear that individual TAFs may be generally required for transcription while others function at a subset of promoters. To complicate the issue further, it is apparent that certain TAFs in both human and yeast systems can be found in large protein complexes distinct from TFIID, such as the SAGA complex (25) and the SWI/SNF complex (10). Taken together, these studies indicate that different TAFs may have distinct functional roles in transcription, yet the nature of the specific functions for a majority of the TAFs remains to be elucidated.
In addition to TAFs, other transcription factors associate with TBP at the core promoter to mediate transcription. One such factor, TFIIA, has been shown to stabilize the interaction between TBP and DNA at the TATA element (reviewed in reference 27). Mutational studies of both TBP and TFIIA demonstrate the importance of the TFIIA-TBP interaction for transcription in vivo (35, 53, 68, 80). TFIIA has been described as a coactivator, since in vitro functions of certain activators are TFIIA dependent (51, 67), and as an antirepressor, because TFIIA can mediate displacement of certain transcriptional inhibitors that act on TBP (4, 5, 22, 32, 42, 52, 56, 58, 69).
A growing body of evidence suggests that the functions of TFIIA and TAFs are connected. DNase I footprinting experiments show that the addition of TFIIA alters the DNA protection pattern of TFIID (14, 15, 51). Consistent with these findings, UV-cross-linking experiments indicate that TFIIA induces a conformational change in TFIID that alters specific TAF interactions with the core promoter (64). Moreover, a set of TFIIA mutations that can form a TBP-TFIIA-DNA complex are defective for forming a complex with TFIID (69). Finally, the three-dimensional structure of the human TFIID-TFIIA-TFIIB complex clearly suggests TFIIA-TAF interactions, since TFIIA maps to a large noncentral lobe of TFIID, with TBP being located more centrally in the structure (1). It is not yet understood how this TFIIA-TAF communication is established or which particular TAFs are involved.
In this report, we investigate the importance of the interactions between TFIIA and yeast TAFs. We demonstrate a direct interaction between TFIIA and TAF40, as well as a direct interaction between TAF40 and TBP. We also find that mutations in TFIIA that impair the TFIIA-TAF40 interaction result in conditional growth phenotypes and defects in transcription in vivo. These results suggest that TAF40 serves as a link between TFIIA and TFIID functions, and they reveal a new role for TAF40 in RNA Pol II transcriptional regulation.
MATERIALS AND METHODS
DNA constructs.
Activation domain (AD) hybrids were cloned into the 2μ LEU2 marked vector, pACT2.2 (19), which contains the ADH1 promoter, a nuclear localization sequence, the hemagglutinin (HA) epitope, and the Gal4 AD (residues 768 to 881). DNA-binding domain (DB) hybrids were created by subcloning from the corresponding AD constructs into the pPC97-TRP vector (85) (CEN, TRP3), which contains the ADH1 promoter, a nuclear localization sequence, and the Gal4 DB (residues 1 to 147). The TAF40 Escherichia coli expression plasmid was created by cloning the TAF40 open reading frame into the pET15b vector using PCR and designed oligonucleotides. The E. coli expression plasmids for Toa2 glutathione S-transferase (GST)-TFIIB have been described elsewhere (80). GST-Toa1 was constructed by subcloning an EcoRI fragment containing the open reading frame of Toa1 into the pGEX1λT vector (Pharmacia); GST-TBP was constructed by PCR of TBP followed by cloning into the GST-2T vector (Pharmacia). TOA2-YCP22 contains TOA2 driven by its native promoter and terminator, which were generated by PCR from genomic DNA. A NcoI site was engineered at the ATG start codon and utilized for inserting six myc epitopes (GEQKLISEEDLN), creating myc-TOA2-YCP22. Site-directed TOA2 mutants were created using oligonucleotide primers containing the desired mutation and PCR. Mutant derivatives were subsequently subcloned into the Gal4-DNA binding domain vector (pPC97-TRP1) and into pET15b (Novagen) lacking the histidine tag. All PCR products were completely sequenced.
Yeast strains.
All strains used in the yeast two-hybrid assay were transformants of MaV103 (85). MaV103 contains the GAL1 promoter (with four Gal4 binding sites) fused to the HIS3 promoter and structural gene; GAL4 and GAL80 are both deleted in the strain. Viability tests of TOA2 mutant derivatives were conducted in ROY100, a derivative of KY114 (relevant genotype MATa ade2-101 leu2::PET56 trp1Δ1 ura3-52), which was created using a two-step gene knockout of the complete open reading frame of the TOA2 gene and contains TOA2 on a 2 μm, URA3-marked plasmid. The plasmid shuffle technique, which involves transforming ROY100 with TRP1-marked TOA2 derivatives, followed by selection for loss of the URA3-marked plasmid by growth on 5-fluoro-orotic acid (5-FOA), was used to test the mutant derivatives for viability and to create Toa2 mutant strains for further characterization.
Yeast two-hybrid assays and phenotypic studies.
Both Gal4 DB and Gal4 AD plasmids were transformed into the yeast strain MaV103 using a standard lithium acetate transformation. The resulting strains were grown in the appropriate selection media, and 10-fold serial dilutions were performed. Cells were spotted onto the appropriate plates, which either contained or lacked 3-aminotriazole (AT), and grown at 30°C for 4 to 7 days. For phenotypic studies, 10-fold serial dilutions of strains were spotted on plates with rich media containing either glucose (yeast-peptone-dextrose [YPD]) or galactose (yeast-peptone-galactose [YPG]), and plates were incubated at either 30 or 38°C.
Protein purification.
TFIIA was purified as described previously (74) by expressing the GST-Toa1 and -Toa2 subunits separately in E. coli, denaturing both in 8 M urea, combining the subunits, and dialyzing out the urea. GST-TBP, GST-TFIIB, and GST were expressed and purified from bacteria as described elsewhere (80). TAF40 was purified with a denaturing and refolding method similar to that used to make recombinant yeast TFIIA. BL21 (DE3) cells containing TAF40 cloned into the bacterial expression plasmid pET15b (His-TAF40) were grown to an optical density at 600 nm (OD600) of 0.6 and induced for 2 h with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). One liter of cells was harvested, washed with 200 ml of buffer A (20 mM Tris-HCl [pH 7.5], 200 mM NaCl), and resuspended in 25 ml of buffer 1 (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol [DTT], 0.05% NP-40), and frozen at −70°C. Cells were thawed and sonicated. The insoluble fraction was collected by spinning at 10,000 × g for 15 min at 4°C. Pellets were resuspended in 30 ml of buffer 2 (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 8 M urea, 5 mM DTT). A 7.5-ml volume of buffer 3 (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1 mM DTT, 10% glycerol) was added slowly, and the solution was cleared by spinning at 10,000 × g for 15 min at 4°C. The supernatant was dialyzed against buffer 3 containing 1 mM imidazole. The dialyzed proteins were cleared by spinning at 10,000 × g for 15 min at 4°C. The soluble material was bound to Ni-nitrilotriacetic acid resin (Qiagen), washed with wash buffer (20 mM Tris [pH 7.5], 100 mM KCl, 10% glycerol, 40 mM imidazole, 1 mM DTT), and eluted with elution buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 10% glycerol, 1 mM DTT, 200 mM imidazole). Elution fractions were dialyzed against buffer containing 20 mM HEPES (pH 7.9), 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 10% glycerol, and 1 mM DTT.
In vitro interaction studies.
Approximately 12.5 pmol of GST-fusion protein or GST alone was incubated with 10 to 20 pmol of His-TAF40 protein in 200 μl of binding buffer (20 mM HEPES [pH 7.9], 20 mM Tris [pH 7.5], 200 mM NaCl, 50 mM KCl, 10 mM MgCl2, 0.025% NP-40, 10% glycerol, 0.5 mM DTT) for 3 h at 4°C. Complexes were recovered by incubation with glutathione Sepharose for 1 h at 4°C in binding buffer with 3% bovine serum albumin. Complexes were washed two times in 400 μl of binding buffer, incubated with sodium dodecyl sulfate (SDS) loading buffer, and boiled, and 10 μl of sample was separated by SDS-polyacrylamide gel electrophoresis (PAGE). Gels were analyzed by immunoblotting with antibodies specific to His-TAF40 (Santa Cruz Biotechnology) or GST (Sigma) and visualized by chemiluminescence detection (Pierce).
Electrophoretic mobility shift assays were performed using a 32P-labeled 45-bp fragment containing the adenovirus early 1B TATA box, as described previously (80). Purified recombinant yeast TBP (5 nM), TFIIA or TFIIB (3 nM), TAF40 (19 to 142 nM), and 100 ng of poly(dG-dC) were incubated at 25°C for 30 min in 20 μl of 20 mM Tris (pH 7.5), 40 mM HEPES (pH 7.9), 100 mM KCl, 1 mM DTT, 0.5 mM phenylmethyl sulfonyl fluoride, and 10% glycerol. Complexes were separated from unbound DNA by 6% nondenaturing acrylamide gel electophoresis in 0.5× Tris-borate-EDTA and quantified by phosphorimaging.
Transcriptional analysis.
Quantitative S1 nuclease analysis was done as described elsewhere (34), with approximately 30 to 50 μg of RNA. For the temperature shift and AT (Sigma) inductions, cells were grown in synthetic complete medium to an OD600 of 0.5 to 1.0. Cells were pre-heat shocked at 38°C for 15 min and incubated at 30°C for 1 h, followed by 38°C for 1 h. AT was added to a concentration of 20 mM, and cells were incubated for an additional hour at 38°C. Total RNA was prepared by hot-phenol extraction and was quantitated at OD260. RNA amounts in each reaction mixture were normalized to the levels obtained from a probe to the intron of the tryptophan tRNA gene (tRNAW).
RESULTS
Yeast TAF40 associates with TFIIA in vivo.
TFIIA interacts with TBP and DNA at the promoter and stabilizes the TBP-DNA interaction. Yet, within a cell, TBP associates with TAFs to form the TFIID complex. We used a yeast two-hybrid assay to investigate the potential interplay between TFIIA and TAFs. Yeast TFIIA is composed of two subunits encoded by the genes TOA1 and TOA2, both of which are required for viability (73). The DB of Ga14 was fused in frame with Toa2 (DB-Toa2), creating the bait for the two-hybrid assay. TAF proteins were fused in frame to the Ga14 AD. Fusion proteins were expressed in a yeast strain with the HIS3 gene under the control of the GAL1 promoter (which contains four Ga14 binding sites). Interactions between Toa2 and the AD-fusion proteins were determined by examining activation of the HIS3 gene. HIS3 gene activation was assayed by growth in the presence of AT, a competitive inhibitor of the HIS3 gene product (28). Strains in which the HIS3 gene is highly expressed, due to interactions between the DB-fusion protein and the AD-fusion protein, will grow on AT.
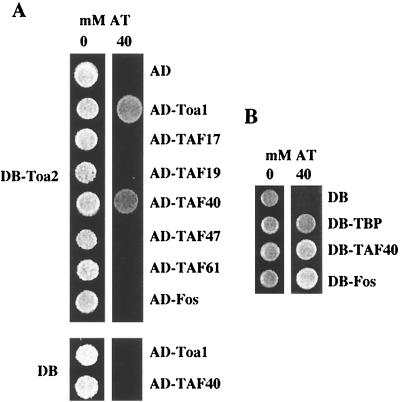
In contrast to a DB fusion to Toa1, which activates transcription (79), expression of DB-Toa2 showed no HIS3 gene activation (Fig. 1A). The difference between the activity of the two subunits is not understood, but it does provide the opportunity to use DB-Toa2 in the two-hybrid assay. Since DB-Toa2 exhibited a strong interaction with AD-Toa1, DB-Toa2 is not defective for subunit interactions with Toa1, and this also suggests that DB-Toa2 interacts with native Toa1 within the cell. We then tested the ability of DB-Toa2 to interact with all 13 of the known essential yeast TAFs found in the TFIID complex (for review, see references 26 and 27). A strong interaction was observed between Toa2 and TAF40. Because two-hybrid interactions can be indirect and TAF40 is a component of the TFIID complex, it was surprising that DB-Toa2 showed no interaction with the other TAF proteins shown in Fig. 1 (TAF17, TAF19, TAF47, and TAF61) or TAF25, TAF48, TAF60, TAF65, TAF67, TAF90, TAF130, and TAF150/TSM1 (data not shown). The lack of interaction was not due to lack of expression, since each of the TAF fusion proteins was easily detectable by immunoblotting of whole-cell extracts with antibodies specific to the HA tag present in the AD vector (data not shown). These results suggest that TAF40 associates with Toa2 within the cell, perhaps directly.
FIG. 1.
TFIIA associates with TAF40 in vivo. (A) Two-hybrid assays were used to demonstrate that TFIIA interacts specifically with Toa1 and TAF40. The indicated Gal4 AD fusion proteins were tested for the ability to interact with a DB fusion of Toa2 (DB-Toa2). Approximately 104 cells were spotted onto synthetic complete plates containing either 0 or 40 mM AT. Growth on AT is indicative of an interaction between the two hybrid proteins. The bottom panel shows that strains containing AD-Toa1 or AD-TAF40 do not grow on AT with the DB vector alone. (B) TAF40 stimulates transcription in an artificial recruitment assay. DB-TBP, DB-40, and DB-Fos each stimulate high levels of transcription when tethered to a promoter via a heterologous DB.
A reciprocal interaction between TAF40 and TFIIA could not be examined because expression of the fusion of TAF40 to the DB resulted in high levels of transcriptional stimulation (Fig. 1B). This indicates that recruitment of TAF40 to a promoter is sufficient to stimulate transcription of that gene. This result is in accord with similar recruitment studies that used other TAFs and TBP (3, 7, 12, 21, 24, 36, 37, 89).
TFIIA has been shown to interact with certain ADs (16, 40, 70). A trivial explanation for the interaction detected is that DB-Toa2 interacts with the AD of AD-TAF40, and the TAF40 domain stimulates transcription. To examine this possibility, the Fos C-terminal AD, which strongly stimulates transcription when bound to the promoter via the Ga14 DB (Fig. 1B), was cloned into the Ga14 AD vector (AD-Fos). Expression of the AD-Fos hybrid protein, which has two tandem functional ADs, did not yield a positive two-hybrid interaction with DB-Toa2 (Fig. 1A). The inability of DB-Toa2 to interact with AD-Fos eliminates the possibility that Toa2 is simply interacting with the Ga14 AD of AD-TAF40 and, in effect, mimicking the activation by DB-TAF40 seen in the artificial recruitment assay.
TAF40 interacts with TFIIA and TBP.
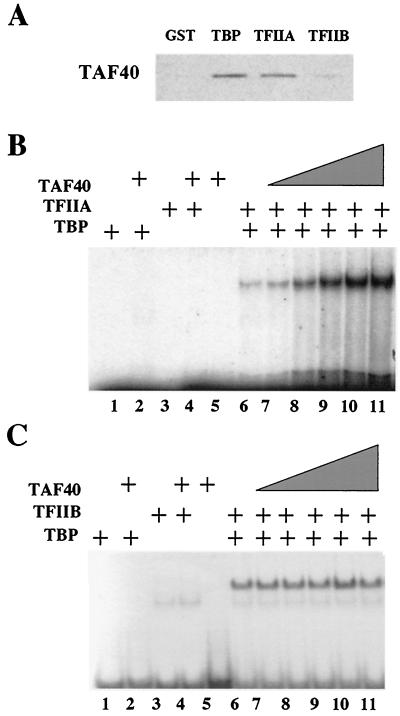
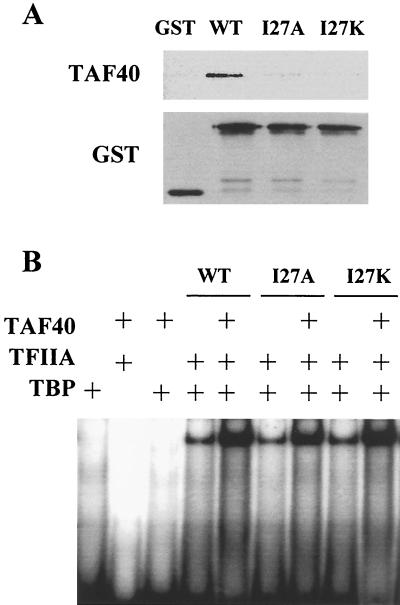
To demonstrate a direct interaction between TAF40 and TFIIA or TBP, recombinant proteins were produced in bacteria and their ability to physically interact was examined using GST pull-down assays. Immunoblot analysis of the isolated complexes revealed that significant amounts of TAF40 interact with the GST-TFIIA (Fig. 2A). In addition, GST-TBP interacts with TAF40 directly. TAF40 does not interact with either GST-TFIIB or GST.
FIG. 2.
TAF40 interacts directly with TFIIA or TBP. (A) To test whether TFIIA interacts directly with TAF40 in vitro, a GST pull-down assay was used. Recombinant GST-TFIIA, GST-TBP, GST-TFIIB, or GST was incubated with recombinant histidine-tagged TAF40 (TAF40). After SDS-PAGE, TAF40 was detected using antibodies specific to the histidine tag of TAF40. Antibodies specific to the GST tag were used to confirm that similar amounts of GST and the GST fusion proteins were utilized (data not shown). (B) TAF40 increases TBP-TFIIA-DNA complex formation on a TATA box. For all reactions, 9 nM radiolabeled adenovirus early 1B TATA box probe was used. Concentrations of TBP and TFIIA were held constant at 5 and 1.5 nM, respectively. Lane 1 contains TBP, lane 2 contains TBP and TAF40 (142 nM), lane 3 contains TFIIA, lane 4 contains TFIIA and TAF40 (142 nM). Lane 5 contains TAF40 (142 nM) alone. Lane 6 contains TBP and TFIIA, and lanes 7 through 11 contain TBP and TFIIA with increasing amounts of TAF40 (19 to 142 nM). (C) TAF40 does not increase the TBP-TFIIB complex formation on a TATA box. All reactions are as in panel B, except that 3 nM TFIIB is used in place of TFIIA.
Electrophoretic mobility gel shift assays were used to test whether the presence of TAF40 affects the TBP-TFIIA-DNA ternary complex. In the absence of magnesium, TBP does not form a stable complex with DNA, and the addition of TFIIA stabilizes the TBP-DNA interaction and shifts the TATA-containing probe (Fig. 2B). Under conditions of subsaturating amounts of TFIIA, the addition of TAF40 resulted in a 20-fold enhancement of the TFIIA-TBP-DNA complex (Fig. 2B). Enhancement of the TBP-TFIIA-DNA complex was not observed when equivalent amounts of bovine serum albumin or buffer were added (data not shown). In addition, TAF40 had no effect on the amount of ternary complex formed with TFIIB (Fig. 2C). Excess TAF40 also resulted in no changes in the DNA-binding properties of TBP, TFIIA, or TFIIB alone, either in the presence or absence of magnesium ions (Fig. 2B and C and data not shown). Thus, enhancement of complex formation by TAF40 is specific to the TBP-TFIIA-DNA ternary complex.
Although TAF40 addition results in a significant enhancement of the TBP-TFIIA-DNA complex, it does not alter the mobility of the complex. This result suggests that either the addition of TAF40 to the complex results in a conformational change that masks the added mass of TAF40, or that TAF40 is not stable to the gel running conditions. We feel that the latter hypothesis is supported by the observation that inclusion of antibodies specific to TAF40 does not result in a shift in the complex size but instead abolishes the ability of TAF40 to enhance TBP-TFIIA-DNA complex formation (data not shown).
Mutations in a hydrophobic patch of TFIIA are defective for interacting with TAF40 in vivo.
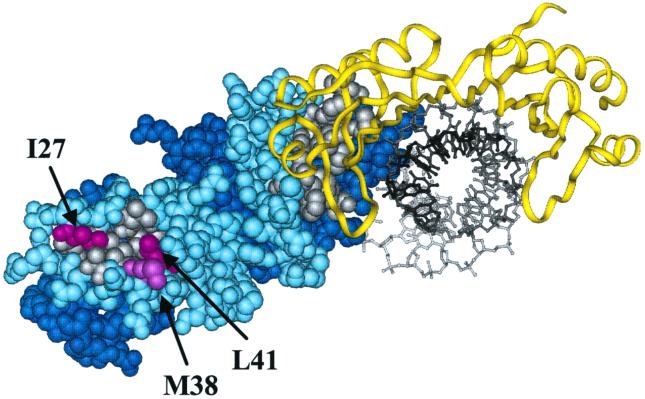
Analysis of the crystal structures of the yeast TFIIA-TBP-DNA ternary complex (23, 81) reveals several striking features. TFIIA consists of two domains, a β domain and a four-helix bundle (4HB) domain (Fig. 3). The β domain makes all of the contacts with TBP and also binds DNA upstream of the TATA element. The 4HB domain of TFIIA projects away from the TBP-TFIIA-DNA complex into solution. In addition, there are two large solvent-exposed patches of hydrophobic residues on TFIIA; one patch is within the β domain and the other is within the 4HB domain. Hydrophobic interactions are important for many protein-protein interactions. In fact, the hydrophobic region on the β domain contacts TBP. We hypothesized that the hydrophobic patch on the 4HB domain may contribute to other TFIIA functions, possibly for interactions with TAF40. Three residues within this hydrophobic patch were targeted for mutational studies: isoleucine at position 27 of Toa2 was changed to alanine (I27A) or lysine (I27K); methionine at position 38 was changed to alanine (M38A) or lysine (M38K); and leucine at position 41 was substituted with either alanine (L41A) or aspartic acid (L41D).
FIG. 3.
Crystal structure of the TFIIA-TBP-DNA complex showing the location of the amino acids replaced in the hydrophobic region of the 4HB. TBP is shown in a yellow ribbon, and DNA is black. TFIIA is shown in the space-filling model, with Toa1 in dark blue and Toa2 in light blue. The two hydrophobic patches on TFIIA are shown in gray; one patch contacts TBP. The amino acids in Toa2 selected for replacement by alanine or radical amino acids are shown in magenta and are indicated by the arrows and the labels. The figure was created with Insight II, using the coordinates of the TBP-TFIIA-DNA structure (81).
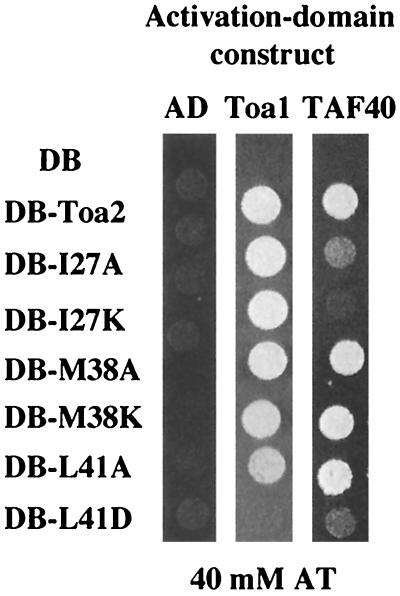
Each of the mutant derivatives was cloned into the DB vector. To determine whether any of the mutations causes a drastic change in the folding ability of Toa2, the derivatives were tested for the ability to interact with AD-Toa1 (Fig. 4). DB-L41D was defective for interacting with Toa1, indicating that this Toa2 derivative is compromised for TFIIA formation. The five remaining derivatives were indistinguishable from wild-type Toa2 with regard to their ability to interact with Toa1.
FIG. 4.
Mutations in Toa2 impair interactions with TAF40. The two-hybrid assay was used to identify mutations in the hydrophobic patch of the TFIIA 4HB that are defective for interaction with TAF40. The AD constructs are indicated across the top, and the DB-Toa2 derivatives are shown along the left side. Approximately 104 cells were spotted onto plates containing 40 mM AT. All strains grew robustly on media lacking AT (data not shown).
We next tested the Toa2 derivatives for defects in interactions with TAF40. I27A, I27K, and L41D showed significantly weakened interactions with TAF40 in the two-hybrid assay (Fig. 4). Since L41D was also defective for interaction with Toa1, the loss of the TAF40 interaction may be the result of global defects in the structure of this protein. In contrast, substitutions at I27 are specifically defective for TAF40 interactions, in that both I27A and I27K are indistinguishable from wild-type Toa2 for Toa1 interactions. The remaining derivatives exhibited interactions with both TAF40 and Toa1 that were comparable to that with wild-type Toa2. These results suggest that the hydrophobic region on the 4HB domain of TFIIA, in particular residue I27 of Toa2, plays an important role in the interaction between TFIIA and TAF40.
Toa2 derivatives impart mutant growth phenotypes.
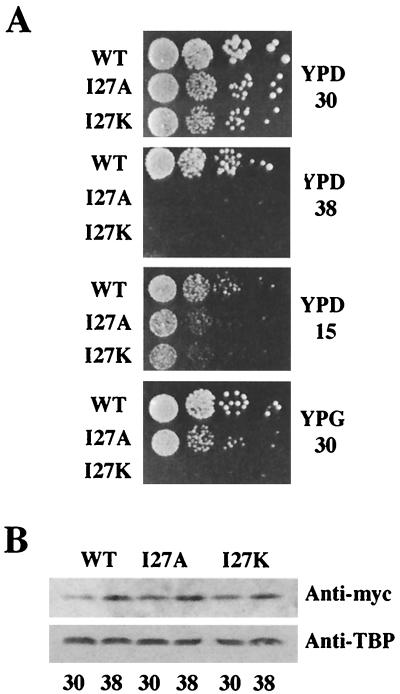
To examine the physiological relevance of the TFIIA-TAF40 interaction, the I27A and I27K Toa2 mutants (under the control of the TOA2 promoter and terminator) were expressed in a TOA2 deletion strain. Both alleles supported cell viability, but each caused a slow-growth phenotype at 30°C and a temperature-sensitive phenotype at 38°C (Fig. 5A). The doubling time for wild-type cells was 2.5 h, whereas I27A- and I27K-containing cells have a doubling time of 3.5 to 4 h at 30°C (data not shown). The slow-growth phenotype at 30°C is consistent with the observation that the TFIIA-TAF40 interaction is disrupted in the two-hybrid assay, which is performed at this same temperature (30°C). Furthermore, the I27K mutant was unable to support growth on galactose-containing medium, suggesting an inability to respond to the Gal4 activator protein. Mutant phenotypes were not the result of a destabilization of Toa2 protein, since the I27A and I27K strains produced amounts of Toa2 protein comparable to that in wild-type Toa2 at 30 and 38°C, as assayed by immunoblot analyses of whole-cell yeast extracts (Fig. 5B). None of the mutant phenotypes could be suppressed by overexpression of TAF40 (data not shown). This suggests that interactions with TAF40 and the I27A and I27K forms of TFIIA are severely compromised, since simple overexpression is not sufficient to counteract the loss of interaction.
FIG. 5.
TAF40-defective Toa2 derivatives confer mutant phenotypes. (A) Strains containing the I27A and I27K mutations of Toa2 were tested for conditional phenotypes in a TOA2 deletion strain. The indicated strains were serially diluted (from 104 to 10 cells) and spotted onto rich media plates containing glucose (YPD) or galactose (YPG) and incubated at either 15, 30, or 38°C. (B) Levels of Toa2, I27A, and I27K proteins are indistinguishable. Strains containing wild-type Toa2 in addition to either myc-tagged wild-type Toa2 (WT), myc-tagged I27A (I27A), or myc-tagged I27K (I27K) were harvested after incubation at 30 or 38°C. Extracts (20 μg) were subjected to SDS-PAGE and immunoblotting with anti-myc antibodies and anti-TBP antibodies (for a load control). The presence of wild-type Toa2 in each of the strains allows for an accurate analysis of the stability of the mutant derivatives, since cell viability is not dependent on their expression.
In vitro analysis of the TFIIA mutants I27A and I27K.
In order to determine the extent to which the mutations at the I27 residue were defective for the interaction with TAF40 in vitro, GST pull-down assays were performed. Immunoblot analysis of the recovered complexes showed that TFIIA recombinant proteins containing the I27A and I27K substitutions were significantly decreased in their ability to interact with TAF40 when compared to wild-type TFIIA (Fig. 6A). Moreover, TAF40 binding by I27K was indistinguishable from the binding observed with GST alone. These results coincide with those of the two-hybrid studies, since in both the two-hybrid assay and the pull-down assay, I27K displayed a more dramatic defect in interacting with TAF40. In contrast, TFIIA substituted with I27A or I27K is fully functional in forming the TBP-TFIIA-DNA complex (Fig. 6B). In addition, both I27A- and I27K-substituted TFIIA were competent for complex enhancement by TAF40 (Fig. 6B). This result is not unexpected, since the direct interaction between TBP and TAF40 (Fig. 2) may compensate for the loss of interaction between TAF40 and TFIIA.
FIG. 6.
: In vitro analysis of the TFIIA mutants I27A and I27K. (A) To test the ability of TFIIA substituted with I27A and I27K to interact with TAF40, a GST pull-down assay was used. Recombinant GST, GST-TFIIA (WT), and TFIIA formed with the I27A (GST-I27A) and I27K (GST-I27K) mutants were incubated with recombinant histidine-tagged TAF40. The complexes were isolated by incubation with GST resin followed by washing. Samples were separated by SDS-PAGE and analyzed by immunoblotting using antibodies specific to the histidine tag of TAF40. Immunoblot analysis was also done using antibodies specific to GST to assay the amounts of GST and GST-fusion proteins recovered. (B) TFIIA substituted with either I27A or I27K is fully functional in forming the TBP-TFIIA-DNA complex, and formation of the complex is enhanced by TAF40. For all reactions, 9 nM radiolabeled adenovirus early 1B TATA box probe was used. Lanes contain TBP (5 nM), WT or mutant TFIIA (1.5 nM), or TAF40 (142 nM) as indicated.
TFIIA mutants are defective for transcription in vivo.
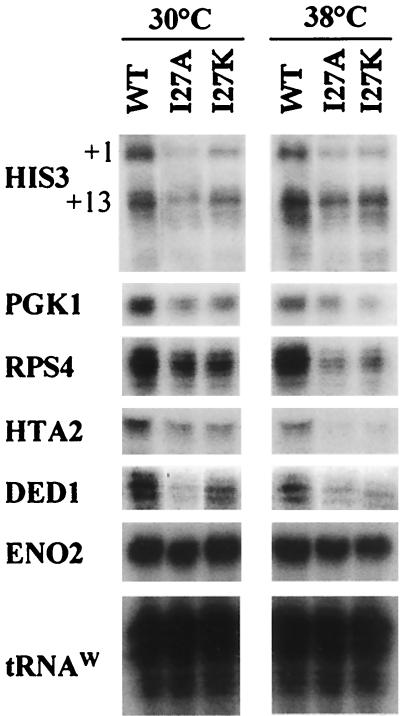
To determine the ramifications of a defect in the TFIIA-TAF40 interaction, we compared wild-type, I27A, and I27K strains for their transcriptional competency. Constitutive transcription of a collection of Pol II-transcribed genes was examined at 30 and 38°C (Fig. 7). When cultured at 30°C, the I27A and I27K strains exhibited a significant reduction in HIS3 gene expression compared to expression in wild-type cells. Levels of transcription of both the +1 and +13 transcripts of HIS3 were decreased. The +1 transcript is generated from a noncanonical promoter element, while the +13 transcribed is derived from a conventional TATA element (13, 33). This suggests that the TAF40-TFIIA interaction is important for transcription from both canonical and noncanonical promoters in vivo. RPS4, DED1, HTA2, and PGK1 mRNA levels also decreased at both 30 and 38°C. The decreases in transcription at the permissive temperature (30°C) are consistent with the fact that the TAF40-TFIIA interaction defect is observed at 30°C in the two-hybrid assay. The transcriptional effects on expression of RPS4, HTA2, and DED1 in strains with defective TFIIA-TAF40 interactions are consistent with similar observations on the expression of these genes in a TAF40 mutant strain (44). In contrast to the genes described above, transcription from the ENO2 gene was unaffected. Thus, requirement for the TFIIA-TAF40 interaction is promoter specific. Promoter-specific dependency on TAF40 is also supported by chromatin immunoprecipitation studies, which showed TAF40 occupancy can vary on transcriptionally active promoters (45).
FIG. 7.
Transcriptional analysis of the TFIIA mutants defective in the TAF40 interaction for a collection of Pol II-transcribed genes. Wild-type (WT) and mutant Toa2 (I27A or I27K) strains were grown to log phase and shifted to the restrictive temperature for 1 h. Thirty to 50 μg of total RNA was hybridized with a 100-fold excess of the indicated probe and treated with S1 nuclease. The HIS3 +1 and +13 initiation sites are indicated. The Pol III-transcribed gene tRNAw served as a loading control.
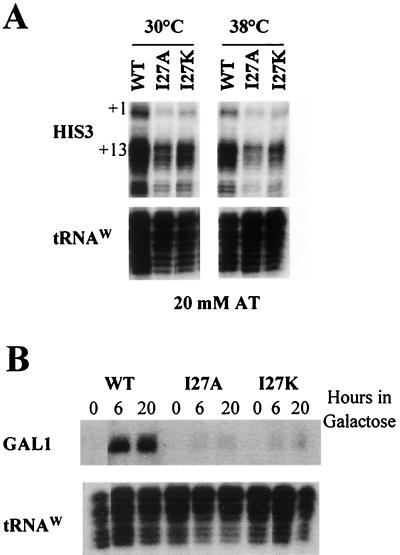
We also tested the ability of the I27A and I27K strains to respond to acidic activators. Gcn4-dependent activation of HIS3 transcription was assayed by growing the cells in AT, a competitive inhibitor of the HIS3 gene product (Fig. 8A). To examine the response at the restrictive temperature, cells were incubated at 38°C for 1 h, AT was added, and the cells were incubated for an additional hour before harvesting. For both the I27A and I27K mutants, activation of HIS3 transcription was decreased compared to that in wild-type cells. Activation by the acidic activator Gal4 was determined by growing cells in galactose-containing medium and assaying for GAL1 transcript levels (Fig. 8B). Both the I27A and I27K mutants displayed a significant decrease in GAL1 transcription. Taken together, these defects in both constitutive and induced transcription for the I27A and I27K derivatives of Toa2 indicate that the TAF40-TFIIA interaction plays an important role in transcription in vivo.
FIG. 8.
Transcriptional analysis of activator-induced genes. (A) Analysis of Gcn4-dependent activation of HIS3 transcription. For assays at 30°C, 20 mM AT was added for 1 h, followed by harvesting of the cells. For assays at 38°C, cells were grown to early log phase, shifted to 38°C for 1 h, and then grown for an additional hour in the presence of 20 mM AT. Total RNA was analyzed for HIS3 and tRNAw expression by S1 analysis. (B) Analysis of Gal4-dependent activation of GAL1 transcription. Strains were grown in raffinose-containing medium to early log phase and then grown for the indicated time in galactose at 30°C. Total RNA (30 to 50 μg) was hybridized with 100-fold excesses of GAL1 and tRNAw probes and then subjected to S1 nuclease digestion.
TFIIA mutants defective for interactions with both TAF40 and TBP are not viable.
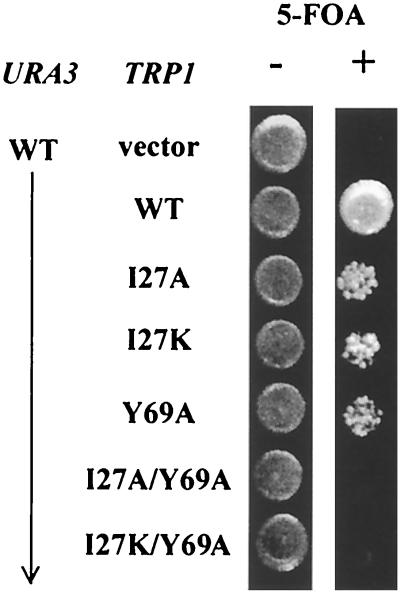
Combining previous work and the results presented above, we conclude that TFIIA interacts with TFIID via TAF40 and TBP. To determine how critical the contacts with TAF40 and TBP are for TFIIA functions in vivo, mutations defective for the TFIIA-TAF40 interaction and the TFIIA-TBP interaction were engineered into the same Toa2 allele. TFIIA substituted with a Y69A mutation in Toa2 is not competent for forming the TBP-TFIIA complex in vitro and results in a temperature-sensitive phenotype and transcriptional defects in vivo (68). We tested the hypothesis that TFIIA interactions with TFIID are essential by combining mutations in Toa2 at either I27A or I27K (which are TAF40 defective) with Y69A (which is TBP defective). While the I27A, I27K, and Y69A single mutants were able to support cell viability, both the I27A/Y69A and I27K/Y69A double mutants were unable to support cell viability in a strain deleted for TOA2 (Fig. 9). Thus, TFIIA interactions with TFIID via TBP or TAF40 are essential for cell survival.
FIG. 9.
TFIIA mutants defective for TAF40 and TBP interaction cannot support cell viability. The growth of strains containing the indicated Toa2 derivative (or vector) was analyzed by spotting approximately 104 cells onto plates deficient for tryptophan and containing (+) or lacking (−) 5-FOA. When wild-type Toa2 (carried on a URA3 plasmid) is shuffled out on 5-FOA, cell viability is dependent on Toa2 functions carried on the TRP1 plasmid. WT, wild type.
DISCUSSION
TFIIA and TAF40 interact directly.
The ability of TFIIA to interact with TBP and stabilize it on a promoter is well characterized, but how TFIIA communicates with TBP in the context of TFIID remains unclear. TFIIA is generally required for transcription in the presence of TAFs, yet it is uncertain how the functions of TFIIA and TAFs are connected. We report here that an important link between TFIIA and TFIID resides in TAF40. The interaction between TFIIA and TAF40 was observed both in vivo and in vitro. Thus, the TFIIA-TAF40 interaction may be directly involved in processes that are TFIIA and TAF dependent.
We also identified a direct interaction between TAF40 and TBP. TAF-TBP interactions have been identified in other organisms (29, 30, 41, 57, 63, 82, 87, 88). In addition, a 100-amino-acid fragment of yeast TAF130/145 has been shown to interact with TBP (6, 42), and the conserved C-terminal domain of yeast TAF68 interacts with TBP (75). However, TAF40 is the first full-length TAF that has been shown to interact directly with yeast TBP in solution. A TBP-TAF40 interaction is consistent with studies that show the human homologue of TAF40, human TAF28, interacts with human TBP (47, 48, 57) and indicates that this interaction is conserved from yeast to humans. Human TAF28 has also been shown to interact with human TAF18, TAF55, TAF100, TAF135, and the viral activator Tax (8, 11, 18, 47, 48, 57). Taken together with our studies, these results suggest that TAF40 may have critical functions in TAF-TAF, TAF-TBP, and TAF-TFIIA interactions.
Mutations in TFIIA affect the interaction with TAF40.
The functional importance for the TFIIA-TAF40 interaction is supported by mutational studies of the Toa2 subunit of TFIIA in the 4HB domain of TFIIA. Mutations at the I27 residue of Toa2 (I27A and I27K) caused a defect in the interaction with TAF40. The loss of interaction is not due to global changes in the structure of the protein, since these Toa2 derivatives interact normally with the other subunit of TFIIA (both in vivo and in vitro), are expressed in cells at levels similar to that in wild-type Toa2, and are functional for TFIIA-TBP interactions in vitro. Although the 4HB of TFIIA is not involved in either TBP or DNA interactions in the crystal structures, a deletion derivative of human TFIIA lacking two of the helices in this domain is not responsive to activators in vitro (54). Taken together with our results, this suggests that the 4HB domain of TFIIA is a functionally important target that facilitates formation of an active transcription initiation complex. Since the isoleucine at position 27 of Toa2 is conserved in yeast, Drosophila, and human TFIIA (23), this surface may play a critical role in the interaction of the higher eukaryotic homologues of TFIIA and TAF40. It is interesting to speculate that an interaction between TFIIA and TAF40 would orient the TAF40-TBP interaction on the N-terminal repeat of TBP. The surface of TBP situated on the same side of the structure as I27 is the precise location of the altered amino acids in a TBP mutant that is defective in TFIID formation in vivo (71).
The I27A- and I27K-substituted derivatives displayed mutant growth phenotypes and defects in transcription in vivo. A complete cessation in transcription was not expected, since the mutants support cell viability. Transcriptional defects observed in previous studies in which TFIIA or TAF40 was inactivated (35, 44, 53, 61) are comparable to those reported here. It is clear that inactivation of TFIIA does not typically cause dramatic transcriptional effects. In our TFIIA mutant strains, it is likely that disruption of TFIIA-TAF40 interactions could be compensated by redundancy in other interactions that contribute to formation of the TBP-TFIIA-TAF40 complex. Important interactions for complex stability may arise from TFIIA-TAF40 contacts, TAF40-TBP contacts, TFIIA-TBP contacts, and interactions between DNA and both TFIIA and TBP. This idea is supported by our in vitro observations in which the Toa2 mutant proteins are functional in the enhancement of the DNA-TBP-TFIIA complex formation by TAF40. Further support for redundant interactions between TFIIA and TFIID is provided by the observation that TFIIA mutants defective for both the TAF40 and TBP interactions are not viable. In addition, the importance of compensatory interactions in the TFIIA-TBP-DNA complex is also demonstrated by recent reports showing that mutations in the region of TFIIA that binds DNA can suppress mutant phenotypes of TBP alleles with DNA binding defects (53). Moreover, mutations in Toa2 at glycine 30 (G30) also suppress the TBP mutant defective for DNA binding and yet, G30 mutants have no effect on in vitro interactions involving TBP, TFIIA, and DNA (53). This result suggests that G30 substitutions can compensate in vivo by increasing interactions with another component of the complex. G30 is located near the hydrophobic patch of Toa2 and directly abuts I27. It is interesting to speculate that mutations at G30 have the potential to alter the TFIIA-TAF40 interaction.
TAF40 may act as a communicator between TFIIA and TFIID.
The association between TFIIA and TAF40 is particularly noteworthy because it supports a growing body of evidence that indicates that transcriptional activity mediated by TAFs is dependent on TFIIA. TFIIA is required for in vitro transcription reactions when TAFs are present (reviewed in reference 27). TFIIA has also been shown to induce a conformational change in the TFIID complex bound to a promoter. Specifically, the presence of TFIIA extends the footprint of TFIID downstream of the transcriptional start site and alters the cross-linking pattern of several TAFs to promoter sequences (14, 15, 20, 64). It has also been shown that an interaction between TFIIA and TFIID results in the generation of a productive form of TFIID that is capable of stably interacting with the promoter (72). Moreover, under certain conditions, TAFs have been shown to inhibit the ability of TBP to bind DNA (42, 43, 63, 83). Yet the addition of TFIIA reverses this TAF inhibition (42, 69).
The identification and characterization of an interaction between TAF40 and TFIIA is a first step in determining the mechanistic requirement for TAF40 in transcription. It is clear that TAF40 performs an essential and nonredundant function in yeast cells, since it is required for cell viability (38) and TAF40 inactivation appears to affect transcription from RNA Pol II promoters in yeast (44, 61). To date, TAF40 appears to be a component specific to the TFIID complex (25, 44, 62, 65), and thus the TFIIA-TAF40 interaction has the potential to serve as a critical link between TFIIA and TFIID functions in vivo.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM56884 to L.A.S.
We are indebted to Kevin Lumb for the Fos AD DNA and for critical reading of the manuscript. We also thank Zarmik Moqtaderi for various TAF DNAs.
REFERENCES
- 1.Andell F, III, Ladurner A G, Inouye C, Tjian R, Nogales E. Three-dimensional structures of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C-A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 3.Apone L M, Virbasius C A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 4.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 5.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 6.Bai Y, Perez G M, Beechem J M, Weil P A. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 8.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A, Davidson I, Moras D. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 9.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 10.Cairns B R, Henry N L, Kornberg R D. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caron C, Mengus G, Dubrowskaya V, Roisin A, Davidson I, Jalinot P. Human TAF(II)28 interacts with the human T cell leukemia virus type I Tax transactivator and promotes its transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3662–3667. doi: 10.1073/pnas.94.8.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Struhl K. Connecting a promoter-bound protein to the TATA-binding protein overrides the need for a transcriptional activation region. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 TATA element: genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 15.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 16.Clemens K E, Piras G, Radonovich M F, Sook Choi K, Duvall J F, DeJong J, Roeder R, Brady J N. Interaction of the human T-cell lymphtropic virus type 1 Tax transactivator with transcription factor IIA. Mol Cell Biol. 1996;16:4656–4664. doi: 10.1128/mcb.16.9.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colgan J, Manley J L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992;6:304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- 18.Dubrovskaya V, Lavigne A, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 19.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 20.Emami K, Jain A, Smale S. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 22.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 23.Geiger J H, Hahn S, Lee S, Sigler P B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Couto E, Klages N, Strubin M. Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc Natl Acad Sci USA. 1997;94:8036–8041. doi: 10.1073/pnas.94.15.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant P A, Schieltz D, Pray-Grant M G, Steger D, Reese J C, Yates III J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 26.Green M R. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- 27.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill D E, Hope I A, Macke J P, Struhl K. Saturation mutagenesis of the yeast HIS3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 29.Hisatake K, Ohta T, Takada R, Guermah M, Yamamoto T, Horikoshi M, Roeder R G. Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors. Proc Natl Acad Sci USA. 1995;92:8195–8199. doi: 10.1073/pnas.92.18.8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann A, Roeder R G. Cloning and characterization of human TAF20/15. J Biol Chem. 1996;271:18194–18202. doi: 10.1074/jbc.271.30.18194. [DOI] [PubMed] [Google Scholar]
- 31.Holstege F C P, Jennings E G, Wyrick C J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 32.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 33.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang J J, Auble D T, Ranish J A, Hahn S. Analysis of yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 37.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 38.Klebanow E R, Poon D, Zhou S, Weil A P. Cloning and characterization of an essential Saccharomyces cerevisiae gene, TAF40, an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1997;272:9436–9442. doi: 10.1074/jbc.272.14.9436. [DOI] [PubMed] [Google Scholar]
- 39.Klein C, Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994;266:280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kokubo T, Gong D-W, Wootton J C, Horikoshi M, Roeder R G, Nakatani Y. Molecular cloning of Drosophila TFIID subunits. Nature. 1994;367:484–487. doi: 10.1038/367484a0. [DOI] [PubMed] [Google Scholar]
- 42.Kokubo T, Swanson M J, Nishikawa J-I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kokubo T, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc Natl Acad Sci USA. 1994;91:3520–3524. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komarnitsky P B, Michel B, Buratowski S. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 1999;13:2484–2489. doi: 10.1101/gad.13.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 46.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 47.Lavigne A, Mengus G, May M, Dubrovskaya V, Tora L, Chambon P, Davidson I. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J Biol Chem. 1996;271:19774–19780. doi: 10.1074/jbc.271.33.19774. [DOI] [PubMed] [Google Scholar]
- 48.Lavigne A C, Gangloff Y-G, Carre L, Mengus G, Birck C, Poch O, Romier C, Moras D, Davidson I. Synergistic transcriptional activation by TATA-binding protein and hTAFII28 requires specific amino acids of hTAFII28 histone fold. Mol Cell Biol. 1999;19:5050–5060. doi: 10.1128/mcb.19.7.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X Y, Bhaumik S R, Green M R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 50.Li X Y, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 51.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 52.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Solution structure of a TBP-TAFII230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 53.Liu Q, Gabriel S E, Roinick K L, Ward R D, Arndt K M. Analysis of TFIIA function in vivo: evidence for a role in TATA-binding protein recruitment and gene-specific activation. Mol Cell Biol. 1999;19:8673–8685. doi: 10.1128/mcb.19.12.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma D, Olave I, Merino A, Reinberg D. Separation of the transcriptional coactivator and anti-repression functions of transcription factor IIA. Proc Natl Acad Sci USA. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macpherson N, Measday V, Moore L, Andrews B. A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics. 2000;154:1561–1576. doi: 10.1093/genetics/154.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 57.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. Cloning and characterization hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 59.Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–672. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 60.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 61.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 62.Natarajan K, Jackson B, Rhee E, Hinnebusch A. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- 63.Nishizawa M, Kokubo T, Horikoshi M, Roeder R G, Nakatani Y. Drosophila TAFII230 and the transcriptional activator VP16 bind competitively to the TATA box-binding domain of the TATA box-binding protein. Proc Natl Acad Sci USA. 1997;94:85–90. doi: 10.1073/pnas.94.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oelgeschlager T, Chiang C, Roeder R. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 65.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X-J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 66.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 67.Ozer J, Bolden A H, Lieberman P M. Transcription factor IIA mutations show activator-specific defects and reveal a IIA function distinct from stimulation of TBP-DNA binding. J Biol Chem. 1996;271:11182–11190. doi: 10.1074/jbc.271.19.11182. [DOI] [PubMed] [Google Scholar]
- 68.Ozer J, Lezina L E, Ewing J, Audi S, Lieberman P M. Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2559–2570. doi: 10.1128/mcb.18.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman P. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–14300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 70.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (γ) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 71.Ranallo R, Struhl K, Stargell L. A TATA-binding protein mutant defective for TFIID complex formation in vivo. Mol Cell Biol. 1999;19:3951–3957. doi: 10.1128/mcb.19.6.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ranish J, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ranish J A, Hahn S. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J Biol Chem. 1991;266:19320–19327. [PubMed] [Google Scholar]
- 74.Ranish J A, Lane W S, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 75.Reese J C, Zhang Z, Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem. 2000;275:17391–17398. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- 76.Sanders S L, Klebanow E R, Weil P A. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J Biol Chem. 1999;274:18847–18850. doi: 10.1074/jbc.274.27.18847. [DOI] [PubMed] [Google Scholar]
- 77.Sanders S L, Weil P A. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 78.Shen W-C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 79.Stargell L A, Moqtaderi Z, Dorris D, Ogg R C, Struhl K. TFIIA has activator-dependent and core promoter functions in vivo. J Biol Chem. 2000;275:12374–12380. doi: 10.1074/jbc.275.17.12374. [DOI] [PubMed] [Google Scholar]
- 80.Stargell L A, Struhl K. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 81.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 82.Tao Y, Guermah M, Martinez E, Oelgeschlager T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder R G. Specific interactions and potential functions of human TAFII100. J Biol Chem. 1997;272:6714–6721. doi: 10.1074/jbc.272.10.6714. [DOI] [PubMed] [Google Scholar]
- 83.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 84.Verrijzer C P, Tijan R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 85.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J D. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;382:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 87.Weinzierl R O J, Dynlacht B D, Tjian R. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993;362:511–517. doi: 10.1038/362511a0. [DOI] [PubMed] [Google Scholar]
- 88.Weinzierl R O J, Ruppert S, Dynlacht B D, Tanese N, Tjian R. Cloning and expression of Drosophila TAFII60 and human TAFII70 reveal conserved interactions with other subunits of TFIID. EMBO J. 1993;12:5303–5309. doi: 10.1002/j.1460-2075.1993.tb06226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]