Abstract
Sneathia is an emerging pathogen implicated in adverse reproductive and perinatal outcomes. Although scarce, emerging data suggest that vaginally residing Sneathia becomes pathogenic following its ascension into the upper urogenital tract, amniotic fluid, placenta, and fetal membranes. The role of Sneathia in women’s health and disease is generally underappreciated because the cultivation of these bacteria is limited by their complex nutritional requirements, slow growth patterns, and anaerobic nature. For this reason, molecular methods are typically required for the detection and differential diagnosis of Sneathia infections. Here, we review the laboratory methods used for the diagnosis of Sneathia infections, the molecular mechanisms underlying its virulence, and its sensitivity to antibiotics. We further review the evidence of Sneathia’s contributions to the pathogenesis of bacterial vaginosis, chorioamnionitis, preterm prelabor rupture of membranes, spontaneous preterm labor, stillbirth, maternal and neonatal sepsis, HIV infection, and cervical cancer. Collectively, growing evidence indicates that Sneathia represents an important yet underappreciated pathogen affecting the development and progression of several adverse clinical conditions diagnosed in pregnant and non-pregnant women, as well as in their neonates.
Keywords: preterm birth, preterm prelabor rupture of membranes (PPROM), bacterial vaginosis, vaginal biofilm, cervical cancer
Introduction
Sneathia is a genus of Gram-negative, rod-shaped, anaerobic, non-motile bacteria recently identified as an important contributor to common obstetric [1–11], neonatal [12–14] and gynecologic pathologies [15–19]. Taxonomically, Sneathia belongs to the family Leptotrichiaceae, which consists of a heterogeneous group of species with diverse metabolic and genetic characteristics [20, 21]. All members of this family are obligate or facultative anaerobes, have fermentative metabolisms, and are non-spore forming [22]. They typically reside on the mucosal surfaces of the oral cavity [23, 24], gastrointestinal tract [23–26] and the urogenital system [23, 24, 27, 28], and several species within this family are documented human pathogens [21, 24, 28, 29].
The genus Sneathia is closely related to that of Leptotrichia. Indeed, Sneathia species were initially classified within the genus Leptotrichia [12]. These two genera share several common properties, such as close 16S rRNA gene sequence similarity and the production of lactic acid [30]. However, compared to Leptotrichia, Sneathia is more fastidious, requires culture media with serum or blood for growth, and, with respect to enzymatic reactions, Sneathia produces β-glucuronidase but not α-glucosidase or β-glucosidase [30]. Based in part on these differences, Sneathia was re-classified as a separate genus in 2001 [30].
At present, the genus Sneathia includes two species (S. amnii and S. sanguinegens), which can be found in the oral cavity [25, 31], gastrointestinal tract [25, 26], and the cervix and/or vagina [27, 28, 32, 33]. In a large cohort study of 736 women, Sneathia was found in 43.3% of vaginal specimens [28]: 76% of the detected Sneathia 16S rRNA gene sequences were from S. amnii, 18% were from S. sanguinegens, and 6% were from the genus Sneathia but could not be assigned to either of these established species [28]. Based on several other reports, Sneathia is present in the cervico-vaginal microbiome of 10–100% of non-pregnant [27, 33–37] and pregnant [8, 9] women, with a mean relative abundance of 1–21% [8, 28, 33, 36].
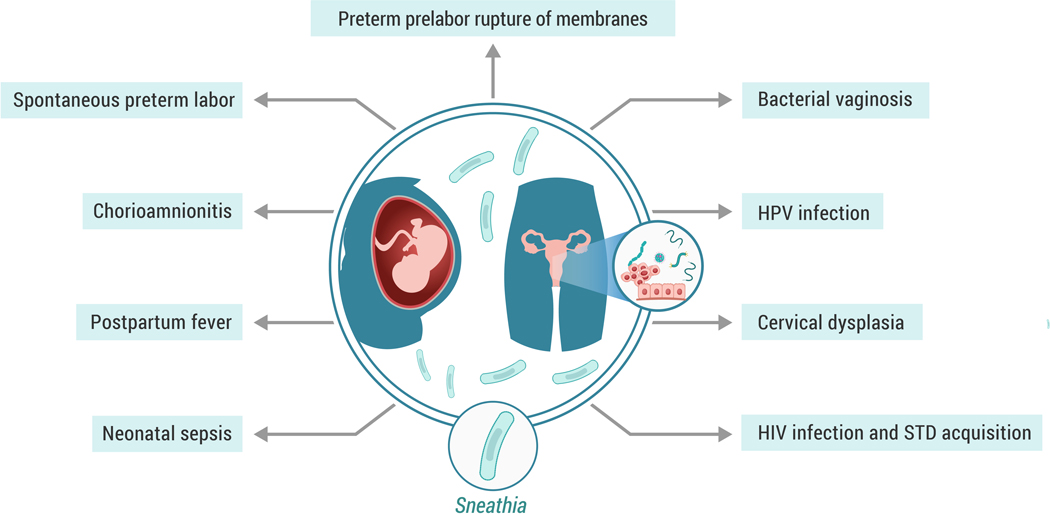
There are indications that Sneathia is a constituent of a non-optimal vaginal microbiome. For instance, Sneathia is posited to be involved in the pathogenesis of bacterial vaginosis [34, 35, 38–42], spontaneous preterm birth [i.e. spontaneous preterm labor and preterm prelabor rupture of membranes (PPROM)] [1–5, 7–10, 43–46], chorioamnionitis [6, 11, 47], stillbirth [48, 49], sexually transmitted diseases [50, 51], and other severe pregnancy complications [13, 52] (see Figure 1). Sneathia species are more often present in the vaginal samples of women with symptoms of bacterial vaginosis than in those from women who are asymptomatic [34, 35, 39, 41]. Indeed, the presence of Sneathia is positively associated with the diagnostic criteria of bacterial vaginosis [36]. A recent study further indicated that Sneathia had among the greatest relative abundance and effect size of the vaginal bacteria associated with preterm birth (delivery at <37 weeks of gestation) [9]. Sneathia is also one of the most frequently detected bacteria in amniotic fluid of patients with PPROM [3, 7] or spontaneous preterm labor [1, 2, 5, 8, 9]. The results of these and other investigations, which are discussed in greater detail below, suggest that Sneathia has the capacity for ascending invasion of the upper reproductive tract during pregnancy. Thus, it is becoming increasingly apparent that Sneathia infections are associated with critically important obstetric and gynecologic pathologies.
Figure 1. Sneathia in female reproductive disease.
During pregnancy, Sneathia spp. are involved in the pathogenesis of preterm prelabor rupture of membranes, spontaneous preterm labor, clinical and histological chorioamnionitis, postpartum fever, as well as neonatal sepsis and bacteremia. In non-pregnant women, the presence of Sneathia spp. in the vaginal fluid is associated with bacterial vaginosis, HPV infection, cervical dysplasia, and increased risk of HIV infection and sexually transmitted diseases (STD).
This review represents the first comprehensive summary of how Sneathia species affect women’s reproductive health and neonatal outcomes. Specifically, we discuss the culture and molecular methods for the isolation and/or identification of Sneathia species, the mechanisms of their virulence, their role in the etiology and pathogenesis of different types of obstetric and gynecologic pathologies, and the antibiotic therapies used to resolve Sneathia infections.
Identification of Sneathia as a novel pathogen
The existence and pathogenic potential of Sneathia species were unrecognized until the development and application of DNA sequencing technologies that allowed for their reliable identification in clinical samples. The first report describing a new infectious agent, Leptotrichia sanguinegens, in the pathogenesis of postpartum and neonatal bacteremia, and its isolation in culture, was provided by Hanff et al in 1995 [12]. The symptoms of the bacterial infection included post-partum fever and elevated white blood cell count, both of which were successfully treated with the antibiotics ampicillin and gentamicin [12]. In 2001, several bacterial isolates from blood and amniotic fluid were found to resemble Leptotrichia sanguinegens. However, the comparison of the DNA nucleotide sequences encoding for the 16S rRNA gene, and detailed analyses of the morphological and metabolic characteristics, showed that these isolates should not be classified within the Leptotrichia genus [30]. They were instead classified as a new genus, Sneathia, named after the British microbiologist Peter Sneath, a pioneer of bacterial systematics [30]. A year later, a novel bacterium with similar characteristics was isolated from amniotic fluid of a woman following intrauterine fetal demise. This bacterium was named Leptotrichia amnionii based on the site of its localization and isolation [48]. In 2004, DNA sequencing indicated that Leptotrichia amnionii should also be re-classified to the Sneathia genus; it was renamed as Sneathia amnii [53].
It should be noted that, until recently, some studies still applied the outdated taxonomic terminology classifying Sneathia as Leptotrichia (examples are shown in Tables 1–4). In this review, we will use the newly accepted taxonomic nomenclature, referring to Leptotrichia sanguinegens and Leptotrichia amnionii as Sneathia sanguinegens and Sneathia amnii, respectively.
Table 1.
Characteristics of Sneathia cultures isolated from patients during pregnancy or postpartum
| Authors | Year | Sample type | Clinical diagnosis | Sneathia species isolated | Number of cases | Culture conditions | Days in culture | Colony morphology |
|---|---|---|---|---|---|---|---|---|
| Hanff et al. [12] | 1995 | Maternal blood | Postpartum fever, neonatal bacteremia | S. sanguinegens (L. sanguinegens) | 4 | Aerobic and anaerobic blood culture bottles; Successful subculture into anaerobic blood agar, chopped-meat glucose medium and chocolate agar supplemented with 20% fresh rabbit, horse, sheep, or human blood | 3 | Pinpoint convex colonies |
| Collins et al. [30] | 2001 | Blood and amniotic fluid | No details of clinical pathology were provided | S. sanguinegens | 1 | Anaerobic blood and chocolate agar plates with additional supplementation of blood or serum | 3 | Pinpoint convex colonies |
| Shukla et al. [48] | 2002 | Amniotic fluid | Intrauterine fetal demise (amniotic fluid was sampled before labor induction) | S. amnii (L. amnionii) | 1 | Anaerobic culture on blood and chocolate agar | 3 | Very small gray colonies, <1 mm in diameter |
| De Martino et al. [54] | 2004 | Maternal blood | Peripartum maternal fever | S. amnii (L. amnionii) (2) and S. sanguinegens (1) | 3 | Anaerobic blood culture vials; Anaerobic subculture on chocolate-polyvitex agar | 2–4 | Not provided |
| Boennelycke et al. [52] | 2007 | Maternal blood | Second trimester spontaneous septic abortion | S. amnii (L. amnionii) | 1 | Anaerobic blood culture bottles | 1 | Not provided |
| Harwich et al. [28] | 2012 | Vaginal fluid | Preterm labor at 26 weeks of gestation | S. amnii | 1 | Anaerobic culture on chocolate agar enhanced with human serum or BHI (brain-heart infusion) agar containing 10% fresh human blood; was able to tolerate transient exposure to air | 2–3 | Flat colonies, ~1 mm in diameter, crystalline on chocolate agar, or mucoid, raised, amorphous, ~2 mm in diameter, on BHI agar supplemented with human blood |
Table 4.
Antibiotic sensitivity and resistance in clinical Sneathia isolates
| Authors | Year | Source of isolation | Bacterial species | Clinical diagnosis | Antibiotic sensitivity | Antibiotic resistance |
|---|---|---|---|---|---|---|
| De Martino et al. [54] | 2004 | Maternal blood | S. amnii (L. amnionii), S. sanguinegens | Intrapartum and postpartum fever | Amoxicillin, amoxicillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cefotaxime, imipenem, chloramphenicol and metronidazole | Vancomycin, intermittent susceptibility to erythromycin |
| Boennelycke et al. [52] | 2007 | Maternal blood | S. amnii (L. amnionii) | Septic abortion at 15 weeks of gestation | Penicillin, metronidazole | Not reported |
| Bachy et al. [166] | 2011 | Joint fluid and synovial biopsy | Sneathia spp. | Elbow septic arthritis | Imipenem, clindamycin, rifampicin, tetracycline and chloramphenicol | Erythromycin, aminoglycosides, fluoroquinolones |
| Harwich et al. [28] | 2012 | Vaginal microbiome | S. amnii | Preterm labor at 26 weeks of gestation | Metronidazole, vancomycin | Nafcillin, tetracycline, ciprofloxacin |
Methods for the detection of Sneathia in clinical samples
Both species of Sneathia are fastidious bacteria with complex and demanding nutritional requirements. Growth media are typically supplemented with blood or serum [12, 28, 30]. Nevertheless, it is incredibly difficult to isolate Sneathia in culture from clinical specimens even when using nutrient-rich, blood-supplemented media. A summary of the data regarding the isolation and culture of Sneathia from clinical specimens is presented in Table 1.
In the first days following inoculation, cultures of Sneathia species progress in their growth through a prolonged lag-phase [12, 28, 30, 48, 54]. Although Sneathia cultures can tolerate transient exposures to aerobic atmospheres [28], and have even shown a limited capacity to grow in such environments [12], Sneathia cultures are typically maintained under anaerobic conditions [12, 28, 30, 48, 52, 54]. Morphological characteristics of S. amnii cultures, as revealed through scanning electron microscopy, include a discernible cell polymorphism with elongated bacilli, short bacilli, and cocci [28]. Cell heterogeneity, including an increased proportion of cocci and the development of bacterial L-forms (i.e. cell-wall deficient morphotypes), is significantly increased in older cultures [28].
As shown in Tables 1 and 2, despite some early reports of the isolation of cultures of both Sneathia species from amniotic fluid [30, 48], more recent attempts to recover Sneathia isolates from the amniotic cavity [2, 5, 7, 55], the chorioamnion [11], and pelvic [19] or vaginal [56] samples were not successful. Thus, it is very difficult to isolate, propagate, and quantify Sneathia in culture in the laboratory. Another explanation for diminished efforts aimed at Sneathia isolation in culture appears to be a wider application of molecular methods that allow for the rapid and specific identification and quantification of Sneathia in clinical specimens, even when it is present at low concentrations.
Table 2.
Identification of Sneathia in upper reproductive tract and neonatal samples using culture-independent methods
| Authors | Year | Gestational age at collection | Clinical pathology | Sample type | Sneathia species identified | Number of cases | Number of cases with Sneathia infections | Was cultivation of Sneathia attempted? | Culture-independent methods used to detect Sneathia | How was Sneathia associated with the clinical pathology? |
|---|---|---|---|---|---|---|---|---|---|---|
| Gardella et al. [1] | 2004 | 22 – 34 weeks | Preterm labor with intact membranes | Amniotic fluid | S. sanguinegens (L. sanguinegens) | 5 | 2 | Yes, not successful | 16S rRNA gene PCR and sequencing | S. sanguinegens was detected in the culture-negative amniotic fluid samples from women with preterm labor with intact membranes and intra-amniotic inflammation. |
| DiGiulio et al. [2] | 2008 | 18 – 35 weeks | Spontaneous preterm labor with intact membranes and preterm birth | Amniotic fluid | S. sanguinegens; Sneathia amnii (L. amnionii) | 113 | 6 | Yes, not successful | 16S rRNA gene end-point PCR and sequencing | S. sanguinegens and S. amnii were detected in the amniotic fluid from women with preterm labor and birth. S. sanguinegens was one of the most common taxa identified. All these cases were diagnosed with intra-amniotic inflammation. |
| DiGiulio et al. [3] | 2010 | 15 – 37 weeks | Preterm prelabor rupture of membranes | Amniotic fluid | S. sanguinegens; S. amnii (L. amnionii) | 204 | 6 | Yes, not successful | 16S rRNA gene and group-specific end-point PCR and sequencing | S. sanguinegens and S. amnii were detected in amniotic fluid from women with PPROM. S. sanguinegens was one of the most common taxa identified. |
| DiGiulio et al. [59] | 2010 | 20 – 40 weeks | Preeclampsia | Amniotic fluid | Sneathia/Leptotrichia spp. | 62 | 3 | No | 16S rRNA gene and group-specific end-point PCR and sequencing | Three out of six patients diagnosed with microbial invasion of the amniotic cavity had a positive PCR for Sneathia/Leptotrichia spp.. |
| Marconi et al. [119] | 2011 | <37 weeks | Preterm labor with intact membranes | Amniotic fluid | S. amnii (L. amnionii) | 40 | 1 | No | 16S rRNA gene PCR and sequencing | S. amnii (L. amnionii) was detected in an amniotic fluid sample from a woman with preterm labor and intact membranes and intra-amniotic inflammation. |
| Kacerovsky et al. [120] | 2013 | 24 – 37 weeks of gestation | Preterm prelabor rupture of membranes | Amniotic fluid | S. sanguinegens; S. amnii (L. amnionii) | 145 | 2 | No | 16S rRNA gene PCR and sequencing | S. sanguinegens and S. amnii (L. amnionii) were detected in the amniotic fluid from women with PPROM. |
| Aujoulat et al. [63] | 2014 | 26 – 29 weeks | Early preterm Infants | Neonatal stool | S. sanguinegens | 30 | 1 | No | 16S rRNA gene PCR and sequencing; PCR-TTGE* | S. sanguinegens (co-colonization with staphylococci and unidentified bacteria) was detected in one neonate’s stool who died at day nine of life due to a non-digestive, unspecified cause. |
| Romero et al. [5] | 2014 | 20 – 35 weeks | Preterm labor with intact membranes | Amniotic fluid | Sneathia spp. | 142 | 2 | Yes, not successful | PCR/ESI-MS** | Sneathia was detected in two amniotic fluid samples from women with preterm labor and intact membranes and with intra-amniotic inflammation and histological chorioamnionitis. |
| Romero et al. [6] | 2015 | 38 – 40 weeks | Clinical chorioamnionitis at term | Amniotic fluid | Sneathia spp. alone and with Gardnerella vaginalis or Ureaplasma spp. | 46 | 3 | Yes, not successful | PCR/ESI-MS** | Sneathia was detected in the amniotic fluid of women with clinical chorioamnionitis at term; polymicrobial invasion was detected in all these samples. |
| Romero et al. [7] | 2015 | 20 – 35 weeks | Preterm prelabor rupture of membranes | Amniotic fluid | Sneathia spp. | 59 | 6 | Yes, not successful | PCR/ESI-MS** | Sneathia was among the most frequent microorganisms detected in amniotic fluid of women with preterm prelabor rupture of membranes. Sneathia positive cases were associated with acute inflammation in the placenta and amniotic cavity. |
| Romero et al. [61] | 2015 | 16 – 32 weeks | Asymptomatic sonographic short cervix ≤25 mm | Amniotic fluid | Sneathia spp. | 231 | 1 | Yes, not successful | PCR/ESI-MS** | Sneathia was detected in one term case (no intra-amniotic inflammation). |
| Carlstein et al. [47] | 2016 | 42 weeks | Postterm pregnancy, prolonged prelabor rupture of membranes, clinical chorioamnionitis | Amniotic fluid | S. sanguinegens | 1 | 1 | Yes, not successful | 16S rRNA gene sequencing | S. sanguinegens was found in amniotic fluid of a woman with clinical chorioamnionitis at term after labor induction with an intracervical balloon. |
| Puri et al. [13] | 2016 | ≤28 weeks | Early preterm infants | Neonatal stool | Sneathia spp. | 106 | 3 | No | 16S rRNA gene sequencing | The preterm infants with chorioamnionitis and funisitis had higher relative abundances of Sneathia in their stool; the presence of Sneathia in stool was associated with a higher risk of sepsis or death. |
| Urushiyama et al. [75] | 2017 | 26 – 37 weeks | Histological chorioamnionitis | Amniotic fluid | S. sanguinegens | 79 | Not reported | No | 16S rRNA gene sequencing | S. sanguinegens was found to be among the top 10 dominant taxa in the group with histological chorioamnionitis stage III. |
| Musilova et al. [44] | 2017 | 24 – 37 weeks | Preterm prelabor rupture of membranes | Amniotic fluid | S. sanguinegens alone and with Ureaplasma spp.; S. amnii (L. amnionii) alone and with Ureaplasma spp. or Chlamydia trachomatis | 479 | 5 | No | 16S rRNA gene sequencing | S. sanguinegens and S. amnii (L. amnionii) were detected in amniotic fluid samples with or without intra-amniotic inflammation from women with PPROM. |
| Musilova et al. [43] | 2017 | 24 – 37 weeks | Preterm prelabor rupture of membranes | Amniotic fluid | S. sanguinegens alone and with Ureaplasma spp. | 287 | 2 | No | 16S rRNA gene sequencing | S. sanguinegens was detected in amniotic fluid samples with intra-amniotic inflammation from women with PPROM. |
| Musilova et al. [46] | 2018 | 34 – 37 weeks | Preterm prelabor rupture of membranes | Amniotic fluid | S. sanguinegens | 159 | 1 | Yes, the results were not specified | 16S rRNA gene sequencing; MALDI-TOF*** was used for isolates | S. sanguinegens was detected in amniotic fluid samples with intra-amniotic inflammation from women with PPROM. |
| Hornychova et al. [45] | 2018 | 24 – 37 weeks | Preterm prelabor rupture of membranes; HPV infection | Amniotic fluid | S. sanguinegens | 100 | 1 | Yes, the results were not specified | 16S rRNA gene sequencing of isolates | S. sanguinegens was detected in amniotic fluid from a woman with PPROM, diagnosed with cervical HPV infection, intra-amniotic inflammation and histological chorioamnionitis and funisitis. |
| Lannon et al. [11] | 2019 | ≥37 weeks | Clinical chorioamnionitis | Vaginal fluid, chorio-decidual space, chorioamnion | Leptotrichia/ Sneathia spp. | 23 | 1 | Yes, not successful | qPCR assay | Leptotrichia/Sneathia spp. were detected simultaneously in the chorioamnion space and vaginal fluid in a woman with mild chorioamnionitis. |
| Vitorino et al. [49] | 2019 | 39 weeks | Neonatal congenital pneumonia (diagnosed postmortem, stillbirth case) from mother with histological acute chorioamnionitis | Neonatal lung, Cerebro-spinal fluid, blood | S. amnii | 1 | 1 | Yes, not successful | 16S rRNA gene PCR and sequencing | S. amnii was attributed as the cause of congenital pneumonia. |
| Romero et al. [55] | 2019 | 20 – 40 weeks | Intra-amniotic inflammation or infection with intact membranes | Amniotic fluid, vaginal fluid | Sneathia spp. | 8 | 4 | Yes, not successful | 16S rRNA gene sequencing | Sneathia was one of the species most commonly shared between paired amniotic fluid and vaginal samples in women with intra-amniotic infection and intact membranes. |
PCR-TTGE - polymerase chain reaction coupled with temporal temperature gradient gel electrophoresis
PCR/ESI-MS - broad-range polymerase chain reaction coupled with electrospray ionization mass spectrometry
MALDI-TOF - matrix-assisted laser desorption ionization time-of-flight—mass spectrometry
Molecular techniques such as polymerase chain reaction (PCR) amplification and 16S rRNA gene sequencing provide alternative means for detecting Sneathia species in clinical samples, even when Sneathia is just one component of mixed microbial communities in these samples. Herein, we will focus on the studies that used PCR and/or 16S rRNA gene sequencing for the identification of Sneathia in perinatal contexts, as summarized in Table 2. PCR has been used for the molecular detection of Leptotrichia/Sneathia spp. in vaginal [11, 34, 57, 58], chorioamnion [11] and amniotic fluid [3, 5–7, 59–61] samples. Targeted sequencing of the whole 16S rRNA gene, or one or more of its variable regions, is another approach recently used for the identification of S. amnii, S. sanguinegens, or Sneathia spp. in general in specimens obtained from the vagina [8–10], upper genital tract [19, 43, 46], and neonatal stool and skin [13, 62, 63]. Currently, publicly available 16S rRNA gene sequence data exist only for two Sneathia type strains that have been identified to the species level, Sneathia amnii SN35 and Sneathia sanguinegens CCUG 41628. The near complete 16S rRNA gene sequences of these isolates are 96.2% similar based on nucleotide matching, with the greatest variability occurring in the V1-V2 regions of the gene. Molecular assays targeting these regions may allow for reliably distinguishing the presence of the two species in clinical samples, but the description of additional isolates from both species is required to determine if there exists consistent differences in the 16S rRNA genes of the two species. Regardless, sequence-based diagnostic tests, whether of the 16S rRNA gene or other gene targets, typically have longer turnaround times than end-point or quantitative real-time PCR, which could potentially delay decision-making in a clinical context [64]. Taken together, these findings indicate a need for the further development of rapid and specific diagnostic laboratory tests for the rapid identification and quantification of individual Sneathia species in clinical specimens.
Genomic characteristics of Sneathia and the molecular mechanisms of Sneathia virulence
Genomic characteristics of Sneathia
The application of modern genomic and molecular biological analyses recently provided significant insights into the pathways underlying the virulence and pathogenesis of Sneathia infections [28, 65]. The chromosomal DNA of members of the bacterial family Leptotrichiaceae is heavily of the A-T type [21, 28]; Leptotrichiaceae genomic DNA has G-C content values of 24–34%, which are low for bacteria [21, 66, 67]. Leptotrichiaceae also has a small genome size that typically varies between 1.2–2.5 mega base pairs (Mbp) [21]; this is only a quarter to half the size of the Escherichia coli genome. Notably, two genera within the family Leptotrichiaceae have uniquely small genomes with high coding density, suggesting that they are only distantly related to other family members. Specifically, it was reported that Sneathia amnii and its close phylogenetic relative Streptobacillus moniliformis [28, 53], a zoonotic pathogen, have the smallest genome sizes in the family Leptotrichiaceae, at 1.34 and 1.67 Mbp, respectively [28, 68]. Ninety-two percent of the S. amnii genome is comprised of protein-coding genes, which is comparable to the coding gene density of the S. moniliformis genome but is higher than that of other members of the family Leptotrichiaceae [28]. This suggests that for S. amnii and S. moniliformis there is a selective cost associated with genome size [28].
Within the genome of S. amnii, genes for DNA replication and cell cycle regulation are abundant, while those for cell signaling, cell motility, and the production of secondary metabolites are rare [28]. S. amnii lacks enzymes required for the synthesis of many amino acids and therefore this bacterium must rely on hosts and other members of the microbiome for these acids and/or their precursors [28]. S. amnii is also limited in its potential carbon and energy sources; it can ferment glucose, maltose, and glycogen [28]. Glycogen, in particular, is prevalent and abundant in the vaginal ecosystem [69]. Therefore, although carbon sources are very limited for S. amnii, the sources that this bacterium can use are reflective of the principal niche it inhabits – the human vagina. This suggests that the reduction in genome size and restricted metabolic capabilities for S. amnii are products of its intimate association with its human host.
To date, S. amnii is the lone Sneathia species to have its genome sequenced, annotated, and published [28].
Molecular mechanisms of Sneathia virulence
Sneathia amnii has strong hemolytic properties, as demonstrated by in vitro tests on both brain-heart infusion (BHI) agar containing 10% human blood and incubation with freshly isolated human erythrocytes [65]. Similarly, S. moniliformis, which is responsible for rat-bite fever, has hemolytic strains [29]. Rat-bite fever normally presents as severe inflammatory responses such as high fever, swollen lymph nodes, and acute systemic inflammation [29]. Unlike Sneathia species, S. moniliformis is not primarily known for obstetric and gynecologic infections, despite occasionally causing them [47, 70, 71].
The vaginal cytokine profiles of non-pregnant women with vaginal S. amnii or S. sanguinegens infections exhibit elevated levels of pro-inflammatory cytokines [72–74], such as IL-8 and IL-1β [72]. Moreover, co-cultures of human vaginal epithelial cells with S. amnii or S. sanguinegens show upregulated secretion of pro-inflammatory cytokines including IL-1α, IL-1β, and IL-8 [72]. Ascending infections by Sneathia spp. during pregnancy are consistently associated with intra-amniotic inflammation [1, 2, 5, 7, 43, 44, 46, 55, 59] and acute histological chorioamnionitis [7, 13, 45, 55, 75]. These data suggest possible involvement in the control of immune responses to Sneathia antigens by vaginal or cervical immune cells, such as antigen-presenting cells [72, 73] (see Figure 2).
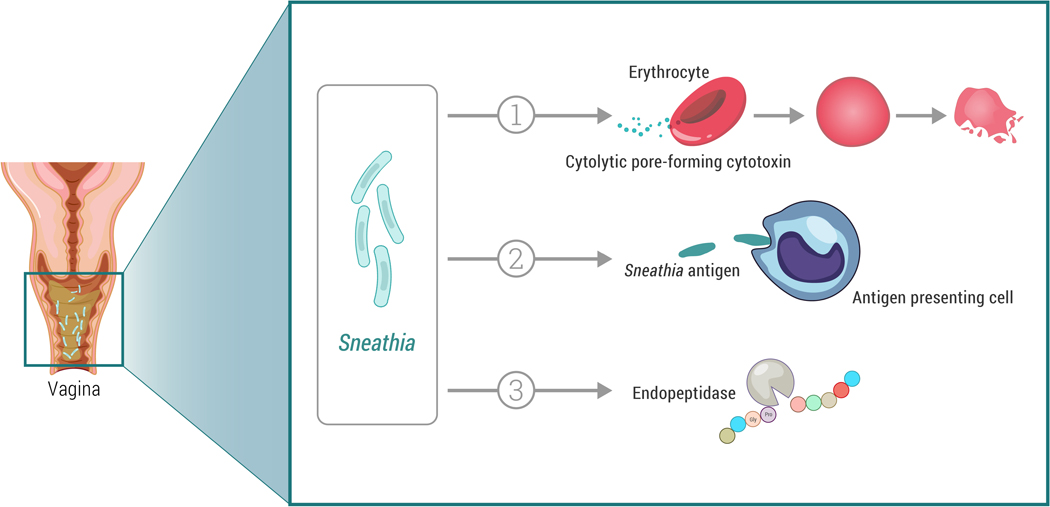
Figure 2. Potential Sneathia virulence factors.
1. Pore-forming cytotoxin CptA (cytopathogenic toxin component A) could damage and/or lyse erythrocytes, vaginal and cervical epithelial cells, and cells of fetal membranes. Preliminary evidence is from in vitro studies of S. amnii [28, 65].
2. Sneathia antigens may be sensed by vaginal and cervical antigen presenting cells and/or other immune cells causing inflammatory responses. Preliminary evidence is from microbial and immune mediator profiling of the human genital tract, transcriptional profiling of cervical antigen presenting cells, and in vitro co-culture studies of human vaginal epithelial cells and S. amnii or S. sanguinegens [72, 73].
3. O-sialoglycoprotein endopeptidase may help Sneathia to transverse the cervix via cervical mucus. Preliminary evidence is from a genomic analysis of S. amnii [28].
A few studies have attempted to elucidate the mechanisms driving Sneathia virulence. For example, it was reported that S. amnii is capable of damaging the fetal membranes, as shown in co-culture experiments with sections of human chorionic tissue from healthy term births [65]. After 18 hours of incubation, histological examination demonstrated that bacterial cells destabilized the surface layers of the tissue and invaded into the fetal membranes, damaging the trophoblast and causing a loss of tissue viability [65]. Sneathia amnii is also capable of adhering to the surface of cervical epithelial cancer cells in culture resulting in the dissociation of intercellular epithelial contacts, cell rounding, and progressive loss of adherence [28]. These effects were evident after just two hours of exposure, indicating a capacity for rapid infectivity by S. amnii.
It was also found that the co-culture of S. amnii with either human amniotic epithelial cells isolated from normal term placentas or chorionic trophoblast cells from a choriocarcinoma cell line led to decreased cell viability and increased degenerative changes [65]. This bacterial cytotoxicity results in the detachment of host cells from each other and from the underlying substrates. The molecular basis of the adhesive capacity of S. amnii cells might be the presence of a gene encoding a putative fibronectin-binding protein, and at least one adhesion homolog [28]. It appears that, because of these binding sites, S. amnii may attach to host cells through fibronectin molecules bound to cell integrins and other extracellular matrix proteins.
An important mechanism underlying S. amnii virulence is the production and release of a pore-forming cytotoxin CptA that destabilizes host cell membranes [65]. As shown in Figure 2 for erythrocytes, it causes the permeabilization of membrane lipid bilayers, leakage of cytoplasmic content, and irreversible injury to host cells [65]. Additionally, S. amnii appears to release hydrolytic enzymes that can cross tissue barriers and membranes. To penetrate the vaginal and cervical epithelial and mucous layers and enter into underlying host tissues, S. amnii may activate a protease capable of cleaving and degrading sialylated proteins [28] (see Figure 2). These data support reports of elevated sialidase activity in vaginal specimens containing Leptotrichia/Sneathia spp. [76]. The S. amnii genome also appears to encode proteins similar to invasins, such as a YadA-like surface protein, and putative internalins that facilitate the crossing of epithelial and connective tissue barriers and gestational membranes [28].
Taken together, these results show that Sneathia (at least S. amnii) virulence and pathogenicity are products of several molecular mechanisms that allow Sneathia to pass tissue and cell barriers and to spread within infected tissues. It is becoming increasingly likely that these characteristics are essential to the invasive ascent of Sneathia from the lower reproductive tract into the fetal membranes, amniotic fluid, and placenta. The putative mechanism of Sneathia’s ascending infection is illustrated in Figure 3. However, our current understanding of detailed cellular and molecular mechanisms underlying Sneathia virulence, particularly in the context of obstetric and gynecologic disease, remain limited.
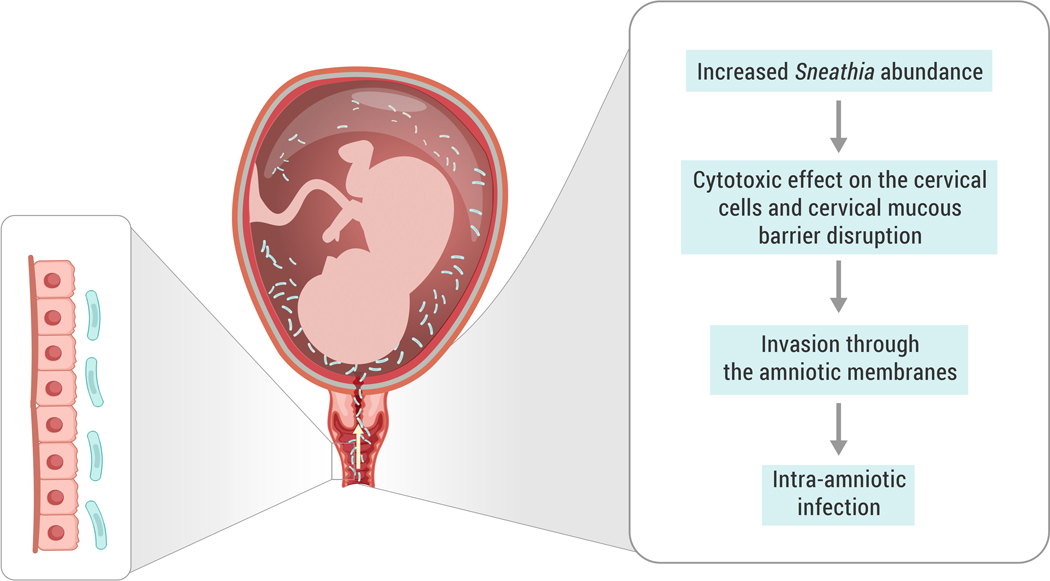
Figure 3. Putative mechanism of Sneathia ascending infection during pregnancy.
Increased abundance of Sneathia in the vaginal microbiome facilitates biofilm formation and/or further spread of Sneathia to the cervical canal. Sneathia’s virulence factors affect the cervical epithelium and help the bacteria spread via cervical mucus. The putative invasins and pore-forming cytotoxin promote penetration of the chorioamniotic membranes and lead to intra-amniotic invasion. Further spread of Sneathia in the amniotic cavity and injury of amniotic cells promotes inflammation, leading to intra-amniotic infection.
Sneathia in female reproductive disease
Bacterial vaginosis
Sneathia spp. have been identified as members of Community State Type IV (CST-IV) of the vaginal microbiome, which is characterized by a diminished presence of Lactobacillus species in the vagina [27]. Although CST-IV is an asymptomatic phenotype in many women, patients diagnosed with bacterial vaginosis have a vaginal microbiome with a CST-IV composition [27, 77]. They also have vaginal secretions with an elevated pH compared to women with Lactobacillus-dominant CSTs [27]. Most importantly, in the context of this review, there is an increased presence of Sneathia spp. in the vaginal microbiome of women with bacterial vaginosis diagnosed by increased Nugent scores [38–40, 78–80], Nugent-Boon scores [81], or Amsel test criteria [34, 78–80].
Bacterial vaginosis is a common clinical condition [82, 83] characterized by a general absence of Lactobacillus species, accompanied by evident vaginal discharge and an unpleasant odor [34, 84, 85]. The specific etiology of bacterial vaginosis remains incompletely understood [86]. Currently, the formation of a bacterial biofilm is considered a primary mechanism underlying the development and progression of bacterial vaginosis [87–89]. A bacterial biofilm is a structured community of bacterial cells secured within a self-produced extracellular matrix that is adherent to an inert surface or biological tissue [90, 91]. Several genera, especially Gardnerella, but also Atopobium, Mobiluncus, Peptostreptococcus, Prevotella, and Sneathia, are associated with both the condition of bacterial vaginosis and biofilm formation [41, 87, 89, 91, 92].
In one association study, an increased relative abundance of S. sanguinegens in the vaginal microbiome was associated with the presence of clue cells (i.e. vaginal epithelial cells with bacteria attached to their surface) and bacterial biofilms in vaginal fluids [41], suggesting that Sneathia spp. may be important contributors to the process of vaginal biofilm formation [41]. Also, the detection of Sneathia spp. in the anal canal of asymptomatic women was reported to correlate with a higher predisposition for and recurrence of bacterial vaginosis [25]. Similarly, Sneathia spp. can inhabit the male urogenital tract [50, 93], and carriage of Sneathia by male partners has been associated with incidences of bacterial vaginosis [94]. Thus, the potential transmission of Sneathia spp. from extra-vaginal reservoirs on the body or the body of sexual partners might contribute to the increased risk of Sneathia colonization of the female genital tract [25]. It is not uncommon for bacterial vaginosis to precede severe obstetric [95–98] and gynecologic [99–101] complications, such as pelvic inflammatory disease [99, 102]. In agreement with these observations, Sneathia spp. have been found in the cervix [18], endometrium [18], and in the surgically removed fallopian tubes and pelvic pus [19, 103] of women diagnosed with pelvic inflammatory diseases. Taken together, these findings provide insights into the pathways underlying the persistence and progression of Sneathia in the female reproductive tract and its potential contribution to the occurrence and recurrence of bacterial vaginosis.
A role for Sneathia in human papilloma virus infection and cervical cancer
Recent data suggest that Sneathia infections in the urogenital tract are associated with increased risk of several other common types of gynecologic disease. Infections with Sneathia spp. have also been found to accompany sexually transmitted diseases [51], including HIV [73, 104], suggesting Sneathia spp. may be opportunistic pathogens. Conversely, Sneathia, with its capacity to disintegrate the stability of the fetal membranes and epithelial cell layers in vitro [28, 65] may potentiate local inflammation and tissue damage [72, 73], thereby increasing the risk of acquisition of HIV [73, 104] and other sexually transmitted infections [51]. That is, Sneathia may actually be a primary agent of disease.
Of special interest are recent reports implicating Sneathia spp. in the pathogenesis of human papilloma viral infections. Cervical cancer is one of the most common types of female malignancies [105, 106], and human papillomavirus (HPV) is considered the principal cause of cervical intraepithelial neoplasia [107] and cervical cancer [105, 108]. HPV infection leads to the development of pre-malignant epithelial lesions in the cervix, which can progress into malignant neoplastic growth and cervical carcinogenesis [109–111]. Several studies of cervical and vaginal microbiomes have revealed a greater relative abundance of Sneathia spp. in specimens obtained from women diagnosed with HPV infection [33, 112–115]. For example, a study of identical female twins showed that the vaginal microbiomes of HPV-infected individuals contained much lower levels of the normally dominant Lactobacillus spp. and significantly elevated levels of Fusobacteria and Sneathia spp. than those of their non-infected genetically identical siblings [112]. Moreover, Sneathia spp. were detected in HPV-infected patients with different types of cervical intraepithelial lesions [15–17], including high squamous intraepithelial lesions that are indicative of chronic HPV infection [15]. Sneathia amnii, at least, can adhere to the surface of malignant cervical epithelial cells [28]. This suggests a potential for toxic products released by adherent Sneathia to alter the characteristics of host tissue and directly mediate effects on the cervical microenvironment.
An analysis of correlations between the severity of cervical neoplasms and variation among vaginal microbiomes found that Sneathia spp. were enriched in all precancerous women with cervical epithelial neoplasia, accompanied by decreased Lactobacillus dominance and abnormally increased vaginal pH [17]. However, the abundance of Sneathia does not appear to be elevated in vaginal specimens received from patients with invasive cervical carcinoma [17]. One potential explanation is that Sneathia is outcompeted by other bacterial species under conditions of changing tissue environment, including possible pH shifts at the advanced stages of tumor growth [17]. This suggests that the activation of Sneathia growth is positively associated with HPV infection, onset of cervical intraepithelial neoplasia, and precancerous neoplastic progression. These findings also suggest that the shift in the composition of cervicovaginal microbiomes of HPV-infected patients may have a potential predictive value for clinical prognosis of cervical intraepithelial neoplastic tumor progression [17].
Sneathia infections and adverse perinatal outcomes
Infections with Sneathia spp. are associated with several adverse perinatal conditions and outcomes such as 1) spontaneous preterm birth, particularly PPROM, 2) clinical chorioamnionitis, 3) septic abortions, 4) neonatal sepsis, 5) intrauterine fetal demise, and 6) maternal intrapartum and postpartum septic complications.
Spontaneous preterm birth
Data summarizing the detection of Sneathia spp. in amniotic fluid in cases of spontaneous preterm birth are presented in Table 2. In these studies, spontaneous preterm labor was defined as regular contractions accompanied by cervical change at less than 37 weeks of gestation. PPROM was defined as spontaneous rupture of the membranes at less than 37 weeks of gestation and at least one hour before the onset of contractions. Births that followed spontaneous preterm labor and PPROM are collectively referred to as spontaneous preterm births [116, 117]. A transabdominal amniocentesis may be performed at the onset of preterm labor and PPROM to diagnose the presence of intra-amniotic inflammation and/or microbial invasion of the amniotic cavity [2, 3, 5, 7, 55, 118].
Among patients with preterm labor and intact membranes, Sneathia was one of the most frequently detected bacterial genera in amniotic fluid (Table 2). Patients diagnosed with Sneathia intra-amniotic infection showed signs of intra-amniotic inflammation [1, 2, 5, 55, 119] and delivered preterm with further histological manifestations of acute chorioamnionitis [2, 5, 55]. Intra-amniotic infection by Sneathia spp. in these cases was detected by using PCR [1, 59], PCR/ESI-MS [5], and 16S rRNA gene sequencing approaches [2, 43, 44, 46, 55, 119, 120].
A recent study showed that Sneathia spp. were found in both the vaginal secretions and amniotic fluid of the same women with preterm labor and intact membranes [55]. The patients examined in this study were diagnosed with intra-amniotic inflammation and subsequently delivered preterm with signs of funisitis and acute chorioamnionitis [55]. This suggests ascension of Sneathia from the vagina into the amniotic cavity and supports the findings presented above regarding the virulence properties of Sneathia [28, 65].
Along with these observations in women with spontaneous preterm labor with intact membranes, Sneathia spp. were among the most common bacteria detected in amniotic fluid of women with PPROM [3, 7]. Moreover, the detection of Sneathia spp. was associated with intra-amniotic infection and adverse pregnancy outcomes [7, 43–46]. Sneathia spp. were also detected in amniotic fluid of an otherwise asymptomatic woman with a sonographic short cervix, although surprisingly in this case the woman delivered at term with no signs of intra-amniotic inflammation [61].
Several studies of the vaginal microbiome have showed a positive correlation between the relative abundance of Sneathia spp. and the occurrence of preterm birth in African American [4, 8, 9] and Caucasian [10] women. In a cohort of primarily African American women with a prior history of preterm birth, women with increased absolute abundance of Sneathia spp. in the vagina in first and early second trimesters were significantly more likely to experience a spontaneous preterm delivery [4]. Moreover, longitudinal studies showed that levels of S. amnii [9] and S. sanguinegens [8] in the vagina were significantly associated with increased risk of preterm birth in cohorts of predominantly African-American women. Similarly, increased levels of S. sanguinegens were evident in the vaginal samples of Caucasian women with preterm labor and birth [10]. Furthermore, in a recent study of a cohort of predominantly African-American women, women who ultimately experienced spontaneous preterm birth exhibited stronger negative associations between S. sanguinegens and the immune mediators CCL26, CCL22, CCL2, CXCL10, and IL-16 in vaginal fluid than did women who ultimately delivered at term [121].
Overall, Sneathia spp. are one of the most frequently detected bacterial taxa in the amniotic cavity in women with spontaneous preterm birth [2, 3, 7]. However, further studies of the purported etiologic and causative links between vaginal and intra-amniotic infections with Sneathia and spontaneous preterm birth are needed to better understand the pathogenic mechanisms involved.
Sneathia infections in the pathogenesis of clinical chorioamnionitis
Clinical chorioamnionitis is one of the most common obstetric pathologies associated with intra-amniotic bacterial infection [122–125]. The diagnostic criteria of clinical chorioamnionitis are based on the studies of Gibbs et al. [126, 127], and refer to the presence of maternal fever associated with clinical signs (i.e., foul-smelling discharge, uterine tenderness, maternal and fetal tachycardia) as well as laboratory abnormalities (i.e., leukocytosis) [6]. However, such clinical criteria do not accurately identify patients with proven intra-amniotic infection (i.e., those with microorganisms detected by culture or molecular microbiologic techniques and an associated intra-amniotic inflammatory response) [128]. In women with clinical chorioamnionitis and proven intra-amniotic infection [6], local and systemic maternal-fetal inflammatory processes are considered to be immune responses to ascending microbial invasion of the amniotic cavity [55, 118, 129–149]. Polymicrobial infections of the amniotic cavity are often observed in patients diagnosed with clinical chorioamnionitis [6, 150–152] and are generally associated with a greater level of inflammatory response than infections with a single microorganism [6]. A key challenge in understanding the role of Sneathia spp. in the pathogenesis of clinical chorioamnionitis and other obstetric pathologies is elucidating their specific contributions under conditions in which they are but one member of a mixed microbial community responsible for polymicrobial infections.
In two studies of women diagnosed with clinical chorioamnionitis at term and polymicrobial invasion of the amniotic cavity, Sneathia spp. were detected in amniotic fluid using PCR/ESI-MS [6] and 16S rRNA gene sequencing [47]. Bacterial cultures from these patients proved to be negative for Sneathia [6, 47], but this may be due to the fastidious culture requirements of this genus [28]. Another study reported polymicrobial invasion that included infection with Sneathia spp. in the chorioamnion and vaginal swabs of the same patient with mild symptoms of clinical chorioamnionitis at term [11]. In this study, Sneathia spp. were identified by 16S rRNA gene PCR. However, attempts to isolate the bacteria in culture were again unsuccessful.
Thus, these findings indicate that Sneathia infections play an important role in the pathogenesis of clinical chorioamnionitis. Further studies examining the mechanisms of virulence of Sneathia spp. and their capacity to initiate maternal-fetal inflammatory responses are warranted.
Sneathia and neonatal sepsis
Neonatal sepsis has been defined as an invasive bacterial infection of the blood and/or the cerebrospinal fluid that occurs in the first three months after birth [125, 153]. The first study suggesting the contribution of S. sanguinegens to the pathogenesis of neonatal bacterial sepsis was published in 1995 [12]. Sneathia infection was identified in blood cultures of two vaginally delivered infants: one of them was born at 35 weeks and the other was born at 41.5 weeks, both were afebrile. The second infant had tachycardia and signs of respiratory distress associated with increased respiratory rate that required positive pressure for respiratory support. The indication for septic workup was a foul smell from the neonates. In another study, stool samples obtained from 106 extremely preterm neonates (i.e. those born earlier than 28 weeks of gestation) revealed that a higher relative abundance of Sneathia spp. during the first week of life was associated with histological chorioamnionitis with funisitis and a higher risk of late onset neonatal sepsis or death [13]. Recently, it was reported that a high relative abundance of Sneathia spp. in the vaginal microbiome during pregnancy was associated with histological chorioamnionitis with funisitis and early onset neonatal sepsis [14]. Taken together, these data indicate that intra-amniotic Sneathia infections during pregnancy have deleterious effects on neonatal life.
Septic abortion, intrauterine fetal demise, and stillbirth
To date, there have been three studies implicating S. amnii in septic complications of pregnancy leading to fetal death. The first report included the isolation of S. amnii in anaerobic culture from maternal blood after a septic abortion at 15 weeks [52]. Another clinical case reported the identification of S. amnii through anaerobic culture of the amniotic fluid of a patient with intrauterine fetal demise [48]. In a third case, S. amnii was identified in the lung tissue of a stillborn infant using 16S rRNA gene sequencing [49]. In this case, histopathological analysis of the placenta and umbilical cord revealed the manifestations of acute chorioamnionitis and funisitis, and rare Gram-negative coccobacilli were detected in the umbilical artery. However, 16S rRNA gene sequencing did not detect S. amnii in the placental tissue [49]. These findings suggest the involvement of Sneathia in the pathogenesis of septic abortion, intrauterine fetal demise, and stillbirth. However, the question as to whether Sneathia is a causative primary agent of infection leading to fetal death needs to be further examined.
Peripartum septic maternal complications
Postpartum infections, including blood stream infections (bacteremia), are a subset of maternal pathological conditions occurring between delivery and the 42nd day postpartum [154]. S. sanguinegens has been isolated in blood culture from a patient diagnosed with postpartum bacteremia [12]. S. sanguinegens and S. amnii were further isolated from the blood cultures of three patients diagnosed with intrapartum and postpartum maternal fever [54]. Two out of three of these patients showed symptoms of fetal distress during delivery; both were infected with S. amnii. However, in all three cases, the newborns did not develop any detectable clinical or laboratory evidence of infection. Most recently, using 16S rRNA gene sequencing, Kotaskova et al. [155] reported that S. sanguinegens was detected in prosthetic valve tissue in a patient with postpartum endocarditis developed following postpartum fever and spontaneous preterm birth. Repeated blood cultures for this patient were negative. The results of these studies suggest that Sneathia infection can spread from the reproductive tract to other organs and systems of the body. Collectively, these findings show that Sneathia species appear to be important infectious agents contributing to the pathogenesis of postpartum bacteremia.
Antibiotic treatment of Sneathia infections
In the previous sections of this review, we discussed several pathological conditions associated with Sneathia infections. Administration of antibiotics is the standard treatment for these infections [156, 157]. To date, literature reporting successful antibiotic therapies for Sneathia infections remain very limited. The current literature is summarized in Table 3.
Table 3.
Use of antibiotics for the treatment of Sneathia infections
| Authors | Year | Sample type | Sneathia species | Clinical diagnosis | Method of Sneathia detection | Antibiotic administration | Effect of antibiotic treatment |
|---|---|---|---|---|---|---|---|
| Mitchell et al. [58] | 2009 | Vaginal secretion | Leptotrichia/Sneathia spp. | Bacterial vaginosis in pregnancy | qPCR assay | Oral or vaginal administration of metronidazole | There were decreases in the concentrations of Leptotrichia/Sneathia in women receiving oral metronidazole, but not in the vaginal treatment subgroup; however, the cure rate and persistence after treatment were not different between these two groups. |
| Ling et al. [158] | 2013 | Vaginal secretion | Sneathia spp. | Bacterial vaginosis | 16S rRNA gene sequencing | Intravaginal administration of metronidazole or intravaginal probiotic (L. crispatus) | There was an equivalent decrease in Sneathia spp. among women treated with intravaginal metronidazole or intravaginal probiotic (Lactobacillus crispatus). |
| Macklaim et al. [161] | 2015 | Vaginal secretion | Sneathia spp. | Bacterial vaginosis | 16S rRNA gene sequencing | Oral administration of tinidazole with or without oral probiotic (Lactobacillus rhamnosus and L. reuteri strains) | The combination of tinidazole with oral probiotic (Lactobacillus rhamnosus and L. reuteri strains), but not tinidazole alone, significantly decreased the relative abundance of Sneathia. |
| Cruciani et al. [162] | 2015 | Vaginal secretion | Sneathia spp. | Bacterial vaginosis | VaginArray (PCR-based microarray tool) | Intravaginal administration of rifaximin | Rifaximin resulted in a significant drop in the absolute abundance of Sneathia in most women. |
| Hilbert et al. [80] | 2016 | Vaginal secretion | Leptotrichia/Sneathia spp. | Bacterial vaginosis | qPCR assay | Oral metronidazole or tinidazole, intravaginal metronidazole suppository, intravaginal metronidazole plus miconazole, clindamycin cream | Leptrotrichia/Sneathia were largely undetectable after 7–10 days in the resolved and recurrent bacterial vaginosis groups, however, it was detected 40–45 days later in both groups. |
| Balkus et al. [37] | 2017 | Vaginal secretion | Leptotrichia/Sneathia spp. | Women with a high-risk of vaginal infections | qPCR assay | Intravaginal administration of metronidazole plus miconazole or placebo | The combination of metronidazole and miconazole significantly reduced the abundance of Leptotrichia/Sneathia compared to the placebo group. |
| Gottschick et al. [41] | 2017 | Vaginal secretion | S. amnii | Bacterial vaginosis | 16S rRNA gene sequencing | Oral administration of metronidazole | The relative abundance of S. amnii was significantly reduced after treatment. |
| Brown et al. [14] | 2018 | Vaginal secretion | S. sanguinegens; Sneathia spp. | Preterm prelabor rupture of membranes (PPROM) | 16S rRNA gene sequencing | Oral administration of erythromycin | The prophylactic oral administration of erythromycin to patients who experienced PPROM with Lactobacillus-dominant vaginal microbiomes led to a shift in the vaginal microbiota and the detection of S. sanguinegens and unclassified Sneathia in the vaginal fluid after treatment. |
Briefly, metronidazole is a first-line therapy for the treatment of bacterial vaginosis; this bactericidal antibiotic targets mainly anaerobic bacteria, including Sneathia [157]. Metronidazole treatment leads to a significant decrease in the absolute [37, 57, 80] and relative abundance [158, 159] of Sneathia spp. in vaginal secretions and/or urine of non-pregnant women, as determined using culture-independent molecular techniques [37, 57, 80, 158, 159]. Among pregnant women with bacterial vaginosis, metronidazole treatment did not significantly affect Sneathia infection when applied vaginally [58]; however, it did resolve Sneathia infection when the antibiotic was taken orally [58]. Furthermore, oral administration of tinidazole (an antibiotic that targets bacterial DNA integrity, repair, and transcription [160]) either alone [80] or in combination with probiotics [161], as well as oral administration of rifaximin [162] (an antibiotic that inhibits bacterial RNA synthesis [163, 164]), appear to effectively decrease the relative and absolute abundance of Sneathia spp. in the vaginal fluid of women with bacterial vaginosis.
A principal challenge of bacterial vaginosis treatment is a high recurrence rate of the infection within one year [165]. Notably, among women successfully treated for bacterial vaginosis with rifaximin, there was a reduction in the prevalence and absolute abundance of Sneathia spp. after seven days, yet there was an increase in Sneathia spp. after 28 days [162]. Similarly, in a separate study of bacterial vaginosis [80], Sneathia spp. were seldom detected at high absolute abundances in the vaginal fluids of women who remained responsive to antibiotic treatment after 7–10 days, yet in women who remained responsive to antibiotic treatment after 40–45 days Sneathia spp. were abundant.
In addition to the studies concerning the clinical effectiveness of the antibiotic therapies presented above, there have been four reports characterizing the sensitivity of Sneathia isolates in culture to different antibiotics [28, 52, 54, 166]. These findings are summarized in Table 4, and suggest that different Sneathia species may differ in their sensitivity to different antibiotics depending on the source of their isolation. For example, multiple isolates of S. amnii and S. sanguinegens cultured from the blood were reported to be resistant to vancomycin [54]; yet, S. amnii isolated from vaginal fluid was sensitive to this antibiotic [28]. This finding may be due to intraspecific variation in the structure of the cell envelope [28], or intraspecific genetic heterogeneity resulting from horizontal gene transfer, including the acquisition of plasmids conferring antibiotic resistance [167, 168].
Further investigation of the molecular mechanisms underlying the sensitivity and resistance of Sneathia to different antibiotics is essential for a better understanding of the pathogenesis and treatment of Sneathia infections. Genomic analysis of different Sneathia isolates may reveal the existence of distinct strains of Sneathia and allow for the development of targeted antibiotic therapies that will help in the management of recurrent bacterial vaginosis and other Sneathia associated pathologies in obstetric and gynecologic clinical practice.
Conclusions and future directions
The data presented in this review suggest a significant role for Sneathia infections in the pathogenesis of PPROM [3, 7, 43–46], spontaneous preterm labor [1, 2, 4, 5, 8–10], chorioamnionitis [6, 11], stillbirth [48, 49], bacterial vaginosis [34, 38–41, 57, 79–81], HIV infection [73, 104], and cervical cancer [15–17]. Several of these pathologies, such as spontaneous preterm birth and intrapartum-related complications associated with bacterial infections, are among the leading causes of death [169–172] and long-term disabilities [173, 174] in children under five years of age.
A growing body of evidence suggests that Sneathia spp. can become opportunistic pathogens following shifts in the vaginal microbiome away from Lactobacillus-dominated communities [14]. Furthermore, although Sneathia spp. are not the only bacteria associated with bacterial vaginosis [34, 38–40, 42, 78–81, 175], they do appear to play a key role in biofilm formation observed in this clinical condition [41]. This capacity may be instrumental in the development and progression of bacterial vaginosis.
Recent results of in vitro and in silico experiments open the door for further elucidation of the mechanisms underlying Sneathia spp. virulence and their capacity for ascending invasion into fetal membranes [28, 65]. The potential capacity of Sneathia spp. for attaching to and damaging host cells using cytolytic [65] and cytotoxic [28] compounds enables their progressive invasion through the vaginal and cervical epithelium into deeper layers of tissue, where they cause tissue injury. Tissue damage might be further enhanced by a pore-forming cytotoxin that destabilizes lipid cell surface membranes leading to leakage of hemoglobin from erythrocytes and the permeabilization of cervical, amniotic and chorionic surface layers [65]. Given the fact that Sneathia spp. are non-motile, these abilities may be instrumental to their ascending invasion through the female urogenital tract (Figure 3).
New evidence demonstrates that the virulence of vaginal bacteria and the progression of the bacterial infection might significantly vary among different strains of the same species [92, 176–178] and be affected by the location of the bacteria within the tissue, as well as the stage and depth of their invasion during the process of infection [179]. For this reason, it is important to examine virulence among conspecific Sneathia strains. Moreover, recent studies of the vaginal microbiome showed that the presence and growth characteristics of different vaginal bacterial species might be affected by the genetic polymorphisms within human populations [18, 180] and the intensity of local host immune responses [72, 181, 182]. Additional genomic [18, 28], metagenomic [183–185], metabolomic [186] and proteomic [187–189] approaches are needed to further elucidate the mechanisms underlying the virulence of Sneathia spp. and strains. Further studies in this field will advance our understanding of the etiological and pathogenic factors involved in Sneathia infections in reproductive and perinatal medicine and serve to evaluate the potential value of Sneathia spp. as prognostic biomarkers of adverse outcomes for both mothers and newborns.
Acknowledgments
Funding details
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS); and, in part, with Federal funds from NICHD/NIH/DHHS under Contract No. HHSN275201300006C. KRT and NG-L were further supported by the Wayne State University Perinatal Research Initiative in Maternal, Perinatal and Child Health. Dr. Romero has contributed to this work as part of his official duties as an employee of the United States Federal Government.
Footnotes
Disclosure of interest
The authors report no conflicts of interest.
References
- 1.Gardella C, et al. , Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor. Am J Perinatol, 2004. 21(6): p. 319–23. [DOI] [PubMed] [Google Scholar]
- 2.DiGiulio DB, et al. , Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One, 2008. 3(8): p. e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiGiulio DB, et al. , Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol, 2010. 64(1): p. 38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DB, et al. , Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery. Paediatr Perinat Epidemiol, 2014. 28(2): p. 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero R, et al. , A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reprod Immunol, 2014. 71(4): p. 330–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero R, et al. , Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J Perinat Med, 2015. 43(1): p. 19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero R, et al. , Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med, 2015. 28(12): p. 1394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elovitz MA, et al. , Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat Commun, 2019. 10(1): p. 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fettweis JM, et al. , The vaginal microbiome and preterm birth. Nat Med, 2019. 25(6): p. 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hocevar K, et al. , Vaginal Microbiome Signature Is Associated With Spontaneous Preterm Delivery. Front Med (Lausanne), 2019. 6: p. 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lannon SMR, et al. , Parallel detection of lactobacillus and bacterial vaginosis-associated bacterial DNA in the chorioamnion and vagina of pregnant women at term. J Matern Fetal Neonatal Med, 2019. 32(16): p. 2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanff PA, et al. , Leptotrichia sanguinegens sp. nov., a new agent of postpartum and neonatal bacteremia. Clin Infect Dis, 1995. 20 Suppl 2: p. S237–9. [DOI] [PubMed] [Google Scholar]
- 13.Puri K, et al. , Association of Chorioamnionitis with Aberrant Neonatal Gut Colonization and Adverse Clinical Outcomes. PLoS One, 2016. 11(9): p. e0162734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown RG, et al. , Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med, 2018. 16(1): p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra A, et al. , Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep, 2015. 5: p. 16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audirac-Chalifour A, et al. , Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS One, 2016. 11(4): p. e0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laniewski P, et al. , Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep, 2018. 8(1): p. 7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BD, et al. , Toll-like receptor variants and cervical Atopobium vaginae infection in women with pelvic inflammatory disease. Am J Reprod Immunol, 2018. 79(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, et al. , Characterization of pelvic and cervical microbiotas from patients with pelvic inflammatory disease. J Med Microbiol, 2018. 67(10): p. 1519–1526. [DOI] [PubMed] [Google Scholar]
- 20.Gupta RS and Sethi M, Phylogeny and molecular signatures for the phylum Fusobacteria and its distinct subclades. Anaerobe, 2014. 28: p. 182–98. [DOI] [PubMed] [Google Scholar]
- 21.Eisenberg T, et al. , Phylogenetic and comparative genomics of the family Leptotrichiaceae and introduction of a novel fingerprinting MLVA for Streptobacillus moniliformis. BMC Genomics, 2016. 17(1): p. 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett KW and Eley A, Fusobacteria: new taxonomy and related diseases. J Med Microbiol, 1993. 39(4): p. 246–54. [DOI] [PubMed] [Google Scholar]
- 23.Eribe ER and Olsen I, Leptotrichia species in human infections. Anaerobe, 2008. 14(3): p. 131–7. [DOI] [PubMed] [Google Scholar]
- 24.Eribe ERK and Olsen I, Leptotrichia species in human infections II. J Oral Microbiol, 2017. 9(1): p. 1368848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrazzo JM, et al. , Extravaginal reservoirs of vaginal bacteria as risk factors for incident bacterial vaginosis. J Infect Dis, 2012. 205(10): p. 1580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon JY, et al. , Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine, 2018. 37: p. 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravel J, et al. , Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A, 2011. 108 Suppl 1: p. 4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harwich MD Jr., et al. , Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics, 2012. 13 Suppl 8: p. S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaastra W, et al. , Rat bite fever. Vet Microbiol, 2009. 133(3): p. 211–28. [DOI] [PubMed] [Google Scholar]
- 30.Collins MD, et al. , Characterization of some strains from human clinical sources which resemble “Leptotrichia sanguinegens”: description of Sneathia sanguinegens sp. nov., gen. nov. Syst Appl Microbiol, 2001. 24(3): p. 358–61. [DOI] [PubMed] [Google Scholar]
- 31.Vesty A, et al. , Oral microbial influences on oral mucositis during radiotherapy treatment of head and neck cancer. Support Care Cancer, 2020. 28(6): p. 2683–2691. [DOI] [PubMed] [Google Scholar]
- 32.Gajer P, et al. , Temporal dynamics of the human vaginal microbiota. Sci Transl Med, 2012. 4(132): p. 132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onywera H, et al. , The cervical microbiota in reproductive-age South African women with and without human papillomavirus infection. Papillomavirus Res, 2019. 7: p. 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredricks DN, Fiedler TL, and Marrazzo JM, Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med, 2005. 353(18): p. 1899–911. [DOI] [PubMed] [Google Scholar]
- 35.Fethers K, et al. , Bacterial vaginosis (BV) candidate bacteria: associations with BV and behavioural practices in sexually-experienced and inexperienced women. PLoS One, 2012. 7(2): p. e30633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srinivasan S, et al. , Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One, 2012. 7(6): p. e37818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balkus JE, et al. , Impact of Periodic Presumptive Treatment for Bacterial Vaginosis on the Vaginal Microbiome among Women Participating in the Preventing Vaginal Infections Trial. J Infect Dis, 2017. 215(5): p. 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamrakar R, et al. , Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis, 2007. 7: p. 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spear GT, et al. , Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis, 2008. 198(8): p. 1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dols JA, et al. , Molecular assessment of bacterial vaginosis by Lactobacillus abundance and species diversity. BMC Infect Dis, 2016. 16: p. 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottschick C, et al. , Treatment of biofilms in bacterial vaginosis by an amphoteric tenside pessary-clinical study and microbiota analysis. Microbiome, 2017. 5(1): p. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noyes N, et al. , Associations between sexual habits, menstrual hygiene practices, demographics and the vaginal microbiome as revealed by Bayesian network analysis. PLoS One, 2018. 13(1): p. e0191625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musilova I, et al. , Maternal serum C-reactive protein concentration and intra-amniotic inflammation in women with preterm prelabor rupture of membranes. PLoS One, 2017. 12(8): p. e0182731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musilova I, et al. , Maternal white blood cell count cannot identify the presence of microbial invasion of the amniotic cavity or intra-amniotic inflammation in women with preterm prelabor rupture of membranes. PLoS One, 2017. 12(12): p. e0189394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hornychova H, et al. , Cervical human papillomavirus infection in women with preterm prelabor rupture of membranes. PLoS One, 2018. 13(11): p. e0207896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musilova I, et al. , Late preterm prelabor rupture of fetal membranes: fetal inflammatory response and neonatal outcome. Pediatr Res, 2018. 83(3): p. 630–637. [DOI] [PubMed] [Google Scholar]
- 47.Carlstein C, Marie Soes L, and Jorgen Christensen J, Aerococcus christensenii as Part of Severe Polymicrobial Chorioamnionitis in a Pregnant Woman. Open Microbiol J, 2016. 10: p. 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shukla SK, et al. , Leptotrichia amnionii sp. nov., a novel bacterium isolated from the amniotic fluid of a woman after intrauterine fetal demise. J Clin Microbiol, 2002. 40(9): p. 3346–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitorino P, et al. , Sneathia amnii and Maternal Chorioamnionitis and Stillbirth, Mozambique. Emerg Infect Dis, 2019. 25(8): p. 1614–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson DE, et al. , Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One, 2010. 5(11): p. e14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brotman RM, et al. , Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex Transm Dis, 2012. 39(10): p. 807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boennelycke M, et al. , Leptotrichia amnionii found in septic abortion in Denmark. Scand J Infect Dis, 2007. 39(4): p. 382–3. [DOI] [PubMed] [Google Scholar]
- 53.Eribe ER, et al. , Genetic diversity of Leptotrichia and description of Leptotrichia goodfellowii sp. nov., Leptotrichia hofstadii sp. nov., Leptotrichia shahii sp. nov. and Leptotrichia wadei sp. nov. Int J Syst Evol Microbiol, 2004. 54(Pt 2): p. 583–92. [DOI] [PubMed] [Google Scholar]
- 54.De Martino SJ, et al. , Peripartum bacteremias due to Leptotrichia amnionii and Sneathia sanguinegens, rare causes of fever during and after delivery. J Clin Microbiol, 2004. 42(12): p. 5940–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romero R, et al. , Evidence that intra-amniotic infections are often the result of an ascending invasion - a molecular microbiological study. J Perinat Med, 2019. 47(9): p. 915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivasan S, et al. , More Easily Cultivated Than Identified: Classical Isolation With Molecular Identification of Vaginal Bacteria. J Infect Dis, 2016. 214 Suppl 1(Suppl 1): p. S21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fredricks DN, et al. , Changes in vaginal bacterial concentrations with intravaginal metronidazole therapy for bacterial vaginosis as assessed by quantitative PCR. J Clin Microbiol, 2009. 47(3): p. 721–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell CM, et al. , Comparison of oral and vaginal metronidazole for treatment of bacterial vaginosis in pregnancy: impact on fastidious bacteria. BMC Infect Dis, 2009. 9: p. 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DiGiulio DB, et al. , Microbial invasion of the amniotic cavity in preeclampsia as assessed by cultivation and sequence-based methods. J Perinat Med, 2010. 38(5): p. 503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiGiulio DB, et al. , Microbial invasion of the amniotic cavity in pregnancies with small-for-gestational-age fetuses. J Perinat Med, 2010. 38(5): p. 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romero R, et al. , Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance. J Matern Fetal Neonatal Med, 2015. 28(11): p. 1343–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez-Bello MG, et al. , Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A, 2010. 107(26): p. 11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aujoulat F, et al. , Temporal dynamics of the very premature infant gut dominant microbiota. BMC Microbiol, 2014. 14: p. 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulkarni P. and Frommolt P, Challenges in the Setup of Large-scale Next-Generation Sequencing Analysis Workflows. Comput Struct Biotechnol J, 2017. 15: p. 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gentile GL, et al. , Identification of a cytopathogenic toxin from Sneathia amnii. J Bacteriol, 2020. [DOI] [PMC free article] [PubMed]
- 66.Hildebrand F, Meyer A, and Eyre-Walker A, Evidence of selection upon genomic GC-content in bacteria. PLoS Genet, 2010. 6(9): p. e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bohlin J, et al. , The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC Genomics, 2017. 18(1): p. 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nolan M, et al. , Complete genome sequence of Streptobacillus moniliformis type strain (9901). Stand Genomic Sci, 2009. 1(3): p. 300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mirmonsef P, et al. , Glycogen Levels in Undiluted Genital Fluid and Their Relationship to Vaginal pH, Estrogen, and Progesterone. PLoS One, 2016. 11(4): p. e0153553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faro S, Walker C, and Pierson RL, Amnionitis with intact amniotic membranes involving Streptobacillus moniliformis. Obstet Gynecol, 1980. 55(3 Suppl): p. 9S–11S. [DOI] [PubMed] [Google Scholar]
- 71.Pins MR, et al. , Isolation of presumptive Streptobacillus moniliformis from abscesses associated with the female genital tract. Clin Infect Dis, 1996. 22(3): p. 471–6. [DOI] [PubMed] [Google Scholar]
- 72.Anahtar MN, et al. , Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity, 2015. 42(5): p. 965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gosmann C, et al. , Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated with Increased HIV Acquisition in Young South African Women. Immunity, 2017. 46(1): p. 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabo MC, et al. , Associations between vaginal bacteria implicated in HIV acquisition risk and proinflammatory cytokines and chemokines. Sex Transm Infect, 2020. 96(1): p. 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Urushiyama D, et al. , Microbiome profile of the amniotic fluid as a predictive biomarker of perinatal outcome. Sci Rep, 2017. 7(1): p. 12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marconi C, et al. , Do Atopobium vaginae, Megasphaera sp. and Leptotrichia sp. change the local innate immune response and sialidase activity in bacterial vaginosis? Sex Transm Infect, 2013. 89(2): p. 167–73. [DOI] [PubMed] [Google Scholar]
- 77.Nelson TM, et al. , Vaginal biogenic amines: biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis? Front Physiol, 2015. 6: p. 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haggerty CL, et al. , Clinical characteristics of bacterial vaginosis among women testing positive for fastidious bacteria. Sex Transm Infect, 2009. 85(4): p. 242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ling Z, et al. , Diversity of cervicovaginal microbiota associated with female lower genital tract infections. Microb Ecol, 2011. 61(3): p. 704–14. [DOI] [PubMed] [Google Scholar]
- 80.Hilbert DW, et al. , Utilization of molecular methods to identify prognostic markers for recurrent bacterial vaginosis. Diagn Microbiol Infect Dis, 2016. 86(2): p. 231–42. [DOI] [PubMed] [Google Scholar]
- 81.Dols JA, et al. , Microarray-based identification of clinically relevant vaginal bacteria in relation to bacterial vaginosis. Am J Obstet Gynecol, 2011. 204(4): p. 305 e1–7. [DOI] [PubMed] [Google Scholar]
- 82.Koumans EH, et al. , The prevalence of bacterial vaginosis in the United States, 2001–2004; associations with symptoms, sexual behaviors, and reproductive health. Sex Transm Dis, 2007. 34(11): p. 864–9. [DOI] [PubMed] [Google Scholar]
- 83.Kenyon C, Colebunders R, and Crucitti T, The global epidemiology of bacterial vaginosis: a systematic review. Am J Obstet Gynecol, 2013. 209(6): p. 505–23. [DOI] [PubMed] [Google Scholar]
- 84.Sobel JD, Bacterial vaginosis. Annu Rev Med, 2000. 51: p. 349–56. [DOI] [PubMed] [Google Scholar]
- 85.van der Veer C, et al. , Accuracy of a commercial multiplex PCR for the diagnosis of bacterial vaginosis. J Med Microbiol, 2018. 67(9): p. 1265–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onderdonk AB, Delaney ML, and Fichorova RN, The Human Microbiome during Bacterial Vaginosis. Clin Microbiol Rev, 2016. 29(2): p. 223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swidsinski A, et al. , Adherent biofilms in bacterial vaginosis. Obstet Gynecol, 2005. 106(5 Pt 1): p. 1013–23. [DOI] [PubMed] [Google Scholar]
- 88.Hardy L, et al. , A fruitful alliance: the synergy between Atopobium vaginae and Gardnerella vaginalis in bacterial vaginosis-associated biofilm. Sex Transm Infect, 2016. 92(7): p. 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jung HS, et al. , Etiology of bacterial vaginosis and polymicrobial biofilm formation. Crit Rev Microbiol, 2017. 43(6): p. 651–667. [DOI] [PubMed] [Google Scholar]
- 90.Costerton JW, Stewart PS, and Greenberg EP, Bacterial biofilms: a common cause of persistent infections. Science, 1999. 284(5418): p. 1318–22. [DOI] [PubMed] [Google Scholar]
- 91.Hardy L, et al. , Bacterial biofilms in the vagina. Res Microbiol, 2017. 168(9–10): p. 865–874. [DOI] [PubMed] [Google Scholar]
- 92.Schwebke JR, Muzny CA, and Josey WE, Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: a conceptual model. J Infect Dis, 2014. 210(3): p. 338–43. [DOI] [PubMed] [Google Scholar]
- 93.Liu CM, et al. , Penile Microbiota and Female Partner Bacterial Vaginosis in Rakai, Uganda. mBio, 2015. 6(3): p. e00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mehta SD, et al. , The Microbiome Composition of a Man’s Penis Predicts Incident Bacterial Vaginosis in His Female Sex Partner With High Accuracy. Frontiers in Cellular and Infection Microbiology, 2020. 10(433). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hillier SL, et al. , Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N Engl J Med, 1995. 333(26): p. 1737–42. [DOI] [PubMed] [Google Scholar]
- 96.Goldenberg RL, et al. , The preterm prediction study: fetal fibronectin, bacterial vaginosis, and peripartum infection. NICHD Maternal Fetal Medicine Units Network. Obstet Gynecol, 1996. 87(5 Pt 1): p. 656–60. [DOI] [PubMed] [Google Scholar]
- 97.Leitich H, et al. , Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol, 2003. 189(1): p. 139–47. [DOI] [PubMed] [Google Scholar]
- 98.Romero R, et al. , Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth. Am J Obstet Gynecol, 2004. 190(6): p. 1509–19. [DOI] [PubMed] [Google Scholar]
- 99.Hillier SL, et al. , Role of bacterial vaginosis-associated microorganisms in endometritis. Am J Obstet Gynecol, 1996. 175(2): p. 435–41. [DOI] [PubMed] [Google Scholar]
- 100.Harmanli OH, et al. , Urinary tract infections in women with bacterial vaginosis. Obstet Gynecol, 2000. 95(5): p. 710–2. [DOI] [PubMed] [Google Scholar]
- 101.Swidsinski A, et al. , Presence of a polymicrobial endometrial biofilm in patients with bacterial vaginosis. PLoS One, 2013. 8(1): p. e53997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Haggerty CL, et al. , Bacterial vaginosis and anaerobic bacteria are associated with endometritis. Clin Infect Dis, 2004. 39(7): p. 990–5. [DOI] [PubMed] [Google Scholar]
- 103.Gundi VA, Desbriere R, and La Scola B, Leptotrichia amnionii and the female reproductive tract. Emerg Infect Dis, 2004. 10(11): p. 2056–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McClelland RS, et al. , Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis, 2018. 18(5): p. 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Munoz N, et al. , Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med, 2003. 348(6): p. 518–27. [DOI] [PubMed] [Google Scholar]
- 106.Cohen PA, et al. , Cervical cancer. Lancet, 2019. 393(10167): p. 169–182. [DOI] [PubMed] [Google Scholar]
- 107.Wentzensen N, Vinokurova S, and von Knebel Doeberitz M, Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res, 2004. 64(11): p. 3878–84. [DOI] [PubMed] [Google Scholar]
- 108.zur Hausen H, Papillomavirus infections--a major cause of human cancers. Biochim Biophys Acta, 1996. 1288(2): p. F55–78. [DOI] [PubMed] [Google Scholar]
- 109.Steenbergen RD, et al. , Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer, 2014. 14(6): p. 395–405. [DOI] [PubMed] [Google Scholar]
- 110.Doorbar J, et al. , Human papillomavirus molecular biology and disease association. Rev Med Virol, 2015. 25 Suppl 1: p. 2–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Egawa N, et al. , Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses, 2015. 7(7): p. 3863–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee JE, et al. , Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One, 2013. 8(5): p. e63514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Di Paola M, et al. , Characterization of cervico-vaginal microbiota in women developing persistent high-risk Human Papillomavirus infection. Sci Rep, 2017. 7(1): p. 10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Y, et al. , Patients With LR-HPV Infection Have a Distinct Vaginal Microbiota in Comparison With Healthy Controls. Front Cell Infect Microbiol, 2019. 9: p. 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen Y, et al. , Association between the vaginal microbiome and high-risk human papillomavirus infection in pregnant Chinese women. BMC Infect Dis, 2019. 19(1): p. 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goldenberg RL, et al. , Epidemiology and causes of preterm birth. Lancet, 2008. 371(9606): p. 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Di Renzo GC, et al. , Guidelines for the management of spontaneous preterm labor: identification of spontaneous preterm labor, diagnosis of preterm premature rupture of membranes, and preventive tools for preterm birth. J Matern Fetal Neonatal Med, 2011. 24(5): p. 659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee SE, et al. , The intensity of the fetal inflammatory response in intraamniotic inflammation with and without microbial invasion of the amniotic cavity. Am J Obstet Gynecol, 2007. 197(3): p. 294 e1–6. [DOI] [PubMed] [Google Scholar]
- 119.Marconi C, et al. , Amniotic fluid interleukin-1 beta and interleukin-6, but not interleukin-8 correlate with microbial invasion of the amniotic cavity in preterm labor. Am J Reprod Immunol, 2011. 65(6): p. 549–56. [DOI] [PubMed] [Google Scholar]
- 120.Kacerovsky M, et al. , Amniotic fluid protein profiles of intraamniotic inflammatory response to Ureaplasma spp. and other bacteria. PLoS One, 2013. 8(3): p. e60399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Florova V, et al. , Vaginal host immune-microbiome interactions in a cohort of primarily African-American women who ultimately underwent spontaneous preterm birth or delivered at term. Cytokine, 2021. 137: p. 155316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Newton ER, Chorioamnionitis and intraamniotic infection. Clin Obstet Gynecol, 1993. 36(4): p. 795–808. [DOI] [PubMed] [Google Scholar]
- 123.Rouse DJ, et al. , The Maternal-Fetal Medicine Units cesarean registry: chorioamnionitis at term and its duration-relationship to outcomes. Am J Obstet Gynecol, 2004. 191(1): p. 211–6. [DOI] [PubMed] [Google Scholar]
- 124.Tita AT and Andrews WW, Diagnosis and management of clinical chorioamnionitis. Clin Perinatol, 2010. 37(2): p. 339–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shane AL, Sanchez PJ, and Stoll BJ, Neonatal sepsis. Lancet, 2017. 390(10104): p. 1770–1780. [DOI] [PubMed] [Google Scholar]
- 126.Gibbs RS, et al. , Quantitative bacteriology of amniotic fluid from women with clinical intraamniotic infection at term. J Infect Dis, 1982. 145(1): p. 1–8. [DOI] [PubMed] [Google Scholar]
- 127.Gibbs RS, et al. , A randomized trial of intrapartum versus immediate postpartum treatment of women with intra-amniotic infection. Obstet Gynecol, 1988. 72(6): p. 823–8. [DOI] [PubMed] [Google Scholar]
- 128.Romero R, et al. , Clinical chorioamnionitis at term III: how well do clinical criteria perform in the identification of proven intra-amniotic infection? J Perinat Med, 2016. 44(1): p. 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gilstrap LC 3rd and Cox SM, Acute chorioamnionitis. Obstet Gynecol Clin North Am, 1989. 16(2): p. 373–9. [PubMed] [Google Scholar]
- 130.Gibbs RS and Duff P, Progress in pathogenesis and management of clinical intraamniotic infection. Am J Obstet Gynecol, 1991. 164(5 Pt 1): p. 1317–26. [DOI] [PubMed] [Google Scholar]
- 131.Willi MJ, et al. , Chorioamnionitis: elevated interleukin-6 and interleukin-8 concentrations in the lower uterine segment. J Perinat Med, 2002. 30(4): p. 292–6. [DOI] [PubMed] [Google Scholar]
- 132.Lee SE, et al. , Funisitis in term pregnancy is associated with microbial invasion of the amniotic cavity and intra-amniotic inflammation. J Matern Fetal Neonatal Med, 2006. 19(11): p. 693–7. [DOI] [PubMed] [Google Scholar]
- 133.Gotsch F, et al. , The fetal inflammatory response syndrome. Clin Obstet Gynecol, 2007. 50(3): p. 652–83. [DOI] [PubMed] [Google Scholar]
- 134.Redline RW, Inflammatory response in acute chorioamnionitis. Semin Fetal Neonatal Med, 2012. 17(1): p. 20–5. [DOI] [PubMed] [Google Scholar]
- 135.Romero R, et al. , Clinical chorioamnionitis at term VI: acute chorioamnionitis and funisitis according to the presence or absence of microorganisms and inflammation in the amniotic cavity. J Perinat Med, 2016. 44(1): p. 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Romero R, et al. , Clinical chorioamnionitis at term V: umbilical cord plasma cytokine profile in the context of a systemic maternal inflammatory response. J Perinat Med, 2016. 44(1): p. 53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Romero R, et al. , Clinical chorioamnionitis at term IV: the maternal plasma cytokine profile. J Perinat Med, 2016. 44(1): p. 77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Romero R, et al. , Clinical chorioamnionitis at term II: the intra-amniotic inflammatory response. J Perinat Med, 2016. 44(1): p. 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chaiyasit N, et al. , Clinical chorioamnionitis at term VIII: a rapid MMP-8 test for the identification of intra-amniotic inflammation. J Perinat Med, 2017. 45(5): p. 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martinez-Varea A, et al. , Clinical chorioamnionitis at term VII: the amniotic fluid cellular immune response. J Perinat Med, 2017. 45(5): p. 523–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gomez-Lopez N, et al. , Amniotic fluid neutrophils can phagocytize bacteria: A mechanism for microbial killing in the amniotic cavity. Am J Reprod Immunol, 2017. 78(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gomez-Lopez N, et al. , Are amniotic fluid neutrophils in women with intraamniotic infection and/or inflammation of fetal or maternal origin? Am J Obstet Gynecol, 2017. 217(6): p. 693.e1–693.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Romero R, et al. , Clinical Chorioamnionitis at Term: New Insights into the Etiology, Microbiology, and the Fetal, Maternal and Amniotic Cavity Inflammatory Responses. Nogyogyaszati Szuleszeti Tovabbkepzo Szemle, 2018. 20(3): p. 103–112. [PMC free article] [PubMed] [Google Scholar]
- 144.Gomez-Lopez N, et al. , Clinical chorioamnionitis at term IX: in vivo evidence of intra-amniotic inflammasome activation. J Perinat Med, 2019. 47(3): p. 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gomez-Lopez N, et al. , Cellular immune responses in amniotic fluid of women with preterm labor and intra-amniotic infection or intra-amniotic inflammation. Am J Reprod Immunol, 2019. 82(5): p. e13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gomez-Lopez N, et al. , The origin of amniotic fluid monocytes/macrophages in women with intra-amniotic inflammation or infection. J Perinat Med, 2019. 47(8): p. 822–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Theis KR, et al. , Microbial burden and inflammasome activation in amniotic fluid of patients with preterm prelabor rupture of membranes. J Perinat Med, 2020. 48(2): p. 115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Galaz J, et al. , Cellular immune responses in amniotic fluid of women with preterm prelabor rupture of membranes. J Perinat Med, 2020. 48(3): p. 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Galaz J, et al. , Cellular immune responses in amniotic fluid of women with preterm clinical chorioamnionitis. Inflamm Res, 2020. 69(2): p. 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Casey BM and Cox SM, Chorioamnionitis and endometritis. Infect Dis Clin North Am, 1997. 11(1): p. 203–22. [DOI] [PubMed] [Google Scholar]
- 151.Duff P, Antibiotic selection in obstetric patients. Infect Dis Clin North Am, 1997. 11(1): p. 1–12. [DOI] [PubMed] [Google Scholar]
- 152.Burnham P, et al. , Separating the signal from the noise in metagenomic cell-free DNA sequencing. Microbiome, 2020. 8(1): p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kuzniewicz MW, et al. , A Quantitative, Risk-Based Approach to the Management of Neonatal Early-Onset Sepsis. JAMA Pediatr, 2017. 171(4): p. 365–371. [DOI] [PubMed] [Google Scholar]
- 154.van Dillen J, et al. , Maternal sepsis: epidemiology, etiology and outcome. Curr Opin Infect Dis, 2010. 23(3): p. 249–54. [DOI] [PubMed] [Google Scholar]
- 155.Kotaskova I, et al. , First report of Sneathia sanguinegens together with Mycoplasma hominis in postpartum prosthetic valve infective endocarditis: a case report. BMC Infect Dis, 2017. 17(1): p. 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Goncalves LF, Chaiworapongsa T, and Romero R, Intrauterine infection and prematurity. Ment Retard Dev Disabil Res Rev, 2002. 8(1): p. 3–13. [DOI] [PubMed] [Google Scholar]
- 157.Lofmark S, Edlund C, and Nord CE, Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin Infect Dis, 2010. 50 Suppl 1: p. S16–23. [DOI] [PubMed] [Google Scholar]
- 158.Ling Z, et al. , The restoration of the vaginal microbiota after treatment for bacterial vaginosis with metronidazole or probiotics. Microb Ecol, 2013. 65(3): p. 773–80. [DOI] [PubMed] [Google Scholar]
- 159.Gottschick C, et al. , The urinary microbiota of men and women and its changes in women during bacterial vaginosis and antibiotic treatment. Microbiome, 2017. 5(1): p. 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lamp KC, et al. , Pharmacokinetics and pharmacodynamics of the nitroimidazole antimicrobials. Clin Pharmacokinet, 1999. 36(5): p. 353–73. [DOI] [PubMed] [Google Scholar]
- 161.Macklaim JM, et al. , Changes in vaginal microbiota following antimicrobial and probiotic therapy. Microb Ecol Health Dis, 2015. 26: p. 27799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cruciani F, et al. , Development of a microarray-based tool to characterize vaginal bacterial fluctuations and application to a novel antibiotic treatment for bacterial vaginosis. Antimicrob Agents Chemother, 2015. 59(5): p. 2825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Scarpignato C. and Pelosini I, Rifaximin, a poorly absorbed antibiotic: pharmacology and clinical potential. Chemotherapy, 2005. 51 Suppl 1: p. 36–66. [DOI] [PubMed] [Google Scholar]
- 164.Rivkin A. and Gim S, Rifaximin: new therapeutic indication and future directions. Clin Ther, 2011. 33(7): p. 812–27. [DOI] [PubMed] [Google Scholar]
- 165.Bradshaw CS, et al. , High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy and factors associated with recurrence. J Infect Dis, 2006. 193(11): p. 1478–86. [DOI] [PubMed] [Google Scholar]
- 166.Bachy B, et al. , Septic arthritis due to a Sneathia species most closely related to Sneathia sanguinegens. J Med Microbiol, 2011. 60(Pt 11): p. 1693–6. [DOI] [PubMed] [Google Scholar]
- 167.Frost LS, et al. , Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol, 2005. 3(9): p. 722–32. [DOI] [PubMed] [Google Scholar]
- 168.Alekshun MN and Levy SB, Molecular mechanisms of antibacterial multidrug resistance. Cell, 2007. 128(6): p. 1037–50. [DOI] [PubMed] [Google Scholar]
- 169.Di Renzo GC, The great obstetrical syndromes. J Matern Fetal Neonatal Med, 2009. 22(8): p. 633–5. [DOI] [PubMed] [Google Scholar]
- 170.Liu L, et al. , Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet, 2015. 385(9966): p. 430–40. [DOI] [PubMed] [Google Scholar]
- 171.Chawanpaiboon S, et al. , Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob Health, 2019. 7(1): p. e37–e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lee AC, Blencowe H, and Lawn JE, Small babies, big numbers: global estimates of preterm birth. Lancet Glob Health, 2019. 7(1): p. e2–e3. [DOI] [PubMed] [Google Scholar]
- 173.Kurjak A, Di Renzo GC, and Stanojevic M, Globalization and perinatal medicine--how do we respond? J Matern Fetal Neonatal Med, 2010. 23(4): p. 286–96. [DOI] [PubMed] [Google Scholar]
- 174.Blencowe H, et al. , Preterm birth-associated neurodevelopmental impairment estimates at regional and global levels for 2010. Pediatr Res, 2013. 74 Suppl 1: p. 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Muzny CA, et al. , Identification of Key Bacteria Involved in the Induction of Incident Bacterial Vaginosis: A Prospective Study. J Infect Dis, 2018. 218(6): p. 966–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Harwich MD Jr., et al. , Drawing the line between commensal and pathogenic Gardnerella vaginalis through genome analysis and virulence studies. BMC Genomics, 2010. 11: p. 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yeoman CJ, et al. , Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS One, 2010. 5(8): p. e12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Castro J, Jefferson KK, and Cerca N, Innate immune components affect growth and virulence traits of bacterial-vaginosis-associated and non-bacterial-vaginosis-associated Gardnerella vaginalis strains similarly. Pathog Dis, 2018. 76(9). [DOI] [PubMed] [Google Scholar]
- 179.Garcia EM, et al. , Interaction of Gardnerella vaginalis and Vaginolysin with the Apical versus Basolateral Face of a Three-Dimensional Model of Vaginal Epithelium. Infect Immun, 2019. 87(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nasioudis D, et al. , Bacterial vaginosis: a critical analysis of current knowledge. BJOG, 2017. 124(1): p. 61–69. [DOI] [PubMed] [Google Scholar]
- 181.Mitchell C, et al. , Hydrogen Peroxide-Producing Lactobacilli Are Associated With Lower Levels of Vaginal Interleukin-1beta, Independent of Bacterial Vaginosis. Sex Transm Dis, 2015. 42(7): p. 358–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Murphy K. and Mitchell CM, The Interplay of Host Immunity, Environment and the Risk of Bacterial Vaginosis and Associated Reproductive Health Outcomes. J Infect Dis, 2016. 214 Suppl 1: p. S29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Niu SY, et al. , Bioinformatics tools for quantitative and functional metagenome and metatranscriptome data analysis in microbes. Brief Bioinform, 2018. 19(6): p. 1415–1429. [DOI] [PubMed] [Google Scholar]
- 184.Berman HL, McLaren MR, and Callahan BJ, Understanding and interpreting community sequencing measurements of the vaginal microbiome. BJOG, 2020. 127(2): p. 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ma B, et al. , A comprehensive non-redundant gene catalog reveals extensive within-community intraspecies diversity in the human vagina. Nat Commun, 2020. 11(1): p. 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Borgogna JC, et al. , The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG, 2020. 127(2): p. 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shaw JL, Smith CR, and Diamandis EP, Proteomic analysis of human cervico-vaginal fluid. J Proteome Res, 2007. 6(7): p. 2859–65. [DOI] [PubMed] [Google Scholar]
- 188.Di Quinzio MK, et al. , Proteomic analysis and characterisation of human cervico-vaginal fluid proteins. Aust N Z J Obstet Gynaecol, 2007. 47(1): p. 9–15. [DOI] [PubMed] [Google Scholar]
- 189.Parry S, et al. , Cervicovaginal fluid proteomic analysis to identify potential biomarkers for preterm birth. Am J Obstet Gynecol, 2020. 222(5): p. 493 e1–493 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]