Abstract
Although polyploid cells were first described nearly two centuries ago, their ability to proliferate has only recently been demonstrated. It also becomes increasingly evident that a subset of tumor cells, polyploid giant cancer cells (PGCCs), play a critical role in the pathophysiology of breast cancer (BC), among other cancer types. In BC, PGCCs can arise in response to therapy-induced stress. Their progeny possess cancer stem cell (CSC) properties and can repopulate the tumor. By modulating the tumor microenvironment (TME), PGCCs promote BC progression, chemoresistance, metastasis, and relapse and ultimately impact the survival of BC patients. Given their pro- tumorigenic roles, PGCCs have been proposed to possess the ability to predict treatment response and patient prognosis in BC. Traditionally, DNA cytometry has been used to detect PGCCs.. The field will further derive benefit from the development of approaches to accurately detect PGCCs and their progeny using robust PGCC biomarkers. In this review, we present the current state of knowledge about the clinical relevance of PGCCs in BC. We also propose to use an artificial intelligence-assisted image analysis pipeline to identify PGCC and map their interactions with other TME components, thereby facilitating the clinical implementation of PGCCs as biomarkers to predict treatment response and survival outcomes in BC patients. Finally, we summarize efforts to therapeutically target PGCCs to prevent chemoresistance and improve clinical outcomes in patients with BC.
Keywords: Polyploid giant cancer cell, Breast cancer, patient prognosis, chemo resistance, artificial intelligence
Introduction
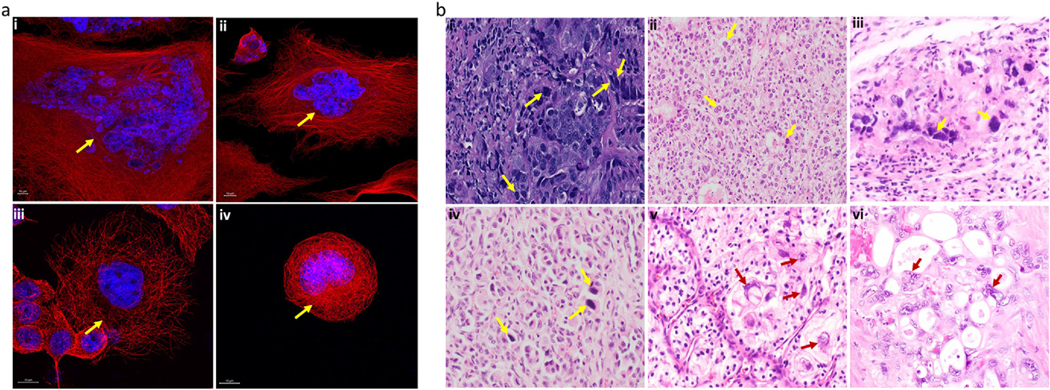
Polyploidy is an important evolutionary feature in plants, animals, and fungi, in which a diploid cell or organism acquires multiple sets of chromosomes. Polyploidy plays a critical role in normal growth and development of cells (e.g., in macrophages, hepatocytes, osteoclasts, cardiomyocytes, and muscle cells), as well as in tissue regeneration [1–4]. On the other hand, unscheduled polyploidy is commonly associated with pathophysiological states, such as chronic hepatitis, several age-related diseases, and cancer [5–8]. Growing evidence strongly suggests the crucial role of a subset of cancer cells known as polyploid giant cancer cells (PGCCs) in tumor heterogeneity and therapy resistance. PGCCs often arise in response to stress, including chemotherapy [2], ionizing radiation [9], and hypoxia [10,11]. They are involved in tumor initiation and progression, therapy resistance, and metastasis in various carcinomas, including breast, ovarian, prostate, and lung carcinomas [12,13]. Figure 1 illustrates representative immunofluorescence and hematoxylin- eosin (H&E)-stained images of PGCCs from various cancer cell lines and tissues.
Fig 1.
Representative images of PGCCs in various cancer types. a Representative immunofluorescence images in (i) MDAMB-231 TNBC cells (ii) PC-3 prostate cancer cells (iii) Panc89 pancreatic cancer cells, and (iv) WIDR colon cancer cells. The arrowheads indicate PGCCs. Red: α-tubulin (microtubules); blue: Hoechst (nuclei). Images were acquired using an LSM 700 confocal microscope (magnification,63X). The yellow arrowheads indicate PGCCs. b Representative H&Estained cancer tissues. Blue: hematoxylin (nuclei); pink: eosin (cytoplasm). Images were acquired under a light microscope (i) PGCCs (yellow arrow) seen with irregular and multiple nuclei at the boundaries of poorly differentiated ductal carcinoma of the breast (magnification, 400X); (ii) hepatocellular carcinoma exhibiting sheets of large clear cells with distinct cell borders and scattered hyperchromatic tumor giant cells, some with multinucleation (yellow arrows) (magnification, 200X, H and E stain); (iii)Embryonal carcinoma of testicles showing PGCCs (yellow arrows)(magnification, 600X, H and E stain); (iv) Well differentiated neuroendocrine tumor with striking nesting of bland tumor cells with eccentric nuclei. Scattered hyperchromatic tumor giant cells are also present (yellow arrows) (magnification, 200X, H and E stain); (v) Clear cell renal cell carcinoma, bizarre tumor cells on the right (red arrows) compared to smaller clear cells on left with low nuclear to cytoplasmic ratio (magnification, 400X) (vi) Clusters of malignant cells with prominent cytoplasmic vacuoles are embedded in desmoplastic stroma. Note the presence of multi nucleated tumor giant cells containing bizarre irregular nuclei (red arrows) with coarse chromatin and prominent nucleoli.
Breast cancer (BC) is the second most common cause of cancer mortality among women [14]. Metastasis and recurrence after surgical removal of the primary tumor are the primary causes of death in BC patients; PGCCs have been implicated in both BC metastasis and recurrence. In this review, we present the current state of knowledge about the clinical relevance of PGCCs in BC and the role of digital pathology and artificial intelligence (AI) in PGCC detection to predict treatment response and survival outcomes. We also discuss methodological obstacles hindering the study of PGCCs, including the lack of robust biomarkers to accurately identify PGCCs and their progeny. Lastly, we describe efforts to therapeutically target PGCCs to prevent chemoresistance and tumor relapse in patients with BC.
PGCCs and breast cancer
The presence of PGCCs in various solid tumors is widely documented [15]. Although PGCCs may be found in premalignant lesions, PGCCs are more frequently observed during the advanced stages of cancer [16] and after treatment [11]. Typically, PGCCs are considered destined for senescence or apoptosis. Due to their presumed inability to proliferate, PGCCs were believed to suppress tumor progression. However, it has become increasingly evident that PGCCs play a crucial role in BC pathophysiology, including tumor relapse, metastasis, and chemoresistance [11,16,17].
PGCCs have been observed in low numbers in chemosensitive cells before treatment; however, PGCC numbers are drastically increased in response to therapy-induced stress [18]. Compared with other cancer cells, PGCCs are more resilient to chemotherapy, as revealed by time-lapse imaging of triple-negative BC (TNBC) MDA-MB-231 cells [18]. A positive correlation between the number of PGCCs and the tumor stage and grade has also been reported [19]. In addition, chemoresistant MDA- MB-231 cells displayed a higher number of PGCCs than parental MDA-MB-231 cells [6]. Notably, the number of PGCCs was significantly associated with the degree of metastasis in patients with BC [16]. A growing amount of evidence also demonstrates a causal role for PGCCs in BC initiation, progression, metastasis, and chemoresistance. Gerashchenko et al. observed that near-triploid DNA content in BC samples (including TNBC) was a marker of poor response to neoadjuvant chemotherapy (NAC) and that these cancers had a fourfold higher proportion of polyploid cells compared with near-diploid cancers [20,21]. Interestingly, our recent analysis of tumors from TNBC patients treated with NAC showed that tumor tissues from patients with residual disease had significantly higher numbers of PGCCs than tumors from patients with pathological complete response (unpublished data), suggesting that PGCCs may underlie chemoresistance to NAC.
Moderate, clinically relevant doses of chemotherapy drugs often lead to senescence instead of apoptosis, accompanied by the formation of PGCCs. Even though most of these PGCCs eventually undergo apoptosis, 10%–20% of PGCCs survive senescence [22–24]. Importantly, surviving PGCCs display stem cell-like properties and can undergo asymmetric division to form progeny cells with near-diploid nuclei. These near-diploid progeny cells can undergo normal mitosis and repopulate the tumor, resulting in tumor relapse. Furthermore, single PGCCs isolated from cancer cell lines formed tumors when injected into immunodeficient mice, underpinning the tumorigenicity of PGCCs [11,24,25] By contrast, injection of hundreds of non-PGCC diploid cancer cells into nude mice failed to produce tumors. The PGCC progeny arising from the newly formed tumor tended to be resistant to chemotherapy and hypoxia [11,24]. Although polyploidy is associated with multidrug resistance [26,27], therapy-induced PGCCs promoted short-term chemoresistance only to the drug they had been exposed to in vitro. We have previously reported that long-term docetaxel exposure in castration-resistant prostate cancer cells resulted in the formation of multinucleated polyploid (MP; another term for PGCC) cells which had tumorigenic potential in nude mice [28]. These MP cells and their progeny were chemoresistant and possessed CSC-like properties (positive for CD44 marker) [28]. In a study by Pirsko et al., the treatment of MDA-MB-231 cells with doxorubicin promoted doxorubicin resistance; intriguingly, sensitivity to other therapeutics (e.g., vinblastine, cisplatin, and 5-fluorouracil) was increased, suggesting that a brief time window may be available for combination therapies to eradicate both non-PGCCs and the chemo resistant PGCCs [29].
The extensive crosstalk between PGCCs and the tumor microenvironment (TME) contributes to the ability of PGCCs to promote chemoresistance and metastasis [12][15]. However, a thorough investigation of PGCCs’ clinical significance in BC is lacking. TNBC (negative for estrogen, progesterone, and HER2 receptors) is a highly heterogeneous BC subtype. TNBC and one of its subtypes, quadruple negative breast cancer (QNBC; TNBC that additionally lacks expression of androgen receptor), are particularly aggressive in women of African descent. Reports suggest that differences in the breast TME between non-Hispanic whites and African Americans may be partly responsible for racial disparities in BC outcomes. The investigation of differences in numbers, size, and the features of PGCCs in conjunction with the TME is an uncharted terrain and may provide further insight into the mechanisms underlying racial disparities in BC.
Life cycle of PGCCs
The primary objective of cancer therapy is the complete elimination of cancer cells through apoptosis and autophagy, which, however, is rarely achieved due to genetic mutations, development of tolerance, or limited drug bioavailability. Evidence strongly suggests that instead of immediately succumbing to death by apoptosis, tumor cells undergo cellular growth arrest referred to as therapy- induced senescence, before eventually undergoing apoptosis [30]. However, therapy-induced stress may cause mitotic slippage in a subset of cells and the formation of PGCCs [31], which remain metabolically active and are capable of escaping senescence [32–34].
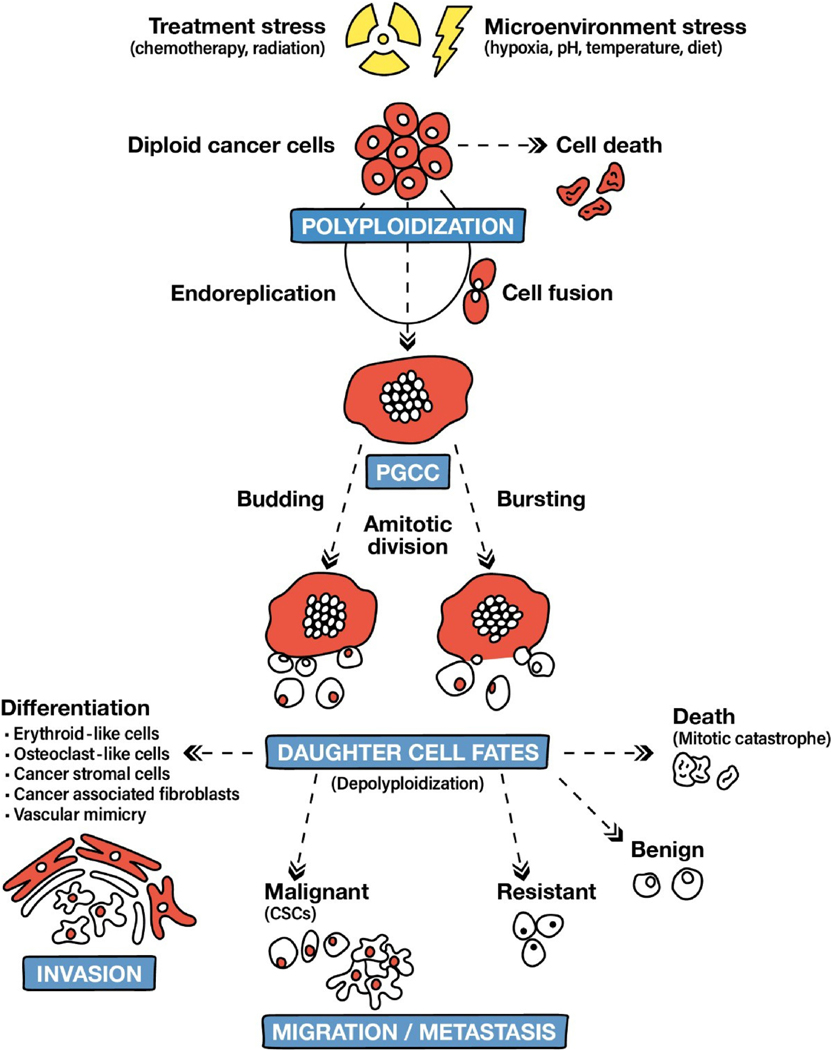
The formation and subsequent proliferation of PGCCs by giant cell division and budding, was initially described as neosis by Sundaram et al., [35], and was further expanded upon by Erenpreisa and Cragg, [36], and later referred to by Niu and colleagues as the “giant cell cycle”. The cycle consists of four phases: initiation, self-renewal, termination, and stability [26]. This is rooted in the embryonic theory of cancer, wherein the division of PGCCs by endoreplication is not unlike the programmed, blastomere-mediated dedifferentiation of germ cells [37] During the initiation phase, diploid cancer cells enter the polyploidy cycle in response to a stressor, bypassing the cell division checkpoints and evading apoptosis. In the self-renewal phase, the polyploid cells continue to undergo endoreplication cycles, while the remaining diploid cells succumb to stress-induced apoptosis. The termination phase involves the depolyploidization of PGCCs to form near-diploid daughter cells. During the stability phase, the newly generated daughter cells undergo normal mitotic division, giving rise to the next generation of near-diploid cancer cells which fuel tumor growth [26].
PGCCs are formed through either cell fusion or endoreplication, with the latter being the more prevalent mechanism of PGCC formation (Figure 2). Formation of PGCCs through cell fusion has been reported in Hodgkin’s lymphoma [38] and glioblastoma [39] cell lines. Despite reports of PGCCs generation through cell fusion, cell fusion in MDA-MB-231 and ovarian cancer cells contributed to only 10%–20% of all PGCCs [11]. These studies involved co-culturing cells that expressed either GFP (green fluorescent protein) or RFP (red fluorescent protein), followed by exposure to drugs or hypoxia to induce PGCC formation. Cell fusion was then quantified by counting cells emitting a merged yellow fluorescence. In contrast to cellular fusion that may give rise to a binucleate cell, endoreplication generally results in the formation of a single large nucleus containing multiple copies of the genome [40]. During endoreplication, cells escape the mechanisms regulating the cell cycle due to alterations in the mitotic cycle or cytokinesis [12]. Cytokinetic failure leads to the formation of binucleate cells, which may fuse to form an enlarged single nucleus. During endoreplication, cells bypass mitosis before metaphase (known as endocycle) or after metaphase (referred to as endomitosis). In contrast to mitotic bypass resulting in a single large spherical nucleus, mitotic failure after metaphase may result in a single lobulated nucleus.
Fig. 2.
Generation and fates of PGCCs and their progeny (adapted from Niu et al. and Chen et al.) [12,41].
Although the mechanisms underlying PGCC formation are under investigation, it is known that endoreplication relies on the disruption of cell cycle checkpoints enforced by cyclin-dependent kinases (CDKs). The p53/p21 and p16/pRB pathways are key regulators of CDKs, playing a pivotal role in the induction of apoptosis or senescence in tumor cells after chemotherapy [42]. Depending upon the extent of DNA damage, p53 activation results in either DNA repair (when the damage is limited) or cell cycle arrest and apoptosis (when the damage is extensive). Severe DNA damage due to chemotherapy or hypoxia results in the upregulation of p53, which inhibits the activity of cyclin/CDK complexes responsible for the assembly of the mitotic apparatus, thereby blocking the transition at the G1/S or G2/M phase of the cell cycle. However, some cells with DNA damage re- enter the cell cycle and exit at the G2 phase, giving rise to polyploid cells [43].
As most solid tumors harbor p53 mutations, DNA damage suppresses cyclin B expression in a p53-independent manner, allowing cancer cells to undergo endoreplication. High expression of the cyclin B binding partner CDK1 has been linked to the formation of PGCCs [44]. In PGCCs, increased expression of CDK1 [44,45] and its downstream effector survivin [46] has been associated with the ability of PGCCs to re-enter the cell cycle and thereby escape therapy-induced senescence. Dysregulation of Aurora kinase B, which interacts with survivin to regulate chromosome segregation, has also been implicated in the formation of polyploid cells [9]. G1/S transition is regulated by the transient expression of the CDK inhibitor p21. In p53-deficient tumors, chronic overexpression of p21 can lead to unfettered cell replication despite the accumulation of DNA damage, leading to the formation of PGCCs [47].
Fate of PGCC progeny
Observations from colony formation assays led to the conclusion that PGCCs either do not proliferate or proliferate very slowly. Even though a fraction of PGCCs may undergo cell death after senescence, the rest are capable of asymmetric division resulting in the formation of aneuploid or diploid cells [30]. PGCCs can produce regular-sized mononucleated cells through neosis, which involves karyokinesis via nuclear budding followed by asymmetric cytokinesis [32,35].We have previously reported neosis in prostate cancer cells [28] in alignment with findings from other groups in multiple human cancer cell lines, including breast [16] and ovarian cancer cell lines [48]. Besides budding, burst-like division has also been reported in some cell lines [11] (Figure 2). These mononucleated daughter cells, occasionally referred to as “Raju cells”, transiently exhibit stem cell-like properties before undergoing cell division or apoptosis. In fact, tumor budding has been associated with lymph node and distant metastasis and recurrence and is an emerging independent prognostic factor [49].
Although a portion of daughter cells die due to mitotic catastrophe, depolyploidization due to meiosis-like division can occur in daughter cells to producing near-diploid cells (Figure 2). These daughter cells can resume mitosis and are pluripotent and stem cell-like, with the ability to self-renew and differentiate into epithelial cancer cells or benign cells. PGCC progeny cells resemble embryonic stem cells and can give rise to any cell type of the three germ layers. They can be induced to differentiate into benign tissues (e.g., bone, cartilage, and adipose tissue) [11] or stromal cells (e.g., endothelial cells, inflammatory cells, fibroblasts, and erythroid cells) [50] (Figure 2). Thus, PGCC progeny cells can become a part of the TME by populating the tumor stroma [51] or contribute to vascular mimicry (VM) [52] channels, increasing the risk of metastasis [51,52]. The PGCC progeny can also differentiate into malignant cells, which are often more chemoresistant and metastatic than parental cells, owing to newly acquired mutations [11,17]. Due to their mesenchymal phenotype, these cells can also migrate to various tissues or organs to form new tumors [16].
Characteristics of PGCCs and potential biomarkers
Cancer stemness
PGCCs express both normal and CSC markers such as octamer-binding transcription factor-4 (OCT4), NANOG, sex-determining region Y-box 2 (SOX2), CD44, and CD133 [11,20,53]. PGCC daughter cells also exhibit stem cell-like properties and express CSC markers, such as the surface glycoproteins CD44 and CD133 [11]. They exist in a highly dedifferentiated state similar to embryonic cells and are more pluripotent than typical CSCs. The PGCC progeny resemble blastomeres during embryonic development and can differentiate into different cell types of the three germ layers (ectoderm, mesoderm, and endoderm) in vitro [26]. They exhibit spatiotemporal expression of embryonic and self-renewal markers, such as NANOG, OCT3, OCT4, aldehyde dehydrogenase-1A (ALDH1A) [29], and SOX-2 [11]. The daughter cells can differentiate into different types of benign cells (e.g., adipocytes, chondrocytes, erythrocytes, and osteocytes) or give rise to carcinomas of different grades. A study by Zhang and colleagues demonstrated that PGCC progeny from the MCF-7 cell line could differentiate into benign stromal cells, including myoepithelial, endothelial, and erythroid cells [50]. Given the role of the tumor stroma in regulating the bioavailability and enzymatic degradation of drugs, PGCCs can play an important role in promoting resistance to various anticancer therapies [54].
Epithelial-to-mesenchymal transition
PGCCs and their progeny often undergo epithelial-to-mesenchymal transition (EMT). EMT involves dramatic cytoskeletal alterations that result in changes in cell adhesion and morphology and is essential for the metastatic spread of cancer cells. Expression of high levels of the mesenchymal markers vimentin, fibronectin, and N-cadherin and low levels of the epithelial markers cytokeratin and E-cadherin have been observed in PGCCs [16] and their progeny [17]. In cancer cells, EMT and the acquisition of stemness phenotypes are closely linked [55]. Similarly, a correlation between the expression of EMT and stemness markers has been reported in PGCC progeny cells. This relationship is exemplified by the induction of EMT in human mammary epithelial cells by the transforming growth factor-β (TGF-β), which also facilitates the generation of cells with stem cell-like properties (CD44 expression and self-renewal ability) [56]. Furthermore, the expression of Twist family members [56,57] and Snail proteins [56] can induce a stem cell-like phenotype in BC cell lines. In BC cell lines, induction of Twist-1 expression and TGF-β exposure are associated with chemoresistance due to the increased expression of ATP-binding cassette (ABC) transporters mediating drug efflux [58]. ABCG2 (ATP-binding cassette superfamily G member 2) overexpression is common in PGCCs and their progeny cells [11], promoting multidrug resistance. Thus, the chemoresistance-metastasis-stemness nexus described is apparent in PGCC daughter cells.
Tumor microenvironment
PGCCs can promote resistance to chemotherapy by modulating the TME. Exposure of chemosensitive BC cell lines to conditioned medium (previously used to culture chemoresistant BC cell lines) induced the expression of B-cell lymphoma-extra-large (Bcl-xL) and other anti-apoptotic proteins and suppressed the expression of pro-apoptotic proteins [6]. PGCC-derived growth factors and cytokines, including vascular endothelial growth factor (VEGF) and macrophage migration inhibitory factor (MIF), were able to promote chemoresistance in chemosensitive cancer cells [6]. MIF [59] and VEGF [60] are implicated in angiogenesis essential for the sustained growth of tumors. In addition, MIF inhibits the key tumor suppressor p53 [59]. Given the role of p53 in the induction of apoptosis or senescence in response to DNA damage, PGCC-derived MIF may allow cancer cells to grow despite the accumulation of DNA damage [59]. PGCCs can also modulate the composition of the TME by recruiting diploid cancer cells from adjacent areas; these diploid cells can eventually become PGCCs. The complexity of the TME contributes to spatial variability in drug exposure within the tumor, resulting in an uneven distribution of PGCCs within the TME. In an in vitro study on PC3 cells treated with a drug gradient to simulate the TME, PGCCs emerged in larger numbers in areas exposed to higher drug doses [61]. Diploid cells from low-dose areas migrated to these high-dose regions and formed PGCCs. It is likely that PGCC-derived factors are partly responsible for the migration of diploid cells to the high-dose regions.
The relationship between PGCCs and the TME is bidirectional, with TME-associated stressors inducing the formation of PGCCs. TME stressors include hypoxia, interstitial pressure, uneven blood flow, and extracellular matrix stiffness [13]. Tumor progression limits oxygen availability to the surrounding tissues and establishes a hypoxic microenvironment, which can promote resistance to chemotherapy, radiotherapy, and immunotherapy. Cobalt chloride (CoCl2)-induced hypoxia in cell lines resulted in the formation of PGCCs [10,16]. One of the common responses to CoCl2-induced hypoxia is the stabilization of the transcription factor hypoxia-inducible factor 1-α (HIF1α), which regulates chemoresistance [62], stemness [63], and EMT [64]. HIF1α stabilization has also been reported in PGCCs after induction of therapy-induced senescence in MDA-MB-231 cells [6]. These PGCCs exhibited elevated levels of reactive oxygen species (ROS), which stabilized HIF1α [6]. They also displayed a lower mitochondrial potential, presumably as a consequence of oxidative stress-induced mitochondrial damage. Furthermore, cellular senescence is closely linked to mitochondrial dysfunction and ROS accumulation [65], consistent with the observation that most PGCCs generated after drug treatment had higher levels of the senescence marker β-galactosidase [6,66]. Interestingly, ROS have been implicated in cancer cell escape from therapy-induced senescence and the formation of polyploid cells. In a study by Mosieniak et al., the ROS scavenger Trolox temporarily reduced the number of polyploid cells generated in HCT-116 cells after doxorubicin treatment [66]. Chen et al. highlighted that PGCC stemness, vasculogenic mimicry, metastasis, and chemoresistance were the net output of the dynamic relationship between PGCCs and the TME [12].
Vascular mimicry
PGCCs and their progeny cells contribute to VM, thereby enabling the survival of aggressive tumors. Similar to angiogenesis, VM also facilitates the supply of nutrients and oxygen to tumors. VM is induced by hypoxia and involves the formation of tumor-derived channels that connect tumor cells with endothelial blood vessels. PGCCs and their progeny can form the walls of VM vessels [19]. Although the role of PGCCs in VM has been mostly studied in human glioma and ovarian cancer, PGCCs and their progeny cells are believed to have a similar role in VM in BC. CSCs, hypoxia, and mesenchymal stem cells are essential for the induction of VM; PGCCs in BC display all these properties, supporting the notion that PGCCs promote VM in BC. Significantly, the PGCC progeny cells in the BC cell lines MCF-7 [50] and BT-549 [67] were capable of differentiating into erythroid cells in vitro. The presence of PGCCs and PGCC-derived erythrocytes in VM vessels has also been demonstrated. Unlike erythroid cells generated from hematopoietic stem cells and mesenchymal stem cells in the bone marrow, PGCC-derived erythroid cells contained high levels of fetal and embryonic hemoglobin [67]. Compared with adult hemoglobin, fetal and embryonic hemoglobin have a higher affinity for oxygen, facilitating the survival of the tumor in a hypoxic environment.
A shifting genetic, epigenetic, and metabolic landscape
The ability of PGCCs to cope with stressors, such as hypoxia and drugs, may be attributed to the multiple copies of each gene, allowing for DNA recombination and variation in epigenetic patterns. PGCCs can epigenetically silence p53 pathway components and thereby escape apoptosis and senescence [68]. Furthermore, PGCCs express high levels of DNA repair genes, including BRCA1 (breast cancer gene-1) and p19Arf, rendering PGCCs resilient to the cytotoxic effects of anticancer treatments [68]. PGCCs also show considerable ploidy level variations after drug treatment [18], contributing to the overall heterogeneity of tumor cells.
Due to these extensive genetic and epigenetic changes, PGCCs significantly differ from non-PGCCs in terms of morphology and mechanical properties. PGCCs generated after paclitaxel treatment of MDA-MB-231 cells exhibited remarkable cytoplasmic and nuclear stiffness accompanied by changes in cytoskeletal organization [18]. The changes in cytoskeletal organization characterized by thick and elongated actin stress fibers contributed to the enhanced migration potentials of these cells. In the same study, PGCCs also exhibited dysregulation of the RhoA-ROCK1 pathway associated with increased malignancy [18].
PGCCs also show a distinct metabolic shift from oxidative phosphorylation to glycolysis [69], allowing PGCCs to survive under hypoxic conditions. A recent study involving the chemoresistant TNBC cell line MDA-MB-436, showed that PGCCs had a higher number of small mitochondria and small lipid droplets than the parental chemosensitive cells. The accumulation of lipid droplets was accompanied by changes in lipid metabolism, as suggested by the transcriptomic profile of the PGCCs. Furthermore, PGCCs from chemoresistant TNBC cells exhibited higher levels of oxidative phosphorylation and reduced levels of glycolysis indicated by a lower lactate secretion [70]. The discrepancies in the utilization of oxidative phosphorylation in PGCCs may have resulted from methodological differences, such as cell density, temperature, and the elapsed time before cell assessment. It is also possible that PGCCs utilize aerobic glycolysis under normoxic conditions in vivo and switch to anaerobic glycolysis in response to hypoxia. The metabolism-regulating kinase mTOR is also upregulated in PGCCs, consistent with elevated glycolysis levels. Thus, mTOR can be used as a PGCC marker. In a study by Mosieniak et al., mTOR was overexpressed in the colorectal carcinoma cell line HCT-116 after doxorubicin treatment, and mTOR upregulation was accompanied by increased numbers of polyploid cells and reversible therapy-induced senescence [66]. By contrast, mTOR was not overexpressed in MCF-7 cells, which showed senescence but no polyploidy after doxorubicin treatment. Importantly, doxorubicin-induced senescence in MCF-7 cells was irreversible, indicating that polyploidization is required for tumor cells to grow after doxorubicin treatment [66].
Isolation and characterization of PGCCs
PGCCs are identified based on their DNA content and morphological features such as cell and nuclear size. Nevertheless, considerable morphological heterogeneity exists among PGCCs. Over the last thirty years, PGCCs have been referred to as multinucleated cells (MN) [71], giant cells [72], endopolyploidy tumor cells [23], and monster cells [73]. Although the distinction between mononucleated and multinucleated PGCCs may be informative, the mechanisms underlying their formation may be similar.
PGCCs are typically isolated from mononuclear cells using size-based filtration techniques such as light scattering in FACS (fluorescence-activated cell sorting) [21] and manually through a nylon filter [24]. More recently, CoCl2-induced hypoxia treatment has been employed as a method to isolate PGCCs; CoCl2 selectively kills diploid tumor cells, thereby allowing for the isolation of PGCCs. To study the molecular mechanisms underlying PGCC formation, Zhang and colleagues isolated PGCCs and evaluated the protein expression patterns in CoCl2 generated PGCCs and in PGCCs that were in the process of budding [74]. For comparison, they also assessed the protein expression patterns in diploid cancer cells using an ITRAQ (Isobaric Tags for Relative and Absolute Quantitation)-based high-throughput approach. Differences were observed in the expression of proteins involved in cell cycle regulation, stem cell generation, hypoxic response, and EMT [74].
Traditionally, quantification of PGCCs in DNA-stained cell samples is based on profiling their DNA content (DNA histograms) through flow and image cytometry. Flow cytometry is widely used owing to its ability for rapid analysis of a large number of cells and precise quantification of S-phase cells [75,76]. Flow cytometry analysis can only be performed on cell suspensions, whereas image cytometry can also be done on histological sections, allowing for a morphological assessment of the cells and, thus, a better characterization of the tumor cell population [76]. Flow cytometry cannot discriminate cancerous cells from normal stromal cells, macrophages, and granulocytes unless cells are specifically labeled. By contrast, examination of cells under a microscope can distinguish cancer cells from normal cells based on their size, shape, and textural features of the nuclei. A four type (I-IV) diagnostic and prognostic classification of BC DNA histograms was first proposed by Auer et al. [77]The Auer classification, however, is unable to discern populations of BC cells that differ in their biological traits. Subsequently, additional parameters (e.g., ploidy balance, proliferation index, DNA index) have been suggested to take into account the phase of the mitotic cycle of individual cells, the growth fraction of the cell population, and the different karyotypes of cells [78–81].
A study by Diaz-Carballo described at least four different morphological patterns in multinucleate cells obtained after etoposide treatment in carcinoma cell lines [82]. The nuclei shape in “spiral” PGCCs was wreath-like in two dimensions and spiral-like in three dimensions. These spiral cells expressed high levels of proliferating cell nuclear antigen (PCNA), indicating a proliferating state. “Monastery” PGCCs had large nuclei surrounded by progeny cells encapsulated in a wall-like structure. Mitochondria tended to accumulate around the wall-like structure and were considered to play a vital role in the formation of this structure. “Pregnant” cells were similar to monastery cells but lacked wall-like structures. Interestingly, the progeny cells surrounding the large nucleus possessed their own cellular membrane and could undergo replication while still embedded in the large parent cell. “Shepherd” cells were relatively rare; they were pear-shaped and had a lobopodium considerably larger than the cell body. Shepherd cells were generally spotted in the vicinity of multiple smaller cells [82]. A study by Zhang et al. reported that PGCCs from MDA-MB-231 and HEY cells had a neuron-like shape; in contrast, PGCCs from the ovarian cell line SK-OV-3 had a more spherical shape without branches [11]. The nomenclature describing the PGCC progeny with distinct morphological features is inconsistent, and the functional significance of PGCCs with different morphologies needs to be clarified.
Currently, our laboratory is investigating whether PGCCs and other TME components have predictive value for treatment response (measured as pathologic complete response versus residual disease) and could serve as a novel biomarker of therapy resistance in BC patients. To this end, we have proposed a digital pathology approach for the analysis of PGCCs and other key TME structures using whole slide images (WSIs) of BC tissue samples, as discussed below.
PGCC and TME mapping using digital pathology
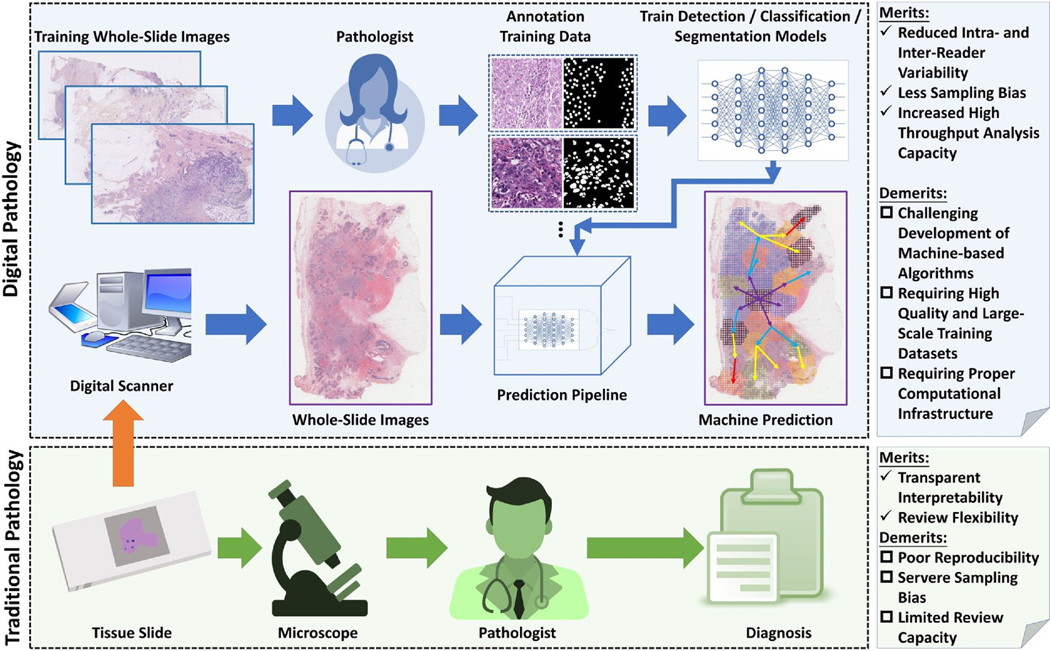
Traditional pathology involves the preparation of tissue slides and their microscopic evaluations by pathologists. Despite proffering certain merits, such as transparent interpretability and review flexibility, traditional pathology is subject to high intra-reader and inter-reader variability, low review and processing capacity, and significant sampling bias. Whole-slide image cytometry which combines whole slide imaging (H&E stained) with DNA image cytometry (Feulgen stained), has also been used for detecting abnormal DNA content (83). It is more objective than histological assessment and cost-effective and simpler to use compared to image cytometry (83). By contrast, digital pathology pipelines supported by high-performance computing can automate slide analysis, allowing for a large number of whole slide pathology images to be analyzed simultaneously. Phenotypic features can be extracted from all tissue domains, providing a more comprehensive tissue characterization. Due to the computational nature of digital pathology pipelines, all derived features are quantitative. Thus, the resulting analysis is highly reproducible, with a strong resistance to inter- and intra-analysis variation and sampling bias [84–86]. However, digital pathology requires the development of new and effective AI algorithms, generation of large-scale annotation data for training, powerful computational facilities, and strong IT support [87].
Given its superior performance, digital pathology is poised to become a widely used tool in cancer diagnosis, treatment response prediction, and patient prognosis [88,89]. Recognizing its tremendous potentials, our research group is actively focused on developing various digital pathology pipelines. One such pipeline to enumerate PGCCs and their various avatars in relation to other TME features is briefly discussed here (Figure 3).
Fig. 3.
A comparison of traditional and digital pathology and a brief overview of the development of a digital pathology pipeline to characterize PGCCs and other TME components.
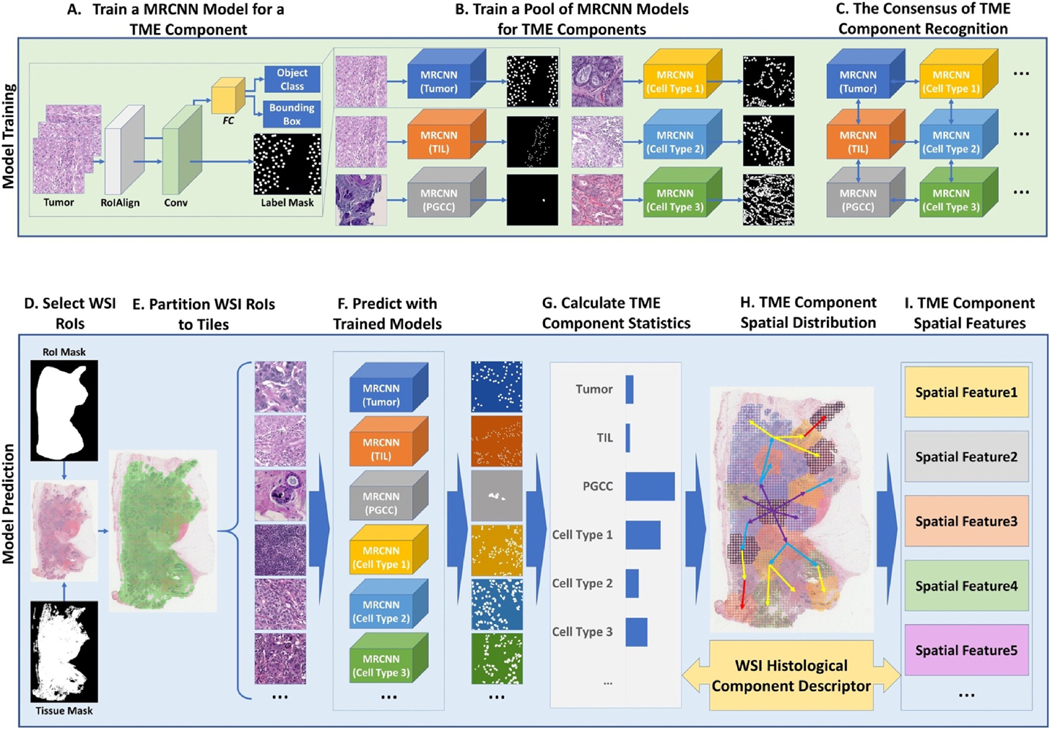
Before computational analysis, physical slides are scanned by high-throughput digital scanners in the data preparation step. The resulting digital microscopy images have high resolutions and capture tissue information at the cellular level. These imaging data are prepared to support TME component detection and segmentation. Deep learning models are trained to automatically detect the locations of TME components, including PGCCs. Points are annotated at each target object center to train deep learning models for detection. Segmentation, on the other hand, is used to delineate the accurate boundaries of cells and other TME components of interest and requires precise boundary annotations for training [90].
Due to its promising performance and generalizability, deep learning has become exceptionally popular for biomedical image processing. Most models derived from this modern AI framework consist of two analysis steps. In the first step, primitive image features from multiple scales are extracted by the convolutional layers with different convolutional kernels. The extracted features are implicitly related to different visual properties, ranging from texture, to color, brightness and scale. This process is followed by the second step, in which a decision is made by the deep learning model to realize the detection, classification, and segmentation task. In order to optimize the resulting performance, a deep learning model can iteratively learn features and update its parameters for decision-making simultaneously during the training stage. Such an end-to-end training pattern makes the deep model training process highly adaptive and effective to achieve optimal performance. For PGCC recognition, for instance, features are computationally extracted to cover multinuclear spatial organization, intensity, contrast, image texture, cell size, and other image primitive properties that are automatically determined by the model during the training process. As these features are derived from multiple scales, they capture the relative position of PGCCs and the surrounding components, including clusters of cells of other populations. The model decision component optimizes weights of features to detect, recognize and segment PGCCs in a way that best converges to the training ground truth. Notably, DNA content differences between diploid cells and polyploid cells (e.g., PGCCs) can be captured by a high-resolution scanner under sufficiently high power. With image processing methods and deep learning-based segmentation and counting algorithms, such differences can contribute to distinguishing PGCCs from other cell clusters.
Mask R-CNN (MRCNN) [91] is an appropriate and widely used deep learning model for cell population analysis using digital images. MRCNN is an extended version of the deep learning architecture Faster R-CNN which was developed based on Fast R-CNN [92,93]. The Fast R-CNN model works in two consecutive stages: 1) region of interest (ROI) proposal, and 2) deep convolutional network for classification and bounding box detection. In the ROI proposal stage, a set of ROIs (e.g., PGCC region candidates) is produced and passed to the second stage of the network for further analyses. In the second stage, the deep convolutional network produces region feature maps that are further processed by the ROI pooling layer, which constructs a feature vector of a fixed length. The feature vector is provided to the fully connected network for classification and detection. By contrast, Faster R-CNN presents a region proposal network (RPN) for the ROI proposal task that is connected with the deep convolutional network. Because of this change in the architecture, the two stages in Fast R-CNN are combined into an end-to-end network, contributing to a significant reduction in the training and prediction time cost. By comparison, MRCNN further enhances Faster R-CNN by replacing the ROI pooling layers with ROI align layers for better segmentation modularity required for segmentation tasks. The MRCNN is implemented [94] with Tensorflow [95] and Keras [96] and uses Feature Pyramid Network (FPN) [97]and a ResNet101 [98] as the convolutional network backbone.
For training, both point and contour features are annotated exhaustively. Human annotators (e.g., pathologists) would annotate all cells of interest (e.g., PGCCs) using a ROI image and assign appropriate class labels to annotated cells. High-quality and definitive annotations covering a full spectrum of representative cells for each cell population are essential to successful machine learning for distinguishing distinct cell populations and other TME components. In the training stage, the point annotations are dilated by image morphological operations to circles as large as the average cell size. The resulting dilated circle masks are used as training labels. The bounding box branches of the MRCNN model are used for point predictions. The center of each resulting bounding box is considered as a cell center. By contrast, a binary mask for each annotated cell is produced by filling an annotated contour with a foreground color. Such binary masks are used as training labels. The contour predictions are made by the segmentation branch of the MRCNN model. The output of the segmentation branch is an array of masks, each capturing the contour information of a segmented cell.
As deep learning methods require a large scale of good quality annotations, an exhaustive annotation is recommended to ensure good prediction performance. In this annotation manner, all the positive samples in a ROI are expected to be annotated. The remaining samples that are not annotated are considered negative ones. However, human annotators may occasionally miss some positive samples by mistake. The impact of these false-negative samples on the machine prediction performance can be alleviated by an enhanced focus on positive samples when it comes to computing the loss function. It is also possible that negative samples are given positive labels in annotation practice. Although this inevitably makes machine algorithms learn features from the wrong samples, the negative effect of few incorrect annotations on algorithm training is limited. This is especially true for the deep learning-based methods that focus on the overall statistical and probability distributions of the sample space. Deep learning model training involves numerous training iterations. The multiple iterations of training and data-updating can help reduce the negative impact of erroneously labeled samples during the training stage. Moreover, the sample annotation process itself can be completed in an iterative manner. After each annotation iteration, the deep learning model can be trained and applied to the training data to identify samples with prediction results different from the human-provided ground truth. These samples can be provided to pathologists for further review and annotation correction. The resulting annotations will be used to update the training data. This process of leveraging trained machine models to identify candidates with incorrect annotations is repeated until the prediction results from the trained model agree with human labels for the vast majority of samples. This method can effectively improve the training data quality by recognizing data outliers due to human annotation mistakes.
For performance enhancement, image color is regulated in the pre-processing step. Given the noticeable stain variation, all images are normalized by color [99] and centralized by subtracting the average RGB value from the training images before they are provided to the model. The resulting images can be augmented by various methods, including left-right flip, up-down flip, rotation, pixel rescaling, and Gaussian blurring [100]. The sizes of the RPN anchors are selected based on the cell sizes. The batch size of each training step is suggested to be as large as possible, contributing to training acceleration. When the batch size is small due to the limited GPU memory, the batch normalization can be skipped. The non-maximum suppression (NMS) is used in the RPN and the detection branch. The thresholds of the intersection over union (IoU) and the minimum confidence score of NMS in RPN and detection need to be set appropriately for enough retained ROI proposals and cell clumps. Additionally, weights of different types of losses need to be properly assigned. The stochastic gradient descent (SGD) optimizer is often used to train a deep learning model with a proper selection of the initial learning rate and the learning momentum. The L2 regularization is usually applied to each layer weight with a small decaying weight. As most cells present relatively regular shape, training can be completed with less than 100 epochs.
Overall, there are two major strategies to construct a deep learning model to analyze multiple TME components. The first approach is to train a single comprehensive deep learning model that can detect, classify, and segment multiple TME components. The second method is to construct a group of deep learning models, each specifically trained for a single TME component, such as PGCCs. In practice, the second strategy usually outperforms the first. However, the downside of the second strategy is that one consensus model has to be in place to harmonize and aggregate outputs from the group of models after training [101]. For each TME component, the overall dataset is divided into training, validation, and testing sets, commonly with a ratio of 70:15:15. After a deep learning model is optimized by training data at each iteration, the refined model makes predictions on the validation set. The validation accuracy is used to control the learning rate reduction, stop the training process, and select the final model. The trained model is then applied to the testing set to confirm the model performance [90].
The development of an analysis pipeline is necessary to apply the deep learning model to WSIs. First, the ROIs are extracted from the WSIs based on the ROI and tissue masks. The ROI masks are generated from manual annotations by domain experts, who highlight the ROIs based on their knowledge. The tissue masks are generated by tissue detection approaches, and each ROI is partitioned into an array of image tiles with appropriate size for further analysis. The trained deep learning models are used to make predictions on these image tiles for detection, classification, and segmentation analysis of different TME components, including PGCCs. Prediction results from image tiles are next spatially aggregated to produce WSI TME component distribution maps. These distribution maps are used to extract statistical and spatial features of the WSI TME components [90]. These features serve as the numerical representation of each WSI to further predict treatment response (Figure 4).
Fig. 4.
A schema of the deep learning model based WSI analysis to characterize PGCCs and other key TME components.
Therapeutic approaches to target PGCCs
The role of PGCCs in the development of chemoresistance, metastasis, and tumor recurrence, as well as their ability to modify the TME, makes PGCCs a promising therapeutic target. Despite increasing efforts to develop anti-PGCC strategies to augment the clinical efficacy of established cancer therapies, anti-PGCC treatments that have shown promise in preclinical studies have not yet been incorporated into clinical practice.
A better understanding of the pathways involved in the formation of PGCCs and their progeny have led to the development of anti-PGCCs therapies with promising preclinical efficacy. Identifying immune and metabolic biomarkers associated with PGCC function may also facilitate the development of therapeutic strategies to target PGCCs. For example, polyploidy induced endoplasmic reticulum (ER) stress and upregulated the ER protein calreticulin on the cell surface, which can be targeted by the immune system [102]. This finding may partly explain why injected PGCCs are eliminated in immunocompetent but not in nude mice [103]. PGCCs are immunogenic, and there is potential for targeting PGCCs using chimeric antigen receptor (CAR) T-cell therapies. However, the dynamic changes in the TME limit the clinical success of immunotherapies [104]. Studies aiming to address these challenges by either genetically modifying CAR T-cells to increase their viability or using them in combination with antibodies are currently underway.
In addition, PGCCs have a higher metabolic rate than normal euploid cells due to their larger cellular and nuclear volume [69,105]. PGCCs frequently show higher levels of glycolysis than non-malignant cells, which rely on oxidative phosphorylation; hence, PGCCs are more susceptible to glycolysis inhibitors. The mTOR signaling pathway plays an essential role in glycolysis, and mTOR inhibitors have proved effective in inducing apoptosis and autophagy in acute myeloid leukemia polyploid cells [106]. A promising candidate is the anti-diabetic drug metformin, which has shown promising anticancer effects in preclinical studies [70]. Metformin exerts its effects partly by activating the AMP- activated protein kinase (AMPK), thereby inhibiting the mTOR pathway. High-throughput screening studies have also identified resveratrol and aspirin as potential anti-PGCC agents, exerting their effects via AMPK activation [107].
Sphingolipid metabolism alterations have been demonstrated in PGCCs and non-PGCCs. In addition to playing a structural role in the plasma membrane, sphingolipids regulate various cellular functions [108]. For instance, the sphingolipids ceramide and sphingosine regulate the cell cycle and cell survival, with the former being pro-apoptotic and the latter being anti-apoptotic. Ceramide is broken down into sphingosine by the lysosomal enzyme acid ceramidase. This lysosomal enzyme is upregulated in PGCCs, and its inhibition disrupted the formation of PGCC progeny [109]. Nevertheless, ceramidase inhibition did not impact the formation of new PGCCs, which could still exert detrimental effects by influencing the TME by releasing trophic factors.
Besides targeting these metabolic pathways, pharmacological approaches targeting key players in the endoreplication cycle are also being investigated. By binding to the endoreplication protein Chk2, mifepristone can block PGCC formation [110]. As undifferentiated cells are more vulnerable to the effects of mifepristone, the use of mifepristone as an anti-PGCC therapy could have some adverse effects.
Despite encouraging findings on the potential use of anti-PGCC treatments in combination with established anticancer drugs, further investigations are required to eliminate the possibility that these combination treatments can give a survival advantage to a small portion of resilient cells, which, upon treatment cessation, will form PGCCs.
Concluding thoughts: the way forward
Although the existence of PGCCs has been known since 1858 (first observed by Rudolph Virchow) [111], their importance in cancer pathophysiology remained underestimated until recently because of their purported inability to proliferate. However, it has become evident that PGCCs can produce progeny cells and promote tumor relapse, chemoresistance, and metastasis by modulating the TME. Their importance in racial disparities in BC outcomes is an attractive area of investigation, which we are actively pursuing. Given the extensive crosstalk between PGCCs and the TME, PGCC formation and proliferation must be contextualized within the framework of their interaction with other TME components. The TME can be described as a dynamic network of diploid cancer cells, PGCCs, PGCC progeny cells, stromal cells, blood vessels, and VM channels, which form the individual nodes of the network. In this network, the weight of each node and the inter-nodal connectivity dynamically change in response to chemotherapy or other stressors. This model can help form hypotheses and enhance our understanding of the changes in PGCCs within the context of their bidirectional relationship with the TME in BC. However, one of the drawbacks of this model emanates from its inability to capture the differences among different zones in the TME.
The pivotal role of PGCCs in BC pathophysiology necessitates the development of approaches to identify these cells and strategies to eradicate them. Currently, the field is hamstrung by a lack of PGCC markers. Digital pathology and AI-assisted methods may help identify robust PGCC markers and overcome the challenges in identifying PGCCs. These techniques can also aid in predicting therapy resistance and survival outcomes. The number of PGCCs in the tumor is correlated with the tumor grade, and the development of anti-PGCCs therapeutics can improve clinical outcomes. The emergence of consensus among the scientific and clinical community regarding the terminology and features of PGCCs will also accelerate scientific progress in the field.
Acknowledgments
Funding
This study was supported by a grant from the National Cancer Institutes of Health (R01CA239120) to RA and from NCI (1U01CA242936) to JK.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C, The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents, The Journal of Clinical Investigation. 119 (2009) 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Davoli T, de Lange T, The causes and consequences of polyploidy in normal development and cancer, Annual Review of Cell and Developmental Biology. 27 (2011) 585–610. [DOI] [PubMed] [Google Scholar]
- [3].Liu Z, Yue S, Chen X, Kubin T, Braun T, Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1, Circulation Research. 106 (2010) 1498. [DOI] [PubMed] [Google Scholar]
- [4].Takegahara N, Kim H, Mizuno H, Sakaue-Sawano A, Miyawaki A, Tomura M, Kanagawa O, Ishii M, Choi Y, Involvement of receptor activator of nuclear factor-κB ligand (RANKL)-induced incomplete cytokinesis in the polyploidization of osteoclasts, Journal of Biological Chemistry. 291 (2016) 3439–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Borradaile NM, Pickering JG, Polyploidy impairs human aortic endothelial cell function and is prevented by nicotinamide phosphoribosyltransferase, American Journal of Physiology-Cell Physiology. 298 (2009) C66–C74. 10.1152/ajpcell.00357.2009. [DOI] [PubMed] [Google Scholar]
- [6].Parekh A, Das S, Parida S, Das CK, Dutta D, Mallick SK, Wu P-H, Kumar BP, Bharti R, Dey G, Multi-nucleated cells use ROS to induce breast cancer chemo-resistance in vitro and in vivo, Oncogene. 37 (2018) 4546–4561. [DOI] [PubMed] [Google Scholar]
- [7].Ikebe H, Takamatsu T, Itoi M, Fujita S, Age-dependent changes in nuclear DNA content and cell size of presumably normal human corneal endothelium, Experimental Eye Research. 43 (1986) 251–258. [DOI] [PubMed] [Google Scholar]
- [8].Gentric G, Maillet V, Paradis V, Couton D, L’hermitte A, Panasyuk G, Fromenty B, Celton-Morizur S, Desdouets C, Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease, The Journal of Clinical Investigation. 125 (2015) 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Erenpreisa J, Ivanov A, Wheatley SP, Kosmacek EA, Ianzini F, Anisimov AP, Mackey M, Davis PJ, Plakhins G, Illidge TM, Endopolyploidy in irradiated p53-deficient tumour cell lines: Persistence of cell division activity in giant cells expressing Aurora B- kinase, Cell Biol Int. 32 (2008) 1044–1056. 10.1016/j.cellbi.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lopez-Sánchez LM, Jimenez C, Valverde A, Hernandez V, Peñarando J, Martinez A, Lopez-Pedrera C, Muñoz-Castañeda JR, Juan R, Aranda E, CoCl 2, a mimic of hypoxia, induces formation of polyploid giant cells with stem characteristics in colon cancer, PloS One. 9 (2014) e99143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J, Generation of cancer stem-like cells through the formation of polyploid giant cancer cells, Oncogene. 33 (2014) 116–128. 10.1038/onc.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen J, Niu N, Zhang J, Qi L, Shen W, Donkena KV, Feng Z, Liu J, Polyploid Giant Cancer Cells (PGCCs): The Evil Roots of Cancer, Curr Cancer Drug Targets. 19 (2019) 360–367. 10.2174/1568009618666180703154233. [DOI] [PubMed] [Google Scholar]
- [13].White-Gilbertson S, Voelkel-Johnson C, Giants and monsters: Unexpected characters in the story of cancer recurrence., Advances in Cancer Research. 148 (2020) 201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A, Global cancer statistics, 2012, CA: A Cancer Journal for Clinicians. 65 (2015) 87–108. [DOI] [PubMed] [Google Scholar]
- [15].Amend SR, Torga G, Lin K-C, Kostecka LG, de Marzo A, Austin RH, Pienta KJ, Polyploid giant cancer cells: Unrecognized actuators of tumorigenesis, metastasis, and resistance, Prostate. 79 (2019) 1489–1497. 10.1002/pros.23877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fei F, Zhang D, Yang Z, Wang S, Wang X, Wu Z, Wu Q, Zhang S, The number of polyploid giant cancer cells and epithelial-mesenchymal transition-related proteins are associated with invasion and metastasis in human breast cancer, J Exp Clin Cancer Res. 34 (2015). 10.1186/s13046-015-0277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang X, EMT-related protein expression in polyploid giant cancer cells and their daughter cells with different passages after triptolide treatment, Medical Oncology. (2019) 9. [DOI] [PubMed] [Google Scholar]
- [18].Xuan B, Ghosh D, Cheney EM, Clifton EM, Dawson MR, Dysregulation in Actin Cytoskeletal Organization Drives Increased Stiffness and Migratory Persistence in Polyploidal Giant Cancer Cells, Sci Rep. 8 (2018). 10.1038/s41598-018-29817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qu Y, Zhang L, Rong Z, He T, Zhang S, Number of glioma polyploid giant cancer cells (PGCCs) associated with vasculogenic mimicry formation and tumor grade in human glioma, J Exp Clin Cancer Res. 32 (2013) 75. 10.1186/1756-9966-32-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gerashchenko BI, Salmina K, Eglitis J, Huna A, Grjunberga V, Erenpreisa J, Disentangling the aneuploidy and senescence paradoxes: A study of triploid breast cancers non-responsive to neoadjuvant therapy, Histochemistry and Cell Biology. 145 (2016) 497–508. [DOI] [PubMed] [Google Scholar]
- [21].Gerashchenko BI, Salmina K, Eglitis J, Erenpreisa J, Probing breast cancer therapeutic responses by DNA content profiling, International Journal of Medicine and Medical Research. 5 (2019) 47–57. [Google Scholar]
- [22].Illidge T, Polyploid giant cells provide a survival mechanism for p53 mutant cells after dna damage, Cell Biology International. 24 (2000) 621–633. 10.1006/cbir.2000.0557. [DOI] [PubMed] [Google Scholar]
- [23].Erenpreisa J, Cragg MS, Three steps to the immortality of cancer cells: senescence, polyploidy and self-renewal, Cancer Cell International. 13 (2013) 92. 10.1186/1475-2867-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Puig P-E, Guilly M-N, Bouchot A, Droin N, Cathelin D, Bouyer F, Favier L, Ghiringhelli F, Kroemer G, Solary E, Martin F, Chauffert B, Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy, Cell Biology International. 32 (2008) 1031–1043. 10.1016/j.cellbi.2008.04.021. [DOI] [PubMed] [Google Scholar]
- [25].Weihua Z, Lin Q, Ramoth AJ, Fan D, Fidler IJ, Formation of solid tumors by a single multinucleated cancer cell, Cancer. 117 (2011) 4092–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Niu N, Mercado-Uribe I, Liu J, Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells, Oncogene. 36 (2017) 4887–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sharma S, Zeng J-Y, Zhuang C-M, Zhou Y-Q, Yao H-P, Hu X, Zhang R, Wang M-H, Small-molecule inhibitor BMS-777607 induces breast cancer cell polyploidy with increased resistance to cytotoxic chemotherapy agents, Mol Cancer Ther. 12 (2013) 725–736. 10.1158/1535-7163.MCT-12-1079. [DOI] [PubMed] [Google Scholar]
- [28].Mittal K, Donthamsetty S, Kaur R, Yang C, Gupta MV, Reid MD, Choi DH, Rida PCG, Aneja R, Multinucleated polyploidy drives resistance to Docetaxel chemotherapy in prostate cancer, British Journal of Cancer. 116 (2017) 1186–1194. 10.1038/bjc.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pirsko V, Čakstiņa I, Nitiša D, Samoviča M, Daneberga Z, Miklaševičs E, Alterations of The Stem-Like Properties in The Breast Cancer Cell Line MDA-MB-231 Induced by Single Pulsed Doxorubicin Treatment, Proceedings of the Latvian Academy of Sciences. Section B. Natural, Exact, and Applied Sciences. 73 (2019) 89–99. 10.2478/prolas-2019-0015. [DOI] [Google Scholar]
- [30].Mirzayans R, Andrais B, Murray D, Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment, Cancers. 10 (2018) 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ogden A, Rida PCG, Knudsen B, Kucuk O, Aneja R, Docetaxel-induced polyploidization may underlie chemoresistance and disease relapse, Cancer Lett. 367 (2015) 89–92. 10.1016/j.canlet.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rajaraman R, Guernsey DL, Rajaraman MM, Rajaraman SR, Stem cells, senescence, neosis and self-renewal in cancer, Cancer Cell International. 6 (2006) 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chitikova ZV, Gordeev SA, Bykova TV, Zubova SG, Pospelov VA, Pospelova TV, Sustained activation of DNA damage response in irradiated apoptosis-resistant cells induces reversible senescence associated with mTOR downregulation and expression of stem cell markers, Cell Cycle. 13 (2014) 1424–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Elmore LW, Di X, Dumur C, Holt SE, Gewirtz DA, Evasion of a single-step, chemotherapy-induced senescence in breast cancer cells: implications for treatment response, Clinical Cancer Research. 11 (2005) 2637–2643. [DOI] [PubMed] [Google Scholar]
- [35].Sundaram M, Guernsey DL, Rajaraman MM, Rajaraman R, Neosis: a novel type of cell division in cancer, Cancer Biol Ther. 3 (2004) 207–218. 10.4161/cbt.3.2.663. [DOI] [PubMed] [Google Scholar]
- [36].Erenpreisa J, Cragg MS, MOS, aneuploidy and the ploidy cycle of cancer cells, Oncogene. 29 (2010) 5447–5451. [DOI] [PubMed] [Google Scholar]
- [37].Liu J, The dualistic origin of human tumors, in: Seminars in Cancer Biology, Elsevier, 2018: pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rengstl B, Newrzela S, Heinrich T, Weiser C, Thalheimer FB, Schmid F, Warner K, Hartmann S, Schroeder T, Küppers R, Rieger MA, Hansmann M-L, Incomplete cytokinesis and re-fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed-Sternberg cells, Proc Natl Acad Sci U S A. 110 (2013) 20729–20734. 10.1073/pnas.1312509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kaur E, Rajendra J, Jadhav S, Shridhar E, Goda JS, Moiyadi A, Dutt S, Radiation-induced homotypic cell fusions of innately resistant glioblastoma cells mediate their sustained survival and recurrence, Carcinogenesis. 36 (2015) 685–695. 10.1093/carcin/bgv050. [DOI] [PubMed] [Google Scholar]
- [40].Erenpreisa J, Kalejs M, Cragg MS, Mitotic catastrophe and endomitosis in tumour cells: an evolutionary key to a molecular solution, Cell Biol Int. 29 (2005) 1012–1018. 10.1016/j.cellbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- [41].Niu N, Zhang J, Zhang N, Mercado-Uribe I, Tao F, Han Z, Pathak S, Multani AS, Kuang J, Yao J, Linking genomic reorganization to tumor initiation via the giant cell cycle, Oncogenesis. 5 (2016) e281–e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bharadwaj D, Mandal M, Senescence in polyploid giant cancer cells: A road that leads to chemoresistance, Cytokine & Growth Factor Reviews. 52 (2020) 68–75. 10.1016/j.cytogfr.2019.11.002. [DOI] [PubMed] [Google Scholar]
- [43].Middleton A, Suman R, O’Toole P, Akopyan K, Lindqvist A, p53-dependent polyploidisation after DNA damage in G2 phase, BioRxiv. (2020) 2020.06.09.141770. 10.1101/2020.06.09.141770. [DOI] [Google Scholar]
- [44].Wang Q, Wu PC, Dong DZ, Ivanova I, Chu E, Zeliadt S, Vesselle H, Wu DY, Polyploidy road to therapy-induced cellular senescence and escape, International Journal of Cancer. 132 (2013) 1505–1515. 10.1002/ijc.27810. [DOI] [PubMed] [Google Scholar]
- [45].Roberson RS, Kussick SJ, Vallieres E, Chen S-YJ, Wu DY, Escape from Therapy-Induced Accelerated Cellular Senescence in p53-Null Lung Cancer Cells and in Human Lung Cancers, Cancer Res. 65 (2005) 2795–2803. 10.1158/0008-5472.CAN-04-1270. [DOI] [PubMed] [Google Scholar]
- [46].Wang Q, Wu PC, Roberson RS, Luk BV, Ivanova I, Chu E, Wu DY, Survivin and escaping in therapy induced cellular senescence, International Journal of Cancer. 128 (2011) 1546–1558. 10.1002/ijc.25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sheikh MS, Rochefort H, Garcia M, Overexpression of p21WAF1/CIP1 induces growth arrest, giant cell formation and apoptosis in human breast carcinoma cell lines., Oncogene. 11 (1995) 1899–1905. [PubMed] [Google Scholar]
- [48].Lv H, Shi Y, Zhang L, Zhang D, Liu G, Yang Z, Li Y, Fei F, Zhang S, Polyploid giant cancer cells with budding and the expression of cyclin E, S-phase kinase-associated protein 2, stathmin associated with the grading and metastasis in serous ovarian tumor, BMC Cancer. 14 (2014) 576. 10.1186/1471-2407-14-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Erenpreisa J, Salmina K, Huna A, Jackson TR, Vazquez-Martin A, Cragg MS, The “virgin birth”, polyploidy, and the origin of cancer, Oncoscience. 2 (2014) 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang S, Uribe I. Mercado, Liu J, Tumor stroma and differentiated cancer cells can be originated directly from polyploid giant cancer cells induced by paclitaxel, International Journal of Cancer. 134 (2014) 508–518. 10.1002/ijc.28319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Khamis ZI, Sahab ZJ, Sang Q-XA, Active roles of tumor stroma in breast cancer metastasis, International Journal of Breast Cancer. 2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sun B, Zhang S, Zhang D, Du J, Guo H, Zhao X, Zhang W, Hao X, Vasculogenic mimicry is associated with high tumor grade, invasion and metastasis, and short survival in patients with hepatocellular carcinoma, Oncology Reports. 16 (2006) 693–698. 10.3892/or.16.4.693. [DOI] [PubMed] [Google Scholar]
- [53].Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, Ivanov A, Jascenko E, Scherthan H, Cragg M, Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells, Experimental Cell Research. 316 (2010) 2099–2112. [DOI] [PubMed] [Google Scholar]
- [54].Valkenburg KC, de Groot AE, Pienta KC, Targeting the tumour stroma to improve cancer therapy, Nat Rev Clin Oncol. 15 (2018) 366–381. 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mallini P, Lennard T, Kirby J, Meeson A, Epithelial-to-mesenchymal transition: what is the impact on breast cancer stem cells and drug resistance, Cancer Treatment Reviews. 40 (2014) 341–348. [DOI] [PubMed] [Google Scholar]
- [56].Mani SA, Guo W, Liao M-J, Eaton E.Ng, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA, The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells, Cell. 133 (2008) 704–715. 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fang X, Cai Y, Liu J, Wang Z, Wu Q, Zhang Z, Yang CJ, Yuan L, Ouyang G, Twist2 contributes to breast cancer progression by promoting an epithelial–mesenchymal transition and cancer stem-like cell self-renewal, Oncogene. 30 (2011) 4707–4720. 10.1038/onc.2011.181. [DOI] [PubMed] [Google Scholar]
- [58].Saxena M, Stephens MA, Pathak H, Rangarajan A, Transcription factors that mediate epithelial–mesenchymal transition lead to multidrug resistance by upregulating ABC transporters, Cell Death & Disease. 2 (2011) e179–e179. 10.1038/cddis.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Guda MR, Rashid MA, Asuthkar S, Jalasutram A, Caniglia JL, Tsung AJ, Velpula KK, Pleiotropic role of macrophage migration inhibitory factor in cancer, Am J Cancer Res. 9 (2019) 2760–2773. [PMC free article] [PubMed] [Google Scholar]
- [60].Toi M, Hoshina S, Takayanagi T, Tominaga T, Association of vascular endothelial growth factor expression with tumor angiogenesis and with early relapse in primary breast cancer, Japanese Journal of Cancer Research. 85 (1994) 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lin K-C, Torga G, Sun Y, Axelrod R, Pienta KJ, Sturm JC, Austin RH, The role of heterogeneous environment and docetaxel gradient in the emergence of polyploid, mesenchymal and resistant prostate cancer cells, Clinical & Experimental Metastasis. 36 (2019) 97–108. [DOI] [PubMed] [Google Scholar]
- [62].Doktorova H, Hrabeta J, Khalil MA, Eckschlager T, Hypoxia-induced chemoresistance in cancer cells: The role of not only HIF-1., Biomedical Papers of the Medical Faculty of Palacky University in Olomouc. 159 (2015). [DOI] [PubMed] [Google Scholar]
- [63].Qin J, Liu Y, Lu Y, Liu M, Li M, Li J, Wu L, Hypoxia-inducible factor 1 alpha promotes cancer stem cells-like properties in human ovarian cancer cells by upregulating SIRT1 expression, Scientific Reports. 7 (2017) 10592. 10.1038/s41598-017-09244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lundgren K, Nordenskjöld B, Landberg G, Hypoxia, Snail and incomplete epithelial– mesenchymal transition in breast cancer, British Journal of Cancer. 101 (2009) 1769–1781. 10.1038/sj.bjc.6605369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sikora E, Bielak-Zmijewska A, Mosieniak G, What is and what is not cell senescence, Postępy Biochemii. (2018) 9. [DOI] [PubMed] [Google Scholar]
- [66].Mosieniak G, Sliwinska MA, Alster O, Strzeszewska A, Sunderland P, Piechota M, Was H, Sikora E, Polyploidy Formation in Doxorubicin-Treated Cancer Cells Can Favor Escape from Senescence, Neoplasia. 17 (2015) 882–893. 10.1016/j.neo.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhang S, Mercado-Uribe I, Liu J, Generation of erythroid cells from fibroblasts and cancer cells in vitro and in vivo, Cancer Lett. 333 (2013) 205–212. 10.1016/j.canlet.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z, Wu X, Wu J, Wang C, Zhong J, Huang Q, Garcia-Aguilar J, Pfeifer GP, Shen B, Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression, Nat Commun. 3 (2012) 815. 10.1038/ncomms1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Coward J, Harding A, Size Does Matter: Why Polyploid Tumor Cells are Critical Drug Targets in the War on Cancer, Front. Oncol 4 (2014). 10.3389/fonc.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M, Buchanan M, Chabot C, Ramanathan A, Balachandran B, Légaré S, Przybytkowski E, Lan C, Krzemien U, Cavallone L, Aleynikova O, Ferrario C, Guilbert M-C, Benlimame N, Saad A, Alaoui-Jamali M, Saragovi HU, Josephy S, O’Flanagan C, Hursting SD, Richard VR, Zahedi RP, Borchers CH, Bareke E, Nabavi S, Tonellato P, Roy J-A, Robidoux A, Marcus EA, Mihalcioiu C, Majewski J, Basik M, A Unique Morphological Phenotype in Chemoresistant Triple-Negative Breast Cancer Reveals Metabolic Reprogramming and PLIN4 Expression as a Molecular Vulnerability, Mol Cancer Res. 17 (2019) 2492–2507. 10.1158/1541-7786.MCR-19-0264. [DOI] [PubMed] [Google Scholar]
- [71].Karna P, Sharp SM, Yates C, Prakash S, Aneja R, EM011 activates a survivin-dependent apoptotic program in human non-small cell lung cancer cells, Molecular Cancer. 8 (2009) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jiang Q, Zhang Q, Wang S, Xie S, Fang W, Liu Z, Liu J, Yao K, A Fraction of CD133+ CNE2 Cells Is Made of Giant Cancer Cells with Morphological Evidence of Asymmetric Mitosis, J. Cancer 6 (2015) 1236–1244. 10.7150/jca.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hawryluk EB, Baran JL, Gerami P, Sepehr A, ‘Monster cell’ melanoma with pulmonary metastasis and cyclin D1 amplification, Journal of Cutaneous Pathology. 40 (2013) 61–65. 10.1111/cup.12024. [DOI] [PubMed] [Google Scholar]
- [74].Zhang S, Mercado-Uribe I, Hanash S, Liu J, iTRAQ-based proteomic analysis of polyploid giant cancer cells and budding progeny cells reveals several distinct pathways for ovarian cancer development, PloS One. 8 (2013) e80120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ormerod MG, Flow cytometry: a practical approach, Oxford University Press, 2000. [Google Scholar]
- [76].Gerashchenko BI, Huna A, Erenpreisa J, Characterization of breast cancer DNA content profiles as a prognostic tool, Experimental Oncology. (2014). [PubMed] [Google Scholar]
- [77].Auer GU, Fallenius AG, Erhardt KY, Sundelin BS, Progression of mammary adenocarcinomas as reflected by nuclear DNA content, Cytometry: The Journal of the International Society for Analytical Cytology. 5 (1984) 420–425. [DOI] [PubMed] [Google Scholar]
- [78].Baisch H, Göhde W, Linden WA, Analysis of PCP-data to determine the fraction of cells in the various phases of cell cycle, Radiation and Environmental Biophysics. 12 (1975) 31–39. [DOI] [PubMed] [Google Scholar]
- [79].Fernö M, Baldetorp B, Borg AAKe, Olsson H, Sigurdsson H, Killander D, Flow cytometric DNA index and S-phase fraction in breast cancer in relation to other prognostic variables and to clinical outcome, Acta Oncologica. 31 (1992) 157–165. [DOI] [PubMed] [Google Scholar]
- [80].Opfermann M, Brugal G, Vassilakos P, Cytometry of breast carcinoma: Significance of ploidy balance and proliferation index, Cytometry: The Journal of the International Society for Analytical Cytology. 8 (1987) 217–224. [DOI] [PubMed] [Google Scholar]
- [81].Barlogie B, W W Göhde Johnston DA, Smallwood L, Schumann J, Drewinko B, Freireich EJ, Determination of ploidy and proliferative characteristics of human solid tumors by pulse cytophotometry, Cancer Res. 38 (1978) 3333–9. [PubMed] [Google Scholar]
- [82].Díaz-Carballo D, Gustmann S, Jastrow H, Acikelli AH, Dammann P, Klein J, Dembinski U, Bardenheuer W, Malak S, Araúzo-Bravo MJ, Schultheis B, Aldinger C, Strumberg D, Atypical Cell Populations Associated with Acquired Resistance to Cytostatics and Cancer Stem Cell Features: The Role of Mitochondria in Nuclear Encapsulation, DNA Cell Biol. 33 (2014) 749–774. 10.1089/dna.2014.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang Y, McManus DT, Arthur K, Johnston BT, Kennedy AJ, Coleman HG, Murray LJ, Hamilton PW, Whole slide image cytometry: a novel method to detect abnormal DNA content in Barrett’s esophagus, Lab Invest. 95 (2015) 1319–30. [DOI] [PubMed] [Google Scholar]
- [84].Aeffner F, Zarella MD, Buchbinder N, Bui MM, Goodman MR, Hartman DJ, Lujan GM, Molani MA, Parwani AV, Lillard K, Turner OC, Vemuri VNP, Yuil-Valdes AG, Bowman D, Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association, J Pathol Inform. 10 (2019) 9. 10.4103/jpi.jpi_82_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Agarwal R, Diaz O, Lladó X, Yap MH, Martí R, Automatic mass detection in mammograms using deep convolutional neural networks, JMI. 6 (2019) 031409. 10.1117/1.JMI.6.3.031409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chang HY, Jung CK, Woo JI, Lee S, Cho J, Kim SW, Kwak T-Y, Artificial Intelligence in Pathology, J Pathol Transl Med. 53 (2019) 1–12. 10.4132/jptm.2018.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Griffin J, Treanor D, Digital pathology in clinical use: where are we now and what is holding us back?, Histopathology. 70 (2017) 134–145. 10.1111/his.12993. [DOI] [PubMed] [Google Scholar]
- [88].Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D, Systematic analysis of breast cancer morphology uncovers stromal features associated with survival, Sci Transl Med. 3 (2011) 108ra113. 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- [89].Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I, Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images, EBioMedicine. 27 (2018) 317–328. 10.1016/j.ebiom.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zarella MD, Bowman D, Aeffner F, Farahani N, Xthona A, Absar SF, Parwani A, Bui M, Hartman DJ, A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association, Arch Pathol Lab Med. 143 (2019) 222–234. 10.5858/arpa.2018-0343-RA. [DOI] [PubMed] [Google Scholar]
- [91].He K, Gkioxari G, Dollár P, Girshick R, Mask r-cnn, in: Proceedings of the IEEE International Conference on Computer Vision, 2017: pp. 2961–2969. [Google Scholar]
- [92].Ren S, He K, Girshick R, Sun J, Faster r-cnn: Towards real-time object detection with region proposal networks, ArXiv Preprint ArXiv:1506.01497. (2015). [DOI] [PubMed] [Google Scholar]
- [93].Girshick R, Fast R-CNN Object detection with Caffe, in: Proceeding of the 13th European Conference on Computer Vision, 2014. [Google Scholar]
- [94].Abdulla W, Mask r-cnn for object detection and instance segmentation on keras and tensorflow, (2017). [Google Scholar]
- [95].Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Tensorflow: A system for large-scale machine learning, in: 12th ${$USENIX$}$ Symposium on Operating Systems Design and Implementation (${$OSDI$}$ 16), 2016: pp. 265–283. [Google Scholar]
- [96].Chollet F, Keras: Deep learning library for theano and tensorflow, URL: Https://Keras.Io/k. 7 (2015) T1. [Google Scholar]
- [97].Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S, Feature pyramid networks for object detection, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017: pp. 2117–2125. [Google Scholar]
- [98].He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016: pp. 770–778. [Google Scholar]
- [99].Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, Steiger K, Schlitter AM, Esposito I, Navab N, Structure-preserving color normalization and sparse stain separation for histological images, IEEE Transactions on Medical Imaging. 35 (2016) 1962–1971. [DOI] [PubMed] [Google Scholar]
- [100].Jung A, imgaug Documentation, (n.d.) 1124. [Google Scholar]
- [101].Al-Milaji Z, Ersoy I, Hafiane A, Palaniappan K, Bunyak F, Integrating segmentation with deep learning for enhanced classification of epithelial and stromal tissues in H&E images, Pattern Recognition Letters. 119 (2019) 214–221. 10.1016/j.patrec.2017.09.015. [DOI] [Google Scholar]
- [102].Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, An immunosurveillance mechanism controls cancer cell ploidy, Science. 337 (2012) 1678–1684. [DOI] [PubMed] [Google Scholar]
- [103].Boilève A, Senovilla L, Vitale I, Lissa D, Martins I, Métivier D, Van Den Brink S, Clevers H, Galluzzi L, Castedo M, Immunosurveillance against tetraploidization-induced colon tumorigenesis, Cell Cycle. 12 (2013) 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Scarfò I, Maus MV, Current approaches to increase CAR T cell potency in solid tumors: targeting the tumor microenvironment, Journal for ImmunoTherapy of Cancer. 5 (2017) 28. 10.1186/s40425-017-0230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Donovan P, Cato K, Legaie R, Jayalath R, Olsson G, Hall B, Olson S, Boros S, Reynolds BA, Harding A, Hyperdiploid tumor cells increase phenotypic heterogeneity within Glioblastoma tumors, Mol. BioSyst 10 (2014) 741–758. 10.1039/C3MB70484J. [DOI] [PubMed] [Google Scholar]
- [106].Liu L-L, Long Z-J, Wang L-X, Zheng F-M, Fang Z-G, Yan M, Xu D-F, Chen J-J, Wang S-W, Lin D-J, Liu Q, Inhibition of mTOR pathway sensitizes acute myeloid leukemia cells to aurora inhibitors by suppression of glycolytic metabolism, Mol Cancer Res. 11 (2013) 1326–1336. 10.1158/1541-7786.MCR-13-0172. [DOI] [PubMed] [Google Scholar]
- [107].Lissa D, Senovilla L, Rello-Varona S, Vitale I, Michaud M, Pietrocola F, Boilève A, Obrist F, Bordenave C, Garcia P, Michels J, Jemaà M, Kepp O, Castedo M, Kroemer G, Resveratrol and aspirin eliminate tetraploid cells for anticancer chemoprevention, PNAS. 111 (2014) 3020–3025. 10.1073/pnas.1318440111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Parveen F, Bender D, Law S-H, Mishra VK, Chen C-C, Ke L-Y, Role of Ceramidases in Sphingolipid Metabolism and Human Diseases, Cells. 8 (2019). 10.3390/cells8121573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gilbertson S. White, Lu P, Jones CM, Chiodini S, Hurley D, Das A, Delaney JR, Norris JS, Johnson C. Voelkel, Tamoxifen is a candidate first-in-class inhibitor of acid ceramidase that reduces amitotic division in polyploid giant cancer cells—Unrecognized players in tumorigenesis, Cancer Medicine. 9 (2020) 3142–3152. 10.1002/cam4.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kapperman HE, Goyeneche AA, Telleria CM, Mifepristone inhibits non-small cell lung carcinoma cellular escape from DNA damaging cisplatin, Cancer Cell International. 18 (2018) 185. 10.1186/s12935-018-0683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Virchow R, Cellular Pathology as Based Upon Physiological and Pathological Histology..., JB Lippincott, 1863. [DOI] [PubMed] [Google Scholar]